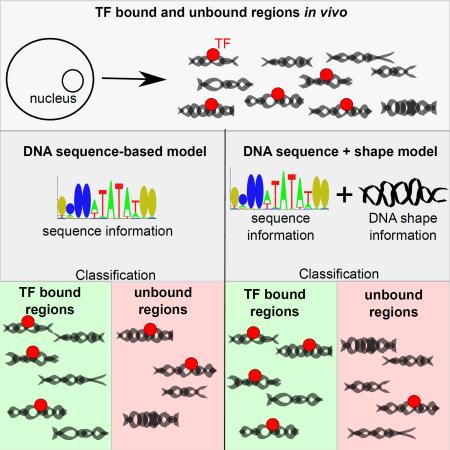
SUMMARY
Interactions of transcription factors (TFs) with DNA comprise a complex interplay between base-specific amino acid contacts and readout of DNA structure. Recent studies highlighted the complementarity of DNA sequence and shape in modeling TF binding in vitro. Here, we provide a comprehensive evaluation of in vivo datasets to assess the predictive power obtained by augmenting various DNA sequence-based models of TF binding sites (TFBSs) with DNA shape features (helix twist, minor groove width, propeller twist, and roll). Results from 400 human ChIP-seq datasets for 76 TFs show that combining DNA shape features with position specific scoring matrix (PSSM) scores improves TFBS predictions. Improvement was also observed using TF flexible models and a machine-learning approach using a binary encoding of nucleotides in lieu of PSSMs. Incorporating DNA shape information is most beneficial for E2F and MADS-domain TF families. Our findings indicate that incorporating DNA sequence and shape information benefits the modeling of TF binding under complex in vivo conditions.
INTRODUCTION
One of many mechanisms that control gene expression, transcriptional regulation involves transcription factors (TFs) as key proteins (Jacob and Monod, 1961; Ptashne and Gann, 1997). Most TFs are sequence-specific DNA binding proteins that recognize specific genome positions through a complex interplay between nucleotide-amino acid contacts (base readout) and readout of DNA structure (shape readout) (Slattery et al., 2014). Deciphering how TFs identify and bind specific target sequences—the TF binding sites (TFBSs)—is a key challenge in understanding transcriptional gene regulation (Dror et al., 2016; Wasserman and Sandelin, 2004; Zambelli et al., 2012).
TFBSs are short and often degenerate sequence motifs. These characteristics make it computationally difficult to model and predict TFBSs at the genomic scale (Badis et al., 2009). Moving beyond initial consensus sequence methods, the classical computational model to describe TFBSs is the position-specific scoring matrix (PSSM), which uses an additive method to summarize frequencies of every nucleotide at each position of the TFBS (Stormo, 2013). These second-generation models, however, do not capture position interdependencies or variable spacing. Therefore, several experimental assays have been designed to unravel characteristics of TF-DNA interactions at the large scale. In vitro high-throughput (HT) binding assays, such as protein binding microarrays (PBMs) (Berger et al., 2006), HT SELEX (Jolma et al., 2010; Zhao et al., 2009), and SELEX-seq (Slattery et al., 2011), expose DNA sequences selected by TFs and reveal their binding preferences. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) represents the in vivo counterpart of these in vitro assays, allowing for the identification of DNA regions bound by a targeted TF at the genomic scale (Johnson et al., 2007).
Large-scale data derived from HT experiments highlight higher-order positional interaction features of TFBSs that cannot be captured by classical PSSMs, even though methods based on these traditional models perform quite well (Weirauch et al., 2013). Recently, computational advances have used experimental assays to construct sophisticated models that capture a broad range of TFBS representations. For instance, PSSMs have been extended to dinucleotides to capture interrelationships within TFBSs (Siddharthan, 2010). Using PBM data, binding energy models include energy parameters to describe contributions of dinucleotides to binding affinity (Zhao et al., 2012). These models describe TF-DNA binding specificity well in cases where PSSMs have performed insufficiently. Utilizing ChIP-seq data, we developed the TF flexible model (TFFM) framework to improve in vivo prediction of TFBSs (Mathelier and Wasserman, 2013). TFFMs capture interdependencies of successive nucleotides within TFBSs and the flexible length of TFBSs within a single hidden Markov model framework.
The abovementioned third-generation methods enable TFBS prediction by representing sequence properties. A parallel approach utilizes the three-dimensional DNA structure, or DNA shape, to capture, at least in part, the interdependencies between nucleotide positions within TFBSs (Gordân et al., 2013; Tsai et al., 2015; Yang and Ramsey, 2015; Zhou et al., 2015). Large-scale DNA structural information can be computed by the DNAshape method (Zhou et al., 2013), which computes four DNA shape features: helix twist (HelT), minor groove width (MGW), propeller twist (ProT), and Roll. Recent studies demonstrated the complementary role of DNA sequence and shape information in determining protein-DNA binding specificity in vitro (Joshi et al., 2007; Rohs et al., 2009; Slattery et al., 2011). For example, the binding specificity of Hox proteins was analyzed using SELEX-seq data to show the direct role of DNA shape features in protein-DNA readout (Abe et al., 2015). Using PBM and SELEX-seq data, we showed that complementing DNA sequence with shape information enhanced prediction of TF binding affinities (Zhou et al., 2015). DNA shape information at regions flanking core binding sites was highly predictive of differential binding derived from BunDLE-seq assays (Levo et al., 2015). While previous works demonstrated that models combining DNA sequence and shape improve quantitative models of TF binding in vitro, we addressed here three key questions: (1) Do more complex in vivo protein-DNA interactions exhibit similar properties; (2) when DNA shape properties are integrated with sequence-based TFBS prediction methods, do we observe an improvement in performance; and (3) do specific TF families benefit more than others from the integration of DNA shape features in TF binding models?
Here, we capitalized on the availability of DNA shape information extracted from GBshape (Chiu et al., 2015), our genome browser database of DNA shape features computed from our DNAshape prediction tool (Zhou et al., 2013), at TF-bound regions derived from ChIP-seq experiments to address the three aforementioned questions.
