Abstract
Background
Stromal tumor infiltrating lymphocytes (TILs) density is an outcome predictor in triple-negative breast cancer (TNBC). Herein we asked whether TILs are related to coding mutation load and to the chemical class of the resulting mutated amino acids, i.e., charged, polar, and hydrophobic mutations.
Methods
We examined paraffin tumors from TNBC patients who had been treated with adjuvant chemotherapy mostly within clinical trials (training cohort, N = 133; validation, N = 190) for phenotype concordance; TILs density; mutation load and types.
Results
Concordance of TNBC phenotypes was 42.1% upon local / central, and 72% upon central / central pathology assessment. TILs were not associated with mutation load, type and class of mutated amino acids. Polar and charged mutation patterns differed between TP53 and PIK3CA (p<0.001). Hydrophobic mutations predicted for early relapse in patients with high nodal burden and <50% TILs tumors (training: HR 3.03, 95%CI 1.11–8.29, p = 0.031; validation: HR 2.90, 95%CI 0.97–8.70, p = 0.057), especially if compared to patients with >50% TILs tumors (training p = 0.003; validation p = 0.015).
Conclusions
TILs density is unrelated to mutation load in TNBC, which may be regarded as an unstable phenotype. If further validated, hydrophobic mutations along with TILs density may help identifying TNBC patients in higher risk for relapse.
Introduction
Triple negative breast cancer (TNBC) is a diagnosis by exclusion corresponding to tumors that are immunohistochemically (IHC) negative for estrogen receptor (ER), progesterone receptor (PgR) and HER2 protein overexpression [1]. TNBC are in fact a variety of diseases under the same umbrella and as such, are characterized by extensive histological, phenotypic and genetic diversity [2]. The majority of TNBC are basal-like breast carcinomas (BLBC). The two terms are not interchangeable although they share a 70–80% overlap. BLBC are diagnosed as an intrinsic breast cancer subtype with widely used classifiers [3], while TNBC are called by ER/PgR/HER2 IHC as a proxy for BLBC [4]. In the clinic, in comparison to patients with other breast cancer subtypes for whom survival has improved or is expected to further improve with the application of disease-specific drugs, survival of patients with TNBC is still generally poor with yet no therapeutic options other than cytotoxic chemotherapy.
At the genomic level, TNBC are characterized by early structural aberrations that persist through clonal evolution and by a high incidence of TP53 mutations [5–9]; TNBC mutation load is more than ten times higher as compared to luminal carcinomas [9] but, with the exception of TP53, mutations and mutated genes are non-recurrent among tumors and may be heterogeneously affected within the same tumor [6, 9]. Research is currently ongoing for translating the huge amount of genomic data into clinical benefit for TNBC patients with no conclusive results yet.
In comparison to luminal subtypes, TNBC are also characterized by relatively higher rates of stromal tumor lymphocytic infiltrates (TILs) that reflect host anti-tumor immune response [10, 11] and confer better prognosis in the adjuvant [12–15] and neoadjuvant settings [16–18]. Irrespectively of their clonal frequency and impact on tumor maintenance, mutations producing neoantigens have been implicated in triggering host immune response against tumors [19]. TNBC are rich in mutations and in TILs, which may indicate interdependence of the two parameters, as has been shown for high mutational loads in other tumors arising in immunogenic environments, e.g., lung cancer, melanoma, cancers within the Lynch syndrome [19, 20]. Such associations have not yet been addressed in tumors arising in non-immunogenic environments, such as in the breast.
Predicting which mutations, provided that they are expressed within mutant epitopes, will result in triggering CD8 mediated cancer cell killing remains a still unmet challenge. The chemical nature of resulting mutant amino acids (charged, polar, hydrophobic) and epitopes might provide a hint in this direction. It was early recognized that preserved mutations during protein evolution preferentially occur for similar amino acid classes [21], because this way self-distraction would not be triggered. Further, polar-to-hydrophobic substitutions were identified as mutations evoking TILs response in patients with melanomas [22]. Supporting the above, hydrophobicity may provide a basis for T cells to recognize alien epitopes as immunogenic [23].
In a first attempt to approach the above issues at the tumor tissue level, herein we asked whether mutational load and the chemical nature of the resulting mutant amino acids in TNBC is associated with TILs density affecting the outcome of patients with operable disease. We hypothesized that mutant amino acid classes might interfere with patient outcome according to TILs density in the tumor. To this end, we examined mutation characteristics and TILs, as well as standard clinico-pathological parameters that affect early TNBC patient outcome, such as nodal status and tumor size, in two independent patient cohorts with informative targeted massively parallel sequencing data (MPS). Patients had mostly been treated within adjuvant clinical trials by the Hellenic Cooperative Oncology Group (HeCOG). Because the cohorts had been selected at different time points we also compared phenotype concordance upon local and central pathology testing; in addition, intra-tumoral (spatial) genetic heterogeneity was examined in different samples from different paraffin block depths from the same tumor.
Materials and Methods
Patients, tumors and samples
In the present retrospective/prospective translational study, two independent series of TN tumor tissues with informative targeted massively parallel sequencing (MPS) data were compared for the presence of variants and mutations, and were examined for patient outcome. Routinely processed TNBC tissues (FFPE) with annotated patient data were retrieved at two different time points from the HeCOG tissue bank and clinical databases; central histological review, phenotypic reassessment, and MPS were implemented in the Laboratory of Molecular Oncology (MOL; Hellenic Foundation for Cancer Research / HeCOG / Aristotle University of Thessaloniki, Thessaloniki, Greece). In total, 365 patients that had been diagnosed as TNBC locally and/or centrally were examined. Cohort (A), which served as the training set, included prospectively collected tumor blocks for biomarker analysis from TNBC patients treated in a series of adjuvant clinical trials by HeCOG, i.e., HE 10/00, HE 10/04A, HE 10/04B, HE 10/05, HE 10/08, and HE 10/10, as described in S1 Table or from patients who were ineligible to enter these trials and were routinely treated in HeCOG affiliated clinical centers. The 133 cases included in this cohort were selected based on tumor phenotype concordance in local and central pathology laboratories. Cohort (B) included tumors that had been characterized as TNBC upon local testing from patients treated only within the HE 10/97, HE 10/00, HE 10/05, and HE 10/08 trials (S1 Table). This cohort included 190 tumors and was used as a validation set. Patient characteristics for the two cohorts are shown in Table 1. For 82 cases in the validation cohort (S2 Table), targeted massively parallel sequencing (MPS) data from at least 2 DNA samples from the same tumor (76 from the same paraffin block; 6 from different blocks) were available [24]; these data were used for studying spatial heterogeneity within tumors. The study outline is summarized in Figure A in S1 File.
