Abstract
Tight junctions are the outermost structures of intercellular junctions and are classified as transmembrane proteins. These factors form selective permeability barriers between cells, act as paracellular transporters and regulate structural and functional polarity of cells. Although tight junctions have been previously studied, comparison of the transcriptional-translational levels of these molecules in canine organs remains to be investigated. In the present study, organ-specific expression of the tight junction proteins, claudin, occludin, junction adhesion molecule A and zona occludens 1 was examined in the canine duodenum, lung, liver and kidney. Results of immunohistochemistry analysis demonstrated that the tight junctions were localized in intestinal villi and glands of the duodenum, bronchiolar epithelia and alveolar walls of the lung, endometrium and myometrium of the hepatocytes, and the distal tubules and glomeruli of the kidney. These results suggest that tight junctions are differently expressed in organs, and therefore may be involved in organ-specific functions to maintain physiological homeostasis.
Keywords: Canis lupus familiaris, tight junction, claudin, junction adhesion molecule-A, zona occludens 1, occludin
Introduction
Intercellular junctions include gap junctions, adhering junctions and tight junctions. Gap junctions chemically and electrically link adjacent cells. Tight junctions build the intercellular barrier. Together, these junctions modulate intracellular and intercellular signaling and transport (1). These junctions perform a number of roles, including acting as barriers against the extracellular environment, regulating cellular permeability and polarity, and serving as intermediates/transducers in cell signaling cascades (2).
Tight junction proteins are commonly classified into three types: Transmembrane, cytoskeletal and cytoplasmic plaque proteins (3). Claudin (CLDN), occludin (OCLN) and junction adhesion molecule A (JAM-A) are transmembrane proteins, whereas zona occludens 1 (ZO-1) is a cytoplasmic plaque protein.
CLDNs are integral transmembrane proteins (4), containing four transmembrane domains and two extracellular loops. The first extracellular loop is long and influences paracellular charge selectivity. The second extracellular loop is shorter and facilitates CLDN-CLDN interactions (5,6). CLDNs contribute to the polarity of epithelial cells and regulate the paracellular selective permeability of ions (7). The expression pattern and function of each CLDN varies depending on the organ (8). OCLN is a primary transmembrane protein consisting of four transmembrane domains, two extracellular loops and three cytoplasmic domains (9). The unique charged structures of the extracellular loops may enable cells to attach to each other (10). OCLN acts as a paracellular barrier, maintains cell surface polarity through interaction with ZO-1 and regulates the permeability of endothelial cells (11,12). Expression of this protein has been reported in numerous organs and tissues, including the brain, testis, epididymis, kidney, liver, lung, stomach, duodenum, ileum, colon, skin and blood vessels (13,14). JAM-A is an integral transmembrane protein with numerous functions, including intercellular junction assembly, paracellular barrier formation, promotion of leukocyte migration, platelet activation and angiogenesis (15). ZO-1 interacts directly with transmembrane factors including CLDNs, OCLN and JAM-A, and is known to organize and regulate the structure of tight junctions (16). In addition, ZO-1 shuttles between the nucleus and plasma membrane (17), modulates paracellular permeability and is involved in the control of gene expression (18). ZO-1 is expressed in numerous organ types (19).
Tight junction proteins are physiologically important and have been associated with numerous diseases. Therefore, a number of investigations have been conducted to examine the expression and regulation of tight junction proteins in animals and humans. However, few studies have been performed using dogs as the experimental model. In particular, comparisons of mRNA and protein expression levels in canine organs remain to be performed. Therefore, the aim of the present study was to measure the expression and localization of CLDNs (CLDN1, 2, 4 and 5), OCLN, JAM-A and ZO-1 in dogs. The organ-specific mRNA and protein expression levels of these factors in canine duodenum, lung, liver and kidney were measured using reverse transcription-polymerase chain reaction (RT-PCR), RT-quantitative PCR (RT-qPCR) and western blot analysis. Localization of proteins in the canine organs was determined by immunohistochemistry.
Materials and methods
Experimental animals
A total of 3 female beagle dogs (age, 3 years; mean weight, 9.5 kg; Central Lab Animal Inc., Seoul, Korea) were sacrificed for the present study. The dogs were fed a commercial diet (Natural Balance Korea, Inc., Suwon, Korea) and tap water and were housed in stainless steel cages in a controlled environment maintained with 12-h light/dark cycles (temperature, 23±2°C; relative humidity, 50±10%; ventilation, 17±1 times/min). The dogs were euthanized by administering 20–30 ml KCl (100 g KCl dissolved in 1 l water) intravenously. A midline incision was made to collect samples from the duodenum, lung, liver and kidney. The Institutional Animal Care and Use Committee of Chungbuk National University (Cheongju, Korea) approved all experimental procedures.