RESULTS
Machine Learning Models Combining DNA Sequence and Shape Features
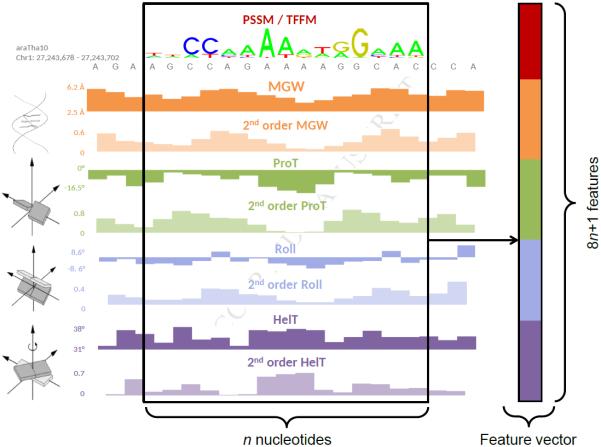
To assess the effects of including DNA structural information in predictions of TFBSs in ChIP-seq datasets, we developed a computational framework combining DNA sequence and shape information to model and predict TFBSs. The availability of numerous ChIP-seq regions enables application of a discriminative supervised machine learning approach (Libbrecht and Noble, 2015). Specifically, a DNA sequence that is considered as a potential TFBS was represented by a (feature) vector that combined 1 to 4n features that encode sequence information and 8n features that capture DNA shape information, where n is DNA sequence length. We encoded DNA sequence information of the putative TFBS using either the PSSM or TFFM score computed from the sequence, or a binary encoding using 4 bits per nucleotide (Zhou et al., 2015). DNA shape-related features are the predicted values of HelT, MGW, ProT, and Roll at each position of the TFBS, extracted from GBshape (Chiu et al., 2015). The vector was further augmented with four second-order shape features that capture structural dependencies at adjacent nucleotide positions (Zhou et al., 2015) (Figure 1). Assuming that each ChIP-seq region contains a TFBS, we constructed a feature vector for the best hit per ChIP-seq peak and background region predicted by a TF binding profile (PSSM or TFFM) to train a classifier. To discriminate between TF bound (ChIP-seq) and unbound (background) regions, we used a gradient boosting classifier, which is an ensemble machine learning classifier that combines multiple weak learners to improve predictive power (Friedman et al., 2001). The gradient boosting classifier was based on decision trees that, given an input feature vector, output the probability of the feature vector to be associated with a ChIP-seq peak or a background region. This approach naturally handles heterogenous features (e.g., DNA sequence and shape information), is robust to outliers, and is able to manage irrelevant input such as noise from ChIP-seq experiments (Friedman et al., 2001).
Figure 1. Feature Vectors of PSSM + DNA Shape and TFFM + DNA Shape Classifiers.
Feature vectors combine sequence scores with respect to the TF binding profile (PSSM or TFFM), the normalized values of four DNA shape features (MGW, ProT, Roll, and HelT), and their normalized product terms at adjacent positions as second-order shape features (Zhou et al., 2015). In 4-bits + DNA shape classifiers, TF binding profile score is replaced by binary 4-bits encoding of the corresponding sequence (Zhou et al., 2015).
Classifiers combining PSSM score, TFFM score, or 4-bits nucleotide encoding with DNA shape features are referred to as PSSM + DNA shape, TFFM + DNA shape, or 4-bits + DNA shape classifiers, respectively. Open-source Python software for generating and using these classifiers is provided at https://github.com/amathelier/DNAshapedTFBS.
Incorporating DNA Shape Features Improves TFBS Prediction in Human In Vivo Datasets
We compiled a set of 400 uniformly processed human ENCODE ChIP-seq datasets for which a JASPAR TF binding profile (Mathelier et al., 2014) was available for the corresponding immunoprecipitated (ChIPed) TF (Data S1). These datasets, covering 76 TFs, were used to compare the predictive powers of three computational models that consider DNA sequence information alone with their DNA shape-augmented classifiers. The first two DNA sequence-based models are PSSMs and TFFMs, which are widely used to score TFBSs in ChIP-seq datasets. The third model, the 4-bits classifier, is a discriminative model that uses a binary encoding of DNA sequence information (Zhou et al., 2015).
Here, the predictive power of a model refers to its ability to discriminate ChIP-seq regions (defined as the 50-bp region surrounding each side of the ChIP-seq peak maximum) from matched background sequences. The 50-bp regions were selected because they are enriched for TFBSs (Hunt et al., 2014; Wilbanks and Facciotti, 2010). To avoid sequence composition biases, we selected each set of background sequences to match either the G+C (%GC) content or dinucleotide composition of the ChIP-seq regions. Unless otherwise indicated, background sequences matching the %GC content of ChIP-seq regions were used in the following results. Predictive powers of PSSM scores and PSSM + DNA shape classifiers were assessed through 10-fold cross-validation (CV). We optimized the PSSMs derived from JASPAR TF binding profiles with the perceptron algorithm using the DiMO tool (Patel and Stormo, 2014) on the constructed foreground and background training sets (range: 495–83,123, median: 15,171, mean: 21,098, standard deviation: 17,220 sequences). Parameters of PSSM + DNA shape classifiers were learned from the same training sets. Vectors used by the classifiers for a ChIP-seq region correspond to the combination of the best PSSM score in the region and the 8n DNA shape feature values computed for this hit. To assess predictive power, we varied the threshold for scores to compute the recall (sensitivity), specificity, and precision values. Areas under the precision and recall curve (AUPRC) and the receiver-operating characteristic curve (AUROC) were computed for each model on each ChIP-seq dataset to evaluate predictive power. Unless otherwise noted, we provide the AUPRC values and the p-values for significance calculated by the Wilcoxon signed-rank test.
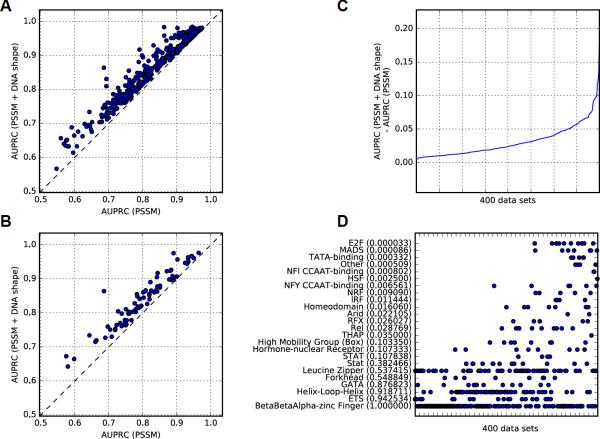
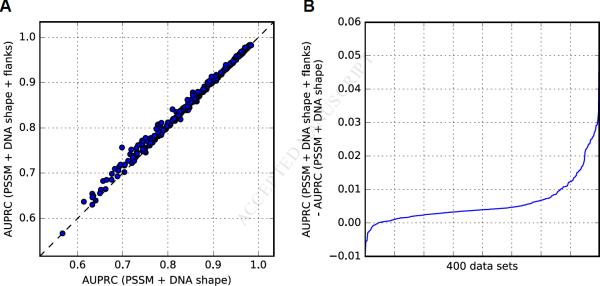
Comparing AUPRC values derived from the PSSM scores or PSSM + DNA shape classifiers, we found that shape-augmented classifiers performed better for all 400 ChIP-seq datasets (p = 2.7 × 10−67; Figure 2A). Considering the median AUPRC values per TF over all ChIP-seq datasets associated with the TF, we observed consistent improvement for all TFs when DNA shape features were incorporated (p = 3.6 × 10−14; Figure 2B). We computed the difference of discriminative power between the two models (Figure 2C) to assess the improvement obtained by using the PSSM + DNA shape classifiers.
Figure 2. Effect of DNA Shape Features on TFBS Predictions in ChIP-seq Data.
(A) AUPRC values obtained for 400 ENCODE human ChIP-seq datasets using PSSM scores (x-axis) or classifiers combining PSSM scores and DNA shape features (y-axis). Dashed line represents equal AUPRC values obtained with both methods.
