Abstract
Osteosarcoma (OS) is the most common primary malignant tumor of bone, showing complex chromosomal rearrangements but with few known consistent changes. Deeper biological understanding is crucial to find new therapies to improve patient survival. We have sequenced the whole exome of two primary tumors (before and after chemotherapy), one metastatic tumor and a matched normal sample from two OS patients, to identify mutations involved in cancer biology. The metastatic samples were also RNA sequenced. By RNA sequencing we identified dysregulated expression levels of drug resistance- and apoptosis-related genes. Two fusion transcripts were identified in one patient (OS111); the first resulted in p53 inactivation by fusing the first exon of TP53 to the fifth exon of FAM45A. The second fusion joined the two first exons of FGFR1 to the second exon of ZNF343. Furthermore, FGFR1 was amplified and highly expressed, representing a potential treatment target in this patient. Whole exome sequencing revealed large intertumor heterogeneity, with surprisingly few shared mutations. Careful evaluation and validation of the data sets revealed a number of artefacts, but one recurrent mutation was validated, a nonsense mutation in CHM (patient OS106), which also was the mutation with the highest expression frequency (53%). The second patient (OS111) had wild-type CHM, but a downregulated expression level. In a panel of 71 clinical samples, we confirmed significant low expression of CHM compared to the controls (p = 0.003). Furthermore, by analyzing public datasets, we identified a significant association between low expression and poor survival in two other cancer types. Together, these results suggest CHM as a candidate tumor suppressor gene that warrants further investigation.
Background
Osteosarcoma (OS) is the most common primary malignant tumor of bone, with an overall incidence rate of 3.3 cases/million [1]. OS is highly aggressive [2, 3] and predominately affects children and adolescents [4]. Modern treatment protocols combine surgery, chemotherapy and sometimes radiotherapy, but the 5-year survival rate remains about 60–70% [5]. Recurrent or metastatic tumors are often multidrug resistant [6, 7], and the 20–25% of patients with metastatic disease at time of diagnosis have a 5-year survival rate at roughly 30% [8]. OS show complex chromosomal rearrangements with multiple gains and losses [9, 10], but few consistent changes are known. The clinical progress has been limited by a poor understanding of the massive, chaotic genetic events observed in this tumor type, and is further complicated by low incidence, limited material due to pre-surgery chemotherapy and routine decalcification of formalin-fixed paraffin-embedded tissue blocks.
By international collaboration, over 2,200 OS patients have been recruited to the EURAMOS-1 trial (European and American Osteosarcoma Studies) [11], the largest OS trial to date. The aim was to evaluate whether giving interferon to good responders or salvage chemotherapy (ifosfamide and etoposide) to the poor responders would improve the outcome compared to the standard treatment. Unfortunately, the event-free survival did not improve and severe side-effects increased [11]. Thus, deeper biological understanding seems crucial to develop new therapies to improve OS treatment.
The rapid advancement of high throughput sequencing allows comprehensive characterization of genomic changes, and by comparing spatial and temporal tumor biopsies the genetic basis for tumor progression, metastasis and treatment effects can be addressed. Furthermore, by sequencing OS tumors, mutations or translocations known in other cancer types may be identified, thereby opening the window for personalized cancer treatment for OS patients. The identification of new, potential treatment targets is especially important for those where the life-expectancy is low and the standard treatment options are few.
In this study, unique triplet progression samples consisting of primary tumors (before and after chemotherapy) and metastasis, of two OS patients have been studied in detail and compared to matched normal samples. We have sequenced the exomes of all samples and the transcriptome of the metastatic samples to study how these tumors relate, and to detect alterations associated with osteosarcoma biology, treatment, drug resistance and progression.
Results
Case descriptions
Both patients were included in the EURAMOS-1 trial at Oslo University Hospital and consented to our associated research project (approval ID from the Regional Ethics Committee of Southern Norway: S-06133). None of the patients experienced local recurrences during the course of their disease.
Case #1: OS106
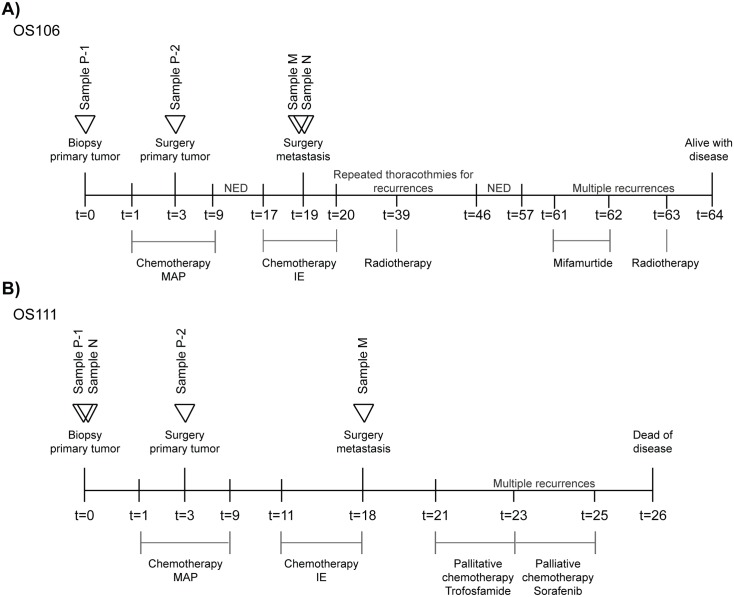
The first patient, a 16 years old male, hereafter referred to as OS106, presented a grade 4 primary OS in the proximal tibia without detectable metastases at diagnosis. The tumor was biopsied (sample P-1) before neoadjuvant treatment with chemotherapy (MAP: methotrexate, doxorubicin and cisplatin), followed by limb-salvage surgery after 2 months. The primary tumor (sample P-2) was removed with wide margins of only 2 mm towards the joint, but with a cut off of 20 mm as the shortest margin in normal soft tissues. The patient showed poor response to chemotherapy, defined in the trial as <90% necrosis [11] and was randomized to continue MAP postoperatively. Metastases in the lungs were discovered 17 months after the initial diagnosis. The patient then received a second line of chemotherapy (IE: ifosfamide and etoposide), followed by surgical removal of the lung metastases 2 months later (one of which is sample M). Surrounding healthy lung tissue, but distant from the tumor, was used as normal control (sample N). The patient progressed, and new lung metastases were detected. Stereotactic radiotherapy was given and four thoracotomies (two times each lung) were conducted to remove all macroscopic metastases. The patient had then no evidence of disease for 11 months, before metastases in the skull and costa were discovered. After treatment with the immunomodulator mifamurtide and radiotherapy, the patient is alive with disease (64 months after diagnosis). A schematic overview of the treatment timeline is presented in Fig 1A.
Fig 1. Schematic timeline of the disease and treatment courses of the two patients for (A): OS106 and (B): OS111.
Time is shown in months. P-1: primary tumor treatment naïve, P-2: primary tumor treatment experienced. N: normal, M: metastasis, MAP: methotrexate, doxorubicin and cisplatin, IE: ifosfamide and etoposide, NED: No evidence of disease.
