Abstract
Background & Aims
Innate immune dysfunction can promote chronic inflammatory diseases of the liver. For example, mice lacking the flagellin receptor Toll-like receptor 5 (TLR5) show microbial dysbiosis and predisposition to high-fat diet (HFD)-induced hepatic steatosis. The extent to which hepatocytes play a direct role in detecting bacterial products in general, or flagellin in particular, is poorly understood. In the present study, we investigated the role of hepatocyte TLR5 in recognizing flagellin, policing bacteria, and protecting against liver disease.
Methods
Mice were engineered to lack TLR5 specifically in hepatocytes (TLR5ΔHep) and analyzed relative to sibling controls (TLR5fl/fl). TLR5 messenger RNA levels, responses to exogenous flagellin, elimination of circulating motile bacteria, and susceptibility of liver injury (concanavalin A, carbon tetrachloride, methionine- and choline-deficient diet, and HFD) were measured.
Results
TLR5ΔHep expressed similar levels of TLR5 as TLR5fl/fl in all organs examined, except in the liver, which showed a 90% reduction in TLR5 levels, indicating that hepatocytes accounted for the major portion of TLR5 expression in this organ. TLR5ΔHep showed impairment in responding to purified flagellin and clearing flagellated bacteria from the liver. Although TLR5ΔHep mice did not differ markedly from sibling controls in concanavalin A or carbon tetrachloride–induced liver injury models, they showed exacerbated disease in response to a methionine- and choline-deficient diet and HFD. Such predisposition of TLR5ΔHep to diet-induced liver pathology was associated with increased expression of proinflammatory cytokines, which was dependent on the Nod-like-receptor C4 inflammasome and rescued by microbiota ablation.
Conclusions
Hepatocyte TLR5 plays a critical role in protecting liver against circulating gut bacteria and against diet-induced liver disease.
Keywords: Innate Immunity, TLR5, Hepatocytes, Inflammation, Steatosis
Abbreviations used in this paper: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CCL4, carbon tetrachloride; CFU, colony-forming unit; ConA, concanavalin A; CXCL, chemokine (C-X-C motif) ligand 1; DC, dendritic cell; Hep, hepatocyte; HFD, high-fat diet; IEC, intestinal epithelial cell; IL, interleukin; KO, knock-out; mRNA, messenger RNA; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NLR, nod-like receptor; NPC, nonparenchymal cell; MCD, methionine- and choline-deficient diet; LPS, lipopolysaccharide; PBS, phosphate-buffered saline; RT-PCR, reverse-transcription polymerase chain reaction; TLR, Toll-like receptor; WT, wild-type
Summary.
Innate immune dysfunction can promote chronic inflammatory diseases of the liver, such as nonalcoholic fatty liver disease. Here, we show a role for hepatocyte Toll-like receptor 5 in detecting flagellin, clearing bacteria from the liver, and protecting against diet-induced hepatic diseases.
The mammalian gastrointestinal tract is inhabited by a complex community of 100 trillion bacteria (1–2 kg in mass), collectively referred to as gut microbiota. Although gut microbiota play an essential role in host metabolism and immune system development,1 failure to manage gut microbiota expeditiously can lead to chronic inflammatory diseases of the intestine such as Crohn's disease and ulcerative colitis.2, 3, 4, 5 A key means by which the host manages its microbiota are the Toll-like receptors (TLRs) and the nod-like receptors (NLRs), which confer the host innate ability to recognize a broad range of microbes. Deficiency in TLR and/or NLR signaling can result in changes in microbiota composition that promote intestinal inflammation and metabolic diseases. For example, mice with engineered deficiencies in TLR5, TLR2, NLRP6, or NLRP3 show altered gut microbiota composition that is associated with features of the metabolic syndrome, which could be transferred to WT mice via co-housing and/or fecal transplant, suggesting a role for microbiota in driving this disorder.6, 7, 8 Such metabolic syndrome included features of nonalcoholic fatty liver disease (NAFLD) when mice were fed high-fat diets (HFD).
A primary mechanism by which altered microbiota might promote NAFLD and other features of metabolic syndrome is by inducing low-grade inflammation, which is a central feature of these disorders. For example, the altered microbiota composition of TLR5 knock-out (KO) mice was associated with higher levels of fecal bioactive lipopolysaccharide (LPS) and flagellin, suggesting that alteration of microbiota may result in inherently greater potential to promote inflammation.9 Although observations that total loss of TLR function alters microbiota composition and promotes inflammation were suggested to be an artifact of mouse husbandry practices,10 the observation that epithelial cell–specific deletion of TLR5 alters microbiota composition relative to TLR5-floxed siblings and results in low-grade inflammation/metabolic syndrome argues against this notion.11 Such inflammation possibly might result from systemic dissemination of gut microbial products and increasing circulating proinflammatory cytokines in response to these products. Moreover, such microbial products might themselves disseminate from the intestine, via portal vein, to the liver and other tissues that do not normally harbor large populations of bacteria, and are thought to be very responsive to these products. Such receiving of intestinal venous blood by the liver has been proposed to result in the need for the liver to serve as a firewall to capture bacteria or their products that breach the intestine,12 but also might be a means by which aberrant microbiota promote NAFLD.13 Indeed, the concept that reduced intestinal barrier function can result in gut microbiota products breaching the intestine, sometimes referred to as leaky gut syndrome, increasingly is thought to play a central role in metabolic disease.14, 15 In support of this notion, detection of LPS in the liver by Kupffer, macrophage-like cells, promotes HFD-induced steatosis in mice.16 However, the extent to which hepatocytes, the predominant cell type in the liver, play a role in recognizing bacterial products other than LPS remains largely undefined. Because hepatocytes share many properties with enterocytes, which are highly responsive to flagellin via TLR5 and normally lack classic LPS-induced TLR4 signaling,17, 18 we reasoned the former pathway might be operable in hepatocytes.
Hence, we generated mice lacking TLR5 in hepatocytes, and examined their phenotypes in assays of innate immunity and models of liver injury and inflammation. Such studies showed a role for hepatocyte TLR5 in detecting bacterial flagellin, clearing bacteria from the liver, and protecting against diet-induced hepatic disease.
Materials and Methods
Generation of Experimental Mice
WT, albumin-CRE, Villin-CRE, and CD11c-CRE mice were purchased from Jackson Laboratories (Bar Harbor, ME). The latter were bred to TLR5fl/fl mice, whose generation recently was described11 to create the TLR5fl/fl, TLR5Δhepatocyte (Hep), TLR5Δintestinal epithelial cell (IEC), and TLR5Δdendritic cell (DC) mice used herein. The global TLR5KO mice used here originally were generated by Dr Shizuo Akira (Osaka University, Osaka, Japan)19 and back-crossed/maintained as previously described.8 Mice lacking NLRC4 (NLRC4KO), generated on a pure C57BL/6J background, were kindly provided by Vishva Dixit (Genentech, Inc, South San Francisco, CA) and used to generate TLR5/NLRC4 double-KO mice (TLR5-NLRC4 DKO), as previously described.20 All animals used in this study were on a C57BL/6J genetic background.
All mice were bred and housed at Georgia State University (Atlanta, GA), under institutionally approved protocols (Institutional Animal Care and Use Committee number A14033). Mice were fed with the standard LabDiet (St. Louis, MO) rodent chow LabDiets 5001 used in this facility. Where indicated, a HFD (60% of calories from fat), a methionine and choline-deficient diet (MCD) (A02082002B). and its associated methionine- and choline-sufficient control diet (A02082003B) were used to feed the mice (HFD, 8 weeks; MCD, 4 weeks, as previously described8, 21). All the experiments using HFD and MCD were performed on female mice, whereas other experiments were performed on either female or male animals. All the figures present values obtained from 1 independent experiment (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5 and 7), except for the HFD feeding experiment (Figures 6 and 8), performed twice.
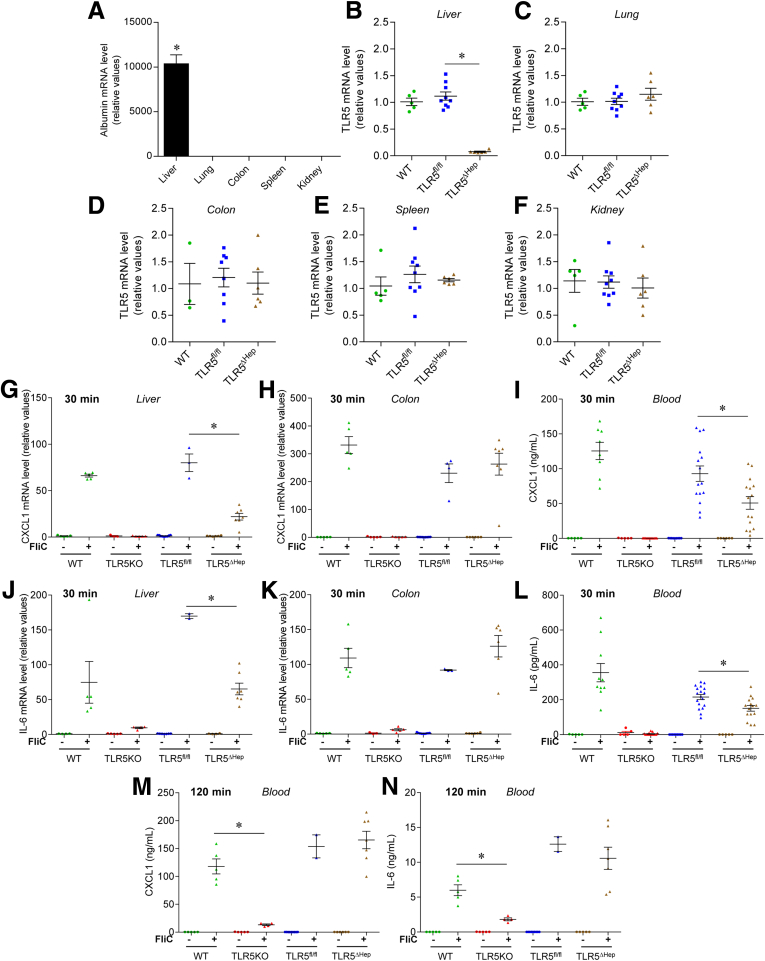
Figure 1.
