Abstract
Aneuploidy is a hallmark of breast cancer; however, our knowledge of how these complex genomic rearrangements evolve during tumorigenesis is limited. In this study we developed a highly multiplexed single-nucleus-sequencing method to investigate copy number evolution in triple-negative breast cancer patients. We sequenced 1000 single cells from 12 patients and identified 1–3 major clonal subpopulations in each tumor that shared a common evolutionary lineage. We also identified a minor subpopulation of non-clonal cells that were classified as: 1) metastable, 2) pseudo-diploid, or 3) chromazemic. Phylogenetic analysis and mathematical modeling suggest that these data are unlikely to be explained by the gradual accumulation of copy number events over time. In contrast, our data challenge the paradigm of gradual evolution, showing that the majority of copy number aberrations are acquired at the earliest stages of tumor evolution, in short punctuated bursts, followed by stable clonal expansions that form the tumor mass.
Introduction
Aneuploidy is pervasive in human cancers1 and is frequently (>90%) detected in breast cancer patients2,3. DNA copy number aberrations (CNAs) often lead to gene dosage effects that promote tumor growth through the overexpression of oncogenes or down-regulation of tumor suppressor genes. However most genomic studies have analyzed a single time-point sample (biopsy or surgery) making it difficult to study the natural progression of chromosome evolution during tumorigenesis. Currently, the prevailing model for copy number evolution posits that CNAs are acquired gradually and sequentially over extended periods of time, leading to successively more malignant stages of cancer4,5. An alternative model is punctuated copy number evolution (PCNE), in which CNAs are acquired in short bursts of crisis, followed by stable clonal expansions that form the tumor mass (Supplementary Fig. 1). Previous work has implicated a punctuated model to explain localized chromosome rearrangements, including chromothripsis6, chromoplexy7 and firestorms8. However, there has been limited data showing that genome-wide aneuploidy arises in a short punctuated burst, at the earliest stages of tumor evolution.
Intratumor heterogeneity provides a window into time, by representing a permanent record of the mutations that occurred during tumor progression. By assuming that mutational complexity increases over time, it is possible to reconstruct the evolutionary history of a tumor9,10 and investigate PCNE. However, most tumors consist of complex mixtures of single cells with different genotypes, complicating such studies. To address this problem, we previously developed a single cell DNA sequencing method called Single-Nucleus-Sequencing (SNS)11,12. We applied this method to sequence single tumor cells from two breast cancer patients, which provided initial evidence for PCNE12. However, these data were limited to two patients, mainly due to the high costs and low throughput associated with SNS. To address this problem, we developed a highly multiplexed single-nucleus-sequencing (HM-SNS) method that can profile 48–96 single cells in parallel.
In this study we applied HM-SNS to investigate the clonal substructure and evolution of CNAs in triple-negative breast cancer (TNBC) patients. TNBCs are a subtype of breast cancer that is characterized by a lack of estrogen receptor (ER), progesterone receptor (PR) and Her2 amplification13. TNBC patients show poor survival and frequently develop resistance to chemotherapy14. The majority of TNBC patients harbor TP53 mutations3 and show complex aneuploid rearrangements2,15. Genomic studies have shown that TNBC patients display a large amount of inter-patient heterogeneity in somatic mutations3, in addition to extensive intra-tumor heterogeneity within the tumor mass16–19. However, most studies of TNBC patients have been limited to bulk tumor analysis, and thus we investigated the clonal substructure of 12 treatment-naïve TNBC patients at single cell genomic resolution (Supplementary Table 1).
RESULTS
Highly-Multiplexed Single-Cell Copy Number Profiling
To profile genome-wide copy number in single cells we developed HM-SNS and applied it to sequence 1000 single cells from 12 TNBC patients (Fig. 1a). Nuclear suspensions were prepared from large (0.6–1cm3) frozen tumor specimens and stained with DAPI for flow-sorting. Single nuclei were gated by ploidy and deposited into individual wells on a 96-well plate for whole-genome-amplification (WGA) using degenerative-oligonucleotide-PCR (DOP-PCR)11,12. After WGA, barcoded libraries were prepared for each single cell and 48–96 libraries were pooled (Online Methods). The pooled libraries were sequenced on the Illumina platform at 76 single-end cycles. Single nuclei were sequenced at sparse coverage depth and copy number profiles were calculated from sequence read depth at 220kb resolution (Online Methods). On average 83 single cells (range 48–120) were sequenced from each TNBC patient (Supplementary Table 2). In each patient we observed a 2N diploid peak (D) and one or more aneuploid peaks that ranged from 1.8 – 4.1N in the flow-sorting histograms (Fig. 1b). Single nuclei were isolated from the aneuploid (A) and diploid (D) peaks, in addition to broadly gating nuclei from all ploidy distributions using universal (U) gates for a subset of tumors.
