Abstract
Biologic research is experiencing a transformation brought about by the ability of programmable nucleases to manipulate the genome. In the recently developed CRISPR/Cas system, short RNA sequences guide the endonuclease Cas9 to any location in the genome, causing a DNA double–strand break (DSB). Repair of DSBs allows the introduction of targeted genetic manipulations with high precision. Cas9–mediated gene editing is simple, scalable, and rapid, and it can be applied to virtually any organism. Here, we summarize the development of modern gene editing techniques and the biology of DSB repair on which these techniques are based. We discuss technical points in applying this technology and review its use in model organisms. Finally, we describe prospects for the use of gene editing to treat human genetic diseases. This technology offers tremendous promise for equipping the nephrology research community to better model and ultimately, treat kidney diseases.
Keywords: molecular genetics, gene therapy, transgenic mouse
“I’m all for the scissors. I believe more in the scissors than I do in the pencil.” – Truman Capote
Gene editing can be defined as the process of permanently altering cellular DNA sequences to introduce a desired mutation. Although manipulating DNA has formed a foundation for modern biomedical research efforts for the last 40 years, remarkable new approaches are revolutionizing this process. Mutations can now be introduced into the genomic DNA of cells from virtually any species. These advances have been made possible using custom nucleases, proteins that can now be targeted to specific sites in the genome, where they can correct mutations, introduce new DNA, activate or suppress transcription, and perform other functions. Increasingly, these exciting and powerful approaches are affecting clinical applications in addition to basic research.
Early approaches to site–specific genome modification relied on homologous recombination, a powerful method to introduce a sequence of interest to a specific DNA locus. This discovery was the subject of a Nobel Prize to Mario R. Capecchi, Sir Martin J. Evans, and Oliver Smithies in 2007, but it is also a relatively inefficient and time-consuming technique requiring considerable operator experience.1,2
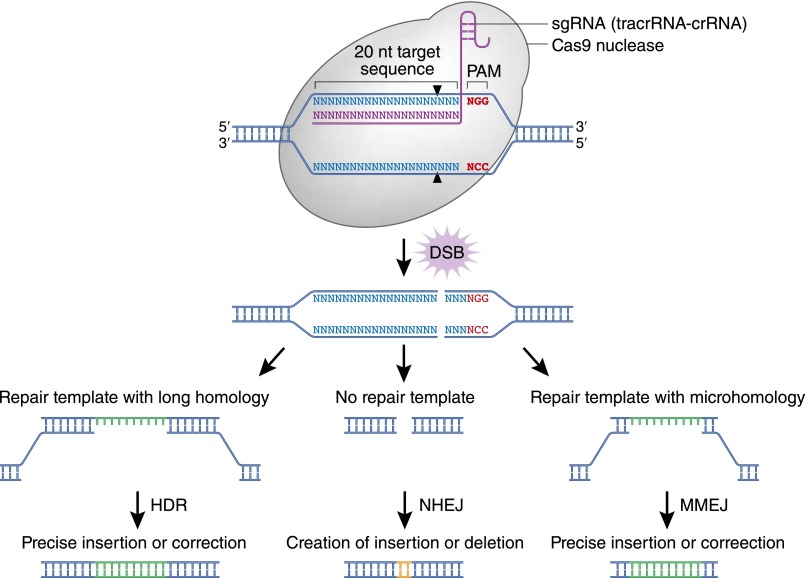
Research into cellular repair mechanisms for DNA double–strand breaks (DSBs) underpins newer approaches to gene editing. Cells have two primary repair pathways: homology-directed repair (HDR) and nonhomologous end joining (NHEJ) (Figure 1). We will discuss a third pathway, microhomology–mediated end joining (MMEJ), below. HDR uses homology regions between the exposed ends of a DSB and a donor DNA template to repair the cut region. By contrast, in the NHEJ pathway, the broken ends of the DSB are ligated back together. Because this process involves processing of the broken ends to generate compatible ends, the repair often results in small deletions or insertions.3 The important concept is that, if one can introduce a DSB at a desired locus, then either mode of repair might come into play, leading to a permanent genetic alteration. As a consequence, investigators have searched intensively for methods of inducing DSBs at a precise location.
Figure 1.