Total RNA extraction, RT-PCR and RT-qPCR
Organs were placed in TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and homogenized with ULTRA-TURRAX® (IKA-Works, Breisgau, Germany), according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed using the first strand Maloney murine leukemia virus reverse transcriptase (Intron Biotechnology, Inc., Seongnam, Korea) and random 9-mer primer (Takara Bio, Inc., Otsu, Japan) to synthesize complementary DNA (cDNA).
β-actin, a housekeeping control gene, served as a loading control and an endogenous reference for normalization of target gene expression levels for RT-PCR and RT-qPCR. The following dog-specific primers were used: CLDN1, sense, 5′-AAG ACG ATG AGG TGC AGA AG-3′ and antisense, 5′-GTG AAG AGA GCC TGA CCA AA-3′; CLDN2, sense, 5′-TGA GAT GCA CTG TCT TCT GC-3′ and antisense, 5′-AGC TTC TCC GAT CTC GAA CT-3′; CLDN4, sense, 5′-TCA TGG TCG TCA GCA TCA-3′ and antisense, 5′-AGT CCC GGA TGA TAT TGT TG-3′; CLDN5 sense, 5′-GGC CAT TGT GCA GAA GAA-3′ and antisense, 5′-TGA CTC AAC AGT CTG TCC TCC T-3′; OCLN sense, 5′-AAG AAC TCT CTC GCC TGG AT-3′ and antisense, 5′-GAT GTG GGA CAA TTT GCT CT-3′; JAM-a, sense, 5′-CTT CGA TCC TGT GTC AGC TT-3′ and antisense, 5′-TCT ATA GGC GAA CCA GAT GC-3′; ZO-1 sense, 5′-GGA GAG GTG TTT CGT GTT GT-3′ and antisense, 5′-ACT GCT CAG CCC TGT TCT TA-3′; β-actin, sense, 5′-AAG TCC AGC TTC TGT TTC CTC-3′ and antisense, 5′-GCA GTG ATC TCC TTC TGC AT-3′.
For RT-PCR, genes were amplified in a 20 μl PCR reaction containing 1 unit of i-StarTaq™ DNA polymerase (Intron Biotechnology, Inc.), 1.5 mM MgCl2, 2 mM dNTP and 20 pmol primers. The cycling conditions were as follows: 30 cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 30 sec. PCR products (10 μl) were separated on a 2.3% agarose gel and stained with ethidium bromide. Gel images were captured under UV illumination using a Gel Doc EQ system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RT-qPCR was performed on 1 μl cDNA using SYBR® (Takara Bio, Inc.) or TaqMan® (Applied Biosystems; Thermo Fisher Scientific, Inc.) probe methods, according to the manufacturer's instructions. The cycling conditions were as follows: A total of 40 cycles of denaturation at 95°C for 15 sec, annealing at 58°C for 15 sec and extension at 72°C for 30 sec. Data for each sample were analyzed by comparing quantification cycle (Cq) values at constant fluorescence intensity. The quantity of transcript was inversely associated with the observed Cq, and for every two-fold dilution of the transcript, the Cq was expected to increase by one increment. Relative expression (R) was calculated using the equation: R=2−ΔΔCq (20).
