Abstract
In this study, quantitative structure activity relationship (QSAR) models for the antioxidant activity of polysaccharides were developed with 50% effective concentration (EC50) as the dependent variable. To establish optimum QSAR models, multiple linear regressions (MLR), support vector machines (SVM) and artificial neural networks (ANN) were used, and 11 molecular descriptors were selected. The optimum QSAR model for predicting EC50 of DPPH-scavenging activity consisted of four major descriptors. MLR model gave EC50 = 0.033Ara-0.041GalA-0.03GlcA-0.025PC+0.484, and MLR fitted the training set with R = 0.807. ANN model gave the improvement of training set (R = 0.96, RMSE = 0.018) and test set (R = 0.933, RMSE = 0.055) which indicated that it was more accurately than SVM and MLR models for predicting the DPPH-scavenging activity of polysaccharides. 67 compounds were used for predicting EC50 of the hydroxyl radicals scavenging activity of polysaccharides. MLR model gave EC50 = 0.12PC+0.083Fuc+0.013Rha-0.02UA+0.372. A comparison of results from models indicated that ANN model (R = 0.944, RMSE = 0.119) was also the best one for predicting the hydroxyl radicals scavenging activity of polysaccharides. MLR and ANN models showed that Ara and GalA appeared critical in determining EC50 of DPPH-scavenging activity, and Fuc, Rha, uronic acid and protein content had a great effect on the hydroxyl radicals scavenging activity of polysaccharides. The antioxidant activity of polysaccharide usually was high in MW range of 4000–100000, and the antioxidant activity could be affected simultaneously by other polysaccharide properties, such as uronic acid and Ara.
Introduction
In our normal metabolism process, oxygen free radicals and non-oxygen free radicals are continuously produced, and lower concentrations of free radical can play a crucial role in regular physiological functions [1–5]. However, many diseases, such as cardiovascular diseases, diabetes, aging and cancer, can be conducted by unregulated overproduction of free radicals [6–8]. Thus, it is essential to develop natural and effective antioxidants [9]. Previously reports revealed that many natural polysaccharides possess potent scavenging activities of free radicals and can be used as potential antioxidants [10–11]. It is always impossible to obtain a large quantity of experimental data because of a lack of perfect data sites, and so the study on relationship between bioactivities and the properties of polysaccharides by model forecast approach was relatively poor [12].
The quantitative structure-activity relationship (QSAR) model, which use relevant molecular physico-chemical properties to predict important treatment responses, is considered as an alternative to the experimental evaluation [13]. It has gained increasingly attention, and a variety of QSAR methods have been developed for water treatment process selection, membrane separation and adsorption etc [14–15].
To date, QSAR models for predicting the bioactivities of polysaccharides have seldom been developed. A study reported the relationship between monosaccharide composition ratio and macrophage stimulatory activity by model forecast approach [12]. To obtain theoretical supports for applications of polysaccharides from natural products, the main aim of this work was to establish reliable soft measurement models to predict performance and study the relationship between polysaccharide properties and antioxidant activities of polysaccharides by QSAR. In our QSAR studies, multiple linear regression (MLR) method, and the nonlinear methods including artificial neural network (ANN) and support vector machine (SVM) were used.
Materials and Methods
Data set
The present study showed that the antioxidant activity of polysaccharide has related with many factors, including monosaccharide composition [16], uronic acid (UA), molecular weight (MW), protein content (PC) and sulfate group content et al [17]. In the data selection, we chose natural purified polysaccharides without sulfate groups to study QSAR models for predicting antioxidant activities of polysaccharides. A various set of polysaccharides and their antioxidant activities were collected from different published papers [18–45]. Antioxidant activities of polysaccharides were represented by the 50% effective concentration (EC50). To set up a more reliable model, we selected 141 compounds. The detailed publication lists with corresponding antioxidant activities and compounds were given. The normalization process was adopted in the distribution of the parameters with 2 as the bottom of the log logarithm, and MW was divided by 10000 in the normalization process.
In models, a training data set was applied to develop the model. A test set, which was never included during their development, was used to validate the predictive power of model [46–47]. The training set and test set were chosen by random distribution.
Descriptors
The structure of polysaccharide was complex and could be represented by variety of descriptors. However, the major composition of polysaccharide was monosaccharide joined together by glycosidic bonds, which was essential to their bioactivities, so we used monosaccharide composition as descriptors. The following descriptors of monosaccharide composition were considered for modeling EC50 values in MLR, ANN and SVM analysis. Descriptors of monosaccharide composition: rhamnose (Rha), arabinose (Ara), mannose (Man), glucose (Glc), galactose (Gal), fucose (Fuc), xylose (Xyl), ribose (Rib), glucuronic acid (GlcA) and galacturonic acid (GalA). Usually, gas chromatography (GC) and high-performance liquid chromatography (HPLC) were performed for the identification and quantification of monosaccharide composition. For HPLC analysis, glucuronic acid (GlcA) and galacturonic acid (GalA) could not be identified. Thus, total uronic acid (UA) could be determined by other methods, such as the sulfuric acid carbazole method, and then UA was also used as a descriptor in our models. The descriptors of PC and MW were also adopted in models. STATISTIC.10 method was used to establish SVM, MLP and ANN models, and the picture was drawn by using RStudio (Version 0.99.902–2009–2016 RStudio, Inc.).
Linear model generation
There were primarily two different approaches for choosing a descriptor subset in MLR, and they were filter and wrapper methods. The procedure of filter method was that setting and filtering descriptors were supposed to generate the top priority subset before training. However, the learning algorithm was wrapped into the selection procedure in the wrapper method [48]. In MLR, we used wrapper as the target learning algorithm. The training data set was applied only for selecting descriptor. At first, we employed a two-dimensional research method. It was a combination of forward and backward search. Then we assessed the selected descriptors on the target learning algorithm. In the learning process, we used 10 fold cross validation method. In stepwise MLR analysis, we selected training descriptor sets and then established a linear model [49].
Artificial neural network and Support vector machines
It was appropriate for artificial neural network (ANN) to model nonlinear relationship. We can find many reviews about ANN research and its application in QSAR studies [49–51]. In this study, we employed multi-layer perceptron (MLP) [52] and three layer reverse Back-Propagation (BP) network. In the back-propagation ANN, we utilized the technique of supervised learning, and the trained network was trained by minimizing the squared error of the network’s output. The first step of training model was to confirm the number of layers and neurons in each player. The second step was to optimize the learning rate as well as momentum parameters. In the input layer, the architecture of the network was composed of eleven neurons, which were the eleven relative descriptors chosen. In the output layer, there was one neuron, i.e. EC50 values of the antioxidant activity. In all the layers, logistic function was applied. In the hidden layer, through changing the number of neurons, we got the lowest RMSE and highest correlation coefficient. We applied 30% of the training data set for verification. The verification was employed to hinder from the over fitting. All of optimization process were taken with 10 fold cross validation [53].
Support vector machines (SVM) was originally developed for the classification problem, and SVM has been used to solve nonlinear regression estimation. Nowadays, SVM has demonstrated much success in QSAR and quantitative structure-property relationship (QSPR) studies [54–57]. We selected support vector machine classifier method (epsilon-SVM) which was most commonly used in QSPR and QSAR studies to optimize the value of kernel parameter g (gamma) [53].
Validation techniques and model performance evaluation
We used a 10 fold cross validation technique. This procedure divided the data set into 10 folds or groups, created the model using 9 of the sets, and tested it on the remaining group. When the procedure was repeated, each of the 10 groups had served as a test group. The root mean square error (RMSE) was calculated, averaged, and then used to evaluate the predictive performance of three models.
Results and Discussion
Models for the DPPH scavenging activity of polysaccharides
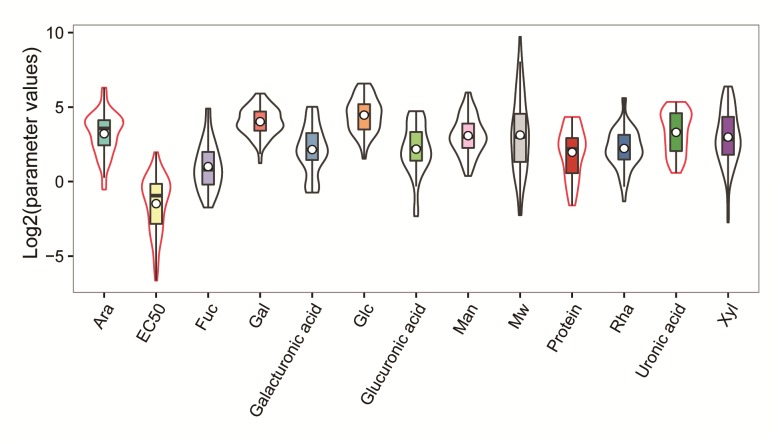
The data was divided into two parts using random classification. One was the training set, the other was the test set. The entire data set including 74 compounds was divided into two clusters. The test set of 22 compounds was chosen randomly from this cluster, and the remaining compounds were used as the train set. Compound number 4, 5, 7, 9, 17, 20, 25, 30, 31, 32, 33, 34, 36, 38, 44, 54, 57, 62, 63, 64, 69,73 were selected as the test set, and the rest of the compounds were the train set. The test set and train set were given in Table 1. The data distribution of parameter was shown in Fig 1, the data distribution was uniform, and no other single variable values was close to EC50 values distribution (-6, 2). The shape of data distribution from EC50 and Ara was similar, which indicated that there was a certain relation between them. In addition to MW, other physical quantities were all the components of polysaccharides, so MW was used to establish the model by itself.
Table 1. Polysaccharides data set with descriptors and EC50 values of the DPPH scavenging activity.