Simplified schematic illustrating DSB repair mechanisms induced by CRISPR/Cas9. The three pathways, HDR, NHEJ, and MMEJ, are used for different gene editing purposes. The black triangles indicate locations of the cut site. NHEJ leads to frequent small insertions or deletions that can lead to disruptive frameshift mutations and premature stop codons. This approach is best suited for generating point mutants or knocking out a gene. Both HDR and MMEJ pathways can be used to introduce longer DNA sequences, but they each require an exogenous donor DNA template. These mechanisms are well suited for introducing precise mutations or knocking in an epitope tag or reporter gene, such as green fluorescent protein. Modified from Sakuma et al.,34 with permission.
The early forms of customized nucleases, such as Zinc Finger Nucleases (ZFNs) and transcription activator–like effector nucleases (TALENs), were engineered to bind specific DNA sequences using knowledge of specific DNA recognition mechanisms shared by transcription factors. For example, zinc finger (ZF) domains in eukaryotic transcription factors4,5 and transcription activator–like effectors (TALEs) from Xanthomonas bacteria6–8 recognize specific DNA sequences. By designing a cDNA to encode tandem repeats of these ZF or TALE modules, synthetic proteins result that target a specific DNA sequence up to 18 bp.9 By fusing the synthetic ZF or TALE with a nuclease, such as Fokl (a nonspecific restriction endonuclease from Flavobacterium okeanokoites), a DSB is induced at the precise location where the ZF or TALE binds.10
Both of these approaches advanced the field considerably, but they also suffer from major limitations. Assembling tandem repeats of ZFs and TALEs is laborious because of the repetitive sequences, and a single ZF nuclease may not be active; therefore, researchers often order several to produce one active nuclease.11 Although TALEs are highly independent of neighboring repeats and not as labor intensive to create, binding sites are limited to sequences that start with a T base,12 and they also require considerable validation.
In contrast to ZFNs and TALENs, the recent clustered, regularly interspaced, short palindromic repeats (CRISPR)–associated endonuclease (Cas) system is considerably simpler to design and use. It is also cheaper. A single ZF costs about $5000, and a single TALEN might be $500 if a laboratory constructs it on their own. By contrast, a single CRISPR/Cas experiment costs only the $30 required to synthesize short oligonucleotides. The CRISPR/Cas technology is based on the bacterial adaptive immunity system entitled CRISPR. This system provides adaptive immunity against foreign DNAs, such as viruses and plasmids. The type 2 CRISPR system first identifies foreign DNA, and then, the Cas cuts this foreign DNA into small pieces. These are subsequently incorporated into a CRISPR locus within the bacterial genome. These loci are then transcribed to generate small RNAs, which will eventually guide the Cas endonuclease to that same foreign DNA sequence, on the basis of RNA-DNA sequence complementarity.13–15 A distinguishing feature of the CRISPR-Cas defense is that it uses RNA-DNA binding to guide the nuclease to specific genetic sequences rather than protein-DNA binding as with ZFNs and TALENs.
Development of the CRISPR/Cas System
The CRISPR/Cas system offers a dramatically simpler target design approach than either ZFN or TALEN methods. In fact, CRISPR technology has become so flexible, powerful, and easy to use that Science magazine named it the 2015 Breakthrough of the Year.16 Of the three types of CRISPR systems identified to date, type 2 has been adapted for development because of its simplicity.17,18 In type 2 systems, the CRISPR array is transcribed as preclustered, regularly interspaced, short palindromic repeats RNA (precrRNA), which hybridizes with target–independent transactivating clustered, regularly interspaced, short palindromic repeats RNAs (tracrRNAs) to form a complex with Cas9 proteins. During crRNA maturation, each crRNA-tracrRNA-Cas9 complex is released as an active DNA nuclease. The complex first scans DNA for a protospacer motif (PAM) and binds to sequences exhibiting complementarity to the 20-nucleotide cRNA that are immediately followed by the PAM.19 The canonical PAM for the most commonly used Streptococcus pyogenes Cas9 is 5′-NGG-3′, where N can be any nucleotide (Figure 1).18
Scientists have simplified the dual tracrRNA-crRNA system by engineering a single guide RNA (sgRNA) that combines tracrRNA-crRNA functions. This leaves a two-component system, in which one can target any DNA sequence simply by introducing the Cas9 nuclease and appropriate sgRNA into target cells.17 An sgRNA target can be any sequence as long as it is immediately followed by the PAM sequence, which is so common that this is not a practical limitation. There are a variety of online resources available to identify genomic loci with unique sequences to minimize off-target effects (Table 1).
