Abstract
Hfq is a global regulator that is involved in environmental adaptation of bacteria and in pathogenicity. To gain insight into the role of Hfq in Vibrio alginolyticus, an hfq deletion mutant was constructed in V. alginolyticus ZJ-T strain and phenotypically characterized. Deletion of hfq led to an alteration of colony morphology and reduced extracellular polysaccharide production, a general impairment of growth in both rich medium and minimal media with different carbon sources or amino acids, enhanced sensitivity to oxidative stress and to several antibiotics. Furthermore, a differential transcriptomic analysis showed significant changes of transcript abundance for 306 protein coding genes, with 179 genes being up regulated and 127 down-regulated. Several of these changes could be related to the observed phenotypes of the mutant. Transcriptomic data also provided evidence for the induction of the extracytoplasmic stress response in absence of Hfq. Altogether, these findings point to broad regulatory functions for Hfq in V. alginolyticus cells, likely to underlie an important role in pathogenicity.
Introduction
Vibrio alginolyticus is a common gram-negative halophilic bacterium that is part of the normal microflora in marine environment worldwide. However, more and more studies suggest that it is also a potential threat to marine animals and humans by causing serious infections [1–3]. A recent report showed that the incidence of diseases caused by vibrios in USA has dramatically increased in the last decade and V. alginolyticus has now been listed as one of the most common pathogens together with Vibrio parahaemolyticus and Vibrio vulnificus [4]. Indeed, V. alginolyticus was proposed to correspond to biotype 2 of V. parahaemolyticus since these two species have almost indistinguishable phenotypes except for the ability to utilize sucrose [5]. The risk of outbreaks caused by V. alginolyticus is underestimated compared with its close relatives. Its pathogenic mechanism, adaptation and epidemic traits remain unknown.
To respond to environmental cues, bacterial cells need to coordinate their gene expression quickly and precisely. Regulatory non-coding small RNAs (sRNAs) are now known to play essential roles in post-transcriptional regulation of gene expression and in virulence [6, 7]. Bacterial sRNAs can regulate target mRNAs positively or negatively, at the translational level or through affecting mRNA stability [8]. Many sRNAs exert their functions by incomplete base pairing with target mRNAs. Such interactions often require the assistance of the RNA chaperone Hfq, an RNA-binding protein belonging to the Sm protein family [8]. Hfq binds to both target mRNA and sRNA and stabilizes the interaction by a mechanism not fully understood [9]. In addition, in case of a negative regulation by the sRNA, Hfq may be involved in the recruitment of RNase E for further degradation of both the mRNA and the sRNA [8, 9]. More rarely, interaction of the sRNA with its target can interfere with RNase E cleavages and stabilize the mRNA (see [10] for a review).
Deletion of hfq was reported to affect motility, biofilm formation, central metabolism, virulence and stress response in a variety of species mostly gram-negative [11–14]. For instance, in Salmonella, the expression of at least 20% genes (about 1,000 genes) is affected by Hfq [12, 13]. Compared to other gram-negative species, especially Escherichia coli and Salmonella sp., still relatively little is known about the role of Hfq and sRNAs in Vibrio sp. In V. cholerae, sRNAs present in multiple copies, such as Qrr or CsrBs, the latters targetting the CsrA protein rather than mRNAs [15] have been found to be important regulators of the quorum sensing response in V. cholerae and V. harveyi [16–18], which itself controls virulence gene expression in pathogenic Vibrio [19, 20]. Hfq was also found to down regulate expression of the alternative extracytoplasmic stress (ECS) RpoE sigma factor in V. cholerae [21]. RpoE itself controls the production of the sRNA VrrA, which represses translation of the outer membrane protein OmpT in an Hfq dependent manner [22]. In V. parahaemolyticus, a Δhfq mutant is more resistant to oxidative stress and has increase production of the thermostable direct hemolysin (TdH), an important virulence factor in this species [23, 24].
In V. alginolyticus, a previous study showed that deletion of hfq decreased motility, biofilm formation and virulence [14]. In this study, we further characterized potential roles of Hfq in V. alginolyticus by phenotypic and transcriptomic analyses of the deletion mutant. Our data show that Hfq participates in diverse cellular processes including colony morphology, nutrient utilization, and envelope composition by modulating the expression of a large variety of genes some of which are likely to be controlled by Hfq-dependent sRNAs.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| V. alginolyticus | ||
| ZJ-T | Apr (ampicillin resistant), translucent/smooth variant of wild strain ZJ51 [25]; isolated from diseased Epinephelus coioides off the Southern China coast | [26] |
| Δhfq-T | Apr; ZJ-T carrying an in-frame deletion of hfq gene | This study |
| hfq+-T | Apr, Cmr (chloramphenicol resistant), Δhfq-T, z:: pNQ705-1-hfq | This study |
| E. coli | ||
| SY327 | Δ(lac pro) argE(Am) rif malA recA56 λpir lysogen; suicide vector pDM4’s intermediate host | [27] |
| S17-1 | thi pro hsdR hsdM+recA RP4-2-Tc::Mu-Km::Tn7 λpir lysogen; donor strain for conjugation | [28] |
| Plasmids | ||
| pDM4 | Cmr; suicide vector with an R6K origin, requiring the Pir protein for its replication, and the sacBR gene of Bacillus subtilis | [29] |
| pNQ705-1 | Cmr; suicide vector with a Pir dependent, R6K origin | [29] |
| pDM4-Δhfq | Cmr; pDM4 containing the mutant allele of hfq | This study |
| pNQ705-1-hfq | Cmr; pNQ705-1 containing the wild-type allele of hfq | This study |
V. alginolyticus was cultured in Tryptic Soy Broth (TSB) (BD, USA) at 30°C. Escherichia coli strains were cultured in Luria–Bertani (LB) (Invitrogen, USA) medium supplemented with appropriate antibiotics at 37°C. For transconjugants selection, TCBS medium (BD, USA) was used with 5 μg.ml-1 chloramphenicol (Cm). To select transconjugants having undergone plasmid excision and allelic exchange, expression of the sacB gene carried by the pDM4 plasmid was induced by adding 10% sucrose to the medium. This gene encodes levansucrase which is lethal for gram-negative bacteria [29]. For colony morphology observation, LB agar plates were used. Unless otherwise indicated, M63 minimum medium was made according to the following recipe: 3 g.L-1 KH2PO4, 7 g.L-1 K2HPO4, 2 g.L-1 (NH4)2SO4, 0.5×10−3 g.L-1 FeSO4, 30 g.L-1 NaCl, 2×10−3 M MgSO4, 5×10−3 g.L-1 thiamine and 4 g.L-1 D-glucose.
Construction of a hfq mutant and complementation strain
An in-frame deletion of the hfq gene, retaining the five first codons of the gene (15 bp) and the stop codon was constructed in V. alginolyticus ZJ-T strain by overlapping extension (SOE) PCR as described by Milton et al [29]. Briefly, two fragments flanking the hfq deletion (Fig 1A), were amplified with two pairs of primers, hfq-A and -B and hfq-C and -D respectively, B and C containing overlapping extensions (S1 Table). These two fragments were further assembled by PCR with primer pair hfq-A and hfq-D, producing a fragment comprising the 321 bp upstream and 249 bp downstream regions of the hfq gene, and including. This fragment was then cloned into the suicide vector pDM4, generating plasmid pDM4-Δhfq (Table 1), using E. coli SY327 as an intermediate host. The recombinant plasmid was transferred by conjugation from strain S17-1 (Table 1) to V. alginolyticus wild-type strain ZJ-T before allelic exchange as described above. The presence of the hfq in-frame deletion was then confirmed by sequencing and the strain named Δhfq-T (Fig 1A, Table 1).
