Abstract
Thrips palmi Karny (Thysanoptera: Thripidae) is the sole vector of Watermelon bud necrosis tospovirus, where the crop loss has been estimated to be around USD 50 million annually. Chemical insecticides are of limited use in the management of T. palmi due to the thigmokinetic behaviour and development of high levels of resistance to insecticides. There is an urgent need to find out an effective futuristic management strategy, where the small RNAs especially microRNAs hold great promise as a key player in the growth and development. miRNAs are a class of short non-coding RNAs involved in regulation of gene expression either by mRNA cleavage or by translational repression. We identified and characterized a total of 77 miRNAs from T. palmi using high-throughput deep sequencing. Functional classifications of the targets for these miRNAs revealed that majority of them are involved in the regulation of transcription and translation, nucleotide binding and signal transduction. We have also validated few of these miRNAs employing stem-loop RT-PCR, qRT-PCR and Northern blot. The present study not only provides an in-depth understanding of the biological and physiological roles of miRNAs in governing gene expression but may also lead as an invaluable tool for the management of thysanopteran insects in the future.
Introduction
MicroRNAs (miRNAs) are a family of small (~18–25 nucleotides (nts)), endogenously initiated, non-coding RNAs (ncRNAs) that primarily regulate gene expression in animals, plants and protozoan in a sequence-specific manner. In mammals, approximately 60% of protein-coding gene activities are under the control of miRNAs and they regulate almost every cellular process investigated [1,2]. miRNAs can regulate gene expression either by translation repression or by degradation of mRNA through deadenylation [3]. The second to seventh nucleotides in the 5' end of the miRNA form the “seed” region that provides the most of the pairing specificity [4,5]. miRNA-mediated regulation plays a key role in cellular and developmental processes such as cell division, cell death, disease, hormone secretion and neural development [6–9]. Lin-4 was the first member of the miRNA family, discovered in Caenorhabditis elegans, which regulates the timing of larval development [10]. Subsequently, many miRNAs have been revealed from wide varieties of organisms including insects [11], plants [12], viruses [13] and vertebrates [14].
The majority of miRNAs are ~22 nts in length and the biogenesis of miRNAs is a multiple step process that is widely conserved among eukaryotes. miRNA biogenesis requires RNAase III-like enzymes, Drosha and Pasha (DGCR8, in vertebrates), for generating pre-miRNA (~70 nts) from the primary miRNA (pri-miRNA) transcript [12] which are translocated to the cytoplasm by exportin-5 [15]. Another class of RNAase III enzyme, Dicer (1 & 2 in insects), produces a ~22 bp miRNA:miRNA* duplex from the previously generated pre-miRNA in the cytoplasm [16,17]. Mature miRNAs are then selectively loaded into the RNA-induced silencing complex that contains Argonaute family proteins [5]. Thus, the mature RISC containing the guide strand recognizes the complementary mRNA and cleaves thereby inhibiting the protein translation [18]. Identification of miRNA involves three main approaches, forward genetics, bioinformatics prediction [19,20] and direct cloning and sequencing [21,22,11]. In the recent past, the high-throughput next generation sequencing (NGS) technology, for example, Illumina platform (www.illumina.com) and Roche 454 pyrosequencing platform (www.454.com) have become robust methods of identifying miRNAs from animals and plants [23–29]. As a result, several miRNAs have been reported in the recent past from various orders of insects such as Diptera, Hymenoptera, Coleoptera, Orthoptera, Lepidoptera, Hemiptera and Homoptera [27].
Thrips (Thysanoptera: Thripidae) are one of the major sucking pests on many crops and nearly 6000 species are currently described [30]. Less than 1% of them are considered as pests of agricultural and horticultural crops, and cause crop damage either by direct feeding or as vectors by transmitting plant pathogenic tospoviruses [31]. Globally, 12 species of thrips have been reported as vectors of tospoviruses [32] and among them, T. palmi is an important polyphagous pest and an extremely successful invasive species that originated in Southeast Asia but has recently spread to large parts of tropical and sub-tropical countries. In India, Watermelon bud necrosis tospovirus is succesfully transmitted by T. palmi adults that acquire the virus during larval stages [33]. The peculiar feature of thrips transmission is that only the nymphs can acquire the virus while the adults can transmit [34]. The rapid parthenogenetic reproduction and the feeding behavior of thrips can cause considerable crop damage. The worldwide annual loss due to tospoviruses is estimated at USD 1 billion and in Asia alone, it is over USD 89 million [34]. Currently, the management of T. palmi includes mainly chemicals such as imidacloprid and pyrethroids against which T. palmi has already developed resistance [35]. Given the paucity of genomic information of T. palmi, it is important to profile the miRNAs for understanding their regulatory role in the biology and the great promise they hold in futuristic pest management. Thus, for the first time, we identified, characterized and validated both conserved and novel miRNAs from T. palmi. Further analysis identified putative target genes for these miRNAs, which will shed more light on the identification of highly specific miRNAs that can be used shortly for thysanopteran pest management.
Materials and Methods
Ethical treatment of animals
No specific permissions were required for these locations/activities, and field studies did not involve endangered or protected species. Ethical approval was not required to work with thrips, the subject species in this study; T. palmi is an invertebrate and not listed on the endangered species list. Thrips are ubiquitous in their natural ranges.
Insect Culture
T. palmi were reared on French bean pods (Phaseolus vulgaris cv. Arka Komal) in plastic containers (10X10 cm) at 30 ± 2°C, 80 ± 10% RH and 16:8 L:D photoperiod as described previously [36]. Total RNA was isolated from whole-body homogenates of sample mix, containing a total of 50 mg of different life stages such as eggs, larvae, pupae and adults of T. palmi using TRIzol reagent (Invitrogen, Carlsbad CA, USA).
Library preparation and sequence data generation
Samples were processed according to Illumina TruSeq™ Small RNA sample preparation guide. Size fractionated small RNA populations (18–28 nts) were extracted, purified and ligated to 3' and 5' adapters using T4 RNA Ligase (Life Technologies, Ambion, USA). Ligated products were reverse transcribed using SuperScript II (Life Technologies, Invitrogen, USA) followed by PCR amplification with 11 cycles and two size selection gels. High-throughput sequencing of the small RNA libraries was performed on Illumina Hiseq2000.
Bioinformatics analysis
The obtained sequence tags were subjected to a primary analysis in which low-quality tags, 3' and 5' adapter contaminants were discarded. The sequencing data were investigated against the Rfam (http://rfam.sanger.ac.uk/) and RepBase (http://www.girinst.org/repbase/) as references to annotate the ncRNAs namely, rRNAs, tRNAs, snRNAs, snoRNAs and repeat-associated small RNAs and degraded fragments of expressed genes (exons and introns) in the remaining sequences. All such mapped reads were removed from the dataset before further analysis. Remaining unique sequences were aligned with the miRNA sequences available in the miRBase (v21, http://www.miRBase.org/) to identify the conserved miRNAs. Novel miRNA candidates were identified by employing the miRDeep2 [37] and miRCat (http://srna-workbench.cmp.uea.ac.uk/tools/mircat/) software. Frankliniella occidentalis genome (http://www.ncbi.nlm.nih.gov/nuccore/644576459) was used as a reference to extract the potential secondary hairpin structures, employing Vienna RNAfold [38].
Homology analysis
Homology analysis was carried out with conserved miRNAs of T. palmi with the miRNAs of other organisms from the miRBase database (Release 21.0) [39]. BLASTn embedded in the miRBase database was used to compare the T. palmi miRNAs with other species, with an E-value of 0.01 to find out more miRNA homologs. The naming of the miRNAs in this study has been done according to Griffith-Jones, et al., 2006. Since these miRNAs were predicted from T. palmi, the prefix for all miRNAs was fixed as ‘tpa’. The rest of the naming convention criteria were in accordance with miRBase [40].
Phylogenetic analysis of microRNA family
All the identified miRNAs were classified into different miRNA precursor families (www.rfam.sanger.ac.uk). Few miRNA families (miR-279, miR-281, miR-1000 and miR-1175) were selected for phylogenetic analysis. RaxML.v.7.0.4 [41] was employed to construct the Maximum Likelihood (ML) tree with 2000 bootstrap replications.
Prediction of miRNA targets
Target identification is crucial for understanding the biological functions of miRNAs. Unlike the plant counterparts, the imperfect complementarity of animal miRNAs with their target sequences on mRNA makes it more difficult to judge the accuracy of prediction [20]. Targets for identified miRNAs were predicted employing miRanda program [42], against the Expressed Sequence Tags (ESTs) and transcriptome (NCBI Accession: PRJNA203209) database of F. occidentalis. An alignment score [43] greater than or equal to 130 and miRNA:mRNA binding energy (Minimum Free Energy (MFE, ΔG)) less than -20 kcal/mol were considered as putative target genes. The targets were further annotated against NCBI-RefSeq invertebrate protein database and Gene Ontology (GO) terms were assigned (using Blast-2-GO) based on the annotation. The circos plot was generated using Circos [44] to visualize the interaction between miRNAs and their targets.
Validation and quantification of T. palmi miRNAs
stem-loop RT-PCR
We were able to validate few of the conserved and novel microRNAs employing Stem-loop RT-PCR primers designed based on the previous reports [22]. Briefly, stem-loop RT primers bind to the 3' portion of miRNA molecules, initiating reverse transcription of the miRNAs. Later, the RT product was amplified using a miRNA specific forward primer and the universal reverse primer.
Reverse transcription quantitative PCR (qRT-PCR)
In the present study, we selected differentially expressed and functionally significant 13 miRNAs (nine conserved and four novel) for qRT-PCR. Briefly, Mir-X-miRNA qRT-PCR SYBR Kit (Clontech Laboratories, Inc., USA) was used for the qRT-PCR reactions, which has a single-step, single-tube reaction to produce the first-strand cDNA, which was then specifically and quantitatively amplified using a miRNA-specific primer and SYBR Advantage qPCR chemistry. All the qRT-PCR assays were conducted according to the MIQE guidelines [45]. U6 snRNA was used as an internal control gene for normalization. qRT-PCR was performed on Light Cycler 480 (Roche, USA) using 1:20 diluted cDNAs and SYBR Advantage Premix (Clontech Laboratories, Mountain View, USA), according to the manufacturer’s instructions. Assays were performed in triplicates for three independent biological experiments and the relative gene expression data were analyzed using 2-ΔΔCT method [46]. The values of these three independent experiments were statistically analyzed using one-way ANOVA to calculate the statistical significance.
miRNA northern blot
Small RNAs were isolated from pooled T. palmi (eggs, larvae, pupae and adults) employing mirVANA miRNA isolation kit (Life Technologies, USA). 5 μg of T. palmi small RNAs were resolved on 15% polyacrylamide gel containing 8M urea. RNA was electro blotted [TransBlot SD Semi-Dry Electrophoretic transfer cell (Bio-Rad, USA)] for 90 minutes at 20V onto HyBond-N+ membrane (GE Healthcare, USA) and immobilized by UV cross-linking (Stratagene, USA). The membrane was hybridized with 5' digoxigenin-labeled locked nucleic acid probes for miRNA detection (100ng/ml, Exiqon) at 37°C overnight. Later the membranes were washed twice in 2x SSC at 37°C for 15 minutes each. Digoxigenin signals were detected with DIG Northern starter kit (Roche) according to the manufacturer’s instructions.