Table 1. Patient and tumor characteristics in the two study cohorts.
| Patients | training | validation | p-value |
|---|---|---|---|
| N | 133 | 190 | |
| Age (years) | |||
| Mean (SD) | 50.7 (12.9) | 52.2 (12.4) | 0.21 |
| Median | 49 | 53 | |
| Min-Max | 28–77 | 21–83 | |
| Ki67 | |||
| Mean (SD) | 53.4 (31.1) | 49.8 (32.6) | 0.30 |
| Median | 52 | 55 | |
| Min-Max | 0–100 | 0–100 | |
| N (%) | N (%) | ||
| Age (years) | |||
| ≤50 | 68 (51.2) | 74 (39.0) | 0.030 |
| >50 | 65 (48.8) | 116 (61.0) | |
| Menopausal status | |||
| Postmenopausal | 64 (48.2) | 103 (54.2) | 0.28 |
| Premenopausal | 69 (51.8) | 87 (45.8) | |
| Tumor size | |||
| ≤2 | 43 (32.6) | 72 (37.8) | 0.33 |
| >2 | 89 (67.4) | 118 (62.2) | |
| Positive lymph nodes | |||
| 0–3 | 91 (70.0) | 122 (64.2) | 0.28 |
| ≥4 | 39 (30.0) | 68 (35.8) | |
| Histological grade | |||
| I | 2 (1.6) | 4 (2.2) | 0.60 |
| II | 20 (15.0) | 36 (19.0) | |
| III | 111 (83.4) | 150 (79.0) | |
| Histological type | |||
| Medullary | 6 (4.6) | 16 (8.4) | 0.29 |
| NST | 112 (84.2) | 148 (77.8) | |
| Other | 15 (11.2) | 26 (13.6) | |
| Surgery (binary) | |||
| MRM | 57 (42.8) | 99 (52.2) | 0.10 |
| Other | 76 (57.2) | 91 (47.8) | |
| Hormonotherapy | |||
| No | 107 (80.4) | 160 (85.1) | 0.27 |
| Yes | 26 (19.6) | 28 (14.9) | |
| Radiotherapy | |||
| No | 34 (25.6) | 44 (23.7) | 0.70 |
| Yes | 99 (74.4) | 142 (76.3) | |
| ER/PgR/HER2 local | |||
| Either positive | 0 (0.0) | 0 (0) | |
| TNBC | 133 (100) | 190 (100) | |
| ER/PgR/HER2 central | |||
| Either positive | 0 | 80 (42.2) | |
| TNBC | 133 (100) | 97 (51.0) | |
| Basal | 0.050 | ||
| Yes | 110 (84.0) | 133 (74.7) | |
| No | 21 (16.0) | 45 (25.3) | |
| Survival data | |||
| Median FU in months | 79 | 70 | |
| N of valid cases | 133 | 190 | |
| Deaths, N | 35 | 39 | 0.070 |
| Event free at 3 years, % | 86,6 | 88,4 | |
| Event free at 5 years, % | 76,7 | 82,9 | |
| Relapse, N | 44 | 47 | 0.051 |
| Event free at 3 years, % | 75,7 | 82,6 | |
| Event free at 5 years, % | 71,2 | 77,2 |
Notes: N: number; MRM: modified radical mastectomy; NST: non-specific type; FU: follow-up.
Patients had provided written consent for the use of their biologic material for research purposes and the study was approved by the Bioethics Committee of the Aristotle University of Thessaloniki School of Medicine (#77/10June2014) and by the Institutional Review Board of Papageorgiou Hospital of Thessaloniki (#725/10May2013). TNBC had been prospectively diagnosed in local pathology laboratories as ER/PgR/HER2 IHC negative.
Central tumor assessment at MOL included histology review; ER/PgR/HER2/Ki67 IHC and HER2 FISH where needed [25] by adapting HER2 evaluation according to the more recent ASCO/CAP guidelines [26]. Briefly, ER and PgR cut-off was 1% positive nuclei; HER2 was considered overexpressed for intense circumscribed staining in >10% tumor cells and HER2 gene amplification for HER2/CEP17 ratio ≥2 or for ≥6 HER2 copies; Ki67 was assessed as % positive nuclei (continuous variable); and, CK5 and EGFR were applied for typing basal-like carcinomas with 1% cut-off for positivity [27]. IHC and FISH were performed on in-house low-density tissue microarrays (TMA) that contained two 1.5mm cores per tumor. TMA H&E sections were also evaluated for tumor cell content (TCC%; tumor nuclei vs. all nuclei). TMAs for the two cohorts were constructed at independent time points but phenotyping was accomplished with the same cut-offs for both series; the evaluated tumor areas in overlapping cases were not identical for the same tumor. Such paired samples were used for the assessment of spatial heterogeneity. Stromal tumor infiltrating lymphocytes (TILs) density was also assessed on whole H&E sections based on Salgado et al [11], as previously described [13].
MPS genotyping
DNA was extracted from TMA cores. The majority of samples (78%) had TCC ≥50% but samples with as low as 15% TCC were also processed. DNA was extracted with magnetic beads (VERSANT Tissue Prep Kit, Siemens Healthcare, Erlangen, Germany); quantity was measured with the Qubit fluorometer (Life Technologies, Paisley, UK); and, DNA amplification performance was evaluated by two different control qPCR assays. Criteria for processing samples for genotyping were ≥2ng/ul DNA amplifiable at Ct≤32 for two different qPCR control assays.
Genotyping was accomplished in an Ion Proton Sequencer; the two custom highly-multiplexed panels [24] targeted coding mutations and single nucleotide polymorphisms (SNPs) previously implicated in breast cancer [5, 7]. The T-panel was used in the training cohort and in 78 out of 82 cases with matched tumor samples; the B-panel was used in the validation cohort and in all matched samples (Figure A in S1 File). Samples were accepted for further evaluation if 90% of amplicons had been read >100 times and if >5 eligible variants were present. Variants obtained from Ion Reporter v.4 were considered ineligible for analysis if non-annotated, if indels with GC-stretches (reading artifacts with semiconductor sequencing); if position coverage <100 and if variant coverage <40. The mean and median reads for the analyzed variant positions were 894.3 and 509 with the T-panel in the training set; 1002.8 and 646 with the B-panel in the validation set.
