Abstract
Background & Aims
Gut dysbiosis is closely involved in the pathogenesis of inflammatory bowel disease (IBD). However, it remains unclear whether IBD-associated gut dysbiosis contributes to disease pathogenesis or is merely secondary to intestinal inflammation. We established a humanized gnotobiotic (hGB) mouse system to assess the functional role of gut dysbiosis associated with 2 types of IBD: Crohn's disease (CD) and ulcerative colitis (UC).
Methods
Germ-free mice were colonized by the gut microbiota isolated from patients with CD and UC, and healthy controls. Microbiome analysis, bacterial functional gene analysis, luminal metabolome analysis, and host gene expression analysis were performed in hGB mice. Moreover, the colitogenic capacity of IBD-associated microbiota was evaluated by colonizing germ-free colitis-prone interleukin 10–deficient mice with dysbiotic patients' microbiota.
Results
Although the microbial composition seen in donor patients' microbiota was not completely reproduced in hGB mice, some dysbiotic features of the CD and UC microbiota (eg, decreased diversity, alteration of bacterial metabolic functions) were recapitulated in hGB mice, suggesting that microbial community alterations, characteristic for IBD, can be reproduced in hGB mice. In addition, colonization by the IBD-associated microbiota induced a proinflammatory gene expression profile in the gut that resembles the immunologic signatures found in CD patients. Furthermore, CD microbiota triggered more severe colitis than healthy control microbiota when colonized in germ-free interleukin 10–deficient mice.
Conclusions
Dysbiosis potentially contributes to the pathogenesis of IBD by augmenting host proinflammatory immune responses. Transcript profiling: GSE73882.
Keywords: Dysbiosis, Microbiota, Crohn's Disease, Ulcerative Colitis
Abbreviations used in this paper: CD, Crohn's disease; CE-TOFMS, capillary electrophoresis time-of-flight mass spectrometry; GB, gnotobiotic; GF, germ-free; hGB, humanized gnotobiotic; IBD, inflammatory bowel disease; IFN, interferon; IL, interleukin; ILC, innate lymphoid cell; IVC, individual ventilated cage; NK, natural killer; OTU, operational taxonomic unit; rRNA, ribosomal RNA; SCFA, short-chain fatty acid; Th, T helper; UC, ulcerative colitis; WT, wild type
Summary.
The dysbiotic feature of the microbiota in inflammatory bowel disease patients (eg, decreased diversity) was reproduced in humanized gnotobiotic mice. The microbiota isolated from patients with Crohn's disease and ulcerative colitis was shown to have altered bacterial function in humanized gnotobiotic mice as shown by bacterial functional gene analysis and luminal metabolome analysis. Host gene expression induced in humanized gnotobiotic mice owing to colonization by the microbiota isolated from patients with Crohn’s disease resembled the core gene expression patterns observed in the intestinal mucosa of Crohn’s disease patients; the microbiota also promoted the development of colitis when used to colonize inflammatory bowel disease–prone interleukin 10–deficient mice.
The resident gut microbiota is essential for numerous vital host physiological processes, including digestion of dietary factors, development of the gut immune system, and colonization resistance against incoming pathogens.1 Not surprisingly, a breakdown of the homeostatic relationship between the host and the microbiota can lead to the development of various intestinal and extraintestinal disorders, including inflammatory bowel disease (IBD).1 IBD comprises 2 major disorders, Crohn’s disease (CD) and ulcerative colitis (UC), characterized by chronic inflammation of the gastrointestinal tract.2, 3 Although the precise etiology of IBD has not yet been defined, it is widely accepted that the gut microbiota is central to the initiation and persistence of disease. Indeed, intestinal inflammation only develops in the presence of a conventional microbiota in most experimental models of IBD, whereas animals housed under germ-free (GF) conditions fail to develop intestinal inflammation.1, 4, 5, 6 In IBD patients, alterations in the gut microbiota have been identified repeatedly and termed dysbiosis.7 However, it remains unclear whether dysbiosis contributes to the pathogenesis of IBD or is merely a secondary factor that develops as a result of gut inflammation.
In support of a pathogenic role of gut dysbiosis in IBD patients, it has been reported that certain pathobionts, such as adherent-invasive Escherichia coli strains, accumulate in patients with CD as a result of gut dysbiosis. These pathobionts are capable of facilitating intestinal inflammation in experimental models.8, 9, 10, 11 Likewise, a potential mechanism is suggested by the observation that perturbations in the metabolic function of the microbiota, caused by dysbiosis, can influence the production of immune-regulatory bacterial byproducts, such as short-chain fatty acids (SCFAs), thereby compromising mucosal defense.12 Furthermore, a recent report showed that gut dysbiosis and an altered host gene expression profile were observed in the ileum of histologically normal tissues in treatment-naive, newly diagnosed patients with CD colitis.13 Thus, it is conceivable that dysbiosis is not simply a result of inflammation. Rather, dysbiotic microbiota is functionally defective and contributes to inflammation. However, as of now, a detailed mechanism connecting IBD-associated dysbiosis and the resultant detrimental host immune responses has not been elucidated.
Alternatively, it has been reported that intestinal inflammation alters the community structure of the microbiota.14, 15, 16 The mucosal inflammatory milieu selectively fuels the growth of facultative anaerobes, including Proteobacteria, at the expense of obligate anaerobes, including Firmicutes and Bacteroidetes. These conditions give rise to blooms of facultative anaerobes, such as E coli.14, 15 The overgrowth of certain bacteria resulting from intestinal inflammation skews the balance of the whole microbial community, leading to lower diversity. Consistently, low diversity and richness of the gut microbial community along with an increased abundance of Enterobacteriaceae, including E coli, and decreased abundance of the phylum Firmicutes, are observed in patients with IBD.11, 12, 17 Thus, gut dysbiosis in IBD may be a secondary manifestation of intestinal inflammation
The gnotobiotic (GB) mouse model, in which GF mice are colonized with selected known populations of bacteria, is a powerful system used to characterize the functions of bacterial populations in vivo.18 GB mice also can be used to evaluate the function of human microbiota in mice.19, 20 Many studies have shown that it is possible to recapitulate the metabolic features of human microbiota in humanized GB (hGB) mice.19, 20 For instance, hGB mice colonized with the microbiota isolated from obese patients, as compared with lean controls, tend to become more obese when they are fed a high-fat diet, indicating that functional features of the disease-associated microbiota can be recapitulated successfully in mice.21 In the present study, we established this model to functionally characterize the dysbiotic microbiota from IBD patients. By using hGB mice colonized with the microbiota isolated from patients with CD and UC, we showed that dysbiosis, present in the donor microbiota, can be recapitulated, at least to some extent, in mice. Certain dysbiotic features (eg, lower community diversity) associated with donor patients' stool samples were reproduced in recipient mice, and the resulting microbial metabolic profiles were different. Likewise, colonization by the IBD-associated dysbiotic microbiota influenced gene expression profiles in the colon, showing that the microbiota in CD and UC has a distinct functional impact on host immunity. Strikingly, we also found that the microbiota from CD patients induced more severe intestinal inflammation when colonized in GF interleukin (IL)10-deficient mice, a mouse model for CD.