Results
Overview of the small RNA Library
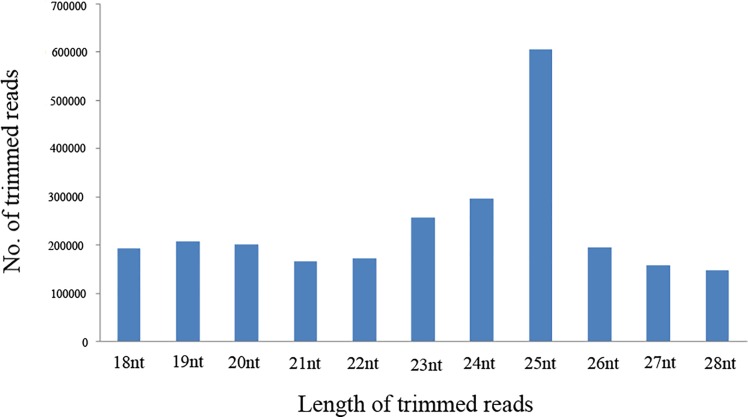
We obtained a dataset of about 14 million reads from the pooled T. palmi small RNA library (egg, larva, pupa and adult) sequenced on Illumina Next Generation Sequencing platform. After various mapping (Table 1), the trimmed high-quality small RNA reads were employed to identify both known and novel miRNAs. Size distribution of the high-quality reads in the library (Fig 1) revealed that the peak was at 25 nts, which was also observed in the pooled library of Plutella xylostella (L.) [47]. A small portion (<5%) of our library consisted of read length of around 26 to 28 nts, which could be putative piwi- interacting RNAs (piRNAs) from T. palmi (S1 Table).
Table 1. Summary Statistics of T. palmi small RNA data analysis.
| Mappable (%) | ||
|---|---|---|
| Raw reads | 14,148,849 | 100 |
| Number of trimmed reads | 5,045,585 | 35.66 |
| Mapped to mRNA | 2,212,874 | 15.63 |
| Repbase | 1,458,987 | 10.31 |
| Rfam mapped | 2,817,765 | 19.87 |
| Total unmappable for miRNA | 2,610,489 | 18.45 |
| miRBase mapped reads | 19,434 | < 1.0 |
| Average length | 23 |
Fig 1. Length distribution of mappable reads obtained from T. palmi deep-sequencing.
Reads with ≥ 18 nt to ≤ 26 nt were considered for miRNA mapping.
Identification of known miRNA
MiRNAs are known to be conserved among different species within a kingdom. Here, in our study, the mappable sequences were aligned to miRNA sequences from miRBase v.21.0. The analysis resulted in a total of 67 conserved miRNAs representing 54 different miRNA families (Table 2), among which the average similarity between the homologs reached 85% and few of them had a similarity to the extent of 95–100% with 1–2 nts or no difference. Analysis of the 54 miRNA families revealed that 15 were found to be exclusively present in arthropod species (Table 3), while 25 miRNA families were vertebrate specific. Seven miRNA families (miR-10, miR-100, miR-71, miR-9, miR-92, miR-15 and miR-281) were found to be highly conserved in the Animal Kingdom (Table 3) during evolution implicating their importance in regulating the gene transcripts involved in the physiological process. Among the known miRNAs, miR-281 and miR-750 were highly expressed with an expression value of 9560 and 5849 respectively (Table 2).
Table 2. Expression value of known miRNAs in T. palmi.
The first column represents miRNA families; the second column represents the number of reads annotated on the particular miRNA family; the third column represents mature miRNA sequences; the fourth column represents the length of mature miRNAs; the fifth column represents miRNAs falling in to related miRNA family.
| miRNA family | Expression value (Reads) | Length (nt) | Name of the miRNA | Sequence (5' - 3') | Homologous miRNA |
|---|---|---|---|---|---|
| miR-750 | 5849 | 23 | tpa-miR-750a | CCAGAUCUAACUCUUCCAGCUCA | isc-miR-750 |
| 1916 | 22 | tpa-miR-750b | CCAGAUCUAACUCUUCCAGCUC | ame-miR-750 | |
| 70 | 25 | tpa-miR-750c | CCAGAUCUAACUCUUCCAUAUGACG | tca-miR-750-3p | |
| 30 | 23 | tpa-miR-750d | UCAGAUCUAACUCUUCCAGUUCU | sme-miR-750-3p | |
| 4 | 22 | tpa-miR-750e | CAGAUCUAACUCUUCCAGCUCA | lgi-miR-750 | |
| miR-92 | 1687 | 22 | tpa-miR-92 | AAUUGCACCCGUCCCGGCCUGA | ame-miR-92b |
| miR-281 | 9560 | 22 | tpa-miR-281 | AAGAGAGCUAUCCGUCGACAGU | dme-miR-281-2-5p |
| miR-6240 | 675 | 26 | tpa-miR-6240 | CCAAAGCAUCGCGAAGGCCCACGGCG | mmu-miR-6240 |
| miR-2779 | 103 | 20 | tpa-miR-2779a | AUAUCCGGCUCGAAGGACCA | bmo-miR-2779 |
| 101 | 18 | tpa-miR-2779b | AUCCGGCUCGAAGGACCA | mse-miR-2779 | |
| miR-2796 | 479 | 23 | tpa-miR-2796 | GUAGGCCGGCGGAAACUACUUGC | ame-miR-2796 |
| miR-7550 | 100 | 18 | tpa-miR-7550 | AUCCGGCUCGAAGGACCA | ipu-miR-7550 |
| miR-993 | 95 | 20 | tpa-miR-993 | GAAGCUCGACUCUACAGGUC | ppc-miR-993 |
| miR-5124 | 85 | 20 | tpa-miR-5124 | GGUCCAGUGACUAAGAGCAU | mmu-miR-5124a |
| miR-9 | 77 | 18 | tpa-miR-9 | UCUUUGGUAUCCUAGCUG | bmo-miR-9c-5p |
| miR-2478 | 56 | 20 | tpa-miR-2478 | GUAUCCCACUUCUGACACCA | bta-miR-2478 |
| miR-1175 | 64 | 22 | tpa-miR-1175-5p | AAGUGGAGUAGUGGUCUCAUCG | aae-miR-1175-5p |
| 6 | 24 | tpa-miR-1175-3p | UGAGAUUCAACUCCUCCAACUUAA | bmo-miR-1175-3p | |
| miR-279 | 32 | 25 | tpa-miR-279a | UGACUAGAUCCAUACUCGUCUAUAG | tca-miR-279d-3p |
| 47 | 22 | tpa-miR-279b | UGACUAGAUCCAUACUCGUCUG | bmo-miR-279c-3p | |
| 24 | 21 | tpa-miR-279c | UGACUAGAUCCAUACUCAGCU | ppc-miR-279 | |
| 4 | 23 | tpa-miR-279d | UGACUAGAUCCAUACUCGUCUGC | mse-miR-279d | |
| miR-3931 | 25 | 23 | tpa-miR-3931 | UACUUUGAGUCGGUACGAAUCCU | isc-miR-3931 |
| miR-10 | 21 | 22 | tpa-miR-10 | AACCCUGUAGACCCGAAUUUGA | gsa-miR-10b-5p |
| miR-5119 | 20 | 19 | tpa-miR-5119 | CAUCUCAUCCUGGGGCUGG | mmu-miR-5119 |
| miR-800 | 18 | 22 | tpa-miR-800 | GCCAAACUCGGAAAUUGUCUGC | cel-miR-800-3p |
| miR-6990 | 18 | 21 | tpa-miR-6990 | CCCAGGGUGAGUCAGGGCUCU | mmu-miR-6990-5p |
| miR-6489 | 15 | 20 | tpa-miR-6489-5p | GGCACCGGACUGGCGCCCUU | mja-miR-6489-5p |
| 6 | 23 | tpa-miR-6489-3p | CGACGGAAAGGUGUCCAAGCUGG | mja-miR-6489-3p | |
| miR-7156 | 15 | 23 | tpa-miR-7156 | UUGUUCUCAAACUGGCUGUCAGA | hsa-miR-7156-5p |
| miR-5108 | 14 | 19 | tpa-miR-5108 | GUAGAGCACUGGAUGGUUU | mmu-miR-5108 |
| miR-9373 | 14 | 21 | tpa-miR-9373 | CAUCGCUCUUGGCCAGCUCGU | dme-miR-9373-3p |
| miR-100 | 13 | 24 | tpa-miR-100 | AACCCGUAGAUUCGAAUUUGUGUU | asu-miR-100b-5p |
| miR-3767 | 12 | 21 | tpa-miR-3767 | UACACAUUAUUUACUACUACU | ame-miR-3767 |
| miR-3049 | 11 | 22 | tpa-miR-3049 | UCCGUCCAACUCCUUUCCGUCU | ame-miR-3049-3p |
| miR-6382 | 11 | 24 | tpa-miR-6382 | UGGAAUGUAAAGAGAGCACACAAG | mmu-miR-6382 |
| miR-998 | 10 | 21 | tpa-miR-998 | UAGCACCAUGGAAUUCAGCUG | api-miR-998 |
| miR-6493 | 3 | 24 | tpa-miR-6493-5p | ACGUCCGGCAGGUUUUACCCCU | mja-miR-6493-5p |
| 9 | 22 | tpa-miR-6493-3p | AGGGGGAAACCGCGCUGAGCGUUA | mja-miR-6493-3p | |
| miR-9198 | 9 | 23 | tpa-miR-9198 | CUUGGCACUGUCAGUGGAUGUGA | efu-miR-9198a |
| miR-2379 | 7 | 22 | tpa-miR-2379 | AGGCUGCUGGAGAAGAUAUUUU | bta-miR-2379 |