For the purposes of the present study, coding mutations and common SNPs (minor allele frequency (MAF) >0.1%) that were targeted with both panels were examined; mutations corresponded to amino acid changing variants in coding regions for which no MAF was reported or, if registered SNPs, with MAF <0.1%. Pathogenicity of mutations was not further addressed, since, despite the numerous bioinformatics tools that have been proposed for this purpose [28], predicting functional implications of tumor mutations is still questionable [29].
Common gene targets with both panels were analyzed in the training and validation sets, and for the assessment of heterogeneity. For avoiding false heterogeneous calls, only positions that had been covered >100 times with both panels were compared from raw Ion Reporter data, as described in [24].
Statistics
TIls were examined as a continuous variable and, for presentation purposes, as a 3-scale variable, 0–5%, 5–50% and >50%, whereby the latter represented lymphocyte-predominant (LP) breast cancer (BC), previously associated with most favorable outcome in TNBC [13].
Classic mutation types (missense, frameshift indels, nonsense) were examined along with the nature of the resulting mutated amino acids. Amino acids were classified as charged (Arg, Lys, Asp, Glu); polar (Gln, Asn, His, Ser, Thr, Tyr, Cys, Met, Trp); and, hydrophobic (Ala, Ile, Leu, Phe, Val, Pro, Gly). These were examined as single markers or in combined patterns that were present in tumors. Venn diagrams were plotted for overlapping features with Oliveros, J.C. (2007–2015) Venny. An interactive tool for comparing lists with Venn's diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html
Classic descriptive statistics were applied (chi-square or Fisher’s exact test for categorical variables; Mann-Whitney or Kruskal-Wallis test for testing categorical against continuous variables). According to patient data origin, follow up periods ranged from 15 years for patients in the HE 10/00 trial down to 5 years in the more recent HE 10/10 trial (S1 Table). For this reason, only disease-free survival (DFS) was assessed in the present study. DFS was measured from the date of diagnosis until verified disease relapse or death, whichever occurred first, or loss from follow-up. Event free patients were censored at the date of last contact. Time-to-event distributions were compared by using Kaplan-Meier curves and log-rank tests. Univariate Cox regression analysis was used for reporting hazard ratios, while the Firth’s correction for monotone likelihood was used in the case of zero or few events. Survival status was updated in June 2014.
Regarding multivariate Cox regression models, the clinicopathological parameters were chosen by backward elimination among the following adjustment factors: age (>50 vs. ≤50 years), menopausal status (post vs. pre), tumor size (>2 cm vs. ≤2 cm), histological grade (I-II vs. III), adjuvant radiotherapy (yes vs. no), and Ki67 as a continuous variable by 5% increments. Positive nodes (≥4 vs. 0–3) and TILs density (LPBC vs. non-LPBC) were also added to models in the case they were not included in any combined variable.
All univariate tests were two-sided, with the significance level at α = 0.05, while significance threshold multivariate models was set at α = 0.15, a level higher than usual in order to control for bias in the estimations. No correction for multiple testing was made due to the exploratory nature of the study.
The analysis was fully compliant with the reporting recommendations for tumor marker prognostic studies [30]. The SAS software was used for statistical analysis (SAS for Windows, version 9.3, SAS Institute Inc., Cary, NC).
Results
Cohort specifications
The phenotypes of the 190 tumors in the validation cohort were TNBC upon local and central testing in 48.9%, locally TNBC / centrally nonTNBC in 42.1%, and locally TNBC without central assessment in 8.9% of the cases (Figure A in S1 File). Discordance concerned ER/PgR status, in accordance with previous reports even between central laboratories [31]. The incidence of basal carcinomas was significantly lower in locally TNBC / centrally nonTNBC (61.2%) as compared to concordant (86%) tumors (p<0.0001). The 82 tumors with ≥2 matched DNA samples that were also examined in the validation cohort had been phenotyped once locally as TNBC, and twice centrally at independent time points; 23 out of these tumors (28%) had one central call as nonTNBC upon heterochronous testing. Patient characteristics in this 82-case subset (S2 Table) did not differ from those in the actual study cohorts (Table 1).
Genotyping results and coding mutation features
Among 2158 variants in all cases tested with the T-panel (133 in the training cohort and 78 in the matched series), 215 were coding mutations (10%) and 1657 were common SNPs (76.1%). Correspondingly, among 2304 variants in the 190 cases tested with the B-panel, 277 were mutations (12%) and 1697 common SNPs (74%). The mean and median numbers of mutations per tumor were similar with both panels (Figure B in S1 File). Coding mutation patterns in the two cohorts, i.e., mutation incidence and gene involvement were as expected for TNBC [5–7, 32]. However, the incidence of TP53 and PIK3CA mutations was significantly different between the training cohort and the locally/centrally discordant subset of the validation cohort (S3 Table). In the 82-case subset with ≥2 matched samples, data from 170 tissue and 12 blood samples were available for analysis (Figure A in S1 File). The results of matched genotype comparisons with the two panels are shown in Figure C in S1 File and in S4 Table. It appeared that (a) SNPs were more frequently conserved in matched samples than coding mutations; (b) coding mutations were common in half of the matched cases only (45% common TP53 mutations); (c) no two tumors had identical genotypes when taking into account both SNPs and coding mutations. Private mutations with either panel for the same tumor were examined in the validation cohort. Thus, the final number of mutations in the training and validation cohort was 149 and 257, respectively; these were distributed in 94/133 (70.7%) and in 125/190 (65.8%) cases.