Case #2: OS111
The second patient, 37 years old female, hereafter referred to as OS111, presented a grade 4 primary OS in the distal femur without detectable metastases at diagnosis. The primary tumor was biopsied (sample P-1) and a blood sample (sample N) was obtained. The patient was enrolled in the EURAMOS-1 trial and received pre- and postoperative chemotherapy (MAP). The response to chemotherapy was good (≥90% necrosis), but the patient had severe side effects of methotrexate. The treatment courses were thus reduced and more delayed than originally scheduled. Two months after the start of chemotherapy, the primary tumor was removed by a limb-salvage procedure. The tumor was extending to the articular surface, but wide margins were obtained with 55 mm in the resected bone and 20 mm in soft tissue (sample P-2). Soon after completion of adjuvant chemotherapy, metastases were detected in both lungs. Chemotherapy (IE) was initiated, and the lung metastases were removed by surgery after 7 months (one of which is sample M). New metastatic lesions were discovered three months after the thoracotomy, and palliative chemotherapy (Ixoten/Trofosfamide followed by Nexavar/Sorafenib) was given. Metastases continued to appear and the patient succumbed 26 months after diagnosis. A schematic overview over the treatment timeline is presented in Fig 1B.
Drug resistance is associated with transcriptional changes
The transcriptomes of both metastatic samples (OS106 M and OS111 M) were sequenced (RNA-seq), as well as four normal, primary osteoblast cultures that were used as controls. In total, OS106 had 1,934 downregulated transcripts (with a log2 fold reduction of <-2) and 8,089 upregulated transcripts (log2 fold increase of >2) compared to the osteoblast average. OS111 had a total of 2,140 downregulated transcripts and 9,350 upregulated transcripts. The transcriptome data and corresponding expression levels (as normalized read counts for each sample) are presented in S1 Table.
Both patients were considered to have acquired multidrug resistant disease. To identify possible causes of drug resistance, we investigated the expression of genes commonly associated with resistance to MAP. In particular, resistance is often caused by increased drug efflux caused by upregulation of members in the ATP-binding cassette (ABC) family of membrane transporters, downregulation of the genes RFC1 and DHFR, mutations in TOP2β and upregulation of the GSTP1 isoenzyme [12–25], but inhibition of apoptosis has also been reported to cause drug resistance [26–29].
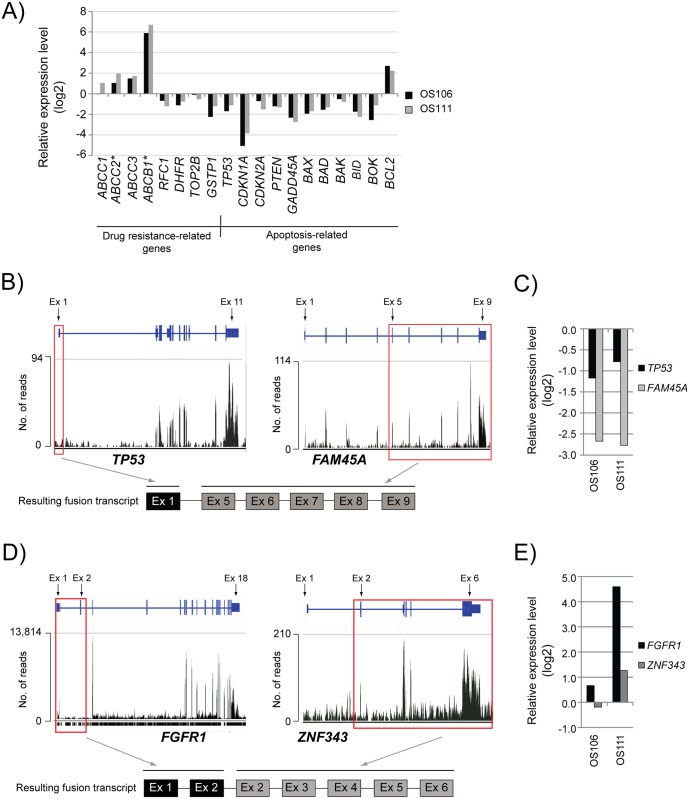
The relative expression levels compared to the average of the osteoblasts are shown in Fig 2A. In OS111, the ABC transporters ABCC1, ABCC2, ABCC3 and ABCB1 (P-glycoprotein or multidrug resistance protein) were upregulated. The genes, except ABCC1, were also upregulated in OS106. However, upon inspection of the normalized read counts, it was apparent that the expression of both ABCC2 and ABCB1 was low in the controls and therefore the relative expression may be overestimated (indicated with an asterisk after the gene name in Fig 2A). The genes RFC1 and DHFR were downregulated in both samples. There was no evidence of mutations in, or overexpression of, TOP2β in either of the samples, and GSTP1 was rather found to be downregulated. We also identified dysregulation of several genes involved in apoptosis. TP53 was downregulated in both patients, as were the tumor suppressors CDKN1A, CDKN2A, PTEN and GADD45A, which can be activated by both p53-dependent and -independent mechanisms. The pro-apoptotic genes BAX, BAD, BAK, BID and BOK were downregulated and the anti-apoptotic gene BCL2 was upregulated (Fig 2A). Overall, gene expression profiles indicate an upregulation of drug resistance mechanisms and inhibition of apoptosis for both patients when compared to the normal controls.
Fig 2. Transcriptome analysis of OS106 and OS111.
(A) The relative expression level of known drug resistance-related genes and apoptosis-related genes. The values are shown as log2 of the expression level ratio, compared to the average of the four normal osteoblasts. Asterisk (*) indicates low number of reads in the osteoblasts. (B) The expression of all exons in TP53 and FAM45A, respectively, shown as total number of reads in patient OS111. Red squares indicate the exons that are fused to form the resulting fusion transcript. ex: exon; no.: number (C) The relative expression level of TP53 and FAM45A. The values are shown as log2 of the expression level ratio, compared to the average of the osteoblasts. The expression of all transcripts are included, both wild-type and aberrant. (D) The expression of all exons in FGFR1 and ZNF343, respectively, shown as total number of reads in patient OS111. Red squares indicate the exons that are fused to form the resulting fusion transcript. (E) The relative expression level of FGFR1 and ZNF343. The values are shown as log2 of the expression level ratio, compared to the average of the osteoblasts. The expression of all transcripts are included, both wild-type and aberrant.
In patient OS111, RNA-seq revealed a fusion transcript where the first exon of TP53 was fused to the fifth exon of FAM45A. The expression profile of all exons in both genes, and an illustration of the corresponding fusion transcript are shown in Fig 2B (see S2 Table for fusion transcript breakpoint sequences). We did not find any evidence supporting the reciprocal fusion transcript. The expression levels (comparing the normalized read counts) of both TP53 and FAM45A were downregulated in both patients compared to the average of the osteoblasts (Fig 2C). However, the fusion transcript was not highly expressed, as we identified a total of 11 paired-end reads spanning the breakpoint, thus additional mechanisms are likely to contribute to the downregulation of TP53 in this patient. For patient OS106, we did not detect any aberrations of TP53 that could explain the reduced expression level, indicating that the downregulation must be caused by mechanisms not detected using WES or RNA-seq.
FGFR1 is upregulated in OS111
A second fusion transcript was detected in OS111 (no fusion transcripts were detected in OS106), fusing FGFR1 to ZNF343. FGFR1 deregulation commonly occurs through gene amplification, point mutation or chromosomal translocations causing a constitutive receptor activation and ligand-independent signaling (reviewed in [30]). However, this particular fusion transcript contained only the first two exons of FGFR1 fused to the second exon of ZNF343 (Fig 2D). The first two exons of FGFR1 encodes only the signal peptide [31], thus this particular fusion transcript does not result in an active FGFR1. We could not identify the reciprocal fusion transcript in our data, but the exons excluded in the fusion transcript (exons 3–18) have a substantial higher expression level than the two included in the fusion (exons 1–2) (Fig 2D). This could suggest high expression from a reciprocal fusion, but producing a transcript where the first exon of ZNF343 is spliced out. On the other hand, many splice variants have been shown for FGFRs, including some lacking the signal peptide sequences [32], thus such transcripts could also originate from a wild-type (wt) allele. In OS111, the expression level of FGFR1 was 24 times higher than the expression level in the osteoblasts (Fig 2E) (comparing normalized read counts).