Table 1.
CRISPR/Cas9 resources
| Resource Name | Description | Web Address |
|---|---|---|
| Benchling-crispr | Highly integrated sgRNA design tool that supports ≥40 reference genomes; automatically annotates genome coordinates with the exon and coding DNA sequence | https://benchling.com/crispr |
| Optimized CRISPR design, Zhang laboratory, Massachusetts Institute of Technology | Input sequence region of interest sequence and software lists candidate sgRNA sequences with scores on the basis of the number of off targets and positions of mismatches; 16 different species | http://crispr.mit.edu/ |
| E-CRISP, Boutros laboratory, German Cancer Research Center | Highly customizable sgRNA calculator including efficiency and possible off–target sites | http://www.e-crisp.org/E-CRISP/ |
| CRISPRdirect, Database Center for Life Sciences, Japan | Search for sgRNAs by gene accession number, genomic location, or sequence; >200 species and sgRNA priority on the basis of the number of off-target sites | http://crispr.dbcls.jp/ |
| The CRISPR page at CNB | A compendium of landmark articles, background information, and links to other CRISPR resources | http://wwwuser.cnb.csic.es/~montoliu/CRISPR/ |
| Google Group | A forum for real-time discussion of all topics related to CRISPR/Cas; currently, 2290 topics and 3959 members | https://groups.google.com/forum/#!forum/crispr |
| Addgene’s CRISPR/Cas9 Plasmids and Resources | Excellent overview of CRISPR/Cas9 and the best place to acquire all plasmids needed for gene editing | https://www.addgene.org/crispr/ |
As of May of 2016.
The sgRNA target sequence is typically synthesized as a partially complementary oligonucleotide and cloned into a plasmid that encodes a cassette for expression of the sgRNA and Cas9. The plasmid may also encode either a fluorescent marker or resistance gene, enabling enrichment of transfected cells. The delivery method varies depending on the cell line: standard transfection methods may be sufficient for easy to transfect cell lines, whereas electroporation or viral delivery can be used for difficult to transfect cell lines. The activity of sgRNAs can be tested by several straightforward methods (Table 2).
Table 2.
Screening of sgRNA activity
| Method | Description |
|---|---|
| Surveyor or T7E1 mismatch cleavage assay | Uses enzyme to cleave mismatched heteroduplex DNA after denaturation and reannealing; cleaved fragments represent mutated DNA on agarose gel, allowing estimation of efficiency |
| Tracking of indels by decomposition (https://tide.nki.nl/) | Online program that determines the ratio of mutated alleles in a background of wild-type alleles; chromatogram from sequencing of the region around the sgRNA site is uploaded, and the algorithm calculates the ratio of mutated alleles |
| Deep sequencing | The most sensitive method: it provides not only efficiency of sgRNA edits but also, detailed sequence–based information on the kinds of mutations that each gRNA can generate |
Basic Knockout and Knock–in CRISPR/Cas Applications
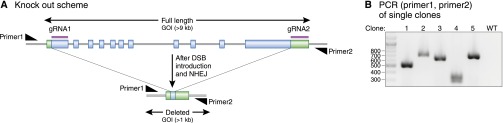
To knock out a gene, an sgRNA can be designed to bind to the 5′ end of the coding region. After a DSB is introduced and repaired through NHEJ, small deletions or insertions will be created, many causing frameshift mutations that result in nonsense-mediated decay. Alternatively, two sgRNAs can be designed at the 5′ and 3′ ends of the gene to delete the whole genic region (Figure 2A). In the former approach, it may be time consuming to screen clones, because DNA from each clone has to be individually sequenced to identify frameshift mutations. In the latter approach, screening can be carried out efficiently by PCR. For example, Figure 2 depicts one such strategy, in which two sgRNAs flank a long genic region, and the screening primers are located upstream and downstream of the sgRNAs. With a 1-minute extension PCR, deleted alleles yield PCR products, whereas the wild-type allele is too large to amplify (Figure 2).