Western blotting
Organs were placed in 50 or 1,000 μl Pro-prep™ Protein Extraction solution (Intron Biotechnology, Inc.) depending on organ volume, and were then homogenized. Total protein (60 μg) was loaded onto 5–12.5% sodium dodecyl sulfate-polyacrylamide gels, electrophoresed, and transferred onto polyvinylidene difluoride membranes (PerkinElmer, Inc., Waltham, MA, USA). After blocking in 5% skim milk for 1 h, membranes were subsequently incubated overnight at 4°C with the following primary antibodies: Mouse anti-CLDN1 (dilution, 1:1,000; cat. no. 37-4900; Invitrogen; Thermo Fisher Scientific, Inc.), mouse anti-CLDN2 (dilution, 1:1,000; cat. no. 32-5600; Invitrogen; Thermo Fisher Scientific, Inc.), mouse anti-CLDN5 (dilution, 1:1,000; cat. no. 35-2500; Invitrogen; Thermo Fisher Scientific, Inc.), rabbit anti-CLDN4 (dilution, 1:500; cat. no. 32-9400; Invitrogen; Thermo Fisher Scientific, Inc.), rabbit anti-OCLN (dilution, 1:1,000; cat. no. 71-1500; Invitrogen; Thermo Fisher Scientific, Inc.), rabbit anti-ZO-1 (dilution, 1:1,000; cat. no. 40-2200; Invitrogen; Thermo Fisher Scientific, Inc.), rabbit anti-JAM-A (dilution, 1:1,000; cat. no. sc-25629; Santa Cruz Biotechnology Inc., Dallas, TX, USA) and rabbit anti-β-actin (dilution, 1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.), diluted in 5% bovine serum albumin (BSA; cat. no. 0903; Amresco, LLC, Solon, OH, USA) solution dissolved in phosphate-buffered saline (PBS). The horseradish peroxidase-conjugated secondary antibodies, poly-clonal anti-mouse (dilution, 1:2,000; cat. no. bs-0330R-HRP; Bioss, Woburn, MA, USA) and polyclonal anti-rabbit (dilution, 1:2,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.), were diluted to 1:3,000 in 2.5% skim milk dissolved in Tris-buffered saline containing Tween 20 and were incubated for 2 h at room temperature. Protein bands were visualized with an Enhanced Chemiluminescence reagent (Santa Cruz Biotechnology, Inc.) and exposed to Biomax™ Light film (Kodak, Rochester, NY, USA) for 1–5 min. Signal specificity was confirmed by blotting without the primary antibody, and bands were normalized to β-actin. Signal intensity for each band was measured using ImageJ software (version 1.50e; National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry
Organ-specific localization of tight junction proteins was investigated by immunohistochemistry. After 1 week of 10% formalin fixation, tissue processing was performed using the Tissue-Tek VIP 5 (Sakura Finetek USA, Inc., Torrance, CA, USA) to embed tissues in paraffin. Embedded blocks were sectioned at 3.5 μm. Each slide was boiled in buffered citrate solution for 20 min to effect antigen retrieval and washed with TBS-T for 5 min. To block endogenous peroxidase activity, slides were placed in 3% hydrogen peroxide for 30 min at room temperature. To prevent non-specific antibody binding, sections were incubated with 10% goat serum (Vector Laboratories, Inc., Burlingame, CA, USA) in phosphate-buffered saline for 1 h at room temperature. The slides were incubated with the primary antibodies listed in the western blotting section diluted 1:200 in 5% BSA dissolved in PBS, overnight at room temperature in a moist chamber. Biotinylated anti-mouse or anti-rabbit secondary antibodies (dilution, 1:400; cat. nos. BA-9200 and BA-1000, respectively; Vector Laboratories, Inc.) were added and incubated at 37°C for 2 h. Slides were washed as described previously. Elite ABC kit (Vector Laboratories, Inc.) was added to slides and incubated at 37°C for 2 h. Diaminobenzidine (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) was used as a chromogen. Sections were counterstained with Harris hematoxylin (Sigma-Aldrich; Merck Millipore).
Statistical analysis
The results of all experiments are presented as the mean ± standard deviation, which was calculated using GraphPad Prism software program, (version 4.0; GraphPad Software, Inc., La Jolla, CA, USA).
Results
Organ-specific mRNA expression levels of CLDN family members
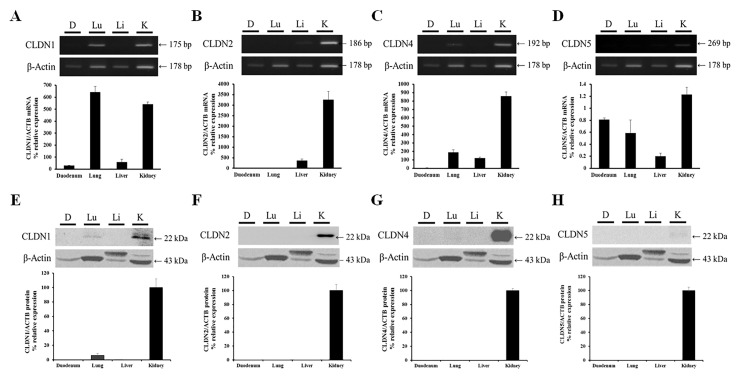
Organ-specific mRNA expression levels of CLDN1, 2, 4 and 5 in duodenum, lung, liver and kidney tissues were analyzed by RT-PCR and RT-qPCR. β-actin served as an internal control. The expression patterns of CLDNs varied among the different organs. CLDN1 was expressed in all organs examined with the greatest expression levels in lung. Expression levels were relatively high in the kidney and low in the liver and duodenum (Fig. 1A). CLDN2 expression levels were high in the kidney, low in liver and undetectable in duodenum and lung (Fig. 1B). CLDN4 was expressed in all the examined organs except for the duodenum; the greatest expression levels were observed in the kidney (Fig. 1C). CLDN5 mRNA was barely detected; however, its expression levels were greatest in the kidney (Fig. 1D).