Figure 1. Highly-multiplexed single nucleus sequencing of TNBC patients.
(a) Highly-multiplexed single nucleus sequencing method. Tumor tissues are dissociated into nuclear suspensions and stained with DAPI for flow-sorting by DNA ploidy. Single nuclei are deposited into 96-well plates and whole-genome-amplified by DOP-PCR. Single cell libraries are barcoded with unique 8bp identifier and 48–96 libraries are pooled together for sparse next-generation sequencing. The sequence reads are demultiplexed using the cell barcodes after sequencing is completed for copy number profile calculations. (b) FACS plots of DAPI intensity showing ploidy distributions for each TNBC patient. Single cells were isolated from different distributions of ploidy that were gated as: D (diploid), A (aneuploidy), or U (universal).
Clonal Substructure and Diversity During Tumor Growth
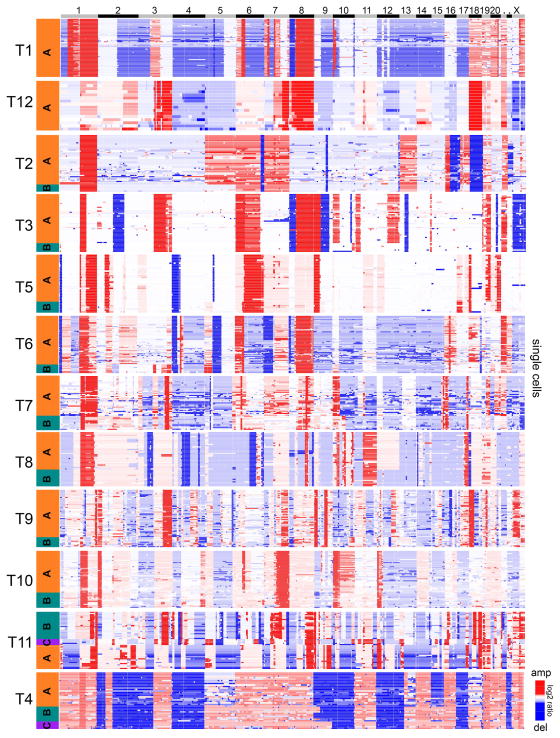
To delineate the clonal substructure of each tumor, we performed 1-dimensional hierarchical clustering of the aneuploid single cell copy number profiles. Clustered heatmaps identified 1–3 major subpopulations of clones (A, B, C) in each tumor (Fig. 2). Within each subpopulation, the single cells shared highly similar copy number profiles (mean pairwise r = 0.87), representing stable clonal expansions that occurred during tumor growth. A similar population substructure was also observed by clustering all of the aneuploid and diploid cells from each TNBC patient, where the diploid cells formed another independent cluster (Supplementary Fig. 2). To quantitatively determine the optimal number of clusters in each tumor, we applied PAMK-medoids clustering20 (Supplementary Fig. 3). The PAMK results were consistent with the hierarchical clustering results in most TNBC patients. Principle component analysis (PCA) was also consistent with the clustering results, by showing that 1–3 major clusters were present in each tumor (Fig. 3a). We quantified the genotype frequencies of the subpopulations, which revealed that some clones achieved higher frequencies in the tumor mass (Fig. 3b). To calculate a global metric of clonal diversity, we computed Shannon diversity indices for each TNBC patient (Online Methods). The diversity indices showed a broad range across the TNBC patient cohort, and corresponded to the number of clonal subpopulations that were present in each tumor (Fig. 3c). These data suggest that most TNBC tumors consisted of 1–3 major clonal subpopulations, and that complex aneuploid tumor profiles were highly stable (clonal stasis) during tumor growth.
Figure 2. Clonal subpopulations identified by clustering aneuploid cells.
Hierarchical 1-dimensional clustering of the single cell aneuploidy copy number profiles from each TNBC patient. The clonal subpopulations (A, B, C) are colored in orange, teal or purple. Single cells are plotted on the Y-axis, while copy number aberrations are plotted in genomic order on the X-axis.