Figure 2.
Schematic illustration of a gene knockout using CRISPR/Cas9 gene editing. (A) Two gRNAs were designed on the coding start region and 3′ untranslated region (UTR), respectively, of gene of interest (GOI) to cut out about 9 kb. The light blue rectangles and green rectangles show coding sequences and UTRs, respectively. The screening primers are located outside of the gRNAs. (B) PCR is performed on DNA isolated from individual clones of cells subject to gene editing. The extension time is purposefully brief, so that the 9-kb WT allele cannot be amplified. Different sizes of deleted alleles are detected, because each clone has different deletion lengths near the DSB.
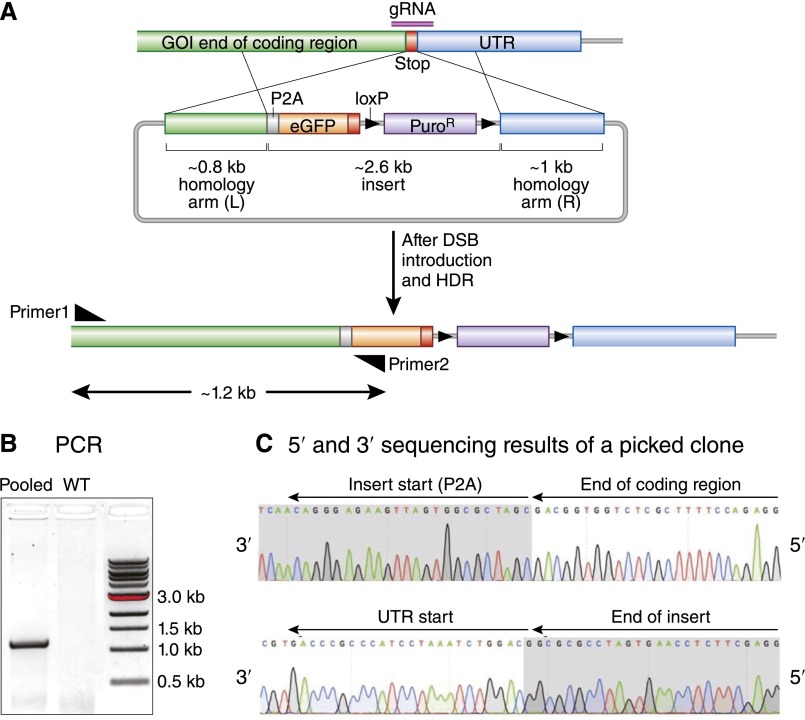
To introduce specific genetic alterations, such as reporters and disease-specific mutations, knock-in strategies may be used. In this case, an additional DNA template must be provided along with the plasmid-encoding Cas9 and the sgRNA. This additional DNA must have two homology arms and the desired new DNA sequence in between them. Insertion of small epitope tags can be accomplished with single-stranded oligonucleotides using homology arms of about 60 nucleotides. Insertion of a larger sequence, such as green fluorescent protein, requires a plasmid donor with longer homology arms of around 0.8 kb. When this donor DNA with homology arms adjacent to the cut site is present, the HDR pathway may be triggered, and the novel sequences located in between the homology arms will be incorporated. Figure 3 shows successful introduction of a 2.6-kb insert using 0.8- to 1-kb homology arms. A donor-specific primer and a primer that anneals outside the region spanned by the homology arms should be used to screen clones with knock-in alleles. Clones with the inserted alleles should give PCR bands, whereas wild-type alleles do not (Figure 3B).
Figure 3.
Illustration of a C–terminal P2A-eGFP–floxed puromycin expression cassette knock-in for gene of interest (GOI) by CRISPR/Cas9 editing. (A) A gRNA was designed to overlap with the stop codon of a GOI, and the donor vector contained homology arms of 800–1000 bp. After HDR, a P2A-eGFP–floxed puromycin expression cassette will be inserted in frame just after the coding region and before the stop codon. Screening primer 1 was designed outside of the left homology arm to prevent detection of the donor plasmid. The insert–specific primer 2 was designed on eGFP. (B) Results of pooled genomic PCR from mouse embryonic stem cells that detects the insertion of the desired sequence into the genome. (C) Sequence results from a single clone indicate precise integration of the desired sequence into the genome. Only the results of the 5′ sequence are shown. UTR, untranslated region.