Generation and characterization of mice lacking TLR5 in the liver. (A) Analysis of albumin mRNA expression by quantitative RT-PCR in multiple organs of WT mice. (B–F) Analysis of TLR5 mRNA expression by quantitative RT-PCR in the (B) liver, (C) lung, (D) colon, (E) spleen, and (F) kidney of WT, TLR5fl/fl, and TLR5ΔHep mice. Results are expressed as relative values compared with the WT group, defined as 1. (G–L) Mice were injected intraperitoneally with 20 μg of purified flagellin (+) or vehicle (200 μL of sterile PBS [-]). Thirty minutes later, serum and organs were isolated. (G and H) Analysis of CXCL1 mRNA expression by quantitative RT-PCR in (G) liver and (H) colonic mucosa of WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (I) Analysis of CXCL1 protein expression level by enzyme-linked immunosorbent assay in sera of WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (J and K) Analysis of IL6 mRNA expression by quantitative RT-PCR in (J) liver and (K) colonic mucosa of WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (L) Analysis of IL6 protein expression level by enzyme-linked immunosorbent assay in the serum of WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (M and N) WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice were injected intraperitoneally with 20 μg of purified flagellin (+) or vehicle (200 μL of sterile PBS [-]), and serum were collected 120 minutes after. (M) Analysis of CXCL1 protein expression level by enzyme-linked immunosorbent assay in the serum of WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (N) Analysis of IL6 protein expression level by enzyme-linked immunosorbent assay in the serum of WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (B–N) Points are from individual mice, with bars representing means ± SEM. (A) Data are the means ± SEM. N = 2–6, except for panels I and L, with N = 5–15. Significance was determined by the Student t test. *P < .05.
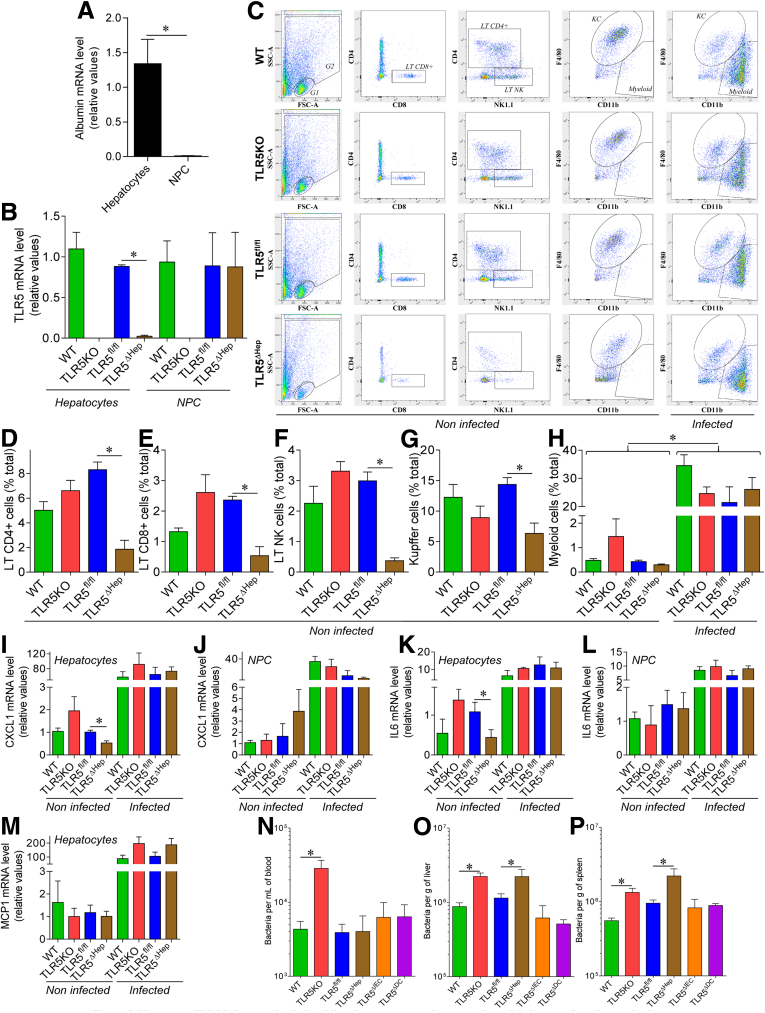
Figure 2.
Hepatocyte TLR5 helps to maintain basal liver immune state and promotes bacterial clearance from liver and spleen. (A and B) Hepatic parenchymal (hepatocytes) and NPCs were isolated from WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (A) Analysis of albumin mRNA expression in WT animals by quantitative RT-PCR. (B) Analysis of TLR5 mRNA expression by quantitative RT-PCR. Results are expressed as relative values compared with (A) hepatocyte or (B) WT groups, defined as 1. (C–P) Eight-week-old WT, TLR5KO, TLR5fl/fl, TLR5ΔHep, TLR5ΔIEC, and TLR5ΔDC mice were administered intravenously 107 live flagellated E coli strain MG1655. Hepatic parenchymal (hepatocytes) and NPCs were isolated from noninfected and infected WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. Liver NPCs were immunophenotyped and quantified by flow cytometry. (C) Representative flow cytometry dot plots. (D) Lymphocyte T CD4+ (LT CD4+), (E) lymphocyte T CD8+ (LT CD8+), (F) lymphocyte T natural killer (LT NK), (G) Kupffer cell, and (H) myeloid cell quantification, expressed as the percentage of total cells, with 20,000 cells being analyzed. Analysis of (I and J) CXCL1, (K and L) IL6, and (M) MCP1 mRNA expression by quantitative RT-PCR in (I, K, and M) purified hepatocytes and (J and L) purified NPC. Results are expressed as relative values compared with noninfected WT and TLR5fl/fl groups, defined as 1. (N–P) Six hours after inoculation, CFUs in (N) blood, (O) liver homogenate, and (P) spleen homogenate were enumerated on selective media. Data are the means ± SEM. No CFUs were detected in mice not administered E coli. N = 5–10. Significance was determined by the Student t test. *P < .05.
Figure 3.
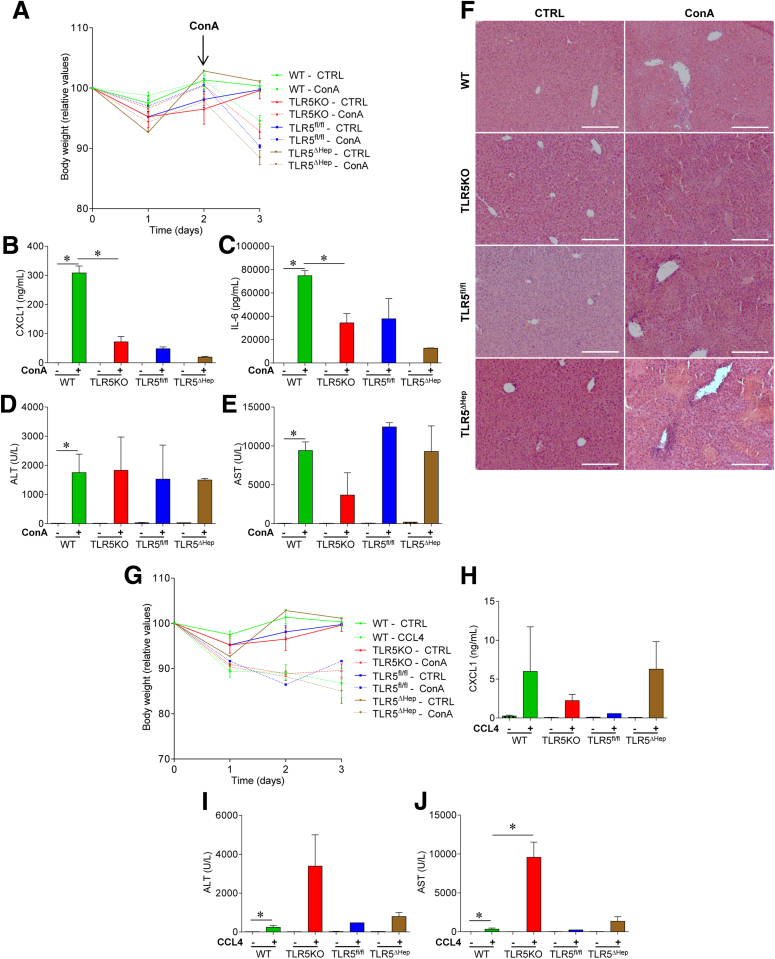
Hepatocyte TLR5 does not play a major role in protecting against acute concanavalin A or CCL4 treatments. (A–F) WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice were injected intravenously with 200 μL of sterile PBS containing (+) or not (-) ConA (15 mg/kg of body weight), and then euthanized 24 hours after injection. (A) Body weight over time. (B) Analysis of CXCL1 protein expression level by enzyme-linked immunosorbent assay in the serum. (C) Analysis of IL6 protein expression level by enzyme-linked immunosorbent assay in the serum. (D) Serum ALT concentrations. (E) Serum AST concentrations. (F) H&E staining of liver sections. (G–J) WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice were injected intraperitoneally with 200 μL of sterile olive oil containing (+) or not (-) CCL4 (1 μL/g of body weight), and euthanized 72 hours after injection. (G) Body weight over time. (H) Analysis of CXCL1 protein expression level by intraperitoneally in the serum. (I) Serum ALT concentrations. (J) Serum AST concentrations. Scale bar: 50 μm. Data are the means ± SEM. N = 3–5. Significance was determined by the Student t test. *P < .05. CTRL, control.
Figure 4.
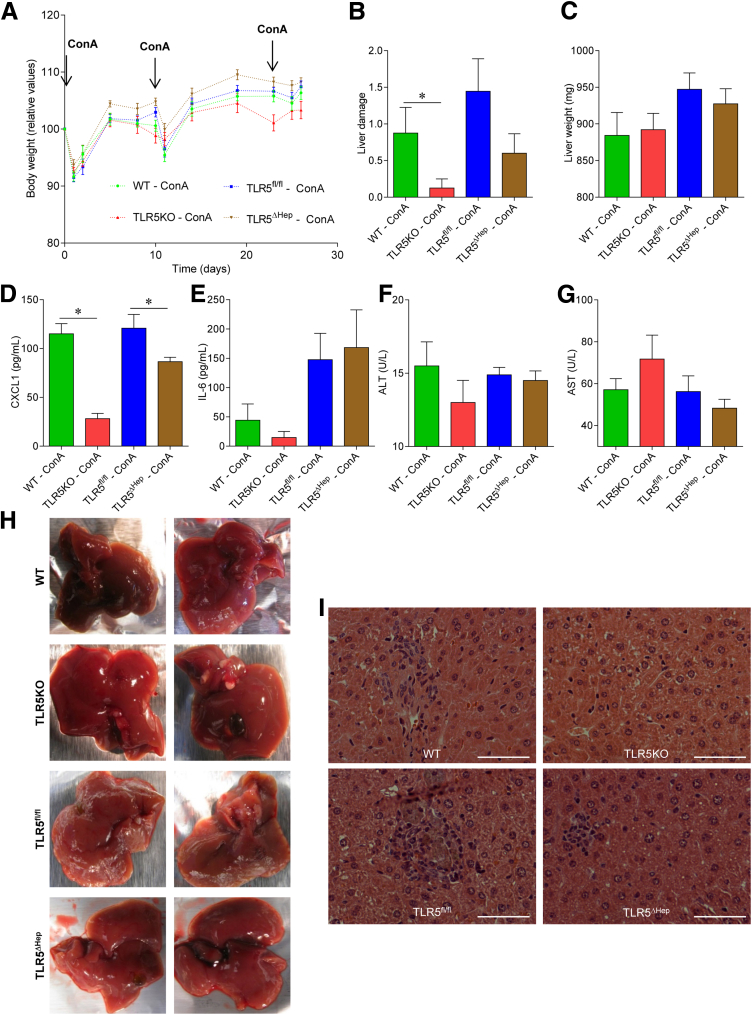
Hepatocyte TLR5 does not play a major role in protecting against chronic concanavalin A treatment. WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice were injected intravenously with 200 μL of sterile PBS containing ConA (5 mg/kg of body weight) every week for 3 weeks, for a total of 3 injections. Forty-eight hours after the last injection, mice were euthanized. (A) Body weight over time, (B) macroscopic liver damage, and (C) liver weight were measured. (D) Analysis of CXCL1 protein expression level by enzyme-linked immunosorbent assay in the serum. (E) Analysis of IL6 protein expression level by enzyme-linked immunosorbent assay in the serum. (F) Serum ALT concentrations. (G) Serum AST concentrations. (H) Macroscopic picture of livers from ConA-treated WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. (I) H&E staining of liver sections from ConA-treated WT, TLR5KO, TLR5fl/fl, and TLR5ΔHep mice. Scale bar: 25 μm. Data are the means ± SEM. N = 3–5. Significance was determined by the Student t test. *P < .05.