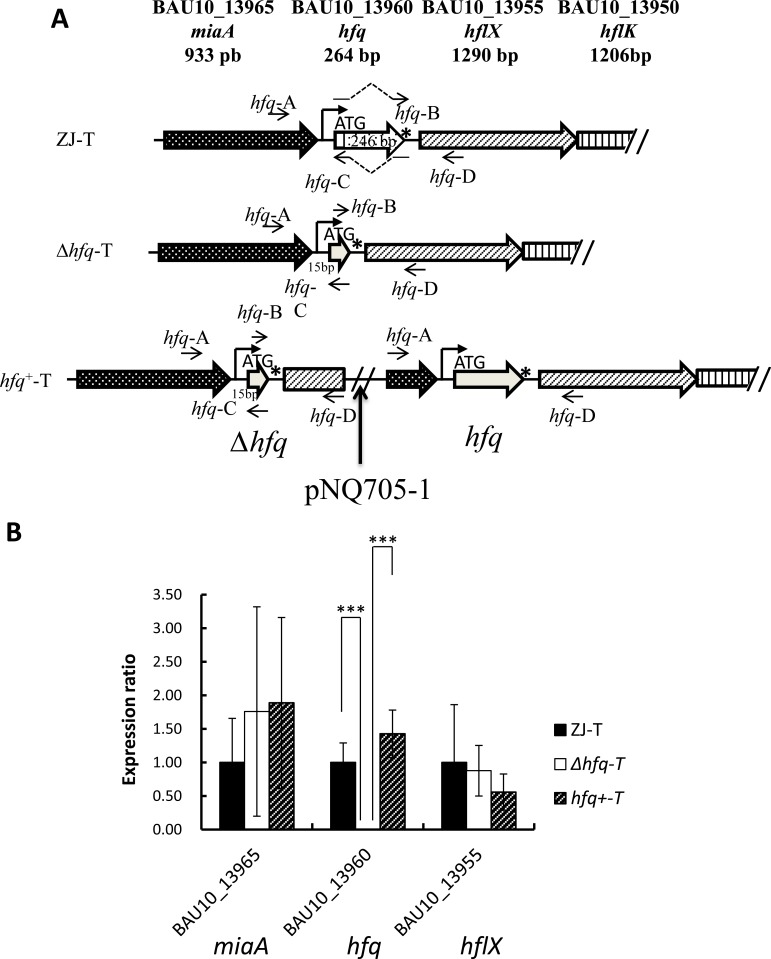
Fig 1.
Structure of the hfq locus (A) and neighboring gene expression in Vibrio alginolyticus ZJ-T and derivative strains (B). A. The hfq gene is downstream of the miaA gene (locus ID = BAU10_13965) and upstream of 3 genes (hflX, K, C) likely to form an operon. A putative Sigma70 promoter identified upstream of hfq is indicated. 246 bps of the hfq ORF, from the 6th codon to the stop codon not included (marked by a star) were deleted as described in Materials and Methods, giving rise to the Δhfq-T strain. This mutation does not affect the potential promoter of the operon, neither the ribosome binding site of the downstream gene. A wild type hfq gene was reinserted at the Δhfq locus of the Δhfq-T strain by insertion of pNQ705-1-hfq by homologous recombination (see Materials and Methods for details) generating a duplication of the locus, corresponding to the fragment inserted in pNQ705-1, from primer hfq-A to primer hfq-D. The depicted situation corresponds to an insertion of the plasmid downstream of the hfq deletion. B. Relative expression of hfq, the upstream gene (miaA) and the downstream gene (hflX) in derivative strains compared to WT. Relative expression (normalized to the WT level for each gene) was determined by qPCR as described in Materials and Methods. Error bars correspond to standard deviations from three biological replicates. Statistically significant differences are indicated (***p < 0.001).
A complementation strain, carrying both the deleted allele and the WT allele, was constructed as described previously [30–32]. Briefly, a PCR fragment containing the hfq gene and its flanking regions (including the hfq gene native promoter, hfq being the first gene of an operon -see Fig 1A) was amplified using the primers hfq-A and hfq-D. The fragment was then inserted into plasmid pNQ705-1 and the recombinant plasmid, pNQ705-1-hfq, was transferred into the hfq mutant by conjugation, leading to integration of pNQ705-1-hfq at the hfq locus by homologous recombination. The resulting strain has two copies of the hfq locus, one mutant, one WT. The presence of an intact hfq gene was confirmed by PCR analysis and sequencing. The complementation strain was named hfq+-T. (Fig 1A, Table 1). Quantitative RT-PCR (qPCR) was used to test a potential effect of the hfq deletion on neighboring gene expression (potential polar effects) as described below.
Quantitative RT-PCR
Overnight cell cultures of ZJ-T, Δhfq-T and hfq+-T were diluted to OD600 = 0.01 in TSB medium and grown to early exponential phase (OD600 = 0.5) and bacterial cells were collected. All reagents were from Takara Bio Inc. Total RNA was isolated by using RNAiso Plus, DNase-treated and 1 μg of RNA was reverse-transcribed with PrimeScriptTM RT reagent Kit. PCR amplification was then carried out on a RotorGene RG-3000 (Qiagen) using SYBR Premix Ex Taq™ with primers specific to the genes to be assayed (S1 Table). Three housekeeping genes (recA, uvrA and gyrA) were used as internal controls. Relative gene expression was calculated by the 2−ΔΔCt method [33] and normalized to the wild type ZJ-T value. Measurements were done in triplicates. Statistical significance was assayed by one-way ANOVA LDS method (*p < 0.05, **p < 0.01, ***p < 0.001).
Colony and cell morphology
To observe colony morphology, overnight cell cultures were serially diluted and 100 μl of each dilution was spread on LB agar plates. Single colonies were observed using the BIO-RAD image system Gel DocTM XR+. Bacterial cell morphology was observed by Transmission Electron Microscopy (Hitachi H-7650 TEM). Briefly, ZJ-T, Δhfq-T and hfq+-T strains were cultured for 8–10 hours in TSB, then placed on copper grids and negatively stained with 3% phosphotungstic acid at pH 7.0 for 2 min.
Extracellular polysaccharide (EPS) quantification
To quantify EPS, alcian blue which stains acidic polysaccharides was used as described by Antony Croxatto et al [34] with modification. Briefly, 5 μl overnight cultures of ZJ-T, Δhfq-T and hfq+-T were spotted on LB agar plate and incubated at 30°C for 24 h. Bacterial spots were scraped from the plate and re-suspended in filtered 4 mM EDTA to a same final OD600 = 1.0. EPS was sheared off the cells by three successive centrifugations at 25,000 g for 30 min followed by re-suspension in the same volume of 4mM EDTA. 1 ml of the supernatant was mixed with 4 ml alcian blue solution and incubated at room temperature for one hour. The solution was centrifuged at 1,500 g for 20 min, and the supernatant was discarded carefully. The pellet was washed in 4 ml ethanol once and re-suspended in 4 ml 10% SDS. The absorbance at 620 nm was recorded. The average quantity of EPS was normalized as OD620/OD600 and one-way ANOVA LDS method was used for statistical analysis of the difference between the wild type, the mutant and the complementation strain with IBM SPSS Statistics 19. p <0.05 was retained as the threshold for statistical significance.
Measurement of bacterial growth
Bacterial strains were grown overnight in TSB medium at 30°C with shaking at 200 rpm. To test growth in rich medium, each culture was brought to OD600 = 1.0 using fresh TSB and then diluted into fresh TSB (1:1000). To investigate the effect of various carbon sources on growth, M63 was modified by replacing D-glucose with 0.4% (w/v) of different carbohydrates, (D-maltose, D-trehalose, D-fructose, N-acetyl-D-glucosamine) or TCA cycle intermediates (sodium citrate, α-ketoglutaric acid, sodium succinate, sodium malonate, oxaloacetic acid and fumaric acid). To probe the effect of amino acid(s) on growth, D-glucose and (NH4)2SO4 in M63 were left out and replaced by the nonpolar amino acids L-alanine (150mM), L-valine (20mM), L-leucine (20mM) or L-isoleucine (20mM), and polar amino acids L-threonine (50mM), L-aspartic acid (10mM), L-lysine (15mM), L-glutamic acid (10mM), L-serine (50mM), L-arginine (50mM), L-histidine hydrochloride (50mM) or L-glutamine (10mM) as both carbon and nitrogen source. Minimal medium assays were carried out as follow: Overnight cultures were collected by centrifugation and washed once with M63 without D-glucose and (NH4)2SO4, then individually re-suspended in the above modified M63 to the same cell density. Cultures (3 replicates in each case) were then incubated at 30°C with continuous shaking at 200 rpm in 96 well plates. OD600 was measured at regular time intervals using the Multiskan Ascent plate reader (Thermo Fisher Scientific).
Stress response assays
Briefly, overnight cultures in TSB of ZJ-T, Δhfq-T and hfq+-T were re-suspended at OD600 = 5.0 (2.5×109 CFU/ml), and then serially diluted 10 fold into fresh TSB medium. 5 μl of each dilution was spotted on TSB agar plates alone or containing 6mM CuSO4 or 0.0015% H2O2 and incubated at 30°C for 24 h.
Antibiotic susceptibility assays
Antibiotics were bought from Hangzhou Binge Microorganism Reagent Co., and disk diffusion assays were carried out as described previously [35] with modifications. Disks with various concentrations of antibiotics were made. 200 μl of overnight bacterial culture was added to 10 ml fresh TSB medium, then mixed gently and plated on TSB agar plates. Disks were placed on the dried plates and incubated at 30°C for 24 h before measuring the inhibition zone. Measurements were done in triplicates. Statistical significance was assayed by one-way ANOVA LDS method as described above (p < 0.05).