Figure 1.
Organ-specific mRNA and protein expression levels of CLDN1, 2, 4 and 5 in the canine duodenum, lung, liver and kidney. mRNA expression levels of (A) CLDN1 were greatest in the lung and kidney, whereas (B) CLDN2 and (C) CLDN4 mRNA was primarily expressed in kidney. (D) CLDN5 mRNA expression levels were low. (E) CLDN1 protein expression levels were high in the kidney, low in the lung and undetectable in duodenum and liver. (F) CLDN2, (G) CLDN4 and (H) CLDN5 protein expression was detected only in the kidney. mRNA and protein expression were normalized to β-actin. Data are presented as the mean ± standard deviation. CLDN, claudin; ACTB, β-actin; D, duodenum; Lu, lung; Li, liver; K, kidney.
Organ-specific protein expression levels and localization of CLDN family members
Organ-specific protein expression levels of CLDN1, 2, 4 and 5 were determined in the duodenum, lung, liver and kidney samples obtained from all three dogs by western blotting (Fig. 1E–H). β-actin served as an internal control. Greater protein expression levels of CLDN1 were detected in the kidney compared with the lung; however, CLDN1 protein was not detected in the duodenum or liver. High protein expression levels of CLDN2, 4 and 5 were detected in the kidney; however, these proteins were not detected in other organs, suggesting that CLDN family members may have tissue-specific roles in the kidney.
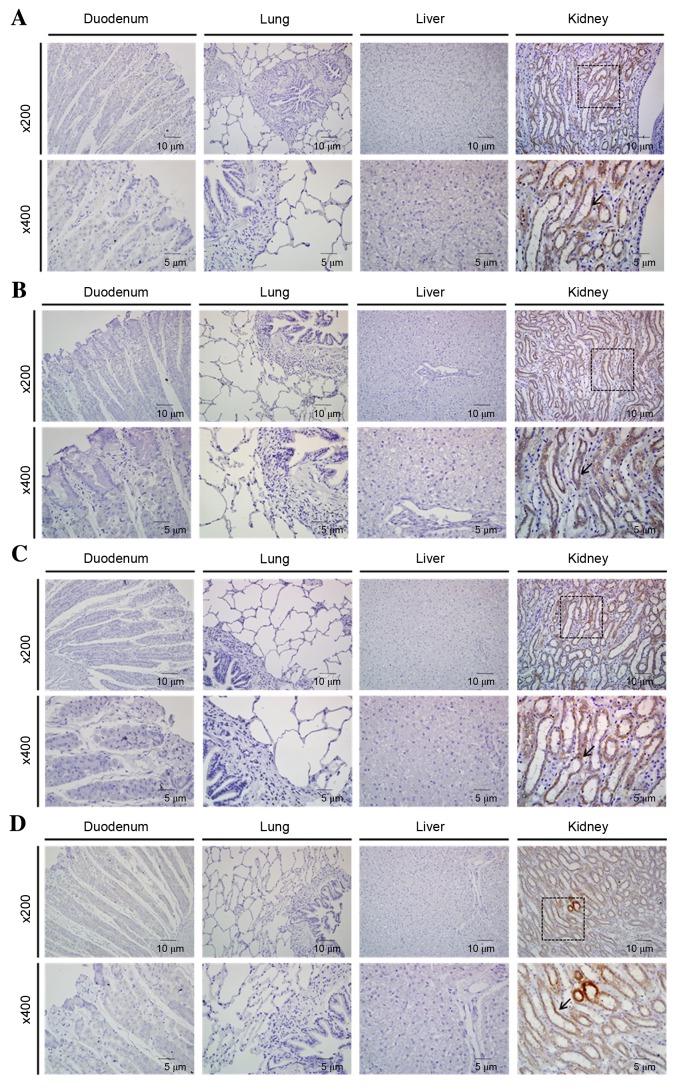
Localization of CLDN1, 2, 4 and 5 in the duodenum, lung, liver and kidney was evaluated by immunohistochemical analysis. CLDN1, 2, 4 and 5 was detected in the kidney tissues, but not within the duodenum, lung or liver tissues (Fig. 2). Within the kidney the localization sites of all CLDNs were identical, with expression detected in the renal distal tubules.