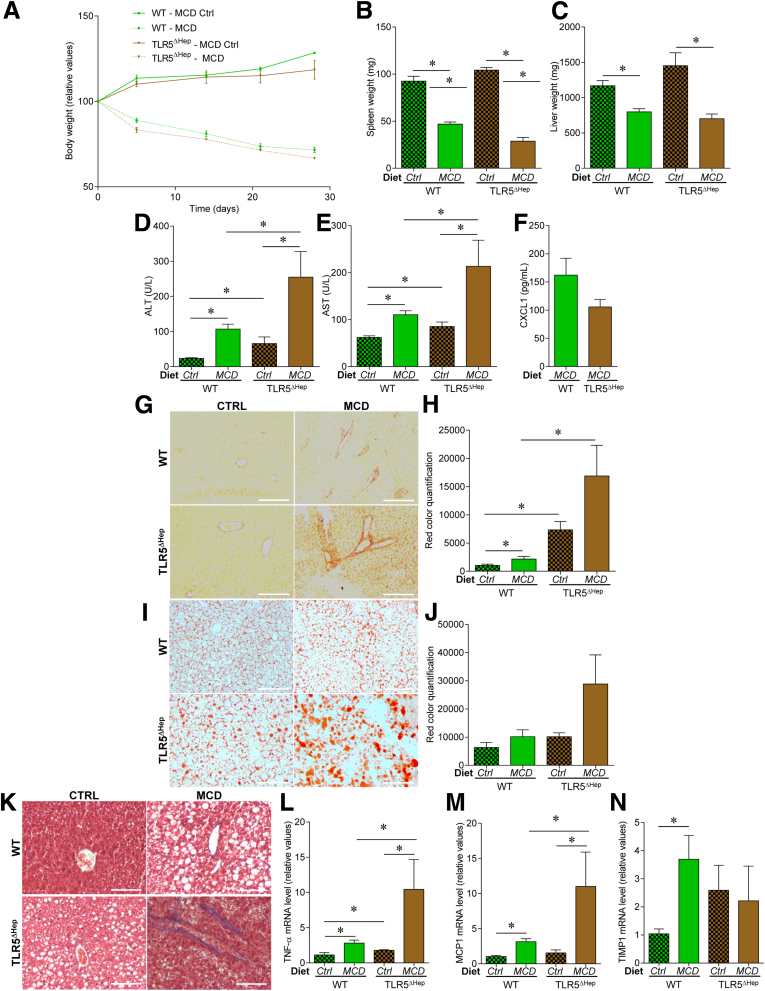
Figure 5.
Liver TLR5 protects against NASH induced by a methionine- and choline-deficient diet. WT and TLR5ΔHep mice were fed with a methionine-and choline-sufficient control diet (Ctrl) or a MCD, for 4 weeks. (A) Body weight over time, (B) spleen weights, (C) liver weights, (D) serum ALT concentrations, (E) serum AST concentrations, and (F) serum CXCL1 concentrations. (G and H) Liver fibrosis estimation using Sirius red staining, with (G) representative images and (H) quantification shown. (I and J) Liver steatosis estimation using Oil Red O staining, with (I) representative images and (J) quantification shown. (K) Liver fibrosis estimation using Masson's trichrome staining, with representative images shown. (L–N) Liver mRNAs were isolated and quantitated for expression of genes involved in inflammation and fibrosis: (L) tumor necrosis factor (TNF)-α, (M) MCP1, and (N) TIMP metallopeptidase inhibitor 1. Results are expressed as relative values compared with WT mice fed with control diet group, defined as 1. Scale bar: 50 μm. Data are the means ± SEM. N = 5. Significance was determined by the Student t test. *P < .05.
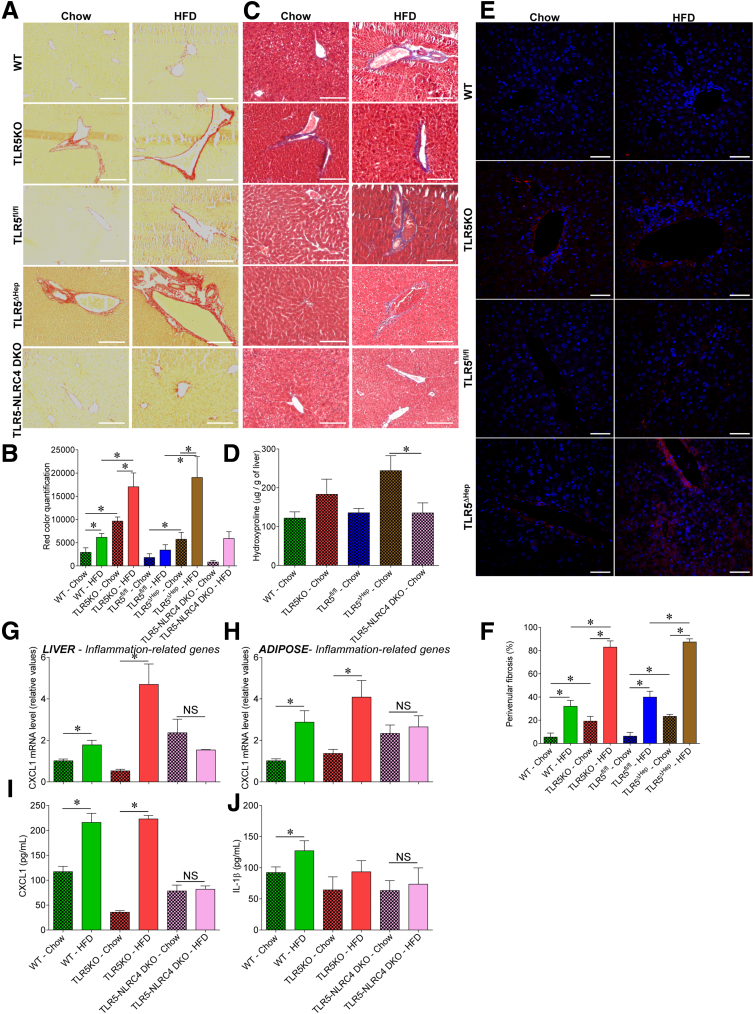
Figure 7.
Liver TLR5 protects against high-fat diet–induced fibrosis. WT, TLR5KO, TLR5fl/fl, TLR5ΔHep, and TLR5-NLRC4 double knock-out (DKO) mice were fed with a regular chow diet or a HFD, comprising 60% fat, for 8 weeks. (A and B) Liver fibrosis estimation using Sirius red staining, with (A) representative images and (B) quantification shown. (C) Liver fibrosis estimation using Masson's trichrome staining, with representative images shown. (D) Hydroxyproline quantification. (E and F) Liver fibrosis estimation using collagen immunostaining and confocal microscopy analysis, with (E) representative images (staining of collagen in red and DNA in blue), and (F) quantification (percentage of vein presenting collagen accumulation, determined through the examination of 10 veins per slide) shown. (G) Liver and (H) adipose tissue mRNA were isolated and quantitated for expression of the CXCL1 gene, involved in inflammation. Results are expressed as relative values compared with WT mice fed with regular chow, defined as 1. (I) Analysis of CXCL1 protein expression level by enzyme-linked immunosorbent assay in the serum. (J) Analysis of IL1β protein expression level by enzyme-linked immunosorbent assay in the serum. Scale bar: 50 μm. Data are the means ± SEM. N = 5. Significance was determined by the Student t test. *P < .05.
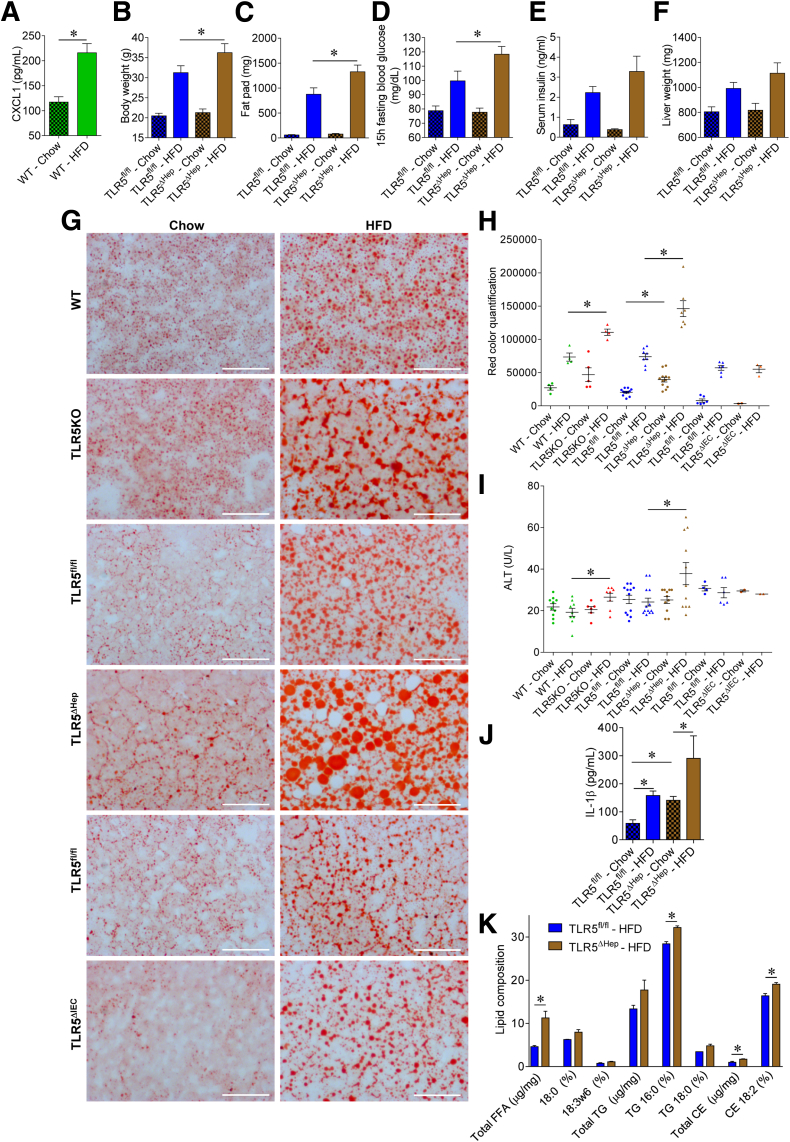
Figure 6.