| No | Namea | Rhab | Arac | Mand | Glce | Galf | Fucg | Xylh | GlcAi | GalAj | UAk | PCl | EC50 | Refs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | S1 (Glu) | 0 | 0 | 22.13 | 28.33 | 13.89 | 0 | 31.48 | 4.03 | 0 | 4.03 | 0 | 1.104 | 18 |
| 2 | S1 (Visco) | 0 | 0 | 63.01 | 10.37 | 16.06 | 0 | 4.7 | 5.87 | 0 | 5.87 | 0 | 0.794 | 18 |
| 3 | GBP50S2 | 46.7 | 0 | 42.2 | 0 | 11.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.412 | 19 |
| 4 | LBP-s80 | 0 | 62.9 | 13.2 | 2.9 | 12.5 | 3.8 | 4.7 | 0 | 0 | 17.07 | 0.69 | 2.97 | 20 |
| 5 | LBP-s75 | 0 | 56.8 | 19.2 | 3.6 | 10.2 | 4.9 | 5.3 | 0 | 0 | 35.69 | 1.3 | 1.98 | 20 |
| 6 | WB1 | 0 | 0 | 12.5 | 59.4 | 9.76 | 11.7 | 6.64 | 0 | 0 | 0 | 0 | 0.12 | 21 |
| 7 | WB2 | 0 | 0 | 7.7 | 26 | 24.7 | 28.4 | 13.2 | 0 | 0 | 0 | 0 | 0.31 | 21 |
| 8 | WB3 | 0 | 0 | 9.7 | 18.6 | 24.5 | 29.9 | 17.3 | 0 | 0 | 0 | 0 | 0.21 | 21 |
| 9 | IOP40 | 4.4 | 4.2 | 9.8 | 40 | 14.5 | 3.3 | 9.6 | 9.7 | 4.6 | 5 | 2.4 | 0.88 | 22 |
| 10 | IOP60 | 9.7 | 5.6 | 8 | 32.2 | 12.6 | 0 | 22.9 | 4.7 | 4.4 | 2.2 | 3.2 | 0.697 | 22 |
| 11 | IOP80 | 11 | 5.6 | 9.7 | 31.3 | 8.5 | 0 | 25.3 | 2.8 | 3.9 | 1.5 | 4.6 | 1.19 | 22 |
| 12 | FUP-1 | 0 | 0 | 0 | 9.81 | 6.78 | 0 | 83.41 | 0 | 0 | 0 | 0 | 0.47 | 23 |
| 13 | CLP-2 | 3.3 | 2.1 | 14.5 | 48 | 28 | 0 | 4.1 | 0 | 0 | 23.59 | 1.48 | 0.86 | 24 |
| 14 | CLP-3 | 0 | 0 | 8.6 | 56 | 29.4 | 0 | 6 | 0 | 0 | 17.06 | 0.95 | 1.27 | 24 |
| 15 | TYAP-1 | 0 | 78.98 | 0 | 5.74 | 10.6 | 0 | 4.68 | 0 | 0 | 0 | 0 | 3.92 | 25 |
| 16 | PV-P1 | 0 | 24.2 | 1.9 | 8.3 | 9.7 | 0 | 55.9 | 0.8 | 3.5 | 3.4 | 1.22 | 0.878 | 26 |
| 17 | PV-P2 | 3.6 | 15.7 | 14.4 | 16 | 21.6 | 0 | 28.7 | 0.3 | 5.4 | 5.7 | 4.22 | 0.169 | 26 |
| 18 | PV-P3 | 6.1 | 16.5 | 16.1 | 11.2 | 13.3 | 0 | 36.8 | 0.2 | 7.9 | 8.1 | 7.09 | 0.048 | 26 |
| 19 | Control-EPS | 6.1 | 14.6 | 20.4 | 20.7 | 24.2 | 0 | 14 | 0 | 0 | 0 | 19.75 | 2.3 | 27 |
| 20 | Control-IPS1 | 3.1 | 7.2 | 28 | 36.7 | 19 | 0 | 6 | 0 | 0 | 0 | 17.57 | 1.08 | 27 |
| 21 | Tween 80-IPS1 | 3.3 | 2.4 | 6.9 | 73.4 | 11.4 | 0 | 2.6 | 0 | 0 | 0 | 16.72 | 0.74 | 27 |
| 22 | Tween 80-IPS2 | 1.4 | 5.2 | 18.3 | 60.9 | 12.1 | 0 | 2.1 | 0 | 0 | 0 | 15.61 | 0.84 | 27 |
| 23 | CPSI | 0 | 0 | 27 | 73 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.23 | 28 |
| 24 | G1 | 10.9 | 1.2 | 6.2 | 52.5 | 14.9 | 0 | 14.3 | 0 | 0 | 4.2 | 6.49 | 0.34 | 29 |
| 25 | G2 | 12.2 | 0.8 | 4.9 | 56 | 16.2 | 0 | 9.9 | 0 | 0 | 7.45 | 5.11 | 0.56 | 29 |
| 26 | G3 | 12.2 | 3.8 | 3.2 | 50.2 | 12.5 | 0 | 18.1 | 0 | 0 | 1.92 | 3.62 | 0.87 | 29 |
| 27 | P1 | 11.4 | 30.3 | 1.5 | 9.2 | 44.4 | 0 | 3.2 | 0 | 0 | 0 | 0 | 0.62 | 30 |
| 28 | P2 | 10.4 | 22.1 | 3.1 | 11.2 | 53.1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.07 | 30 |
| 29 | CP | 1.2 | 15.6 | 7.5 | 28.2 | 24.7 | 0 | 5.4 | 4.8 | 12.6 | 17.4 | 7.57 | 0.09 | 31 |
| 30 | SCG | 0 | 19.93 | 4.43 | 15.37 | 60.27 | 0 | 0 | 0 | 0 | 0 | 0 | 0.7 | 32 |
| 31 | PNMP2 | 0 | 5.78 | 28.62 | 14.42 | 41.57 | 7.24 | 2.37 | 0 | 0 | 0 | 0 | 0.3297 | 16 |
| 32 | PNMP3 | 0 | 3.45 | 26.58 | 21.55 | 36.42 | 8.44 | 3.56 | 0 | 0 | 0 | 0 | 0.1516 | 16 |
| 33 | GLP60 | 0 | 0 | 3.2 | 85.9 | 8.2 | 1.5 | 0 | 0 | 0 | 0 | 0 | 0.97 | 8 |
| 34 | GLP80 | 0 | 0 | 9.4 | 79.4 | 5.4 | 1.1 | 0 | 0 | 0 | 0 | 0 | 0.72 | 8 |
| 35 | GLP | 0 | 0 | 4.8 | 86.5 | 6.1 | 1.2 | 0 | 0 | 0 | 0 | 0 | 0.9 | 8 |
| 36 | LLPs-D | 6.83 | 2.73 | 9.2 | 19.23 | 58.19 | 0.57 | 3.25 | 0 | 0 | 0 | 0 | 0.38 | 33 |
| 37 | LLPs-L | 5.03 | 19.39 | 6.07 | 22.82 | 37.45 | 7.04 | 2.21 | 0 | 0 | 0 | 0 | 0.99 | 33 |
| 38 | SMWP-1 | 0 | 0 | 27 | 34 | 11 | 0 | 28 | 0 | 0 | 0 | 0.53 | 0.13 | 34 |
| 39 | EAP40-1 | 2.63 | 0 | 36 | 46.79 | 14.58 | 0 | 0 | 0 | 0 | 0 | 0.33 | 0.28 | 35 |
| 40 | EAP60-1 | 3.37 | 2.28 | 2.89 | 43.61 | 37.67 | 0 | 10.18 | 0 | 0 | 0 | 0.48 | 0.52 | 35 |
| 41 | CMP-1 | 4.2 | 0 | 0 | 95.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.15 | 36 |
| 42 | GPA1 | 0.4 | 21.2 | 10.6 | 13.8 | 27.5 | 2.2 | 0 | 14.8 | 9.5 | 23.04 | 3.75 | 0.08 | 37 |
| 43 | GPA2 | 0.8 | 15.6 | 8.2 | 18 | 21.4 | 1.6 | 1.6 | 18 | 14.8 | 32.79 | 4.38 | 0.06 | 37 |
| 44 | GPA3 | 3.8 | 7.5 | 6.3 | 34.3 | 16.3 | 1.3 | 3.1 | 24.3 | 3.1 | 27.01 | 5.53 | 0.03 | 37 |
| 45 | Ac-CP1 | 2.5 | 16.4 | 5.6 | 17.6 | 27.5 | 0 | 1.8 | 2.6 | 26 | 15.78 | 7.25 | 0.06448 | 38 |
| 46 | Ac-CP2 | 2.8 | 15.3 | 6 | 14.3 | 26.2 | 0 | 2 | 2.9 | 30.5 | 25.99 | 6.93 | 0.07829 | 38 |
| 47 | Ac-CP3 | 2.2 | 15.6 | 5.6 | 9.8 | 29.9 | 0 | 1.1 | 3.4 | 32.4 | 27.43 | 7.09 | 0.07804 | 38 |
| 48 | CP | 1.2 | 15.6 | 7.5 | 28.1 | 24.8 | 0 | 5.4 | 4.8 | 12.6 | 16.14 | 7.57 | 0.09837 | 38 |
| 49 | WFPs | 5.2 | 18.5 | 3.5 | 15.9 | 21.3 | 0 | 7.4 | 3.3 | 24.9 | 28.1 | 0 | 0.007 | 39 |
| 50 | APs-2-1 | 4.6 | 8 | 0 | 32.3 | 24.2 | 0 | 21.1 | 0 | 9.8 | 9.8 | 1.9 | 0.4545 | 40 |
| 51 | APs-3-1 | 1.5 | 2.8 | 0 | 35.1 | 34 | 0 | 16.7 | 2 | 7.9 | 7.9 | 1.3 | 0.2243 | 40 |
| 52 | PTPS-3 | 6.82 | 26.22 | 13.83 | 10.23 | 39.34 | 3.21 | 0.35 | 0 | 0 | 40.66 | 13.27 | 1.72 | 41 |
| 53 | PTPS-5 | 15.98 | 20.84 | 15.29 | 6.08 | 40.33 | 1.68 | 0.15 | 0 | 0 | 40.44 | 19.96 | 1.45 | 41 |
| 54 | PSS-EPS | 8.2 | 7.7 | 24 | 35.3 | 15.4 | 0 | 9.4 | 0 | 0 | 0 | 20.19 | 1.497 | 42 |
| 55 | UKLOxa | 5.5 | 10.2 | 6.1 | 11.3 | 28.4 | 0.3 | 7.2 | 26.5 | 4.5 | 31 | 0 | 0.0546 | 43 |
| 56 | UKLK1 | 3.6 | 6.1 | 5.8 | 10.9 | 9.4 | 1.8 | 5.03 | 6.5 | 2.9 | 9.4 | 0 | 0.136 | 43 |
| 57 | UKLK4 | 2.5 | 6.6 | 2.6 | 8.3 | 6.6 | 1.1 | 64.4 | 7.1 | 0.8 | 7.9 | 0 | 0.6023 | 43 |
| 58 | UKSOxa | 5.6 | 9.9 | 11.8 | 18.7 | 16.9 | 0.4 | 9.4 | 24.3 | 3 | 27.3 | 0 | 0.0165 | 43 |
| 59 | UKSOxa-PG | 8.6 | 15.7 | 7.1 | 12.4 | 26.1 | 0.5 | 15.4 | 10.2 | 4 | 14.2 | 0 | 0.3751 | 43 |
| 60 | UKSK1 | 3.5 | 4 | 1.9 | 6.2 | 3.6 | 0.7 | 76.5 | 2.8 | 0.8 | 2.6 | 0 | 0.177 | 43 |
| 61 | UKSK4 | 3.6 | 8.7 | 3.7 | 8.3 | 6.2 | 0.9 | 66 | 1.9 | 0.7 | 2.6 | 0 | 0.0038 | 43 |
| 62 | PMBOxa | 8.8 | 11.9 | 9.5 | 23.8 | 18.3 | 0.7 | 8.4 | 16.3 | 2.3 | 18.6 | 0 | 0.0217 | 43 |
| 63 | PMBOxa-PG | 6.7 | 12.4 | 10.3 | 26.6 | 25.5 | 0.8 | 6.7 | 8 | 3 | 11 | 0 | 0.144 | 43 |
| 64 | PMBK1 | 4.8 | 13.3 | 4.8 | 17.2 | 13.7 | 1.5 | 37.5 | 5.3 | 1.9 | 7.2 | 0 | 0.3143 | 43 |
| 65 | PMBK4 | 2.4 | 18.4 | 2.3 | 9.9 | 8.9 | 2.6 | 51.9 | 3 | 0.6 | 3.6 | 0 | 0.6547 | 43 |
| 66 | AMBOxa | 10.9 | 14.5 | 4.2 | 25.9 | 14.9 | 0.5 | 6.1 | 16.8 | 6.2 | 23 | 0 | 0.0184 | 43 |
| 67 | AMBOxa-PG | 17.4 | 22.6 | 4 | 5.9 | 16.6 | 0.6 | 7.1 | 20.8 | 5 | 25.8 | 0 | 0.3533 | 43 |
| 68 | AMBK1 | 2 | 4.6 | 1.9 | 32.2 | 10.7 | 1.8 | 44.2 | 1.5 | 1.1 | 2.6 | 0 | 0.1093 | 43 |
| 69 | AMBK4 | 3.2 | 27.2 | 1.3 | 17.1 | 6.5 | 2.1 | 40.6 | 1.3 | 0.7 | 2 | 0 | 1.4203 | 43 |
| 70 | PS1 | 0.79 | 0.69 | 60.51 | 32.66 | 2.35 | 2.98 | 0 | 0 | 0 | 0 | 0 | 1.21 | 44 |
| 71 | PS2 | 10.96 | 5.81 | 36.16 | 26.92 | 14.55 | 4.52 | 1.04 | 0 | 0 | 0 | 0 | 0.73 | 44 |
| 72 | PS3 | 48.55 | 10.73 | 7.35 | 11.41 | 13.85 | 4.62 | 3.45 | 0 | 0 | 0 | 0 | 0.67 | 44 |
| 73 | WKCP-N | 0 | 2.22 | 0 | 91.95 | 5.83 | 0 | 0 | 0 | 0 | 0 | 0 | 0.61 | 45 |
| 74 | WKHP-N | 0 | 12.9 | 0 | 73.71 | 10 | 0 | 1.34 | 2.45 | 0 | 3.2 | 0 | 1.08 | 45 |
aname from reference
brhamnose
carabinose
dmannose
eglucose
f galactose
gfucose
hxylose
iglucuronic acid
jgalacturonic acid
kuronic acid
lprotein content
Fig 1. Data distribution of parameter.