Figure 3. Clonal composition and diversity of TNBC tumors.
(a) Principal Component Analysis of single cell copy number profiles sequenced from each TNBC tumor. Copy number profiles are colored by hierarchical clustering analysis and labeled as follows: diploid (D) or aneuploid tumor subpopulations (A, B, C). (b) Percentage of subclone genotypes in each tumor. (c) Shannon diversity index of copy number profiles from each tumor, with dotted lines indicating low, intermediate and high diversity groups.
Divergent Subpopulations in Polyclonal Tumors
Polyclonal tumors shared most CNAs between the subpopulations, but also differed by a few discrete subclonal events that emerged in the later stages of tumor evolution. The subclonal CNAs distinguished the clones and often resulted in the amplification of oncogenes and deletion of tumor suppressors. In several cases, the subclonal CNAs were associated with increased genotype frequencies of the clones in the tumor mass, suggesting that they may have provided a fitness advantage. To further investigate this possibility, we calculated clonal frequencies (cf) in the polyclonal tumors (Online Methods, Supplementary Table 3). For instance, in tumor T3, two major clonal subpopulations (A, B) were identified, in which clone A acquired additional amplifications of chromosome 10p and 12q (Fig. 4a). The 10p amplification increased the copy number of GATA3, while the 12q amplification increased the copy number of MDM2 in addition to several other genes. These amplifications were associated with an increased frequency of clone A (cf = 0.85) compared to clone B (cf = 0.15). In another polygenomic tumor (T2) we identified two major clonal subpopulations (A, B) that differed by a broad amplification on chromosome 5 that encompassed 14 cancer genes, including MAP3K1, ERBB2IP and PIK3R1 (Fig. 4b). This amplification was associated with an increased frequency of clone A (cf=0.87) relative to clone B (cf=0.13). Similar subclonal CNAs were found in other TNBC patients (T5 and T8) and often were associated with increased genotype frequencies in the clones that harbored the new CNAs (Supplementary Fig. 4). These data show that in addition to stable clonal expansions, TNBC patients can continue to acquire single CNAs in the later stages of tumor progression, and that these events are associated with the increased prevalence of new subpopulations.
Figure 4. Divergent subpopulations in polyclonal tumors.
Clustered heatmaps of single cell aneuploid copy number profiles in polyclonal tumors. (a) Tumor T3 heatmap with two subpopulations (A, B) identified. Subpopulation A (orange cluster) diverged from subpopulation B (teal cluster) by acquiring additional amplifications on chromosomes 10p and 12q, resulting in the amplification of GATA3 and MDM2 in addition to many other genes. (b) Tumor T2 heatmap with two subpopulations (A, B) identified. Subpopulation A (orange) diverged from the B subpopulation (teal) by the amplification of chromosome 5, containing many cancer genes including MAP3K1, ERBB2IP and PIK3R1.
Non-Clonal Copy Number Profiles in Tumors
While most cancer cells clustered into 1–3 major clonal subpopulations, we also identified a minor fraction (<10%) of non-clonal single cell copy number profiles in each tumor. On average the non-clonal copy number profiles occurred at 7.4 ± 0.8% (SEM) in the aneuploid fractions, 7.9 ± 1.4% (SEM) in the diploid fractions and 5.9 ± 1.0% (SEM) in the adjacent normal tissue cells (Fig. 5a–d, Supplementary Table 4). Based on the patterns of the CNA profiles, we identified three major classes of non-clonal cells: 1) metastable tumor cells, 2) pseudodiploid cells, and 3) chromazemic cells (Fig. 5e–h).
Figure 5. Non-clonal copy number profiles in tumors and normal breast tissues.
(a) Percentage of non-clonal cells in each tumor. (b) Percentage of non-clonal metastable aneuploid cells in the aneuploid fractions of each tumor. (c) Percentage of non-clonal pseudo-diploid cells in the diploid fractions of each tumor (d) Percentage of pseudodiploid cells in matched normal breast tissues from four TNBC patients (T3, T5, T8 and T10). (e) Examples of two metastable aneuploid cells (upper panels) compared to the copy number profiles of the major aneuploid subpopulations (lower panels). (f) Example of a pseudo-diploid cell isolated from diploid fractions of tumors. (g) Example of a pseudo-diploid cell isolated from matched normal breast tissues. (h) Examples of four chromazemic cells with large homozygous deletions of whole chromosomes or chromosome arms.