Liver TLR5 protects against high-fat diet–induced metabolic syndrome and steatosis. (A) Analysis of CXCL1 protein expression level by enzyme-linked immunosorbent assay in sera of WT mice fed with a regular chow diet or a HFD, comprising 60% fat, for 8 weeks. (B–K) TLR5fl/fl and TLR5ΔHep mice were fed with a regular chow diet or a HFD, comprising 60% fat, for 8 weeks. (B) Body weight, (C) fat-pad weights, (D) 15-hour fasting blood glucose concentration, (E) 5-hour fasting insulinemia, and (F) liver weights were measured. (G and H) Liver lipid staining using Oil Red O, with (G) representative images and (H) quantification shown. (I) Serum ALT concentrations. (J) Serum IL1β concentrations. (K) Lipid composition in the liver. FFA, free-fatty acid; TG, triglyceride; CE, cholesterol ester. Scale bar: 50 μm. Data are the means ± SEM. (H and I) Points are from individual mice, with bar representing means ± SEM. N = 5–15. Significance was determined by the Student t test. *P < .05.
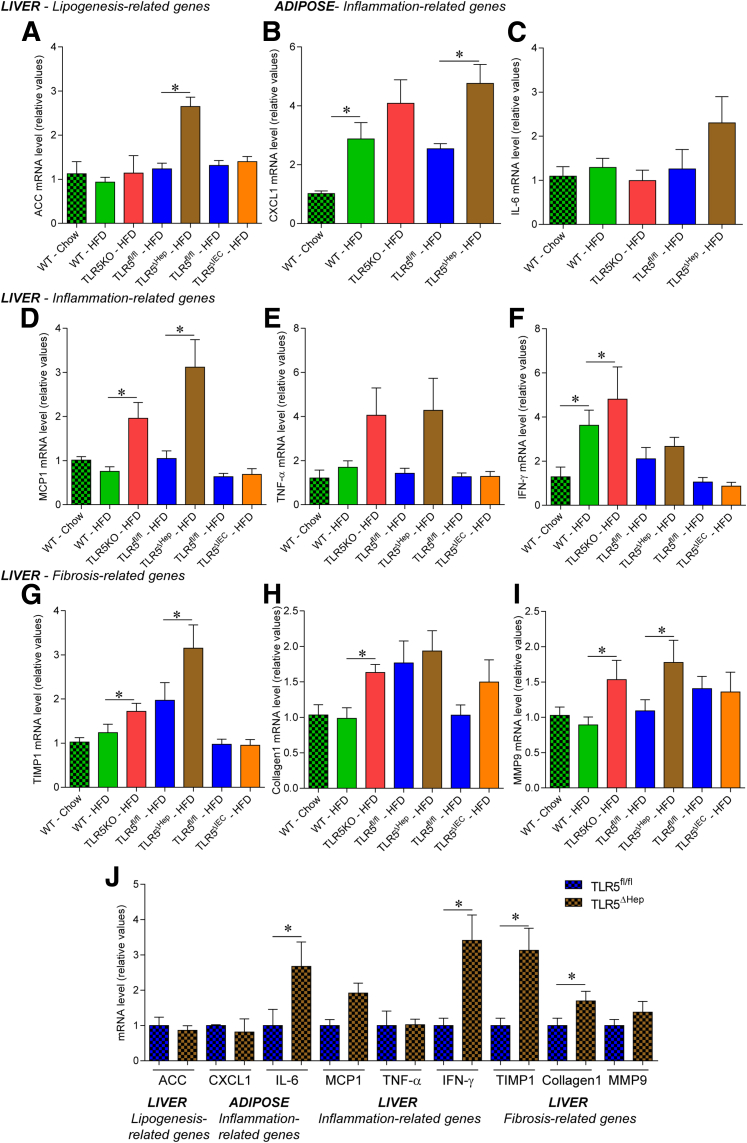
Figure 8.
Loss of hepatocyte TLR5 potentiates high-fat diet–induced proinflammatory gene expression in liver. WT, TLR5KO, TLR5fl/fl, TLR5ΔHep, and TLR5ΔIEC mice were fed with a regular chow diet or a HFD, comprising 60% fat, for 8 weeks. (A) Liver mRNAs were isolated and quantitated for expression of the acetyl-coenzyme A carboxylase gene, involved in lipogenesis. (B and C) Adipose tissue mRNAs were isolated and quantitated for expression of genes involved in inflammation: (B) CXCL1 and (C) IL6. (D–I) Liver mRNAs were isolated and quantitated for expression of genes involved in inflammation and fibrosis: (D) MCP1, (E) tumor necrosis factor (TNF)-α, (F) interferon (IFN)-γ, (G) TIMP1, (H) collagen 1, and (I) matrix metalloprotease 9. Results are expressed as relative values compared with (A–I) WT or (J) TLR5fl/fl mice fed with regular chow, defined as 1. Data are the means ± SEM. N = 5–15. Significance was determined by the Student t test. *P < .05.
Flagellin Treatment
Flagellin was isolated and purified by high-performance liquid chromatography, as previously described.22 Six-week-old mice were injected intraperitoneally with 20 μg of purified flagellin or vehicle (phosphate-buffered saline [PBS]) as a control. Blood was collected at 30 and 120 minutes and hemolysis-free serum was generated by centrifugation using serum separator tubes (Becton Dickinson, Franklin Lakes, NJ). Mice then were euthanized, and organs (liver, lung, colon, spleen, and kidney) were collected and stored at -80°C for further analysis.
Cytokine Analysis
Serum CXCL1 (chemokine CXC motif ligand 1), interleukin (IL)1β, and IL6 concentrations were determined using Duoset cytokine enzyme-linked immunosorbent assay kits (R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions. Serum insulin concentration was determined using the EZRMI-13K rat/mouse insulin enzyme-linked immunosorbent assay kit (Millipore, Billerica, MA) according to the manufacturer’s instructions.
Quantitative Reverse-Transcription Polymerase Chain Reaction
Total RNAs were isolated from liver, lung, spleen, abdominal fat tissue, and colon using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Messenger RNAs (mRNAs) were purified using the RNeasy mini kit RNA cleanup procedure (Qiagen, Valenica, CA). Quantitative reverse-transcription polymerase chain reactions (RT-PCRs) were performed using the iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad, Hercules, CA) in a CFX96 apparatus (Bio-Rad) with specific mouse oligonucleotides (Table 1). Results were normalized to the 36B4 (housekeeping gene).
Table 1.
Primers Used in This Study
| Gene name | Oligonucleotide sequence (5’-3’) |
|---|---|
| 16S | 515F GTGCCAGCMGCCGCGGTAA 806R GGACTACHVGGGTWTCTAAT |
| 36B4 | TCCAGGCTTTGGGCATCA CTTTATTCAGCTGCACATCACTCAGA |
| Albumin | GAAGACCCCAGTGAGTGAGC GCTTCACCAGCTCAGCAAGA |
| TLR5 | ATGGATGCTGAGTTCCCCCA AAAGGCTATCCTGCCGTCTG |
| CXCL1 | TTGTGCGAAAAGAAGTGCAG TACAAACACAGCCTCCCACA |
| IL6 | GTGGCTAAGGACCAAGACCA GGTTTGCCGAGTAGACCTCA |
| MCP1 | GCTGGAGCATCCACGTGTT TGGGATCATCTTGCTGGTGAA |
| TNF-α | CGAGTGACAAGCCTGTAGCC CATGCCGTTGGCCAGGA |
| IFN-γ | AGCAAGGCGAAAAAGGATGC TCATTGAATGCTTGGCGCTG |
| TIMP1 | ACAGACAGCCTTCTGCAACT CGCTGGTATAAGGTGGTCTCG |
| Collagen-1 | CTGGACTTCCTGGTCCTCCT CCATAGGACATCTGGGAAGCA |
| MMP9 | TAGCTACCTCGAGGGCTTCC GCCTTGGGTCAGGCTTAGAG |
| ACC | AGCAGATCCGCAGCTTG ACCTCTGCTCGCTGAGTGC |
ACC, acetyl-coenzyme A carboxylase; CXCL1, chemokine (C-X-C motif) ligand 1; IFN, interferon; IL6, interleukin 6; MCP1, monocyte chimioattractant; MMP, matrix metalloprotease 9; TIMP1, TIMP metallopeptidase inhibitor 1; TLR 5, toll like receptor 5; TNF, tumor necrosis factor.
Intravenous Injection of Bacteria
As previously described,12 live Escherichia coli (flagellated commensal strain MG1655) was administered (107 colony forming units [CFU], intravenously) via tail vein. Six hours after injection, blood was collected and hemolysis-free serum was generated by centrifugation using serum separator tubes (Becton Dickinson). Serum then was diluted serially, plated on a Luria Bertani agar plate, and incubated for 12 hours at 37°C. CFUs were counted and expressed as bacteria number per milliliters of blood. In parallel, spleen and liver were collected, homogenized in sterile PBS, serially diluted, plated on a Luria Bertani agar plate, and incubated for 12 hours at 37°C. Bacteria then were enumerated and results are expressed as bacteria per gram of tissue.
Isolation of Hepatic Parenchymal and Nonparenchymal Cells
Liver perfusion was performed as previously described.23, 24 Briefly, after liver perfusion with perfusion buffer (Hank's balanced salt solution, 5 mmol/L HEPES, 0.5 mmol/L EDTA; Sigma, St. Louis, MO) via inferior vena cava, collagenase solution was perfused (Hank's balanced salt solution, 5 mmol/L HEPES, 0.5 mmol/L CaCl2, 0.5 mg/mL collagenase; Sigma). Digested livers were homogenized and passed through a 100-μm cell strainer. After centrifugation at 50g for 3 minutes, the supernatant was collected and nonparenchymal cells (NPC) were purified using 40% iodixanol solution (Progen, Heidelberg, Germany). The pelleted parenchymal cells (hepatocytes) were washed once.
Total RNAs were isolated from purified hepatocytes and NPC using TRIzol (Invitrogen) according to the manufacturer’s instructions, and quantitative RT-PCRs were performed as described earlier.