MLR results
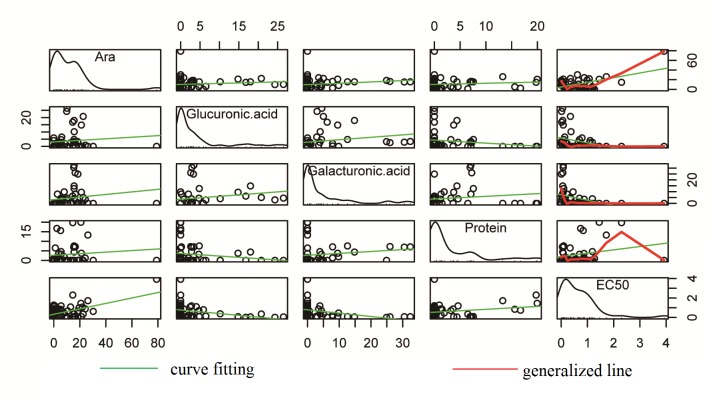
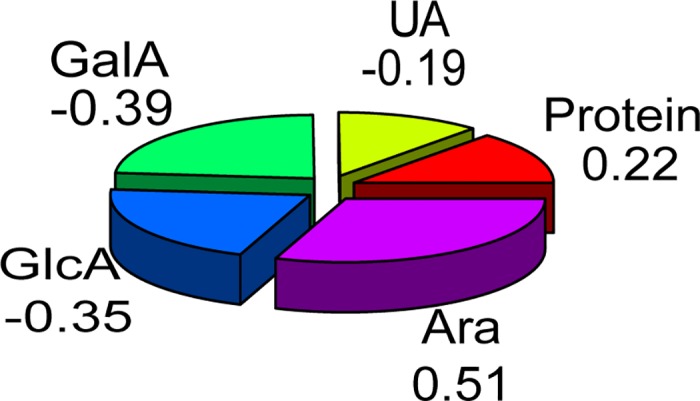
In this study, the training data set of 52 compounds was used. A stepwise linear regression analysis was used to determine the relationship between the dependent variable of EC50 and the independent variables of uronic acid (UA), protein content (PC) and monosaccharide compositions (Rha, Ara, Man, Glc, Gal, Fuc, Xyl, GlcA and GalA). To achieve this goal, regression analysis was implemented by using the forward stepwise. In stepwise regression procedures, the first was to choose the most correlated independent variable, and then to select independent variable which was most correlated with the remaining variance in the dependent variable. This procedure was to increase the additional independent variable with R-squared (R2) which was not changing until a significance of at least 80%. Accordingly, the variables of Ara, GalA, GlcA and PC were included in the regression model. The relationship between the matrix of parameters and EC50 was shown in Fig 2. One variable data was used as the abscissa, another variable data was used as ordinate, and all points had been portrayed by the matrix scatter plot. From the diagonal we can see that the distribution of the data was all similar in shape. Fig 3 showed the correlation between model parameters and EC50, and the proportion of Ara, GalA and GlcA accounted 0.51, 0.39 and 0.35, respectively, which indicated that they had the most effect on EC50. In Fig 3, we can see that EC50 had a positive correlation with Ara and PC, and it has negative correlation with GalA and GlcA, which was consistent with the model given in equation. The regression Eq 1, which could be obtained through the statistical analysis, was as follows. Because the effect of UA on EC50 was little, UA was not added to the model equation. The linear model selected four major relevant descriptors, and gave a stable model with R = 0.807 and RMSE = 0.423.
Fig 2. Correlation between the matrix of parameters and EC50 value of the DPPH scavenging activity.
Fig 3. Proportion of the parameters effecting on EC50 value of the DPPH scavenging activity.
| (1) |
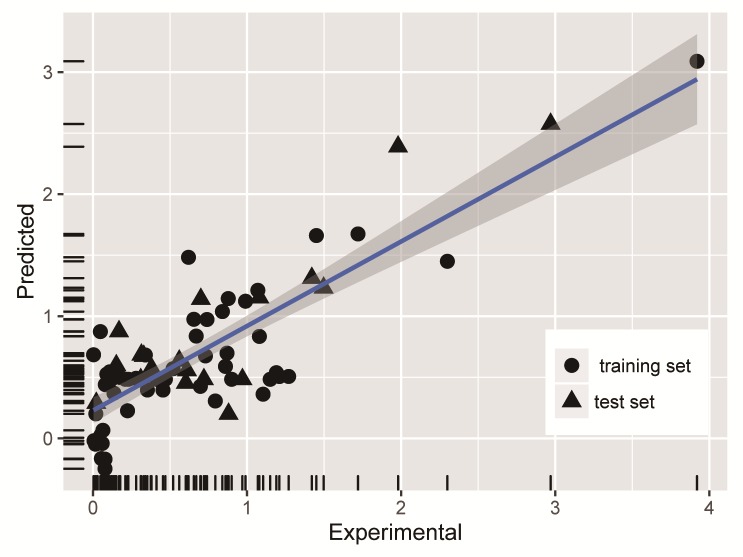
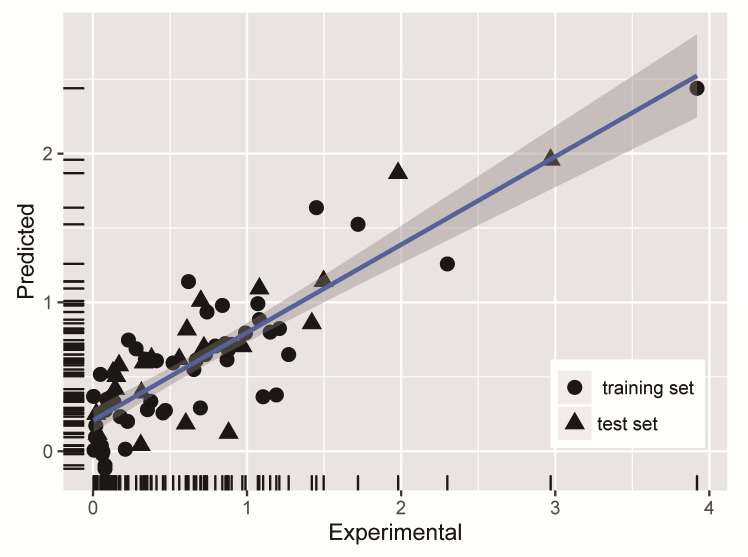
In the model, R value was 0.807 (p <0.001), fit indicators of the model were acceptable, the model was coincided with the data structure, and Ara, GalA, GlcA, PC and EC50 were significant correlation. The predicted EC50 values of the training and test set by using the MLR equation were given in the Table 2. Predicted values and experimental values of EC50 in two sets of data were plotted and shown in Fig 4. Most of the data were distributed from 0 to 1.5, and there were some predicted and negative values existing in the left lower corner. The experimental values of these negative values were between 0 and 0.2, which could be accepted. Experimental values and predicted points were distributed in two sides of the curve fitting, and most point of test set distributed among the prediction set, which illustrated that the establishment of training set used for the multiple regression model was very good to predict the numerical value of test set. The above linear model was applied to predict the 22 test data set, and these test data were never used in model building. The result showed R = 0.872, RMSE = 0.361 and p = 1.245E-7, which showed that there was a significant correlation. Multiple linear regressions (MLR) established the relationship between the dependent variable of EC50 and the independent variable of polysaccharide properties. The results showed that the statistics for MLR equation were good, and it also offered some views about the polysaccharide properties influences on DPPH-scavenging activity of polysaccharides.
Table 2. Experimental and predicted values of EC50 for the DPPH-scavenging activity of polysaccharides using MLR, ANN and SVM models.
| No | Namea | Exp | MLR | SVM | ANN | |||
|---|---|---|---|---|---|---|---|---|
| Predict | Residue | Predict | Residue | Predict | residue | |||
| 1 | S1 (Glu) | 1.104 | 0.361582 | 0.742418 | 0.366201 | 0.737799 | 0.953944 | 0.150056 |
| 2 | S1 (Visco) | 0.794 | 0.305859 | 0.488141 | 0.706948 | 0.087052 | 0.956870 | -0.162870 |
| 3 | GBP50S2 | 0.412 | 0.483627 | -0.071627 | 0.608142 | -0.196142 | 0.376554 | 0.035446 |
| 4 | LBP-s80 | 2.97 | 2.575070 | 0.394930 | 1.958210 | 1.011790 | 3.430658 | -0.460658 |
| 5 | LBP-s75 | 1.98 | 2.388860 | -0.408860 | 1.868621 | 0.111379 | 2.222439 | -0.242439 |
| 6 | WB1 | 0.12 | 0.483627 | -0.363627 | 0.367456 | -0.247456 | 0.395514 | -0.275514 |
| 7 | WB2 | 0.31 | 0.483630 | -0.173630 | 0.040901 | 0.269099 | 0.181648 | 0.128352 |
| 8 | WB3 | 0.21 | 0.483627 | -0.273627 | 0.013756 | 0.196244 | 0.138245 | 0.071755 |
| 9 | IOP40 | 0.88 | 0.200140 | 0.679860 | 0.123591 | 0.756409 | 0.508640 | 0.371360 |
| 10 | IOP60 | 0.697 | 0.425514 | 0.271486 | 0.291228 | 0.405772 | 0.814391 | -0.117391 |
| 11 | IOP80 | 1.19 | 0.537777 | 0.652223 | 0.378394 | 0.811606 | 0.955650 | 0.234350 |
| 12 | FUP-1 | 0.47 | 0.483627 | -0.013627 | 0.274257 | 0.195743 | 0.145075 | 0.324925 |
| 13 | CLP-2 | 0.86 | 0.589228 | 0.270772 | 0.723493 | 0.136507 | 0.863565 | -0.003565 |
| 14 | CLP-3 | 1.27 | 0.506954 | 0.763046 | 0.649755 | 0.620245 | 1.104942 | 0.165058 |
| 15 | TYAP-1 | 3.92 | 3.088460 | 0.831540 | 2.438504 | 1.481496 | 3.770749 | 0.149251 |
| 16 | PV-P1 | 0.878 | 1.145072 | -0.267072 | 0.681146 | 0.196854 | 0.592526 | 0.285474 |
| 17 | PV-P2 | 0.169 | 0.876230 | -0.707230 | 0.577149 | -0.408149 | 0.551492 | -0.382492 |
| 18 | PV-P3 | 0.048 | 0.874380 | -0.826380 | 0.516508 | -0.468508 | 0.321103 | -0.273103 |
| 19 | Control-EPS | 2.3 | 1.450109 | 0.849891 | 1.258621 | 1.041379 | 2.311131 | -0.011131 |
| 20 | Control-IPS1 | 1.08 | 1.152520 | -0.072520 | 1.093392 | -0.013392 | 1.781373 | -0.701373 |
| 21 | Tween 80-IPS1 | 0.74 | 0.973340 | -0.233340 | 0.935877 | -0.195877 | 0.419414 | 0.320586 |
| 22 | Tween 80-IPS2 | 0.84 | 1.038431 | -0.198431 | 0.979781 | -0.139781 | 0.776261 | 0.063739 |
| 23 | CPSI | 0.23 | 0.483627 | -0.253627 | 0.747002 | -0.517002 | 0.339270 | -0.109270 |
| 24 | G1 | 0.34 | 0.682566 | -0.342566 | 0.618879 | -0.278879 | 0.678410 | -0.338410 |
| 25 | G2 | 0.56 | 0.635490 | -0.075490 | 0.619038 | -0.059038 | 0.912242 | -0.352242 |
| 26 | G3 | 0.87 | 0.697843 | 0.172157 | 0.614961 | 0.255039 | 0.920268 | -0.050268 |
| 27 | P1 | 0.62 | 1.482949 | -0.862949 | 1.139900 | -0.519900 | 0.912532 | -0.292532 |
| 28 | P2 | 1.07 | 1.212505 | -0.142505 | 0.991519 | 0.078481 | 0.774189 | 0.295811 |
| 29 | CP | 0.09 | 0.525923 | -0.435923 | 0.348891 | -0.258891 | 0.112901 | -0.022901 |
| 30 | SCG | 0.7 | 1.140940 | -0.440940 | 1.009837 | -0.309837 | 0.745078 | -0.045078 |
| 31 | PNMP2 | 0.3297 | 0.674260 | -0.344560 | 0.595812 | -0.266112 | 0.513151 | -0.183451 |
| 32 | PNMP3 | 0.1516 | 0.597410 | -0.445810 | 0.504217 | -0.352617 | 0.423722 | -0.272122 |
| 33 | GLP60 | 0.97 | 0.483630 | 0.486370 | 0.704868 | 0.265132 | 0.650600 | 0.319400 |
| 34 | GLP80 | 0.72 | 0.483630 | 0.236370 | 0.697402 | 0.022598 | 0.519819 | 0.200181 |
| 35 | GLP | 0.9 | 0.483627 | 0.416373 | 0.718195 | 0.181805 | 0.627824 | 0.272176 |
| 36 | LLPs-D | 0.38 | 0.573660 | -0.193660 | 0.623545 | -0.243545 | 0.573738 | -0.193738 |
| 37 | LLPs-L | 0.99 | 1.123127 | -0.133127 | 0.794887 | 0.195113 | 0.608499 | 0.381501 |
| 38 | SMWP-1 | 0.13 | 0.496640 | -0.366640 | 0.528723 | -0.398723 | 0.605999 | -0.475999 |
| 39 | EAP40-1 | 0.28 | 0.491730 | -0.211730 | 0.688481 | -0.408481 | 0.330165 | -0.050165 |
| 40 | EAP60-1 | 0.52 | 0.570610 | -0.050610 | 0.593314 | -0.073314 | 0.667831 | -0.147831 |
| 41 | CMP-1 | 1.15 | 0.483627 | 0.666373 | 0.800434 | 0.349566 | 0.948406 | 0.201594 |
| 42 | GPA1 | 0.08 | 0.440122 | -0.360122 | 0.275073 | -0.195073 | 0.109224 | -0.029224 |