| miR-1000 | 21 | 22 | tpa-miR-1000a | AUAUUGUCCUGUCACAGCAGUA | api-miR-1000 |
| 14 | 21 | tpa-miR-1000b | AUAUUGUCCUGUCACAGCAGU | bmo-miR-1000 | |
| miR-3086 | 6 | 22 | tpa-miR-3086 | CCCAAUGAGCCUACAGUCUAAG | mmu-miR-3086-3p |
| miR-71 | 5 | 25 | tpa-miR-71 | UGAAAGACCUGUUGGUAGUGAGACG | ppc-miR-71a |
| miR-2944 | 5 | 23 | tpa-miR-2944 | UAUCACAGCCGUAGUUGCCUUAC | tca-miR-2944b-3p |
| miR-4724 | 5 | 21 | tpa-miR-4724 | GUACCUUCUGGUUCAGCUAGU | hsa-miR-4724-3p |
| miR-7475 | 5 | 20 | tpa-miR-7475 | CCGCCGCCGCCGCGCCCUCC | gga-miR-7475-5p |
| miR-8316 | 5 | 19 | tpa-miR-8316 | AUGGUGUCCAGGUCGUCGC | ppc-miR-8316-3p |
| miR-412 | 4 | 20 | tpa-miR-412a | UGGUCGACCAGCUGGAAAGU | rno-miR-412-5p |
| 7 | 23 | tpa-miR-412b | UGGUCGACCAGCUGGAAAGUAAU | cgr-miR-412-5p | |
| miR-1726 | 4 | 22 | tpa-miR-1726 | AAGCUUGUUGGGUUUGGUUUGU | gga-miR-1726 |
| miR-965 | 4 | 22 | tpa-miR-965 | UAAGCGUAUAGCUUUUCCCCUU | hme-miR-965 |
| miR-6787 | 4 | 22 | tpa-miR-6787 | UGGCGGGGGUAGAGCUGGCUGC | hsa-miR-6787-5p |
| miR-454 | 3 | 22 | tpa-miR-454 | ACCCUAUCAAUAUUGUCUCUGC | hsa-miR-454-5p |
| miR-1723 | 3 | 23 | tpa-miR-1723 | UGGGAGCGGAAUGUGCAGCCUCA | gga-miR-1723 |
| miR-786 | 3 | 23 | tpa-miR-786 | UAAUGCCCUUGCUGAGAUUCCAU | crm-miR-786 |
| miR-3344 | 3 | 25 | tpa-miR-3344 | UUGCAAGAAGGACUCAGCCAGCGAG | bmo-miR-3344 |
| miR-4045 | 3 | 22 | tpa-miR-4045 | CCACAAUGAAAGUAGAUGUCCG | cin-miR-4045-5p |
| miR-3878 | 3 | 19 | tpa-miR-3878 | UGGACGGAGAACUAAUUGU | tca-miR-3878-5p |
| miR-4459 | 3 | 22 | tpa-miR-4459 | CCAGGAGGCGGAGGAGGUGGAG | hsa-miR-4459 |
| miR-4638 | 3 | 23 | tpa-miR-4638 | CCUGGACACCGCUCAGCCGGCCG | hsa-miR-4638-3p |
| miR-5625 | 3 | 21 | tpa-miR-5625 | CCCGGAAGUUCUUGAGUAGGA | mmu-miR-5625-5p |
| miR-7258 | 3 | 20 | tpa-miR-7258 | AAAAGGACUUGACUGCAGCA | mdo-miR-7258-5p |
| miR-7690 | 3 | 23 | tpa-miR-7690 | AAUCAUCCGGGAGUUGGGAAAGA | cbn-miR-7690 |
| miR-15 | 3 | 23 | tpa-miR-15 | CGUAGCAGCACGUCAUGGUUUGU | ssa-miR-15a-5p |
| miR-8511 | 3 | 18 | tpa-miR-8511 | UCAGUCUUUUCCUCUUUC | pxy-miR-8511 |
Table 3. Homology analysis of T. palmi miRNA homologs.
| tpa-miR | Insects | Other Arthropods | Other Invertbrates | Vertebrates | Note |
|---|---|---|---|---|---|
| tpa-miR-10 | √ | √ | √ | √ | Highly conserved |
| tpa-miR-100 | √ | √ | √ | √ | Highly conserved |
| tpa-miR-1000 | √ | — | — | — | Insect specific |
| tpa-miR-1175 | √ | — | √ | — | Invertebrate specific |
| tpa-miR-2796 | √ | — | — | — | Insect specific |
| tpa-miR-279 | √ | √ | √ | — | Invertebrate specific |
| tpa-miR-71 | √ | √ | √ | √ | Highly conserved |
| tpa-miR-750 | √ | √ | √ | — | Invertebrate specific |
| tpa-miR-965 | √ | √ | — | — | Arthropod specific |
| tpa-miR-993 | √ | √ | √ | — | Invertebrate specific |
| tpa-miR-998 | √ | — | — | — | Insect specific |
| tpa-miR-9 | √ | √ | √ | √ | Highly conserved |
| tpa-miR-2779 | √ | — | — | — | Insect specific |
| tpa-miR-92 | √ | √ | √ | √ | Highly conserved |
| tpa-miR-15 | — | — | √ | √ | Highly conserved |
| tpa-miR-1723 | — | — | — | √ | Vertebrate specific |
| tpa-miR-1726 | — | — | — | √ | Vertebrate specific |
| tpa-miR-2379 | — | — | — | √ | Vertebrate specific |
| tpa-miR-2478 | — | — | — | √ | Vertebrate specific |
| tpa-miR-2944 | √ | — | — | Insect specific | |
| tpa-miR-3049 | √ | — | — | — | Insect specific |
| tpa-miR-3086 | — | — | — | √ | Vertebrate specific |
| tpa-miR-3344 | √ | — | — | — | Insect specific |
| tpa-miR-3767 | √ | — | — | — | Insect specific |
| tpa-miR-3878 | √ | — | — | — | Insect specific |
| tpa-miR-3931 | — | √ | — | — | Arthropod specific |
| tpa-miR-4045 | — | — | — | √ | Vertebrate specific |
| tpa-miR-412 | — | — | — | √ | Vertebrate specific |
| tpa-miR-4459 | — | — | — | √ | Vertebrate specific |
| tpa-miR-454 | — | — | — | √ | Vertebrate specific |
| tpa-miR-4638 | — | — | — | √ | Vertebrate specific |
| tpa-miR-4724 | — | — | — | √ | Vertebrate specific |
| tpa-miR-5108 | — | — | — | √ | Vertebrate specific |
| tpa-miR-5119 | — | — | — | √ | Vertebrate specific |
| tpa-miR-5124 | — | — | — | √ | Vertebrate specific |
| tpa-miR-5625 | — | — | — | √ | Vertebrate specific |
| tpa-miR-6240 | — | — | — | √ | Vertebrate specific |
| tpa-miR-6382 | — | — | — | √ | Vertebrate specific |
| tpa-miR-6489 | — | √ | — | — | Arthropod specific |
| tpa-miR-6493 | — | √ | — | — | Arthropod specific |
| tpa-miR-6787 | — | — | — | √ | Vertebrate specific |
| tpa-miR-6990 | — | — | — | √ | Vertebrate specific |
| tpa-miR-7156 | — | — | — | √ | Vertebrate specific |
| tpa-miR-7258 | — | — | — | √ | Vertebrate specific |
| tpa-miR-7475 | — | — | — | √ | Vertebrate specific |
| tpa-miR-7550 | — | — | — | √ | Vertebrate specific |
| tpa-miR-7690 | — | — | √ | — | Invertebrate specific |
| tpa-miR-786 | — | — | √ | — | Invertebrate specific |
| tpa-miR-800 | — | — | √ | — | Invertebrate specific |
| tpa-miR-8316 | — | — | √ | — | Invertebrate specific |
| tpa-miR-8511 | √ | — | — | — | Insect specific |
| tpa-miR-9198 | — | — | — | √ | Vertebrate specific |
| tpa-miR-9373 | √ | — | — | — | Insect specific |
| tpa-miR-281 | √ | √ | √ | √ | Highly conserved |
Identification of miRNA-star strands
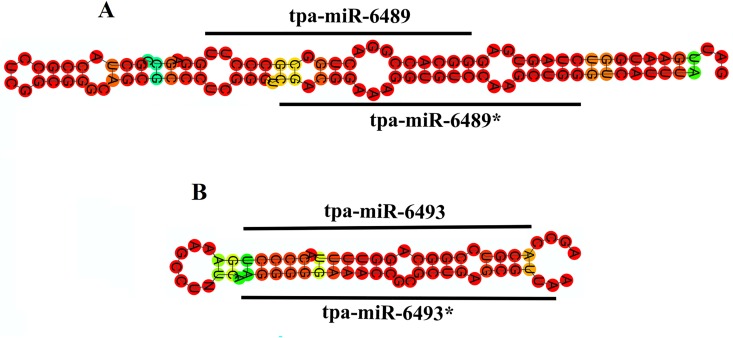
In most of the cases, once the mature miRNA strand is loaded into RISC, its star strand will be degraded soon after being exported to the cytosol. However, our analysis revealed that two T. palmi miRNA star (miRNA*) strands namely, miR-6489* and miR-6493* were obtained from our library for their corresponding mature miRNAs (Fig 2). The expression values (Number of reads) of miR-6489* were lower than that of their corresponding miRNAs, whereas, for miR-6493* it was three times higher (Table 2).
Fig 2. Stem loop structures of two miRNAs and their star strands.
(A) The secondary structure of tpa-miR-6489 and tpa-miR-6489*. (B) The secondary structure of tpa-miR-6493 and tpa-miR-6493*. Both miRNAs and its star reads were marked by black bars. The secondary structure was predicted by employing RNA fold WebServer (www.rna.tbi.univie.ac.at/lgi-bin/RNAfold.cgi).