Copy number analyses of all tumors were performed based on the whole exome data. For OS111, we found a large amplification on chromosome 8 including FGFR1 in the P-1 and P-2 (in S1 Fig, copy number variation for all samples along chromosome 8:35,000,000–43,000,000 is shown). For sample M, of which the transcriptome data was obtained, there was no evidence of amplification, thus other mechanisms besides amplification must be causing the overexpression of FGFR1 in the metastatic sample. Patient OS106 showed no amplification corresponding to the location of FGFR1 (S1 Fig) and had an expression level comparable to the expression of the osteoblasts (1.6 fold) (Fig 2E).
The expression level of the fusion partner ZNF343 was also upregulated in OS111 (2.4 fold of that in the osteoblasts), whereas the level in OS106 was at a similar level as the controls (0.8 fold) (Fig 2E). It is possible that the fusion between FGFR1 and ZNF343 in OS111 causes upregulation of ZNF343. This is further supported by the increase in expression of the exons included in the fusion transcript (exons 2–6), compared to exon 1 in ZNF343 (Fig 2D).
Whole exome sequencing analysis
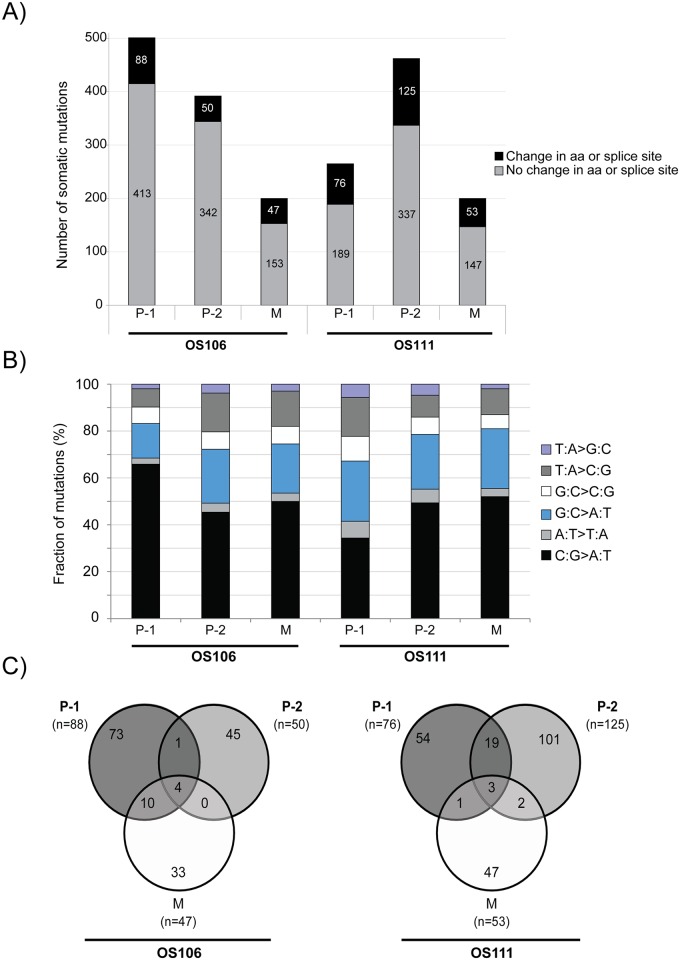
The whole exomes of three progression samples (P-1, P-2 and M) and their corresponding normal sample (N) from both patients (OS106 and OS111) were sequenced (WES). Each tumor sample was compared to their respective normal sample using the validated Norwegian Cancer Genomics Consortium (NCGC, cancergenomics.no/en/) variant-calling pipeline [33], identifying between 200 and 500 somatic mutations per tumor (Fig 3A). Mutations in the introns were most frequent, followed by missense mutations, silent mutations and mutations in the untranslated regions (UTRs), respectively (see S2 Fig for an overview of the frequencies of different mutation types).
Fig 3. WES analysis of OS106 and OS111.
(A) The total number of somatic mutations identified in each sample for both patients. Black represents nonsynonymous mutations in the coding sequence and/or mutations in the splice sites, whereas grey represents all other mutations. The respective numbers of mutations are indicated in the bars. aa: amino acids, P-1: primary tumor treatment naïve, P-2: primary tumor after treatment, M: metastasis. (B) The mutation spectrum showing the proportions of transitions and transversions in percent (adding up to 100%). (C) Venn diagram showing the distribution of mutations (only nonsynonymous mutations in the coding sequence and mutations in the splice sites) between the matched tumor samples in each patient. The total number of mutations in each tumor is shown in parenthesis.
Analysis of the mutation spectra showed that C:G>A:T transversions were dominant for all samples, ranging from 34% to 66% frequency (Fig 3B). The second highest mutation rate was G:C>A:T transition, ranging from 15% to 26% frequency (Fig 3B). Both predominant mutation types have been associated with cisplatin DNA damage [34, 35]. In both patients the mutation spectrum of the treated sample (P-2) was more similar to the metastatic sample (M), than to the treatment naïve sample (P-1), further indicating that many of the mutations were induced by chemotherapy.
WES reveals high intertumor heterogeneity
An overview of all somatic mutations and their corresponding coverage by WES and RNA-seq is shown in S3 Table. For further analysis, only the nonsynonymous mutations in the coding regions and the mutations in the splice sites were included. We focused on the recurrent mutations, present in at least two tumors in at least one of the patients. An overview over the number of mutations in each sample and the number of shared mutations between the tumors of each patient is shown in Fig 3C. However, some of the recurrent mutations were initially called from read sequences that mapped to more than one place in the genome with either identical or very high sequence similarity. The mutations called from such reads were discarded, even though they were reported in the COSMIC and/or the dbSNP databases. These were mutations in the genes ARMC4 (p.D425Y), MUC4 (p.A4217E, p.L4230P, p.H4205Q), TIMM23B (splice site, chr10: 51374435 A>G), DDX11 (p.A607P, p.P368S), CDC27 (p.F26S and p.L27P) and FANCD2 (splice site, chr3: 10106408 C>T), demonstrating that care must be taken when evaluating mutation data. Two recurrent mutations (in CHM and ZNF197) were identified from unique read sequences. Both mutations were only present in patient OS106, and were validated by RNA-seq in the metastatic sample (expression frequency of 53% and 52% for the mutations in CHM and ZNF197, respectively). The mutation in CHM was further validated by targeted resequencing, but we were unable to design specific primers for the mutation in ZNF197. However, since the mutation in ZNF197 results in one basic amino acid residue replacing another (p.K803R) we anticipate limited functional consequences. An overview of the coverage by WES, RNA-seq and targeted resequencing is shown in S4 Table.