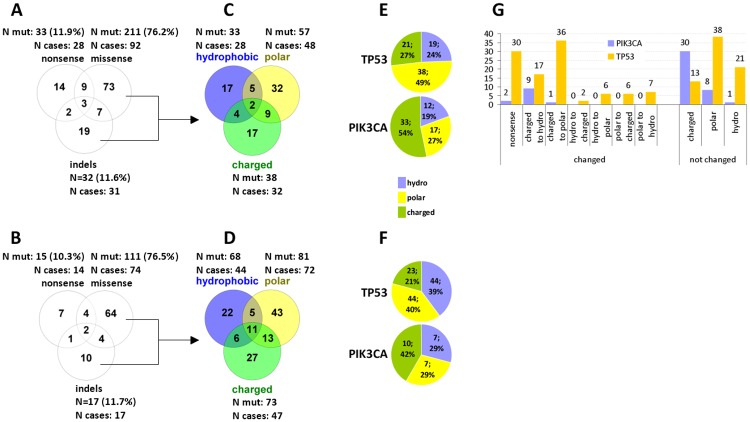
The type of mutations (missense, indels, nonsense) was strikingly similar, and the distribution of resulting charged, hydrophobic and polar amino acids was not statistically significantly different in the two cohorts (S5 Table). Multiple mutations per tumor were mostly missense (Fig 1A and 1B), which was particularly prominent in the hypermutated cases (22 and 60 mutations per case) in the validation cohort. Relatively few tumors had multiple types of mutations (11/94 [11.7%] and 18/127 [14.2%] in the training and validation cohort, respectively). In comparison, the rate of combinations of resulting mutated amino acid types was more than double in both cohorts (Fig 1C and 1D). The distribution of mutated amino acid types differed in TP53 and PIK3CA (Fig 1E and 1F); mutations in TP53 were more frequently polar and the mutated amino acid class was more frequently changed in comparison to PIK3CA mutations that resulted in charged amino acids without class change (Fisher’s exact p<0.0001) (Fig 1G).
Fig 1. Coding mutation types and amino acid classes in TNBC.
A, C, E: training cohort. B, D, F: validation cohort. The distribution of classic mutation types (A and B) and of mutated amino acid classes (C and D) did not differ between the two cohorts. Mutated amino acid classes in TP53 and PIK3CA showed the same distribution in the two cohorts but differed significantly for the two genes (E and F). The difference concerned both changed and not changed amino acid classes as compared to the reference (G, combined data for the two cohorts).
Mutation load and types of mutations are not associated with TILs density
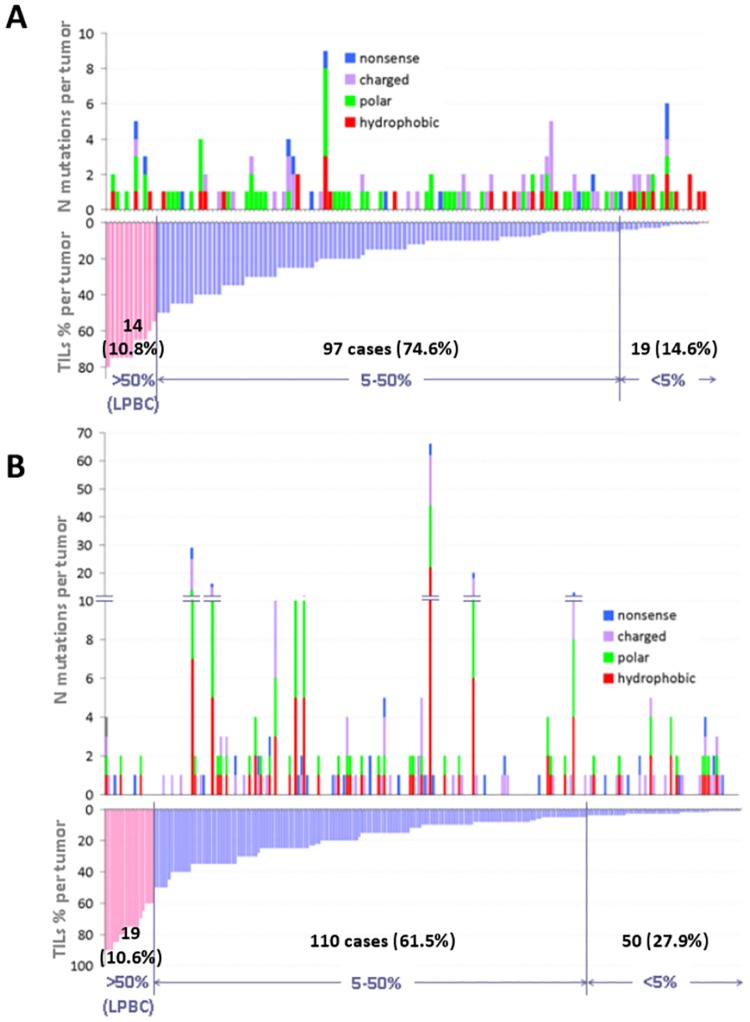
It was possible to assess TILs in 130 and 179 tumors only, in the training and validation cohort, respectively, due to absence of adequate stroma in the available tumor sections. The rate of tumors with >50% TILs (LPBC) was constant, but the rate of tumors with <5% TILs, which was used as a lower cut-off for presentation purposes, was higher in the validation cohort (Fig 2A and 2B). Other than expected, LPBC had similar or lower mutation load and mutated genes than tumors with <5% TILs. In the training cohort, tumors with <5% TILs had significantly higher mutation load (mean 1.4 vs. 1.02) and numbers of mutated genes (mean 1.2 vs. 0.8) as compared to tumors with higher TILs (Mann-Whitney p = 0.0347 and p = 0.0302, respectively). No corresponding association was demonstrated in the validation cohort. Comparison of continuous TILs against the same mutation parameters did not yield significant associations either. In both cohorts, the rate of mutant tumors in LPBC was lower than the overall such rate. Because TP53 mutations were by far the most abundant in both TNBC cohorts, and also because unique mutation combinations with or without TP53 were observed, single gene comparisons against TILs would not yield meaningful results and were not applied. Classic types of mutations were not associated with TILs density, while nonsense mutations were also observed in LPBC.
Fig 2. Coding mutations and mutated amino acid classes in association with TILs density.
A: Training cohort, N cases = 130; B: Validation cohort, N cases = 179. The rate of LPBC was constant in the two cohorts, but the rate of tumors with <5% TILs was significantly higher in the validation cohort. The mutation load in LP tumors was similar to or lower than that observed in tumors with <5% TILs. No difference was observed in the distribution of mutation types according to TILs density.
The by definition lymphocyte-rich medullary carcinomas were particularly poor in mutations of any class; other than expected, hydrophobic mutations were more frequent in tumors with low TILs, which reached statistical significance in the training cohort (S6 Table). Single mutated amino acid classes were rare in LPBC, especially with regards to hydrophobic mutations. In the validation cohort, polar mutations were associated with larger tumor size (p<0.001), which was not significant in the training cohort. No further associations between mutated amino acid classes and clinicopathological parameters were observed.
