Abstract
The expression and functions of microRNA (miR)-411 have been investigated in several types of cancer. However, until now, miR-411 in human breast cancer has not been examined. The present study investigated the expression, biological functions and molecular mechanisms of miR-411 in human breast cancer, discussing whether it offers potential as a therapeutic biomarker for breast cancer in the future. The expression levels of miR-411 in human breast cancer tissues and cells were measured using reverse transcription-quantitative polymerase chain reaction analysis. Following transfection with miR-411 mimics, an MTT assay, cell migration and invasion assay, western blot analysis and luciferase assay were performed in human breast cancer cell lines. According to the results, it was found that miR-411 was significantly downregulated in breast cancer, and associated with lymph node metastasis and histological grade. Additionally, it was observed that miR-411 suppressed cell growth, migration and invasion in the breast cancer cells. The present study also provided the first evidence, to the best of our knowledge, that miR-411 was likely to directly target specificity protein 1 in breast cancer. These findings indicated that miR-411 may be used a therapeutic biomarker for the treatment of breast cancer in the future.
Keywords: breast cancer, microRNA-411, specificity protein 1, specificity protein 1, therapy
Introduction
Breast cancer, the second most common type of malignancy worldwide, is the most frequent type of cancer among women and accounts for 29% of all new cancer diagnoses in women (1,2). Every year, ~1,300,000 women are newly diagnosed with breast cancer, with a mortality rate of almost 40,000 (3). Breast cancer is a heterogeneous and complex disease. The carcinogenesis of breast cancer involves genetic alterations, epigenetic alterations and environmental factors (4,5). Currently, the primary therapeutic strategies for patients with early breast cancer are surgery, followed by hormonal therapy, chemotherapeutic approaches, radiotherapy and biological therapies. However, for patients with advanced disease, it remains a prevalent and life threatening malignancy (6,7). Although several oncogenic and tumor-suppressive genes have been demonstrated to have important functions in the carcinogenesis and progression of breast cancer, the molecular mechanisms underlying this process remain to be fully elucidated. Therefore, it is important to understand these molecular mechanisms and to develop novel targeted therapies for breast cancer.
In breast cancer, changes in the expression of microRNAs (miRNAs) have been demonstrated (8–10). miRNAs are a group of non protein-coding, signal-stranded, small RNAs, which are ~19–25 nucleotides in length (11). miRNAs function as negative regulators of the expression of target mRNAs, and are involved in cell proliferation, differentiation, cell cycle, survival and all fundamental cellular processes implicated in carcinogenesis (12). Accumulating evidence suggests that over one third of all human genes may be targeted by miRNAs (13). miRNAs inhibit the translation or degradation of target mRNAs through binding to the complementary 3′ untranslated region of target mRNAs by base-pairing (14). There is also increasing evidence to suggest that specific miRNAs can be downregulated or upregulated in the different types of tumor (15–17). The upregulation of miRNAs can result in the degradation of tumor suppressor genes, whereas the downregulation of miRNAs can lead to an increase in the expression of oncogens (18). Therefore, identification of the targets of miRNAs is important to understand the functions of miRNAs in tumorigenesis and cancer development. It also suggests that miRNAs may be a target for breast cancer therapy.
The expression and functions of miR-411 have been investigated in several types of cancer. However, until now, miR-411 has not been investigated in human breast cancer. In the present study, the expression, biological functions and molecular mechanisms of miR-411 in human breast cancer were examined. The results showed that the expression of miR-411 was significantly decreased in human breast cancer, and was associated with lymph node metastasis and histological grade. In addition, miR-411 suppressed cell proliferation, migration and invasion by directly targeting specificity protein 1 (SP1). These findings have therapeutic implications, which may be exploited for the treatment of human breast cancer.
Materials and methods
Clinical specimens
The present study was approved by the Protection of Human Subjects Committee of The Second People's Hospital of Shenzhen (Shenzhen, China). Written informed consent was obtained from all patients (age range, 26–75 years; stage range, 50 cases of stage I–II, 64 cases of stage III) involved in the study. In total, 114 pairs of human breast cancer tissues and their corresponding adjacent non-neoplastic tissues were obtained with patients' approval. The tissue samples were snap frozen in liquid nitrogen at the time of surgery and stored at −80°C in a refrigerator.
Cell culture and transfection
The human breast cancer cell lines, MCF-7, MDA-MB-231 and SKBR3, and normal mammary epithelial cell line, MCF-10A, were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin (Gibco; Thermo; Fisher Scientific, Inc.) and 100 ug/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.) in a humidified 5% CO2 cell incubator at 37°C.