(B) Median AUPRC values over all ChIP-seq datasets associated with each TF (one point per TF), obtained using PSSM scores (x-axis) or PSSM + DNA shape classifiers (y-axis). Dashed line represents equal AUPRC values obtained with both methods.
(C) Predictive power improvement obtained when considering DNA shape features (y-axis) as the difference between AUPRC values obtained with PSSM + DNA shape classifiers and PSSM scores. Larger difference corresponds to stronger improvement. Datasets (x-axis) are ranked by increasing difference values.
(D) For each TF family (y-axis), an associated dataset is represented at the corresponding x-coordinate where the dataset appears in B. Names of TF families are given on y-axis, with significant Mann–Whitney U test p-values in parentheses (not corrected for multiple hypothesis testing). See also Data S2–S4 and S7.
Using the same analyses, we found that the predictive power of the TFFM + DNA shape classifiers was better than that of the TFFMs for 396/400 ChIP-seq datasets (p = 4.4 × 10−67; Data S2). Classifiers performed strictly better than TFFMs for all TFs when we considered the median AUPRC values per TF (p = 3.6 × 10−14; Data S2).
Finally, we compared the 4-bits and 4-bits + DNA shape classifiers, which were trained and tested on sequences of the highest-scoring hit per ChIP-seq region derived from the PSSMs. DNA shape-augmented classifiers performed consistently better than 4-bits classifiers for 365/400 ChIP-seq datasets (p = 2.7 × 10−57) and 70/76 TFs (p = 1.3 × 10−12) when considering the median AUPRC values (Data S2).
We confirmed the improvement in discriminative power of the models incorporating DNA shape features by considering background sequences matching the dinucleotide composition of ChIP-seq regions (Data S3) and TF-bound regions recurrently found in multiple ChIP-seq datasets for the same TF (Data S4).
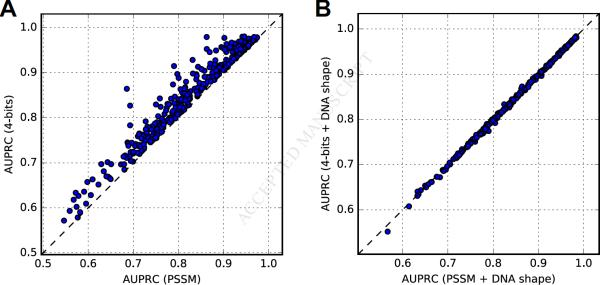
The relative improvement obtained when incorporating DNA shape information varied depending on the baseline DNA sequence-based approach. Unsurprisingly, the 4-bits + DNA shape classifiers exhibited a smaller improvement over the 4-bits classifiers compared to the shape-based improvements obtained with PSSMs and TFFMs. The higher baseline performance of the 4-bits method is consistent with the superiority of discriminative over generative models to distinguish bound from unbound regions in ChIP-seq (Libbrecht and Noble, 2015) (Figure 3A and Data S5). Nonetheless, PSSM + DNA shape classifiers performed consistently better than 4-bits + DNA shape classifiers for 344/400 datasets (p = 7.7× 10−43; Figure 3B) and 64/76 TFs (p = 1.0 × 10−8; Data S5).
Figure 3. Predictive Power of PSSM and 4-bit Approaches for TFBSs in ChIP-seq Regions.
AUPRC values obtained for 400 human ENCODE ChIP-seq datasets, obtained by considering (A) PSSM scores (x-axis) and 4-bits classifiers (y-axis), or (B) PSSM + DNA shape (x-axis) and 4-bits + DNA shape (y-axis) classifiers. See also Data S5.
Although 4-bits classifiers outperformed PSSM scores, the higher discriminative power of PSSM + DNA shape compared to 4-bits + DNA shape classifiers reinforces the capacity of DNA shape features to improve TFBS predictions in ChIP-seq datasets. Importantly, the combination of sequence information (captured by PSSMs, TFFMs, or 4-bits classifiers) with DNA shape properties performed better than generative (PSSM and TFFM) and discriminative (4-bits classifier) approaches modeling DNA sequence, indicating that DNA shape provides additional information.
Although the utility of DNA shape to predict TFBSs was reported before (Abe et al., 2015; Yang and Ramsey, 2015; Yang et al., 2014; Zhou et al., 2015), we provide evidence, from an extensive collection of 400 human in vivo datasets for 76 TFs, that this observation is generalizable and relevant to noisy environments and data (Fan and Struhl, 2009; Hunt et al., 2014; Hunt and Wasserman, 2014; Jain et al., 2015; Park et al., 2013; Teytelman et al., 2013).
DNA Shape at Genomic Flanking Regions Improves TFBS Predictions In Vivo
Sequences immediately flanking TFBSs were previously shown to contribute to TF binding specificity (Gordân et al., 2013), which is determined, in part, by DNA shape outside the core binding sites (Afek et al., 2014; Barozzi et al., 2014; Dror et al., 2015). We extended our DNA shape-augmented models to consider eight DNA shape features at 15-bp-long regions 5' and 3' of the TFBSs, as in Barozzi et al. (2014).
Augmenting DNA shape-based classifiers with additional DNA shape information from flanking sequences improved the discriminatory power of classifiers trained using 10-fold CV for 378 (~94%), 373 (~93%), and 375 (~94%) datasets compared to PSSM + DNA shape, TFFM + DNA shape, and 4-bits + DNA shape classifiers, respectively (Figure 4 and Data S6). Our findings agree with results from in vitro studies of the role of flanking regions in TF-DNA binding (Dror et al., 2016; Gordân et al., 2013; Levo et al., 2015).
Figure 4. Predictive Power of DNA Shape Features at TFBS Flanking Regions.
(A) AUPRC values obtained for 400 human ENCODE ChIP-seq datasets when using classifiers combining PSSM scores and DNA shape features at core TFBSs (x-axis), or classifiers combining PSSM scores and DNA shape features at both core TFBSs and surrounding 15 bp on each side (y-axis). Dashed line represents equal AUPRC values for both methods.
(B) AUPRC value differences (y-axis) between flank-augmented classifiers and PSSM + DNA shape classifiers (x-axis). Datasets (x-axis) are ranked by increasing difference values. See also Data S6.
E2F and MADS-domain TF Families Benefit Most from DNA Shape Information
Next, motivated by the observation that DNA structural information improves the prediction of TFBSs for some ChIP-seq datasets more than others (Figure 2C and Data S2, S3), we investigated whether predictions for certain TF families with similar DNA-binding domains specifically benefit from incorporating DNA shape information.