| 43 | GPA2 | 0.06 | -0.041681 | 0.101681 | -0.019261 | 0.079261 | 0.043427 | 0.016573 |
| 44 | GPA3 | 0.03 | 0.004720 | 0.025280 | 0.116442 | -0.086442 | 0.087376 | -0.057376 |
| 45 | Ac-CP1 | 0.06448 | 0.065797 | -0.001317 | -0.003399 | 0.067879 | 0.022510 | 0.041970 |
| 46 | Ac-CP2 | 0.07829 | -0.170541 | 0.248831 | -0.094824 | 0.173114 | 0.017536 | 0.060754 |
| 47 | Ac-CP3 | 0.07804 | -0.249175 | 0.327215 | -0.117474 | 0.195514 | 0.017260 | 0.060780 |
| 48 | CP | 0.09837 | 0.525923 | -0.427553 | 0.343823 | -0.245453 | 0.125297 | -0.026927 |
| 49 | WFPs | 0.007 | -0.019404 | 0.026404 | 0.007187 | -0.000187 | 0.016533 | -0.009533 |
| 50 | APs-2-1 | 0.4545 | 0.395343 | 0.059157 | 0.257761 | 0.196739 | 0.074447 | 0.380053 |
| 51 | APs-3-1 | 0.2243 | 0.225857 | -0.001557 | 0.200810 | 0.023490 | 0.147594 | 0.076706 |
| 52 | PTPS-3 | 1.72 | 1.674231 | 0.045769 | 1.524571 | 0.195429 | 1.532898 | 0.187102 |
| 53 | PTPS-5 | 1.45 | 1.661066 | -0.211066 | 1.636562 | -0.186562 | 1.546828 | -0.096828 |
| 54 | PSS-EPS | 1.497 | 1.233340 | 0.263660 | 1.142511 | 0.354489 | 1.906331 | -0.409331 |
| 55 | UKLOxa | 0.0546 | -0.165611 | 0.220211 | 0.036641 | 0.017959 | 0.072630 | -0.018030 |
| 56 | UKLK1 | 0.136 | 0.369956 | -0.233956 | 0.332098 | -0.196098 | 0.164552 | -0.028552 |
| 57 | UKLK4 | 0.6023 | 0.453730 | 0.148570 | 0.185824 | 0.416476 | 0.087298 | 0.515002 |
| 58 | UKSOxa | 0.0165 | -0.047842 | 0.064342 | 0.094666 | -0.078166 | 0.070345 | -0.053845 |
| 59 | UKSOxa-PG | 0.3751 | 0.529760 | -0.154660 | 0.335927 | 0.039173 | 0.159281 | 0.215819 |
| 60 | UKSK1 | 0.177 | 0.498201 | -0.321201 | 0.232651 | -0.055651 | 0.102440 | 0.074560 |
| 61 | UKSK4 | 0.0038 | 0.684537 | -0.680737 | 0.368651 | -0.364851 | 0.231762 | -0.227962 |
| 62 | PMBOxa | 0.0217 | 0.288880 | -0.267180 | 0.250147 | -0.228447 | 0.105432 | -0.083732 |
| 63 | PMBOxa-PG | 0.144 | 0.528240 | -0.384240 | 0.417461 | -0.273461 | 0.559193 | -0.415193 |
| 64 | PMBK1 | 0.3143 | 0.684450 | -0.370150 | 0.393583 | -0.079283 | 0.306324 | 0.007976 |
| 65 | PMBK4 | 0.6547 | 0.975208 | -0.320508 | 0.549260 | 0.105440 | 0.626754 | 0.027946 |
| 66 | AMBOxa | 0.0184 | 0.200785 | -0.182385 | 0.172081 | -0.153681 | 0.053022 | -0.034622 |
| 67 | AMBOxa-PG | 0.3533 | 0.395625 | -0.042325 | 0.279658 | 0.073642 | 0.083612 | 0.269688 |
| 68 | AMBK1 | 0.1093 | 0.545151 | -0.435851 | 0.353599 | -0.244299 | 0.546221 | -0.436921 |
| 69 | AMBK4 | 1.4203 | 1.312850 | 0.107450 | 0.859646 | 0.560654 | 1.691224 | -0.270924 |
| 70 | PS1 | 1.21 | 0.506384 | 0.703616 | 0.824544 | 0.385456 | 1.219299 | -0.009299 |
| 71 | PS2 | 0.73 | 0.675246 | 0.054754 | 0.649997 | 0.080003 | 0.467911 | 0.262089 |
| 72 | PS3 | 0.67 | 0.837512 | -0.167512 | 0.612208 | 0.057792 | 0.702244 | -0.032244 |
| 73 | WKCP-N | 0.61 | 0.556840 | 0.053160 | 0.816849 | -0.206849 | 0.776698 | -0.166698 |
| 74 | WKHP-N | 1.08 | 0.834885 | 0.245115 | 0.884773 | 0.195227 | 1.335107 | -0.255107 |
aname from reference
Fig 4. A comparison of experimental vs predicted EC50 using MLR method.
ANN results
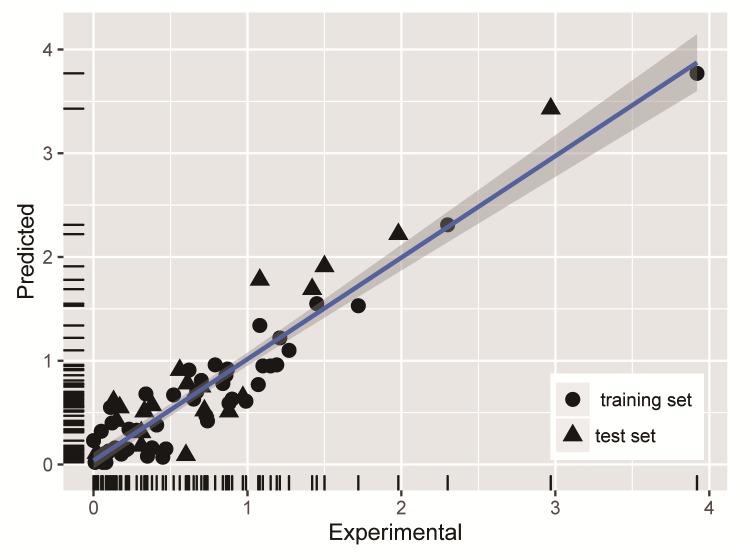
Polysaccharide properties were considered as the input layer node in neural networks, and EC50 values of the DPPH-scavenging activity was the output layer node. Numbers of nodes had a great influence on the test results. The optimization was done with 10 fold cross validation, and 30% of test data were used for validation. Selected parameters of the number of neurons in the hidden layer were optimized by changing from 4 to 14, and it was worthy to mention that the initial value of 7 selected was optimal. The selected network adopted Broyden Fletcher Goldfard Shanno (BFGS) algorithm which was still seen as the best Quasi-Newton algorithm. When the entire training data was trained in the network with the optimized parameters, it gave R = 0.96 and RMSE = 0.018. The experimental and predicted values of EC50 for the train data using the ANN model were plotted and shown in Fig 5. The experimental value was abscissa, the point distribution of the prediction value for the y-coordinate was on both sides of the curve fitting from 0 to 1.5, and the point distribution was uniform and closed to each other. According to the view of point, the density of horizontal and vertical coordinates and the fitting effect were perfect. The predicted values of EC50 for the train and test data were given in the Table 2. The test set was used for prediction and gave R = 0.933 and RMSE = 0.055.
Fig 5. A comparison of experimental vs predicted EC50 using ANN method.
SVM results
We selected radial basis function (RBF) kernel for function modeling in SVM, the best parameter C, g and ε were selected by using 10 fold cross validation, a SVM model was obtained by training the whole training set, and then the model was used for the test set. By varying the parameter values in the training set systematically, we optimized SVM parameters, and calculated RMSE of the model. The parameter value which gave the lowest RMSE was selected. The regularization parameter C controlled the alternate use between maximizing the margin and minimizing the training error. If the value of C was too small, then there was not sufficient stress on fitting the training data. To have a stable learning procedure, a large value of C should be set up first [57]. To discover an optimal value of C, the RMSE of SVM model with different C values was calculated. Then, this value C = 9 was selected as the optimal value. We achieved the selected parameters (g = 0.091, ε = 0.1, C = 9) and the final training running in the whole training set, and EC50 of the DPPH-scavenging activity was predicted. The predicted EC50 on the basis of this model was plotted and shown in Fig 6 and Table 2. The statistical parameters of this model were R = 0.851 and RMSE = 0.151 for the training set, and the test set was used for prediction and gave R = 0.865 and RMSE = 0.144.
Fig 6. A comparison of experimental vs predicted EC50 using SVM method.
Comparison of MLR, ANN and SVM models
The statistical parameters obtained from the investigative models for train and test set were shown in Table 3. The error estimates were applied to model performance evaluation, and RMSE were lower for nonlinear models (SVM, ANN) generated by the machine learning methods than that by multiple linear regression. The correlation coefficients (R) given by SVM and ANN models were also higher than that by multiple linear regression. The above results indicated that the performances of nonlinear models SVM and ANN were better than that of a linear MLR model for the prediction of DPPH-scavenging activity of polysaccharides. The comparison of the nonlinear models demonstrated that ANN model accurately predicted the relationship between polysaccharide properties and the DPPH-scavenging activity for the train data set, and this was obviously evident from a lower RMSE (0.018) and a higher R (0.96) value. While ANN model was also the best one in the prediction of the test set.
Table 3. Comparison of MLR, ANN and SVM models for the DPPH scavenging activity of polysaccharides.
| Method | Parameters | Training set | Test set |
|---|---|---|---|
| MLR | R | 0.807 | 0.872 |
| RMSE | 0.423 | 0.361 | |
| ANN | R | 0.96 | 0.933 |
| RMSE | 0.018 | 0.055 | |
| SVM | R | 0.851 | 0.865 |
| RMSE | 0.151 | 0.144 |
Effect of MW on the scavenging activity of DPPH radical
Molecular weight was seen as an important indicator of the antioxidant activity of polysaccharides [20], so a single study was used to evaluate the relationship of MW and antioxidant activity of polysaccharides. Due to the relatively large difference in MW of polysaccharide from 2250 to 538500 (Table 4), MW was normalized before the analysis, the size of MW was taken with a base-8 of log, and the data was shown in Table 4 [58–66].