Identification of novel miRNAs
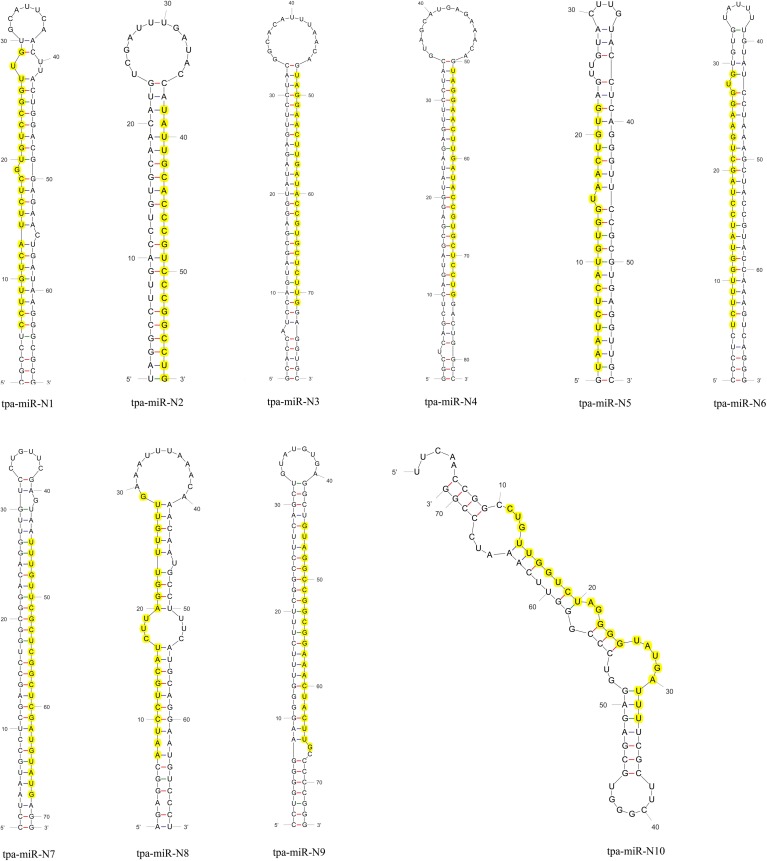
We utilized the genomic sequence assembly of F. occidentalis (Thripidae: Thysanoptera) to identify the novel miRNAs, as no genomic information is available for T. palmi. Miranalyzer pipeline identified a total of 10 novel miRNAs from T. palmi for the first time (Table 4), with their predicted precursor secondary structures (Fig 3). The complete details of the mature miRNAs and their corresponding pre-miRNAs have been given in Table 4. The length of the novel miRNAs ranged from 21–24 nucleotides with a preference of Uracil (60%) followed by Cytosine (20%) at the 5' end. Among these ten miRNAs, five were located in the 5' arm while the other five arose from 3' arm (Table 4, Fig 3).
Table 4. Details of T. palmi novel miRNAs obtained from the current study.
Information regarding mature and precursor sequences, start and end position, orientation, expression values, MFE value and (A+U) content were given.
| miRNAs | Sequence | Locus | Scaffold | start | end | Hairpin Sequence | Start | End | Orientation | Abundance | miRNA length | MFE | Hairpin (A+U)% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tpa-miR-N1 | CCUUGUCAUUCUCGUGUCCGGUUG | KL024066 | Scaffold373 | 148612 | 148635 | CGCCUCCUUGUCAUUCUCGUGUCCGGUUGUGCAUUCAACUUACUGGACGGAGAACUGAUAAGGGCGCG | 554 | 577 | Negative | 10 | 24 | -33.5 | 45.59 |
| tpa-miR-N2 | UAUUGCACCCGUCCCGGCCUG | KL023851 | Scaffold158 | 512267 | 512247 | UAGGCCUUGACCUGUGCAACAUGUCGAUUUGAUACCAUAUUGCACCCGUCCCGGCCUG | 1724 | 1744 | Negative | 7 | 21 | -20.8 | 46.55 |
| tpa-miR-N3 | UAGGAACUUGAUACCGUGCUCUUG | KL023856 | Scaffold163 | 605332 | 605309 | GCACCAUCCAGUAGCGAGGUAUAGAGUUCCUACGGCACAUUUAACAGUAGGAACUUGAUACCGUGCUCUUGGAGGUGC | 2469 | 2492 | Negative | 4007 | 24 | -41.22 | 50 |
| tpa-miR-N4 | UAGGAACUUGAUACCGUGCUCCUG | KL023856 | Scaffold163 | 531005 | 531028 | GGCUCAGCUCAGUAGCGAGGUAUAGAGUUCCUACGUAGCAUGAGAAACAGUAGGAACUUGAUACCGUGCUCCUGGACUGGCC | 4132 | 4155 | Positive | 1026 | 24 | -40.8 | 47.56 |
| tpa-miR-N5 | UAAUCUCAUGUGGUAACUGUG | KL023731 | Scaffold38 | 137500 | 137520 | GUAAUCUCAUGUGGUAACUGUGAGUUGUACUUGUACCUCAGGGUUCCGCGUGAGGUUGC | 5735 | 5755 | Positive | 21 | 21 | -31.3 | 50.85 |
| tpa-miR-N6 | UCUUUGGUAUCCUAGCUGAAGGUG | KL023728 | Scaffold35 | 1566837 | 1566860 | CCCUCUCUUUGGUAUCCUAGCUGAAGGUGUGUGUAUUUUGUAUCCUAAAGCUACCGUACCAAAGUCAGGG | 13396 | 13419 | Positive | 13 | 24 | -26.8 | 54.29 |
| tpa-miR-N7 | UUUGUUCGCUCGGCUCGAUGUAUG | KL023727 | Scaffold34 | 1931509 | 1931532 | CCUAAUGCCUCGAGCCUGGCGGACAGGUUGUCCUGUUCGAGUAAUUUGUUCGCUCGGCUCGAUGUAUGAGG | 1925 | 1948 | Positive | 380 | 24 | -37.9 | 45.07 |
| tpa-miR-N8 | AAUCCUGCAUCUUAGGUUUGUUG | KL023694 | Scaffold1 | 2310295 | 2310273 | AGAGGCAAUCCUGCAUCUUAGGUUUGUUGAAAUUUAAACAAACAAUGCCUUUCAUGCAGGAAUGUCCCU | 5827 | 5849 | Negative | 9 | 23 | -25 | 60.87 |
| tpa-miR-N9 | GUAGGCCGGCGGAAACUACUUG | KL023695 | Scaffold2 | 1399443 | 1399464 | CCUGGGGAAGGGGUUUCUUUCGGCCUUCAGCUGUAUGUGAGGCUGUAGGCCGGCGGAAACUACUUGCCCCCGGG | 2544 | 2565 | Positive | 9 | 22 | -44.6 | 37.84 |
| tpa-miR-N10 | CUGUUGGUCUAGGGGUAUGAUUU | KL023751 | Scaffold58 | 1266 | 1245 | UUCAACCGGCCUGUUGGUCUAGGGGUAUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGG | 1197 | 1281 | Positive | 37 | 23 | -21.5 | 42.3 |
Fig 3. Various hairpin secondary structures of the ten novel pre-miRNAs of T. palmi.
The mature miRNAs are indicated by yellow shades. The secondary structure was predicted using RNA fold WebServer.
Abundance of novel miRNAs
The novel miRNAs identified from T. palmi varied in their expression values in the library. Among the novel miRNAs, tpa-miR-N3 (4007 copies), tpa-miR-N4 (1026 copies) and tpa-miR-N7 (380 copies) had the highest abundance compared to the remaining novel miRNAs (Table 4). Whereas, few other novel miRNAs namely, tpa-miR-N1, tpa-miR-N2, tpa-miR-N6, tpa-miR-N8 and tpa-miR-N9 were found to be very minimal (≤ 15 copies). The length and Minimum Free Energy (MFE) for these novel pre-miRNAs ranged from 58–82 nts and -20.8 to -44.6 kcal/mol respectively. The (A+U) % of the novel pre-miRNAs was in the range of 37.84% to 60.87% (Table 4).
Sequence and phylogenetic analysis
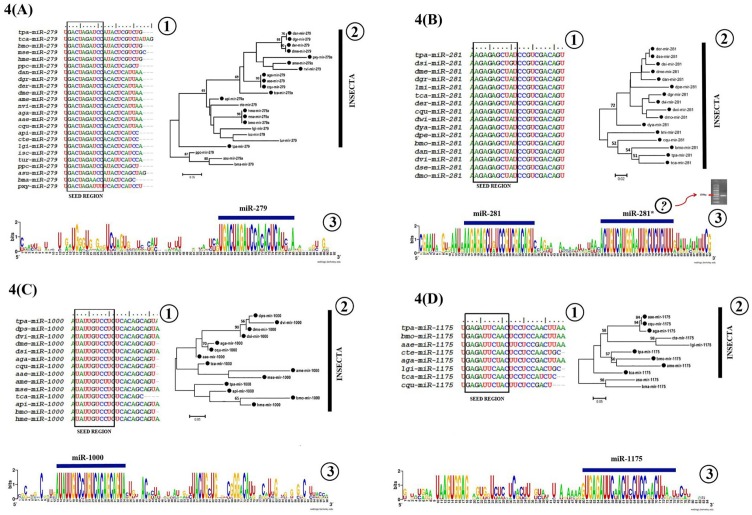
Sequence and phylogenetic analyses revealed that some of the known miRNAs were expressed in a wide range of insect species and are highly conserved (Table 3, Fig 4A1–4D1 and Fig 4A3–4D3). Mature miRNAs are highly conserved among various species within the Kingdom and are considered to be the evolutionarily conserved regulators of the gene expression [48]. The phylogenetic trees for miR-1000, miR-1175, miR-281 and miR-279 revealed that T. palmi miRNAs grouped with the closely related species of insects (Fig 4A2–4D2). However, few miRNAs (miR-1000, miR-2796, miR-965, miR-998, miR-2779, etc.) are highly specific to few species (Table 3). Fig 4A–4D revealed that T. palmi miRNAs are well conserved, particularly in the seed region compared to the homologous miRNAs from other species.
Fig 4.
(A) to (D). Homology, phylogeny and weblogo analysis of T. palmi miRNAs. 4(A) to (D) 1. Homology in the seed region of the T. palmi miRNA with respect to its counterpart from other insect species. Sequence conservation of the T. palmi mature miRNAs including the seed region over a wide range of insects. The first three letters of each miRNAs indicating the name of the species. 4(A) to (D) 2. Phylogenetic trees (ML tree, RaxML.v.7.0.4) of four families of precursor miRNA sequences from various members of the animal kingdom. 4(A) to (D) 3. T. palmi pre-miRNAs weblogo. The pre-miRNA sequence logos for the T. palmi, in which mature miRNA is indicated by blue bars. Each logo consists of stacks of symbols, one for each nucleotide position in the sequence. The height indicates the sequence conservation at that nucleotide position and the height of symbols within the stack indicates the relative frequency of each nucleotide at that position. Fig 4(B) 3 indicated the possible presence of miR-281* which was not evident from the NGS raw reads. However, the presence of miR-281* has been validated by stem-loop RT-PCR.