Metastable tumor cells are aneuploid cancer cells that share highly similar copy number profiles with the major subpopulations, but have evolved additional gains or losses of single chromosomes or arms (Fig. 5e). In tumor T3 we identified 53 single aneuploid tumor cells that shared a common copy number profile, and 6 unique metastable tumor cells with non-clonal amplifications and deletions. One metastable tumor cell from T3 showed an additional amplification of chromosome 5p compared to the major aneuploid tumor cells (Fig. 5e–h, left panel). In tumor T6 we identified 79 single tumor cells that shared a common copy number profile and 6 unique metastable tumor cells with non-clonal CNAs. One metastable tumor cell with an additional amplification of chromosome 18p is shown in comparison to the major aneuploid tumor subpopulation (Fig. 5e, right panel). Metastable tumor cells acquired single CNAs in the later stages of tumor evolution, but represent evolutionary ‘dead-ends’ that did not undergo further expansion to achieve prevalence in the tumor mass.
Pseudodiploid cells are single cells with flat 2N copy number profiles that have acquired additional gains or losses of single chromosomes or arms at random genomic locations (Fig. 5f–g). While most CNAs were randomly distributed, one exception was a frequent (23%) loss of the X chromosome in multiple cells from different patients (p<0.0001, one-tailed t-test) (Supplementary Table 5). To determine if non-clonal diploid cells were due to a tumor field effect, we also profiled normal breast tissues and found that 5.9% of cells also had non-clonal profiles (Fig. 5d, Fig. 5g). These data suggest that random copy number gains and losses occur during normal mitosis, and are unlikely to be associated with a tumorigenic field effect (Supplementary Table 6).
Chromazemic cells (-zemia = ‘damage’ or ‘loss’) are non-clonal cells with large homozygous deletions of whole chromosomes or chromosome arms that occur at random locations in the genome (Fig. 5h). These cells are unlikely to be viable, due to the large homozygous deletions of chromosomes. Chromazemic cells may be the byproduct of asymmetric cell divisions or possibly dying cells and are found in diploid fractions, normal tissues and aneuploid fractions.
Punctuated Copy Number Evolution
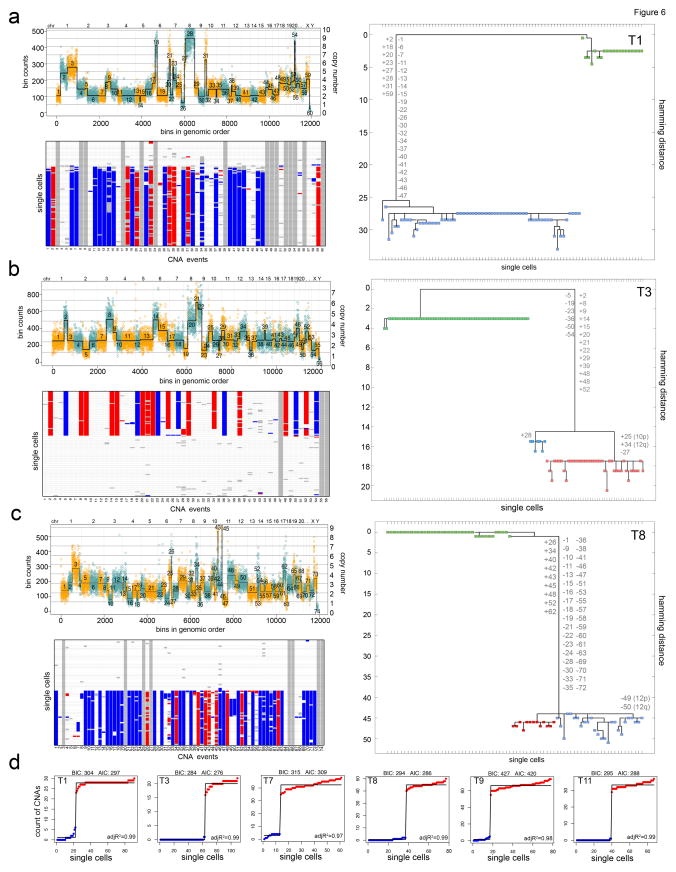
To trace tumor evolution, we constructed phylogenetic trees from the single cell copy number data. Intratumor heterogeneity provides a permanent record of the mutations that occurred during tumor growth, enabling lineages to be reconstructed by assuming that mutational complexity increases with time9,10. Copy number segmentation was performed using a multi-sample breakpoint algorithm21 to identify common chromosome breakpoints that occur across single cells within each tumor. We then calculated a trinary event matrix to treat all large and small CNA events equally for phylogenetic analysis using maximum parsimony (MP) (Online Methods). The resulting MP trees show that each tumor evolved a long root branch of founder (‘truncal’) CNAs that were acquired concurrently in the early stages of tumor evolution and maintained stably in the clones during tumor growth (Fig. 6a–c). Evidence of gradual intermediate branching was not observed as cells progressed from diploid to aneuploid genomes. Although some TNBC tumors showed clear evidence of divergent subclones in the later stages of tumor evolution, these clones typically only diverged by a few (N=1–3) CNAs, compared to the many (N=24–132) CNAs that were acquired in early punctuated bursts. Another important characteristic of the phylogenetic trees is that they show that all cancer cells share a common evolutionary origin in each tumor, suggesting that they evolved from a single normal cell in the breast tissue, not multiple initiating cells.