CHM is a candidate tumor suppressor gene
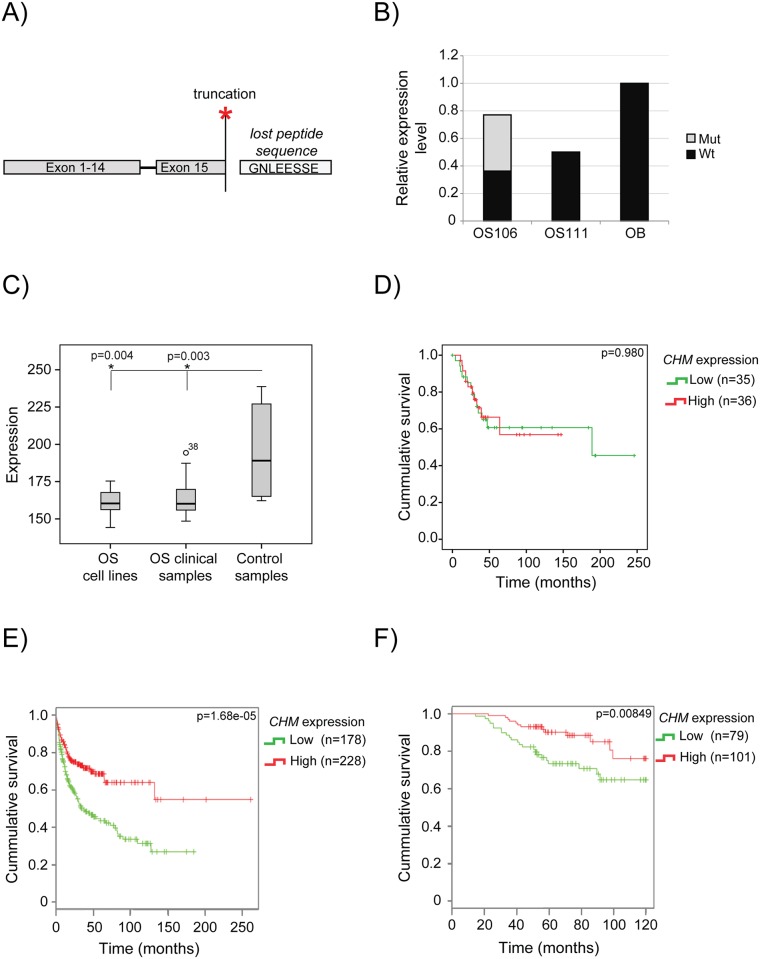
The identified nonsense mutation in CHM (p.G646*), causing a truncation in the last exon (Fig 4A), was present in the treatment naïve primary tumor and in the metastasis of patient OS106. The expression level was similar to the average level in the four, normal osteoblasts, but 53% of the reads carried the mutation (Fig 4B). No mutation was detected in OS111, but the expression level of wt CHM was half that observed in osteoblasts (Fig 4B). We further compared the expression level of CHM in a panel of OS cell lines (n = 19) and OS clinical samples (n = 71) to control samples (four bone samples and two osteoblastic cultures (n = 6)). Gene expression profiling of these samples has previously been performed as a part of the EuroBoNeT network of excellence [36–38] (summary of the clinical data can be found in S5 Table). The expression level of CHM was significantly downregulated both in the cell lines and clinical samples compared to the control samples (Fig 4C). We also investigated whether the expression level of CHM in the clinical samples was correlated with survival. Of the 71 clinical samples, only one had an expression level above the median of the control samples. Thus, the survival analysis were performed using median split (Fig 4D), where the samples are stratified according to whether their respective expression level was above or below the median expression level of the clinical samples, and not the median level of the controls which was used in the expression level analysis (Fig 4C). No significant association between expression level and survival was found (Fig 4D). Using DRUGSURV, a publicly available database where expression profiling and clinical information from several studies are available [39], we investigated the expression of CHM in other cancer types. The top two hits were from studies on diffuse large B cell lymphoma [40] (GEO accession GSE10846) and breast cancer [41] (GEO accession GSE24450), and both showed that poor survival was significantly associated with low expression of CHM (Fig 4E and 4D, respectively). Together, these data suggest CHM as a candidate tumor suppressor gene in cancer, but the potential mechanism has to be further elucidated.
Fig 4. CHM analysis.
(A) The identified nonsense mutation p.G646* in CHM results in the loss of the last eight amino acids. (B) The relative expression level of CHM in OS106 and OS111 compared to the average of the four osteoblasts. The nonsense mutation was only identified in OS106 at a frequency of 53% revealed by the RNA-seq (illustrated with grey bars). Mut: nonsense mutation, wt: wild-type. C) Expression level of CHM in osteosarcoma cell lines (n = 19), osteosarcoma clinical samples (n = 71) and normal control samples (n = 6). The difference was statistical significant with p-values of 0.004 and 0.003, for cell lines and clinical samples, respectively (Mann-Whitney U test) (D) Survival analysis of CHM expression in 71 clinical OS samples. The samples are sorted according to low (below median) or high (above the median) expression level. The difference was not significant with a p-value of 0.980 (Log Rank/Mantel-Cox test). (E) Survival plot obtained from DRUGSURV showing expression of CHM in patients with large diffuse B-cell lymphoma. Low expression was significantly associated with poor survival, p = 1.68e-05 (Chi-square 18.5 on 1 degree of freedom). (F) Survival plot obtained from DRUGSURV showing expression of CHM in patients with breast cancer. Low expression was significantly associated with poor survival, p = 8.49e-03 (Chi-square 6.9 on 1 degree of freedom).
Discussion
Osteosarcomas are rare tumors and it is difficult to obtain multiple sets of progression samples. By comparing matched progression tumors, novel insights into cancer biology may be revealed. Comparing the matched tumor progression genomes showed a high degree of intertumor heterogeneity. Other studies of matched tumor samples have shown a substantially higher level of concordance of shared mutations than what we found in this study, both by WGS [42, 43], WES [44] and targeted resequencing of smaller gene panels [45]. However, studies performed on multiple spatial biopsies in renal-cell carcinomas have identified large intratumor heterogeneity, with less than 40% shared mutations [46]. OS have exceptionally unstable genomes [47], which is probably causing the large heterogeneity seen in this study. In addition, two months of chemotherapy are separating samples P-1 and P-2, and further chemotherapy for the metastatic lesion. Not only have the tumors had time to acquire additional mutations, but also been under a strong selection pressure, which could increase the intertumor heterogeneity through e.g. branched evolution [48]. Not surprisingly, our data indicate that chemotherapy impacts the mutation burden in osteosarcoma. This also raises the question whether studies on treated tumors may include large amounts of mutational noise that is unrelated to the mechanisms of tumor development.
Both patients were considered to have acquired multidrug resistant disease. We found dysregulated expression levels of many of the genes reported to be involved in resistance to MAP [12–25]. Several of the ABC transporters were found to be upregulated, including ABCB1. Upregulation of ABCB1, also known as P-glycoprotein or multidrug resistance protein, is well known to confer drug resistance in OS [21, 23, 49–52]. ABCB1 was highly upregulated in both patients, albeit the expression levels in the controls were low and consequently the relative expression level may be overestimated. In addition, we found several apoptosis-related genes to be dysregulated, also known to confer drug resistance [26–29]. Interestingly, we identified a fusion transcript joining the first exon of TP53 to the fifth exon of FAM45A in OS111, most likely causing an inactivation of p53. Inactivation of TP53 in osteosarcoma by rearrangements (with a hotspot in intron 1) has previously been reported by us and others [53–55], and may well be the cause of the fusion transcript in this patient, although intronic translocation sites cannot be confirmed by WES. However, also frequent trans-splicing of RNAs have been reported in OS cell lines [53]. The translocation in TP53 appears to be an osteosarcoma-specific way to inactivate p53, and seems to be relatively frequent [53–55].