TILs density and hydrophobic mutations on patient DFS
Patient follow up period and the number of events in both cohorts is shown in Table 1. The effect of clinicopathological characteristics on DFS is shown in S7 Table. High nodal burden was the only parameter significantly associated with poor DFS in both cohorts. Mutations in single repeatedly affected individual genes were not associated with patient outcome (S8 Table). Risk probability analysis with respect to TILs density revealed an overall higher risk for relapse in the training as compared to the validation cohort but the effect of TILs on DFS was in the same direction (Figure D in S1 File).
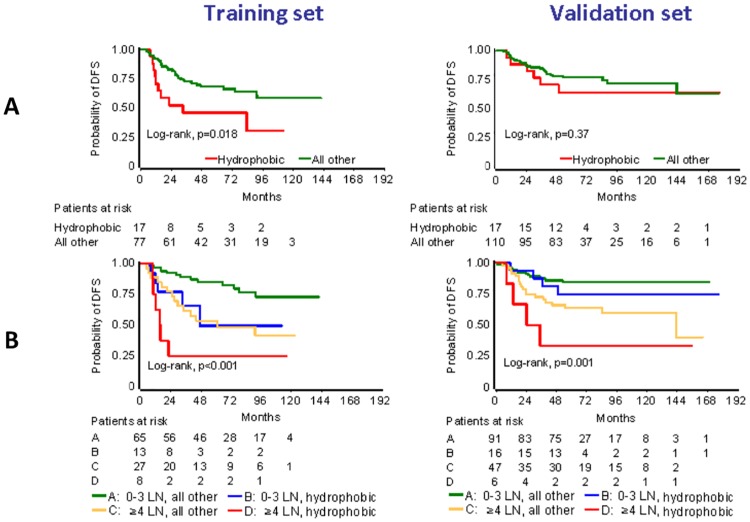
Because of the rare incidence of single mutated amino acid classes in LPBC (Fig 2, S6 Table), the relevant analysis was applied for non-LPBC. The presence of charged and polar mutations, either as single parameters or as combined mutated amino acid classes, had no effect on patient outcome in both cohorts. In the training cohort, the presence of only hydrophobic mutations was strongly associated with poor DFS, whereby 9/10 relapses in the corresponding 17 patients occurred within the first 3 years; in the validation cohort, statistical significance was not reached for the same parameter but again, 5/6 relapses were observed within the first 3 years (Fig 3A; S8 Table). Based on this analysis, a first multivariate model was created including hydrophobic mutations and all clinicopathological parameters (patient age and menopausal status, nodal status, tumor size, histology, grade, radiotherapy). High nodal status was the only common independent unfavorable parameter in both cohorts. Resulting hydrophobic mutations remained significant in the training cohort, while a similar trend was observed in the validation cohort when continuous TILs were included in the model (Table 2A and 2B).
Fig 3. Hydrophobic mutations adversely affect patient DFS.
Analysis was conducted in non-LPBC (tumors with <50% TILs). LN in B: nodal status. Patients with tumors with hydrophobic mutations performed worse in every context, in both cohorts.
Table 2. Basic parameter adjustments in the two cohorts.
| A. TILs as a categorical variable were not retained in the model | |||||
| TRAINING COHORT | N patients | N events | HR | 95%CI | Wald's p |
| Menopausal status | |||||
| post vs. pre | 61 vs. 69 | 18 vs. 25 | 0.6 | 0.32–1.10 | 0.10 |
| Size | |||||
| >2 vs. ≤2 cm | 87 vs. 43 | 32 vs. 11 | 1.8 | 0.90–3.58 | 0.097 |
| Radiotherapy | |||||
| yes vs. no | 99 vs. 31 | 31 vs. 12 | 0.44 | 0.21–0.90 | 0.026 |
| Mutated amino acid class | |||||
| hydrophobic vs. all other | 26 vs. 104 | 12 vs. 31 | 2.31 | 1.17–4.58 | 0.017 |
| Number of positive nodes | |||||
| ≥4 vs. 0–3 | 39 vs. 91 | 21 vs. 22 | 3.88 | 2.01–7.50 | <0.001 |
| VALIDATION COHORT | |||||
| Histo grade | |||||
| III vs. I-II | 150 vs. 40 | 32 vs. 15 | 0.59 | 0.32–1.09 | 0.094 |
| Mutated amino acid class | |||||
| hydrophobic vs. all other | 24 vs. 166 | 8 vs. 39 | 1.69 | 0.78–3.65 | 0.18 |
| Number of positive nodes | |||||
| ≥4 vs. 0–3 | 68 vs. 122 | 28 vs. 19 | 2.75 | 1.52–4.97 | <0.001 |
| B. TILs as a continuous variable were favourable in the validation cohort only | |||||
| TRAINING COHORT | N patients | N events | HR | 95%CI | Wald's p |
| Menopausal status | |||||
| post vs. pre | 61 vs. 69 | 18 vs. 25 | 0.6 | 0.32–1.10 | 0.10 |
| Size | |||||
| >2 vs. ≤2 cm | 87 vs. 43 | 32 vs. 11 | 1.8 | 0.90–3.58 | 0.097 |
| Radiotherapy | |||||
| yes vs. no | 99 vs. 31 | 31 vs. 12 | 0.44 | 0.21–0.90 | 0.026 |
| Mutated amino acid class | |||||
| hydrophobic vs. all other | 26 vs. 104 | 12 vs. 31 | 2.31 | 1.17–4.58 | 0.017 |
| Number of positive nodes | |||||
| ≥4 vs. 0–3 | 39 vs. 91 | 21 vs. 22 | 3.88 | 2.01–7.50 | <0.001 |
| VALIDATION COHORT | |||||
| Histo grade | |||||
| III vs. I-II | 126 vs. 37 | 24 vs. 13 | 0.57 | 0.27–1.19 | 0.13 |
| Mutated amino acid class | |||||
| hydrophobic vs. all other | 23 vs. 140 | 8 vs. 29 | 2.07 | 0.94–4.59 | 0.072 |
| Number of positive nodes | |||||
| ≥4 vs. 0–3 | 52 vs. 111 | 19 vs. 18 | 2.46 | 1.26–4.79 | 0.008 |
| TILs continuous (incr. by 5%) | - | - | 0.86 | 0.76–0.98 | 0.024 |
| Ki67 (incr. by 5%) | - | - | 1.06 | 1.00–1.12 | 0.053 |
Based on these results, we partitioned non-LPBC into high and low nodal status; patients with hydrophobic mutations had unfavorable outcome in both cohorts, particularly pronounced in patients with high nodal status (Fig 3B). Distinguishing low and high TILs in non-LPBC (<5% and 5–50% TILs) further supported the unfavorable effect of resulting hydrophobic mutations in the 5–50% TILs subset, in both cohorts (univariate Cox results in S8 Table). This analysis was not possible in patients with <5% TILs, because only two tumors had hydrophobic mutations in this subset in the validation cohort.