The mature miR-411 mimics, negative control (NC) and the luciferase reporter plasmid were synthesized and verified by GenePharma (Shanghai, China). The cells were seeded into a 6-well plate and cultured in RPMI-1640 medium without antibiotics. When the cell density reached 30–40%, tansfections were performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from homogenized tissues and cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RT process was performed using an M-MLV Reverse Transcription system (Promega Corporation, Madison, WI, USA) in a 25 µl volume. The temperature protocol for RT was as follows: 95°C for 2 min; 20 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 2 min; and 72°C for 5 min. Following RT, qPCR was performed using the reagents of the SYBR green I mix (Takara Bio, Dalian, China) in a 20 µl reaction volume, according to the manufacturer's protocol. The reaction system contained 10 µl SYBR Green I mix, 2 µl cDNA, 2 µl forward primer, 2 µl reverse primer (Guangzhou RiboBio Co., Ltd., Guangzhou, China) and 4 µl double-distilled water. The temperature protocol for the reaction was as follows: 95°C for 10 min; 40 cycles at 95°C for 15 sec and 60°C for 1 min. Each sample was analyzed in triplicate. U6 small RNA was used as an internal control. Gene expression was quantified using the 2−ΔΔCq method (19).
MTT assay
The assessment of cell proliferation was performed using an MTT assay, according to the manufacturer's protocol. Following transfection for 24 h, the transfected cells were seeded into 96-well plates at a density of 3,000 cells/well. The MTT assay was performed every 24 h until 96 h. A 20 µl volume of MTT solution (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added into each well. Following incubation for 4 h at 37°C, the MTT solution was removed and the formazan precipitates were dissolved in 200 µl DMSO. The A490 of each well was measured using a plate reader. The suppression rate was calculated using the following formula: Suppression rate = (1 − A490miR-411 / A490NC) × 100%. All experiments were performed in triplicate.
Cell migration and invasion assays
The migration and invasion potentials of the cells were evaluated using Transwell chambers with an 8-µm pore polycarbonate membrane (Corning Costar, Cambridge, MA, USA). For the invasion assay, the Transwell chambers were coated with Matrigel (BD Biosciences, San Jose, CA, USA). Following transfection for 24 h, 5×104 of the transfected cells in 300 µl serum-free medium were added into the upper chamber. A 500 µl volume of medium containing 20% FBS was then added to the lower chamber as a chemoattractant. Following 12 h incubation for the migration assay 24 h incubation for the invasion assay, cells, which had not migrated or invaded through the pores were carefully removed using a cotton swab. The chambers were then fixed with 100% methanol and stained with 0.5% crystal violet (Beyotime Institute of Biotechnology, Haimen, China). The migrated or invaded cells were counted under a light microscope (CKX41; Olympus Corporation, Tokyo, Japan).
Western blot analysis
The rabbit anti-human monoclonal SP1 (1:1,000; cat. no. 9389) and rabbit anti-human monoclonal GADPH (1:1,000; cat. no. 2118) primary antibodies used in the present study were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The cells were washed and lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology) 72 h post-transfection. Protein concentration was determined using the bicinchoninc acid protein assay kit (Beyotime Institute of Biotechnology). Equal quantities of protein (20 µg) were separated by 10% SDS-PAGE and electroblotted onto a PVDF membrane (EMD Millipore, Billerica, MA, USA). The membrane was blocked in phosphate-buffered saline (PBS) containing 0.1% Tween-20 (Beyotime Institute of Biotechnology) and 5% non-fat dry milk. The membrane was then probed with a primary antibodies for overnight incubation at 4°C. Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:2,000; cat. no. BS13278; Bioworld Technology, Inc., St. Louis Park, MN, USA) was then added for 1 h at room temperature, followed by incubation at 1:1,000 dilution in Tris-buffered saline with Tween-20. Finally, the protein bands were detected using ECL solution (Pierce Biotechnology, Inc., Rockford, IL, USA) and images were captured using a FluorChem imaging system (Alpha Innotech, San Leandro, CA, USA). GADPH was used as a loading control.
Luciferase assay
A luciferase assay was performed to determine whether SP1 was a direct target of miR-411. The cells were seeded into 12-well plates at 50% confluence. After 24 h at 37°C in an atmosphere containing 5% CO2, the cells were transfected with the luciferase reporter plasmid, miR-411 mimics or NC using Lipofectamine 2000 reagent, performed in three independent experiments. At 48 h post-transfection, the activities of firefly and Renilla luciferase were examined using a Dual-Luciferase Reporter Assay system (Promega Corporation). The activity of firefly luciferase was normalized to the activity of Renilla luciferase, respectively.