Fluorescence-Activated Cell Sorter–Based Characterization of Hepatic Nonparenchymal Cells
Purified NPC were treated with red blood cell lysis buffer, and cell preparations were stained with the following antibody cocktail: CD8a-Pacific Blue (clone 53-6-7; Thermo Fisher Scientific, Waltham, MA), CD4-PerCP Cy5.5 (clone RMA 4-5; eBioscience, San Diego, CA), CD11b–fluorescein isothiocyanate (clone MI-70; eBioscience), NK1.1-PE-Cy7 (clone PKI 36; eBioscience), Tie2-PE (clone TEK4; eBioscience), and F4/80-APC (clone BM8; eBioscience). Incubation was performed at 4°C for 20 minutes, followed by fixation at 37°C for 10 minutes in 4% formaldehyde. A total of 20,000 cells were examined with a BD LRS Fortessa (BD Biosciences, San Jose, CA), and data were analyzed with FlowJo software version 10 (Ashland, OR). The Live/Dead Yellow staining kit was used to confirm cell viability (Thermo Fisher Scientific). Two gates were designed using forward scatter-A vs side scatter-A plots (Figure 2C), as previously described.23 G1 was used further to analyze lymphocytes T CD4+ (LT CD4+), CD8+ (LT CD8+), and natural killer (LT NK) using CD4, CD8a, and NK1.1 markers. G2 was sorted further using Tie2 and CD11b markers, with CD11bint/high Tie2int/low subsequently used to analyze Kupffer and myeloid cells using CD11b and F4/80 markers.
Bacterial Load Quantification by Quantitative RT-PCR
For quantification of hepatic bacterial load, total RNAs were isolated from liver using TRIzol (Invitrogen) according to the manufacturer’s instructions, and subsequently purified using the RNeasy Mini Kit (Qiagen). RNAs then were subjected to quantitative PCR using the iScript One-Step RT-PCR Kit with SYBR Green with universal 16S ribosomal RNA primers 515F and 806R (Table 1). Results were normalized to the housekeeping 36B4 gene.
Assessment of Basal Phenotypes
Mice were weighed after weaning and every week thereafter. Body weight data shown is expressed as a percentage compared with the initial body weight (day 0), defined as 100%. Eight weeks after weaning (11 weeks old), mice were fasted for 5 hours, at which time blood was collected by retro-orbital capillary plexus. Mice then were euthanized, and colon length, colon weight, spleen weight, liver, and adipose tissue weights were measured.
Concanavalin A–Induced Hepatitis
Eight-week-old mice were injected intravenously (tail vein) with Concanavalin A (ConA) (Sigma), 15 mg/kg of body weight, diluted in sterile pyrogen-free PBS, as previously described.25 Twenty-four hours after injection, blood was collected by retro-orbital capillary plexus and hemolysis-free serum was generated by centrifugation of blood using serum separator tubes (Becton Dickinson). Mice then were euthanized, and liver weight was measured and collected for downstream analysis. Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were analyzed at the Comparative Clinical Pathology Services (Columbia, MO) under the supervision of a board-certified veterinary clinical pathologist (Charles E. Wiedmeyer). A liver damage score from 1 to 4 was attributed macroscopically according to abnormal liver surface and to the extent of lesions. For chronic exposure to ConA, 8-week-old mice were injected intravenously (tail vein) with ConA (5 mg/kg of body weight, diluted in sterile pyrogen-free PBS; Sigma) every week for 3 weeks, for a total of 3 injections. Forty-eight hours after the last injection, serum and tissues were collected and analyzed, as described earlier.
Carbon Tetrachloride–Induced Hepatitis
Eight-week-old mice were injected intraperitoneally with 200 μL of sterile olive oil containing (+) or not (-) carbon tetrachloride (CCL4) (1 μL/g of body weight), as previously described.26 Seventy-two hours after injection, blood was collected by retro-orbital capillary plexus and hemolysis-free serum was generated by centrifugation of blood using serum separator tubes (Becton Dickinson). Serum ALT and AST levels were analyzed at the Missouri State University metabolic services core.
Serum ALT and AST Quantification
Serum ALT and AST levels were analyzed at the Comparative Clinical Pathology Services (Columbia, MO) under the supervision of a board-certified veterinary clinical pathologist (Charles E. Wiedmeyer). Results are expressed as units per liter.
H&E Staining
After euthanasia, mice livers were fixed in 10% buffered formalin for 24 hours at room temperature, transferred to Ethanol 70°, and then embedded in paraffin. Tissues were sectioned at 5-μm thickness and subjected to H&E staining.
Oil Red Staining
After euthanasia, mice livers were frozen in optimum cutting temperature and stored at -80°C until analysis. Tissues were sectioned using a cryostat at -20°C at 8-μm thickness and stained with Oil Red O using a previously described protocol.27 Briefly, 1.9 g of Oil Red O (Sigma) was dissolved in 300 mL of isopropyl alcohol. After homogenization, 200 mL of distilled water was added and the solution was incubated at 4°C for 30 minutes. After filtering (0.45 μm), solution was used to stain OCT liver sections for 6 minutes. Finally, sections were washed using running tap water for at least 30 minutes and mounted using a sterile glycerol 40% solution. Photoshop CS6 edition (Adobe, San Jose, CA) was used to quantify the red staining specifically, using the same settings and threshold for all of the slides.
Determination of Liver Lipid Contents
After overnight fasting, mice were euthanized and livers were collected, snap-frozen, and subsequently stored at -80°C. Frozen liver tissues were used for lipid extraction by the Vanderbilt Mouse Metabolic Phenotyping Center Lipid Core (DK59637). After extraction, free fatty acid, triglyceride, and cholesterol ester composition were determined.
Determination of Liver Fibrosis
After euthanasia, mouse livers were fixed in 10% buffered formalin for 24 hours at room temperature, transferred to Ethanol 70°, and then embedded in paraffin. Tissues were sectioned at 5-μm thickness and subjected to collagen immunostaining (primary antibody: anti-mouse collagen type I, AB765P; Millipore; secondary antibody: anti-rabbit IgG cy5) with DNA staining using Hoechst. Observations were performed with a Zeiss LSM 700 confocal microscope (Zeiss Microscopy, Peabody, MA) with Zen 2011 software version 7.1. This software was used to determine the percentage of veins presenting with collagen accumulation, determined through the examination of 10 veins per slide. Fibrosis also was evaluated using Masson trichrome (kit ab150686; Abcam, Cambridge, MA) and Sirius red (kit ab15068; Abcam) staining, performed on 5-μm thickness paraffin liver sections and following the manufacturer's instructions. Photoshop CS6 edition was used to quantify the red staining specifically, using the same settings and threshold for all of the slides.
Determination of Hepatic Hydroxyproline Content
Liver was homogenized in distilled water to a final concentration of 100 mg/mL. A total of 100 μL of 12 mol/L HCl was added to 100 μL of liver suspension, and samples were incubated at 120°C for 3 hours. Hydrolyzed liver (50 μL) was added to a 96-well plate and evaporated at 60°C. Chloramine T/Oxidation Buffer Mixture (Sigma) was added and samples were incubated at room temperature for 5 minutes. Dimethylaminobenzaldehyde reagent was added, samples were incubated at 60°C for 90 minutes, and absorbance was measured at 560 nm. Purified hydroxyproline was used as a standard.
Overnight Fasting Blood Glucose Measurement
Mice were placed in a clean cage and fasted overnight for 15 hours. Blood glucose concentration was determined using a Nova Max Plus Glucose Meter (Billerica, MA) and expressed as milligrams per deciliters.
Antibiotic Treatment
Four-week-old mice were placed on broad-spectrum antibiotics ampicillin (1.0 g/L) and neomycin (0.5 g/L) in drinking water for 14 weeks, as previously described.8
Statistical Analysis
N designate total sample number. Significance was determined using the Student t test or 1-way analysis of variance using GraphPad Prism software (version 6.04; La Jolla, CA). Differences were noted as significant at a P value of .05 or less. The Kolmogorov–Smirnov test was used to verify that all data were distributed normally.
Results
Generation and Characterization of Mice Lacking Hepatic TLR5
Mice lacking the flagellin receptor, TLR5, are prone to develop a NAFLD-like phenotype when maintained on a compositionally defined HFD, possibly as a consequence of low-grade intestinal inflammation and/or a direct loss of liver cell TLR5.8 In accord with the latter possibility, cultured hepatocytes express TLR5 and respond to flagellin.28 Moreover, we recently observed that loss of gut epithelial cell TLR5 resulted in low-grade inflammation and some aspects of metabolic syndrome but did not predispose the liver to HFD-induced nonalcoholic steatohepatitis (NASH), suggesting a potential role for hepatocyte TLR5 in protecting the liver in this disease model.11 Hence, we sought to generate mice that lack TLR5 specifically in hepatocytes and, subsequently, investigate their phenotype. C57BL/6 mice in which exon 1 of the TLR5 gene was flanked by loxP sites11 were crossed to mice engineered to express CRE recombinase under the control of the albumin promoter, allowing subsequent use of a breeding scheme in which all dams maintained WT TLR5 (TLR5fl/fl) function while, on average, 50% of each litter would lack TLR5 in albumin-expressing cells (TLR5ΔHep). In WT mice, as expected, albumin mRNA was highly expressed in the liver and undetectable in other tissues where TLR5 is known to be expressed, including lung, colon, spleen, and kidney, suggesting our approach would provide highly specific deletion of TLR5 in liver (Figure 1A). In accord, TLR5ΔHep, which were born at the expected Mendelian ratios and lacked obvious abnormalities, showed WT levels of TLR5 in all tissues examined, but had a more than 93% reduction in TLR5 mRNA in the liver relative to both WT and TLR5fl/fl mice, showing that depletion of TLR5 was indeed specific to the liver and that hepatocytes likely accounted for the major portion of liver TLR5 expression (Figure 1B–F and Table 2). To investigate this notion, we isolated and purified hepatocytes and NPCs, followed by TLR5 mRNA level expression analysis. As expected, albumin was found to be highly expressed in the hepatocyte fraction compared with the NPC fraction (Figure 2A), and this approach showed a strong 97% depletion of TLR5 mRNA in hepatocytes from TLR5ΔHep mice compared with TLR5fl/fl animals, with TLR5 mRNA expression being unaffected in NPC (Figure 2B), thus confirming that the deletion of TLR5 was highly specific to hepatocytes.
Table 2.