Transcriptome sequencing and bioinformatic analysis
Two single colonies from strains ZJ-T and Δhfq-T were used to inoculate two independent cultures in TSB medium, which were grown at 30°C to early exponential phase (OD600 = 0.5). Equal amount of cells were harvested from each sample and total RNA was isolated using RNAiso Plus (Takara Bio Inc.) according to the manufacturer's instructions. The quality and quantity of total RNA was verified by measuring the OD260/280 and OD260/230 with a Nanodrop 2000 spectrophotometer, and agarose gel electrophoresis. The whole process was repeated once and four RNA samples from each strain were pooled for RNA-seq analysis. Libraries derived from the mixed RNA samples were sequenced on an Illumina HiSeqTM 2000 at the Beijing Genomics Institute using standard Illumina protocols.
Primary sequencing data produced by Illumina HiSeqTM 2000 were subjected to quality control. Filtered quality reads were mapped to the reference genome sequence of V. alginolyticus ZJ-T (GenBank accession numbers CP016224 and CP016225 for Chromosome 1 and 2 respectively) using the SOAP2 aligner [36]. To identify differentially expressed transcripts, the mapped sequence reads were normalized as Reads Per Kilobase per Million (RPKM) for each annotated feature in the genome as described previously [37] and the expression ratio (fold change) between mutant and wild type was computed. The significance of differential gene expression (p-value) was then calculated by the method of Audic and Claverie [38]. In addition a False Discovery Rate (FDR) was calculated by the method of Benjamini and Yekutieli [39]. Differentially expressed genes (expression ratio ≥2, p-value <0.1 and FDR < 0.001) were classified into different categories based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and ontologies. Raw sequence reads were deposited in the SRA databank under the accession number SRP076968.
Outer membrane protein analysis
Extraction of total outer membrane proteins (OMPs) was conducted as previously described [40]. Briefly, overnight cultures in TSB of ZJ-T, Δhfq-T and hfq+-T were diluted to OD600 = 0.01 in 15 ml TSB medium and incubated at 30°C to OD600 = 0.5. The cultures were centrifuged at 1,000 g for 10 min and the pellets were re-suspended in 13.5 ml ddH2O. The cells were broken by sonication followed by cell debris removal by centrifugation at 12,000 g for 10 min at 4°C. The supernatants were transferred to new tubes containing 1.5 ml of 20% sodium sarcosine, to solubilize inner membrane proteins without affecting OMPs. After incubation at room temperature for 30 min, followed by centrifugation at 240,000 g for 1 h at 4°C in an Beckman Coulter Optima XPN-100 Ultracentrifuge, the pellets were washed with ice-cold ddH2O and re-centrifuged in the same condition before being re-suspended in 1×SDS sample buffer. 15 μl of protein preparation was separated by SDS–PAGE (12%). The gel was stained with Coomassie brilliant blue and photographed with a gelDoc imager (Bio-Rad, USA).
Results
Construction of an hfq null mutant and complementation
An in-frame deletion of the hfq gene, from the sixth codon to the last codon (not including the stop codon) was constructed as described in Materials and Methods. In addition, we constructed a complementation strain, where the WT hfq gene was reintroduced in the chromosome, generating a partial tandem duplication of the locus, containing both the deleted and the WT version of the gene (Fig 1A). Since hfq is the first gene of a four-gene operon, it was important to verify that neither the deletion nor the reintroduction of the wild-type gene had a polar effect on the neighboring genes. Hence, we checked by qPCR expression of all three genes (upstream, hfq and downstream gene) in the three strains: WT, Δhfq and the complementation strain. As shown in Fig 1B, a significant difference of expression was observed only in the case of hfq: expectedly expression could not be detected in the mutant and was restored in the complementation strain.
Hfq regulates colony morphology and synthesis of EPS in V. alginolyticus
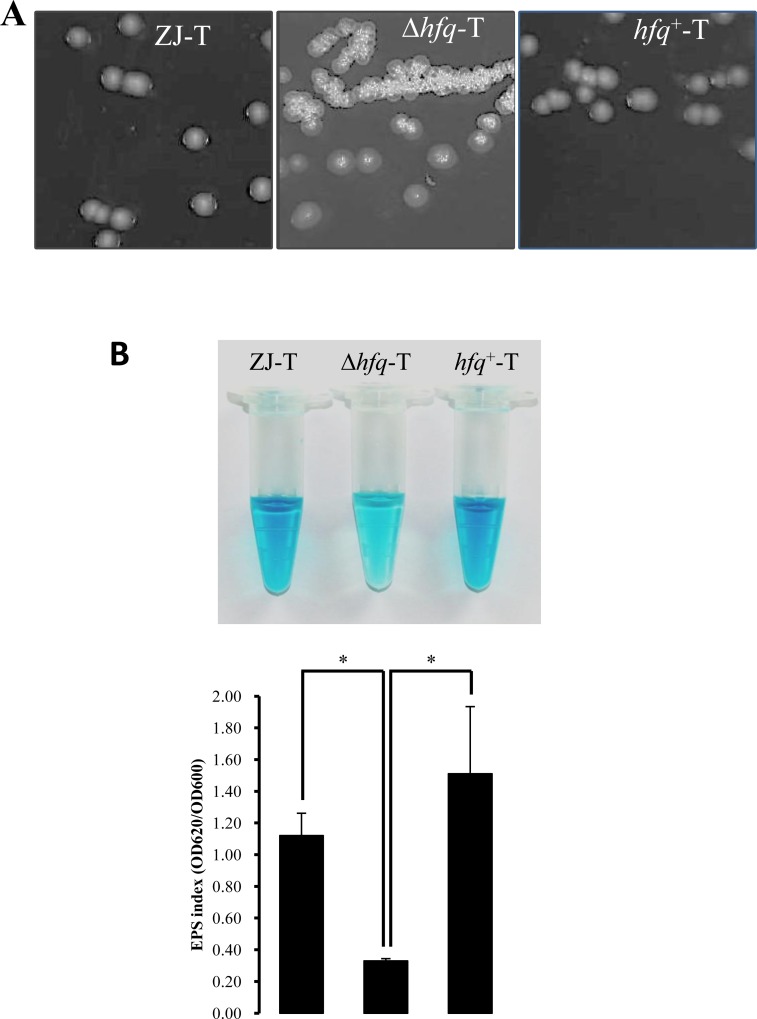
V. alginolyticus wild type ZJ-T strain forms translucent (Tr) and smooth (S) colonies on LB agar plates [26]. In contrast, we observed that the hfq mutant forms opaque (Op) and rugose (R) colonies and that reintroducing the WT hfq allele in strain hfq+-T reverted colony morphology to Tr-S (Fig 2A), indicating a role of Hfq in the morphology change. In V. cholerae, colony morphology changes from Tr to Op have been associated with changes in production of extracellular polysaccharides (Vps) [41, 42]. In Vibrio vulnificus, expression of a Group 1 capsular polysaccharide (CPS) operon was positively associated with an Op phenotype [43]. Whereas the transition from Tr to Op requires increased CPS production, transition from S to R does not [41, 44]. V. vulnificus carries another exopolysaccharide gene cluster, named brp, which was found to be important for rugose colony formation [44]. We quantified by alcian blue the EPS produced by cells grown on LB agar plates. As shown in Fig 2B, EPS staining by alcian blue was significantly decreased in the hfq mutant, compared to the wild type and the complementation strain, whereas transmission electron microscopy revealed no visible change in cell shape or size: all three strains carried similar 3–4 polar flagella and were surrounded by a thick layer of extracellular matrix (S1 Fig).
Fig 2. hfq deletion affects colony morphology and extracellular polysaccharide production.
A. Colony morphology was observed with a BIO-RAD Gel DocTM XR+ imager. hfq deletion led to a colony morphology change from translucent and smooth to opaque and rugose. B. Amounts of extracellular polysaccharides of ZJ-T, Δhfq-T and hfq+-T were assayed in triplicates with alcian blue (top) and quantified by spectroscopy (bottom). Error bars correspond to standard deviations. Statistically significant differences are indicated (* p < 0.05)
Deletion of hfq affects carbon and nitrogen metabolism
When the bacteria grew both in TSB medium and in minimal medium supplemented with D-glucose, D-maltose, D-trehalose, D-fructose or N-acetyl-D-glucosamine as sole carbon source, the lag phase of Δhfq-T was 2 hours longer than that of ZJ-T, and the growth rate of the mutant decreased in exponential phase, as well as total growth. When intermediates of the TCA cycle were supplied as sole carbon source, no differences between WT and mutant were observed in terms of duration of lag phase, but the hfq mutant displayed a significant decrease in growth yield. These phenotypes were reverted in the complementation strain (S2 Fig, Table 2). In addition, none of the strains were able to grow on sodium malonate (S2 Fig, Table 2).
Table 2. Effect of hfq deletion on growth rate and total growth in rich medium and minimal medium M63 supplemented with various carbon sources.