Statistical analysis
Data are presented as the mean ± standard deviation, and were compared using SPSS 17 software (SPSS, Inc., Chicago, IL, USA) using the Student's t-test or one-way analysis of variance followed by least significant difference test. The associations between miR-411 expression and clinicopathological factors were analyzed using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-411 is downregulated in breast cancer tissues and cell lines
The expression levels of miR-411 in breast cancer tissues and their corresponding adjacent non-neoplastic tissues were measured using RT-qPCR. As shown in Fig. 1A, miR-411 was significantly downregulated in the breast cancer tissues, compared with the corresponding adjacent non-neoplastic tissues (P<0.05).
Figure 1.
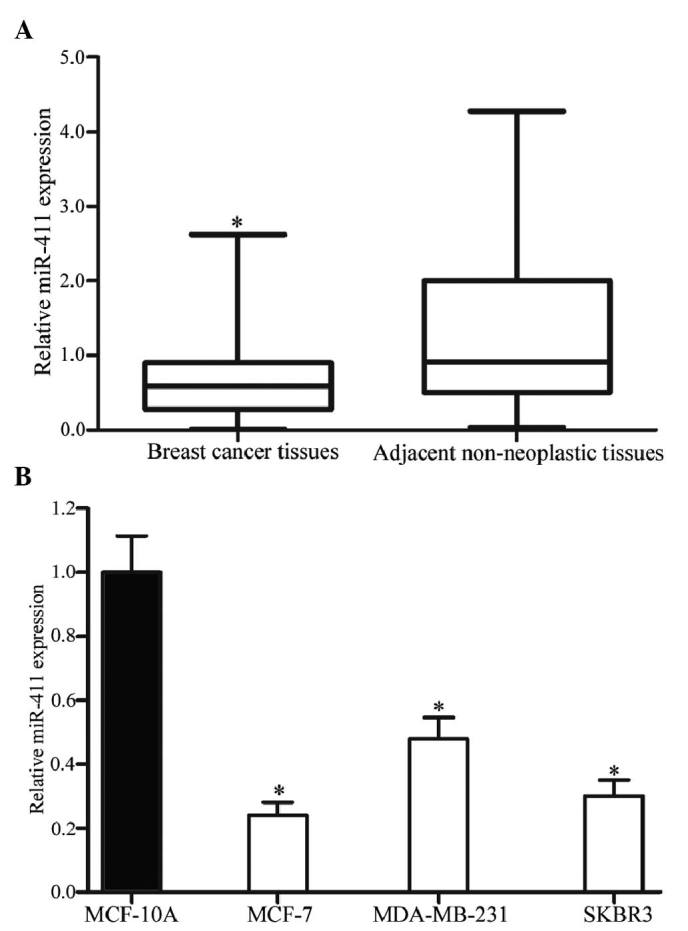
Expression levels of miR-411 in breast cancer tissues and cell lines. (A) Levels of miR-411 in the breast cancer tissues were significantly lower than in their corresponding adjacent non-neoplastic tissues. (B) Levels of miR-411 were also lower in the MCF-7, MDA-MB-231 and SKBR3 cell lines, compared with the MCF-10A cell line. Expression levels of miR-411 were determined by reverse transcription-quantitative polymerase chain reaction analysis and normalized to U6. Data are presented as the mean ± standard deviation. *P<0.05, compared with their respective controls. miR, microRNA.
In addition, the expression levels of miR-411 were detected in breast cancer cell lines and the normal mammary epithelial cell line, MCF-10A. As shown in Fig. 1B, downregulation in the levels of miR-411 were also observed in the MCF-7, MDA-MB-231 and SKBR3 cells, compared with the MCF-10A cells (P<0.05). In these three cell lines, the expression levels in the MCF-7 and SKBR3 cells were lowest, compared with that in the MDA-MB-231 cells. Therefore, MCF-7 and SKBR3 were selected for use in the present study to examine the functions of miR-411 in breast cancer.
Association between the expression of miR-411 and clinicopathological features in patients with breast cancer
To further assess the association between the expression of miR-411 and clinicopathological features in patients with breast cancer, statistical analysis was performed. As shown in Table I, a low expression level of miR-411 was closely associated with lymph node metastasis (P=0.034) and histological grade (P=0.003). However, no correlation was found between the expression of miR-411 and other clinicopathological factors, including age, tumor diameter, stage, and the expression of human epidermal growth factor receptor 2, Ki-67 or P53 (P>0.05).
Table I.