Figure 2.
Organ-specific localization of CLDN1, 2, 4 and 5 in canine duodenum, lung, liver and kidney, as detected by immunohistochemistry. (A) CLDN1, (B) CLDN2, (C) CLDN4 and (D) CLDN5 were detected only in the kidney. Boxes in the upper panels (magnification, ×200) are areas magnified in the lower panels (magnification, ×400). Organs for which no boxes are indicated are those that revealed no detectable staining. Arrows indicate positive staining.
Organ-specific mRNA expression levels of OCLN, JAM-A and ZO-1
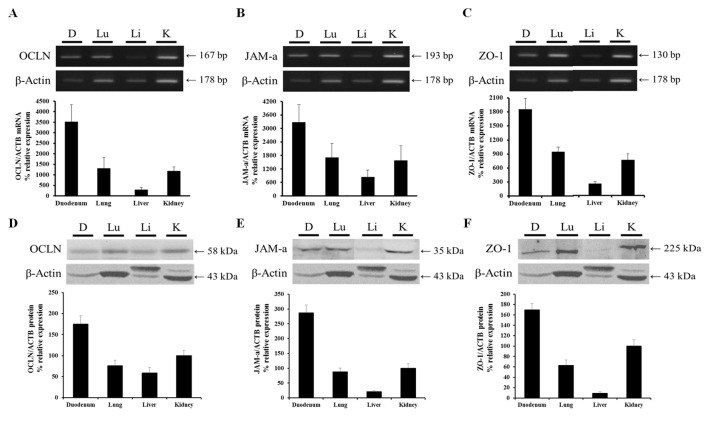
RT-PCR and qPCR results demonstrated that OCLN mRNA expression levels were greatest in the duodenum compared with the other organs. OCLN expression levels were moderate in the lung and kidney and low in the liver (Fig. 3A). JAM-A mRNA expression levels were greatest in the duodenum followed (in descending order) by the lung, kidney and liver (Fig. 3B). ZO-1 mRNA expression levels were greater in the duodenum compared with the other organs. As for OCLN and JAM-A, ZO-1 expression levels were greatest in the duodenum followed (in descending order) by the lung, kidney and liver (Fig. 3C).
Figure 3.
Organ-specific mRNA and protein expression levels of OCLN, JAM-A and ZO-1 in canine duodenum, lung, liver and kidney. mRNA expression levels of (A) OCLN, (B) JAM-A and (C) ZO-1 were greatest in the duodenum, moderate in the lung and kidney, and low in the liver. Protein expression levels of (D) OCLN, (E) JAM-A and (F) ZO-1 followed a similar pattern to mRNA expression levels. mRNA and protein expression were normalized to β-actin. Data are presented as the mean ± standard deviation. OCLN, occludin; JAM-A, junction adhesion molecule A; ZO-1, zona occludens 1; ACTB, β-actin; D, duodenum; Lu, lung; Li, liver; K, kidney.
Organ-specific protein expression levels of OCLN, JAM-A and ZO-1
Western blotting identified that OCLN was expressed in all the examined organs, with a pattern of expression similar to that of mRNA. Protein expression levels were greatest in the duodenum followed (in descending order) by the kidney, lung and liver (Fig. 3D). JAM-A protein was expressed in all the examined organs with the greatest levels observed in the duodenum. The expression levels in the kidney and lung were similar and relatively high compared with the liver, which were in agreement with the JAM-A mRNA expression levels (Fig. 3E). ZO-1 protein was expressed in all the examined organs, and the pattern of protein expression was similar to that of mRNA. The ZO-1 protein expression levels were greatest in the duodenum, moderate in the kidney and lung and low in the liver (Fig. 3F).