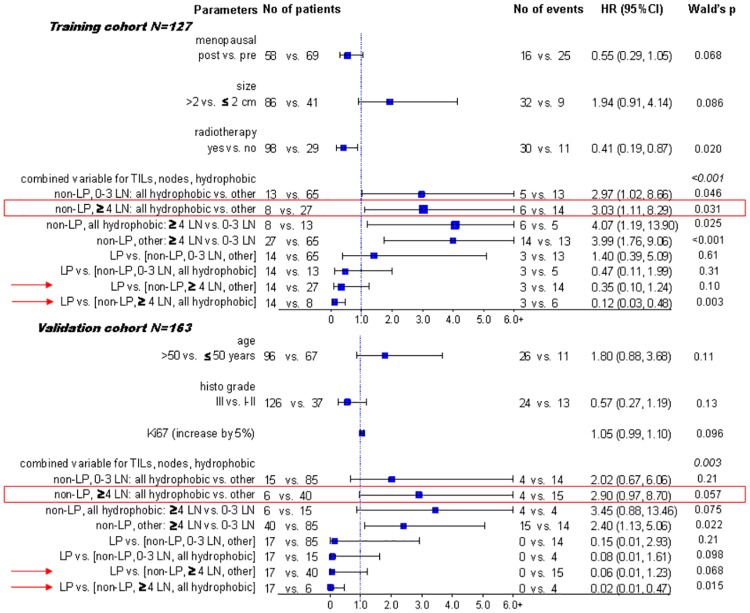
The combined variable including TILs, nodal status and resulting hydrophobic mutations was adjusted for significance with the remaining clinicopathologic variables (Fig 4). The variable retained its significance in both sets, at a first glance because it included nodal status and TILs. Importantly though, all hazard ratios in all subsets were in the same direction, whether significant, marginally significant or non-significant, while the resulting hydrophobic mutations were independently unfavorable prognostic factors in patients with high nodal status, in both the training and validation cohorts. In addition, in both cohorts, LPBC were independently favorable in patients with high nodal burden, especially when compared to patients with the same nodal status and hydrophobic mutations, while only a trend in the same direction was noticed for all other comparisons.
Fig 4. Multivariate analysis results.
The unfavorable effect of hydrophobic mutations in patients with high nodal burden (red boxes), especially when compared to LP-TNBC (arrows) was demonstrated in both the training and validation cohorts.
Discussion
This study confirmed the described complexity and plurality of TNBC mutation profiles [5, 6, 8, 9] in two independent cohorts that were tested with different MPS panels despite that relatively few genes were analyzed. The study also confirmed the favorable prognostic role of increasing TILs density in TNBC in the adjuvant setting [12, 14]. LPBC was highlighted in our previous report [13] as a potentially distinct subgroup of breast tumors with favorable prognosis. Although it has been suggested that LPBC should not be distinguished as a separate category in breast cancer [10], the same effect at the level of relative risk prediction was shown here for the independent training cohort that was not previously analyzed.
Other than described for tumors arising upon exposure to carcinogens or in immunogenic environments [19, 20], and although some TNBC may have mutation load as high as melanomas [33], high TILs density by any approach (continuous or categorical) was not accompanied by higher mutation load or higher numbers of mutated genes, and it was not associated with any structural type of coding mutation the way these are usually studied (missense, indels, nonsense), in neither of the two TNBC cohorts. For the genes analyzed, LP tumors had similar or lower mutation load, as compared to tumors with very low or absent TILs; in the majority of LP tumors no mutations were identified. High TILs in these cases may have been triggered by mutant or chimeric proteins not targeted with the panels; these may also have accumulated as a response to infectious agents the presence of which has been demonstrated in TNBC [34]. Clinically important in the same context is the opposite, that low TILs tumors had mutation load as tumors with higher TILs and LP. This indicates host inefficiency to recognize tumor cells with aberrant proteins and may thus explain the overall worse outcome of such patients.
The novelty here is the investigation of the chemical nature of the resulting mutated amino acids in TNBC genotypes. Although the entire mutation load of the tumors cannot be examined with targeted panels, the presented patterns may be considered representative since the genes mostly affected in TNBC were analyzed. We observed a similar distribution of mutated charged, polar and hydrophobic amino acids in the two cohorts, and similar rates of co-mutation combinations. It will be interesting to define whether these patterns are TNBC-specific or preserved among different tumor types. The small number of mutations in most individual genes did not allow for statistical comparisons. However, mutated amino acid features significantly differed in the most commonly mutated genes, TP53 and PIK3CA. The pattern observed for PIK3CA, i.e., preservation of the charged amino acid class upon mutation may be associated with the low TILs density observed in luminal tumors, where this gene is abundantly mutated [13–15]; the high incidence of changed class and the relatively low rate of preserved charged mutations in TP53 may explain the immunogenicity attributed to mutations of this gene [35, 36].
Among charged, polar and hydrophobic mutated amino acids, only the latter appeared to have an impact on patient outcome. If we accept that hydrophobic epitopes serve in self/non-self discrimination by T cell response [23], and also that increased hydrophobic residues destabilize proteins and turn them prone for proteasome degradation [37], tumors with such mutations would be expected to have dense lymphocytic infiltrations in the normal context. The opposite was observed, in line with tumors being in the escape phase at the time of diagnosis [38, 39]; very low representation of hydrophobic mutations in LP and higher incidence of the same mutations in low TILs tumors. This confirms the inefficiency of host immune response and is compatible with the poor prognosis of such patients. Importantly, we have shown that hydrophobic mutations were associated with poor prognosis in the major subset of early TNBC patients with up to 50% TILs density in their tumors, an effect that was particularly prominent in patients with high nodal status. TNBC patients with high nodal status and non-LP tumors harboring hydrophobic mutations did not benefit from chemotherapy; in fact, this was the only patient subset in both study cohorts with a clear prognostic disadvantage when compared to patients with LP tumors. TILs have been proposed as a marker predictive for response to immune check-point inhibitors [10] and may also be used as triggers for the paused immune system along with conventional therapies [40]. Whether hydrophobic mutations in combination with TILs density may serve as a marker of disabled host immune response is an important clinically relevant question with implications in strategies for the sensitization of the immune system.