Association between the expression of miR-411 and clinicopathological factors.
| Clinical feature | Cases (n) | miR-411 expression
|
P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age | 0.257 | |||
| <50 years | 59 | 37 | 22 | |
| ≥50 years | 55 | 28 | 27 | |
| Tumor diameter | 0.230 | |||
| <2 cm | 76 | 40 | 36 | |
| ≥2 cm | 38 | 25 | 13 | |
| Lymph node metastasis | 0.034 | |||
| Negative | 46 | 32 | 14 | |
| Positive | 68 | 33 | 35 | |
| Stage | 0.349 | |||
| I–II | 50 | 26 | 24 | |
| III | 64 | 39 | 25 | |
| Histological grade | 0.003 | |||
| I–II | 72 | 49 | 23 | |
| III | 42 | 16 | 26 | |
| HER2 expression | 0.456 | |||
| Negative | 56 | 34 | 22 | |
| Positive | 58 | 31 | 27 | |
| Ki-67 expression | 0.848 | |||
| Negative | 46 | 27 | 19 | |
| Positive | 68 | 38 | 30 | |
| P53 expression | 0.835 | |||
| Negative | 33 | 18 | 15 | |
| Positive | 81 | 47 | 34 | |
miR, microRNA; HER2, human epidermal growth factor receptor 2.
miR-411 is upregulated in breast cancer cells following transfection with miR-411 mimics
In order to investigate the effects of miR-411 on breast cancer, the present study transfected the miR-411 mimic into the MCF-7 and SKBR3 cells. RT-qPCR was performed to assess transfection efficiency. At 72 h post-transfection, miR-411 was significantly overexpressed in the cells transfected with miR-411 mimics, compared with the cells transfected with NC (Fig. 2; P<0.05).
Figure 2.

Effect of miR-411 mimic transfection on the expression of miR-411. Transfection of miR-411 mimics into MCF-7 and SKBR3 cells significantly increased the expression of miR-411. Data are presented as the mean ± standard deviation. *P<0.05, compared with their respective NC. miR, microRNA; NC, negative control.
miR-411 suppresses the proliferation of breast cancer cells
An MTT assay was performed to examine the effect of miR-411 on cell proliferation. It was revealed that the upregulation of miR-411 significantly inhibited cell proliferation in the MCF-7 and SKBR3 cells (Fig. 3; P<0.05). At 96 h post-transfection, the inhibitory rates were 28.55±4.34% in the MCF-7 cells and 33.09±5.17% in the SKBR3 cells. These results verified that miR-411 functioned as a tumor suppressor in human breast cancer.
Figure 3.
Cell proliferation. MTT assays revealed significantly inhibited proliferation in miR-411-transfected cells, compared with NC cells of the MCF-7 and SKBR3 cell lines. Data are presented as the mean ± standard deviation. *P<0.05, compared with respective NC. miR, microRNA; NC, negative control.
miR-411 decreases the migration and invasion of breast cancer cells
To investigate the effect of miR-411 on cell motility, cell migration and invasion assays were performed. As shown in Fig. 4A and B, the migration and invasive abilities of the MCF-7 and SKBR3 cells transfected with miR-411 were significantly downregulated, compared with those of the cells transfected with the NC (P<0.05). These results indicated that miR-411 decreased the migration and invasive abilities of the breast cancer cells.
Figure 4.
Overexpression of miR-411 inhibits the migration and invasion of breast cancer cells. (A) miR-411 decreased the migration abilities of the MCF-7 and SKBR3 cells. (B) miR-411 decreased the invasive abilities of the MCF-7 and SKBR3 cells. Magnification, ×200. Data are presented as the mean ± standard deviation. *P<0.05, compared with their respective NC. miR, microRNA; NC, negative control.