Expression Analysis of TLR5 mRNA in Multiple Organs in TLR5fl/fl and TLR5ΔHep Mice
| Tissue | Albumin expression (relative values) | TLR5 fold decrease expression in TLR5ΔHep mice compared with TLR5fl/fl mice | P |
|---|---|---|---|
| Liver | 10,364.08 | 14.50 | <.0001 |
| Lung | 0.17 | 0.88 | .2644 |
| Colon | 0.01 | 1.19 | .4752 |
| Spleen | 0.08 | 1.09 | .5866 |
| Kidney | 2.85 | 1.11 | .5991 |
Liver TLR5 Mediates Flagellin-Induced Gene Expression
We first investigated the consequences of hepatocyte-specific deletion of TLR5 by measuring responses to systemic administration of purified flagellin. Mice were injected intraperitoneally with flagellin, and various tissues were isolated 30 minutes later, at which time responses were likely to reflect direct responses from cells stimulated by flagellin rather than cells indirectly stimulated by flagellin-induced cytokines. In WT mice, liver levels of CXCL1 (mouse homologue of IL8) were highly induced in response to flagellin (Figure 1G–I). In contrast, no such increase was observed in mice with complete loss of TLR5, showing the strong TLR5-dependence of this response. Relative to their TLR5fl/fl siblings, TLR5ΔHep showed a 75% reduction in flagellin-induced CXCL1 levels in the liver, indicating a direct role for hepatocyte TLR5 in mediating a rapid response to flagellin, while also suggesting that nonhepatocytes (eg, Kupffer cells) also might be capable of direct TLR5-mediated recognition of flagellin. A similar pattern of results was seen when IL6 was used as the readout for flagellin responsiveness (Figure 1J–L). In contrast, via both readouts, rapid responsiveness to flagellin in the colon was similar in TLR5fl/fl and TLR5ΔHep, indicating that the decreased responsiveness observed in the liver was specific for that organ (Figure 1G–L). However, the levels of flagellin-induced CXCL1 and IL6 in serum were reduced significantly, albeit modestly, in TLR5ΔHep relative to their TLR5fl/fl siblings at the 30-minute time point, but not at the 120-minute time point, which may reflect secondary cytokine production (Figure 1I, and L–N), suggesting that hepatocytes contribute significantly to circulating cytokines in response to purified flagellin.
Liver TLR5 Promotes Clearance of Hepatic Bacteria
We next investigated the extent to which loss of hepatocyte TLR5 might alter populations of nonparenchymal cells in the liver. First, we performed flow cytometric analysis of the main hepatic immune cell populations and observed that, relative to TLR5fl/fl mice, TLR5ΔHep mice showed significantly decreased levels of CD4+, CD8+, and natural killer lymphocytes, as well as Kupffer cells (Figure 2C–G). Such reduction correlated with reduced hepatocyte expression of cytokines CXCL1 and IL6, which may have influenced immune cell recruitment (Figure 2I–M). However, such alterations were not observed in global TLR5KO mice, suggesting that a broad range of compensatory factors likely can overcome any deficiency of cell recruitment in even modest inflammatory conditions. Hence, we next considered the ability of TLR5ΔHep mice to effectively orchestrate innate immunity upon infection.
It recently was shown that the liver protects the host from the gut microbiota by serving as a “firewall” that clears commensal gut bacteria, breaching the gut mucosa and entering the blood stream via the portal vein.12 Although such liver clearance is presumed to be mediated largely by Kupffer cells, which are macrophage-like cells, we hypothesized a potential role for hepatocyte TLR5 in clearing flagellated commensal bacteria. To investigate this possibility, mice (WT, TLR5KO, TLR5fl/fl, TLR5ΔHep, TLR5ΔIEC, and TLR5ΔDC) were administered the commensal, live, flagellated, E coli strain MG1655 by intravenous injection. Such infection lead to a dramatic recruitment of myeloid cells, as well as an increase in proinflammatory cytokines (CXCL1, IL6, and monocyte chimoattractant protein 1 (MCP1)) in both hepatocytes and NPC, and none of those phenotypes were altered by TLR5 deletion, either global or liver specific (Figure 2H–M). However, relative to their TLR5fl/fl siblings, TLR5ΔHep showed a 3-fold increase in CFUs in the liver and spleen, whereas CFUs in blood were not affected significantly (Figure 2N–P). Mice with a complete deficiency of TLR5 showed a similar increase of CFUs in liver, spleen, and also in the circulating blood, whereas mice lacking TLR5 expression by intestinal epithelial cells (TLR5ΔIEC) or by dendritic cells (TLR5ΔDC) harbor CFUs in blood, liver, and spleen not significantly affected compared with TLR5fl/fl control mice. Thus, although clearance of systemically administered bacteria likely involves multiple cell types and signaling pathways, hepatocyte TLR5 plays a key role in the efficient orchestration of immune responses that keep bacterial loads in check in the liver and, subsequently, in the spleen.
Minimal Role for TLR5 in Experimental Models of Liver Injury
A standard and widely used model for liver injury in rodents is via administration of ConA, which nonspecifically activates immune cells, eventuating in liver injury.29 High doses of ConA have been used to model acute liver injury, and we also designed a new approach by using lower but multiple doses of ConA to induce chronic rather than acute liver damage. It has been suggested that the gut microbiota can function as a rheostat of the immune system, and consequently affect the severity of liver disease. Hence, we subjected TLR5fl/fl and sibling TLR5ΔHep mice, to acute (Figure 3A–F) and chronic (Figure 4) ConA treatment. The extent of the resulting liver injury appeared consistent with what was described previously for WT C57BL/6 mice and, more importantly, was not statistically different between TLR5fl/fl and TLR5ΔHep mice. CCL4 administration was used as another model of liver injury, and, again, only a modest and not significant increase in liver injury was observed in TLR5KO and TLR5ΔHep mice compared with WT and TLR5fl/fl mice, respectively (Figure 3G–J). Overall, these data suggest that hepatocyte TLR5 does not play a major role in classic models of liver injury.
Liver TLR5 Protects Against NASH Induced by a Methionine- and Choline-Deficient Diet
In light of the increasing appreciation of the role of innate immunity in mediating NASH, we next examined the role of hepatocyte TLR5 in a widely used experimental model of this disorder. Specifically, WT and TLR5ΔHep mice were placed on a MCD diet, which is known to drive steatohepatitis within weeks of its administration.21, 30 As expected, WT mice fed with the MCD diet showed marked weight loss and developed liver disease within 4 weeks, as indicated by an increase in both ALT and AST levels (Figure 5A–F). Importantly, when fed with the control diet (methionine and choline sufficient), both ALT and AST levels were increased significantly, albeit modestly, in TLR5ΔHep compared with WT mice (Figure 5D and E). When exposed to the MCD diet, TLR5ΔHep mice showed a similar degree of wasting, but showed exacerbated increases in ALT and AST levels compared with WT animals (Figure 5A, D, and E). This increase in diet-induced liver injury in TLR5ΔHep mice correlated with a decrease in the circulating CXCL1 cytokine level (Figure 5F), suggesting that the enhanced disease in TLR5ΔHep mice reflected an absence of TLR5-mediated immune cell recruitment that normally would prevent and/or remediate liver damage that results from methionine and choline deficiency. We next analyzed fibrosis and steatosis development, and identified exacerbated collagen fiber deposition and liver lipid droplet accumulation in TLR5ΔHep compared with WT mice, both at the basal level (control diet) and after MCD diet treatment (Figure 5G–K). Inflammation-related (tumor necrosis factor-α and MCP1) and fibrosis-related (TIMP metallopeptidase inhibitor 1) gene expression were found be increased in the liver of TLR5ΔHep compared with WT mice at the basal level, with further exacerbation after MCD diet treatment (Figure 5L–N). Altogether, these results show a protective role played by liver TLR5 against NASH.
Liver TLR5 Protects Against High-Fat Diet–Induced Steatosis and Fibrosis
Mice with complete deficiency of TLR5 were observed previously to develop gut bacteria dysbiosis, low-grade inflammation, and features of metabolic syndrome including increased adiposity and dysglycemia.8 Such metabolic abnormalities were more pronounced on a HFD and included hepatic steatosis.8 Most features of TLR5-deficient metabolic syndrome were recapitulated by specific deletion of TLR5 from intestinal epithelial cells (IEC), suggesting the disorder was driven by the inability of TLR5-deficient IECs to manage the gut microbiota.11 However, such TLR5ΔIEC mice did not show increased steatosis, relative to their sibling control mice, even when maintained on a HFD, suggesting that this important feature of their disease might involve loss of TLR5 on liver cells. Moreover, we observed that HFD feeding leads, in WT mice, to a substantial increase in circulating CXCL1 levels, suggesting that the TLR5 signaling pathway, and its associated cytokines, may play a central role in response to HFD challenge (Figure 6A). Hence, to examine the possibility that hepatocyte TLR5 might protect against steatosis, and/or other aspects of metabolic syndrome, we examined a range of parameters in WT, TLR5fl/fl, and TLR5ΔHep mice on a normal chow and HFD. When maintained on a standard mouse chow diet, TLR5ΔHep mice lacked features of low-grade inflammation (no colomegaly or splenomegaly) and metabolic syndrome (no modification of body mass, fat-pad mass, or fasting blood glucose) that were shown by mice with a complete or an IEC TLR5 deficiency (Figure 6B–F).8, 11 However, relative to their TLR5fl/fl sibling controls, TLR5ΔHep showed a significant increase, albeit modest, in liver lipid droplet levels when fed with a regular chow diet, as shown by levels of Oil Red O staining (Figure 6G and H). In accord with our previous study, although HFD treatment leads to moderate steatosis in WT animals, complete loss of TLR5 resulted in exacerbated levels of steatosis in response to HFD8 that correlated with increased levels of liver injury markers, as reflected by levels of serum ALT (Figure 6I). Such increased susceptibility to HFD-induced liver dysfunction was recapitulated completely by loss of TLR5 specifically in hepatocytes in that, compared with their TLR5fl/fl siblings, TLR5ΔHep showed marked increased in both levels of steatosis and serum ALT in response to HFD (Figure 6G–I). Such liver abnormalities correlated with increased levels of the proinflammatory cytokine IL1β (Figure 6J), whose increased expression is known to drive much of the gut pathology shown by mice with complete deficiency of TLR5.31, 32 Such increased levels of IL1β in TLR5-deficient mice were normalized by the global deletion of the NLRC4 gene, which mediates intracellular detection of flagellin, resulting in inflammasome activation (Figure 7J). The absence of NLRC4 also prevented HFD-induced liver and adipose tissue inflammation (Figure 7G–J) compared with TLR5KO animals, suggesting that increased NLRC4 activation mediated the exacerbated liver HFD-induced injury upon loss of hepatocyte TLR5. The increased lipid accumulation in HFD-treated TLR5ΔHep relative to TLR5fl/fl animals was seen in several lipid species, including free fatty acids, triglycerides, and cholesterol esters (Figure 6K). Such increased lipid incorporation correlated with increased expression of the hepatic acetyl-coenzyme A carboxylase gene, whose activity regulates the availability of substrates for fatty acid synthesis33 in the liver of TLR5ΔHep compared with TLR5fl/fl mice (Figure 8A). Analysis of fibrosis showed increased collagen fiber deposition at the basal level in TLR5KO and TLR5ΔHep compared with WT and TLR5fl/fl mice, respectively, as shown by increased collagen fiber deposition, hepatic hydroxyproline (major component of collagen) concentration, and mild perivenular fibrosis (Figure 7A–F). After HFD treatment, TLR5ΔHep and TLR5KO mice developed more profound fibrosis compared with WT and TLR5fl/fl controls (Figure 7A–F). The absence of NLRC4 was sufficient to prevent the exacerbated fibrosis observed in TLR5KO animals, further supporting the role played by NLRC4 in liver injury observed after the loss of TLR5 (Figure 7A–F). The development of hepatic steatosis and fibrosis also was supported by the observation of gene expression alteration in liver and adipose tissue in response to HFD treatment, and further exacerbated in TLR5KO and TLR5ΔHep animals, including increases in genes that mediate inflammation (CXCL1, IL6, MCP1, tumor necrosis factor-α, and interferon-γ) and fibrosis (TIMP metallopeptidase inhibitor 1, collagen 1, and matrix metalloprotease 9) (Figure 8). Importantly, some of those markers were found to be increased significantly in TLR5ΔHep animals compared with TLR5fl/fl controls, even under basal conditions (ie, chow-feeding) (Figure 8J), further supporting a role for liver TLR5 in protecting against liver disease.