Target Prediction
Targets were predicted for known and novel miRNAs of T. palmi employing miRanda on a scale of 0–7 to indicate the stringency of miRNA-target pairing with the smaller numbers representing higher stringency. ESTs and transcriptome of F. occidentalis were used as a reference for target searches with a cut-off score 140.
Targets for known miRNAs
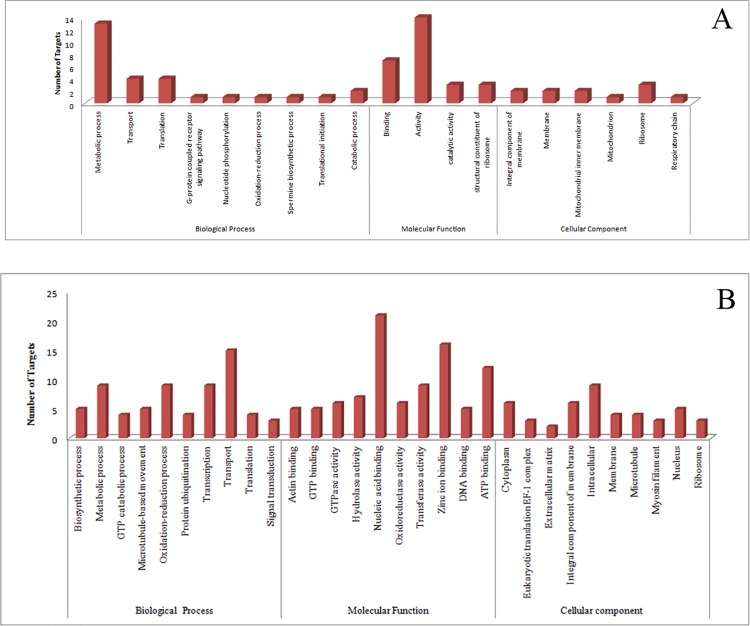
All 67 known miRNAs were searched for targets against ESTs and transcriptome sequences of F. occidentalis. Out of the known 67 miRNAs, 20 and 40 known miRNAs were found to have targets in ESTs and transcriptome respectively (S2 and S3 Tables). The enrichment analysis (Blast-2-GO) was performed employing gene ontology (GO) terms for genes targeted by miRNAs (Fig 5A and 5B). For those targets in the ESTs, three motifs were over-represented in GO-BP (biological process) category like ‘metabolic process’, ‘transport’ and ‘translation’. The GO-MF (molecular function) category was over-represented by the motif ‘activity’ and ‘binding’ (Fig 5A). On the other hand, GO-terms enrichment analysis of miRNA targets in the transcriptome yielded motifs for ‘transport’, ‘metabolic process’ and ‘oxidation-reduction process’ in GO-BP category; while, GO-MF category was over-represented with motifs for ‘nucleic acid binding’, ‘zinc-ion binding’ and ‘ATP binding’ (Fig 5B). Complete details of the Blast-2-GO analysis have been provided in S4 and S5 Tables.
Fig 5.
Gene Ontology (GO) classification of the putative target genes for the T. palmi miRNAs against ESTs (A) and transcriptome (B) sequences of F. occidentalis. GO terms were assigned to each target gene based on the annotation and were summarized into three main GO categories viz. (i) biological process (BP) (ii) molecular function (MF) and (iii) cellular component (CC). Only top ten subcategories are presented in the case of GO for transcriptome sequences.
Targets for novel miRNA
Ten novel miRNAs were searched for their targets in the F. occidentalis transcriptome. A total of 33 miRNA-target pairs were obtained (S6 Table) and further Blast-2-GO analysis yielded ‘regulation of transcription’ and ‘binding’ as GO-BP and GO-MF category respectively (S7 Table). Complete details of the miRNA targets and Blast-2-GO analysis have been provided in S7 Table.
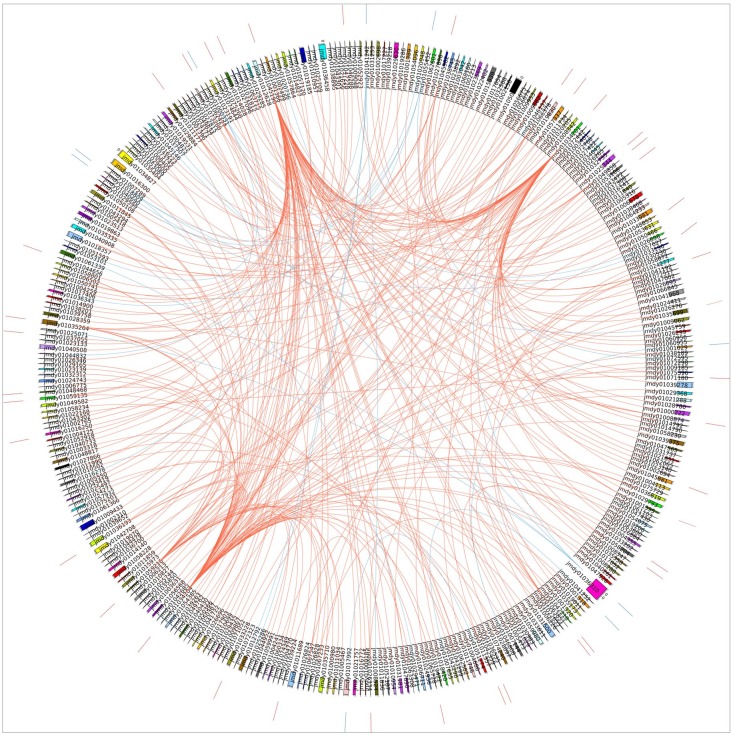
The synteny analysis of the T. palmi miRNAs and their targets were performed by employing circos44. In brief, the Blast analysis was performed using T. palmi miRNA sequences (known and novel) against F. occidentalis scaffolds (Largest 300). The positions of miRNAs were identified and their targets represented in the Circos plot (Fig 6).
Fig 6. The synteny analyses using Circos (Krzywinski et al. 2009).
Map of the Western Flower Thrips, F. occidentalis scaffolds linking T. palmi miRNAs and their targets prepared using Circos. The outer circle represents the highlights of 10 novel miRNA in blue and 40 known miRNA represented in red colour. The inner lines in red colour represent known miRNAs and their targets (839 targets) and blue lines represent 11 novel miRNAs and their targets (33 targets) across 300 scaffolds of F. occidentalis genome.
Validation of T. palmi microRNAs
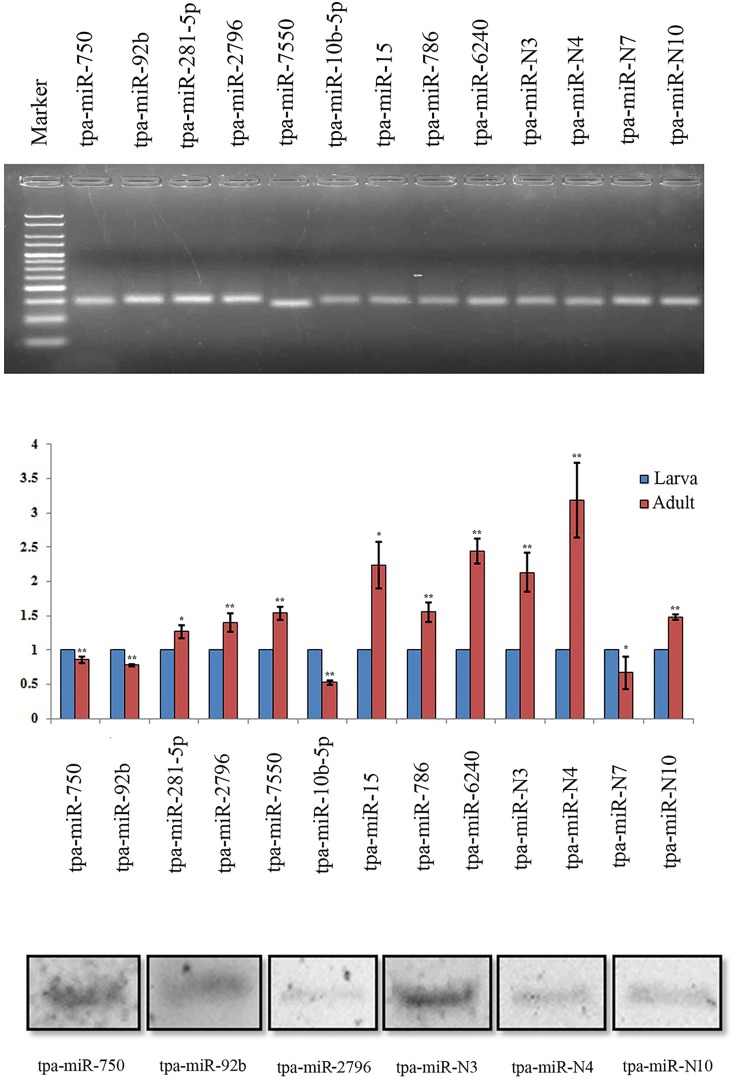
The present study revealed the novel miRNAs from T. palmi (Tables 2 and 4). However, further validation of these miRNAs was performed by (i) stem-loop end-point reverse transcriptase PCR (RT-PCR) (ii) real-time quantitative reverse transcriptase PCR (qRT-PCR) and (iii) small RNA Northern blots. Using stem-loop end-point RT-PCR, we have validated 9 conserved (tpa-miR-750, tpa-miR-92b, tpa-miR-281-5p, tpa-miR-2796, tpa-miR-10b-5p, tpa-miR-786, tpa-miR-6240, tpa-miR-7550 and tpa-mir-15) and 4 novel miRNAs (tpa-miR-N3, tpa-miR-N4, tpa-miR-N7, tpa-miR-N10) from T. palmi using the primer sets as described (Table 5). All of these miRNAs were amplified with an approximate product size of 75 bp (Fig 7A).