Materials and Methods
Donor Stool Sample Preparation
Stool samples were obtained from patients with CD and UC and healthy control subjects according to the University of Michigan Institutional Review Board–approved protocol (IBD databank, HUM00041845). Written informed consent forms were obtained before sample collection. Donor patients and control subjects were not treated with any antibiotics for at least 3 months before sample collection, and had no history of intestinal bacterial infections such as Clostridium difficile, or other infections such as hepatitis B virus, hepatitis C virus, or human immunodeficiency virus. Patients had been histologically and endoscopically diagnosed with CD or UC. Patients with ostomy were excluded. Collected stool samples were stored at -80°C until use. Before inoculation, stool samples were diluted 1:10 with pre-reduced phosphate-buffered saline under anaerobic conditions. Diluted stool samples then were passed through a 100-μm cell strainer and orally inoculated (100 μL per mouse) into GF C57BL/6 mice.
Mice
GF C57BL/6 mice were housed in the Germ-Free Animal Facility at the University of Michigan. GF mice were maintained in flexible film isolators and were checked weekly for GF status by aerobic and anaerobic culture. The absence of the microbiota was verified by microscopic analysis of stained cecal contents that detects any unculturable contamination. Eight- to 16-week-old female and male mice were used for experiments. For the generation of hGB mice, stool samples, which were obtained from control and IBD donors (as described earlier), were orally inoculated into 2–5 recipient GF C57BL/6 mice per donor. The hGB mice were housed in positive-pressure individual ventilated cages (IVCs) (ISOcage P; Techniplast, West Chester, PA) per condition to prevent cross-contamination among the different groups and maintain gnotobiotic conditions.22, 23 All mice were fed a sterilized rodent breeder diet 5013 (LabDiet, St. Louis, MO). All animals were handled in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the University of Michigan.
Bacterial DNA Isolation
Stool samples were obtained from individual donor or hGB mice (stool samples from multiple mice colonized with the same microbiota and housed in the same IVC were pooled). Genomic DNA was extracted by using a modified protocol of the Qiagen DNeasy Blood and Tissue kit (Valencia, CA). These modifications included the following: (1) adding a bead-beating step using UltraClean fecal DNA bead tubes (Mo Bio Laboratories, Inc, West Carlsbad, CA) that were shaken using a Mini-Beadbeater-16 (BioSpec Products, Inc, Bartlesville, OK) for 1.5 minutes; (2) increasing the amount of buffer ATL used in the initial steps of the protocol (from 180 to 360 μL); (3) increasing the volume of proteinase K used (from 20 to 40 μL); and (4) decreasing the amount of buffer AE used to elute the DNA at the end of the protocol (from 200 to 85 μL).
Bacterial 16S rRNA Sequencing
Samples were submitted to the University of Michigan Medical School Host Microbiome Initiative and processed using the MiSeq Illumina sequencing platform. 16S ribosomal RNA (rRNA) gene libraries were constructed using primers specific to the V4 region.
Operational Taxonomic Unit Assignment and Diversity Measurements
Sequences were curated using the community-supported software program mothur (v.1.33)24 and by following the steps outlined in the MiSeq SOP (http://www.mothur.org/wiki/MiSeq_SOP).25 Sequences were assigned to operational taxonomic units (OTUs) using a cut-off value of 0.03 and classified against the Ribosomal Database Project 16S rRNA gene training set (version 9) using a naive Bayesian approach with an 80% confidence threshold. Curated OTU sequence data were converted to relative abundance ± SEM. Within-community diversity (α-diversity) was calculated using the Shannon diversity index (H’) and OTU richness. Between-community diversity (β-diversity) was determined using the Yue and Clayton dissimilarity distance metric. Nonmetric multidimensional scaling was used to ordinate the β-diversity data. An analysis of molecular variation was used to test for significant differences in the community structure using 10,000 permutations. The functional aspect of the bacterial community was investigated using the OTU-based bacterial signaling analysis phylogenetic investigation of communities by reconstruction of unobserved states.26 To test which functional pathways were differentially abundant, biologically consistent, and had the greatest effect size, we used linear discriminant analysis effect size.27
Capillary Electrophoresis Time-of-Flight Mass Spectrometry–Based Metabolome Analysis
Capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS)–based metabolome analysis was conducted as described previously with some modifications.28 In brief, fecal samples were lyophilized by using a VD-800R lyophilizer (TAITEC, Saitama, Japan) for 24 hours. Freeze-dried feces were disrupted with 3.0-mm Zirconia Beads (Biomedical Science, Tokyo, Japan) by vigorous shaking (1500 rpm for 10 min) using Shake Master (Biomedical Science). Fecal metabolites were extracted by the methanol:chloroform:water extraction protocol. CE-TOFMS experiments were performed using the Agilent CE System, the Agilent G3250AA LC/MSD TOF System, the Agilent 1100 Series Binary HPLC Pump, the G1603A Agilent CE-MS adapter, and the G1607A Agilent CE-ESI-MS SprayerKit (all Agilent Technologies, Santa Clara, CA).
Host Gene Expression Analysis
Colonic tissues were harvested from hGB mice 14 days after microbiota colonization. RNA was extracted using the ENZA Total RNA Kit (Omega Biotek, Norcross, GA) according to the manufacturer’s protocol. RNA integrity number was measured using a bioanalyzer instrument (Agilent Technologies) and ranged from 9.2 to 9.6, with 28S/18S ratios between 1.8 and 1.9. Target-labeled complementary RNA was hybridized to a GeneChip Mouse Gene ST 2.1 array (Affymetrix, Santa Clara, CA). Data were normalized with the robust multi-array average procedure using the affy package of Bioconductor implemented in the R statistical language.
Induction of Colitis in IL10-Deficient Mice
Fecal samples were obtained from HC and CD hGB repository mice and were used to prepare the donor microbiota samples. Fecal samples isolated from hGB mice (>14 days after reconstitution) were inoculated into GF Il10-/- mice. GB mice colonized with microbiota from a single donor were housed in an IVC to prevent cross-contamination among groups. Three weeks after fecal transplantation using the microbiota from hGB mice donors, the reconstituted Il10-/--hGB mice were killed, and ceca and colons were collected. Colonic length and weight were measured and then fixed with 4% paraformaldehyde. Fixed tissues were processed for H&E staining. Histologic assessment was performed by a pathologist in a blinded fashion at the Unit for Laboratory Animal Medicine in vivo Animal Core. For histology scoring, a 4-point scale was used to denote the severity of inflammation (0, none; 1, minimal multifocal inflammation [few foci]; 2, moderate multifocal inflammation [numerous foci]; 3, severe multifocal coalescing inflammation; and 4, same as a score of 3 with abscesses or extensive mural involvement), the edema scores (0, none; 1, mild focal or multifocal edema, minimal submucosal expansion (<2×); 2, moderate focal or multifocal edema, moderate submucosal expansion (2–3×); 3, severe multifocal to coalescing inflammation; and 4, same as a score of 3 with diffuse submucosal expansion), and the epithelial score (0, none; 1, mild, multifocal, superficial damage; 2, moderate, multifocal, superficial damage; 3, evere, multifocal to coalescing mucosal damage ± pseudomembrane ± ulcer; 4, same as a score of 3 with significant pseudomembrane or ulcer formation). Each variable then was summed to obtain the overall score.