Values correspond to the mean of three independent cultures. Significant statistical differences between strains and WT are indicated (*p < 0.05, **p < 0.01, ***p <0.001). Growth rates are expressed as generation times.
| Medium | Generation time (minutes) | Total growth (OD600) | ||||
|---|---|---|---|---|---|---|
| ZJ-T | Δhfq-T | hfq+-T | ZJ-T | Δhfq-T | hfq+-T | |
| TSB | 101.15±3.14 | 160.47±1.80 (***)a | 90.70±3.16 | 1.27±0.02 | 1.15±0.02 (***) | 1.28±0.01 |
| M63+ d-glucose | 144.46±3.17 | 195.71±5.24 (***) | 120.19±9.20 | 0.53±0.02 | 0.45±0.01 (***) | 0.53±0.00 |
| M63+d-maltose | 154.81±6.97 | 228.33±8.46 (***) | 126.85±6.76 | 0.56±0.01 | 0.49±0.00 (***) | 0.57±0.01 |
| M63+d-trehalose | 156.55±8.98 | 283.61±10.06 (***) | 128.33±7.86 | 0.56±0.00 | 0.48±0.00 (***) | 0.58±0.00 |
| M63+d-fructose | 119.48±5.44 | 160.42±7.64 (***) | 101.79±8.96 | 0.53±0.00 | 0.45±0.01 (***) | 0.54±0.00 |
| M63+ | ||||||
| N-acetyl-d-glucosamine | 145.79±7.29 | 188.58±5.25 (***) | 123.97±9.91 | 0.5±0.00 | 0.40±0.00 (***) | 0.49±0.00 |
| Sodium citrate | 459.47±40.41 | 881.25±50.39 (***) | 418.03±49.48 | 0.29±0.02 | 0.21±0.00 (***) | 0.29±0.03 |
| α-ketoglutaric acid | 118.37±2.28 | 189.04±5.73 (***) | 94.59±6.08 | 0.47±0.00 | 0.38±0.00 (***) | 0.47±0.00 |
| Sodium succinate | 162.74±8.19 | 156.12±7.60 | 137.71±3.75 | 0.25±0.00 | 0.21±0.00 (***) | 0.25±0.00 |
| Sodium malonate | - | - | - | - | - | - |
| Oxaloacetic acid | 91.18±0.67 | 86.65±2.41 | 71.24±0.59 | 0.24±0.00 | 0.21±0.00 (***) | 0.24±0.00 |
| Fumaric acid | 176.60±4.54 | 144.38±2.26 (***) | 150.06±4.40 | 0.42±0.00 | 0.38±0.00 (***) | 0.43±0.00 |
a *p < 0.05
**p < 0.01
***p <0.001.
To check the impact of hfq on the metabolism of amino acids, bacterial growth was measured in M63 with no added (NH4)2SO4 or D-glucose and supplemented with individual amino acids as both carbon and nitrogen sources. As shown in S3 Fig and in Table 3, deletion of hfq affected the ability of V. alginolyticus to grow on L-threonine or L-arginine, which were the preferred amino acids for the wild type strain, suggesting that hfq is essential for the utilization of these amino acids. In addition, absence of hfq affected cell growth on L-alanine, L-serine, L-histidine or L-leucine (S3 Fig, Table 3). Other tested amino acids could not support growth of either wild type or mutant strains (S3 Fig, Table 3).
Table 3. Effect of hfq deletion on growth rates and total growth in minimal medium M63 supplemented with various amino acids as both carbon and nitrogen sources.
Values correspond to the mean of three independent cultures. Significant statistical differences between strains and WT are indicated (*p < 0.05, **p < 0.01, ***p <0.001).
| Amino acid | Generation time (minutes) | Total growth (OD600) | ||||
|---|---|---|---|---|---|---|
| ZJ-T | Δhfq-T | hfq+-T | ZJ-T | Δhfq-T | hfq+-T | |
| L-alanine | 138.77±12.22 | 208.42±10.22 (***)a | 110.72±2.93 | 0.57±0.01 | 0.41±0.01 (***) | 0.59±0.01 |
| L-threonine | 249.71±10.75 | - (***) | 200.69±14.13 | 0.50±0.01 | -(***) | 0.53±0.01 |
| L-aspartic acid | - | - | - | - | - | - |
| L-lysine | - | - | - | - | - | - |
| L-glutamic acid | - | - | - | - | - | - |
| L-serine | 201.47±10.91 | 617.52±28.50 (***) | 170.73±2.43 | 0.39±0.01 | 0.10±0.01 (***) | 0.39±0.01 |
| L-arginine | 1616.96±93.16 | - (***) | 1686.25±165.43 | 0.29±0.05 | - (***) | 0.25±0.03 |
| L-histidine hydrochloride | 364.42±12.62 | 358.56±5.12 | 335.16±9.80 | 0.81±0.01 | 0.69±0.00 (***) | 0.82±0.01 |
| L-glutamine | - | - | - | - | - | - |
| L-leucine | 1578.13±146.45 | 3727.20±263.49 (***) | 1595.48±59.43 | 0.17±0.00 | 0.06±0.00 (***) | 0.20±0.04 |
| L-valine | - | - | - | - | - | - |
| L-isoleucine | - | - | - | - | - | - |
a *p < 0.05
**p < 0.01
***p <0.001.
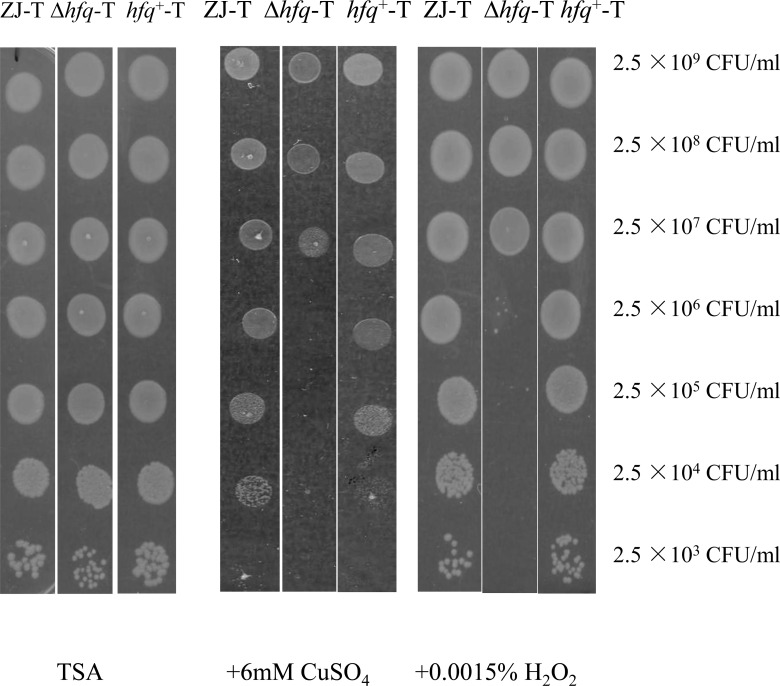
Deletion of hfq increased susceptibility of V. alginolyticus to copper and radical oxygen
Excessive amount of cupric ions are toxic to bacterial cells. Therefore, bacteria, including V. alginolyticus, have developed complex mechanisms to respond to this stressor [45, 46]. As shown in Fig 3, we observed a 1000 fold difference in survival rates between the hfq mutant and the WT on TSB agar plate in the presence of 6mM CuSO4. Excess of copper is toxic because cupric ions catalyze the formation of highly reactive hydroxyl radicals, generating a strong oxidative stress as does hydrogen peroxide. We found that the hfq mutant was also more sensitive to 0.0015% H2O2 (Fig 3). Our results indicate that hfq plays a vital role in responding to oxidative stress generated by radical oxygen.
Fig 3. Δhfq-T showed increased sensitivity to CuSO4 and H2O2.
Serial dilution of cultures of ZJ-T, Δhfq-T and hfq+-T were spotted on TSB agar plates (left panel), in presence of 6 mM CuSO4 (middle panel) or 0.0015% H2O2 (right panel). The picture is representative of the results of at least ten experiments.
Deletion of hfq altered antibiotic resistance of V. alginolyticus
Twenty-five antibiotic agents were used to determine the impact of hfq on antibiotic resistance. As shown in Table 4, all the strains show similar levels of resistance to lincomycin, clindamycin, ampicillin, oxacillin, piperacillin, penicillin-G, acetylspiramycin and amoxicillin. However, Δhfq-T strain was more sensitive, compared to the wild type and the complemented strains, to vancomycin and fosfomycin, both of which inhibit the synthesis of cell wall, and kanamycin, tobramycin as well as neomycin, which belong to the aminoglycoside family of antibiotics and inhibit protein synthesis.