SP1 is a direct target gene of miR-411 in vitro
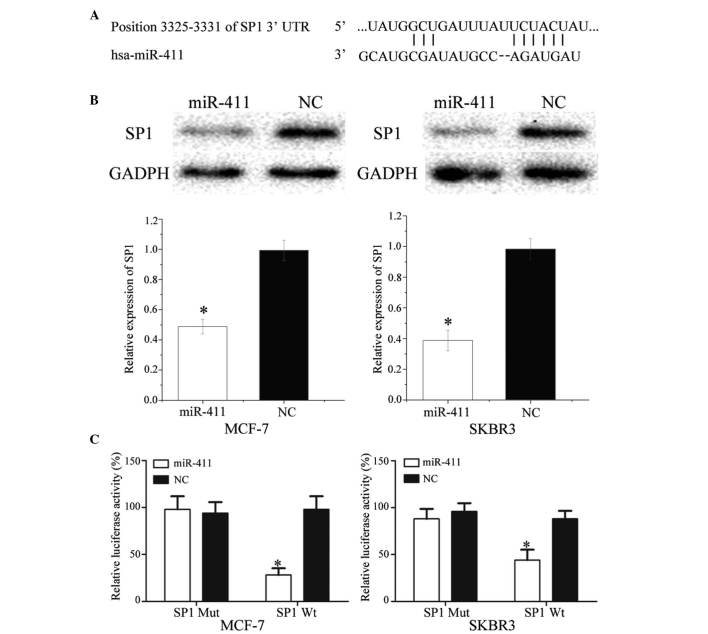
To identify the target of miR-411, TargetScan was used. SP1 was predicted to be a target of miR-411 (Fig. 5A). To verify whether miR-411 directly targets SP1, western blot analysis was performed to determine whether SP1 was downregulated at the protein level in breast cancer cells following transfection with miR-411. As shown in Fig. 5B, SP1 was significantly downregulated in the MCF-7 and SKBR3 cells following transfection with miR-411 (P<0.05).
Figure 5.
SP1 is a direct target gene of miR-411 in vitro. (A) TargetScan revealed that SP1 mRNA contained an miR-411 seed match at position 3,325–3,331 of the SP1 3′UTR. (B) Western blot analysis revealed that SP1 was significantly downregulated in MCF-7 and SKBR3 cells following transfection with miR-411. (C) Overexpression of miR-411 significantly inhibited the luciferase activity of SP1 Wt, but not SP1 Mut in the MCF-7 and SKBR3 cells. Data are presented as the mean ± standard deviation. *P<0.05, compared with their respective NC. SP1, specificity protein 1; miR, microRNA; Wt, wild-type; Mut, mutant; NC, negative control; 3′ UTR, 3′ untranslated region.
Luciferase assays were also performed to determine whether miR-411 directly targeted SP1. As shown in Fig. 5C, miR-411 significantly inhibited the luciferase activity of SP1 WT, but not SP1 Mut, in the MCF-7 and SKBR3 cells (P<0.05). Taken together, these results indicated SP1 was a direct target gene of miR-411 in vitro.
Discussion
In the last decade, accumulating evidence has suggested that the aberrant expression of miRNAs is a characteristic of malignancies, including breast cancer (8,20,21). miR-411 is located at the 14q32 region (22). In hepatocellular cell carcinoma (HCC), miR-411 has been found to be overexpressed in HCC tissues, compared with adjacent non-neoplastic tissues, and its expression was also found to be significantly upregulated in eight HCC cell lines, compared with the human hepatic cell line, LO2. Harafuji et al (23) reported that miR-411 was upregulated in primary and immortalized myoblasts of facioscapulohumeral muscular dystrophy (FSHD), compared with control myoblasts. However, the present study demonstrated that miR-411 was downregulated in breast cancer tumor samples and cell lines. These conflicting results suggest that the expression of miR-411 in cancer is tissue-type dependent, and the present study expanded current knowledge of the expression of miR-411 in cancer.
Previous studies have demonstrated that miRNAs are important in tumorigenesis and development (24). Studies have also indicated that the expression of miRNAs is significantly associated with the clinical recurrence and progression of metastasis (25,26). In HCC, miR-411 enhances cell growth and has been confirmed as an oncogene through the regulation of ITCH (27). In FSHD, miR-411 has been found to inhibit myogenic factors and may have an important function in myogenesis (23). However, there are no previous reports on the functions of miR-411 in breast cancer. In the present study, it was found that miR-411 suppressed breast cancer cell proliferation, migration and invasion. These results indicated that miR-411 has important functions in the tumorigenesis and development of breast cancer, and may have clinical implications in the treatment of breast caner.
The identification of miR-411 target genes is essential for understanding its functions in the carcinogenesis and progression of breast cancer. It is also important for investigating novel targeted therapies for breast cancer. In the present study, a molecular link between miR-411 and SP1 was identified. SP1, a member of the Sp/Krüppel-like factor transcription factor family, was the first transcription factor to be cloned from mammalian cells in 1983 (28). SP1 regulates gene expression through binding to GC-rich sequences (29). It has been found to be upregulated in several types of cancer, including breast cancer (30), gastric cancer (31), hepatocellular carcinoma (32), thyroid cancer (33), colorectal cancer (34), pancreatic cancer (35) and lung cancer (36). Accumulated evidence has indicated that SP1 is critical in a variety of physiological processes, including cell proliferation, apoptosis, cell cycle, differentiation, tumor metastasis and tumor development (37–40). The elevated expression of SP1 has also been found to be associated with poor prognosis in gastric cancer and breast cancer (41,42). These findings suggest that selecting SP1 as a therapeutic approach is practicable for breast cancer.