Quantification of Flagellin
Flagellin levels in feces were measured by using HEK-Blue mTLR5 cells (Invivogen, San Diego, CA) according to a previously reported method.29 Fecal samples were obtained from hGB mice and homogenized for 10 seconds using a bead beater (Biospec Products, Bartlesville, OK). Homogenized fecal samples were centrifuged at 8000 × g for 2 minutes and the supernatants were harvested. HEK-Blue mTLR5 cells were suspended in the HEK-Blue detection medium and stimulated with serially diluted homogenate fecal supernatants. Stimulated cells were incubated for 6 hours at 37°C and reporter activity (alkaline phosphatase activity) was measured at 640 nm. Purified flagellin (from Salmonella typhimurium; Invitrogen) was used as a standard.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism software version 5.0 (GraphPad Software, Inc, San Diego, CA). Differences between 2 groups were evaluated using the Student t test (parametric) or the Mann–Whitney U test (nonparametric). For the comparison of more than 3 groups, statistical analysis was performed using 1-way analysis of variance (parametric) or the Kruskal–Wallis test (nonparametric), followed by the Bonferroni correction for parametric samples, or the Dunn test for nonparametric samples as a post hoc test. P values less than .05 were considered significant.
Results
Gut Dysbiosis Associated With IBD Is Reproduced in hGB Mice
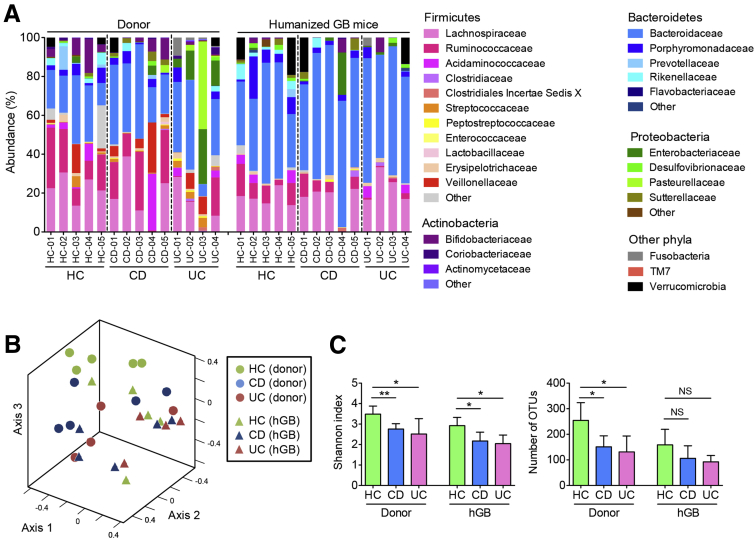
To directly test the function of IBD-associated dysbiosis, we first attempted to establish an hGB mouse model by inoculating GF mice with IBD-associated dysbiotic microbiota. Stool samples were obtained from 5 healthy controls (HC-01–HC-05), 5 CD patients (CD-01–CD-05), and 4 UC patients (UC-01–UC-04), and they were used to orally inoculate GF C57BL/6 mice. Mice were kept under GF conditions for 2 weeks to allow for complete reconstitution with the human microbiota.30 To analyze the microbial composition before and after humanization of the GF mice, donor stool samples and stool samples from recipient hGB mice were collected and analyzed by 16S rRNA sequencing (Figure 1). Compared with HC donor microbiota, the UC donors, in particular, showed a lower abundance of Firmicutes, a greater abundance of Proteobacteria, and more variability in the abundances of predominant bacterial taxa among individual samples, consistent with data from larger cohorts (Figure 2A and Supplementary Table 1). Interestingly, apparent changes in the CD donor microbiota were less obvious (Figure 2A and Supplementary Table 1). As shown in Figure 2B, the microbiota from HC, CD, and UC stool donors tends to fall into different clusters, which is shown in the nonmetric multidimensional scaling plot. After reconstitution in GF mice, these 3 groups still showed different community structures (Figure 2B). Because there was substantial variability within each group, the between-group differences were not statistically significant (Figure 2B). The α-diversity of the CD and UC donor communities was significantly lower compared with the HC donors as measured by the Shannon diversity index and OTU richness (Figure 2C). These dysbiotic features were noted in the recipient mice as well, and α-diversity (Shannon diversity index) was significantly lower in hGB mice colonized with the microbiota from patients with CD and UC (Figure 2C). Even though a similar decrease in microbial richness could be observed in UC and CD hGB mice after reconstitution, the difference was not statistically significant (Figure 2C). These results suggested that hGB mice colonized with the microbiota from IBD patients show dysbiosis, at least to some extent, as observed in the original stool samples.
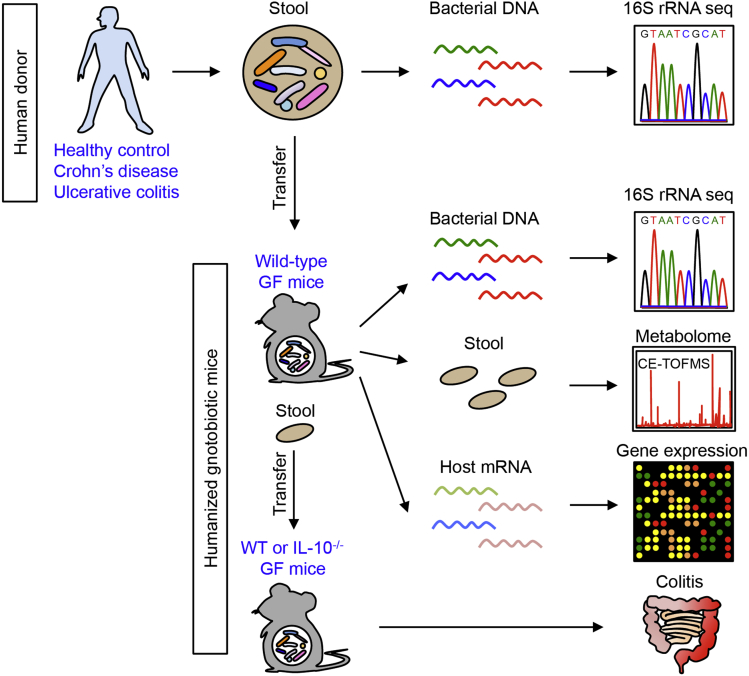
Figure 1.
Analysis of functional dysbiosis in IBD. Stool samples were obtained from healthy control subjects and patients with IBD (Crohn's disease and ulcerative colitis). Intestinal bacteria then were inoculated into GF WT mice (hGB mice). After 2 weeks of reconstitution, gut microbiome (16S rRNA sequencing), luminal metabolome (CE-TOF/MS), and host gene expression profile (DNA microarray) in hGB mice were analyzed. For evaluation of the colitogenic capacity of the microbiota, established hGB mice were used as stool donors. Stool samples obtained from HC- or IBD-hGB mice were transplanted into either GF WT B6 mice or GF Il10-/-mice. After 3 weeks of reconstitution, intestinal inflammation was examined. mRNA, messenger RNA.
Figure 2.
IBD-associated dysbiosis is recapitulated in humanized gnotobiotic mice. (A) Stool samples were obtained from patients with CD, UC, and HCs, and then inoculated into GF mice. After 2 weeks of reconstitution, stool samples from hGB mice were collected and bacterial 16S rRNA sequences were analyzed. Sample IDs of human donors correspond to those of donor-derived hGB mice. (B) Microbial community structures were analyzed by using the Yue and Clayton dissimilarity distance metric (θYC) and are shown in a nonmetric multidimensional scaling plot. (C) Shannon index (α-diversity) and number of OTUs (richness) for each group. Data are presented as means ± SD. *P < .05 by Student t test. ** P < .01.