The second fusion transcript detected in OS111, joined exons 1–2 of FGFR1 to exons 2–6 of ZNF343. The two first exons of FGFR1 encode the signal peptide [31], which directs the location of FGFR1 to the membrane. Thus, if translated, it is possible that the fusion protein is transported to the cell membrane. However, to our knowledge, there are no publications describing the function of ZNF343, thus making the potential function for such a fusion protein unclear. Interestingly, it has been shown that FGFR1 without the signal peptide accumulates in the cytosol and the nucleus [56], and that nuclear FGFR1 has been associated with invasive cancer cells [57–59]. The expression levels of the FGFR1 exons excluded in the FGFR1-ZNF343 fusion transcript (exons 3–18) were substantially higher than the ones included (exons 1–2). However, from our data we cannot conclude whether it contains the reciprocal fused allele (ZNF343-FGFR1) and produces a 5’ truncated FGFR1 transcript, or a splice variant of a wt allele. Due to limited material, we were not able to investigate the subcellular localization of FGFR1 in the tumor samples.
FGFR1 is a high-priority therapeutic target, and there are both specific tyrosine kinase inhibitors and monoclonal antibodies targeting FGFRs [60]. FGFR1 has been shown to be amplified in ~19% of OS tumors with poor response to chemotherapy [61]. Although OS111 had good response to the chemotherapy, drugs targeting FGFRs could have been of value for second-line treatment. Due to limited material, we were unable to establish cell lines or patient-derived xenografts, and no functional studies were possible. However, cancer cell lines with FGFR1 amplification have been shown to be sensitive to an FGFR inhibitor (NVP-BGJ398) [62]. Interestingly, this was especially true for the osteosarcoma, breast and lung cancer cell lines [62]. Furthermore, pharmacological blockade of FGFR1 has also been shown to decrease lung metastases in an osteosarcoma animal model [63]. Thus, there are indications of possible treatment opportunities of OS patients with FGFR1 amplification and/or overexpression.
Although still not strongly supported in OS, we propose CHM (choroideremia (Rab escort protein 1)) to be a candidate tumor suppressor gene. CHM encodes the protein Rab proteins geranylgeranyl transferase component A 1, hereafter referred to as REP-1. REP-1 recognizes newly synthesized Rab proteins and presents them to geranylgeranyl transferases for prenylation. Functioning as a chaperone, REP-1 keeps the hydrophobic geranylgeranylated Rab soluble and delivers it to the appropriate membrane [64], thought to rely on membrane receptors that recognize the complex between REP-1 and specific Rabs. Thus, REP-1 is important for the functionality of Rabs [65], which regulate intracellular vesicular transport [66]. Mutations in CHM are known to cause choroideremia; an X-linked form of progressive blindness caused by degeneration the retinal pigment epithelium and the two underlying cell layers. A second isoform called REP-2, encoded by the gene CHML, is believed to compensate for the loss of REP-1 in most human tissues, except the eye [67], which could explain why choroideremia patients seems otherwise unaffected. However, it has been shown that some Rab proteins are most efficiently prenylated by REP-1, e.g. Rab3A and Rab3D [67]. Furthermore, increased pH levels in lysosomes, reduced rates of proteolytic degradation and altered secretion of cytokines have been shown in monocytes and fibroblasts isolated from choroideremia patients compared to healthy controls [68]. This indicates that loss-of-function mutations in CHM is not necessarily fully compensated by REP-2, and could potentially affect intracellular transportation in cell types besides in the eye.
The identified nonsense mutation causes a truncation of the protein, losing the last 8 amino acids (of 654). To investigate the functional effects of the truncation was beyond the scope of this study. Three out of the eight amino acids are negatively charged (sequence: GNLEESSE), indicating a possible function for the C-terminal protein sequence. However, we found the expression level to be significantly downregulated in both OS cell lines and clinical samples compared to the control samples, which supports the assumption of an inhibitory role for CHM in osteosarcoma biology. We did not find a significant association between expression of CHM and survival in the osteosarcoma samples, which could be due to the overall low expression level in this sample cohort, but by using DRUGSURV, we found that low expression level of CHM in two other cancer types, large diffuse B cell lymphoma and breast cancer, were significantly associated with poor survival. It is possible that the level of CHM in OS is consistently low, and thus affects survival, but that almost all samples in our cohort had lower levels than the best stratum in the other comparisons. We propose, to our knowledge, for the first time a role for CHM in cancer biology, although more thorough studies are necessary to investigate the potential tumor suppressive function of REP-1.
Materials and Methods
Material
The patients included in the study consented to our research project in written. Our research project was approved by the Regional Ethics Committee of Southern Norway, with approval ID number: S-06133.
Tumor tissue was either surgical biopsy (P-1) or surgical specimens (P-2 and M). All were immediately snap-frozen in liquid nitrogen. Corresponding normal control samples (N) were either a blood sample (OS111) or healthy lung tissue (OS106).
Commercially available primary osteoblast cultures isolated from human femur and tibia of different donors (n = 4) (Cambrex BioScience) were used as normal control for the RNA-seq. The osteoblast cells were maintained in medium provided by the manufacturer, split when reaching 80% confluency and harvested when enough cells for RNA isolation were obtained.
Nucleic acid isolation
DNA was isolated using Promega Wizard Genomic DNA purification isolation kit according to the manufacturer’s instructions and stored at 4°C. RNA was isolated using Qiagen miRNeasy isolation kit according to the manufacturer’s instructions and stored at -80°C. The integrity and quality of the DNA and RNA were assessed using an Agilent Technologies TapeStation and 2100 Bioanalyzer, respectively.
Whole exome sequencing and analysis
The exome sequencing libraries of all tumor samples and matching normal was performed using the Agilent SureSelect All Exome v5 platform. One microgram of total genomic DNA was processed, as described in [69]. The libraries were sequenced paired-end (2 x 100 bp) on a HiSeq2500 (Illumina) using TruSeq SBS v3 chemistry. Real-time analysis and base calling were conducted by Illumina’s software packages HSC2.0.2/RTA1.17.21.3. Raw reads were processed using the Illumina CASAVA (v. 1.8.2) to demultiplex data and filter out the low-quality reads.
Initially reads were analyzed and quality checked using FastQC [70]. Alignment of reads to reference genome was performed by Novoalign [70]. Multiple mapping reads, PCR duplicates and not proper pair reads were removed using in-house scripts. Using GATK package, we performed local realignment around indels and base-quality recalibration [71]. Somatic single nucleotide variants were identified using MuTect [72]. Small InDel and raw copy number regions from exome were detected using VarScan 2 [73]. Copy number segments from raw data were identified using DNA copy package [70].
Transcriptome sequencing and analysis
The RNA sequencing libraries were made using the Illumina TruSeq Stranded Total RNA with Ribo-Zero Gold kit, starting from one microgram high quality total RNA and processed according to the manufacturer’s instructions. The resulting libraries were sequenced paired end (2 x 100 bp) on a HiSeq2500.
Differential expression analysis from RNA-seq data performed by using DESeq package [74]. Fusion transcript analysis was performed as described in [53].
Data deposits
Because of Norwegian legal regulations, the ethical approval for this study and the consent signed by the patient, we are not able to deposit our sequencing dataset in a public repository. We will provide access to the data if requested by cancer researchers. For access, please contact the corresponding author Dr. Meza-Zepeda (leonardo.meza-zepeda@rr-research.no).
The gene expression data used in the CHM analysis can be retrieved from GEO. The accession numbers are GSE36004 (cell lines, osteoblasts and bone samples) and GSE30699 (clinical samples).
Targeted resequencing
Validation of the mutations in ZNF197 and CHM was performed by targeted resequencing. Primers were designed to amplify the mutations of interest with an amplicon length of ~140 bp. In addition, a tail complementary to the Barcoded Illumina Primers (Universal forward and indexed reverse primers) was added to the primers. The primer sequences are found in S6 Table.