Using JASPAR (Mathelier et al., 2014), we extracted TF family information of DNA binding domains associated with the 400 human ChIP-seq experiments. In aggregate, we analyzed profiles derived from DNA binding domains associated with 24 TF families, which were associated with TFs in the JASPAR database using a classification scheme (Fulton et al., 2009) (Data S1). Comparing the predictive powers of DNA shape-augmented classifiers to those of DNA sequence-based approaches, we assessed the enrichment of a TF family for larger AUPRC or AUROC difference values using the one-sided Mann–Whitney U test. Predictive power comparisons were performed considering %GC- and dinucleotide-matched background sets. Depending on the DNA sequence-based approach (PSSM, TFFM, or 4-bits), background type (%GC- or dinucleotide-matched), and assessment method (AUPRC or AUROC), we observed enrichment (p < 4.17 × 10−4, with Bonferroni correction for desired α < 0.01) for different TF families (Figure 2D and Data S1, S2). Taken together, the results across all three DNA sequence-based approaches suggest that the E2F and MADS-domain TF datasets benefited the most from inclusion of DNA shape information (Figure 2D and Data S1, S2). It is noteworthy that results for the E2F and MADS-domain TF datasets were not derived from a single TF, but were consistent over several TFs from the same family. Namely, the 10 E2F-associated ChIP-seq datasets were derived from E2F1 (3 datasets), E2F4 (4 datasets), and E2F6 (3 datasets), and seven MADS-domain–associated ChIP-seq datasets were derived from MEF2A (2 datasets), MEF2C (1 dataset), and SRF (4 datasets).
To confirm the results obtained when incorporating DNA shape features for MADS-domain TFs, we considered an independent set of seven Arabidopsis thaliana TFs (Heyndrickx et al., 2014) for which we had JASPAR TF binding profiles (Data S1). Similarly to human MADS-domain TF results (Data S7), we observed improved discriminative power when DNA structural information was considered for plant MADS-domain TFs with PSSM + DNA shape and TFFM + DNA shape models (Data S7). Only the two smallest datasets (94 sequences for FLC and 54 for SVP in training sets) showed decreased discriminative power with DNA shape-augmented classifiers. For one of the three approaches, the 4-bits + DNA shape classifiers, we found no improvement in predictive power for plant MADS-domain TF datasets compared to the 4-bits classifiers (Data S7).
Taken together, we observed that, among all studied protein families, TFs from the E2F and MADS-domain families benefited the most from inclusion of DNA shape information in the classifiers when compared to the three DNA-sequence-based models.
Structural Analyses of E2F and MADS-domain TF Binding Specificities DNA shape readout contributes to E2F-DNA binding
Given our observed improvement in predictive power based on DNA shape information, we next characterized the specific DNA shape features that contributed the most to the improvement in predictive accuracy for the family of E2F TFs. We extracted the importance of each feature learned by the DNA shape-augmented classifiers by combining DNA sequence and shape information for the E2F TFs. To consider the same DNA sequence-based model per TF for all associated ChIP-seq datasets, we selected PSSMs derived from the corresponding JASPAR TF binding profiles. To simplify interpretation, we considered classifiers based on sequence and first-order DNA shape features (Zhou et al., 2015). Figures S1–S2 plot the average feature importance measures obtained over the 10-fold CV training for all ChIP-seq datasets associated with the TFs.
Although the PSSM score was consistently the most important feature (Figure S1), several DNA shape features at different positions were important for TFBS predictions. A commonality among the three E2Fs was the contribution of ProT (and, to lesser extents, HelT and MGW) at proximal flanks of the TFBSs (see first and last positions of heat maps in Figure S2). Nucleotides immediately flanking the E2F TFBSs contributed to the DNA binding specificity of this TF family.
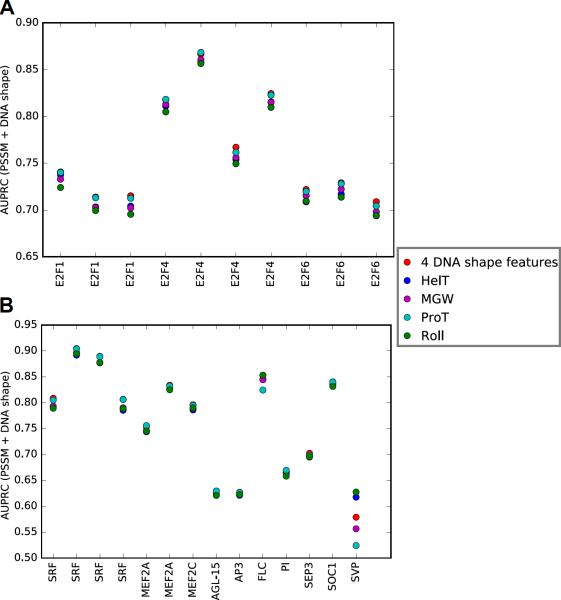
Comparing the predictive powers of classifiers combining sequence with either a single or four DNA shape features, we confirmed the importance of ProT for improving predictive power for the 10 ChIP-seq datasets associated with the three E2Fs (Figures 5A and S3A).
Figure 5. Use of a Single DNA Shape Feature Category for E2F and MADS-box TFBS Recognition in ChIP-seq.
AUPRC values (y-axis) for E2F (A) and MADS-domain (B) TF datasets (x-axis), obtained by using all four first-order DNA shape features or a single feature category along with sequence features in the PSSM + DNA shape classifiers. See also Figure S3.
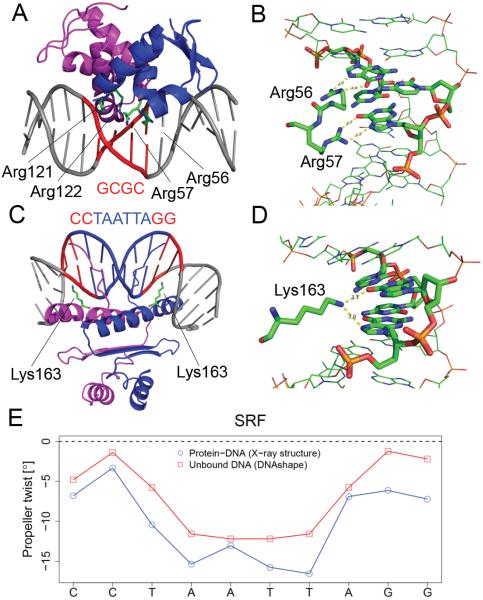
Analysis of the co-crystal structure of the E2F4 TF in complex with DNA and its cofactor DP2 (PDB ID 1CF7) (Zheng et al., 1999) revealed that the RRXYD motifs of the E2F4 and DP2 heterodimer form a compact structural assembly that contacts the major groove (Figure 6A, B). Guanidinium groups of each of the four arginine residues engage in base readout through bidentate hydrogen bonds with guanine bases of the core binding site. This intricate system of eight hydrogen bonds enables readout of structural features and recognition of functional groups of the guanine bases. The angular orientation of the arginine side chains stabilized by other amino acids selects for rotational parameters of the contacted G/C bp through hydrogen bond geometries. This observation is reflected by the importance given to rotational parameters, such as ProT, in our models. Similar results were reported in a recent study of diverse protein families (Dror et al., 2015).
Figure 6. Structural Analysis of E2F and MADS-domain TFs in Complex with DNA.
(A) Co-crystal structure (PDB ID 1CF7) of E2F4 (blue) and DP2 (magenta) forming a heterodimer that binds to core motif GCGC (red).
(B) Detailed view of hydrogen bonds between arginines and guanines in major groove.