Microbial Metabolic Pathways Are Perturbed in IBD
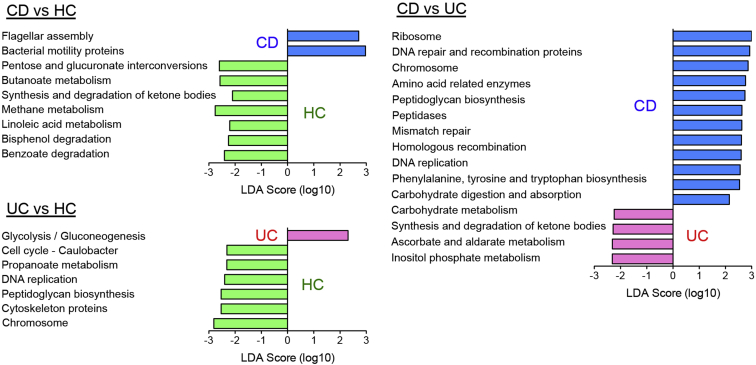
To assess for the presence of a potential functional defect in the IBD-associated dysbiotic microbiota recapitulated in GB mice, we next predicted the functions of the microbial communities by using the phylogenetic investigation of communities by reconstitution of unobserved states algorithm (Phylogenetic Investigation of Communities by Reconstitution of Unobserved States).26 The predicted functional pathways significantly affected by dysbiosis in patients with CD or UC were identified in hGB recipient mice by using the linear discriminant analysis effect size approach.27 As shown in Figure 3, genes related to flagellar assembly and bacterial motility proteins were significantly over-represented and genes involved in certain metabolic pathways, such as carbohydrate and bile acid metabolism, were under-represented in CD hGB mice compared with HC hGB mice. In UC hGB mice, glycolysis/gluconeogenesis-related genes were over-represented compared with HC hGB mice, whereas genes associated with bacterial homeostasis (eg, DNA replication, peptidoglycan synthesis) and certain metabolic pathways (eg, propanoate metabolism) were significantly reduced (Figure 3). There were also significant differences in predicted metabolic function between CD hGB and UC hGB mice. Certain metabolic genes were over-represented in CD but not in UC (Figure 3). These results suggested that dysbiosis in IBD is predicted to compromise the metabolic function of the microbiota. Furthermore, CD-associated and UC-associated dysbiosis result in distinct functional alterations of the microbiota. We also analyzed the functional profile of the microbial communities present in human donor stool samples based on 16S rRNA sequence results (Supplementary Figure 1). There also were significant differences in predicted metabolic function among HC, CD, and UC donor samples (Supplementary Figure 1). However, the predicted functionality differences observed in the microbiota of HC, CD, and UC donor samples did not fully correlate with the differences observed upon reconstitution of hGB mice (Figure 3 and Supplementary Figure 1).
Figure 3.
Microbial functional gene pathways in hGB mice. The abundance of Kyoto Encyclopedia of Genes and Genomes metabolic gene pathways was analyzed by phylogenetic investigation of communities by reconstruction of unobserved states based on 16S rRNA sequencing data in Figure 2. Significantly altered pathway genes in 3 groups (HC-, CD-, and UC-hGB mice) were identified by linear discriminant analysis effect size. The linear discriminant analysis (LDA) score is shown.
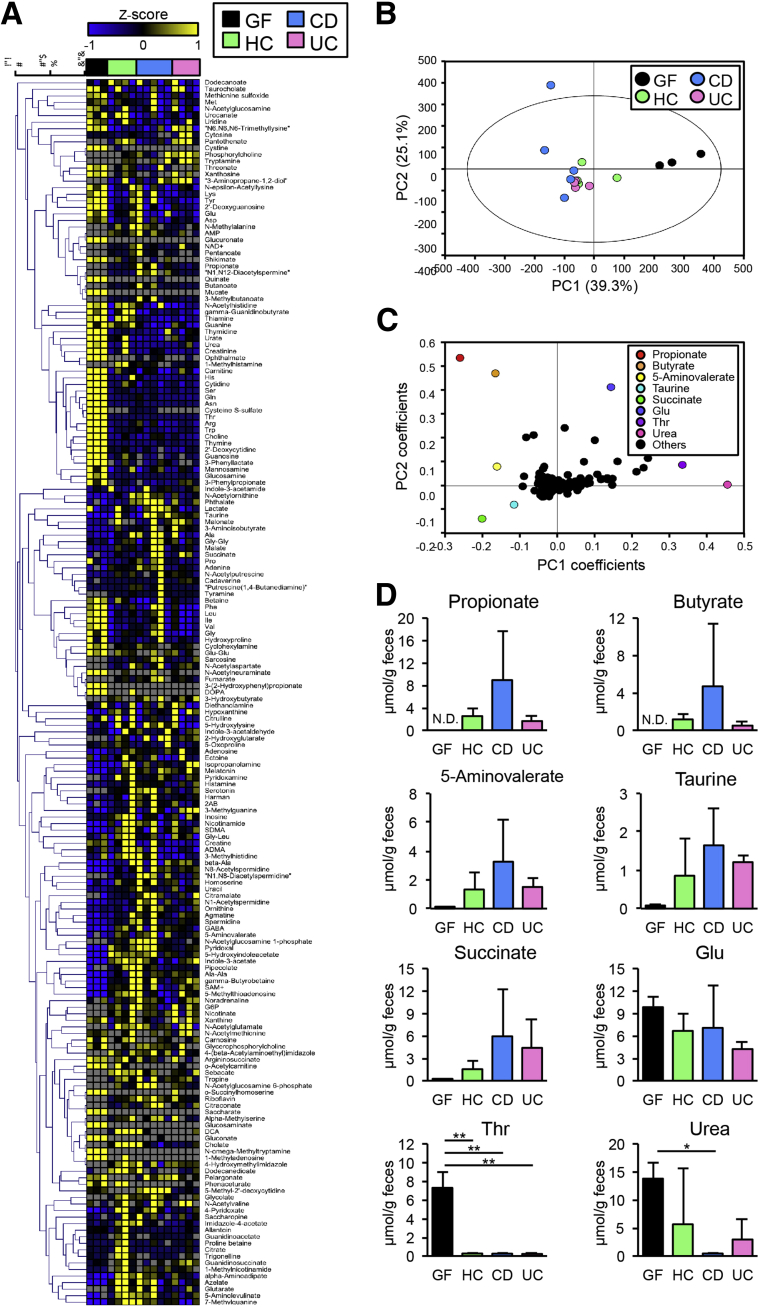
To confirm the result of the predictive functional gene analysis, we next analyzed the metabolome of hGB mice colonized with IBD and control microbiota using CE-TOFMS. As shown in Figure 4A and Supplementary Table 2, hGB mice colonized with IBD microbiota showed distinct metabolic profiles compared with hGB mice reconstituted with HC microbiota. Principal component analysis showed that metabolome of humanized hGB mice is distinct from that of GF controls, confirming that colonization of human microbiota alters the luminal metabolome (Figure 4B). The metabolome in CD hGB seemed to cluster differently from HC and UC (Figure 4B). Orthogonal partial least-squares discriminate analysis of the CE-TOFMS–based metabolome data indicated that the amount of luminal SCFAs, such as propionate and butyrate, succinate, 5-aminovalerate, and taurine, was higher in hGB mice than in GF mice, indicating that generation of these metabolites are microbiota-dependent (Figure 4C and D). CD hGB mice showed a trend toward an increase in SCFAs compared with HC hGB mice, whereas the amount of SCFAs in UC hGB mice was similar to HC hGB (Figure 4D). The amount of certain metabolites, including threonine and urea, was lower after bacterial colonization, suggesting that these metabolites are consumed and/or degraded by the microbiota (Figure 4C and D). These results show that the metabolic functions of the HC and IBD microbiota of hGB mice are distinct, and this suggests a possible mechanism by which IBD dysbiosis may influence disease susceptibility in the host.