The sequences were amplified using nested PCR. The procedure and reagents were identical to the set-up described in RainDance ThunderBolts Cancer Panel Assay Manual, with the exception of the custom-designed primers used in the 1st PCR round and the annealing temperatures were set according to the melting temperatures of the respective primers. Furthermore, droplets were not generated, and thus the droplet stabilizer was not used.
The sequencing of the resulting libraries was performed on MiSeq (Illumina) using TruSeq SBS v3 chemistry. Analysis was performed using the PCR amplicon workflow using BWA for alignment and GATK for variant calling using the MiSeq Reporter software (Illumina).
Supporting Information
The copy number along chromosome 8:35,000,000–43,000,000 of each tumor sample, with emphasis on FGFR1.
(TIF)
An overview over the different mutations types for all tumor samples.
(TIF)
The transcriptome data and corresponding gene expression levels (shown as normalized read counts) for each sample (tumors and controls).
(XLSX)
An overview over the identified fusion transcripts in patient OS111.
(XLSX)
An overview over all identified somatic mutations and corresponding expression level in all samples.
(XLSX)
A detailed overview over the two recurrent mutations.
(XLSX)
A summary of the clinical data of the 71 patients included in the CHM analysis.
(XLSX)
An overview over the primer sequences used for the targeted resequencing.
(XLSX)
Acknowledgments
We would like to acknowledge Dr. Heidi Maria Namløs (Department of Tumor Biology, Oslo University Hospital) for critical reading of the manuscript, the personnel at the Genomics Core Facility at Oslo University Hospital (oslo.genomics.no) for technical assistance with the sequencing and Prof. Janne E. Reseland (Department of Biomaterials, Oslo University Hospital) for providing normal osteoblasts. The work was supported by the Norwegian Research Council (grant 218241) and the Norwegian Cancer Society (grant PR-2007-0163).
Data Availability
Because of Norwegian legal regulations, the ethical approval for this study and the consent signed by the patient, we are not able to deposit our sequencing dataset in a public repository. We will, however, provide access to the data if requested by cancer researchers. We would like to emphasize that all relevant data (i.e. all called somatic variants and gene expression levels) are available in the Supplementary Information, but we are not allowed to openly publish the complete exome of patients. The gene expression data used for the CHM analysis can be found at GEO. The GEO accession numbers GSE36004 (cell lines, osteoblasts and bone samples) and GSE30699 (clinical samples) are provided under “Data deposits” in Materials and Methods. Otherwise, all relevant data can be found within the paper and its Supporting Information Files. For data related queries and requests, please contact the corresponding author Dr. Meza-Zepeda (leonardo.meza-zepeda@rr-research.no).
Funding Statement
TB, ØSB and OM were financed by Oslo University and Oslo University Hospital. SL, JS, CSRC and LAMZ were financed by Oslo University Hospital. The work was further supported by the Norwegian Research Council (grant 218241) and the Norwegian Cancer Society (grant PR-2007-0163). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Berner K, Johannesen TB, Berner A, Haugland HK, Bjerkehagen B, Bohler PJ, et al. Time-trends on incidence and survival in a nationwide and unselected cohort of patients with skeletal osteosarcoma. Acta Oncol. 2015;54(1):25–33. Epub 2014/06/25. 10.3109/0284186X.2014.923934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher CDM, Bridge J.A., Hogendoorn P., Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. Fourth Edition WHO, editor.: Lyon: IARC Press; 2013. [Google Scholar]
- 3.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254 Epub 2012/06/12. 10.1155/2012/627254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorfman HD, Czerniak B. Bone cancers. Cancer. 1995;75(1 Suppl):203–10. . [DOI] [PubMed] [Google Scholar]
- 5.Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–90. Epub 2002/02/01. 10.1200/JCO.20.3.776 . [DOI] [PubMed] [Google Scholar]
- 6.Berner K, Hall KS, Monge OR, Weedon-Fekjaer H, Zaikova O, Bruland OS. Prognostic factors and treatment results of high-grade osteosarcoma in norway: a scope beyond the "classical" patient. Sarcoma. 2015;2015:516843 10.1155/2015/516843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz CL, Gorlick R, Teot L, Krailo M, Chen Z, Goorin A, et al. Multiple drug resistance in osteogenic sarcoma: INT0133 from the Children's Oncology Group. J Clin Oncol. 2007;25(15):2057–62. Epub 2007/05/22. 10.1200/JCO.2006.07.7776 . [DOI] [PubMed] [Google Scholar]
- 8.Kager L, Zoubek A, Potschger U, Kastner U, Flege S, Kempf-Bielack B, et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J Clin Oncol. 2003;21(10):2011–8. Epub 2003/05/14. 10.1200/JCO.2003.08.132 . [DOI] [PubMed] [Google Scholar]
- 9.Mackall CL, Meltzer PS, Helman LJ. Focus on sarcomas. Cancer Cell. 2002;2(3):175–8. 10.1016/S1535-6108(02)00132-0 . [DOI] [PubMed] [Google Scholar]
- 10.Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3(9):685–94. 10.1038/nrc1168 . [DOI] [PubMed] [Google Scholar]
- 11.Whelan JS, Bielack SS, Marina N, Smeland S, Jovic G, Hook JM, et al. EURAMOS-1, an international randomised study for osteosarcoma: results from pre-randomisation treatment. Ann Oncol. 2015;26(2):407–14. Epub 2014/11/26. 10.1093/annonc/mdu526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assaraf YG. Molecular basis of antifolate resistance. Cancer Metastasis Rev. 2007;26(1):153–81. Epub 2007/03/03. 10.1007/s10555-007-9049-z . [DOI] [PubMed] [Google Scholar]
- 13.de Anta JM, Perez-Castro AJ, Freire R, Mayol X. The DNA damage checkpoint is activated during residual tumour cell survival to methotrexate treatment as an initial step of acquired drug resistance. Anticancer Drugs. 2006;17(10):1171–7. Epub 2006/11/01. 10.1097/01.cad.0000236311.73703.d2 . [DOI] [PubMed] [Google Scholar]
- 14.Gorlick R, Goker E, Trippett T, Waltham M, Banerjee D, Bertino JR. Intrinsic and acquired resistance to methotrexate in acute leukemia. N Engl J Med. 1996;335(14):1041–8. Epub 1996/10/03. 10.1056/NEJM199610033351408 . [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Healey JH, Meyers PA, Ladanyi M, Huvos AG, Bertino JR, et al. Mechanisms of methotrexate resistance in osteosarcoma. Clin Cancer Res. 1999;5(3):621–7. Epub 1999/04/01. . [PubMed] [Google Scholar]
- 16.Chen ZS, Lee K, Walther S, Raftogianis RB, Kuwano M, Zeng H, et al. Analysis of methotrexate and folate transport by multidrug resistance protein 4 (ABCC4): MRP4 is a component of the methotrexate efflux system. Cancer Res. 2002;62(11):3144–50. Epub 2002/05/31. . [PubMed] [Google Scholar]
- 17.Kruh GD, Zeng H, Rea PA, Liu G, Chen ZS, Lee K, et al. MRP subfamily transporters and resistance to anticancer agents. J Bioenerg Biomembr. 2001;33(6):493–501. Epub 2002/01/24. . [DOI] [PubMed] [Google Scholar]
- 18.Zeng H, Chen ZS, Belinsky MG, Rea PA, Kruh GD. Transport of methotrexate (MTX) and folates by multidrug resistance protein (MRP) 3 and MRP1: effect of polyglutamylation on MTX transport. Cancer Res. 2001;61(19):7225–32. Epub 2001/10/05. . [PubMed] [Google Scholar]