(C) Co-crystal structure (PDB ID 1SRS) of MADS-domain SRF homodimer in complex with core motif CCTAATTAGG.
(D) Detailed view of hydrogen bonds between lysine and guanines in major groove.
(E) ProT in bound (blue; calculated from X-ray structure) and unbound (red; predicted by DNAshape) target site.
MADS-domain TFs recognize position-specific DNA shape
As described above for E2Fs, we considered DNA shape features individually in the DNA shape-augmented classifiers for the MADS-domain TF ChIP-seq datasets. Figure 5B highlights the importance of ProT for improving the discriminative power of models associated with human and plant MADS-domain TFs. The FLC and SVP datasets, which had small numbers of training sequences, were the only ones for which ProT was not the most important shape feature. Inclusion of a single DNA shape category ensured that DNA shape features did not compensate for each other when considering all four shape features due to dependencies among different features. Previous work showed the important role of MGW at the A-tract of the MADS-box for DNA-binding (Muiño et al., 2014). Our models captured this importance, although ProT remained the most important shape feature.
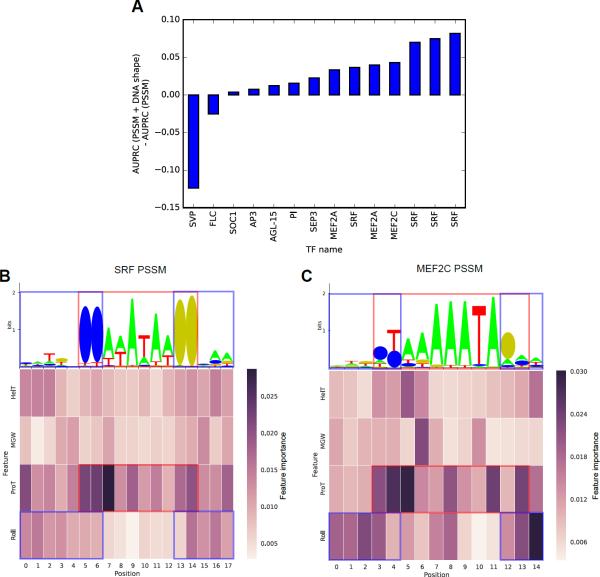
We extracted feature importance measures at each position within the TFBSs learned by the PSSM + DNA shape classifiers for human and plant MADS-domain TFs (Figures S4–S6). The most important DNA shape features for discriminating ChIP-seq bound sites from background genomic regions were ProT and Roll at specific positions within the MADS-box TFBSs (in agreement with Figure 5B). This observation was consistent across all of the human MADS-domain TFs, whereas the signal was more diffuse for plant MADS-domain TFs. DNA shape-augmented classifiers associated with human MADS-domain TF ChIP-seq datasets obtained the strongest discriminative improvements over sequence-based models (Figure 7A).
Figure 7. Feature Importance Measures for MADS-box Recognition in ChIP-seq Datasets.
(A) AUPRC improvements (y-axis) for human and plant MADS-domain TF ChIP-seq datasets (x-axis provides TF names) when using PSSM + DNA shape features vs. PSSM scores.
(B–C) Weblogos derived from JASPAR TF binding profile associated with (B) SRF (MA0083.2) and (C) MEF2C (MA0497.1) TFs are provided in top panels. Heat maps illustrating average feature importance values (y-axis) at each position (x-axis) of TFBSs in the classifiers trained for 10-fold CV analysis of ChIP-seq datasets are provided in bottom panels. Only feature importance measures associated with first-order DNA shape features are considered. Color scale for heat map is given on the right of the heat map. Red boxes highlight core MADS-box motif (CCW6GG). Blue boxes highlight edges of motifs. See also Figures S1, S2, and S4–S6.
As an example, we plotted feature importance measures and sequence logos of the TF binding profiles associated with SRF and MEF2C (Figure 7B, C). These two TFs were associated with datasets that showed the strongest improvements in discriminative power when incorporating DNA shape features for the classification of bound vs. unbound sites (Figure 7A). ProT features seemed to contribute to the binding of the core CArG-box (CCW6GG) (red squares in Figures 7B, C and S7), whereas Roll features were highlighted at the edges of the MADS-box core motif (blue squares in Figures 7B, C and S7).
Structural analyses of various complexes of MADS-domain TFs with their DNA target sites suggested that protein residues recognize specific DNA conformations. Comparison of the feature importance measures for different structural features of the DNA binding sites of human MADS-domain proteins indicated a contribution of ProT to binding specificity (Figures 6, 7, S3B, and S7). Human SRF ChIP-seq datasets were consistently associated with strongest improvement in discriminative power when considering the DNA shape-augmented classifiers to predict TFBSs in TF-bound regions (Figure 7A). Hence, we analyzed the co-crystal structures available for complexes of human SRF and MEF2.
The SRF homodimer uses lysine residues to form base-specific hydrogen bonds with C/G bp in the CArG motif (Figure 6C, D). DNA bends around the protein, and intrinsic structural features are likely responsible for this deformation. As our models indicate a contribution of ProT, we compared ProT observed in the co-crystal structure of the SRF-DNA complex (PDB ID 1SRS) (Pellegrini et al., 1995) with the prediction of ProT for the unbound motif using our DNAshape method (Zhou et al., 2013). The ProT pattern in the unbound target site resembled the pattern in the protein-DNA complex (Figure 6E).
This observation suggests that ProT is intrinsic to the binding site and likely selected by the TF. A less negative ProT within the two adjacent G/C bp at each flank of the motif optimizes the geometry of hydrogen bonds between the lysine side chains and bases (Figure 6D). This preference is coupled to the sequence and selection of functional groups of the bases, whereas structural features enhance the energetics of these specific contacts.
DISCUSSION
Here, we used a machine learning classifier-based approach to demonstrate that combining DNA sequence information with DNA shape features improves discrimination between TF-bound sites in vivo and background genomic regions. These in vivo analyses complement our previous in vitro studies showing that inclusion of DNA shape properties can improve the accuracy of TF binding site prediction (Abe et al., 2015; Yang et al., 2014; Zhou et al., 2015).
A possible limitation of our approach is that we only considered DNA shape features at the best TFBS per ChIP-seq region, derived from PSSM or TFFM scores. Although the site with the highest score represents the best candidate in a ChIP-seq region, another site harboring a lower score could potentially represent a more appropriate DNA-shape readout (Zentner et al., 2015). In our analyses, we used three baseline approaches (PSSM, TFFM, and 4-bits) representing two types of models (generative and discriminative), two background types (%GC- and dinucleotide-matched), and two assessment measures (AUPRC and AUROC). Direct comparisons between the models stressed the higher predictive power of discriminative models using ChIP-seq datasets, in agreement with the literature (Libbrecht and Noble, 2015).
Our computational analyses of 400 human ChIP-seq datasets revealed that when DNA shape features were incorporated in the models, the E2F and MADS-domain TF families showed the largest improvement in TFBS prediction accuracies. TF families with the strongest predictive power improvements varied depending on the baseline model, background type, and assessment measure considered. These results highlight the importance of considering multiple background types and assessment measures when comparing models.