Table 5. List of Universal Reverse primer, Stem-loop RT primers and forward primers employed in small RNA validation.
| Sl. No. | Oligo Name | Oligo Sequence (5' to 3') |
|---|---|---|
| 1 | Universal Reverse | ATCCAGTGCAGGGTCCGAGG |
| 2 | RT/tpa-miR-750 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAGCTG |
| 3 | F/tpa-miR-750 | GCGGCGGCCAGATCTAACTCTTC |
| 4 | RT/tpa-miR-92b | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAGGC |
| 5 | F/tpa-miR-92b | GCGGCGGAATTGCACCCGTCCC |
| 6 | RT/tpa-miR-281-5p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACTGTC |
| 7 | F/tpa-miR-281-5p | GCGGCGGAAGAGAGCTATCCGTC |
| 8 | RT/tpa-miR-2796 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAAGT |
| 9 | F/tpa-miR-2796 | GCGGCGGGTAGGCCGGCGGAAAC |
| 10 | RT/tpa-miR-7550 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGTCC |
| 11 | F/tpa-miR-7550 | GCGGCGGATCCGGCTCGAAGGAC |
| 12 | RT/tpa-miR-10b-5p | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCAAAT |
| 13 | F/tpa-miR-10b-5p | GCGGCGGAACCCTGTAGACCCG |
| 14 | RT/tpa-miR-15 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAC |
| 15 | F/tpa-miR-15 | GCGGCGGCGTAGCAGCACGTCATG |
| 16 | RT/tpa-miR-8511 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGAAAGA |
| 17 | F/tpa-miR-8511 | GCGGCGGTCAGTCTTTTCCTC |
| 18 | RT/tpa-miR-6240 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGCCGT |
| 19 | F/tpa-miR-6240 | GCGGCGGCCAAAGCATCGCGAAG |
| 20 | RT/tpa-miR-N3 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAGAG |
| 21 | F/tpa-miR-N3 | GCGGCGGTAGGAACTTGATACCG |
| 22 | RT/tpa-miR-N4 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGGAG |
| 23 | F/tpa-miR-N4 | GCGGCGGTAGGAACTTGATACCG |
| 24 | RT/tpa-miR-N7 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCATACA |
| 25 | F/tpa-miR-N7 | GCGGCGGTTTGTTCGCTCGGCTC |
| 26 | RT/tpa-miR-N10 | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATCA |
| 27 | F/tpa-miR-N10 | GCGGCGGCTGTTGGTCTAGGGG |
| 28 | RT/tpa-miR-281* | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAGAG |
| 29 | F/tpa-miR-281* | GCGGCGGACUGUCAUGGAAUUGC |
Fig 7. Validation of selected conserved and novel miRNAs from T. palmi.
(A) Stem-loop RT-PCR analyses of nine conserved and four novel miRNAs from T. palmi. The products were resolved on 3% agarose gel in 1X TBE stained with ethidium bromide. HyperLadder™ 25bp (Bioline, USA) employed as a marker. (B) Stem-loop RT-qPCR analysis of spatiotemporally expressed T. palmi miRNAs in larva and adults. ‘*’ and ‘**’ means a statistically significant difference at level p < 0.05 and p < 0.001 respectively for these miRNAs in the larva and adult T. palmi. The error bars indicate standard deviation for three biological replications. (C) Small RNA Northern blot validation. Both conserved (tpa-miR-750, tpa-miR-92b, tpa-miR-2796) and novel (tpa-miR-N3, tpa-miR-N4 and tpa-miR-N10) miRNAs were validated by small RNA northern analysis employing small RNA isolated from pooled T. palmi.
Our study also quantified the expression level of the above-mentioned 13 miRNAs from T. palmi larvae and adults using qRT-PCR (Table 6, Fig 7B). Results suggested that the miRNA expression was higher in larval stages compared to adults in four microRNAs namely tpa-miR-750, tpa-miR-92b, tpa-miR-10b-5p and tpa-miR-N7 (Fig 7B). Finally, we also validated 3 conserved (tpa-miR-750, tpa-miR-92b and tpa-miR-2796) and three novel miRNAs (tpa-miR-N3, tpa-miR-N4 and tpa-miR-N10) employing a more sensitive small RNA Northern blot technique (Fig 7C).
Table 6. MicroRNA specific primers employed in the RT-qPCR.
| miRNA | Sequence (5' - 3') |
|---|---|
| tpa-miR-750 Forward | CCAGATCTAACTCTTCCAGCTC |
| tpa-miR-92b Forward | AATTGCACCCGTCCCGGCCTGA |
| tpa-miR-281-5p Forward | AAGAGAGCTATCCGTCGACAGT |
| tpa-miR-2796 Forward | GTAGGCCGGCGGAAACTACTTGC |
| tpa-miR-7550 Forward | ATCCGGCTCGAAGGACCA |
| tpa-miR-10b-5p Forward | AACCCTGTAGACCCGAATTTGA |
| tpa-miR-15 Forward | CGTAGCAGCACGTCATGGTTTGT |
| tpa-miR-8511 Forward | TCAGTCTTTTCCTCTTTC |
| tpa-miR-6240 Forward | CCAAAGCATCGCGAAGGCCCACGGCG |
| tpa-miR-N3 Forward | TAGGAACTTGATACCGTGCTCTTG |
| tpa-miR-N4 Forward | TAGGAACTTGATACCGTGCTCCTG |
| tpa-miR-N7 Forward | TTTGTTCGCTCGGCTCGATGTATG |
| tpa-miR-N10 Forward | CTGTTGGTCTAGGGGTATGATTT |
Discussion
Illumina deep sequencing approach for identification of microRNAs
With the advent of the next generation sequencing technologies, miRNAs have been discovered at an accelerated pace. Presently miRNAs are known from more than 25 insect species, which includes 12 Drosophila species [49]. Among them, the most recent ones are from Plutella xylostella [47], Spodoptera frugiperda [50], etc. Several miRNAs have been reported from various orders of insects such as Diptera, Hymenoptera, Coleoptera, Orthoptera, Lepidoptera, Hemiptera and Homoptera [27], and for the first time, we report the small RNAs from a thysanopteran insect, T. palmi. In this regard, small RNA library was prepared from the pooled samples of different developmental stages of T. palmi and then Illumina (sequencing-by-synthesis) sequencing technology was used to identify miRNAs from the library. The Illumina sequencing approach is one of the high throughput technologies by which miRNAs of any organisms can be identified [51–60,24]. Size distributions of the high-quality reads varied from 18–28 nts in the library. The peak was at 25 nt which was at par with the previous studies [47,60,61]. According to Bartel (2004), the average miRNA length is 22 nt in animals and study conducted showed that the average length is 23 nt.
The unique read distributes of 26–28 nts with a relative lower abundance are common for many small RNA libraries [62–64] indicating the presence of piRNAs. Piwi RNAs (piRNAs) are the class of small RNAs mediating chromatin modifications [65] which are derived mainly from retro-transposons and other repetitive elements with high sequence diversity [62,65,66]. Thus, the results indicated that T. palmi genome encodes not only miRNAs but also other small RNAs such as piRNAs (S1 Table) in a lower proportion that might be involved in the trans-generational epigenetic inheritance [67].
Homology-based predictions of miRNAs
The identification of small RNAs (especially miRNAs) based on genomic information has been reported previously in several insects [60,62]. In this regard, we report the identification and characterization of miRNAs from T. palmi based on Illumina small RNA sequencing. We employed F. occidentalis genome sequences as a reference for T. palmi since the complete genome for T. palmi is still not available. However, a large proportion (93.12%) of the T. palmi sRNA sequences could be mapped on to F. occidentalis genome. This higher percentage of mapping was possible because T. palmi & F. occidentalis (reference genome) belong to the same family, Thripidae. Mapping onto a whole genome sequence also helped in elucidating the sample proportion of small ncRNAs such as tRNA, rRNA, snoRNA or snRNA [68]. All of these sequences were annotated by aligning the reads with Rfam database, which indicated the efficiency of deep sequencing in identifying small RNAs. Our results indicated that there is a rich small RNA world evident in T. palmi.
Our study revealed 67 conserved and ten novel miRNAs from T. palmi for the first time. The (A+U) content of the pre-miRNAs should be in the range of 30–70% [69], as those with higher (A+U) content bind more strongly to proteins [20,70]. In this regard, the (A+U) content of the novel pre-miRNAs ranged from 37.84% (tpa-miR-N9) to 60.87% (tpa-miR-N8). In our dataset, the most abundant miRNAs were tpa-miR-281 (from the conserved miRNAs) and tpa-miR-N3 (from the novel miRNAs) with a total of 9560 and 4007 number of reads respectively.
miRNAs are known to be conserved among different species within a kingdom and are evolutionarily conserved regulators of gene expression [20,71]. Our homology and phylogenetic analysis revealed that insect miRNAs are known to be well-conserved, despite considerable diversity in the genome (Fig 4A–4D). In most of the cases, detection of miRNA*s is difficult with the available methods as these molecules are liable to degrade soon after being exported to the cytosol [27]. However, in our study during the process of identifying conserved miRNAs, two miRNAs for instance, miR-6489* and miR-6493* that matched to the same precursor sequences with their mismatched complementary mature miRNAs were also detected. The weblogo sequences analysis revealed the likely presence of miR-281* which was not identified in the raw reads of NGS. The presence of miR-281* was identified by BLASTN option in miRBase and further confirmed by stem-loop RT PCR (Table 5, Fig 4B3). The absence of miR-281* could be due to the faster degradation as compared to miR-281.
Possible roles of T. palmi miRNAs
Although thousands of small RNAs have been discovered in the recent past, [39,40,47,50,72] the primary challenge is to fully identify the spatiotemporally expressed microRNAs and to determine their individual functions. The majority of the microRNAs have been identified through either computational prediction or cloning and sequencing [73]. In this study, we employed Illumina next generation sequencing approach to identify miRNAs from T. palmi. Currently, there are several mature and precursor microRNAs deposited in the miRBase [39]. In this connection, we identified a total of 77 miRNAs from T. palmi using high throughput sequencing. The current study is the first report of miRNA profiling from a Thrips species employing deep sequencing approach. This approach is far superior to the other approaches of miRNA identification, as it can discover novel microRNAs [74].
The analysis of the expression value (read numbers) revealed that the highest expression was for miR-281 (9560 reads). Recent studies have proved that microRNA-281 regulate the expression of ecdysone receptor (EcR) isoform B, in the silkworm, Bombyx mori [75]. Thus, miR-281 may be involved in development and metamorphosis of T. palmi by regulating the genes involved in the ecdysone cascade. The second highest expression was for miR-750 with an expression value of 7869. RNAi studies proved that the putative JH receptor ultraspiracle (USP) [76,77] is a likely target of miR-750. Thus, it indicates that miR-750 may be involved in hormone signaling, immunity and stress response by regulating the vitellogenin (Vg) gene in T. palmi [78].