Organ-specific localization of OCLN, JAM-A and ZO-1
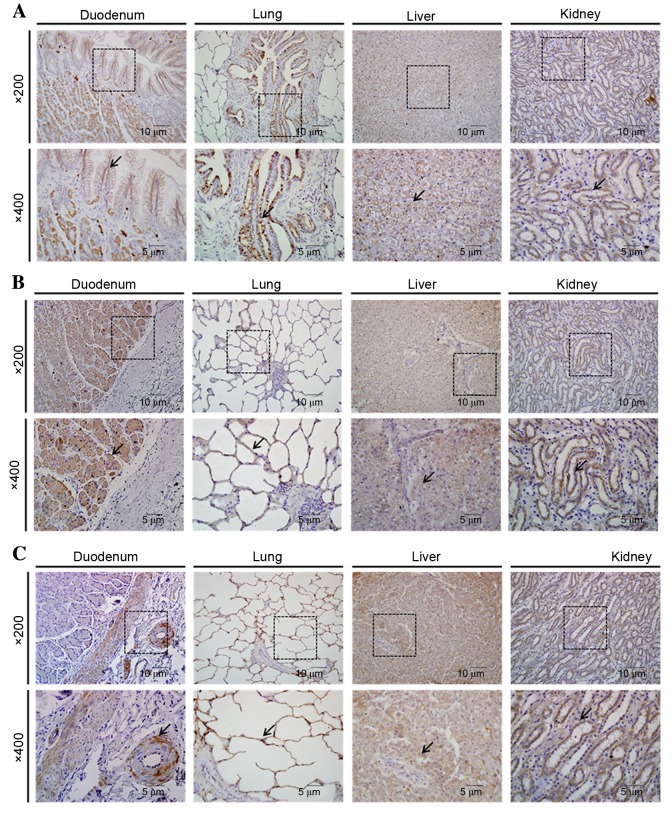
OCLN-specific immunohistochemical staining was observed in all the examined organs. This protein was observed in intestinal villi of the duodenum, bronchiolar epithelium of the lung, connective tissue stroma of the hepatocytes and renal distal tubules (Fig. 4A). JAM-A immunohistochemical staining was observed in all the examined organs, in intestinal villi of the duodenum, alveolar walls of the lung, hepatocytes and in renal distal tubules (Fig. 4B). Immunohistochemistry indicated that ZO-1 was localized in muscularis mucosae and submucosal glands of the duodenum, alveolar walls of the lung, hepatocytes, and distal tubules and glomeruli of the kidney (Fig. 4C).
Figure 4.
Organ-specific localization of OCLN, JAM-A and ZO-1 in canine duodenum, lung, liver, and kidney, as detected by immunohistochemistry. (A) OCLN, (B) JAM-A and (C) ZO-1 immunostaining was detected in all organs examined. Boxes in the upper panels (magnification, ×200) are areas magnified in the lower panels (magnification, ×400). Arrows indicate positive staining. OCLN, occludin; JAM-A, junction adhesion molecule A; ZO-1, zona occludens 1.
Discussion
The present study examined the organ-specific expression and localization of transmembrane proteins (CLDNs, OCLN and JAM-A) and a cytoplasmic plaque protein (ZO-1) in canine organs. Organ-specific variations in transelectrical resistance and paracellular ionic selectivity among epithelia have been demonstrated to be determined by the differential expression and distribution of CLDNs (21). The results of the present study demonstrated that CLDN mRNA and protein were highly expressed in the kidney and commonly localized to the renal distal tubules. This expression and distribution pattern in canine organs differs from those in other animals (22) and humans (23). Compared with humans, low expression levels of CLDN 1, 2, 4 and 5 have been observed in the duodenum, lung and liver of mice (22). The gatekeeper function of CLDNs determines the renal epithelial tight junction paracellular permeability; therefore, CLDNs are associated with renal physiology and pathology. Abnormal expression of CLDNs induces mineral imbalances, impedes recovery from ischemic acute renal injury and disrupts acid/base homeostasis in humans and rats (24). Based on data from the literature and the results of the present study, CLDNs may be critical for maintaining renal physiology in dogs.