SP1 has been found to be regulated by multiple miRNAs in several types of cancer. For example, in HCC, miR-324-5p inhibits cell invasion by targeting SP1 (43). In addition, in gastric cancer, miR-145, miR-133a, miR-133b, miR-22 and miR-335 act as tumor suppressors via the direct inhibition of SP1 (31,44,45). In non-small cell lung cancer, miR-27b decreases cell proliferation and invasion through the regulation of SP1 (46). In addition, in human glioblastoma, miR-377 suppresses cell growth and invasion by inhibiting the activity of SP1 (47). In ovarian cancer, miR-145 enhances cell chemosensitivity to paclitaxel through targeting SP1. In esophageal carcinoma, miR-429 targets SP1 to inhibit cell invasion and enhance cell apoptosis (48). In prostate cancer, miR-330 decreases cell migration and invasion through the downregulation of SP1 (49). Studies have also revealed that miRNAs regulate the expression of SP1 (50–52). For example, miR-200b functions as a tumor suppressor by directly targeting SP1. In the present study, it was demonstrated that miR-411 has a suppressive role in breast cancer through the regulation of SP1. Therefore, miR-411/SP1-based targeted therapy may offer potential as a novel treatment for breast cancer.
In conclusion, the present study showed that miR-411 was downregulated in breast cancer, and was associated with lymph node metastasis and histological grade. It was also demonstrated that miR-411 inhibited breast cancer cell proliferation, migration and invasion by directly targeting SP1. Therefore, miR-411 may offer potential as a targeted therapy for breast cancer. Future investigations are required to address whether the potential of miR-411 may be fully realized in breast cancer therapy.
Acknowledgments
This study was supported by the National Natural Science Fund (grant no. 30500599), the Guangdong Natural Science Fund Project (grant no.≈9151503102000019), the Guangdong province Science and Technology Plan Project (grant no. 2013021800097), the Shenzhen Science and Technology Plan (Medical Treatment and Public Health) Key Project (grant no. 201201028) and the Shenzhen Technical Research and Development Fund Project (grant nos. CXZZ20140414170821163, GIHZ20130412153906740, NJCYJ20120615085512920, JCYJ20120613171430264 and ZYC201006180477A).
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Mangolini A, Ferracin M, Zanzi MV, Saccenti E, Ebnaof SO, Poma VV, Sanz JM, Passaro A, Pedriali M, Frassoldati A, et al. Diagnostic and prognostic microRNAs in the serum of breast cancer patients measured by droplet digital PCR. Biomark Res. 2015;3:12. doi: 10.1186/s40364-015-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann MW, Niederacher D, Schnürch HG, Gusterson BA, Bender HG. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J Mol Med (Berl) 1997;75:429–439. doi: 10.1007/s001090050128. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura S, Yagata H, Ohno S, Yamaguchi H, Iwata H, Tsunoda N, Ito Y, Tokudome N, Toi M, Kuroi K, Suzuki E. Multi-center study evaluating circulating tumor cells as a surrogate for response to treatment and overall survival in metastatic breast cancer. Breast Cancer. 2010;17:199–204. doi: 10.1007/s12282-009-0139-3. [DOI] [PubMed] [Google Scholar]
- 7.Gong Y, He T, Yang L, Yang G, Chen Y, Zhang X. The role of miR-100 in regulating apoptosis of breast cancer cells. Sci Rep. 2015;5:11650. doi: 10.1038/srep11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chávez J, Trevino V, Rodríguez-Padilla C, Reséndez-Pérez D. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9:53–59. doi: 10.1111/j.1743-7563.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Zhu Q, Tang L. MiR-99a antitumor activity in human breast cancer cells through targeting of mTOR expression. PLoS One. 2014;9:e92099. doi: 10.1371/journal.pone.0092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu C, Li J, Wang X, Song L. Unregulated miR-96 induces cell proliferation in human breast cancer by downregulating transcriptional factor FOXO3a. PLoS One. 2010;5:e15797. doi: 10.1371/journal.pone.0015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JC, Gundara JS, Glover A, Serpell J, Sidhu SB. MicroRNA expression profiles in the management of papillary thyroid cancer. Oncologist. 2014;19:1141–1147. doi: 10.1634/theoncologist.2014-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96(Suppl):R40–R44. [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Abdel-Mageed AB, Mondal D, Kandil E. MicroRNA expression profiles in differentiated thyroid cancer, a review. Int J Clin Exp Med. 2013;6:74–80. [PMC free article] [PubMed] [Google Scholar]