Another interesting microRNA obtained in the current study was miR-92 with an expression value of 1687. Previous studies had shown that miR-92 regulates Mef2, the key transcription factor for muscle development and differentiation in Drosophila [79]. An insect-specific microRNA, miR-2796 was identified in T. palmi with an expression value of 479. miR-2796 was found to be the most abundant microRNA in honey bee brain [80]. Additionally, miR-2796 bound to the coding sequence (CDs) of PLC-epsilon gene in Apis and Tribolium, affecting the mRNA stability by splicing rather than the normal canonical translational repression [81]. However, interestingly both miR-2796 and PLC-epsilon gene were missing from the genus Drosophila, even though it was found in other dipterans [80].
Our analyses revealed miR-993 were identified only in invertebrates (Table 3). miR-993 belongs to the miR-100/10 family, and both miR-993 and miR-10 are derived from the ancient miR-100 through duplication and arm-switching [60,82]. miR100/10 family members could regulate the expression of relevant Hox-genes, thus may play a crucial role in insect development [83]. Rest of the insect-specific miRNAs identified in T. palmi may play an important role in insect-specific features, such as metamorphosis, parthenogenesis and biogenesis of pheromones [84]. Whereas, the other invertebrate and vertebrate-specific miRNAs (Table 3) identified from T. palmi required special attention, as their nonexistence in other species of insects could be due to the absence of genomic information for most of those insects [60]. These specific microRNAs may be involved in some special biological processes that distinguish thysanopteran insects from others.
Developmental roles of T. palmi miRNAs
The expression profile of miRNA varies among different developmental stages [85,86]. In the present study, the developmental expression profiles (larval and adult stage) of microRNAs namely, miR-750, miR-92b, miR-281-5p, miR-2796, miR-7550, miR-10b-5p, miR-15, miR-786, miR-6240, tpa-miR-N3, tpa-miR-N4, tpa-miR-N7 and tpa-miR-N10 were investigated by qRT-PCR (Fig 7B). The higher expression of miR-750, miR-92b, miR-10b-5p and tpa-miR-N7 in T. palmi larvae reflected their possible involvement in insect-specific features such as metamorphosis, whereas, the high levels of miR-281-5p, miR-2796, miR-7550, miR-15, miR-786, miR-6240, tpa-miR-N3, tpa-miR-N4 and tpa-miR-N10 in the adult stage indicated their role in the adult development, parthenogenesis and sexual reproduction.
Target prediction is crucial to understand the biological role of a particular miRNA. Unlike their plant counterparts, the imperfect complementarity of animal miRNAs to their target mRNA sequences makes it more difficult to judge the accuracy of prediction. MicroRNAs can bring about mRNA cleavage or translational repression of target mRNAs by binding to 3' UTRs, 5' UTRs and even to coding regions [87]. However, animal miRNAs primarily target the 3' UTRs; and therefore, we limited our target search to (i) expressed sequence tags (ESTs) and (ii) transcriptomic sequences of F. occidentalis. The predicted targets were annotated against GO database and the targeted genes included transcription factors, signal transduction, hormone pathways, molting and even metabolism. Therefore, all these conserved and novel miRNAs identified from T. palmi could play a vital role in diverse biological processes, thus undoubtedly participating in the regulation of thrips growth and development.
In summary, results from this study add to our growing pool of miRNA database and is the first report on such analysis in a thysanopteran insect, T. palmi. Deep sequencing of small RNAs has facilitated the identification of miRNAs from T. palmi. Sixty-seven conserved and ten novel miRNAs that were identified with high confidence and sufficient evidence are the contributions from our study. Most of the T. palmi miRNAs were homologous to insects as compared to the vertebrates. Sequence and phylogenetic analyses revealed that most of the T. palmi miRNAs are highly conserved in various species, making miRNAs, a hallmark of evolutionarily conserved regulators of gene expression. To harmonize the data, and to provide more useful biological insights, we also carried out in silico analysis for identifying potential targets for these miRNAs. Unlike the plant counterparts, the imperfect complementarity of metazoan miRNAs to the target has been found to be sufficient to promote the RNA silencing, as in the case of Drosophila and Bactrocera [88]. Our results indicated that the list of putative mRNA targets was very extensive (S2, S3 and S6 Tables), even with stringent parameters applied to miRanda. Our results suggested that most of the putative target genes for T. palmi miRNAs were associated with several KEGG pathways like metabolic process, transport, translation, signal pathways and oxidative phosphorylation. However, further wet lab experiments are still required for the validation of these targets in understanding the biology of this insect. Expression levels of few miRNAs were also validated by both qRT-PCR and Northern analysis.
Several miRNAs were identified and characterized from animals and plants and among them, very few were further explored for various applications by disrupting specific pathways targeted by these miRNAs. This can be achieved by employing the artificial microRNAs (amiRs) [89]. Recent studies successfully demonstrated the use of amiRNAs for targeting the reporter and the endogenous genes in animals and plants. Identification and expression of a few essential insect-specific gene(s) in plants, can target and degrade an invading insect’s genes, consequently confer insect resistance [90]. Results from our study indicated few miRNAs have been predicted to be involved in the adult development process, which can be further utilized in gene functional studies through RNAi-based approach or in developing miRNA mimics both for feeding and in planta expression [29,88,91] as novel pest management strategies based on gene silencing and insect transgenesis.
Supporting Information
(DOCX)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We thank Sonal Dsouza, Communication and Programme Assistant, Bioversity International for language editing that greatly improved the manuscript. We also thank the three anonymous reviewers for their careful reading and useful suggestions and comments on an earlier version of the manuscript. Our sincere thanks are due to The Director, IIHR, Bangalore for providing necessary facilities. This work is a part of the Ph. D Thesis of the senior author K. B. Rebijith.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Out Reach Program on Management of Sucking Pests (ORP-SP) of Horticultural Crops’ funded by Indian Council for Agricultural Research (ICAR), New Delhi (ICAR-ORP-SP-2012-2017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19: 92–105. 10.1101/gr.082701.108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010;79: 351–79. 10.1146/annurev-biochem-060308-103103 . [DOI] [PubMed] [Google Scholar]
- 3.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21: 452–460. 10.1016/j.ceb.2009.04.009 . [DOI] [PubMed] [Google Scholar]
- 4.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5: R65 10.1186/gb-2004-5-9-r65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136: 215–233. 10.1016/j.cell.2009.01.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431: 350–355. 10.1038/nature02871 . [DOI] [PubMed] [Google Scholar]
- 7.Singh J, Nagaraju J. In silico prediction and characterization of microRNAs from red flour beetle (Tribolium castaneum). Insect Mol. Biol. 2008;17: 427–436. 10.1111/j.1365-2583.2008.00816.x . [DOI] [PubMed] [Google Scholar]
- 8.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3(12): e215 10.1371/journal.pgen.0030215 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nohata N, Hanazawa T, Kinoshita T, Okamoto Y, Seki N. MicroRNAs function as tumor suppressors or oncogenes: Aberrant expression of microRNAs in head and neck squamous cell carcinoma. Auris Nasus Larynx. 2013;40(2): 143–149. 10.1016/j.anl.2012.07.001 . [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75: 843–854. 10.1016/0092-8674(93)90529-y . [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294: 853–58 (2001). 10.1126/science.1064921 . [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics biogenesis mechanism and function. Cell. 2004;116: 281–297. . [DOI] [PubMed] [Google Scholar]
- 13.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38 (Suppl.): S25–S30. 10.1038/ng1793 . [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299: 1540 10.1126/science.1080372 . [DOI] [PubMed] [Google Scholar]
- 15.Lund E, Güttinger S, Calado A, Dahlber JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303: 95–98. 10.1126/science.1090599 . [DOI] [PubMed] [Google Scholar]
- 16.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432: 231–235. 10.1038/nature03049 . [DOI] [PubMed] [Google Scholar]
- 17.Ghosh Z, Chakrabarti J, Mallick B. “miRNomics’—the bioinformatics of microRNA genes”. Biochem Biophys Res Commun. 2007;363: 6–11. 10.1016/j.bbrc.2007.08.030 . [DOI] [PubMed] [Google Scholar]
- 18.Gu L, Knipple DC. Recent advances in RNA interference research in insects: Implications for future insect pest management strategies. Crop Prot. 2013;45: 36–40. 10.1016/j.cropro.2012.10.004 [DOI] [Google Scholar]
- 19.Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46: 243–259. 10.1111/j.1365-313X.2006.02697.x . [DOI] [PubMed] [Google Scholar]
- 20.Rebijith KB, Asokan R, Krishna V, Ranjitha HH, Krishna Kumar NK, Ramamurthy VV. In Silico Prediction and Characterization of MicroRNAs from Aphis gossypii (Hemiptera: Aphididae). Ann Entomol Soc Am. 2014;107(2): 521–31. 10.1603/an12158 [DOI] [Google Scholar]
- 21.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294: 862–864. 10.1126/science.1065329 . [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem–loop RT-PCR. Nucleic Acids Res. 2005;33: e179 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song QX, Liu YF, Hu XY, Zhang WK, Ma B, Chen SY, et al. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011;11: 5 10.1186/1471-2229-11-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Liu Y, Xin X, Kim TS, Cabeza EA, Ren J, et al. Evidence for Positive Selection on a Number of MicroRNA Regulatory Interactions during Recent Human Evolution. PLoS Genet. 2012;8(3): e1002578 10.1371/journal.pgen.1002578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peláez P, Trejo MS, Iniguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes J, et al. Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom. 2012;13: 83 10.1186/1471-2164-13-83 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang F, Li L, Liu L, Li H, Zhang Y, Yao Y, et al. High-throughput sequencing discovery of conserved and novel microRNAs in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol Genet Genomics. 2012;287: 555–63. 10.1007/s00438-012-0699-3 [DOI] [PubMed] [Google Scholar]
- 27.Wu W, Ren Q, Li C, Wang Y, Sang M, Zhang Y, et al. Characterization and Comparative Profiling of MicroRNAs in a Sexual Dimorphism Insect, Eupolyphaga sinensis Walker. PLoS ONE. 2013;8(4): e59016 10.1371/journal.pone.0059016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.28Calla B, Geib SM. MicroRNAs in the oriental fruit fly, Bactrocera dorsalis: extending Drosophilid miRNA conservation to the Tephritidae. BMC Genom. 2015;16: 740 10.1186/s12864-015-1835-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandety RS, Sharif A, Kamita SG, Ramasamy A, Falk B. W. Identification of Novel and Conserved microRNAs in Homalodisca vitripennis, the Glassy-Winged Sharpshooter by Expression Profiling. PLoS ONE. 2015;10(10): e0139771 10.1371/journal.pone.0139771 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mound LA, Morris DC. The insect order Thysanoptera: classification versus systematics. Zootaxa. 2007;1668: 395–411. [Google Scholar]
- 31.German TL, Ullman DE, Moyer JW. Tospoviruses: diagnosis, molecular biology, phylogeny, and vector relationships. Annu Rev Phytopathol. 1992;30: 315–348. 10.1146/annurev.py.30.090192.001531 [DOI] [PubMed] [Google Scholar]
- 32.Mound LA. Thysanoptera: diversity and interacttions. Ann RevEntomol.2005;50: 247–269. 10.1146/annurev.ento.49.061802.123318 . [DOI] [PubMed] [Google Scholar]
- 33.Rebijith KB. Molecular approaches in identification, diversity and management of important insect vectors, Thrips palmi Karny (Thysanoptera) and Aphis gossypii Glover (Hemiptera). Ph.D Thesis, Kuvempu University. 2015.