In contrast to CLDNs, mRNA and protein expression of OCLN, JAM-A and ZO-1 were detected in all the examined organs. Expression patterns of three factors were similar. As for CLDNs, the expression and distribution patterns of OCLN, JAM-A and ZO-1 were different compared with other species (25). For instance, high expression levels of ZO-1 and JAM-A have been observed in the livers of mice (25), whereas, in the present study, low expression levels of these factors were observed in canine livers when compared with other organs. Expression of OCLN, JAM-A and ZO-1 was greater in canine duodenum compared with other organs, and was localized to the intestinal villi and submucosal gland. Numerous groups have demonstrated that tight junction molecules act as epithelial barriers and regulators of inflammatory responses in the intestine. Tight junctions regulate mucosal permeability and enteric pathogens disrupt intestinal physiological functions in various intestinal inflammatory diseases, including inflammatory bowel disease and Crohn's disease (26). As these diseases are observed in dogs, tight junctions may be associated with canine intestinal physiology and pathology. In support of this, a previous study demonstrated that CLDN2 expression in the superficial colonic epithelium is upregulated in dogs with idiopathic colitis (27). OCLN, JAM-A and ZO-1 mRNA and protein expression was observed in the lung, where tight junctions form the alveolar membrane barrier. In addition, tight junctions of the alveolar epithelial and capillary endothelial cells maintain alveolar wall permeability (28). In a previous study, reduction of OCLN expression induced by acute lung injury led to reduced alveolar membrane integrity and elevated alveolar membrane permeability (25). Furthermore, tight junctions have been revealed to help prevent microbial invasion and interact with dendritic cells in pulmonary epithelia (29). Renal expression of OCLN, JAM-A and ZO-1 was localized to the distal tubules and glomeruli. In the kidney, tight junctions of the renal epithelia are widely distributed in renal tubules, and have a transepithelial potential difference and various morphological and functional characteristics. In general, tight junctions regulate the transfer of various solutes and water, and inhibit the interaction between apical and basolateral plasmolemmal domains. It has been determined that ZO-1 acts as a coxsackievirus and adenovirus receptor in glomerular podocytes, and serves as a barrier that controls the movement of macromolecules and ions (30). Thus, tight junctions are important in viral infection of the kidney and renal disease development. The mRNA and protein expression levels of OCLN, JAM-A and ZO-1 in the liver were observed to be relatively low compared with the other examined organs. Hepatocytes closely interact with each other through intercellular junctions to form hepatocytic plates and maintain liver function. Tight junctions of hepatocytes tightly encircle the bile canaliculi, and the cells form a blood-biliary barrier and maintain epithelial polarity (24). In addition, tight junction molecules bind to hepatitis C virus to mediate viral infection and fatty liver-associated disease. In addition, dysfunction and downregulation of tight junctions is associated with primary and metastatic tumor development (31).
In conclusion, the results of the present study demonstrated that the canine tight junction proteins, CLDNs, OCLN, JAM-A and ZO-1 were expressed and regulated in a tissue-specific manner. The data suggest that these proteins may perform unique physiological roles in tissues where they are highly expressed. Therefore, these observations may serve as a basis for further studies of tight junction proteins and their roles in canine physiological or pathological conditions.
Acknowledgments
The present study was supported by the National Research Foundation of Korea (grant no. NRF-2013R1A2A2A05004582) and the Korea Foundation for the Advancement of Science & Creativity, funded by the Korean government.
References
- 1.Green KJ, Getsios S, Troyanovsky S, Godsel LM. Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb Perspect Biol. 2010;2:a000125. doi: 10.1101/cshperspect.a000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin TA, Jiang WG. Tight junctions and their role in cancer metastasis. Histol Histopathol. 2001;16:1183–1195. doi: 10.14670/HH-16.1183. [DOI] [PubMed] [Google Scholar]
- 3.Gumbiner BM. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/S0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 4.Tamura A, Yamazaki Y, Hayashi D, Suzuki K, Sentani K, Yasui W, Tsukita S. Claudin-based paracellular proton barrier in the stomach. Ann N Y Acad Sci. 2012;1258:108–114. doi: 10.1111/j.1749-6632.2012.06570.x. [DOI] [PubMed] [Google Scholar]