The increased accumulation of lipids, and associated changes in gene expression, observed in mice lacking hepatocyte TLR5 also manifested systemically in that HFD-treated TLR5ΔHep mice showed modest but significant increases in body weight, fat-pad mass, and fasting blood glucose/insulinemia, relative to TLR5fl/fl control mice (Figure 6B–D). Thus, loss of hepatocyte TLR5 predisposed mice to HFD-induced hepatofibrosis and steatosis that promoted other aspects of the metabolic syndrome.
Antibiotic Treatment Eliminates High-Fat Diet–Induced Steatosis That Resulted From the Loss of Hepatocyte TLR5
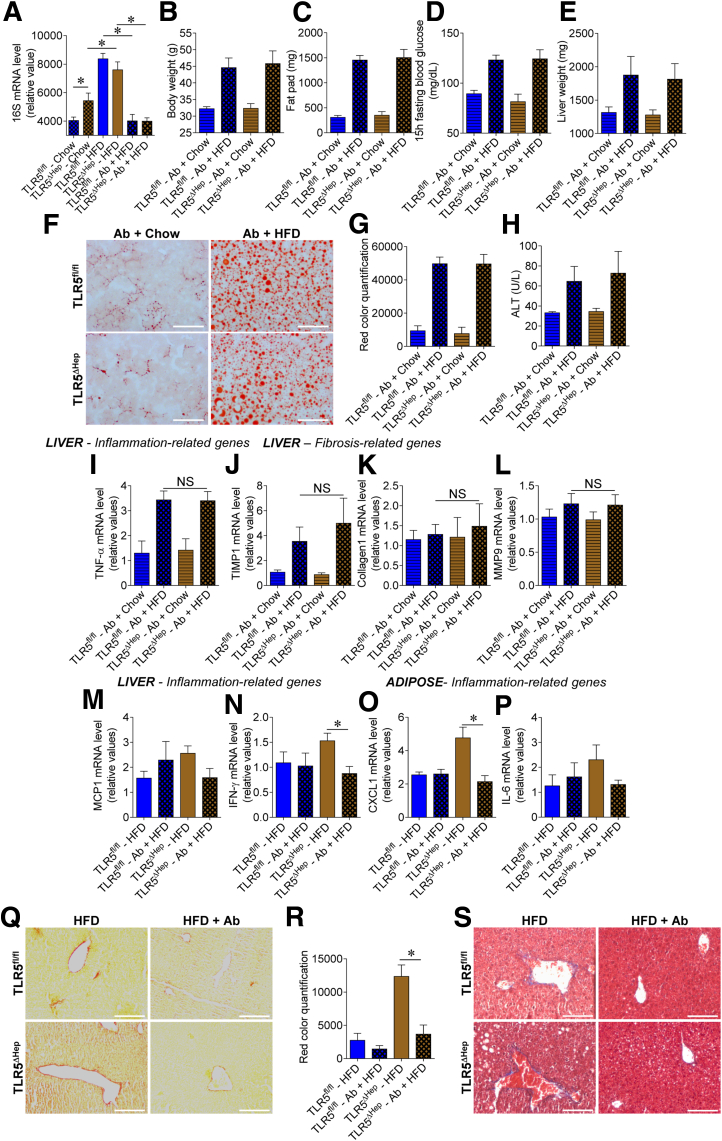
Gut epithelial TLR5 protection against gut inflammation is thought to involve this receptor's role in mediating rapid recruitment of immune cells that afford expedient clearance of bacteria that breach the mucosa.11 Extending this logic to the liver, in conjunction with the general hypothesis that gut bacteria and/or their products that transit to the liver promote hepatic disease,14, 34, 35 suggests that hepatocyte TLR5 might protect against liver disease by promoting efficient clearance of bacteria that cross the gut epithelial barrier and translocate to the liver. In accord with this hypothesis, we observed that, relative to control TLR5fl/fl siblings, TLR5ΔHep mice showed increased levels of bacterial DNA in the liver when fed with a chow diet (Figure 9A). Moreover, HFD treatment leads to an increased bacterial load in the liver of TLR5fl/fl and TLR5ΔHep mice compared with chow control groups (Figure 9A), showing that HFD feeding is associated with a leaky gut syndrome,14, 15 which might explain, at least in part, the uncontrolled liver inflammation in the context of TLR5 deficiency. To test this hypothesis, we maintained TLR5fl/fl and TLR5ΔHep mice on a broad-spectrum antibiotic cocktail while being fed chow or HFD. We observed a significant decrease in bacterial DNA detected in the liver, indicating such antibiotic treatment ablates the HFD-induced increase in liver bacteria in both genotypes (Figure 9A). Preventing the increase in liver bacterial loads is associated with elimination of the differences in metabolic and liver steatosis phenotype that was previously observed between TLR5fl/fl and TLR5ΔHep mice (Figure 9B–H). Moreover, as previously described,36 broad-spectrum antibiotic treatment reduced steatosis development, with an approximately 30% decrease in lipid accumulation after HFD treatment. Antibiotic treatment also reduced HFD-induced increases in liver and adipose gene expression related to inflammation (Figure 9I–P). Antibiotic treatment also abolished the increased fibrosis in TLR5ΔHep compared with TLR5fl/fl mice (Figure 9Q–S). Together, these results indicate that antibiotic treatment abrogated the increased disease severity in TLR5ΔHep compared with TLR5fl/fl animals that otherwise resulted after HFD feeding (Figure 9B–L). Moreover, antibiotic treatment prevented the severe steatosis observed in TLR5ΔHep-fed HFD (Figure 9M–S). Altogether, these results suggest that hepatocyte TLR5 protect against steatosis and inter-related aspects of the metabolic syndrome through a mechanism that likely involves efficient clearance of bacteria that translocate from the gut to the liver.
Figure 9.
Antibiotic treatment prevents the exacerbation of high-fat diet–induced metabolic syndrome, steatosis, and fibrosis that resulted from loss of hepatocyte TLR5. TLR5fl/fl and TLR5ΔHep mice were fed with a regular chow diet or a HFD, comprising 60% fat, for 8 weeks, with or without broad-spectrum antibiotics (ampicillin/neomycin), administered via drinking water. (A) Analysis of 16S mRNA expression by quantitative RT-PCR in the liver of TLR5fl/fl and TLR5ΔHep mice. (B) Body weight, (C) fat-pad weights, (D) 15-hour fasting blood glucose concentration, and (E) liver weights were measured. (F and G) Liver lipid staining using Oil Red O with (F) representative images and (G) quantification shown. (H) Serum ALT concentrations. (I–L) Liver mRNAs were isolated and quantitated for expression of genes involved in inflammation and fibrosis: (I) tumor necrosis factor (TNF)-α, (J) TIMP1, (K) collagen 1, and (L) matrix metalloprotease 9. (M and N) Liver mRNAs were isolated and quantitated for expression of genes involved in inflammation: (M) MCP1 and (N) interferon (IFN)-γ. (O and P) Adipose tissue mRNAs were isolated and quantitated for expression of genes involved in inflammation: (O) CXCL1 and (P) IL6. Results are expressed as relative values compared with TLR5fl/fl mice fed a high-fat diet, defined as 1. (Q and R) Liver fibrosis estimation using Sirius red staining with (Q) representative images and (R) quantification shown. (S) Liver fibrosis estimation using Masson's trichrome staining with representative images shown. Scale bar: 50 μm. Data are the means ± SEM. N = 3–5. Significance was determined by the Student t test. *P < .05.
Discussion
The rapid ascension of NAFLD, and the inter-related diseases it drives, is a major public health problem that warrants better understanding of its pathophysiology. Although increased caloric consumption in general, and saturated fats in particular, is likely a key driver of fatty liver disease, the extent of HFD promotion of NAFLD is driven by a range of genetic and nongenetic determinants. Gut microbiota increasingly are viewed as acting in concert with a HFD to promote NAFLD. Specifically, it is hypothesized that gut microbiota-derived LPS translocate, via the portal vein, to the liver where it activates proinflammatory gene expression through TLR4 expressed on Kupffer cells37 and promotes the onset of disease.16 Accordingly, mice engineered to lack TLR4, globally or in bone marrow–derived cells, are protected from HFD-induced steatosis.38 Herein, we describe that, in contrast, TLR5-mediated detection of bacterial flagellin is mediated by hepatocytes and protects mice from HFD-induced NAFLD. Hence, activation of TLRs in the liver is not always pathologic but, rather, can play an important role in protecting the liver against chronic inflammatory diseases.
Absence of TLR5 on hepatocytes delayed liver clearance of flagellated bacteria, and such liver-mediated clearance of bacteria, termed “firewall function,” is thought to play an important role in protecting against bacteria that breach the intestine.12 Hence, considering that HFD can reduce gut barrier function by reducing tight junction protein expression36 and results in increased bacterial translocation to the liver, we hypothesize that hepatocyte TLR5 may protect against HFD-induced steatosis likely by facilitating rapid clearance of gut-translocating bacteria, which effectively reduces the extent/duration of proinflammatory gene expression in general, and TLR4 activation in particular, as a result of the reduced bacterial loads. Such increased loads of flagellated bacteria likely would result in activation of the flagellin-responsive inflammasome NLRC4, which plays a pivotal role in driving inflammation in mice with a total absence of TLR5,31 leading to increased IL1β production, which is thought to play an important role in liver injury and fibrosis.39, 40, 41 In support of this scenario, we observed that the exacerbated steatosis correlated with enhanced proinflammatory gene expression in general, and increased IL1β cytokine production in particular, whereas the deletion of NLRC4 prevented HFD-induced IL1β and, moreover, abrogated NAFLD in TLR5-deficient mice, although future study will be needed to determine if NLRC4 activation is occurring in the liver. Although increased levels of liver bacterial DNA were observed at basal conditions in TLR5ΔHep compared with TLR5fl/fl animals, in accordance with the concept that the initial impact of an innate immune deficiency will be an increase in bacterial loads, we did not observe any difference in liver bacterial loads after HFD treatment. This observation was reminiscent of our results seen in mice with complete deficiency of TLR5 in that, relative to WT control mice, noncolitic TLR5KO mice harbor a higher load of adherent gut bacteria, whereas TLR5KO mice with overt colitis show a lower level of total adherent gut bacteria,42 perhaps reflecting that an increased bacterial load predisposes to an inflammatory response, which then serves as a compensatory means to keep bacteria in check. Hence, we view the increased level of bacteria in the liver of TLR5ΔHep mice maintained on normal chow to reflect their discrete innate immune deficiency, but that the liver inflammation that ensued upon prolonged high-fat diet feeding served to limit the bacterial load in this organ while promoting liver disease. However, at present, it is also very reasonable to speculate that the ability of hepatocyte TLR5 to protect against diet-induced liver disease is independent of its role in promoting bacterial clearance. Indeed, the ability of TLR5 activation to suppress apoptosis43, 44 may underlie its protection against NAFLD and NASH, resulting from high-fat or MCD diets.