Figure 6. Punctuated copy number evolution and phylogenetic trees.
(a–c) Multi-cell segmentation (upper panels), trinary event matrices (lower panels), where white = 0, red = 1, and blue = −1 and maximum parsimony trees (right panels) from 3 TNBC patients: (a) T1, (b) T3 and (c) T8. Maximum parsimony trees are rooted by the diploid cells and non-clonal profiles were excluded from the analysis. Copy number events with non-integer values were filtered from all cells prior to tree construction and are shown in grey. (d) Linear and step fitting of sorted single cell CNA count data from 6 TNBC patients. Adj R2, BIC and AIC metric are also displayed for each fit.
To further investigate whether the single cell data was consistent with PCNE we performed linear (gradual) and multi-step (punctuated) fitting of the sorted CNA count data from the single cells in each tumor (Online Methods). The 1-step fit resulted in higher correlation values (adj R2 = 0.977) compared to the linear fitting (adj R2 = 0.704) and was statistically significant (p = 2.125e-9, one-tailed t-test) (Fig. 6d, Supplementary Fig. 5). Similarly, better BIC and AIC values were obtained for all tumors when step-fitting was applied. These data support PCNE, by showing that a large number of CNA events increased drastically within a short period of time during tumor evolution.
Absence of Gradual Intermediate Cells in Ungated Fractions
One possible explanation for the absence of gradual intermediate copy number profiles in the tumor mass is that the gating of ploidy distributions by FACS was too narrow and therefore may have missed intermediate cells that occur in between the ploidy peaks (Fig. 1b). To investigate this possibility, we performed universal gating (U) to sample broadly across all of the ploidy distributions in 4/12 TNBC patients and flow-sorted additional single nuclei for HM-SNS. Hierarchical clustering was performed using the narrowly gated and universally gated nuclei data and heatmaps were constructed to compare the clonal substructure (Supplementary Fig. 6). Clustering analyses showed similar population substructure in the universal (U) and ploidy-gated (A, D) populations of tumor cells from each patient, with no evidence of additional intermediate copy number profiles in the universal gates, suggesting that if intermediate profiles exist and persist in the tumor mass, they are very rare. These data are consistent with the cell counts in the FACS histograms, which show no evidence of intermediate density between the aneuploid and diploid populations, with the exception of minor S-phase populations (Fig. 1b).