Whereas most bioinformatics tools rely on PSSMs for TFBS predictions, our findings imply that the field should consider more sophisticated modeling methods. For TFs where only a small number of experimentally derived TF-bound regions are available, traditional PSSMs represent a reasonable alternative, as exemplified by the FLC and SVP plant TFs in our study. We envision that future tools relying on TFBS predictions will incorporate the most appropriate model for each TF or TF family.
With the increasing trove of whole-genome sequencing data, the identification of variants altering gene regulation through disruption of TF-DNA interactions has become an important challenge. Recent approaches have focused on allelic differences in PSSM scores to predict the functional impact of variants within TFBSs (Chen et al., 2016; Mathelier et al., 2015). Our study confirmed that some TFs critically rely on DNA-shape readout for TFBS recognition. Future work will be required to assess how the incorporation of DNA structural properties can help to predict the impact of variants disrupting TFBSs.
METHODS AND RESOURCES
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests will be fulfilled by the corresponding authors Remo Rohs (rohs@usc.edu) and Wyeth W. Wasserman (wyeth@cmmt.ubc.ca).
METHOD DETAILS
ChIP-seq Datasets and TF Binding Profiles
We retrieved uniformly processed human ENCODE ChIP-seq datasets (Dunham et al., 2012) as narrowPeak-formatted files from the UCSC genome browser at http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgTfbsUniform/ (as of April 12th 2013). We associated JASPAR TF binding profiles (Mathelier et al., 2014) with ChIPed TFs wherever possible. Using this approach, we obtained 400 ChIP-seq datasets associated with 76 JASPAR profiles (Data S1).
We retrieved A. thaliana MADS-domain TF ChIP-seq peak positions studied in Heyndrickx et al. (2014) from the bed-formatted file at http://bioinformatics.psb.ugent.be/cig_data/RegNet/. We specifically considered seven MADS-box ChIP-seq datasets for which a JASPAR TF binding profile was available (Data S1).
ChIP-seq Peak Sequences
For human ENCODE ChIP-seq peaks, we analyzed 50-bp regions on each side of the peak maximum provided in the narrowPeak-formatted files. Sequences were extracted using the getfasta subcommand of bedtools (Quinlan and Hall, 2010) from the hg19 version of the human genome from Ensembl release 63 (Cunningham et al., 2015).
For plant TFs, we considered 50 bp on each side of the middle point of the ChIP-seq peaks because the peak maximum position was not provided. Sequences were extracted using the getfasta subcommand of bedtools (Quinlan and Hall, 2010) from the TAIR10 version of the A. thaliana genome from Ensembl plant release 26 (Cunningham et al., 2015).
Recurrent ChIP-seq Peak Regions
We obtained the plots in Data S4 using ChIP-seq peaks found in recurrently ChIPed genomic regions between multiple ChIP-seq experiments for the same TF. Using bedtools, we merged genomic regions where at least two ChIP-seq peaks overlapped among all ChIP-seq experiments associated with the TF. For each merged region, we randomly selected one of the overlapping ChIP-seq peaks. The corresponding set of ChIP-seq peaks was used as the set of foreground sequences in the 10-fold CV.
10-fold CV datasets
For each ChIP-seq dataset, we constructed 10 training (Ti for i є [0, 9]) and 10 testing (Fi for i є [0, 9]) datasets, where Ti is 9 times the size of Fi. For each training or testing set, we constructed a background dataset (Bti or Bfi, respectively) using the BiasAway tool (Hunt et al., 2014). Background sequences were obtained by two methods: (i) by randomly selecting the same number of sequences as in Ti or Fi and matching the same %GC composition distribution from a set of genomic background sequences; and (ii) by shuffling sequences in Ti or Fi while matching the same dinucleotide composition.
Genomic background sequences associated with the human ENCODE ChIP-seq datasets were retrieved from mappable regions of the human genome derived from the ENCODE CrgMappability 36-mer track (ftp://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/wgEncodeCrgMapabilityAlign36mer.bigWig). We retained mappable regions > 200 bp, which were subsequently split into 100-bp segments. Genomic background sequences associated with plant ChIP-seq datasets were selected from 1,000,000 random sequences with lengths matched to the considered ChIP-seq regions. Plant sequences were extracted from the TAIR10 genome sequence using the random subcommand of bedtools (Quinlan and Hall, 2010).
Discriminative Power Assessment
PSSM- and TFFM-based Model Assessments
We assessed the capacity of models to discriminate ChIP-seq peak regions from background sequences using a 10-fold CV methodology with datasets Ti, Fi, Bti, and Bfi. The PSSM + DNA shape approach was assessed as follows. For all matching training (Ti and Bti) and testing (Fi and Bfi) datasets, each composed of ChIP-seq/foreground and background sequences, we:
Optimized the JASPAR binding profile using the DiMO tool (Patel and Stormo, 2014) on the training sequences;
Constructed PSSM from the DiMO-optimized profile using the motifs Biopython module;
Applied PSSM to training sequences (from Ti and Bti), and extracted eight DNA shape features at each best site (i.e., with highest PSSM score) per training sequence;
Constructed, for each site, a vector combining PSSM score with normalized values of the eight DNA shape parameters at each position;
Trained a gradient boosting classifier using the Python scikit-learn module (Pedregosa et al., 2011) with vectors associated with the ChIP-seq (Ti) and background (Bti) sequences;
Applied PSSM to testing sequences (from Fi and Bfi), and extracted the eight DNA shape features at each best site obtained with the PSSM scores;
Constructed corresponding feature vectors, as in 4;
Applied the classifier to vectors to obtain the probability of the sequence being a foreground sequence for each testing sequence; and
Combined all testing sequence probabilities, and computed AUPRC and AUROC values using the Python scikit-learn module (Pedregosa et al., 2011).
To evaluate the TFFM + DNA shape approach on the same sets of training and testing sequences, we:
Initialized a TFFM with the DiMO-optimized profile (described above) using the Python TFFM framework (Mathelier and Wasserman, 2013);
Trained the TFFM on foreground training sequences (from Ti); and
Applied steps 3–9 above using scores from the trained TFFM in place of PSSM. Predictive powers of the PSSMs and TFFMs were obtained using the same 10-fold CV strategy by solely considering the DiMO-optimized PSSM and trained TFFM scores of the best hit in each sequence.
4-bits-based Classifier Assessment
To evaluate the 4-bits + DNA shape classifiers using the testing and training sets described above, we:
Applied the DiMO-optimized PSSM to training sequences (from Ti and Bti), and extracted DNA sequence and eight DNA shape features at each best site per training sequence;
Constructed, for each site, a vector combining the encoded DNA sequence (A is encoded as 1000, T as 0100, G as 0010, and C as 0001) with the normalized values of the eight DNA shape parameters at each position (Zhou et al., 2015);
Trained a gradient boosting classifier using the Python scikit-learn module (Pedregosa et al., 2011) with vectors associated with the ChIP-seq (Ti) and background (Bti) sequences;
Applied the DiMO-trained PSSM to the testing sequences (from Fi and Bfi), and extracted DNA sequence and eight DNA shape features at each best site obtained with the PSSM scores;
Constructed the corresponding feature vectors, as in 2;
Applied the classifier to vectors to obtain the probability of the sequence being a foreground sequence for each testing sequence; and
Combined all testing sequence probabilities, and computed AUPRC and AUROC values using the Python scikit-learn module (Pedregosa et al., 2011).