Table 4. Antibiotic resistance of ZJ-T, △hfq-T and hfq+-T.
| Antibiotics | Concentration | Size of inhibition zone (mm) | p-value | ||
|---|---|---|---|---|---|
| (μg/per disk) | ZJ-T | Δhfq-T | hfq+-T | (Δhfq-T vs ZJ-T) a | |
| lincomycin | 2 | 0 | 0 | 0 | 1.00 |
| vancomycin | 30 | 8.80±1.30 | 13.40±0.89 | 8.40±1.52 | 0.00 |
| clindamycin | 2 | 0 | 0 | 0 | 1.00 |
| streptomycin | 300 | 12.60±1.52 | 14.80±1.92 | 12.67±2.08 | 0.24 |
| ampicillin | 10 | 0 | 0 | 0 | 1.00 |
| oxacillin | 1 | 0 | 0 | 0 | 1.00 |
| piperacillin | 100 | 0 | 0 | 0 | 1.00 |
| kanamycin | 30 | 11.80±0.84 | 14.40±1.52 | 11.40±1.14 | 0.00 |
| penicillin-G | 10 | 0 | 0 | 0 | 1.00 |
| cefazolin | 30 | 13.50±3.54 | 14.50±3.54 | 14.50±3.54 | 0.796 |
| tobramycin | 10 | 11.00±2.00 | 14.67±0.58 | 11.00±1.73 | 0.03 |
| neomycin | 30 | 14.67±1.15 | 17.00±1.00 | 14.67±1.15 | 0.04 |
| novobiocin | 30 | 19.00±3.46 | 21.00±3.61 | 18.33±2.31 | 0.34 |
| erythromycin | 15 | 16.00±1.00 | 17.67±3.06 | 16.33±1.53 | 0.36 |
| medemycin | 30 | 11.00±1.41 | 11.50±0.71 | 11.00±1.41 | 0.71 |
| polymyxin B | 300 | 13.50±2.12 | 14.00±1.41 | 12.50±2.12 | 0.81 |
| acetylspiramycin | 30 | 0 | 0 | 0 | 1.00 |
| spectinomycin | 100 | 12.50±0.71 | 11.00±1.41 | 10.50±2.12 | 0.40 |
| cefixime | 5 | 16.67±4.73 | 19.35±1.15 | 18.33±4.04 | 0.41 |
| amoxicillin | 10 | 0 | 0 | 0 | 1.00 |
| azithromycin | 15 | 17.00±1.00 | 19.67±2.52 | 18.00±1.00 | 0.11 |
| clarithromycin | 15 | 13.00±2.00 | 13.00±2.83 | 11.50±3.54 | 1.00 |
| teicoplanin | 30 | 8.50±0.71 | 10.00±0.00 | 8.50±0.71 | 0.08 |
| fosfomycin | 200 | 17.00±1.73 | 22.67±0.58 | 15.00±1.00 | 0.00 |
| gentamicin | 120 | 19.50±0.71 | 18.50±0.71 | 16.50±0.71 | 0.25 |
a There is no significant difference (p>0.05) between the wild type ZJ-T and the complemented strain hfq+-T.
Global effects of Hfq on V. alginolyticus transcriptome
Transcriptome sequencing of both the WT and the Δhfq strain by Illumina in early exponential phase (see Materials and Methods) was used to identify genes potentially regulated by hfq in V. alginolyticus. Of the 4710 protein coding genes annotated in the V. alginolyticus ZJ-T strain genome, 306 genes (approximately 6.5%) had their expression significantly affected in the mutant with our criteria (fold change ≥ 2, p-value ≤ 0.1 and a FDR ≤ 0.001) (S2 Table). Amongst them, 226 genes could be assigned to 24 KEGG functional categories based on their annotations.
Since Hfq typically regulates targets via the action of sRNAs, and that in some cases, interaction between the sRNA, Hfq and its target leads to degradation of both the target and the sRNA, one could expect some sRNA levels to be affected by the absence of Hfq. Accordingly, we identified 16 V. alginolyticus known sRNAs by sequence comparison with other vibrios and the sRNA database Rfam (http://rfam.xfam.org/)(S3 Table), and determine their expression levels in our transcriptomes. None of them filled out our criteria of statistical significance to be included in the list of differentially expressed genes, although some of them did show changes of expression, such as the qrr2-5 genes whose expression was undetectable in the hfq mutant (S3 Table).
Several categories of differentially expressed genes could be related to the phenotypes described in this study and are discussed below.
(1) Carbon uptake and central metabolism. As shown in S2 Table, 55 out of the 226 genes with predicted functions are involved in carbon utilization and energy conversion (Carbohydrate metabolism, Other carbon source utilization, Central and energy metabolism, and Carbon transport). Interestingly, the relative transcript abundances of the PTS system components II for trehalose, fructose, sucrose and mannitol were all decreased in the hfq mutant. In addition, genes involved in maltodextrin uptake (malGEFK) and conversion (malQ, glgP, glgX) as well as the regulatory gene malT were down regulated by 2–7 fold. Finally, the glycerol uptake facilitator genes glpF and glpK were also down regulated.
Conversely, many genes involved in glycolysis, TCA cycle and electron transport chains were up regulated in the hfq mutant. As shown in S2 Table, phosphoenolpyruvate carboxykinase (PckA) and phosphoenolpyruvate synthase (Pps), which convert oxaloacetate and pyruvate respectively to phosphoenolpyruvate, increased, whereas pyruvate kinase (Pyk) that catalyzes the reverse reaction of Pps was down regulated in the mutant. Other TCA cycle enzyme-encoding genes whose expression increased were fumA, fumC, sucD and sucC.
(2) Amino acids transport and metabolism. Growth of the hfq mutant on several amino acids was strongly affected compared to the WT (S3 Fig, Table 3). Our transcriptomic data suggest an increase of the biosynthesis pathways for glutamate in the mutant. Both genes for glutamate synthase (gltD and gltB), which synthesizes glutamate from glutamine, were up regulated in the hfq deletion background, and gamma-glutamyltranspeptidase (encoded by ggt), which converts glutathione to glutamate, was up regulated 3.7 fold. The valine/leucine/isoleucine biosynthetic pathway was also enhanced with an increase of 3-isopropylmalate dehydrogenase (leuBC) and ketol-acid reductoisomerase (ilvC). Conversely, genes such as fadB, puuB and ilvD, involved in valine/leucine/isoleucine degradation were down regulated.
Three genes encoding polar amino acid transporters of the general L-amino acid permease family (ABC transporters) showed decreased expression in the mutant. In Rhizobium leguminosarum, such ABC transporter, named Aap, has been shown to have a broad amino-acid specificity, importing not only polar amino acids such as glutamate, aspartate or histidine, but also more hydrophobic amino acids such as leucine and methionine [47]. V. alginolyticus encodes two such polar amino acid permeases, the two corresponding ATPase subunits being less expressed in the hfq mutant (aapP1 and aapP2) as well as one polar amino acid transport system substrate-binding protein. Interestingly, the Aap transporter could also export its substrates, for instance glutamate [47].
(3) Motility and chemotaxis. Motility is driven by polar flagella in most Vibrio species in liquid environment and is required for infection [48, 49]. In V. alginolyticus, hfq was found to be required for motility [14], although we could not detect any obvious defect in flagella by TEM (S1 Fig). In contrast, our transcriptomic study provided a molecular basis for this phenotype since, as shown in S2 Table, 11 genes involved in motility or chemotaxis were down regulated by at least two fold, such as the four fliC genes, that encode flagellin. In addition, two motor proteins MotA and MotB that provide driving forces for motility were down regulated in the mutant as well as three of the four methyl-accepting chemotaxis proteins (MCP) and the MCP methyltransferase CheB/CheR fusion protein [49].
Finally, in Vibrio species, four to five sRNAs according to species, named Qrrs, are important mediators of Quorum sensing. At low cell density, Qrrs are made and block the translation of the master regulator gene hapR mRNA, in an Hfq dependent manner [16]. At high cell density, no Qrrs are made leading to expression of HapR. HapR is both a negative and positive transcriptional regulator and controls a number of genes, varying according to species, which are responding to the QS pathway [17]. We identified five Qrrs in V. alginolyticus, which showed very low expression in our transcriptomes (S3 Table). However, we could not detect any expression of Qrr2-4 in the Δhfq mutant. Consistent with this, we found that the hapR gene (BAU10_12125) was de-repressed 2.6 fold in the mutant.