Figure 4.
Fecal metabolome profiles of GF and hGB mice. (A) A heat map showing the quantified metabolic profiles. All concentrations of quantified metabolites were transformed into Z-scores and clustered according to Euclidean distance. Gray areas in the heat map indicate that respective metabolites were not detected. (B) The principal component analysis of the metabolome data. The ellipse denotes the 95% significance limit of the model, as defined by the Hotelling t test. (C) A loading scatter plot of the principal component analysis. (D) The bar graphs showing the concentration of propionate, butyrate, 5-aminovalerate, taurine, succinate, glutamate, threonine, and urea in murine feces, respectively. Data are presented as means ± SEM. *P < .05, ** P < .01. ND, not detected by 1-way analysis of variance, followed by the Tukey post hoc test. PC, principal component.
Dysbiotic Microbiota in IBD Influences Differentially Regulated Mucosal Gene Expression Patterns
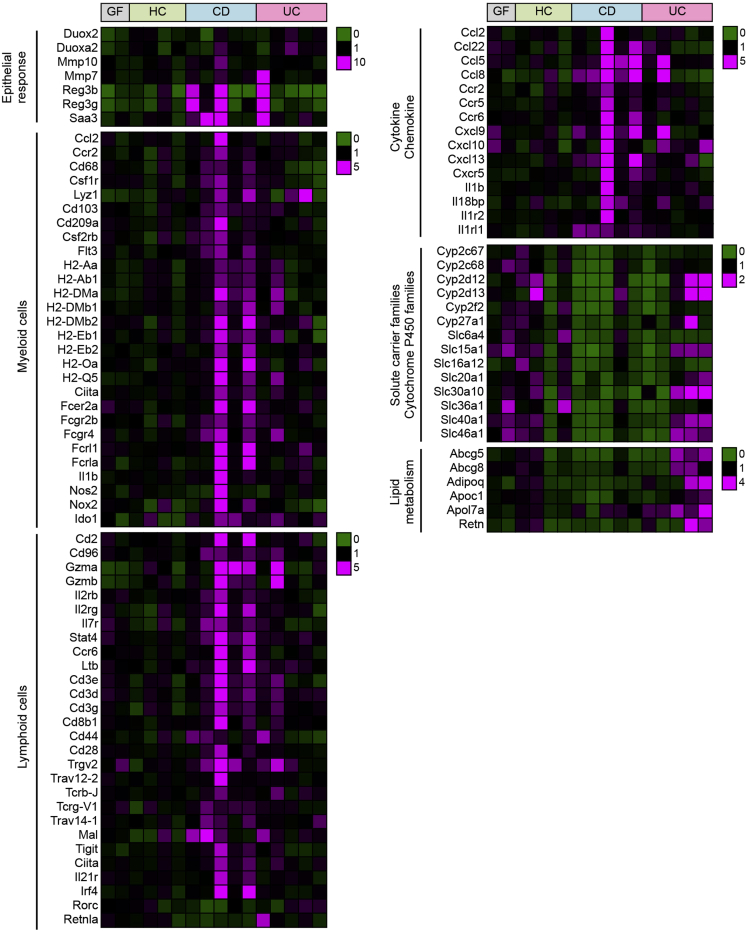
To explore the functional impact of dysbiotic microbiota in IBD patients on host immune responses, we next analyzed gene expression profiles in the colonic mucosa of hGB mice colonized with HC, CD, and UC microbiota. Colonization of HC microbiota induced expression of genes associated with the epithelial response to microbes (eg, Reg3b/3g, Cldn4, Duox2, Duoxa2, and Saa3) and immunoglobulin-related genes in GF mice (Figure 5 and Supplementary Table 3), suggesting that human microbiota can promote mucosal maturation in mice. Colonization of CD microbiota triggered stronger induction of certain epithelial response genes (eg, Reg3b/3g, Mmp10) than HC microbiota (Figure 5 and Supplementary Table 3). Likewise, colonization of CD microbiota promoted expression of gene markers for macrophages (major histocompatibility complex class II genes, Fc receptor genes, Ccl2/Ccr2, Csf1r, Cd68, Lyz1), dendritic cells (major histocompatibility complex class II genes, FcR genes, Csf2rb, Flt3, Cd209a, Cd103), natural killer (NK) cells (Gzma/b, Cd2, Cd96, Il2rb/g, Stat4), group3 innate lymphoid cells (ILCs) (Ltb, Il2rg, Ccr6, Il7r, Ciita), T-helper (Th)1 cells (Stat4, Ciita), and Th17 cells (Saa3, Irf4, Ccr6, Il21r, Stat4) (Figure 5 and Supplementary Table 3). Furthermore, many cytokines, chemokines, and their receptors (Il1b, il1r2, il1rl1, il18bp, Ccl2/5/8/22, Cxcl9/10/13, Ccr2/5/6, Cxcr5) were up-regulated in hGB mice colonized with CD microbiota (Figure 5 and Supplementary Table 3). In contrast, certain genes related to solute carrier families (Slc6a4/15a1/16a12/20a1/30a10/36a1/40a1/46a1) and cytochrome P450 families (Cyp2c67/2c68/2d12/2d13/2f2/27a1) were under-represented in CD hGB compared with HC hGB (Figure 5 and Supplementary Table 3). Gene expression patterns in UC hGB mice were different from those in CD hGB mice. Similar to CD, colonization with UC microbiota also showed enhanced expression of certain epithelial response genes, such as Saa3 and Duoxa2, compared with HC microbiota (Figure 5 and Supplementary Table 3). However, many genes found to be increased in CD hGB mice were not up-regulated in UC hGB mice (Figure 5 and Supplementary Table 3). On the other hand, certain genes, including genes related to lipid metabolism (Retn, Abcg5/8, Adipoq, Apoc1, Apol7a) and some Th17-related genes (Rorc, Retnla, Ccl20), were expressed at greater levels in UC hGB than in CD hGB mice (Figure 5 and Supplementary Table 3).
Figure 5.
Host gene expression in the colonic mucosa of hGB mice. Host gene expression induced by colonization of human microbiota (HC, CD, and UC). A heat map of selected genes, which were expressed differently in CD-hGB or UC-hGB mice compared with HC-hGB mice, is shown. The color range indicates the fold expression of genes compared with the average expression in HC-hGB mice.