It is best practice to design sgRNAs and donor homology arms in such a manner that the homology arm sequences do not contain the full sgRNA target sequences to ensure that the plasmid donor and repaired locus will not be subject to mutagenic NHEJ outcomes. This can be accomplished by mutating the PAM sequence of the donor plasmid.
New Applications of CRISPR Using Modified Cas9
Although the CRISPR/Cas9 system produces DSBs efficiently at target sites, it can also cause off-target mutations at unintended sites.20 This is because sgRNAs can tolerate mismatched nucleotides and even noncanonical PAMs. Recent developments, however, have significantly improved targeting specificity. One solution involves a nickase version of Cas9 containing a D10A or N863A mutation to inactivate one of two conserved nuclease domains.21,22 These nickase mutants use two sgRNAs on opposite strands to make paired single–strand breaks. Off-target nicks caused by single nickases are less mutagenic than DSBs, because nicks are repaired with high fidelity.23 The newest addition is enhanced specificity SpCas9 nucleases, which contains triple mutations to neutralize positive charges in the nontarget strand groove, weakening off–target strand binding. SpCas9 is the simplest solution available and outperforms another recent strategy—the truncated sgRNA strategy, which uses sgRNAs shortened by two or three nucleotides to increase sensitivity to mismatches.24,25
CRISPR/Cas9 can also be used to recruit effector domains to turn gene transcription off or on (CRISPRi or CRISPRa). Catalytically dead (dCas9) variants, which lack nuclease activity but retain DNA binding capacity, can repress transcription sterically, either by blocking transcription initiation or elongation, when the sgRNA directs dCas9 binding to a promoter or exon. Transcriptional repression (CRISPRi) can also be achieved by fusing dCas9 to a transcriptional repressor domain, such as Krüppel–associated box repressor, and targeting the sgRNA near a transcriptional start site.26
Activation of target gene transcription (CRISPRa) is also readily accomplished using dCas9. Fusion of an artificial repeat of the VP64 transcriptional activation domain to dCas9 is sufficient to recruit RNA polymerase and initiate transcription at a site determined by dCas9 binding (for example, near a transcriptional start site).27 This system has recently been improved by fusing several different transcriptional activation domains to dCas9 in tandem. A tripartite construct consisting of the activation domains VP64, p65, and Rta, when fused to dCas9, showed 22- to 320-fold greater activation of endogenous target genes. This dCas9-VP64, p65, and Rta approach also allowed multiplexed activation of up to four genes in the same cell simply through introduction of a collection of guide RNAs targeting those four genes.28 Finally, dCas9 fused to the catalytic domain of the human acetyltransferase p300 has been reported to accomplish gene–specific epigenome editing. The fusion protein catalyzes histone H3 lysine 27 acetylation around genomic sites targeted by sgRNAs.29
CRISPR/Cas technology has also been used successfully for large–scale, high–throughput functional screening. For example, by using a lentiviral knockout sgRNA library, a group has identified genes essential for the intoxication of cells by anthrax and diphtheria toxins.30 Their method involved lentiviral infection of human cultured cells with the sgRNA library to establish library cell lines followed by toxin screening. Surviving cells were then analyzed for their individual sgRNA sequence by deep sequencing, thereby identifying the gene knockout that protected the cell from death by either anthrax or diphtheria.
Genome Engineering in Model Organisms
CRISPR/Cas9 technology can be readily applied to genome modification in model organisms. Genetically modified mice, for example, are used routinely for analysis of gene function, but generating mouse lines with one or multiple targeted mutations remains expensive and time consuming. The fastest timeline for creating a mouse knockout by standard homologous recombination is about 1 year, and this does not include time required to construct the targeting plasmid. With a CRISPR approach, injection of an sgRNA and Cas9 mRNA or protein into the one-cell embryo creates germline knockout mice in one generation—4 weeks.31 Moreover, multiple genes can be targeted at once, which is impossible using standard approaches.32
One-step generation of mouse knock-ins is also possible, but it is less efficient than creating knockouts. Insertion of small epitope tags can be accomplished by coinjecting an sgRNA, Cas9, and single-stranded oligonucleotide containing the epitope tag and homology arms of about 60 nucleotides. Larger insertions, which require plasmid donors with longer homology arms as described in the previous section, are considerably more challenging to accomplish in a one-step procedure, despite early reports to the contrary.33 Some investigators have resorted to creating the knock-ins in mouse embryonic stem cells using CRISPR/Cas9 followed by traditional blastocyst injection.