Hfq regulates OMP synthesis in V. alginolyticus
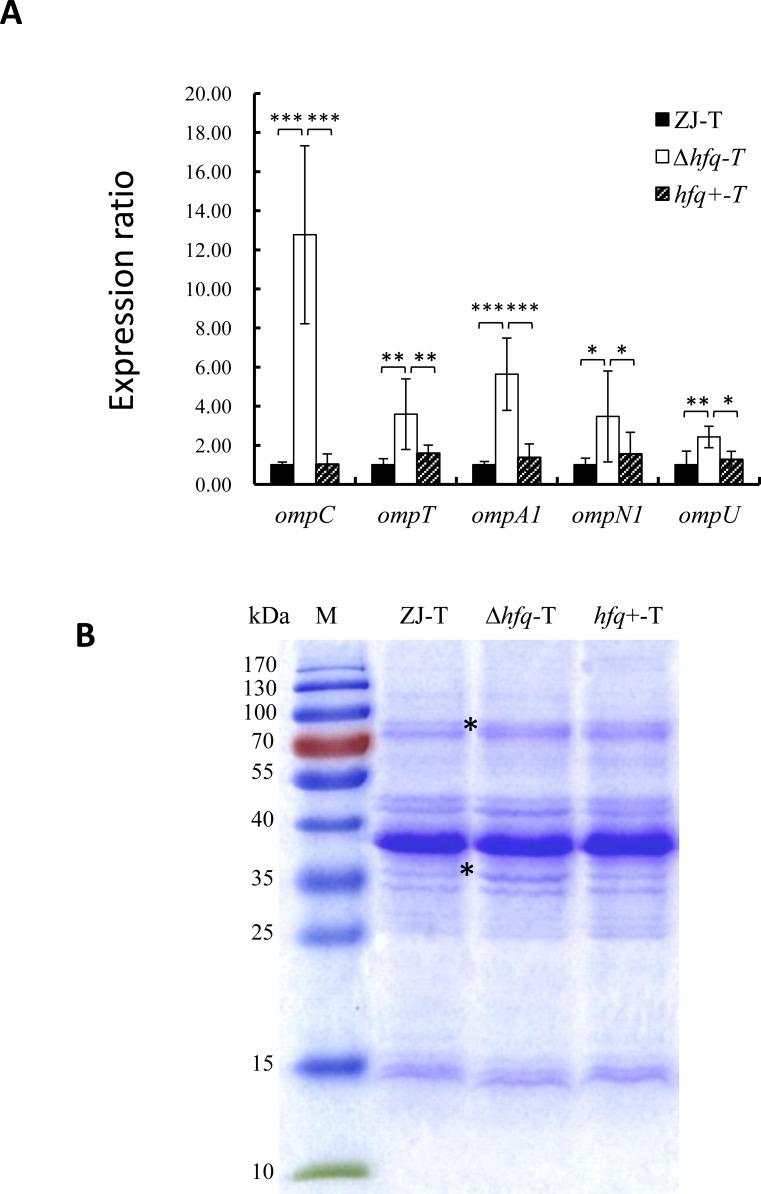
Most outer membrane β-barrel proteins (OMPs) including OmpC, OmpT, OmpA, OmpN and OmpU were significantly up regulated in the hfq mutant, whereas second copies of ompN and ompA were down regulated (S2 Table). Indeed, ompC was the second most up regulated (65.8 X) gene in our data set, (although its expression level in the WT was 2000 fold less than this of ompN1/ ompN2 together) and ompT was the fourth. This up-regulation of several OMP genes was further confirmed by qPCR (Fig 4A) and suggests that the outer membrane composition of V. alginolyticus is remodeled in absence of hfq. To further test this possibility, OMPs were extracted from strains ZJ-T, Δhfq-T and hfq+-T and were analyzed by SDS-PAGE. As shown in Fig 4B, a major band was observed at around the 38 kDa position, consistent with the high level of expression (data not shown) and the sizes of OmpN1 and OmpN2. The main difference between the WT and the mutant was the increase of a minor band corresponding to a size around 35 kDa, that could correspond to the less expressed OmpC. Finally, another minor band also increased in the 75 kDa region, which could correspond to one or several outer-membrane-localized iron receptor proteins in this size range (BAU10_23175, BAU10_12590, BAU10_23170), which were up regulated from 4 to 10 fold in our study (S2 Table).
Fig 4. Hfq regulates outer membrane protein expression.
A. Relative expression of OMP genes in derivative strains compared to WT. Relative expression (normalized to the WT level for each gene) was determined by qPCR as described in Materials and Methods. Error bars correspond to standard deviations from three biological replicates. Statistically significant differences are indicated (*p < 0.05, **p < 0.01, ***p <0.001). B. SDS-PAGE analysis of the outer membrane fraction from ZJ-T, Δhfq-T and hfq+-T. M: Molecular size markers. Apparent sizes of markers are indicated on the left. The positions of two bands that increase in the hfq mutant are indicated by stars.
Discussion
It has been reported that approximately 50% of bacterial genomes carry homologs of hfq [50]. As an RNA molecular chaperone, Hfq exerts its function at the post-transcriptional level mainly by being required for the action of up to hundreds of regulatory non-coding sRNAs, the majority of them still uncharacterized. Hence, loss of Hfq often results in pleiotropic phenotypes in many bacteria [12, 13, 35, 51–55], vibrios not being exceptions [14, 21, 23]. In V. alginolyticus, Hfq had been previously shown to be required for motility, biofilm formation and resistance to various stress such as osmotic stress, ethanol and iron starvation [14]. The mutant was also found to be attenuated in virulence to zebra fish [14]. In here, we describe some additional phenotypes of the hfq mutant: colony morphology changes (Fig 2A), decrease in EPS production (Fig 2B), impaired growth on a range of carbon and nitrogen sources (Tables 2 and 3) and increased sensitivity to copper and hydrogen peroxide (Fig 3). In addition, a differential transcriptomic study showed that deletion of hfq led to altered expression of more than three hundred genes, with several of them that could be related to some of the phenotypes of the mutant.
A first obvious phenotype caused by hfq deletion is the colony appearance that becomes rugose and opaque instead of smooth and translucent in the mutant, with the complementation strain becoming S/Tr again (Fig 2A) indicating that the phenotype is due to the lack of Hfq and not to a polar effect of the deletion and/or secondary mutations in the mutant. The colony morphology of Vibrio species often depends on the amount and the nature of extracellular polysaccharides produced by the cells [56, 57]. Opaque colony formation has been linked to an increase of expression of one of two EPS gene clusters in V. cholerae and V. parahaemolyticus [56, 58], a cluster that is absent in V. alginolyticus. In V. vulnificus, another cluster, named brp was shown to be important for rugose colony appearance [44]. These findings underscore the complexity of the relationship between colony aspect and exopolysaccharides. In our case, the opaque/rugose mutant had globally less EPS than the WT. Although, no genes linked to EPS production or secretion were identified as significantly affected by the hfq deletion in our transcriptomic study (S2 Table), we could identify two cluster of EPS genes that were differentially expressed. In the first one, out of eleven genes, 4 were up regulated 2–4 fold in the mutant, whereas in the second one, 5 out of 10 genes were down regulated 2–4 fold (data not shown). However, these genes were not included in our list in S2 Table because of an FDR higher than our cutoff of 0.001. Clearly, more studies will be required to understand the molecular basis of colony appearance in V. alginolyticus.
Deficiency in carbon and amino acids uptake and utilization caused by loss of hfq were extensively reported in both gram-positive and gram-negative bacteria [35, 52–54]. In this study, we found that a V. alginolyticus hfq mutant displayed several growth defects compared to the WT strain: in rich medium, it displayed an increased lag phase and reached a lower OD600 (S2 Fig) possibly reflecting a reduction in growth yield. Such reduction was also observed when the cells were growing on PTS sugars such as glucose, trehalose, fructose or N-acetyl glucosamine, as well as non-PTS sugars such as maltose (S2 Fig). Consistent with such phenotypes, we found that genes encoding PTS sugar transporter component II and genes involved in maltose transport and utilization had decreased expression. The mutant was also less efficient in using TCA cycle intermediates as carbon sources (S2 Fig) and, at the same time, a large portion of genes involved in glycolysis, TCA cycle and electron transfer chain had increased expression (S2 Table) in contrast to prior study in other bacterial species [54, 59]. The hfq mutant was also less able than the WT to grow on a variety of amino acids, both polar and hydrophobic. Decreased expression of two broad-specificity amino-acid ABC transporters (S2 Table) could contribute to these phenotypes. Finally, the mutant has increased biosynthesis of branched amino acids and glutamate, suggesting an accumulation of glutamate in the mutant. Reduction of the glutamate efflux by the homologues of the Aap transporter would also contribute to such accumulation. All together, deletion of hfq led to a general decrease of V. alginolyticus capability to utilize externally provided compounds, and an increase of central metabolism.
In keeping with decreased motility, and despite the fact we could not observe any apparent change in flagellar structure (S1 Fig), as many as 11 motility and chemotaxis associated genes (including several methyl accepting chemotaxis proteins, S2 Table) had reduced expression in the mutant, suggesting that motility reduction is linked to a decrease in chemotactic signaling and energy production, rather than to a direct regulation of flagellum biogenesis, although fliC genes, which encode flagellin, the subunit of the flagellum, were also less expressed. Decreased motility and EPS production may in turn contribute to less biofilm formation [57, 60].