A main issue highlighted here is about the TNBC phenotype itself. Discordant ER/PgR and HER2 status has repeatedly been addressed for calling luminal and HER2 positive tumors even between reference laboratories [31], but how often TNBC is miscalled or discordantly called remains unknown. As shown here, IHC-determined basal-like tumors are more consistently triple-negative than non-basal tumors, which has not been previously reported; this is nevertheless compatible with the highest concordance reported between IHC and PAM50 for this phenotype [41]. Phenotype discordance in various depths of the same paraffin block may be the result of technical inconsistency (preanalytical, analytical, IHC scoring and interpretation) but it also may be the result of intra-tumoral heterogeneity [6, 8, 9] and, as shown in preclinical models, of the multipotency of tumor stem cells [42]. In the present series discordance in calling TNBC reflected discordant hormone receptor status. This seems in line with TNBC relating more to the Luminal B subtype at the genomic and gene expression level than to any other breast cancer subtype [43]. In the clinic, TNBC means that the patient will not receive hormonal therapy. If we accept that the TNBC phenotype is heterogeneous or transient within the same tumor, the old practice of treating non-basal TNBC with anti-estrogens should perhaps be reconsidered.
Genomic heterogeneity was revealed as more pronounced for coding mutations than for common polymorphisms. From the technical point, the present comparison of matched samples represents one of the largest series tested with two different panels. As shown, non-coding areas in genes without known alterations in breast cancer such as TERT were fairly preserved in matched samples; by contrast, polymorphisms in genes like MDM2, CDKN2A, EGFR, GATA3 and mutations in frequently affected genes such as in TP53 were frequently not recapitulated. Genomic heterogeneity has been repeatedly demonstrated in TNBC, as reviewed by Ng et al [44] and the present data are further confirmatory in the respect. However, despite the stringent criteria that we applied for accepting variants for inter-sample comparisons with two different MPS panels, technical reasons cannot be excluded for the high degree of the observed genomic heterogeneity in this tumor subset [24].
In conclusion, this study shows that the level of TILs in TNBC is not related to the tumor mutational load and provides evidence suggesting that it may be worthy assessing the chemical nature of mutated amino acids for understanding the functionality of the host immune system according to the type of mutant residues present in the tumor. As in all studies dealing with LP breast cancer, a drawback in the present study is the small number of subsets that emerge for analysis. It appears, however, that LP TNBC is not driven by immunogenic mutations and that tumors with very low TILs may be heavily mutated but not recognized by the host. These findings were confirmed in two independent data sets and patient cohorts and may have implications in the rationale and design of trials involving interventions with immunomodulators. In this context, further validation and elucidation of the evidenced aspects of mutated amino acids seems worthy pursuing.
Supporting Information
Figure A, REMARK diagram for the study cohorts; Figure B, Coding mutations and SNPs in the study cohorts, as assessed with the two panels; Figure C, TNBC genotypes in 82 cases with multiple tumor samples; Figure D, Probability of DFS for patients in each TNBC cohort according to TILs density.
(DOC)
(PDF)
(PDF)
(PDF)
(PDF)
(XLS)
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors wish to thank Mrs. Emily Daskalaki for excellent technical assistance with MPS and Ms. Maria Moschoni and Mrs. Stella Dallidou for secretarial assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by an internal Hellenic Cooperative Oncology Group (HeCOG)translational research grant (HE TRANS_BR). The funders played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This study was also partly supported by the Greek General Secretary for Research and Technology (GSRT) Program, Research in Excellence II, funded by 75 % from the European Union and the Operational Program "Education & Lifelong Learning" ESPA-THALIS#266 of the Ministry of Education, Lifelong Learning & Religious Affairs. Health Data Specialists Ltd provided support in the form of salaries for author ZA but did not have any additional role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The specific role of this author is articulated in the ‘author contributions’ section. Health Data Specialists Ltd did not provide financial support in any other form.
References
- 1.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nature reviews Clinical oncology. 2010;7(12):683–92. Epub 2010/09/30. 10.1038/nrclinonc.2010.154 . [DOI] [PubMed] [Google Scholar]
- 2.Turner NC, Reis-Filho JS. Tackling the diversity of triple-negative breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19(23):6380–8. Epub 2013/12/04. 10.1158/1078-0432.CCR-13-0915 . [DOI] [PubMed] [Google Scholar]
- 3.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(8):1160–7. Epub 2009/02/11. 10.1200/JCO.2008.18.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linn SC, Van 't Veer LJ. Clinical relevance of the triple-negative breast cancer concept: genetic basis and clinical utility of the concept. Eur J Cancer. 2009;45 Suppl 1:11–26. Epub 2009/09/25. 10.1016/S0959-8049(09)70012-7 . [DOI] [PubMed] [Google Scholar]
- 5.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. Epub 2012/09/25. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–9. Epub 2012/04/13. 10.1038/nature10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–4. 10.1038/nature11017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lips EH, Michaut M, Hoogstraat M, Mulder L, Besselink NJ, Koudijs MJ, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast cancer research: BCR. 2015;17(1):134 10.1186/s13058-015-0642-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–60. Epub 2014/08/01. 10.1038/nature13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nature reviews Clinical oncology. 2015. Epub 2015/12/17. 10.1038/nrclinonc.2015.215 . [DOI] [PubMed] [Google Scholar]
- 11.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2015;26(2):259–71. Epub 2014/09/13. 10.1093/annonc/mdu450 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic Value of Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancers From Two Phase III Randomized Adjuvant Breast Cancer Trials: ECOG 2197 and ECOG 1199. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2014. 10.1200/JCO.2013.55.0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotoula V, Chatzopoulos K, Lakis S, Alexopoulou Z, Timotheadou E, Zagouri F, et al. Tumors with high-density tumor infiltrating lymphocytes constitute a favorable entity in breast cancer: a pooled analysis of four prospective adjuvant trials. Oncotarget. 2015. Epub 2015/10/28. 10.18632/oncotarget.6231 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2014;25(8):1544–50. Epub 2014/03/13. 10.1093/annonc/mdu112 . [DOI] [PubMed] [Google Scholar]