Although the precise mechanism by which activation of hepatocyte TLR5 promotes bacterial clearance from the liver has not yet been defined, it may involve increases in hepatocyte expression of antimicrobial peptides and cytokines, promoting recruitment and activation of phagocytes that mediate bacterial killing. In addition, activation of hepatocyte TLR5 in response to gut-translocating flagellin may induce cytoprotective gene expression in hepatocytes that allows them to better withstand the stressful environment that can result from lipotoxicity and/or proinflammatory gene expression.45 These potential mechanisms are analogous to mechanisms by which TLR5 protects the intestine. Specifically, TLR5 signaling directly confers epithelia with enhanced ability to survive a range of challenges46, 47 whereas loss of intestinal epithelial TLR5 results in delayed clearance of flagellated bacteria, eventuating in chronic inflammation, which can take the form of colitis or low-grade inflammation that is associated with parameters of the metabolic syndrome.11 Such colitis does not occur in the absence of TLR4,11 thus, perhaps, the liver and intestine both follow the paradigm that a discrete deficiency in TLR signaling can result in bacteria expansion and subsequently enhanced activation of other pathways of innate immune activation that promote chronic inflammation.
The notion that hepatocytes are the key liver cell type that responds to flagellin is in accord with recent studies showing that hepatocytes rapidly and directly respond to a flagellin analog but not to LPS.28 Accordingly, activation of liver TLR5 has been proposed to play a central role in flagellin's radioprotective and anticancer properties.28 The notion that TLR5 may play a role in chronic liver disease is supported by the recent clinical study that found that flagellin-induced IL6 production is impaired in cirrhosis patients.48 Together with our findings, these results support the general theme that activation of innate immune signaling in nonimmune cells often plays an important role in homeostasis and, moreover, underscores the concept that broad inhibition of inflammatory signaling is unlikely to be a successful means to treat chronic inflammatory diseases of the gut. Rather, approaches to better manage the gut microbiota so as to avoid excessive activation of TLR signaling on immune and nonimmune cells might offer greater long-term therapeutic potential. Collectively, our results show that hepatocytes are direct responders to microbial products and, moreover, play an important role in protecting the liver against microbiota-driven chronic inflammatory diseases.
Acknowledgments
The authors thank Celine Thomas for outstanding technical assistance, the Vanderbilt Mouse Metabolic Phenotyping Center Lipid Core (DK59637) for hepatic lipid analysis, Kyle Flannigan and Debby Walthall for help with flow cytometry analysis, and Vishal Singh for helpful discussion and technical advice.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants DK099071 and DK083890 (A.T.G.) and DK097865 (M.V.K.). Benoit Chassaing is a recipient of the Career Development Award from the Crohn’s and Colitis Foundation of America.
References
- 1.Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 2.Chassaing B., Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 3.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Sartor R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 5.Cader M.Z., Kaser A. Recent advances in inflammatory bowel disease: mucosal immune cells in intestinal inflammation. Gut. 2013;62:1653–1664. doi: 10.1136/gutjnl-2012-303955. [DOI] [PubMed] [Google Scholar]
- 6.Caricilli A.M., Picardi P.K., de Abreu L.L. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol. 2011;9:e1001212. doi: 10.1371/journal.pbio.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Henao-Mejia J., Elinav E., Jin C. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijay-Kumar M., Aitken J.D., Carvalho F.A. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chassaing B., Koren O., Carvalho F.A. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069–1080. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ubeda C., Lipuma L., Gobourne A. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chassaing B., Ley R.E., Gewirtz A.T. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology. 2014;147:1363–1377 e17. doi: 10.1053/j.gastro.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balmer M.L., Slack E., de Gottardi A. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6:237ra66. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- 13.Chassaing B., Etienne-Mesmin L., Gewirtz A.T. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59:328–339. doi: 10.1002/hep.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cani P.D., Amar J., Iglesias M.A. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 15.Pierre N., Deldicque L., Barbe C. Toll-like receptor 4 knockout mice are protected against endoplasmic reticulum stress induced by a high-fat diet. PLoS One. 2013;8:e65061. doi: 10.1371/journal.pone.0065061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W., Metlakunta A., Dedousis N. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59:347–357. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho F.A., Aitken J.D., Vijay-Kumar M. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Ann Rev Physiol. 2012;74:177–198. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 18.Abreu M.T. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 19.Uematsu S., Jang M.H., Chevrier N. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 20.Vijay-Kumar M., Carvalho F.A., Aitken J.D. TLR5 or NLRC4 is necessary and sufficient for promotion of humoral immunity by flagellin. Eur J Immunol. 2010;40:3528–3534. doi: 10.1002/eji.201040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahai A., Malladi P., Melin-Aldana H. Upregulation of osteopontin expression is involved in the development of nonalcoholic steatohepatitis in a dietary murine model. Am J Physiol Gastrointest Liver Physiol. 2004;287:G264–G273. doi: 10.1152/ajpgi.00002.2004. [DOI] [PubMed] [Google Scholar]
- 22.Gewirtz A.T., Navas T.A., Lyons S. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 23.Mohar I., Brempelis K.J., Murray S.A. Isolation of non-parenchymal cells from the mouse liver. Methods Mol Biol. 2015;1325:3–17. doi: 10.1007/978-1-4939-2815-6_1. [DOI] [PubMed] [Google Scholar]
- 24.Conrad E., Resch T.K., Gogesch P. Protection against RNA-induced liver damage by myeloid cells requires type I interferon and IL-1 receptor antagonist in mice. Hepatology. 2014;59:1555–1563. doi: 10.1002/hep.26915. [DOI] [PubMed] [Google Scholar]
- 25.Jiang N., Zhang X., Zheng X. A novel in vivo siRNA delivery system specifically targeting liver cells for protection of ConA-induced fulminant hepatitis. PLoS One. 2012;7:e44138. doi: 10.1371/journal.pone.0044138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorn C., Heilmann J., Hellerbrand C. Protective effect of xanthohumol on toxin-induced liver inflammation and fibrosis. Int J Clin Exp Pathol. 2012;5:29–36. [PMC free article] [PubMed] [Google Scholar]
- 27.Mehlem A., Hagberg C.E., Muhl L. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–1154. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 28.Burdelya L.G., Brackett C.M., Kojouharov B. Central role of liver in anticancer and radioprotective activities of Toll-like receptor 5 agonist. Proc Natl Acad Sci U S A. 2013;110:E1857–E1866. doi: 10.1073/pnas.1222805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H.X., Liu M., Weng S.Y. Immune mechanisms of concanavalin A model of autoimmune hepatitis. World J Gastroenterol. 2012;18:119–125. doi: 10.3748/wjg.v18.i2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinclair E.M., Yusta B., Streutker C. Glucagon receptor signaling is essential for control of murine hepatocyte survival. Gastroenterology. 2008;135:2096–2106. doi: 10.1053/j.gastro.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 31.Carvalho F.A., Nalbantoglu I., Ortega-Fernandez S. Interleukin-1beta (IL-1beta) promotes susceptibility of Toll-like receptor 5 (TLR5) deficient mice to colitis. Gut. 2012;61:373–384. doi: 10.1136/gut.2011.240556. [DOI] [PubMed] [Google Scholar]
- 32.Vijay-Kumar M., Sanders C.J., Taylor R.T. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005;62:1784–1803. doi: 10.1007/s00018-005-5121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Roy T., Llopis M., Lepage P. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 35.Musso G., Gambino R., Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care. 2010;33:2277–2284. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cani P.D., Bibiloni R., Knauf C. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 37.Rivera C.A., Adegboyega P., van Rooijen N. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47:571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saberi M., Woods N.B., de Luca C. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab. 2009;10:419–429. doi: 10.1016/j.cmet.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gieling R.G., Wallace K., Han Y.P. Interleukin-1 participates in the progression from liver injury to fibrosis. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1324–G1331. doi: 10.1152/ajpgi.90564.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeSantis D.A., Ko C.W., Liu Y. Alcohol-induced liver injury is modulated by Nlrp3 and Nlrc4 inflammasomes in mice. Mediators Inflamm. 2013;2013:751374. doi: 10.1155/2013/751374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szabo G., Petrasek J. Inflammasome activation and function in liver disease. Nat Rev Gastroenterol Hepatol. 2015;12:387–400. doi: 10.1038/nrgastro.2015.94. [DOI] [PubMed] [Google Scholar]
- 42.Carvalho F.A., Koren O., Goodrich J.K. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng H., Wu H., Sloane V. Flagellin/TLR5 responses in epithelia reveal intertwined activation of inflammatory and apoptotic pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G96–G108. doi: 10.1152/ajpgi.00273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salamone G.V., Petracca Y., Fuxman Bass J.I. Flagellin delays spontaneous human neutrophil apoptosis. Lab Invest. 2010;90:1049–1059. doi: 10.1038/labinvest.2010.77. [DOI] [PubMed] [Google Scholar]
- 45.Vijay-Kumar M., Wu H., Jones R. Flagellin suppresses epithelial apoptosis and limits disease during enteric infection. Am J Pathol. 2006;169:1686–1700. doi: 10.2353/ajpath.2006.060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vijay-Kumar M., Aitken J.D., Sanders C.J. Flagellin treatment protects against chemicals, bacteria, viruses, and radiation. J Immunol. 2008;180:8280–8285. doi: 10.4049/jimmunol.180.12.8280. [DOI] [PubMed] [Google Scholar]
- 47.Zhang B., Chassaing B., Shi Z. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346:861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alazawi W., Spyrou A., Lahiri R. PMO-141 flagellin-induced IL-6 production is selectively impaired in patients with cirrhosis. Gut. 2012;61:A130. [Google Scholar]