- 34.Whitefield AE, Ullman DE, German TL. Tospovirus-Thrips interaction. Ann Rev Phytopathol. 2005;43: 459–89. 10.1146/annurev.phyto.43.040204.140017 . [DOI] [PubMed] [Google Scholar]
- 35.Murai T. Current status of onion thrips, Thrips tabaci, as a pest in Japan. Plant Protect. 2003;57: 53–55. [Google Scholar]
- 36.Rebijith KB, Asokan R, Krishna Kumar NK, Krishna V, Ramamurthy VV. Development of species-specific markers and Molecular differences in mtDNA of Thrips palmi Karny and Scirtothrips dorsalis Hood (Thripidae: Thysanoptera), vectors of tospoviruses (Bunyaviridae) in India. Entomol News. 2011;122(3): 201–213. 10.3157/021.122.0301 [DOI] [Google Scholar]
- 37.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40: 37–52. 10.1093/nar/gkr688 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31: 3429–3431. 10.1093/nar/gkg599 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36 (Database issue): D154–158. 10.1093/nar/gkm952 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34 (Database Issue): D140–D144. 10.1093/nar/gkj112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. The RAxML 704 manual. Available: http://icwwwepflch/~stamatak/ (2008).
- 42.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2004;5(1): R1 10.1186/gb-2003-5-1-r1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol.1981;147: 195–197. 10.1016/0022-2836(81)90087-5 [DOI] [PubMed] [Google Scholar]
- 44.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9): 1639–1645. 10.1101/gr.092759.109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines-minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55: 611–622. 10.1373/clinchem.2008.112797 . [DOI] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4): 402–408. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 47.Liang P, Feng B, Zhou X, Gao X. Identification and Developmental Profiling of microRNAs in Diamondback Moth, Plutellaxylostella (L.). PLoS ONE. 2013;8(11): e78787 10.1371/journal.pone.0078787 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15: 336–360. 10.1038/sj.cr.7290302 . [DOI] [PubMed] [Google Scholar]
- 49.Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, Hannon GJ, et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17(12): 1865–1879. 10.1101/gr.6593807 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kakumani PK, Chinnappan M, Singh AK, Malhotra P, Mukherjee SK, Bhatnagar RK. Identification and Characteristics of microRNAs from Army Worm, Spodoptera frugiperda Cell Line Sf21. PLoS ONE. 2015;10(2): e0116988 10.1371/journal.pone.0116988 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burnside J, Ouyang M, Anderson A, Bernberg E, Lu C, Meyers BC, et al. Deep sequencing of chicken microRNAs. BMC Genomics.2008;9: 185 10.1186/1471-2164-9-185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koh W, Sheng CT, Tan B, Lee QY, Kuznetsov V, Kiang LS, et al. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genom. 2010;11 (Suppl.1): S5–S6, 10.1186/1471-2164-11-S1-S6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inukai S, De-Lencastre A, Turner M, Slack F. Novel MicroRNAs Differentially Expressed during Aging in the Mouse Brain. PLoS ONE. 2012;7(7): e40028 10.1371/journal.pone.0040028 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, et al. Evolution of the human-specific microRNA miR-941. Nat Commun. 2012;3:1145 10.1038/ncomms2146 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji Z, Wang G, Xie Z, Zhang C, Wang J. Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology. Mol Biol Rep. 2012;39: 9361–9371. 10.1007/s11033-012-1779-5 . [DOI] [PubMed] [Google Scholar]
- 56.Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, et al. MicroRNA-34a Inhibits the Proliferation and Metastasis of Osteosarcoma Cells Both In Vitro and In Vivo. PLoS ONE. 2012;7(3): e33778 10.1371/journal.pone.0033778 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avesson L, Reimegar J, Wagner EG, Söderbom F. MicroRNAs in Amoebozoa: Deep sequencing of the small RNA population in the social amoeba, Dictyostelium discoideum reveals developmentally regulated microRNAs. RNA. 2012;18: 1771–1782. 10.1261/rna.033175.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22(1): 107–26. 10.1038/cr.2011.158 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol. 2012;42: 381–395. 10.1016/j.ibmb.2012.01.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge X, Zhang Y, Jiang J, Zhong Y, Yang X, Li Z, et al. Identification of MicroRNAs in Helicoverpa armigera and Spodoptera litura Based on Deep Sequencing and Homology Analysis. Int. J. Biol. Sci. 2013;9(1): 1–15. 10.7150/ijbs.5249 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sattar S, Addo-Quaye C, Song Y, Anstead JA, Sunkar R, Thompson GA. Expression of small RNA in Aphis gossypii and its potential role in the resistance interaction with melon. PLoS ONE. 2012;7: e48579 10.1371/journal.pone.0048579 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6): 792–806. 10.1016/j.molcel.2013.08.017 . [DOI] [PubMed] [Google Scholar]
- 63.Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arrese EL, Soulages JL, et al. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genom. 2010;20: 52 10.1186/1471-2164-11-52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surridge AK Lopez-Gomollon S, Moxon S, Maroja LS, Rathjen T, Nadeau NJ, et al. Characterization and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene. BMC Genom. 2011;12: 62 10.1186/1471-2164-12-62 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505: 353–359. 10.1038/nature12987 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature Rev. Mol. Cell Biol. 2011;12: 246–258. 10.1038/nrm3089 . [DOI] [PubMed] [Google Scholar]
- 67.Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141(18): 3458–3471. 10.1242/dev.094037 . [DOI] [PubMed] [Google Scholar]
- 68.Cordero P, Lucks JB, Das R. An RNA Mapping Database for curating RNA structure mapping experiments. Bioinformatics. 2012;28: 3006–3008. 10.1093/bioinformatics/bts554 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flores-jasso CF, Arenas-Huertero C, Reyes JL, Contreras-Cubas C, Covarrubias A, Vaca L. “First step in pre-miRNAs processing by human Dicer”. Acta Pharmacol Sin. 2009;30: 1177–85. 10.1038/aps.2009.108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta R, Soni N, Patnaik P, Sood I, Singh R, Rawal K, et al. High AU content: a signature of up regulated miRNA in cardiac diseases. Bioinformation. 2010;5: 132–135. 10.6026/97320630005132 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Zhou X, Ge X, Jiang J, Li M, Jia S, et al. Insect-Specific microRNA Involved in the Development of the Silkworm Bombyx mori. PLoS ONE. 2009;4: e4677 10.1371/journal.pone.0004677 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39 (supplement): D152 10.1093/nar/gkq1027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian G, Yin XY, Luo H. Sequencing bias: Comparison of different protocols of microRNA library construction. BMC Biotechnol. 2010;10: 64 10.1186/1472-6750-10-64 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Ann Rev Microbiol. 2010;64: 123–141. 10.1146/annurev.micro.112408.134243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems Nat. Biotechnol. 2013;31(3): 233–239. 10.1038/nbt.2508 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones G, Sharp PA. Ultraspiracle: an invertebrate nuclear receptor for juvenile hormones. Proc Natl Acad Sci U S A. 1997;94: 13499–13503. 10.1073/pnas.94.25.13499 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barchuk AR, Maleszka R, Simões ZL. Apis mellifera ultraspiracle: cDNA sequence and rapid up-regulation by juvenile hormone. Insect Mol Biol. 2004;13: 459–467. 10.1111/j.0962-1075.2004.00506.x . [DOI] [PubMed] [Google Scholar]
- 78.Nunes FMF, Ihle KE, Mutti NS, Simões ZLP, Amdam GV. The gene vitellogenin affects microRNA regulation in honey bee (Apis mellifera) fat body and brain. J. Exp. Biol. 2013;216: 3724–3732. 10.1242/jeb.089243 . [DOI] [PubMed] [Google Scholar]
- 79.Chen Z, Liang S, Zhao Y, Han Z. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development. 2012;139: 3543–3552 10.1242/dev.082719 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greenberg JK, Xia J, Zhou X, Thatcher SR, Gu X, Ament SA, et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes, Brain Behav. 2012;11: 660–70. 10.1111/j.1601-183X.2012.00782.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nature Rev Genet. 2011;12: 99–110. 10.1038/nrg2936 . [DOI] [PubMed] [Google Scholar]
- 82.Griffiths-Jones S, Hui JH, Marco A, Ronshaugen M. MicroRNA evolution by arm switching. EMBO Rep. 2011;12: 172–177. 10.1038/embor.2010.191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE. 2008;3: e 1396 10.1371/journal.pone.0001396 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28: 598–613. 10.1016/j.molcel.2007.09.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu G, Zhou H, Wang Q, Auersperg N, Peng C. Activin receptor-like kinase 7 induces apoptosis through up-regulation of Bax and down-regulation of Xiap in normal and malignant ovarian epithelial cell lines. Mol Cancer Res. 2006;4: 235–246. 10.1158/1541-7786.MCR-05-0174 . [DOI] [PubMed] [Google Scholar]
- 86.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137: 273–282. 10.1016/j.cell.2009.01.058 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104: 9667–9672. 10.1073/pnas.0703820104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jayachandran B, Hussain M, Asgari S. An insect trypsin-like serine protease as a target of miRNA: utilization of miRNA mimics and inhibitors by oral feeding. Insect Biochem Mol Biol. 2013;43(4): 398–406. 10.1016/j.ibmb.2012.10.004 . [DOI] [PubMed] [Google Scholar]
- 89.Miska E. A., How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15: 563–568. 10.1016/j.gde.2005.08.005 . [DOI] [PubMed] [Google Scholar]
- 90.Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18(12): 2024–33. 10.1101/gr.080200.108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agrawal N, Sachdev B, Rodrigues J, Sowjanya Sree K, Bhatnagar RK. Development associated profiling of chitinase and microRNA of Helicoverpa armigera identified chitinase repressive microRNA. Scientific Reports. 2013;3: 2292 10.1038/srep02292 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.