Although gene editing with CRISPR/Cas9 has been described in myriad other model organisms, including rat, zebrafish, frog, monkey, mosquito, pig, plants, and more, there still remains room to improve gene knock–in efficiency and applicability to organisms with a low HDR frequency. An intriguing recent development, called CRISPR/Cas9 precise integration into target chromosome, takes advantage of a separate DNA repair mechanism called MMEJ to facilitate knock-in. In contrast to the conventional HDR, the MMEJ process requires only short homologous sequences of 5–25 bp. This method features a donor plasmid with short homology arms, and the whole template region is flanked by sgRNA target sites. Therefore, Cas9 generates DSBs in both the genomic DNA and the donor vector, exposing the microhomology sequences that are then used for MMEJ–mediated knock-in of the insert.34 The precise integration into target chromosome system has been reported to have about 2.5-fold higher efficiency than HDR–mediated knock-ins in cultured cells and has been successfully used in silkworm, zebrafish, and frog. It certainly seems to be a promising technique that needs to be tested in mouse embryos.35 Additional studies into MMEJ repair mechanisms might improve the methodology further to enable precise and efficient knock-in of large insertions in whole-animal applications.
Recent reports also document the first successful application of gene editing approaches to the study of kidney disease. Freedman et al.36 recently reported protocols for the directed differentiation of human pluripotent stem cells (hPSCs) to kidney organoids. Using CRISPR, they knocked out the podocalyxin gene in hPSC.36 Podocalyxin regulates tight junctions and is expressed in podocytes in mature kidney. After differentiation of wild–type or podocalyxin knockout hPSC to kidney organoids, Freedman et al.36 reported disappearance of protein tracks of the other podocyte proteins synaptopodin and ZO-1 along with a decrease in the gap width between podocyte-like cells.36 This proof of principle experiment establishes a novel platform for investigation of kidney genes in hPSCs.
Freedman et al.36 also generated biallelic loss of function mutations in the polycystic kidney disease 1 (PKD1) and PKD2 genes that are responsible for development of human PKD. After differentiation of these lines and wild-type cells into kidney organoids followed by extended culture, Freedman et al.36 observed large cyst–like structures apparently emanating from the tubular compartment in organoids. Although these were uncommon at 6% of organoids, this seems to be a highly promising new approach to model cystogenesis in PKD in vitro.
Gene Therapy Protocols Using Genome Editing Technologies
One of the fastest growing applications of CRISPR/Cas9 approaches is in gene therapy to cure disease. Sickle cell anemia, for example, is caused by a single-nucleotide mutation in the β-globin gene, which encodes a protein subunit of hemoglobin (Hb) found in red blood cells. The mutation does not alter the ability of Hb to carry oxygen, but under hypoxic conditions, it does promote polymerization of intracellular Hb molecules, which in turn, reduces elasticity of the red blood cell and causes shape change called sickling. The now rigid blood cells block small capillaries, causing occlusion and ischemia.37
One strategy to cure sickle cell anemia is to activate expression of fetal Hb, HbF, which does not require the β-globin subunit and therefore, does not cause sickling. HbF expression is normally downregulated after 6 weeks of age by a transcriptional repressor encoded by the BCL11A gene.38 A group from the Dana Farber Cancer Institute used a CRISPR/Cas9 to perform saturation mutagenesis on the human enhancer (a noncoding element) that regulates BCL11A expression. The group identified sgRNAs that caused complete knockout of BCL11A, with subsequent upregulation of HbF expression.39 This important work provides a framework for therapeutic gene editing in hematopoietic stem cells to restore HbF expression and ameliorate the sickle cell phenotype. The particular enhancer targeted is specific to erythroid elements, thus preserving BCL11A expression in nonerythroid tissues and reducing side effects.