- 19.Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Rev Anticancer Ther. 2006;6(7):1075–85. Epub 2006/07/13. 10.1586/14737140.6.7.1075 . [DOI] [PubMed] [Google Scholar]
- 20.Bramwell VH. Osteosarcomas and other cancers of bone. Curr Opin Oncol. 2000;12(4):330–6. Epub 2000/07/11. 10.1097/00001622-200007000-00009 . [DOI] [PubMed] [Google Scholar]
- 21.Hornicek FJ, Gebhardt MC, Wolfe MW, Kharrazi FD, Takeshita H, Parekh SG, et al. P-glycoprotein levels predict poor outcome in patients with osteosarcoma. Clin Orthop Relat Res. 2000;(373):11–7. Epub 2000/05/16. 10.1097/00003086-200004000-00003 . [DOI] [PubMed] [Google Scholar]
- 22.Serra M, Scotlandi K, Reverter-Branchat G, Ferrari S, Manara MC, Benini S, et al. Value of P-glycoprotein and clinicopathologic factors as the basis for new treatment strategies in high-grade osteosarcoma of the extremities. J Clin Oncol. 2003;21(3):536–42. Epub 2003/02/01. 10.1200/JCO.2003.03.144 . [DOI] [PubMed] [Google Scholar]
- 23.Chan HS, Grogan TM, Haddad G, DeBoer G, Ling V. P-glycoprotein expression: critical determinant in the response to osteosarcoma chemotherapy. J Natl Cancer Inst. 1997;89(22):1706–15. Epub 1997/12/09. 10.1093/jnci/89.22.1706 . [DOI] [PubMed] [Google Scholar]
- 24.Pasello M, Michelacci F, Scionti I, Hattinger CM, Zuntini M, Caccuri AM, et al. Overcoming glutathione S-transferase P1-related cisplatin resistance in osteosarcoma. Cancer Res. 2008;68(16):6661–8. Epub 2008/08/15. 10.1158/0008-5472.CAN-07-5840 . [DOI] [PubMed] [Google Scholar]
- 25.Pasello M, Manara MC, Michelacci F, Fanelli M, Hattinger CM, Nicoletti G, et al. Targeting glutathione-S transferase enzymes in musculoskeletal sarcomas: a promising therapeutic strategy. Analytical cellular pathology (Amsterdam). 2011;34(3):131–45. Epub 2011/06/16. 10.3233/ACP-2011-012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osmak M, Brozovic A, Ambriovic-Ristov A, Hadzija M, Pivcevic B, Smital T. Inhibition of apoptosis is the cause of resistance to doxorubicin in human breast adenocarcinoma cells. Neoplasma. 1998;45(4):223–30. Epub 1999/01/16. . [PubMed] [Google Scholar]
- 27.Bauer JA, Kumar B, Cordell KG, Prince ME, Tran HH, Wolf GT, et al. Targeting apoptosis to overcome cisplatin resistance: a translational study in head and neck cancer. Int J Radiat Oncol Biol Phys. 2007;69(2 Suppl):S106–8. Epub 2007/10/18. 10.1016/j.ijrobp.2007.05.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.PosthumaDeBoer J, van Royen BJ, Helder MN. Mechanisms of therapy resistance in osteosarcoma: a review. Oncology Discovery. 2013;1(1):8 10.7243/2052-6199-1-8 [DOI] [Google Scholar]
- 29.Yang C, Yang S, Wood KB, Hornicek FJ, Schwab JH, Fondren G, et al. Multidrug resistant osteosarcoma cell lines exhibit deficiency of GADD45alpha expression. Apoptosis: an international journal on programmed cell death. 2009;14(1):124–33. Epub 2008/12/05. 10.1007/s10495-008-0282-x . [DOI] [PubMed] [Google Scholar]
- 30.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–29. Epub 2010/01/23. 10.1038/nrc2780 . [DOI] [PubMed] [Google Scholar]
- 31.Hou JZ, Kan MK, McKeehan K, McBride G, Adams P, McKeehan WL. Fibroblast growth factor receptors from liver vary in three structural domains. Science. 1991;251(4994):665–8. Epub 1991/02/08. 10.1126/science.1846977 . [DOI] [PubMed] [Google Scholar]
- 32.Holzmann K, Grunt T, Heinzle C, Sampl S, Steinhoff H, Reichmann N, et al. Alternative Splicing of Fibroblast Growth Factor Receptor IgIII Loops in Cancer. J Nucleic Acids. 2012;2012:950508 10.1155/2012/950508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alioto TS, Buchhalter I, Derdak S, Hutter B, Eldridge MD, Hovig E, et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat Commun. 2015;6:10001 10.1038/ncomms10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta T, Ohmae S, Yamaya K, Kanemichi Y, Tokishita S, Yamagata H. Characterization of the mutational specificity of DNA cross-linking mutagens by the Lac+ reversion assay with Escherichia coli. Teratog Carcinog Mutagen. 2001;21(4):275–82. Epub 2001/06/15. . [DOI] [PubMed] [Google Scholar]
- 35.Meier B, Cooke SL, Weiss J, Bailly AP, Alexandrov LB, Marshall J, et al. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome Res. 2014;24(10):1624–36. Epub 2014/07/18. 10.1101/gr.175547.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuijjer ML, Namlos HM, Hauben EI, Machado I, Kresse SH, Serra M, et al. mRNA expression profiles of primary high-grade central osteosarcoma are preserved in cell lines and xenografts. BMC medical genomics. 2011;4:66 Epub 2011/09/22. 10.1186/1755-8794-4-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namlos HM, Meza-Zepeda LA, Baroy T, Ostensen IH, Kresse SH, Kuijjer ML, et al. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7(10):e48086 Epub 2012/11/08. 10.1371/journal.pone.0048086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buddingh EP, Kuijjer ML, Duim RA, Burger H, Agelopoulos K, Myklebost O, et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17(8):2110–9. Epub 2011/03/05. 10.1158/1078-0432.CCR-10-2047 . [DOI] [PubMed] [Google Scholar]
- 39.Amelio I, Gostev M, Knight RA, Willis AE, Melino G, Antonov AV. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell death & disease. 2014;5:e1051 Epub 2014/02/08. 10.1038/cddis.2014.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou D, Xie WZ, Hu KY, Huang WJ, Wei GQ, He JS, et al. Prognostic values of various clinical factors and genetic subtypes for diffuse large B-cell lymphoma patients: a retrospective analysis of 227 cases. Asian Pacific journal of cancer prevention: APJCP. 2013;14(2):929–34. Epub 2013/04/30. 10.7314/APJCP.2013.14.2.929 . [DOI] [PubMed] [Google Scholar]
- 41.Heikkinen T, Greco D, Pelttari LM, Tommiska J, Vahteristo P, Heikkila P, et al. Variants on the promoter region of PTEN affect breast cancer progression and patient survival. Breast cancer research: BCR. 2011;13(6):R130 Epub 2011/12/17. 10.1186/bcr3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie T, Cho YB, Wang K, Huang D, Hong HK, Choi YL, et al. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics. 2014;104(4):234–41. 10.1016/j.ygeno.2014.07.012 . [DOI] [PubMed] [Google Scholar]
- 43.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–10. 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Shi Y, Lalonde E, Li L, Cavallone L, Ferenczy A, et al. Exome profiling of primary, metastatic and recurrent ovarian carcinomas in a BRCA1-positive patient. BMC Cancer. 2013;13:146 Epub 2013/03/26. 10.1186/1471-2407-13-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vignot S, Frampton GM, Soria JC, Yelensky R, Commo F, Brambilla C, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol. 2013;31(17):2167–72. Epub 2013/05/01. 10.1200/JCO.2012.47.7737 . [DOI] [PubMed] [Google Scholar]
- 46.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. 10.1056/NEJMoa1113205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuijjer ML, Hogendoorn PC, Cleton-Jansen AM. Genome-wide analyses on high-grade osteosarcoma: making sense of a genomically most unstable tumor. Int J Cancer. 2013;133(11):2512–21. 10.1002/ijc.28124 . [DOI] [PubMed] [Google Scholar]