- 15.Xia M, Li H, Wang JJ, Zeng HJ, Wang SH. MiR-99a suppress proliferation, migration and invasion through regulating insulin-like growth factor 1 receptor in breast cancer. Eur Rev Med Pharmacol Sci. 2016;20:1755–1763. [PubMed] [Google Scholar]
- 16.Wu D, Niu X, Pan H, Zhou Y, Qu P, Zhou J. MicroRNA-335 is downregulated in bladder cancer and inhibits cell growth, migration and invasion via targeting ROCK1. Mol Med Rep. 2016;13:4379–4385. doi: 10.3892/mmr.2016.5055. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Bo L, Lu W, Zhou G, Chen Q. MicroRNA-148b targets Rho-associated protein kinase 1 to inhibit cell proliferation, migration and invasion in hepatocellular carcinoma. Mol Med Rep. 2016;13:477–482. doi: 10.3892/mmr.2015.4500. [DOI] [PubMed] [Google Scholar]
- 18.Marini F, Luzi E, Brandi ML. MicroRNA role in thyroid cancer development. J Thyroid Res. 2011;2011:407123. doi: 10.4061/2011/407123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Abedi N, Mohammadi-Yeganeh S, Koochaki A, Karami F, Paryan M. miR-141 as potential suppressor of β-catenin in breast cancer. Tumour Biol. 2015;36:9895–9901. doi: 10.1007/s13277-015-3738-y. [DOI] [PubMed] [Google Scholar]
- 21.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 22.Nadal E, Zhong J, Lin J, Reddy RM, Ramnath N, Orringer MB, Chang AC, Beer DG, Chen G. A MicroRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin Cancer Res. 2014;20:3107–3117. doi: 10.1158/1078-0432.CCR-13-3348. [DOI] [PubMed] [Google Scholar]
- 23.Harafuji N, Schneiderat P, Walter MC, Chen YW. miR-411 is up-regulated in FSHD myoblasts and suppresses myogenic factors. Orphanet J Rare Dis. 2013;8:55. doi: 10.1186/1750-1172-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi B, Piazza GA, Su X, Xi Y. MicroRNA and cancer chemoprevention. Cancer Prev Res (Phila) 2013;6:401–409. doi: 10.1158/1940-6207.CAPR-13-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang JX, Song W, Chen ZH, Wei JH, Liao YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: A microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 26.Heinzelmann J, Unrein A, Wickmann U, Baumgart S, Stapf M, Szendroi A, Grimm MO, Gajda MR, Wunderlich H, Junker K. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: A comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21:1046–1054. doi: 10.1245/s10434-013-3361-3. [DOI] [PubMed] [Google Scholar]
- 27.Xia K, Zhang Y, Cao S, Wu Y, Guo W, Yuan W, Zhang S. miR-411 regulated ITCH expression and promoted cell proliferation in human hepatocellular carcinoma cells. Biomed Pharmacother. 2015;70:158–163. doi: 10.1016/j.biopha.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 29.Kadonaga JT, Courey AJ, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 30.Yue L, Li L, Liu F, Hu N, Zhang W, Bai X, Li Y, Zhang Y, Fu L, Zhang X, Ye L. The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis. 2013;34:927–935. doi: 10.1093/carcin/bgs399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang X, Jiang L, Sun Z, Miao Z, Xu H. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin P, Zhao C, Li Z, Mei C, Yao W, Liu Y, Li N, Qi J, Wang L, Shi Y, et al. Sp1 is involved in regulation of cystathionine gamma-lyase gene expression and biological function by PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell Signal. 2012;24:1229–1240. doi: 10.1016/j.cellsig.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Bonofiglio D, Qi H, Gabriele S, Catalano S, Aquila S, Belmonte M, Ando S. Peroxisome proliferator-activated receptor gamma inhibits follicular and anaplastic thyroid carcinoma cells growth by upregulating p21Cip1/WAF1 gene in a Sp1-dependent manner. Endocr Relat Cancer. 2008;15:545–557. doi: 10.1677/ERC-07-0272. [DOI] [PubMed] [Google Scholar]