Another disease that seems to be amenable to CRISPR–based gene therapy is Duchenne muscular dystrophy (DMD). This muscle disease mainly affects boys who develop progressive skeletal muscle and heart degeneration and die in their mid-20s. DMD is caused by genetic defects in the gene dystrophin, a component of the muscle cell cytoskeleton. It has been known for some time that manipulations causing the transcriptional machinery to skip the exon containing the mutation result in truncated dystrophin protein that still functions adequately enough to reverse the muscle pathology.40
Three independent groups very recently reported CRISPR strategies to achieve exon skipping in the dystrophin gene, resulting in improved muscle function in mouse models of DMD.41–43 These groups (Olson and coworkers,41 Gersbach and coworkers,41 and Wagers and coworkers43) all used adenoassociated viruses (AAVs) to deliver the CRISPR/Cas9 components. AAVs are nonintegrating viruses that are safe for human use, and they efficiently transduce skeletal and heart muscle. Investigators designed novel AAVs that encoded the Cas9 endonuclease along with paired sgRNAs that flank the mutated exon. Simple intravenous injection of the custom AAV resulted in efficient excision of the mutant exon, restoring myofiber and cardiomyocyte function. Wagers and coworkers43 also showed that excision occurred in muscle stem cells, providing hope that this approach may provide a durable cure if adapted to humans.
Potential Applications to Kidney Disease
Applications of CRISPR–mediated gene editing to the treatment of kidney disease can be envisioned. For example, the shortage of transplantable organs remains a critical roadblock to the treatment of ESRD. Xenotransplatation of porcine kidneys has been considered a viable solution to this problem, but concerns about transmission of porcine endogenous retroviruses (PERVs) to humans has limited progress in this field. PERVs cannot be eliminated by biosecure breeding, because these elements are encoded within the pig genome. Recently, Church and coworkers44 used a CRISPR approach to eradicate a gene critical to PERV function at 62 independent sites within the genome of a pig cell line during a single round of CRISPR. This result provides a proof of principle that the same approach could be used to create pig embryos with inactivated PERV sequences and subsequently, cloned pigs with kidneys free from retroviruses.
PKD represents another potential application for therapeutic gene editing. Most patients develop kidney failure from small mutations in either the PKD1 or PKD2 gene. These types of mutations are amenable to repair by the HDR approach and would only require a relatively short oligonucleotide donor template. Two possible therapeutic approaches might be possible. On the one hand, fibroblasts from an affected patient could be isolated and induced to become pluripotent cells (a standard protocol now). The PKD mutation could be corrected by CRISPR at this point, and the pluripotent cells could then be induced to differentiate into a transplantable kidney, which would have the benefit of being immunologically identical to the patient.36 Unfortunately, it is not clear when or even if a transplantable kidney can be grown in the laboratory.
An alternative approach would be to deliver Cas9, a gRNA, and the donor template DNA directly to kidney tubular epithelia in patients affected with PKD and correct the gene defect in all of the nephrons of that patient. Here, the major limitation is that there is no clear method to deliver this DNA to the kidney. The AAV approach seems to be the most promising and is already in clinical trials in other organs; however, there is no evidence of any AAV serotype capable of infecting kidney epithelia.
Ethical Considerations
The ability to edit the genome with relative ease has led to an intense debate about whether and how this technology might be used to modify the genomes of human embryos. Although in utero or preimplantation genetic screening—for Down syndrome, for example—has existed for years, CRISPR/Cas9 could be used to make changes that are inherited, crossing what some consider an ethical boundary. Some scientists have argued that germline genome editing should be banned because of unpredictable effects on future generations, the impossibility of informed consent, and the possible use of gene editing for nontherapeutic purposes (appearance, for example).45 Others argue that such a ban would reflect a lack of imagination and lack of faith in society to properly regulate a field with enormous potential benefit to patients suffering from incurable disease.46
As a result of these concerns, a group of 18 prominent leaders in the field met in 2015 in Napa, California to initiate a discussion on the scientific, medical, legal, and ethical implications of CRISPR/Cas9 gene editing. The group called for a moratorium on the clinical use of germline gene editing until a diverse group of citizens and stakeholders examines the technology and evaluates its risks and potential benefits.47
Conclusions
Over just a few years, the scientific community has witnessed the development of a remarkably powerful, flexible, and exciting new tool for studying and manipulating gene function. We have highlighted the development of gene editing and focused on CRISPR/Cas9-based approaches, currently the most flexible gene manipulation available. These technologies offer the rapid, inexpensive, and efficient targeting of one or multiple genes and are easily accessible for any laboratory environment. These approaches offer powerful new tools to those studying kidney disease and are sure to play an increasingly important role in both kidney investigation and therapeutics in the future.