Table 4. MW and EC50 values of the DPPH scavenging activity.
| Namea | EC50 | Mw | Refs | Name | EC50 | Mw | Refs | Name | EC50 | Mw | Refs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PS | 6.20 | 225000 | 58 | CLP-3 | 1.27 | 60143 | 24 | CMP-1 | 1.15 | 4300 | 36 |
| PSPO-1a | 1.43 | 18000 | 59 | TYAP-1 | 3.92 | 115000 | 25 | PPM | 1.80 | 22000 | 64 |
| LBP-80 | 5.33 | 70600 | 20 | TYAP-2 | 4.11 | 479000 | 25 | PPE | 3.06 | 38000 | 64 |
| LBP-s75 | 1.98 | 71700 | 20 | TYAP-3 | 2.64 | 403000 | 25 | GPA1 | 0.08 | 19600 | 37 |
| LBP-s50 | 4.96 | 538500 | 20 | PS1-1 | 6.81 | 67400 | 62 | GPA2 | 0.06 | 10600 | 37 |
| BSFP-1 | 7.40 | 13300 | 60 | PS1-2 | 4.56 | 15400 | 62 | GPA3 | 0.03 | 6700 | 37 |
| WB2 | 0.31 | 28000 | 21 | PS2-1 | 2.53 | 12100 | 62 | AAP-2A | 0.15 | 2252 | 65 |
| WB3 | 0.21 | 19000 | 21 | PNMP1 | 0.72 | 28400 | 16 | RNLP I | 0.20 | 14900 | 66 |
| SP1 | 3.20 | 9192 | 61 | PNMP2 | 0.33 | 31500 | 16 | WKCP-N | 0.61 | 9600 | 45 |
| FUP-1 | 0.47 | 41000 | 23 | PNMP3 | 0.15 | 26100 | 16 | WKHP-N | 1.08 | 113400 | 45 |
| CLP-1 | 1.69 | 78754 | 24 | AAP | 3.29 | 27700 | 63 | WKHP-A | 3.34 | 169600 | 45 |
| CLP-2 | 0.86 | 51257 | 24 | EAP80-2 | 1.32 | 65313 | 35 |
aname from reference
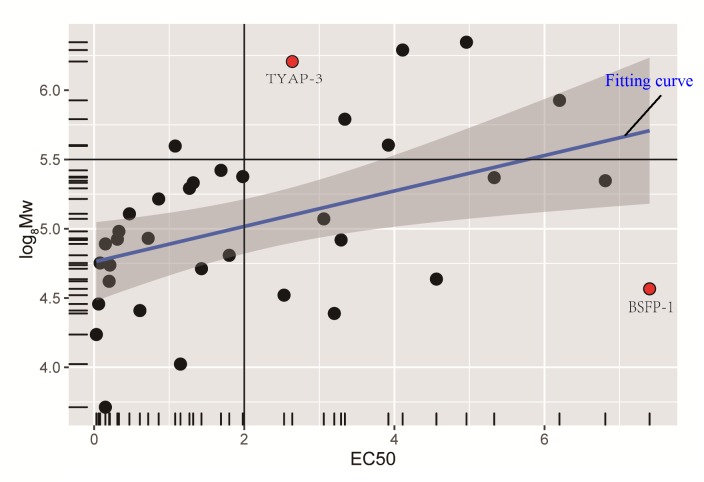
We used EC50 values as the horizontal coordinate and established the correlation between EC50 and MW. As shown in Fig 7, the value of EC50 decreased with the decrease of MW, which indicated that the smaller MW could have the stronger DPPH free radical scavenging activity. This result was in accord with those reported in the literature [20, 59]. In Fig 7, it could also be found that there were some points which did not conform to the rules, such as TYAP-3 and BSFP-1. BSFP-1 had the smaller MW and a relatively larger EC50 value [60], which may be because BSFP-1 had no UA. TYAP-3 had larger MW, but its EC50 value was smaller. The reason may be that the content of Ara accounted for 45.82% in TYAP-3 [25]. Fig 7 showed that when the value of EC50 arranged from 0 to 2, the value of Y axis was 0–5.5, which indicated that MW was between 4000 and 100000.
Fig 7. Correlation scatter plots of EC50 and MW.
According to the above results, we could conclude that the antioxidant activity of polysaccharide usually was higher in MW range of 4000–100000. However, MW was not the only factor, and the antioxidant activity could be affected by other polysaccharide properties, such as UA and Ara.
Models for the hydroxyl radicals scavenging activity of polysaccharides
To make relationship models of monosaccharide composition and the hydroxyl radicals scavenging activity, the entire data set including 67 compounds was divided into two clusters [67–82]. The test set and the train set were given in Table 5.
Table 5. Polysaccharides data set with descriptors and their EC50 values of the hydroxyl radicals scavenging activity.
| No | Namea | Rhab | Arac | Mand | Glce | Galf | Fucg | Xylh | GlcAi | GalAj | UAk | PCl | EC50 | Refs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PS-SI | 0 | 27.3 | 18.2 | 9.1 | 45.4 | 0 | 0 | 0 | 0 | 0 | 0.5 | 0.21 | 67 |
| 2 | CBP-1 | 0 | 0 | 35.9 | 12.8 | 51.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0.638 | 68 |
| 3 | GBP50S2 | 46.7 | 0 | 42.2 | 0 | 11.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.482 | 19 |
| 4 | pMTPS-3 | 0 | 17.3 | 41.6 | 28.3 | 12.6 | 0 | 0 | 0 | 0 | 0 | 0 | 1.9 | 69 |
| 5 | USEP40-1 | 7.95 | 8.42 | 37.34 | 17.94 | 28.36 | 0 | 0 | 0 | 0 | 0 | 0 | 0.376 | 70 |
| 6 | USEP70-1 | 14.47 | 7.78 | 37.27 | 21.85 | 18.64 | 0 | 0 | 0 | 0 | 0 | 0 | 0.524 | 70 |
| 7 | IOP40 | 4.4 | 4.2 | 9.8 | 40 | 14.5 | 3.3 | 9.6 | 9.7 | 4.6 | 5 | 2.4 | 0.58 | 22 |
| 8 | IOP60 | 9.7 | 5.6 | 8 | 32.2 | 12.6 | 0 | 22.9 | 4.7 | 4.4 | 2.2 | 3.2 | 0.46 | 22 |
| 9 | CLP-1 | 0 | 3.6 | 7.9 | 60.2 | 26.4 | 0 | 1.9 | 0 | 0 | 15.84 | 1.43 | 3.68 | 24 |
| 10 | CLP-2 | 3.3 | 2.1 | 14.5 | 48 | 28 | 0 | 4.1 | 0 | 0 | 23.59 | 1.48 | 1.29 | 24 |
| 11 | CLP-3 | 0 | 0 | 8.6 | 56 | 29.4 | 0 | 6 | 0 | 0 | 17.06 | 0.95 | 2.8 | 24 |
| 12 | GPS-2 | 44.7 | 20.9 | 0 | 3.6 | 10.8 | 0 | 19.9 | 0 | 0 | 0 | 0 | 0.069 | 71 |
| 13 | P70-1 | 0 | 0 | 56 | 18 | 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0.548 | 65 |
| 14 | PS1-1 | 0 | 0 | 89.5 | 7.3 | 3.2 | 0 | 0 | 0 | 0 | 0 | 1.67 | 1.14 | 62 |
| 15 | PS1-2 | 0 | 0 | 71.1 | 3.7 | 25.2 | 0 | 0 | 0 | 0 | 0 | 1.86 | 0.48 | 62 |
| 16 | PS2-1 | 0 | 0 | 52.7 | 28 | 16.9 | 0 | 0 | 0 | 2.4 | 0 | 3.85 | 0.36 | 62 |
| 17 | O.ficus-indica -p | 15.3 | 45.5 | 0 | 39.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.6318 | 72 |
| 18 | G1 | 10.9 | 1.2 | 6.2 | 52.5 | 14.9 | 0 | 14.3 | 0 | 0 | 4.2 | 6.49 | 1.88 | 29 |
| 19 | G2 | 12.2 | 0.8 | 4.9 | 56 | 16.2 | 0 | 9.9 | 0 | 0 | 7.45 | 5.11 | 1.41 | 29 |
| 20 | P1 | 11.4 | 30.3 | 1.5 | 9.2 | 44.4 | 0 | 3.2 | 0 | 0 | 0 | 0 | 2.38 | 30 |
| 21 | P2 | 10.4 | 22.1 | 3.1 | 11.2 | 53.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.98 | 30 |
| 22 | CP | 1.2 | 15.6 | 7.5 | 28.2 | 24.7 | 0 | 5.4 | 4.8 | 12.6 | 0 | 7.57 | 0.37 | 31 |
| 23 | SSP II-a | 8.94 | 38.74 | 0 | 2.18 | 31.47 | 0 | 0 | 2.33 | 16.34 | 0 | 0 | 0.7782 | 73 |
| 24 | PNMP2 | 0 | 5.78 | 28.62 | 14.42 | 41.57 | 7.24 | 2.37 | 0 | 0 | 0 | 0 | 0.7117 | 74 |
| 25 | PNMP3 | 0 | 3.45 | 26.58 | 21.55 | 36.42 | 8.44 | 3.56 | 0 | 0 | 0 | 0 | 0.4336 | 74 |
| 26 | LLPs-D | 6.83 | 2.73 | 9.2 | 19.23 | 58.19 | 0.57 | 3.25 | 0 | 0 | 0 | 0 | 0.61 | 33 |
| 27 | LLPs-L | 5.03 | 19.39 | 6.07 | 22.82 | 37.45 | 7.04 | 2.21 | 0 | 0 | 0 | 0 | 0.92 | 33 |
| 28 | SMWP-1 | 0 | 0 | 27 | 34 | 11 | 0 | 28 | 0 | 0 | 0 | 0.53 | 1.08 | 34 |
| 29 | GRMP1 | 0 | 0 | 0 | 31.5 | 0 | 0 | 68.5 | 0 | 0 | 0 | 0 | 0.1472 | 16 |
| 30 | EAP40-1 | 2.63 | 0 | 36 | 46.79 | 14.58 | 0 | 0 | 0 | 0 | 0 | 0.33 | 0.95 | 35 |
| 31 | EAP60-1 | 3.37 | 2.28 | 2.89 | 43.61 | 37.67 | 0 | 10.18 | 0 | 0 | 0 | 0.48 | 1.49 | 35 |
| 32 | EAP80-2 | 1.22 | 0 | 6.73 | 21.64 | 55.56 | 10.39 | 4.46 | 0 | 0 | 0 | 0.14 | 1.84 | 35 |
| 33 | PS-2 | 4.17 | 17.33 | 18.65 | 35.14 | 19.11 | 0 | 5.59 | 0 | 0 | 0 | 0 | 0.89 | 75 |
| 34 | EUPS-2 | 8.83 | 15.77 | 12.39 | 43.94 | 11.15 | 0 | 7.92 | 0 | 0 | 0 | 0 | 1.36 | 75 |
| 35 | CMP-1 | 4.2 | 0 | 0 | 95.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.65 | 36 |
| 36 | EPS-1 | 0 | 0 | 10.6 | 84 | 5.4 | 0 | 0 | 0 | 0 | 0 | 19.86 | 4.84 | 76 |
| 37 | EPS-2 | 0 | 0 | 32.7 | 57.3 | 10.9 | 0 | 0 | 0 | 0 | 0 | 20.3 | 2.69 | 76 |
| 38 | IPS-1 | 0 | 0 | 59 | 8.5 | 32.6 | 0 | 0 | 0 | 0 | 0 | 33.97 | 1.32 | 76 |
| 39 | IPS-2 | 0 | 0 | 42.2 | 19.8 | 38 | 0 | 0 | 0 | 0 | 0 | 20.38 | 1.58 | 76 |
| 40 | IPS-3 | 0 | 0 | 27.2 | 72.8 | 0 | 0 | 0 | 0 | 0 | 0 | 1.9 | 1.91 | 76 |
| 41 | PPM | 0 | 0 | 69.1 | 7.8 | 23.1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.99 | 64 |
| 42 | GPA1 | 0.4 | 21.2 | 10.6 | 13.8 | 27.5 | 2.2 | 0 | 14.8 | 9.5 | 23.04 | 3.75 | 0.22 | 37 |
| 43 | GPA2 | 0.8 | 15.6 | 8.2 | 18 | 21.4 | 1.6 | 1.6 | 18 | 14.8 | 32.79 | 4.38 | 0.21 | 37 |
| 44 | GPA3 | 3.8 | 7.5 | 6.3 | 34.3 | 16.3 | 1.3 | 3.1 | 24.3 | 3.1 | 27.01 | 5.53 | 0.2 | 37 |