The hfq mutant also displayed increased sensitivity to vancomycin, tobramycin, kanamycin, neomycin and fosfomycin, amongst 25 tested antibiotics (Table 4). These antibiotics do not belong to the same family (3 are aminoglycosides, fosfomycin is a phosphonic acid, and vancomycin is a glycopeptide), they don't have the same targets neither the same entrance mechanism in the cell. Hence, it is unlikely that Hfq plays a role in the resistance to these antibiotics through a unique mechanism. However, one contributing factor could be the role of Hfq in controlling the abundance of OMPs. Hydrophilic antibiotics are thought to pass the outer membrane barrier through porins. The fact that a majority of OMPs were significantly up regulated in the hfq mutant could contribute to the increased sensitivity to at least some of these antibiotics [61, 62]. Such changes could also contribute to the observed ECS response, as attested by the induction of the ECS sigma factor RpoE, as well as member of its regulon, such as degQ, rseA, or smpA [63](S2 Table).
In E. coli, level of OMPs is regulated by many Hfq dependent sRNAs [64]. In V. cholerae, two sRNAs that regulate OMP expression have been identified, VrrA and MicX. VrrA is transcribed by the ECS sigma factor RpoE, and represses the production of OmpT in an Hfq dependent way [22]. MicX was shown to repress the ompC gene in an hfq independent manner, whereas the primary transcript of MicX is processed by RNaseE in an hfq dependent fashion to a shorter transcript [65]. However, we were not able to identify significant changes through our transcriptomic approach neither of VrrA nor MicX levels. Further studies will be required to determine if these sRNAs or others, not yet identified, play a role in the observed changes.
Another sigma factor which, in E. coli, is regulated by many sRNAs is the stationary phase and stress response sigma factor RpoS [66, 67]. Maybe surprisingly, rpoS expression was not affected in the mutant. One possible reason could be the absence of significant expression of rpoS in our conditions (early exponential phase) preventing the detection of a significant difference between the wild type and the mutant.
Finally, in vibrios, conserved Hfq dependent sRNAs named qrr (four to five copies according to species) are negative regulators at low cell density of hapR, which encodes the master regulator of the quorum sensing system [17]. As could be expected, V. alginolyticus hapR expression increased 2.6 fold in the mutant (S2 Table). This increase of HapR in exponential phase, possibly linked to the loss of regulation by Qrrs in the hfq mutant could also contribute to the observed decrease of EPS and biofilm formation since in vibrios, HapR is a negative regulator of these processes [68].
In summary, our study once again points at the importance of posttranscriptional regulations mediated by sRNAs with the assistance of the RNA chaperone Hfq for tuning a wide range of functions in bacterial cells [69]. Undoubtedly, numerous sRNAs remain to be discovered in V. alginolyticus to account for the pleiotropic phenotypes of the hfq mutant.
Supporting Information
Magnification; ZJ-T: 2 500 x, Δhfq-T: 4 000 x and hfq+-T: 2 500 x .tif file. Magnification was adjusted for each strain to make a single cell and its flagella fill out the field of vision.
(TIF)
ZJ-T, Δhfq-T and hfq+-T were cultured in triplicates in TSB or M63 minimal medium supplemented with 0.4% (w/v) of the indicated saccharide or TCA cycle intermediates as sole carbon source. (Squares: ZJ-T, circles: Δhfq-T, triangles: hfq+-T). Indicated values correspond to the mean of three measurements, and error bars to standard deviations from three biological replicates
(TIF)
ZJ-T, Δhfq-T and hfq+-T were cultured in triplicates in M63 minimal medium minus D-glucose and (NH4)2SO4 and supplemented with the indicated amino acid as both carbon and nitrogen source. (squares: ZJ-T, circles: Δhfq-T, triangles: hfq+-T). Indicated values correspond to the mean of three measurements, and error bars to standard deviations from three biological replicates
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank the Public Equipment Service Center, SCSIO for giving us access to the Optima XPN-100 Ultracentrifuge and to Dr. Yongli Gao for kindly operating the centrifuge for us. We are also grateful to the anonymous reviewers for helpful comments and suggestions to improve the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31272697), http://www.nsfc.gov.cn/ to CC, The National Natural Science Foundation of China (41276163), http://www.nsfc.gov.cn/ to ZZ, The Project of Science and Technology New Star of Zhujiang in Guangzhou city (2013J2200094), http://www.gzsi.gov.cn/ to ZZ and the Science and Technology Planning Project of Guangdong Province, China (2014B030301064), http://www.gdstc.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Citil BE, Derin S, Sankur F, Sahan M, Citil MU. Vibrio alginolyticus Associated Chronic Myringitis Acquired in Mediterranean Waters of Turkey. Case reports in infectious diseases. 2015;2015:187212 10.1155/2015/187212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee DY, Moon SY, Lee SO, Yang HY, Lee HJ, Lee MS. Septic shock due to Vibrio alginolyticus in a cirrhotic patient: the first case in Korea. Yonsei medical journal. 2008;49(2):329–32. 10.3349/ymj.2008.49.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui R, Alam M, Naser M, Otomo Y, Yasmin M, Nessa J, et al. Prevalence of Vibrio alginolyticus in Sediment Samples of River and Coastal Areas of Bangladesh. 2012.
- 4.Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;54 Suppl 5:S391–5. 10.1093/cid/cis243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SAKAZAKI R. Proposal of Vibrio alginolyticus for the biotype 2 of Vibrio parahaemolyticus. Japanese Journal of Medical Science and Biology. 1968;21(5):359–62. 10.7883/yoken1952.21.359 [DOI] [PubMed] [Google Scholar]
- 6.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: regulators of bacterial virulence. Nature Reviews Microbiology. 2010;8(12):857–66. 10.1038/nrmicro2457 [DOI] [PubMed] [Google Scholar]
- 7.Liu JM, Camilli A. A broadening world of bacterial small RNAs. Current opinion in microbiology. 2010;13(1):18–23. 10.1016/j.mib.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. Journal of Biological Chemistry. 2013;288(12):7996–8003. 10.1074/jbc.R112.441386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10(4):610–8. 10.4161/rna.24201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner EG, Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet. 2015;90:133–208. 10.1016/bs.adgen.2015.05.001 . [DOI] [PubMed] [Google Scholar]
- 11.Sonnleitner E, Schuster M, Sorger‐Domenigg T, Greenberg EP, Bläsi U. Hfq‐dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Molecular microbiology. 2006;59(5):1542–58. 10.1111/j.1365-2958.2006.05032.x [DOI] [PubMed] [Google Scholar]
- 12.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4(8):e1000163 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, et al. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One. 2009;4(3):e4809 10.1371/journal.pone.0004809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Wang Q, Liu Q, Cao X, Shi C, Zhang Y. Roles of Hfq in the stress adaptation and virulence in fish pathogen Vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Applied microbiology and biotechnology. 2011;91(2):353–64. Epub 10.1007/s00253-011-3286-3 [DOI] [PubMed] [Google Scholar]
- 15.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10(2):156–63. 10.1016/j.mib.2007.03.007 . [DOI] [PubMed] [Google Scholar]
- 16.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118(1):69–82. 10.1016/j.cell.2004.06.009 [DOI] [PubMed] [Google Scholar]
- 17.Nguyen A, Jacq A. Small RNAs in the Vibrionaceae: an ocean still to be explored. Wiley Interdisciplinary Reviews: RNA. 2014;5(3):381–92. 10.1002/wrna.1218 [DOI] [PubMed] [Google Scholar]
- 18.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58(4):1186–202. 10.1111/j.1365-2958.2005.04902.x . [DOI] [PubMed] [Google Scholar]
- 19.Bardill JP, Hammer BK. Non-coding sRNAs regulate virulence in the bacterial pathogen Vibrio cholerae. RNA biology. 2012;9(4):392–401. 10.4161/rna.19975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bardill JP, Zhao X, Hammer BK. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol Microbiol. 2011;80(5):1381–94. 10.1111/j.1365-2958.2011.07655.x . [DOI] [PubMed] [Google Scholar]
- 21.1106 Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. 2004;53(1):345–54. 10.1111/j.1365-2958.2004.04142.x . [DOI] [PubMed] [Google Scholar]
- 22.Song T, Sabharwal D, Wai SN. VrrA mediates Hfq-dependent regulation of OmpT synthesis in Vibrio cholerae. J Mol Biol. 2010;400(4):682–8. 10.1016/j.jmb.2010.05.061 . [DOI] [PubMed] [Google Scholar]
- 23.Nakano M, Takahashi A, Su Z, Harada N, Mawatari K, Nakaya Y. Hfq regulates the expression of the thermostable direct hemolysin gene in Vibrio parahaemolyticus. BMC microbiology. 2008;8:155 10.1186/1471-2180-8-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su Z, Nakano M, Koga T, Lian X, Hamamoto A, Shimohata T, et al. Hfq regulates anti-oxidative ability in Vibrio parahaemolyticus. J Gen Appl Microbiol. 2010;56(3):181–6. 10.2323/jgam.56.181 . [DOI] [PubMed] [Google Scholar]
- 25.Xie ZY, Hu CQ, Chen C, Zhang LP, Ren CH. Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Lett Appl Microbiol. 2005;41(2):202–7. 10.1111/j.1472-765X.2005.01688.x . [DOI] [PubMed] [Google Scholar]