Microbiota of CD Patients Confers Increased Susceptibility to Experimental Colitis
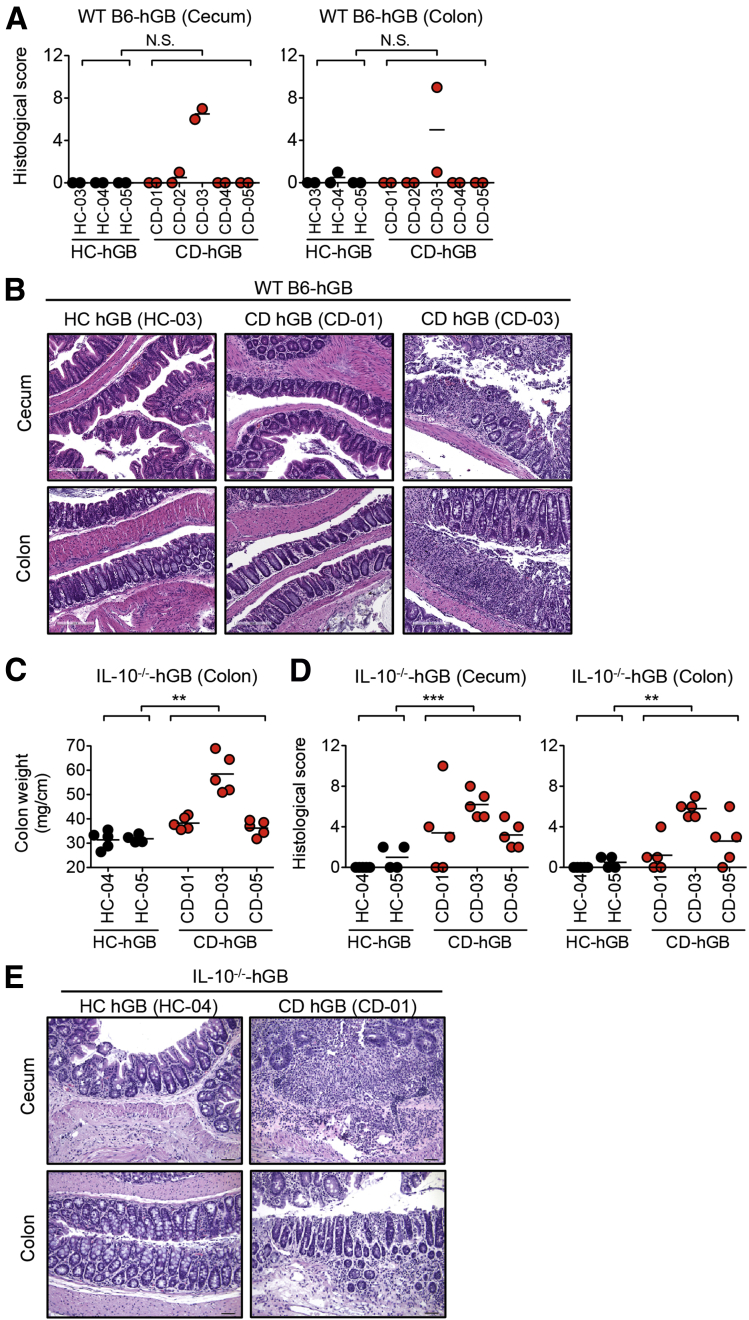
Although colonization by the dysbiotic microbiota isolated from CD patients elicited proinflammatory immune responses (eg, Th17, Th1, IL1β signaling) (Figure 5), none of the microbiota samples triggered overt intestinal pathology, with the exception of 1 patient (CD-03) (Figure 6A and B). This suggests that in most cases CD-associated dysbiosis does not immediately lead to pathogenesis under these conditions. Notably, the microbiota derived from patient CD-03 had the strongest inductive effect on proinflammatory genes when used to colonize GF wild-type (WT) B6 mice (Figure 5), indicating that this patient’s microbiota may harbor pathogenic bacteria. We next assessed the colitogenic capacity of CD dysbiotic microbiota in colitis-prone mice. Upon colonization of GF IL10-/- mice, the HC microbiota did not induce any overt signs of intestinal inflammation (ie, colon thickening or intestinal pathology) (Figure 6C–E). In contrast, the CD microbiota elicited the development of severe colitis in GF IL10-/- mice. IL10-/- mice colonized with CD microbiota showed significantly increased colon weight and overt intestinal pathology (Figure 6C–E). These results show that the intestinal bacteria from CD patients can induce the development of colitis in IBD-prone mice.
Figure 6.
CD-associated microbiota promotes the development of colitis in IBD-prone mice. (A and B) Stool samples were isolated from HC-hGB mice (HC-03, HC-04, and HC-05) and CD-hGB mice (CD-01, CD-02, CD-03, CD-04, and CD-05), and then inoculated into GF WT B6 mice. After 3 weeks of reconstitution, cecal and colonic tissues were harvested. (A) Histologic score of colon. Each dot indicates an individual mouse. NS, not significant by Mann–Whitney U test. (B) A representative histologic image of WT B6-HC-hGB and CD-hGB mice. Scale bar: 200 μm. (C–E) Stool samples were isolated from HC-hGB mice and CD-hGB mice and then inoculated into GF Il10-/- mice. After 3 weeks of reconstitution, cecal and colonic tissues were harvested. (C) Colonic weights. Each dot indicates an individual mouse. **P < .01 by Mann–Whitney U test. (D) Histologic score of colon. Each dot indicates an individual mouse. **P < .01 by Mann–Whitney U test. ***P < .001. (E) A representative histologic image of colonic tissues isolated from IL10-/--HC-hGB and CD-hGB mice. Scale bar: 50 μm.
Discussion
New and provocative results have been published as of late, supporting a potential pathogenic role for gut microbial dysbiosis in the manifestation of IBD. Studies have reported that changes in microbial composition and host gene expression profiles of ileal tissue samples that do not show histologic inflammation from newly diagnosed, treatment-naive CD colitis patients, suggest a possible causal role for IBD-associated dysbiosis.11, 13 A potential mechanism for dysbiosis-mediated host responses is suggested by recent functional analyses of the microbiota in IBD, which showed that certain metabolic functions of the microbiota are perturbed in IBD. For example, perturbed carbohydrate metabolism suggests the compromised formation of SCFAs. Because SCFAs play crucial roles in the development of regulatory T cells and enhancement of the epithelial barrier function,31, 32, 33 dysbiosis in IBD may compromise host regulatory immune responses. Also, amino acid metabolism is significantly altered in the dysbiotic microbiota associated with IBD.12, 34 In the clinical setting of IBD, there are significant inherent challenges in deciphering whether dysbiosis contributes to disease pathogenesis or is merely a secondary change associated with the disease process. Also, it is not possible to exclude that these data could be significantly affected by environmental and host genetic factors, including unappreciated inflammation. Therefore, alternative approaches are required to unravel the true impact of IBD-associated dysbiosis on host responses to rule out any extrinsic factors and focus squarely on host–microbial interaction. To this end, we have used the GB mouse system to re-create IBD-associated dysbiosis in mice.
As reported previously,19, 20 we showed that the microbiota harvested from human subjects, including IBD patients, can stably colonize the gastrointestinal tract of mice. However, the dysbiotic communities present in the stool samples of IBD donors were not completely recapitulated in hGB mice in our study. For example, although an increased abundance of Proteobacteria was observed in IBD donor stool samples, this phenomenon was not fully reproduced in hGB mice. Because intestinal inflammation is required for Proteobacteria to bloom in the gut, the lack of complete reproducibility may be owing to different levels of inflammation between IBD patients (donors) and recipient mice. Likewise, the predicted functionality differences in the donor group of microbiota samples did not fully correlate with hGB mice because this analysis is based on 16S rRNA sequencing results (Figure 3 and Supplementary Figure 1). Given that IBD patients do harbor genetic defects in the host response to microbes, it is possible that these defects foster dysbiosis that is not maintained in the murine gut. Likewise, other features related to diet, environment, and host factors presumably shape the microbiota after reconstitution in mice and represent a limitation of this model. Nonetheless, certain features of the dysbiotic microbiota, including decreased bacterial diversity and richness, also were observed after reconstitution in hGB mice. Ultimately, we did note lower bacterial diversity, altered bacterial metabolite levels, differential host response profiles, and an apparent difference in the potential to develop inflammation after transfer of IBD-associated dysbiotic microbiota, showing the potential utility of this model for assessing various aspects of the host-microbiota relationship that may be critical for the pathogenesis of IBD.