| 45 | RCP-II | 9.8 | 21.3 | 0 | 7.9 | 33.8 | 0 | 9.3 | 0 | 17.9 | 23.6 | 0 | 0.96 | 77 |
| 46 | AAP-2A | 8 | 25.7 | 0 | 49.3 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0.022 | 65 |
| 47 | TPC | 0 | 12.7 | 0 | 11.2 | 5.4 | 0 | 33.8 | 27.1 | 0 | 0 | 0 | 0.101 | 78 |
| 48 | TPC-1 | 0 | 21.2 | 16 | 26.3 | 6.4 | 0 | 17.3 | 0 | 0 | 30 | 2.8 | 0.184 | 79 |
| 49 | TPC-2 | 0 | 26.4 | 13.9 | 37.5 | 0 | 0 | 0 | 0 | 0 | 47.6 | 3.8 | 0.158 | 79 |
| 50 | TPC-3 | 0 | 37.2 | 0 | 14.9 | 8.3 | 0 | 23.1 | 0 | 0 | 51.8 | 4 | 0.093 | 79 |
| 51 | GO-2 | 24.2 | 0 | 0 | 25.8 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 1.13 | 80 |
| 52 | GO-3 | 24.5 | 0 | 0 | 14.1 | 0 | 0 | 0 | 61.4 | 0 | 0 | 0 | 0.93 | 80 |
| 53 | GO-4 | 22.4 | 0 | 0 | 0.7 | 0 | 0 | 0 | 76.9 | 0 | 0 | 0 | 0.7 | 80 |
| 54 | RNLP I | 10.1 | 51.7 | 3.5 | 22.3 | 8.8 | 0 | 3.6 | 0 | 0 | 6.71 | 0 | 1.74 | 66 |
| 55 | PSCK2-2 | 4 | 12.4 | 43.3 | 0 | 0 | 36.4 | 0 | 0 | 4 | 24.7 | 0 | 1.5 | 81 |
| 56 | PSCK2-3 | 5 | 11.3 | 45.7 | 2.5 | 0 | 35.5 | 0 | 0 | 0 | 6.64 | 0 | 4.8 | 81 |
| 57 | APs-1-1 | 1.4 | 8.1 | 0 | 68.2 | 0 | 0 | 22.3 | 0 | 0 | 0 | 3.1 | 0.2092 | 40 |
| 58 | APs-2-1 | 4.6 | 8 | 0 | 32.3 | 24.2 | 0 | 21.1 | 0 | 9.8 | 0 | 1.9 | 0.1967 | 40 |
| 59 | APs-3-1 | 1.5 | 2.8 | 0 | 35.1 | 34 | 0 | 16.7 | 2 | 7.9 | 0 | 1.3 | 0.1715 | 40 |
| 60 | WSEPS | 0 | 14.5 | 0 | 31.9 | 40.6 | 0 | 0 | 13 | 0 | 0 | 0 | 0.07 | 82 |
| 61 | CT-EPS | 11.4 | 7.4 | 19 | 40 | 13.5 | 0 | 8.7 | 0 | 0 | 0 | 14.87 | 1.62 | 42 |
| 62 | PSS-EPS | 8.2 | 7.7 | 24 | 35.3 | 15.4 | 0 | 9.4 | 0 | 0 | 0 | 20.19 | 1.119 | 42 |
| 63 | PSS-DEPS | 3.3 | 5.6 | 25.5 | 31.5 | 29.8 | 0 | 4.3 | 0 | 0 | 0 | 26.47 | 3.522 | 42 |
| 64 | CT-IPS | 2.1 | 6.2 | 18 | 59.7 | 9 | 0 | 5 | 0 | 0 | 0 | 25.06 | 8.828 | 42 |
| 65 | PSS-IPS | 1.7 | 6.9 | 8.6 | 73.1 | 5 | 0 | 4.7 | 0 | 0 | 0 | 10.82 | 0.779 | 42 |
| 66 | PS2 | 10.96 | 5.81 | 36.16 | 26.92 | 14.55 | 4.52 | 1.04 | 0 | 0 | 0 | 0 | 0.98 | 44 |
| 67 | PS3 | 48.55 | 10.73 | 7.35 | 11.41 | 13.85 | 4.62 | 3.45 | 0 | 0 | 0 | 0 | 0.66 | 44 |
aname from reference
brhamnose
carabinose
dmannose
eglucose
f galactose
gfucose
hxylose
iglucuronic acid
jgalacturonic acid
kuronic acid
lprotein content
We selected five relevant descriptors in MLR model, and a stable model EC50 = 0.12PC+0.083Fuc+0.013Rha-0.02UA+0.372 (R = 0.664, RMSE = 1.149, F = 8.268, p<5.17E-5) was given. According to the model, PC, Fuc, Rha and UA had significant correlation with EC50 of the hydroxyl radicals scavenging activity, and the relevant correlation coefficient was shown in Table 6.
Table 6. Correlation matrix showing inter-correlation among various parameters and EC50 of the hydroxyl radicals scavenging activity.
| EC50 | PC | Fuc | Rha | UA | |
|---|---|---|---|---|---|
| EC50 | 1.000000 | ||||
| PCa | 0.515359 | 1.000000 | |||
| Fucb | 0.270504 | -0.134435 | 1.000000 | ||
| Rhac | -0.093930 | -0.125825 | -0.017084 | 1.000000 | |
| UAd | -0.126494 | -0.028296 | 0.167403 | -0.180576 | 1.000000 |
aprotein content
bfucose
crhamnose
duronic acid
The statistical parameters of MLR, ANN and SVM models for the train set and the test set were shown in Table 7. According to a lower RMSE and a higher R value, the results indicated that nonlinear model ANN was better than models obtained from MLR and SVM for the prediction of hydroxyl radicals scavenging activity of polysaccharides.
Table 7. Comparison of MLR, ANN and SVM models for the hydroxyl radicals scavenging activity of polysaccharides.
| Method | Parameters | Training set | Test set |
|---|---|---|---|
| MLR | R | 0.664 | 0.523 |
| RMSE | 1.149 | 1.117 | |
| ANN | R | 0.944 | 0.857 |
| RMSE | 0.119 | 0.257 | |
| SVM | R | 0.836 | 0.767 |
| RMSE | 0.751 | 0.645 |
Sensitivity analysis from ANN
According to two ANN models, the results of sensitivity analysis were shown in Table 8. The higher sensitivity coefficient indicated that this descriptor had the more influence upon the antioxidant activity of polysaccharides. The results indicated that Ara and GalA had a great effect on DPPH-scavenging activity, and PC, UA and GalA had a great effect on hydroxyl radicals scavenging activity of polysaccharides, which was consistent with the results from MLR.
Table 8. Sensitivity analysis from ANN models.
| Sensitivity coefficients | Composition | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Araa | GalAb | PCc | GlcAd | UAe | Glcf | Xylg | Manh | Gali | Fucj | Rhak | |
| DPPH-scavenging activity | 6.48 | 3.4 | 3.25 | 2.98 | 2.61 | 1.52 | 1.29 | 1.23 | 1.11 | 1.11 | 0.97 |
| Hydroxyl radicals scavenging activity | 1.85 | 4.76 | 7.37 | 1.21 | 3.78 | 3.24 | 1.08 | 1.49 | 2.9 | 3.44 | 1.35 |
aarabinose
bgalacturonic acid
cprotein content
dglucuronic acid
euronic acid
fglucose
gxylose
hmannose
igalactose
jfucose
krhamnose
Conclusions
To establish quantitative structure-activity relationship (QSAR) models for antioxidant activity of polysaccharides, MLR, SVM and ANN methods were used, and polysaccharide properties (UA, PC, monosaccharide compositions, MW) as descriptors were selected. MLR models for predicting EC50 of DPPH-scavenging activity and hydroxyl radicals scavenging activity of polysaccharides consisted of four major descriptors, and the models were EC50 = 0.033Ara- 0.041GalA- 0.03GlcA- 0.025PC +0.484 and EC50 = 0.12PC +0.083Fuc +0.013Rha -0.02UA+0.372, respectively. A comparison of results from models indicated that the ANN model with R = 0.96 and RMSE = 0.018 predicted more accurately the DPPH-scavenging activity of polysaccharides than SVM and MLR models. ANN model (R = 0.933, RMSE = 0.055) was also the best one for predicting the hydroxyl radicals scavenging activity of polysaccharides. According to MLR and ANN models, Ara and GalA were most critical in determining the DPPH-scavenging activity of polysaccharides, and PC, UA and GalA had a great effect on hydroxyl radicals scavenging activity of polysaccharides. The polysaccharide of MW 4000–100000 usually owned higher DPPH-scavenging activity, but the antioxidant activity could simultaneously be affected by other polysaccharide properties. These results may provide some new insights in the complex study of polysaccharide structure and bioactivities, and we can simply predict the antioxidant activity of polysaccharide by using the established models after determining the monosaccharide composition ratios and MW.
It is worth noting that the highly GalA-containing polysaccharide could exhibit significantly antioxidant activity, which might be because they owned the functional group–COOH. It has been reported that the functional groups such as–COOH, CH3CO–and–SH were generally recognized as good electron or hydrogen donors that might be related to the antioxidant activity of polysaccharides [5]. The antioxidant activity of polysaccharide was also found to correlate to complex structure such as glycosidic linkages, branch ratios, and microstructure etc, polysaccharide properties is not enough for fine detailed structure of polysaccharide, and the research on more precise structure-function relationships remained to be explored.