- 15.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31(7):860–7. Epub 2013/01/24. 10.1200/JCO.2011.41.0902 . [DOI] [PubMed] [Google Scholar]
- 16.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(1):105–13. Epub 2009/11/18. 10.1200/JCO.2009.23.7370 . [DOI] [PubMed] [Google Scholar]
- 17.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33(9):983–91. Epub 2014/12/24. 10.1200/JCO.2014.58.1967 . [DOI] [PubMed] [Google Scholar]
- 18.Dieci MV, Criscitiello C, Goubar A, Viale G, Conte P, Guarneri V, et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: a retrospective multicenter study. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2014;25(3):611–8. Epub 2014/01/10. 10.1093/annonc/mdt556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin SD, Coukos G, Holt RA, Nelson BH. Targeting the undruggable: immunotherapy meets personalized oncology in the genomic era. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2015;26(12):2367–74. Epub 2015/09/16. 10.1093/annonc/mdv382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. The Journal of clinical investigation. 2015;125(9):3413–21. Epub 2015/08/11. 10.1172/JCI80008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185(4154):862–4. 10.1126/science.185.4154.862 [DOI] [PubMed] [Google Scholar]
- 22.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. The Journal of experimental medicine. 1996;183(3):1185–92. Epub 1996/03/01. 10.1084/jem.183.3.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(14):E1754–62. Epub 2015/04/02. 10.1073/pnas.1500973112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotoula V, Lyberopoulou A, Papadopoulou K, Charalambous E, Alexopoulou Z, Gakou C, et al. Evaluation of two highly-multiplexed custom panels for massively parallel semiconductor sequencing on paraffin DNA. PLOS ONE. 2015;10(6):e0128818 Epub 2015/06/04. 10.1371/journal.pone.0128818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fountzilas G, Dafni U, Bobos M, Batistatou A, Kotoula V, Trihia H, et al. Differential response of immunohistochemically defined breast cancer subtypes to anthracycline-based adjuvant chemotherapy with or without paclitaxel. PLOS ONE. 2012;7(6):e37946 Epub 2012/06/09. 10.1371/journal.pone.0037946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31(31):3997–4013. 10.1200/JCO.2013.50.9984 . [DOI] [PubMed] [Google Scholar]
- 27.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(2):157–67. Epub 2010/11/16. 10.1038/modpathol.2010.200 . [DOI] [PubMed] [Google Scholar]
- 28.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Human mutation. 2011;32(4):358–68. Epub 2011/03/18. 10.1002/humu.21445 . [DOI] [PubMed] [Google Scholar]
- 29.Martelotto LG, Ng CK, De Filippo MR, Zhang Y, Piscuoglio S, Lim RS, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome biology. 2014;15(10):484 Epub 2014/10/29. 10.1186/s13059-014-0484-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast cancer research and treatment. 2006;100(2):229–35. 10.1007/s10549-006-9242-8 . [DOI] [PubMed] [Google Scholar]
- 31.McCullough AE, Dell'orto P, Reinholz MM, Gelber RD, Dueck AC, Russo L, et al. Central pathology laboratory review of HER2 and ER in early breast cancer: an ALTTO trial [BIG 2-06/NCCTG N063D (Alliance)] ring study. Breast cancer research and treatment. 2014;143(3):485–92. Epub 2014/01/08. 10.1007/s10549-013-2827-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kriegsmann M, Endris V, Wolf T, Pfarr N, Stenzinger A, Loibl S, et al. Mutational profiles in triple-negative breast cancer defined by ultradeep multigene sequencing show high rates of PI3K pathway alterations and clinically relevant entity subgroup specific differences. Oncotarget. 2014;5(20):9952–65. Epub 2014/10/10. 10.18632/oncotarget.2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nature genetics. 2013;45(10):1127–33. Epub 2013/09/28. 10.1038/ng.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee S, Wei Z, Tan F, Peck KN, Shih N, Feldman M, et al. Distinct microbiological signatures associated with triple negative breast cancer. Scientific reports. 2015;5:15162 Epub 2015/10/16. 10.1038/srep15162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumai T, Ishibashi K, Oikawa K, Matsuda Y, Aoki N, Kimura S, et al. Induction of tumor-reactive T helper responses by a posttranslational modified epitope from tumor protein p53. Cancer immunology, immunotherapy: CII. 2014;63(5):469–78. Epub 2014/03/19. 10.1007/s00262-014-1533-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Met O, Balslev E, Flyger H, Svane IM. High immunogenic potential of p53 mRNA-transfected dendritic cells in patients with primary breast cancer. Breast cancer research and treatment. 2011;125(2):395–406. Epub 2010/03/26. 10.1007/s10549-010-0844-9 . [DOI] [PubMed] [Google Scholar]
- 37.Strub C, Alies C, Lougarre A, Ladurantie C, Czaplicki J, Fournier D. Mutation of exposed hydrophobic amino acids to arginine to increase protein stability. BMC biochemistry. 2004;5:9 Epub 2004/07/15. 10.1186/1471-2091-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. Epub 2015/04/04. 10.1126/science.aaa4971 . [DOI] [PubMed] [Google Scholar]
- 39.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–70. Epub 2011/03/26. 10.1126/science.1203486 . [DOI] [PubMed] [Google Scholar]
- 40.Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015. Epub 2015/10/31. 10.1158/1078-0432.CCR-15-1125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheang MC, Martin M, Nielsen TO, Prat A, Voduc D, Rodriguez-Lescure A, et al. Defining breast cancer intrinsic subtypes by quantitative receptor expression. The oncologist. 2015;20(5):474–82. Epub 2015/04/25. 10.1634/theoncologist.2014-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bao L, Cardiff RD, Steinbach P, Messer KS, Ellies LG. Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast cancer research: BCR. 2015;17(1):137 Epub 2015/10/16. 10.1186/s13058-015-0615-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng CK, Martelotto LG, Gauthier A, Wen HC, Piscuoglio S, Lim RS, et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome biology. 2015;16:107 Epub 2015/05/23. 10.1186/s13059-015-0657-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng CK, Schultheis AM, Bidard FC, Weigelt B, Reis-Filho JS. Breast cancer genomics from microarrays to massively parallel sequencing: paradigms and new insights. Journal of the National Cancer Institute. 2015;107(5). Epub 2015/02/26. 10.1093/jnci/djv015 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure A, REMARK diagram for the study cohorts; Figure B, Coding mutations and SNPs in the study cohorts, as assessed with the two panels; Figure C, TNBC genotypes in 82 cases with multiple tumor samples; Figure D, Probability of DFS for patients in each TNBC cohort according to TILs density.
(DOC)
(PDF)
(PDF)
(PDF)
(PDF)
(XLS)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.