Disclosures
None.
Acknowledgments
The authors thank Beno Freedman for helpful discussions.
Work in the laboratory of B.D.H. is supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases grants DK107274, DK103740, and DK103050; Established Investigator Award EIA14650059 of the American Heart Association; and Biogen Idec.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS: Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317: 230–234, 1985 [DOI] [PubMed] [Google Scholar]
- 2.Thomas KR, Folger KR, Capecchi MR: High frequency targeting of genes to specific sites in the mammalian genome. Cell 44: 419–428, 1986 [DOI] [PubMed] [Google Scholar]
- 3.Pardo B, Gómez-González B, Aguilera A: DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell Mol Life Sci 66: 1039–1056, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller J, McLachlan AD, Klug A: Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4: 1609–1614, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavletich NP, Pabo CO: Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science 252: 809–817, 1991 [DOI] [PubMed] [Google Scholar]
- 6.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U: Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512, 2009 [DOI] [PubMed] [Google Scholar]
- 7.Moscou MJ, Bogdanove AJ: A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Boch J, Bonas U: Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu Rev Phytopathol 48: 419–436, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Segal DJ, Ghiara JB, Barbas CF 3rd: Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci U S A 94: 5525–5530, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF: Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhakta MS, Henry IM, Ousterout DG, Das KT, Lockwood SH, Meckler JF, Wallen MC, Zykovich A, Yu Y, Leo H, Xu L, Gersbach CA, Segal DJ: Highly active zinc-finger nucleases by extended modular assembly. Genome Res 23: 530–538, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL: The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P: CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J: Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marraffini LA, Sontheimer EJ: CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Travis J: Making the cut. Science 350: 1456–1457, 2015 [DOI] [PubMed] [Google Scholar]
- 17.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E: A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasiunas G, Barrangou R, Horvath P, Siksnys V: Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109: E2579–E2586, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E: CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD: High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O: Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F: Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dianov GL, Hübscher U: Mammalian base excision repair: The forgotten archangel. Nucleic Acids Res 41: 3483–3490, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F: Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK: Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA: Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK: CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10: 977–979, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM: Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12: 326–328, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA: Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W: High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509: 487–491, 2014 [DOI] [PubMed] [Google Scholar]
- 31.Inui M, Miyado M, Igarashi M, Tamano M, Kubo A, Yamashita S, Asahara H, Fukami M, Takada S: Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep 4: 5396, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R: One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R: One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T: MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc 11: 118–133, 2016 [DOI] [PubMed] [Google Scholar]
- 35.McVey M, Lee SE: MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet 24: 529–538, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV: Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6: 8715, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frenette PS, Atweh GF: Sickle cell disease: Old discoveries, new concepts, and future promise. J Clin Invest 117: 850–858, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, Voit RA, Yuan GC, Porteus MH, Stamatoyannopoulos JA, Lettre G, Orkin SH: An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342: 253–257, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE: BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527: 192–197, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, Dickson G, Wood MJ, Wilton SD, Straub V, Kole R, Shrewsbury SB, Sewry C, Morgan JE, Bushby K, Muntoni F: Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: An open-label, phase 2, dose-escalation study. Lancet 378: 595–605, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN: Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA: In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ: In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Güell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, Cortazio R, Wilkinson RA, Fishman JA, Church G: Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350: 1101–1104, 2015 [DOI] [PubMed] [Google Scholar]
- 45.Lanphier E, Urnov F, Haecker SE, Werner M, Smolenski J: Don’t edit the human germ line. Nature 519: 410–411, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Check Hayden E: Should you edit your children’s genes? Nature 530: 402–405, 2016 [DOI] [PubMed] [Google Scholar]
- 47.Baltimore D, Berg P, Botchan M, Carroll D, Charo RA, Church G, Corn JE, Daley GQ, Doudna JA, Fenner M, Greely HT, Jinek M, Martin GS, Penhoet E, Puck J, Sternberg SH, Weissman JS, Yamamoto KR: Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science 348: 36–38, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]