Acknowledgments
This work was supported by Promotion Program for Young and Middle-aged Teacher in Science and Technology Research of Huaqiao University (ZQN-PY316), National Nature Science Foundation of China (31201314), and Cultivation Project Funds for Postgraduates Innovation Ability of Huaqiao University.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by Promotion Program for Young and Middle-aged Teacher in Science and Technology Research of Huaqiao University (ZQN-PY316), National Nature Science Foundation of China (31201314), and Cultivation Project Funds for Postgraduates Innovation Ability of Huaqiao University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sun YX, Kennedy JF. Antioxidant activities of different polysaccharide conjugates (CRPs) isolated from the fruiting bodies of Chroogomphis rutilus (Schaeff.: Fr.) O. K. Miller. Carbohydr Polym. 2010; 82: 510–514. 10.1016/j.carbpol.2010.05.010 [DOI] [Google Scholar]
- 2.Liu J, Luo JG, Ye H, Zeng XX. Preparation, antioxidant and antitumor activities in vitro of different derivatives of levan from endophytic bacterium Paenibacillus polymyxa EJS-3. Food Chem Toxicol. 2012; 50: 767–772. 10.1016/j.fct.2011.11.016 [DOI] [PubMed] [Google Scholar]
- 3.Li SQ, Shah NP. Antioxidant and antibacterial activities of sulphated polysaccharides from Pleurotus eryngii and Streptococcus thermophilus ASCC 1275. Food Chem. 2014; 165: 262–270. 10.1016/j.foodchem.2014.05.110 [DOI] [PubMed] [Google Scholar]
- 4.Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007; 39: 44–84. 10.1016/j.biocel.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 5.Wang K, Li W, Rui X, Li T, Chen X, Jiang M, et al. Chemical modification, characterization and bioactivity of a released exopolysaccharide (r-EPS1) from Lactobacillus plantarum 70810. Glycoconj J. 2015; 32: 17–27. 10.1007/s10719-014-9567-1 [DOI] [PubMed] [Google Scholar]
- 6.Zhang K, Ding W, Sun J, Zhang B, Lu F, Lai R, et al. Antioxidant and antitumor activities of 4-arylcoumarins and 4-aryl-3,4- dihydrocoumarins. Biochimie. 2014; 107: 203–210. 10.1016/j.biochi.2014.03.014 [DOI] [PubMed] [Google Scholar]
- 7.Zhu BW, Wang LS, Zhou DY, Li DM, Sun LM, Yang JF, et al. Tada M. Antioxidant activity of sulphated polysaccharide conjugates from abalone (Haliotis discus hannai Ino). Eur Food Res Technol. 2008; 227: 1663–1668. [Google Scholar]
- 8.Kan YJ, Chen TQ, Wu YB, Wu JG, Wu JZ. Antioxidant activity of polysaccharide extracted from Ganoderma lucidum using response surface methodology. Int J Biol Macroml. 2015; 72: 151–157. 10.1016/j.ijbiomac.2014.07.056 [DOI] [PubMed] [Google Scholar]
- 9.Nandita S, Rajini PS. Free radical scavenging activity of an aqueous extract of potato peel. Food Chem. 2004; 85: 611–616. 10.1016/j.foodchem.2003.07.003 [DOI] [Google Scholar]
- 10.Chen Y, Zhang H, Wang YX, Nie SP, Li C, Xie MY. Acetylation and carboxymethylation of the polysaccharide from Ganoderma atrum and their antioxidant and immunomodulating activities. Food Chem. 2014; 156: 279–288. 10.1016/j.foodchem.2014.01.111 [DOI] [PubMed] [Google Scholar]
- 11.Zhang ZS, Zhang QB, Wang J, Zhang H, Niu XZ, Li PC. Preparation of the different derivatives of the low-molecular-weight porphyran from Porphyra haitanensis and their antioxidant activities in vitro. Int J Biol Macromol. 2009; 45: 22–26. 10.1016/j.ijbiomac.2009.03.009 [DOI] [PubMed] [Google Scholar]
- 12.Lo TCT, Jiang YH, Chao ALJ, Chang CA. Use of statistical methods to find the polysaccharide structural characteristics and the relationships between monosaccharide composition ratio and macrophage stimulatory activity of regionally different strains of Lentinula edodes. Analytica Chinica Acta. 2007; 584:50–56. 10.1016/j.aca.2006.10.051 [DOI] [PubMed] [Google Scholar]
- 13.Jia L, Shen Z, Guo W, Zhang Y, Zhu H, Ji W, Fan M. QSAR models for oxidative degradation of organic pollutants in the Fenton process. J Taiwan Inst Chem E. 2015; 46: 140–147. 10.1016/j.jtice.2014.09.014 [DOI] [Google Scholar]
- 14.Yangali-Quintanilla V, Sadmani A, McConville M, Kennedy M, Amy G. A QSAR model for predicting rejection of emerging contaminants (pharmaceuticals, endocrine disruptors) by nanofiltration membranes. Water Res. 2010; 44: 373–384. 10.1016/j.watres.2009.06.054 [DOI] [PubMed] [Google Scholar]
- 15.De Ridder D, Villacorte L, Verliefde A, Verberk J, Heijman S, Amy G, et al. Modeling equilibrium adsorption of organic micropollutants onto activated carbon. Water Res 2010; 44: 3077–3086. 10.1016/j.watres.2010.02.034 [DOI] [PubMed] [Google Scholar]
- 16.Gao CJ, Wang YH, Wang CY, Wang ZY. Antioxidant and immunological activity in vitro of polysaccharides from Gomphidius rutilus mycelium. Carbohydrate Polymers.2013; 92: 2187–2192. 10.1016/j.carbpol.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 17.Shao P, Chen M, Pei YP, Sun PL. In intro antioxidant activities of different sulfated polysaccharides from chlorophytan seaweeds Ulva fasciata. International Journal of Biological Macromolecules. 2013; 59: 295–300. 10.1016/j.ijbiomac.2013.04.048 [DOI] [PubMed] [Google Scholar]
- 18.Fleita D, El-Sayed M, Rifaat D. Evaluation of the antioxidant activity of enzymatically-hydrolyzed sulfated polysaccharides extracted from red algae; Pterocladia capillacea. LWT—Food Science and Technology. 2015; 63: 1236–1244. 10.1016/j.lwt.2015.04.024 [DOI] [Google Scholar]
- 19.Yuan F, Yu RM, Yin Y, Shen JR, Dong QF, Zhong L, et al. Structure characterization and antioxidant activity of a novel polysaccharide isolated from Ginkgo biloba. Int J Biol Macromol. 2010; 46: 436–439. 10.1016/j.ijbiomac.2010.02.002 [DOI] [PubMed] [Google Scholar]
- 20.Zhang M, Wang F, Liu R, Tang XL, Zhang Q, Zhang ZS. Effects of superfine grinding on physicochemical and antioxidant properties of Lycium barbarum polysaccharides. LWT—Food Science and Technology. 2014; 58: 594–601. 10.1016/j.lwt.2014.04.020 [DOI] [Google Scholar]
- 21.Mao JW, Yin J, Ge Q, Jiang ZL, Gong JY. In vitro antioxidant activities of polysaccharides extracted from Moso Bamboo-Leaf. Int J Biol Macromol. 2013; 55: 1–5. 10.1016/j.ijbiomac.2012.12.027 [DOI] [PubMed] [Google Scholar]
- 22.Du XJ, Mu HM, Zhou S, Zhang Y, Liu XL. Chemical analysis and antioxidant activity of polysaccharides extracted from Inonotus obliquus sclerotia. Int J Biol Macroml. 2013; 62: 691–696. 10.1016/j.ijbiomac.2013 [DOI] [PubMed] [Google Scholar]
- 23.Liu CH, Chang JK, Zhang L, Zhang J, Li SY. Purification and antioxidant activity of a polysaccharide from bulbs of Fritillaria ussuriensis Maxim. Int J Biol Macromo. 2012; l 50: 1075–1080. 10.1016/j.ijbiomac.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 24.Feng SL, Cheng HR, Fu Liang, Ding CB, Zhang Li, et al. Ultrasonic-assisted extraction and antioxidant activities of polysaccharides from Camellia oleifera leaves. Int J Biol Macroml. 2014; 68: 7–12. 10.1016/j.ijbiomac.2014.04.026 [DOI] [PubMed] [Google Scholar]
- 25.Dou J, Meng YH, Liu L, Li J, Ren DY, et al. Purification, characterization and antioxidant activities ofpolysaccharides from thinned-young apple. International Journal of Biological Macromolecules. 2015; 72: 31–40. 10.1016/j.ijbiomac.2014.07.053 [DOI] [PubMed] [Google Scholar]
- 26.Li C, Huang Q, Fu X, Yue XJ, Liu RH, You LJ. Characterization, antioxidant and immunomodulatory activities of polysaccharides from Prunella vulgaris Linn. Int J Biol Macromol. 2015; 75: 298–305. 10.1016/j.ijbiomac.2015.01.010 [DOI] [PubMed] [Google Scholar]
- 27.Xu XQ, Quan LL, Shen MW. Effect of chemicals on production, composition and antioxidantactivity of polysaccharides of Inonotus obliquus. International Journal of Biological Macromolecules. 2015; 77: 143–150. 10.1016/j.ijbiomac.2015.03.013 [DOI] [PubMed] [Google Scholar]
- 28.Asker MMS, Ahmed YM, Ramadan MF. Chemical characteristics and antioxidant activity of exopolysaccharide fractions from Microbacterium terregens. Carbohydrate Polymers. 2009; 77: 563–567. 10.1016/j.carbpol.2009.01.037 [DOI] [Google Scholar]
- 29.Zhang ZS, Wang XM, Zhang JJ, Zhao MX. Potential antioxidant activities in vitro of polysaccharides extracted from ginger (Zingiber officinale). Carbohydrate Polymers. 2011; 86: 448–452. 10.1016/j.carbpol.2011.04.062 [DOI] [Google Scholar]
- 30.Zhang ZS, Wang XM, Han ZP, Zhao MX, Yin L. Purification, antioxidant and moisture-preserving activities of polysaccharides from papaya. Carbohydrate Polymers. 2012; 87: 2332–2337. 10.1016/j.carbpol.2011.10.067 [DOI] [Google Scholar]
- 31.Xie JH, Wang ZJ, Shen MY, Nie SP, Gong B, Li HS, et al. Sulfated modification, characterization and antioxidant activities of polysaccharide from Cyclocarya paliurus. Food Hydrocolloids. 2016; 53: 7–15. 10.1016/j.foodhyd.2015.02.018 [DOI] [Google Scholar]
- 32.Lina FB, Miguel AC, José AT, Solange IM. Characterization of polysaccharides extracted from spent coffeegrounds by alkali pretreatment. Carbohydrate Polymers. 2015; 127: 347–354. 10.1016/j.carbpol.2015.03.047 [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Tu ZC, Wang H, Kou Y, Wen QH, et al. Response surface optimization and physicochemical properties of polysaccharides from Nelumbo nucifera leaves. Int J Biol Macromol. 2015; 74: 103–110. 10.1016/j.ijbiomac.2014.11.020 [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y, Wang L, Zhang L, Wang T, Zhou Y, et al. Optimization of extraction and antioxidant activity of polysaccharides from Salvia miltiorrhiza Bunge residue. Int J Biol Macromo. 2015; 79: 533–541. 10.1016/j.ijbiomac.2015.05.024 [DOI] [PubMed] [Google Scholar]
- 35.Cheng HR, Feng SL, Shen SA, Zhang L, Yang RW, Zhou YH, et al. Extraction, antioxidant and antimicrobial activities of Epimedium acuminatum Franch. polysaccharide. Carbohydr Polym. 2013; 96: 101–108. 10.1016/j.carbpol.2013.03.072 [DOI] [PubMed] [Google Scholar]
- 36.Jing YS, Cui XL, Chen ZY, Huang LJ, Song LY, et al. Elucidation and biological activities of a new polysaccharide from cultured Cordyceps militaris. Carbohydrate Polymers. 2014; 102: 288–296. 10.1016/j.carbpol.2013.11.061 [DOI] [PubMed] [Google Scholar]
- 37.Li B, Zhang XY, Wang MZ, Jiao LL. Characterization and antioxidant activities of acidic polysaccharides from Gynostemma pentaphyllum (Thunb.) Markino. Carbohydr Polym. 2015; 127: 209–214. 10.1016/j.carbpol.2015.03.069 [DOI] [PubMed] [Google Scholar]
- 38.Xie JH, Zhang F, Wang ZJ, Shen MY, Nie SP, Xie MY. Preparation, characterization and antioxidant activities of acetylated polysaccharides from Cyclocarya paliurus leaves. Carbohydrate Polymers. 2015; 133: 596–604. 10.1016/j.carbpol.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 39.He NW, Yang XB, Jiao YD, Tian LM, Zhao Y. Characterisation of antioxidant and antiproliferative acidic polysaccharides from Chinese wolfberry fruits. Food Chemistry. 2012; 133: 978–989. 10.1016/j.foodchem.2012.02.018 [DOI] [Google Scholar]