We obtained predictive powers of the 4-bits classifiers using the same 10-fold CV strategy by solely considering the encoded DNA sequence at the best hit in each ChIP-seq region.
DNA Shape Features
We retrieved values for four DNA shape features (HelT, MGW, ProT, and Roll) and their corresponding second-order shape features (Zhou et al., 2015) for Homo sapiens and A. thaliana genomes from the GBshape genome browser (Chiu et al., 2015), considering 10-fold CV using the %GC-matched background sequences as genomic sequences. Feature values at each best site in the training and testing sequences were extracted from the corresponding bigWig files using the extract subcommand of the bwtool software (Pohl and Beato, 2014). For 10-fold CV using the dinucleotide-matched background sequences, DNA shape features were computed with the DNAshapeR tool (Chiu et al., 2016) using a customized second-order shape feature branch of the method, provided at https://github.com/TsuPeiChiu/DNAshapeR/tree/2nd-order. Values of the four DNA shape features and corresponding second-order features were normalized independently by the equation normvalue = (value − minvalue)/(maxvalue − minvalue), where normvalue is the normalized value to compute, value is the DNA shape feature value, and minvalue (maxvalue) corresponds to the minimum (maximum) possible value for the DNA shape feature.
Gradient Boosting Classifiers
We used the GradientBoostingClassifier class implemented in the Python scikit-learn module (Pedregosa et al., 2011) to construct, train, and apply gradient boosting classifiers to DNA sequences. Features used in the classifiers to describe a DNA sequence are vectors composed of a 4-bits encoded DNA sequence, PSSM score, or TFFM score and the eight DNA shape features at each nucleotide position of the DNA sequence.
Scanning DNA Sequences with TFFMs and PSSMs
Position frequency matrices (PFMs) corresponding to JASPAR binding profiles were retrieved from the JASPAR database using the jaspar BioPython module (Mathelier et al., 2014). The same module was used to convert the DiMO-optimized PFMs to PSSMs using the default background distribution of nucleotides. The module was used to convert the PFMs from JASPAR to PSSMs using the JASPAR pseudocount computation described in the module and default background distribution of nucleotides when assessing feature importance measures. Corresponding PSSMs were used to scan DNA sequences and extract the best hit per sequence. When applying a trained TFFM to a DNA sequence, the best hit was retrieved from all positions on both strands using the TFFM framework (Mathelier and Wasserman, 2013). First-order hidden Markov model-based TFFMs were used in this study.
TF Family Annotation
We retrieved the family assignment for each TF dataset from the JASPAR database (Mathelier et al., 2014) using the associated binding profile identifiers. This annotation resulted in the inclusion of 76 TFs representing 24 TF families (Data S1).
Feature Importance Measures
We considered gradient boosting classifiers used in 10-fold CV when considering sequence and first-order shape features. To consider the same PSSM per TF for all associated ChIP-seq datasets, we did not apply the DiMO-optimization step and only considered PSSMs derived from the corresponding JASPAR TF binding profiles. We extracted feature importance measures using the Python scikit-learn module (Pedregosa et al., 2011). Each measure was averaged over all classifiers used in the 10-fold CV for all datasets associated with a TF. Feature importance measures represent how discriminative a feature is in the underlying decision trees; they do not correspond to feature weights. These measures were visualized using the heat map function of the Python seaborn module [doi:10.5281/zenodo.19108].
QUANTIFICATION AND STATISTICAL ANALYSIS
Wilcoxon Signed-Rank Tests
We assessed the significance for the improvement of predictive power when comparing two models using the Wilcoxon signed-rank tests. The wilcoxon function of the scipy.stats Python module was used in Ipython (Perez and Granger, 2007) to compute p-values.
Mann–Whitney U Tests
We assessed enrichments for significant improvement of the discriminative power associated with TF families using one-sided Mann–Whitney U tests. The wilcox.testR function was used in IPython (Perez and Granger, 2007) through the Python rpy2 module (http://rpy.sourceforge.net/). We corrected Mann–Whitney U test p-values for multiple testing by Bonferroni correction. Significant enrichment was defined when p < 4.17 × 10−4, with Bonferroni correction for desired α < 0.01.
DATA AND SOFTWARE AVAILABILITY
Plot Reproducibility
All output data associated with the 10-fold CV analyses, as well as the IPython notebooks (Perez and Granger, 2007) used to produce associated figures and compute Wilcoxon signed-rank and Mann–Whitney U test p-values are provided at https://github.com/amathelier/DNAshapedTFBS_notebooks. Notebooks can be launched by using binder (mybinder.org) at http://mybinder.org/repo/amathelier/DNAshapedTFBS_notebooks. The script feature_importance_heatmap.py producing a heat map associated with a single classifier or set of classifiers is provided at http://github.com/amathelier/DNAshapedTFBS.
DNAshaped Python module
The DNAshaped Python module to train and apply the classifiers for ChIP-seq data can be found at http://github.com/amathelier/DNAshapedTFBS.
Supplementary Material
Highlights.
Considering DNA shape features improved the prediction of TF binding in vivo.
DNA shape at flanking regions of binding sites refined the prediction of TF binding.
Larger improvements were observed for the E2F and MADS-domain TF families.
Propeller twist at specific nucleotide positions of the MADS-box contributed most.
ACKNOWLEDGMENTS
We thank Miroslav Hatas for systems support and Dora Pak for management support, François Parcy, Oriol Fornés Crespo, and Chih-Yu Chen for comments on the manuscript, and all members of the Wasserman and Rohs labs for insightful discussions. AM and WWW were supported by the Genome Canada Large Scale Applied Research Grant 174CDE. Funding was provided by the Child and Family Research Institute, Vancouver, and the British Columbia Children's Hospital Foundation to AM and WWW. AM was also supported by funding from the Norwegian Research Council, Helse Sor-Ost, and the University of Oslo through the Centre for Molecular Medicine Norway (NCMM), which is a part of the Nordic European Molecular Biology Laboratory partnership for Molecular Medicine. RR was supported by the National Institutes of Health (grants R01GM106056 and U01GM103804) and the Alfred P. Sloan Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS AM, WWW, and RR conceived and designed the project. AM implemented and performed the experiments. AM, WWW, and RR analyzed and interpreted the results. AM implemented the DNAshapedTFBS tool. BX performed experiments and analyzed data related to the 4-bits-based classifiers, with contributions from TPC. TPC also generated tools for DNA shape analysis. LY analyzed and interpreted structural data. AM, WWW, and RR wrote the manuscript.