- 34.Pathi S, Jutooru I, Chadalapaka G, Nair V, Lee SO, Safe S. Aspirin inhibits colon cancer cell and tumor growth and downregulates specificity protein (Sp) transcription factors. PLoS One. 2012;7:e48208. doi: 10.1371/journal.pone.0048208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C, Xie K. Crosstalk of Sp1 and Stat3 signaling in pancreatic cancer pathogenesis. Cytokine Growth Factor Rev. 2012;23:25–35. doi: 10.1016/j.cytogfr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YT, Chuang JY, Shen MR, Yang WB, Chang WC, Hung JJ. Sumoylation of specificity protein 1 augments its degradation by changing the localization and increasing the specificity protein 1 proteolytic process. J Mol Biol. 2008;380:869–885. doi: 10.1016/j.jmb.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 37.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 38.Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 39.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Guan X, Zhang J, Jia Z, Wei D, Li Q, Yao J, Xie K. Targeted inhibition of Sp1-mediated transcription for antiangiogenic therapy of metastatic human gastric cancer in orthotopic nude mouse models. Int J Oncol. 2008;33:161–167. [PubMed] [Google Scholar]
- 41.Zhang J, Zhu ZG, Ji J, Yuan F, Yu YY, Liu BY, Lin YZ. Transcription factor Sp1 expression in gastric cancer and its relationship to long-term prognosis. World J Gastroenterol. 2005;11:2213–2217. doi: 10.3748/wjg.v11.i15.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright C, Angus B, Napier J, Wetherall M, Udagawa Y, Sainsbury JR, Johnston S, Carpenter F, Horne CH. Prognostic factors in breast cancer: Immunohistochemical staining for SP1 and NCRC 11 related to survival, tumour epidermal growth factor receptor and oestrogen receptor status. J Pathol. 1987;153:325–331. doi: 10.1002/path.1711530406. [DOI] [PubMed] [Google Scholar]
- 43.Cao L, Xie B, Yang X, Liang H, Jiang X, Zhang D, Xue P, Chen D, Shao Z. MiR-324-5p suppresses hepatocellular carcinoma cell invasion by counteracting ECM degradation through post-transcriptionally downregulating ETS1 and SP1. PLoS One. 2015;10:e0133074. doi: 10.1371/journal.pone.0133074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo MM, Hu LH, Wang YQ, Chen P, Huang JG, Lu N, He JH, Liao CG. miR-22 is down-regulated in gastric cancer, and its overexpression inhibits cell migration and invasion via targeting transcription factor Sp1. Med Oncol. 2013;30:542. doi: 10.1007/s12032-013-0542-7. [DOI] [PubMed] [Google Scholar]
- 45.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 46.Jiang J, Lv X, Fan L, Huang G, Zhan Y, Wang M, Lu H. MicroRNA-27b suppresses growth and invasion of NSCLC cells by targeting Sp1. Tumour Biol. 2014;35:10019–10023. doi: 10.1007/s13277-014-2294-1. [DOI] [PubMed] [Google Scholar]
- 47.Zhang R, Luo H, Wang S, Chen W, Chen Z, Wang HW, Chen Y, Yang J, Zhang X, Wu W, et al. MicroRNA-377 inhibited proliferation and invasion of human glioblastoma cells by directly targeting specificity protein 1. Neuro Oncol. 2014;16:1510–1522. doi: 10.1093/neuonc/nou111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Li M, Zang W, Ma Y, Wang N, Li P, Wang T, Zhao G. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) 2013;36:385–394. doi: 10.1007/s13402-013-0144-6. [DOI] [PubMed] [Google Scholar]
- 49.Mao Y, Chen H, Lin Y, Xu X, Hu Z, Zhu Y, Wu J, Xu X, Zheng X, Xie L. microRNA-330 inhibits cell motility by downregulating Sp1 in prostate cancer cells. Oncol Rep. 2013;30:327–333. doi: 10.3892/or.2013.2452. [DOI] [PubMed] [Google Scholar]
- 50.Cui F, Wang S, Lao I, Zhou C, Kong H, Bayaxi N, Li J, Chen Q, Zhu T, Zhu H. miR-375 inhibits the invasion and metastasis of colorectal cancer via targeting SP1 and regulating EMT-associated genes. Oncol Rep. 2016;36:487–493. doi: 10.3892/or.2016.4834. [DOI] [PubMed] [Google Scholar]
- 51.Kang M, Xiao J, Wang J, Zhou P, Wei T, Zhao T, Wang R. MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 2016;5:1163–1173. doi: 10.1002/cam4.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X, Chen L, Wang W, Meng FB, Zhao RT, Chen Y. MicroRNA-150 inhibits cell invasion and migration and is downregulated in human osteosarcoma. Cytogenet Genome Res. 2015;146:124–135. doi: 10.1159/000437379. [DOI] [PubMed] [Google Scholar]