Beyond bacterial community analysis, focusing on the functional changes of the microbiota caused by dysbiosis in these experiments has yielded more thought-provoking results. A gene-based predictive analysis showed an increase in genes involved in the pathogenic features of bacteria, including flagellar assembly and bacterial motility proteins, in CD hGB mice.35 Consistent with the gene-based predictive analysis, the actual flagellin load in feces tended to be higher in CD hGB mice compared with HC hGB (Supplementary Figure 2A). Notably, the higher production of flagellin by the CD microbiota became more obvious when the microbiota was used to colonize GF Il10-/- mice (Supplementary Figure 2B). This result suggests that the CD microbiota contains bacteria that have the potential to express flagellin, but flagellin expression is not turned on under physiological conditions. In certain microenvironments, such as during intestinal inflammation and/or owing to genetic variation, these bacteria start producing flagellin and may exacerbate intestinal inflammation. Consistent with this notion, a previous study showed that the immune environment regulates the production of flagellin by the commensal microbiota.29 Thus, the CD microbiota may become more pathogenic when it is exposed to certain stimuli. In addition to flagellin production, the gene-based predictive analysis showed that there are differences in the metabolic profile of IBD hGB and HC hGB mouse microbiotas. An increased level of gene expression related to bacterial metabolic functions, likely crucial for survival and colonization under normal conditions, was observed in HC hGB. In UC hGB mice, expression of propanoate metabolism genes was decreased significantly compared with HC hGB mice, suggesting that the UC microbiota has a defect in the generation of SCFAs. Consistent with the gene-based predictive analysis, the actual amount of luminal SCFAs tended to be lower in UC hGB mice compared with HC hGB mice. In contrast, succinate levels tended to be higher in UC hGB mice. Because certain bacteria can generate SCFAs from succinate,36 this pathway appears to be compromised in the UC microbiota. Indeed, expression of genes related to succinate metabolism (propanoate metabolism genes) was significantly lower in UC hGB mice. In CD hGB, the gene-based predictive analysis showed that the expression of butanoate (butyrate) metabolism pathway genes was significantly lower than HC hGB mice. However, one surprising result shown by the metabolome analysis was that the luminal levels of SCFAs (propionate and butyrate) tended to be higher in CD hGB mice compared with HC hGB mice. Given that this evidence of microbiota dysfunction is consistent with the previous larger cohort study that analyzed the function of IBD microbiota directly in patients,12, 34 it is conceivable that dysbiosis, reproduced in hGB mice, resembled, at least to some extent, the functional abnormalities found in the microbiota of IBD patients. Moreover, it is noteworthy that the bacterial functions associated with CD and UC seem to be distinct because both types of IBD showed similar levels of reduction in bacterial diversity and richness. These results suggest that the functional impact of the dysbiotic microbiota on host immunity might be different in CD compared with UC.
Gut dysbiosis and altered host gene expression may precede inflammation as reported in histologically normal ileal tissues from newly diagnosed patients with CD colitis.11, 13 This evidence supports a primary role for IBD-associated dysbiosis, but cannot rule out a primary effect of altered host gene expression driving subclinical inflammation and secondary dysbiosis. In the present study, we have shown that hGB mice colonized with the CD microbiota showed a stronger expression of epithelial host–defense responses (eg, Reg3b/g and Saa3) compared with hGB mice colonized with the HC microbiota. These genes are known to be induced in response to the attachment of bacteria to the intestinal epithelium, implying that the CD microbiota contains potential pathobionts residing near the intestinal epithelium.37 Therefore, dysbiosis may promote altered gene expression owing to altered host-microbe interactions. Consistent with this finding, previous studies have reported that potentially pathogenic, intestinal epithelium-adhering bacteria accumulate in CD patients.8, 38 In addition to epithelial responses, our data showed alterations in downstream gene expression associated with gut leukocytes. A variety of genes associated with myeloid and lymphoid cells and their activation were increased after reconstitution with IBD compared with HC microbiota. Of note, genes related to proinflammatory features of mononuclear phagocytes, such as IL1β, Nox2, and inducible nitric oxide synthase were induced in response to colonization by the pathogenic microbiota in CD. Furthermore, the gene expression profile in CD hGB mice showed activation of both innate (NK, ILCs) and adaptive (Th1, Th17) lymphocytes. Because accumulating evidence indicates that microbiota-induced IL1β is a key cytokine that promotes differentiation and activation of Th17 cells as well as group 3 ILCs,39 pathogenic microbiota in CD patients may elicit these lymphocyte responses through activation of a mucosal IL1β signaling pathway (Il1b, il1r2). Interferon (IFN)-γ is a cytokine that is believed to be involved in the pathogenesis of CD,13 and although it was not detected by the gene expression analysis per se, genes induced by IFN-γ (Ifi205, Ciita, Ido, Nos2) were over-represented in CD hGB. Thus, IFN-γ signaling also might be activated by the dysbiotic microbiota in CD patients. Expression of other genes, such as those related to solute carrier families and cytochrome P450 families, were decreased in CD hGB compared with HC hGB. It is noteworthy that these immunologic features, both up-regulation and down-regulation of genes, observed in CD hGB mice resemble core gene expression patterns in intestinal mucosa of newly diagnosed, treatment-naive patients with CD.13 Thus, our data support the possibility that immunologic alterations in CD patients are driven by abnormal gut microbiota.
As was the case with the luminal metabolites, we noted differences in the host gene expression profiles in UC vs CD hGB mice. Although host genes induced by colonization of UC microbiota showed some overlap with those induced by CD microbiota, genes related to mononuclear phagocyte development and activation, functions of NK cells, Th1, and ILC3s were not up-regulated in UC hGB. In contrast, in UC hGB mice, expression of genes related to Th17 immunity was greater than that observed in CD hGB mice, although the microbiota from both types of diseases provoked Th17 responses. Thus, our results have shown that the dysbiotic microbiota associated with CD and UC differently affect host immunity.
As further evidence of the negative impact of the dysbiotic microbiota on host immune regulation, we assessed the colitogenic capacity of the dysbiotic microbiota. Notably, only 1 of 5 CD microbiota samples induced the development of inflammation in WT GF recipient mice, despite the increase of core proinflammatory gene expression (Figure 6). This indicates that IBD-associated dysbiosis is not sufficient enough to trigger colitis in most cases. On the other hand, CD-associated microbiota, but not HC-associated microbiota, was capable of inducing colitis in IBD-prone mice. This suggests that additional factors (ie, genetic susceptibility) might be required to potentiate the colitogenic capacity of IBD-associated dysbiotic microbiota. The ability to transfer colitogenic microbiota has been shown previously using mouse models,40 but the present study is an important demonstration that the IBD patient-derived microbiota possesses colitogenic capacity. Thus, the dysbiotic microbiota in CD patients seems to be pathogenic, and may contribute to the manifestation of intestinal inflammation. Furthermore, IBD hGB mice may serve as an effective tool to investigate the mechanisms by which the dysbiotic microbiota promotes aberrant host responses.