- 48.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–13. 10.1038/nature10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caronia D, Patino-Garcia A, Perez-Martinez A, Pita G, Moreno LT, Zalacain-Diez M, et al. Effect of ABCB1 and ABCC3 polymorphisms on osteosarcoma survival after chemotherapy: a pharmacogenetic study. PLoS One. 2011;6(10):e26091 Epub 2011/10/22. 10.1371/journal.pone.0026091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brambilla D, Zamboni S, Federici C, Lugini L, Lozupone F, De Milito A, et al. P-glycoprotein binds to ezrin at amino acid residues 149–242 in the FERM domain and plays a key role in the multidrug resistance of human osteosarcoma. Int J Cancer. 2012;130(12):2824–34. Epub 2011/07/23. 10.1002/ijc.26285 . [DOI] [PubMed] [Google Scholar]
- 51.Pakos EE, Ioannidis JP. The association of P-glycoprotein with response to chemotherapy and clinical outcome in patients with osteosarcoma. A meta-analysis. Cancer. 2003;98(3):581–9. Epub 2003/07/25. 10.1002/cncr.11546 . [DOI] [PubMed] [Google Scholar]
- 52.Scotlandi K, Serra M, Manara MC, Lollini PL, Maurici D, Del Bufalo D, et al. Pre-treatment of human osteosarcoma cells with N-methylformamide enhances P-glycoprotein expression and resistance to doxorubicin. Int J Cancer. 1994;58(1):95–101. 10.1002/ijc.2910580116 . [DOI] [PubMed] [Google Scholar]
- 53.Lorenz S, Baroy T, Sun J, Nome T, Vodak D, Bryne JC, et al. Unscrambling the genomic chaos of osteosarcoma reveals extensive transcript fusion, recurrent rearrangements and frequent novel TP53 aberrations. Oncotarget. 2016;7(5):5273–88. 10.18632/oncotarget.6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribi S, Baumhoer D, Lee K, Edison, Teo AS, Madan B, et al. TP53 intron 1 hotspot rearrangements are specific to sporadic osteosarcoma and can cause Li-Fraumeni syndrome. Oncotarget. 2015;6(10):7727–40. Epub 2015/03/13. 10.18632/oncotarget.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell reports. 2014;7(1):104–12. 10.1016/j.celrep.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers JM, Martins GG, Ostrowski J, Stachowiak MK. Nuclear trafficking of FGFR1: a role for the transmembrane domain. J Cell Biochem. 2003;88(6):1273–91. Epub 2003/03/21. 10.1002/jcb.10476 . [DOI] [PubMed] [Google Scholar]
- 57.Nguyen PT, Tsunematsu T, Yanagisawa S, Kudo Y, Miyauchi M, Kamata N, et al. The FGFR1 inhibitor PD173074 induces mesenchymal-epithelial transition through the transcription factor AP-1. Br J Cancer. 2013;109(8):2248–58. 10.1038/bjc.2013.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chioni AM, Grose R. FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J Cell Biol. 2012;197(6):801–17. Epub 2012/06/06. 10.1083/jcb.201108077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, et al. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO molecular medicine. 2014;6(4):467–81. Epub 2014/02/08. 10.1002/emmm.201302698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25(3):552–63. Epub 2013/11/23. 10.1093/annonc/mdt419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernanda Amary M, Ye H, Berisha F, Khatri B, Forbes G, Lehovsky K, et al. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo-adjuvant chemotherapy. Cancer medicine. 2014;3(4):980–7. Epub 2014/05/28. 10.1002/cam4.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guagnano V, Kauffmann A, Wohrle S, Stamm C, Ito M, Barys L, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer discovery. 2012;2(12):1118–33. 10.1158/2159-8290.CD-12-0210 . [DOI] [PubMed] [Google Scholar]
- 63.Weekes D, Kashima TG, Zandueta C, Perurena N, Thomas DP, Sunters A, et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35(22):2852–61. 10.1038/onc.2015.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra MC, Zerial M. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J. 1994;13(22):5262–73. Epub 1994/11/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62(15):1657–70. Epub 2005/06/01. 10.1007/s00018-005-4486-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horgan CP, McCaffrey MW. Rab GTPases and microtubule motors. Biochem Soc Trans. 2011;39(5):1202–6. Epub 2011/09/23. 10.1042/BST0391202 . [DOI] [PubMed] [Google Scholar]
- 67.Cremers FP, Armstrong SA, Seabra MC, Brown MS, Goldstein JL. REP-2, a Rab escort protein encoded by the choroideremia-like gene. J Biol Chem. 1994;269(3):2111–7. Epub 1994/01/21. . [PubMed] [Google Scholar]
- 68.Strunnikova NV, Barb J, Sergeev YV, Thiagarajasubramanian A, Silvin C, Munson PJ, et al. Loss-of-function mutations in Rab escort protein 1 (REP-1) affect intracellular transport in fibroblasts and monocytes of choroideremia patients. PLoS One. 2009;4(12):e8402 Epub 2009/12/23. 10.1371/journal.pone.0008402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chilamakuri CS, Lorenz S, Madoui M-A, Vodak D, Sun J, Hovig E, et al. Performance comparison of four exome capture systems for deep sequencing. BMC Genomics. 2014;15(1):449 10.1186/1471-2164-15-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Available from: http://master.bioconductor.org/packages/release/bioc/html/DNAcopy.html.
- 71.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9. Epub 2013/02/12. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. Epub 2012/02/04. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106 Epub 2010/10/29. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The copy number along chromosome 8:35,000,000–43,000,000 of each tumor sample, with emphasis on FGFR1.
(TIF)
An overview over the different mutations types for all tumor samples.
(TIF)
The transcriptome data and corresponding gene expression levels (shown as normalized read counts) for each sample (tumors and controls).
(XLSX)
An overview over the identified fusion transcripts in patient OS111.
(XLSX)
An overview over all identified somatic mutations and corresponding expression level in all samples.
(XLSX)
A detailed overview over the two recurrent mutations.
(XLSX)
A summary of the clinical data of the 71 patients included in the CHM analysis.
(XLSX)
An overview over the primer sequences used for the targeted resequencing.
(XLSX)
Data Availability Statement
Because of Norwegian legal regulations, the ethical approval for this study and the consent signed by the patient, we are not able to deposit our sequencing dataset in a public repository. We will, however, provide access to the data if requested by cancer researchers. We would like to emphasize that all relevant data (i.e. all called somatic variants and gene expression levels) are available in the Supplementary Information, but we are not allowed to openly publish the complete exome of patients. The gene expression data used for the CHM analysis can be found at GEO. The GEO accession numbers GSE36004 (cell lines, osteoblasts and bone samples) and GSE30699 (clinical samples) are provided under “Data deposits” in Materials and Methods. Otherwise, all relevant data can be found within the paper and its Supporting Information Files. For data related queries and requests, please contact the corresponding author Dr. Meza-Zepeda (leonardo.meza-zepeda@rr-research.no).