References
- Abe N, Dror I, Yang L, Slattery M, Zhou T, Bussemaker HJ, Rohs R, Mann RS. Deconvolving the Recognition of DNA Shape from Sequence. Cell. 2015;161:307–318. doi: 10.1016/j.cell.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afek A, Schipper JL, Horton J, Gordân R, Lukatsky DB. Protein–DNA binding in the absence of specific base-pair recognition. Proc. Natl. Acad. Sci. USA. 2014;111:17140–17145. doi: 10.1073/pnas.1410569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen, et al. Diversity and Complexity in DNA Recognition by Transcription Factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. Coregulation of Transcription Factor Binding and Nucleosome Occupancy through DNA Features of Mammalian Enhancers. Molecular Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nature Biotechnology. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rozowsky J, Galeev TR, Harmanci A, Kitchen R, Bedford J, Abyzov A, Kong Y, Regan L, Gerstein M. A uniform survey of allele-specific binding and expression over 1000-Genomes-Project individuals. Nat Commun. 2016;7:11101. doi: 10.1038/ncomms11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T-P, Comoglio F, Zhou T, Yang L, Paro R, Rohs R. DNAshapeR: an R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics. 2016;32:1211–1213. doi: 10.1093/bioinformatics/btv735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T-P, Yang L, Zhou T, Main BJ, Parker SCJ, Nuzhdin SV, Tullius TD, Rohs R. GBshape: a genome browser database for DNA shape annotations. Nucleic Acids Res. 2015;43:D103–D109. doi: 10.1093/nar/gku977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I, Golan T, Levy C, Rohs R, Mandel-Gutfreund Y. A widespread role of the motif environment on transcription factor binding across diverse protein families. Genome Res. 2015;25:1286–1280. doi: 10.1101/gr.184671.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror I, Rohs R, Mandel-Gutfreund Y. How motif environment influences transcription factor search dynamics: Finding a needle in a haystack. BioEssays. 2016;38:605–612. doi: 10.1002/bies.201600005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Struhl K. Where does mediator bind in vivo? PLoS ONE. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. The elements of statistical learning. Springer series in statistics Springer; Berlin: 2001. [Google Scholar]
- Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordân R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic Regions Flanking E-Box Binding Sites Influence DNA Binding Specificity of bHLH Transcription Factors through DNA Shape. Cell Reports. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx KS, Velde JV, de, Wang C, Weigel D, Vandepoele K. A Functional and Evolutionary Perspective on Transcription Factor Binding in Arabidopsis thaliana. Plant Cell. 2014;26:3894–3910. doi: 10.1105/tpc.114.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RW, Mathelier A, Peso L, del, Wasserman WW. Improving analysis of transcription factor binding sites within ChIP-Seq data based on topological motif enrichment. BMC Genomics. 2014;15:472. doi: 10.1186/1471-2164-15-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RW, Wasserman WW. Non-targeted transcription factors motifs are a systemic component of ChIP-seq datasets. Genome Biology. 2014;15:412. doi: 10.1186/s13059-014-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jain D, Baldi S, Zabel A, Straub T, Becker PB. Active promoters give rise to false positive “Phantom Peaks” in ChIP-seq experiments. Nucleic Acids Research. 2015;43:6959–6968. doi: 10.1093/nar/gkv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. J. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Research. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional Specificity of a Hox Protein Mediated by the Recognition of Minor Groove Structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, Zalckvar E, Sharon E, Machado ACD, Kalma Y, Lotan-Pompan M, Weinberger A, Yakhini Z, Rohs R, Segal E. Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 2015;25:1018–1029. doi: 10.1101/gr.185033.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht MW, Noble WS. Machine learning applications in genetics and genomics. Nat Rev Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Shi W, Wasserman WW. Identification of altered cis-regulatory elements in human disease. Trends in Genetics. 2015;31:67–76. doi: 10.1016/j.tig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Wasserman WW. The next generation of transcription factor binding site prediction. PLoS computational biology. 2013;9:e1003214. doi: 10.1371/journal.pcbi.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen C. -y., Chou A, Ienasescu H, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Research. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiño JM, Smaczniak C, Angenent GC, Kaufmann K, Dijk ADJ, van Structural determinants of DNA recognition by plant MADS-domain transcription factors. Nucleic Acids Res. 2014;42:2138–2146. doi: 10.1093/nar/gkt1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Lee Y, Bhupindersingh G, Iyer VR. Widespread Misinterpretable ChIP-seq Bias in Yeast. PLOS ONE. 2013;8:e83506. doi: 10.1371/journal.pone.0083506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RY, Stormo GD. Discriminative motif optimization based on perceptron training. Bioinformatics. 2014;30:941–948. doi: 10.1093/bioinformatics/btt748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. Scikit-learn: Machine learning in Python. The Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- Pellegrini L, Tan S, Richmond TJ. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- Perez F, Granger BE. IPython: A System for Interactive Scientific Computing. Computing in Science and Engineering. 2007;9:21–29. [Google Scholar]
- Pohl A, Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;30:1618–1619. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein–DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharthan R. Dinucleotide Weight Matrices for Predicting Transcription Factor Binding Sites: Generalizing the Position Weight Matrix. PLoS ONE. 2010;5:e9722. doi: 10.1371/journal.pone.0009722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, Mann RS. Cofactor Binding Evokes Latent Differences in DNA Binding Specificity between Hox Proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. Absence of a simple code: how transcription factors read the genome. Trends in Biochemical Sciences. 2014;39:381–399. doi: 10.1016/j.tibs.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo GD. Modeling the specificity of protein-DNA interactions. Quant Biol. 2013;1:115–130. doi: 10.1007/s40484-013-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, Oudenaarden A, van Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai ZT-Y, Shiu S-H, Tsai H-K. Contribution of Sequence Motif, Chromatin State, and DNA Structure Features to Predictive Models of Transcription Factor Binding in Yeast. PLoS Comput Biol. 2015;11:e1004418. doi: 10.1371/journal.pcbi.1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature Reviews Genetics. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nature Biotechnology. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ramsey SA. A DNA shape-based regulatory score improves position-weight matrix-based recognition of transcription factor binding sites. Bioinformatics. 2015;31:3445–3450. doi: 10.1093/bioinformatics/btv391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhou T, Dror I, Mathelier A, Wasserman WW, Gordan R, Rohs R. TFBSshape: a motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Research. 2014;42:D148–D155. doi: 10.1093/nar/gkt1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Motif discovery and transcription factor binding sites before and after the next-generation sequencing era. Briefings in Bioinformatics. 2012;14:225–237. doi: 10.1093/bib/bbs016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Kasinathan S, Xin B, Rohs R, Henikoff S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nature Communications. 2015;6:8733. doi: 10.1038/ncomms9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Computational Biology. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ruan S, Pandey M, Stormo GD. Improved Models for Transcription Factor Binding Site Identification Using Nonindependent Interactions. Genetics. 2012;191:781–790. doi: 10.1534/genetics.112.138685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Fraenkel E, Pabo CO, Pavletich NP. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 1999;13:666–674. doi: 10.1101/gad.13.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Shen N, Yang L, Abe N, Horton J, Mann RS, Bussemaker HJ, Gordân R, Rohs R. Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. USA. 2015;112:4654–4659. doi: 10.1073/pnas.1422023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Research. 2013;41:W56–W62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.