It is noteworthy that the microbiota isolated from patients with UC failed to induce the development of colitis even in GF IL10-/- mice (Supplementary Figure 3). Consistently, there were no obvious changes in proinflammatory gene expression in UC hGB mice compared with CD hGB mice (Figure 5). However, in this study, we only tested the colitogenic capacity of the microbiota isolated from 3 UC patients. Therefore, it is possible that the dysbiotic microbiota in certain populations of UC patients is capable of inducing the development of colitis-like, CD-associated microbiota. Moreover, although the majority of IBD-associated dysbiotic microbiota is not immediately pathogenic under experimental conditions (3 weeks after colonization), it is possible that longer exposure to the pathogenic microbiota can lead to the development of intestinal inflammation even in WT recipients. Further investigation is needed to better characterize the colitogenic capacity of IBD-associated dysbiotic microbiota.
Although the humanized GB mouse system is considered to be the gold standard method for evaluation of function of the human microbiota in vivo, there are potential pitfalls to consider. The concept of species-specific microbiota has been illuminated in experiments showing that maturation of the gut-associated immune system is abrogated in GF mice colonized with human- or rat-derived microbiota as compared with mouse-derived microbiota.41 Furthermore, hGB mice in the aforementioned study were unable to combat infectious pathogens effectively. In contrast, other reports have shown that human microbiota sufficiently promotes murine immune responses related to the induction of colitis.37, 38, 42 One explanation for this discrepancy lies in the sites used for analysis. The former study characterized host immune responses after colonization with a human microbiota in the small intestine. In the latter studies, the colon of hGB mice was used to assess host immune responses. Consistent with this notion, it has been reported that immune regulation by the resident microbiota in the small and large intestine is different.43, 44 Our results have confirmed that colonization with a human microbiota induces host immune responses in the colonic mucosa of hGB mice. It is noteworthy that host immune activation by the species-mismatched microbiota seems to be reduced compared with colonization by species-specific microbiota, although CD microbiota was able to induce colitis development in IL10-deficient mice. Thus, although human microbiota can elicit immune development at least in the colon of gnotobiotic mice, the immune responses induced by the species-mismatched microbiota may be weaker than those induced by the species-matched microbiota.
In this study, we have shown that the hGB mouse system is a useful model that can be used to investigate the functional role of IBD-associated dysbiotic microbiota. Dysbiosis alters downstream host immune responses that could serve a contributory role in disease persistence and flares. Taken together, these data suggest that dysbiosis in IBD patients is not merely a secondary change of intestinal inflammation. Although it still remains unclear what drives dysbiosis in IBD, gut dysbiosis is a key player in the vicious cycle of intestinal inflammation in IBD.
Acknowledgment
The authors thank Dr Kathryn A. Eaton, Sara Poe, Chriss Vowles, and Natalie Anderson at the University of Michigan Germ-free Animal Core for assistance with germ-free experiments, Anna Romans for procurement of human stool samples, Ingrid L. Bergin at the Unit for Laboratory Animal Medicine In Vivo Animal Core at the University of Michigan for assistance with pathology interpretation and images, and the University of Michigan Medical School Host Microbiome Initiative for support.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by Japan Society for the Promotion of Science Postdoctoral Fellow for Research Abroad (H.N.-K. and S.K.); the Crohn’s and Colitis Foundation of America, a Young Investigator Grant from the Global Probiotics Council, and the Michigan Gastrointestinal Research Center DK034933 (N.K.), and National Institutes of Health grant DK087708-01 (J.Y.K.).
Supplementary Material
References
- 1.Kamada N., Seo S.U., Chen G.Y. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 2.Sartor R.B. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 3.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Taurog J.D., Richardson J.A., Croft J.T. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sellon R.K., Tonkonogy S., Schultz M. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng T., Wang L., Schoeb T.R. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamboli C.P., Neut C., Desreumaux P. Dysbiosis in inflammatory bowel disease. Gut. 2004;53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darfeuille-Michaud A., Boudeau J., Bulois P. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 9.Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn's disease. Int J Med Microbiol. 2002;292:185–193. doi: 10.1078/1438-4221-00201. [DOI] [PubMed] [Google Scholar]
- 10.Barnich N., Darfeuille-Michaud A. Adherent-invasive Escherichia coli and Crohn's disease. Curr Opin Gastroenterol. 2007;23:16–20. doi: 10.1097/MOG.0b013e3280105a38. [DOI] [PubMed] [Google Scholar]
- 11.Gevers D., Kugathasan S., Denson L.A. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan X.C., Tickle T.L., Sokol H. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haberman Y., Tickle T.L., Dexheimer P.J. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupp C., Robertson M.L., Wickham M.E. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Winter S.E., Winter M.G., Xavier M.N. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamada N., Chen G.Y., Inohara N. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic A.D., Xavier R.J., Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seedorf H., Griffin N.W., Ridaura V.K. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faith J.J., Rey F.E., O'Donnell D. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094–1098. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faith J.J., Ahern P.P., Ridaura V.K. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh P.J., Ridaura V.K., Faith J.J. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecht G., Bar-Nathan C., Milite G. A simple cage-autonomous method for the maintenance of the barrier status of germ-free mice during experimentation. Lab Anim. 2014;48:292–297. doi: 10.1177/0023677214544728. [DOI] [PubMed] [Google Scholar]
- 23.Paik J., Pershutkina O., Meeker S. Potential for using a hermetically-sealed, positive-pressured isocage system for studies involving germ-free mice outside a flexible-film isolator. Gut Microbes. 2015;6:255–265. doi: 10.1080/19490976.2015.1064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss P.D., Westcott S.L., Ryabin T. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozich J.J., Westcott S.L., Baxter N.T. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langille M.G., Zaneveld J., Caporaso J.G. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segata N., Izard J., Waldron L. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishima E., Fukuda S., Shima H. Alteration of the intestinal environment by lubiprostone is associated with amelioration of adenine-induced CKD. J Am Soc Nephrol. 2015;26:1787–1794. doi: 10.1681/ASN.2014060530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cullender T.C., Chassaing B., Janzon A. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridaura V.K., Faith J.J., Rey F.E. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furusawa Y., Obata Y., Fukuda S. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 32.Smith P.M., Howitt M.R., Panikov N. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arpaia N., Campbell C., Fan X. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davenport M., Poles J., Leung J.M. Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:723–731. doi: 10.1097/MIB.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duck L.W., Walter M.R., Novak J. Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe Y., Nagai F., Morotomi M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol. 2012;78:511–518. doi: 10.1128/AEM.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atarashi K., Tanoue T., Ando M. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palm N.W., de Zoete M.R., Cullen T.W. Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw M.H., Kamada N., Kim Y.G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elinav E., Strowig T., Kau A.L. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung H., Pamp S.J., Hill J.A. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eun C.S., Mishima Y., Wohlgemuth S. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10-/- mice. Infect Immun. 2014;82:2239–2246. doi: 10.1128/IAI.01513-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atarashi K., Tanoue T., Shima T. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sano T., Huang W., Hall J.A. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.