- 40.Chen RZ, Tan L, Jin CG, Lu J, Tian L, Chang QQ, et al. Extraction, isolation, characterization and antioxidant activity of polysaccharides from Astragalus membranaceus. Industrial Crops and Products. 2015; 77: 434–443. 10.1016/j.indcrop.2015.08.006 [DOI] [Google Scholar]
- 41.Xu P, Wu J, Zhang Y, Chen H, Wang YF. Physicochemical characterization of puerh tea polysaccharides and their antioxidant and α-glycosidase inhibition. Journal of Functional Foods. 2014; 6: 545–554. 10.1016/j.jff.2013.11.021 [DOI] [Google Scholar]
- 42.Xu XQ, Hu Y, Zhu LH. The capability of Inonotus obliquus for lignocellulosic biomass degradation in peanut shell and for simultaneous production of bioactive polysaccharides and polyphenols in submerged fermentation. J TaiWan Inst Chem E. 2014; 45: 2851–2858. 10.1016/j.jtice.2014.08.029 [DOI] [Google Scholar]
- 43.Mengome LE, Voxeur A, Akue JP, Lerouge P. Screening of antioxidant activities of polysaccharides extracts from endemic plants in Gabon. Bioactive Carbohydrates and Dietary Fibre. 2014; 3: 77–88. 10.1016/j.bcdf.2014.02.001 [DOI] [Google Scholar]
- 44.Kang MC, Kim SY, Kim YT, Kim EA, Lee SH, et al. In vitro and in vivo antioxidant activities of polysaccharide purified from aloe vera (Aloe barbadensis) gel. Carbohydr Polym. 2014; 99: 365–371 10.1016/j.carbpol.2013.07.091 [DOI] [PubMed] [Google Scholar]
- 45.Gao TT, Ma S, Song JY, Bi HT, Tao YD. Antioxidant and immunological activities of water-soluble polysaccharides from Aconitum kusnezoffii Reichb. Int J Biol Macromol. 2011; 49: 580–586. 10.1016/j.ijbiomac.2011.06.017 [DOI] [PubMed] [Google Scholar]
- 46.Tropsha A, Gramatica P, Gombar VK. The importance of being earnest: Validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb SCI. 2003; 22: 69–77. 10.1002/qsar.200390007 [DOI] [Google Scholar]
- 47.Gramatica P. Principles of QSAR models validation: Internal and external. QSAR Comb SCI. 2007; 26: 694–701. 10.1002/qsar.200610151 [DOI] [Google Scholar]
- 48.Witten IH. Frank E. Data Mining: Practical Machine Learning Tools and Techniques, second ed. Morgan Kaufmann, San Francisco, 2005. [Google Scholar]
- 49.Bruno L, Vijay KA, Padmaka VK. Prediction of intrinsic solubility of generic drugs using MLR, ANN and SVM analyses. Eur J Med Chem. 2010; 45: 4018–4025. 10.1016/j.ejmech.2010.05.059 [DOI] [PubMed] [Google Scholar]
- 50.Zupan J. Introduction to artificial neural network (ANN) methods: What they are and how to use them. Acta Chim Slov. 1994; 41: 327–352. [Google Scholar]
- 51.Peterson KL. Artificial neural networks and their use in chemistry. Rev Comp Ch. 2000; 16: 53–140. 10.1002/9780470125939.ch2 [DOI] [Google Scholar]
- 52.Wythoff BJ. Backpropagation neural networks: A tutorial. Chemometr Intell Lab Syst. 1993; 18: 115–155. [Google Scholar]
- 53.Cortes C, Vapnik V. Support vector networks. Mach Learn. 1995; 20: 273–293. [Google Scholar]
- 54.Burges CA. Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discov. 1998; 2: 121–167. [Google Scholar]
- 55.Doucet JP, Barbault F, Xia H, Panaye A, Curr BF. Nonlinear SVM approaches to QSPR/QSAR studies and drug design. Comput. -Aided Drug Des. 2007; 3: 263–289. 10.2174/157340907782799372 [DOI] [Google Scholar]
- 56.Li H, Liang Y, Xu Q. Support vector machines and its applications in chemistry. Chemometr Intell Lab. 2009; 95: 188–198. 10.1016/j.chemolab.2008.10.007 [DOI] [Google Scholar]
- 57.Wang WJ, Xu ZB, Lu WZ, Zhang XY. Determination of the spread parameter in the gaussian kernel for classification and regression neurocomputingig. Neurocomputing. 2003; 55: 643–646. 10.1016/S0925-2312(02)00632-X [DOI] [Google Scholar]
- 58.Kousik M, Eshita K, Saikat M, Sanjoy KG, Debsankar D, et al. Structural characterization and study of immunoenhancing and antioxidant property of a novel polysaccharide isolated from the aqueous extract of a somatichybrid mushroom of Pleurotus florida and Calocybe indica variety APK2. International Journal of Biological Macromolecules.2011; 48: 304–310. 10.1016/j.ijbiomac.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 59.Zhang YX, Dai L, Kong XW, Chen LW. Characterization and in vitro antioxidant activities of polysaccharides from Pleurotus ostreatus. International Journal of Biological Macromolecules. 2012; 51: 259–265. 10.1016/j.ijbiomac.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 60.Ding X, Hou YL, Hou WR. Structure elucidation and antioxidant activity of a novel polysaccharide isolated from Boletus speciosus Forst. International Journal of Biological Macromolecules. 2012; 50: 613–618. 10.1016/j.ijbiomac.2012.01.021 [DOI] [PubMed] [Google Scholar]
- 61.Zhang XH, Liu L, Lin CW. Structural features, antioxidant and immunological activity of a new polysaccharide (SP1) from sisal residue. International Journal of Biological Macromolecules. 2013; 59: 184–191. 10.1016/j.ijbiomac.2013.04.052 [DOI] [PubMed] [Google Scholar]
- 62.Sun HH, Mao WJ, Chen Y, Guo SD, Li HY, et al. Isolation, chemical characteristics and antioxidant properties of the polysaccharides from marine fungus Penicillium sp. F23-2. Carbohydrate Polymers. 2009; 78: 117–124. 10.1016/j.carbpol.2009.04.017 [DOI] [Google Scholar]
- 63.Zeng WC, Zhang Z, Gao H, Jia LR, Chen WY. Characterization of antioxidant polysaccharides from Auricularia auricular using microwave-assisted extraction. Carbohydrate Polymers. 2012; 89: 694–700. 10.1016/j.carbpol.2012.03.078 [DOI] [PubMed] [Google Scholar]
- 64.Jiang P, Yuan L, Cai DL, Jiao LL, Zhang LP. Characterization and antioxidant activities of the polysaccharidesfrom mycelium of Phellinus pini and culture medium. Carbohydrate Polymers. 2015; 117: 600–604 10.1016/j.carbpol.2014.10.013 [DOI] [PubMed] [Google Scholar]
- 65.Pu XY, Ma XL, Liu L, Ren J, Li HB, Li XY, et al. Structural characterization and antioxidant activity in vitro of polysaccharides from angelica and astragalus. Carbohydrate Polymers. 2016; 137: 154–164. 10.1016/j.carbpol.2015.10.053 [DOI] [PubMed] [Google Scholar]
- 66.Xu YQ, Cai F, Yu ZY, Zhang L, Li XG, et al. Optimisation of pressurised water extraction of polysaccharides from blackcurrant and its antioxidant activity. Food Chemistry. 2016; 194: 650–658. 10.1016/j.foodchem.2015.08.061 [DOI] [PubMed] [Google Scholar]
- 67.Aguirre MJ, Isaacs M, Matsuhiro B, Mendoza L, Zúñiga EA. Characterization of a neutral polysaccharide with antioxidant capacity from red wine. Carbohydrate Research. 2009; 344: 1095–1101. 10.1016/j.carres.2009.03.024 [DOI] [PubMed] [Google Scholar]
- 68.Yu RM, Yin Y, Yang W, Ma WL, Yang L, et al. Structural elucidation and biological activity of a novel polysaccharide by alkaline extraction from cultured Cordyceps militaris. Carbohydrate Polymers. 2009; 75: 166–171. 10.1016/j.carbpol.2008.07.023 [DOI] [Google Scholar]
- 69.He L, Ji PF, Gong XG, Li WQ, Cheng JW. Physico-chemical characterization, antioxidant and anticancer activities in vitro of a novel polysaccharide from Melia toosendan Sieb. Et Zucc fruit. International Journal of Biological Macromolecules. 2011; 49: 422–427. 10.1016/j.ijbiomac.2011.05.028 [DOI] [PubMed] [Google Scholar]
- 70.Xiao J, Sun J, Yao LY, Zhao QS, Wang LW, et al. Physicochemical characteristics of ultrasonic extracted polysaccharides from cordyceps cephalosporium mycelia. Int J Biol Macroml. 2012; 51: 64–69. 10.1016/j.ijbiomac.2012.04.029 [DOI] [PubMed] [Google Scholar]
- 71.Zhang JJ, Meng GY, Zhai GY, Yang YH, Zhao HJ, et al. Extraction, characterization and antioxidant activity of polysaccharides of spent mushroom compost of Ganoderma lucidum. Int J Biol Macroml. 2016; 82: 432–439. 10.1016/j.ijbiomac.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 72.Zhong XK, Jin X, Lai FY, Lin QS, Jiang JG. Chemical analysis and antioxidant activities in vitro of polysaccharide extracted from Opuntia ficus indica Mill. cultivated in China. Carbohydrate Polymers. 2010; 82: 722–727. 10.1016/j.carbpol.2010.05.042 [DOI] [Google Scholar]
- 73.Feng K, Chen W, Sun LW, Liu JZ, Zhao YX, et al. Optimization extraction, preliminary characterization and antioxidantactivity in vitro of polysaccharides from Stachys sieboldii Miq. Tubers. Carbohydrate Polymers. 2015; 125: 45–52. 10.1016/j.carbpol.2015.02.026 [DOI] [PubMed] [Google Scholar]
- 74.Wang ZY, Zhou F, Quan Y. Antioxidant and immunological activity in vitro of polysaccharides from Phellinus nigricans mycelia. Int J Biol Macromo. 2014; 64: 139–143. 10.1016/j.ijbiomac.2013.11.038 [DOI] [PubMed] [Google Scholar]
- 75.Chen SH, Chen HX, Tian JG, Wang YW, Xing LS. Enzymolysis-ultrasonic assisted extraction, chemical characteristicsand bioactivities of polysaccharides from corn silk. Carbohydrate Polymers. 2014; 101: 332–341. 10.1016/j.carbpol.2013.09.046 [DOI] [PubMed] [Google Scholar]
- 76.Meng L, Sun SS, Li R, Shen ZP, Wang P, Jiang XL. Antioxidant activity of polysaccharides produced by Hirsutella sp. andrelation with their chemical characteristics. Carbohydrate Polymers. 2015; 117: 452–457. 10.1016/j.carbpol.2014.09.076 [DOI] [PubMed] [Google Scholar]
- 77.Yu ZY, Liu L, Xu YQ, Wang L, Teng X, et al. Characterization and biological activities of a novel polysaccharide isolated from raspberry (Rubus idaeus L.) fruits. Carbohydrate Polymers. 2015; 132: 180–186. 10.1016/j.carbpol.2015.06.068 [DOI] [PubMed] [Google Scholar]
- 78.Chen HX, Zhang M, Xie BJ. Components and antioxidant activity of polysaccharide conjugate from green tea. Food Chemistry. 2005; 90: 17–21. 10.1016/j.foodchem.2004.03.001 [DOI] [Google Scholar]
- 79.Chen HX, Zhang M, Qu ZS, Xie BJ. Antioxidant activities of different fractions of polysaccharide conjugates from green tea (Camellia Sinensis). Food Chemistry. 2008; 106: 559–563. 10.1016/j.foodchem.2007.06.040 [DOI] [Google Scholar]
- 80.Redouan E, Emmanuel P, Michelle P, Bernard C, Josiane C, et al. Evaluation of antioxidant capacity of ulvan-like polymer obtained by regioselective oxidation of gellan exopolysaccharide. Food Chemistry. 2011; 127: 976–983. 10.1016/j.foodchem.2011.01.067 [DOI] [PubMed] [Google Scholar]
- 81.Zhang GY, Yin QS, Han T, Zhao YX, Su JJ. Purification and antioxidant effect of novel fungal polysaccharides from the stroma of Cordyceps kyushuensis. Industrial Crops and Products. 2015; 69: 485–491. 10.1016/j.indcrop.2015.03.006 [DOI] [Google Scholar]
- 82.Wang HG, Jiang XL, Mu HJ, Liang XT, Guan HS. Structure and protective effect of exopolysaccharide from P. Agglomerans strain KFS-9 against UV radiation. Microbiological Research. 2007; 162: 124–129. 10.1016/j.micres.2006.01.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.