- 5.González-Mariscal L, Betanzos A, Nava P, Jaramillo BE. Tight junction proteins. Prog Biophys Mol Biol. 2003;81:1–44. doi: 10.1016/S0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 6.Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life. 2009;61:431–437. doi: 10.1002/iub.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–F876. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukita S, Furuse M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann NY Acad Sci. 2000;915:129–135. doi: 10.1111/j.1749-6632.2000.tb05235.x. [DOI] [PubMed] [Google Scholar]
- 9.Tsukita S, Furuse M. Occludin and claudins in tight-junction strands: Leading or supporting players? Trends Cell Biol. 1999;9:268–273. doi: 10.1016/S0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- 10.Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Itoh M, Yonemura S, Furuse M, Tsukita S. Interspecies diversity of the occludin sequence: cDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balda MS, Whitney JA, Flores C, González S, Cereijido M, Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, Lynch RD, Schneeberger EE. Occludin is a functional component of the tight junction. J Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- 13.Gye MC. Expression of occludin in canine testis and epididymis. Reprod Domest Anim. 2004;39:43–47. doi: 10.1046/j.1439-0531.2003.00474.x. [DOI] [PubMed] [Google Scholar]
- 14.Kimura Y, Shiozaki H, Hirao M, Maeno Y, Doki Y, Inoue M, Monden T, Ando-Akatsuka Y, Furuse M, Tsukita S, Monden M. Expression of occludin, tight-junction-associated protein, in human digestive tract. Am J Pathol. 1997;151:45–54. [PMC free article] [PubMed] [Google Scholar]
- 15.Mandell KJ, Parkos CA. The JAM family of proteins. Adv Drug Deliv Rev. 2005;57:857–867. doi: 10.1016/j.addr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Niessen CM. Tight junctions/adherens junctions: Basic structure and function. J Invest Dermatol. 2007;127:2525–2532. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 17.Islas S, Vega J, Ponce L, Gonzalez-Mariscal L. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp Cell Res. 2002;274:138–148. doi: 10.1006/excr.2001.5457. [DOI] [PubMed] [Google Scholar]
- 18.Balda MS, Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000;19:2024–2033. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Ohta H, Yamaguchi T, Rajapakshage BK, Murakami M, Sasaki N, Nakamura K, Hwang SJ, Yamasaki M, Takiguchi M. Expression and subcellular localization of apical junction proteins in canine duodenal and colonic mucosa. Am J Vet Res. 2011;72:1046–1051. doi: 10.2460/ajvr.72.8.1046. [DOI] [PubMed] [Google Scholar]
- 22.Hwang I, Yang H, Kang HS, Ahn CH, Lee GS, Hong EJ, An BS, Jeung EB. Spatial expression of claudin family members in various organs of mice. Mol Med Rep. 2014;9:1806–1812. doi: 10.3892/mmr.2014.2031. [DOI] [PubMed] [Google Scholar]
- 23.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: Expression in normal and neoplastic tissues. BMC Cancer. 2006;6:186. doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balkovetz DF. Tight junction claudins and the kidney in sickness and in health. Biochim Biophys Acta. 2009;1788:858–863. doi: 10.1016/j.bbamem.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Hwang I, An BS, Yang H, Kang HS, Jung EM, Jeung EB. Tissue-specific expression of occludin, zona occludens-1, and junction adhesion molecule A in the duodenum, ileum, colon, kidney, liver, lung, brain, and skeletal muscle of C57BL mice. J Physiol Pharmacol. 2013;64:11–18. [PubMed] [Google Scholar]
- 26.Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA, Williams IR, Koval M, Peatman E, Campbell JA, et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med. 2007;204:3067–3076. doi: 10.1084/jem.20071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridyard AE, Brown JK, Rhind SM, Else RW, Simpson JW, Miller HR. Apical junction complex protein expression in the canine colon: Differential expression of claudin-2 in the colonic mucosa in dogs with idiopathic colitis. J Histochem Cytochem. 2007;55:1049–1058. doi: 10.1369/jhc.7A7211.2007. [DOI] [PubMed] [Google Scholar]
- 28.Hartsock A, Nelson WJ. Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta. 2008;1778:660–669. doi: 10.1016/j.bbamem.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambrecht BN, Hammad H. Biology of lung dendritic cells at the origin of asthma. Immunity. 2009;31:412–424. doi: 10.1016/j.immuni.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Nagai M, Yaoita E, Yoshida Y, Kuwano R, Nameta M, Ohshiro K, Isome M, Fujinaka H, Suzuki S, Suzuki J, et al. Coxsackievirus and adenovirus receptor, a tight junction membrane protein, is expressed in glomerular podocytes in the kidney. Lab Invest. 2003;83:901–911. doi: 10.1097/01.LAB.0000073307.82991.CC. [DOI] [PubMed] [Google Scholar]
- 31.Lee NP, Luk JM. Hepatic tight junctions: From viral entry to cancer metastasis. World J Gastroenterol. 2010;16:289–295. doi: 10.3748/wjg.v16.i3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]