- 26.Chang C, Jin X, Chaoqun H. Phenotypic and genetic differences between opaque and translucent colonies of Vibrio alginolyticus. Biofouling. 2009;25(6):525–31. 10.1080/08927010902964578 [DOI] [PubMed] [Google Scholar]
- 27.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. Journal of bacteriology. 1988;170(6):2575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon R, Priefer U, A. P. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotechnology. 1983;1:784–91. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 29.Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. Journal of bacteriology. 1996;178(5):1310–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J-j, Chen C, Zhang L-p, Hu C-q. Cloning, identification, and characterization of the rpoS-like sigma factor rpoX from Vibrio alginolyticus. BioMed Research International. 2009;2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber B, Croxatto A, Chen C, Milton DL. RpoS induces expression of the Vibrio anguillarum quorum-sensing regulator VanT. Microbiology. 2008;154(Pt 3):767–80. 10.1099/mic.0.2007/014167-0 . [DOI] [PubMed] [Google Scholar]
- 32.Zhao Z, Zhang L, Ren C, Zhao J, Chen C, Jiang X, et al. Autophagy is induced by the type III secretion system of Vibrio alginolyticus in several mammalian cell lines. Archives of microbiology. 2011;193(1):53–61. 10.1007/s00203-010-0646-9 [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 34.Croxatto A, Lauritz J, Chen C, Milton DL. Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environmental microbiology. 2007;9(2):370–82. 10.1111/j.1462-2920.2006.01147.x [DOI] [PubMed] [Google Scholar]
- 35.Boudry P, Gracia C, Monot M, Caillet J, Saujet L, Hajnsdorf E, et al. Pleiotropic role of the RNA chaperone protein Hfq in the human pathogen Clostridium difficile. Journal of bacteriology. 2014;196(18):3234–48. 10.1128/JB.01923-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–7. 10.1093/bioinformatics/btp336 [DOI] [PubMed] [Google Scholar]
- 37.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 38.Audic S, Claverie J-M. The significance of digital gene expression profiles. Genome research. 1997;7(10):986–95. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–88. . [Google Scholar]
- 40.Chen C, Wang QB, Liu ZH, Zhao JJ, Jiang X, Sun HY, et al. Characterization of role of the toxR gene in the physiology and pathogenicity of Vibrio alginolyticus. Antonie van Leeuwenhoek. 2012;101(2):281–8. 10.1007/s10482-011-9632-8 . [DOI] [PubMed] [Google Scholar]
- 41.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proceedings of the National Academy of Sciences. 1999;96(7):4028–33. 10.1073/pnas.96.7.4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fong JC, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189(6):2319–30. 10.1128/JB.01569-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatzidaki-Livanis M, Jones MK, Wright AC. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. Journal of bacteriology. 2006;188(5):1987–98. 10.1128/JB.188.5.1987-1998.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y, Rowe-Magnus DA. Identification of a c-di-GMP-regulated polysaccharide locus governing stress resistance and biofilm and rugose colony formation in Vibrio vulnificus. Infect Immun. 2010;78(3):1390–402. 10.1128/IAI.01188-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon AS, Harwood VJ, Sayyar S. Growth, copper-tolerant cells, and extracellular protein production in copper-stressed chemostat cultures of Vibrio alginolyticus. Appl Environ Microbiol. 1993;59(1):60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harwood VJ, Gordon A. Regulation of extracellular copper-binding proteins in copper-resistant and copper-sensitive mutants of Vibrio alginolyticus. Applied and environmental microbiology. 1994;60(6):1749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walshaw DL, Poole PS. The general l‐amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Molecular microbiology. 1996;21(6):1239–52. [DOI] [PubMed] [Google Scholar]
- 48.Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. International Journal of Medical Microbiology. 2002;291(8):605–14. 10.1078/1438-4221-00173 [DOI] [PubMed] [Google Scholar]
- 49.Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS microbiology reviews. 2003;27(4):505–23. 10.1016/s0168-6445(03)00064-0 [DOI] [PubMed] [Google Scholar]
- 50.Feliciano JR, Grilo AM, Guerreiro SI, Sousa SA, Leitão JH. Hfq: a multifaceted RNA chaperone involved in virulence. Future microbiology. 2016;11(1):137–51. 10.2217/fmb.15.128 [DOI] [PubMed] [Google Scholar]
- 51.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proceedings of the National Academy of Sciences. 2011;108(3):1110–5. 10.1073/pnas.1010082108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui M, Wang T, Xu J, Ke Y, Du X, Yuan X, et al. Impact of Hfq on global gene expression and intracellular survival in Brucella melitensis. PloS one. 2013;8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang M-K, Lu M-C, Liu L-C, Lin C-T, Lai Y-C. Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae. PLoS One. 2011;6(7):e22248 10.1371/journal.pone.0022248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Möller P, Overlöper A, Förstner KU, Wen T-N, Sharma CM, Lai E-M, et al. Profound impact of Hfq on nutrient acquisition, metabolism and motility in the plant pathogen Agrobacterium tumefaciens. 2014. 10.1371/journal.pone.0110427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arce-Rodriguez A, Calles B, Nikel PI, de Lorenzo V. The RNA chaperone Hfq enables the environmental stress tolerance super-phenotype of Pseudomonas putida. Environmental microbiology. 2015. 10.1111/1462-2920.13052 . [DOI] [PubMed] [Google Scholar]
- 56.Enos-Berlage JL, McCarter LL. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. Journal of Bacteriology. 2000;182(19):5513–20. 10.1128/jb.182.19.5513-5520.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends in microbiology. 2009;17(3):109–18. 10.1016/j.tim.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali A, Johnson JA, Franco AA, Metzger DJ, Connell TD, Morris JG, et al. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infection and immunity. 2000;68(4):1967–74. 10.1128/iai.68.4.1967-1974.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kakoschke T, Kakoschke S, Magistro G, Schubert S, Borath M, Heesemann J, et al. The RNA chaperone Hfq impacts growth, metabolism and production of virulence factors in Yersinia enterocolitica. PLoS One. 2014;9(1):e86113 10.1371/journal.pone.0086113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davey ME, O'Toole G A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–67. 10.1128/mmbr.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin J, Huang S, Zhang Q. Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes and infection. 2002;4(3):325–31. 10.1016/s1286-4579(02)01545-9 [DOI] [PubMed] [Google Scholar]
- 62.Kumar A, Schweizer HP. Bacterial resistance to antibiotics: active efflux and reduced uptake. Advanced drug delivery reviews. 2005;57(10):1486–513. 10.1016/j.addr.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 63.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, et al. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 2009;5(9):e1000651 10.1371/journal.pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Current opinion in microbiology. 2006;9(6):605–11. 10.1016/j.mib.2006.10.006 [DOI] [PubMed] [Google Scholar]
- 65.Davis BM, Waldor MK. RNase E-dependent processing stabilizes MicX, a Vibrio cholerae sRNA. Mol Microbiol. 2007;65(2):373–85. 10.1111/j.1365-2958.2007.05796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henderson CA, Vincent HA, Casamento A, Stone CM, Phillips JO, Cary PD, et al. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA. 2013;19(8):1089–104. 10.1261/rna.034595.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Updegrove TB, Wartell RM. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA• mRNA duplex formation in rpoS riboregulation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2011;1809(10):532–40. 10.1016/j.bbagrm.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 68.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Molecular microbiology. 2003;50(1):101–4. 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- 69.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nature Reviews Microbiology. 2011;9(8):578–89. 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Magnification; ZJ-T: 2 500 x, Δhfq-T: 4 000 x and hfq+-T: 2 500 x .tif file. Magnification was adjusted for each strain to make a single cell and its flagella fill out the field of vision.
(TIF)
ZJ-T, Δhfq-T and hfq+-T were cultured in triplicates in TSB or M63 minimal medium supplemented with 0.4% (w/v) of the indicated saccharide or TCA cycle intermediates as sole carbon source. (Squares: ZJ-T, circles: Δhfq-T, triangles: hfq+-T). Indicated values correspond to the mean of three measurements, and error bars to standard deviations from three biological replicates
(TIF)
ZJ-T, Δhfq-T and hfq+-T were cultured in triplicates in M63 minimal medium minus D-glucose and (NH4)2SO4 and supplemented with the indicated amino acid as both carbon and nitrogen source. (squares: ZJ-T, circles: Δhfq-T, triangles: hfq+-T). Indicated values correspond to the mean of three measurements, and error bars to standard deviations from three biological replicates
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.