Mathematical Modeling of Gradual and Punctuated Evolution
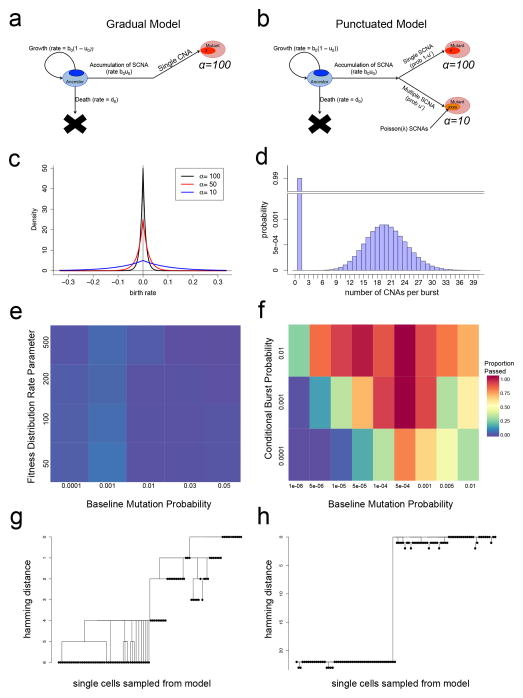
To further investigate alternative scenarios such as punctuated and gradual evolution in silico, we developed a multi-type stochastic branching process model of tumor growth. In this model, during each time-step a cell can divide to produce: 1) two daughter cells that are identical to the mother cell, 2) no cells (death), or 3) one daughter identical to the mother cell and one daughter with a new CNA whose fitness advantage is selected from a mutational fitness distribution22. In the gradual model, each cell division event may lead to the accumulation of a new CNA at a constant rate (Fig. 7a) corresponding to the baseline mutation rate for single copy number changes (Fig. 7c). In the punctuated model (Fig. 7b), each cell division event may either result in the accumulation of a single CNA or, at a different rate, a burst of multiple somatic CNAs whose number is chosen from a Poisson distribution (Fig. 7d). We implemented both models as exact stochastic computer simulations initiating with a single diploid ancestral tumor cell and continued each instantiation of the model until the total number of cells was equivalent to the total number of cells in each TNBC patient. From each simulation, we sampled 100 single cells at random and constructed phylogenetic trees (Supplementary Fig. 7). We then performed AMOVA23 to investigate the topologies of the resulting phylogenies. Permutation testing was applied to obtain p-values for each sample based on the gradual (Fig. 7e) and the punctuated model (Fig. 7f) and to test whether these models were able to recapitulate the tree topologies obtained from the TNBC patient data (Online Methods). We investigated a wide range of parameter values by searching through a total of 162 combinations of parameters. In the trees resulting from the gradual model, we found evidence of many intermediate subpopulations, suggesting that selective sweeps are unlikely to occur in later stages of tumor evolution, even when clones with high fitness values emerge (Fig. 7g, Supplementary Fig. 7b). In contrast, the punctuated simulations resulted in tree structures with long root nodes between the ancestral diploid and aneuploid subpopulations, which are consistent with our single cell data (Fig. 7h, Supplementary Fig. 7a). We then investigated alternative scenarios for gradual simulations (epistasis, cancer stem cells, increasing mutation rates, fixed fitness distributions and mutation rate from a distributions) to test their ability to recapitulate the data (Supplementary Notes, Online Methods). In total, we investigated these alternative scenarios for a total of 2,097 parameter combinations. However, under all of these scenarios, the resulting trees sampled from 100 single cells displayed evidence of many intermediate branching clones (Supplementary Fig. 7b). Collectively, these modeling data support PCNE and suggest that selective sweeps occurring in later stages of tumor growth are unlikely to explain the presence of highly clonal subpopulations.
Figure 7. Mathematical modeling of punctuated and gradual tumor evolution.
(a) Gradual model of multi-type stochastic birth-death-mutation process. (b) Punctuated model of multi-type stochastic birth-death-mutation process with a Poisson mutation burst probability distribution. (c) Fitness distributions with varying shape parameter values (alpha) that are used for sampling as new clones emerge during the binary branching process in the gradual or punctuated models. (d) Poisson probability distribution for multiple CNA events occurring in the punctuated model with a single atom (i.e. point mass) at 1 for single CNA events. (e) Heatmap of AMOVA analysis for different fitness distributions and mutation rates in the gradual model. (f) Heatmap of AMOVA analysis for different burst and mutation probabilities in the punctuated model. Colors indicate the proportion of simulations passing the ‘minimal punctuated criteria’, with p-values < 0.05 in AMOVA permutation and > 90% samples having root nodes with at least 5 CNAs to construct a tree. (g) Tree constructed from sampling 100 random single cells from simulated data generated from the gradual model. (h) Tree constructed from random sampling of 100 single cells from the simulated data from the punctuated model.
Inter-tumor Heterogeneity Between TNBC Patients
In addition to investigating intra-tumor heterogeneity we also compared copy number differences between TNBC patients. Consensus profiles were calculated to represent the bulk tumor populations from each TNBC patient by aggregating the single cell aneuploid copy number profiles (Online Methods). Frequency plots were calculated using all TNBC patients to identify common amplifications and deletions that are recurrent in the patient cohort (Fig. 8a). This analysis identified frequent amplifications on chromosome 1q (MDM4), 3q (PIK3CA), 6p (CCND3), 8q (MYC) and 18 (BCL2, SMAD4), while frequent deletions included chromosome 4p (FGFR3), 5q (PIK3R1), 8p (DBC2), 9p (NR4A3), 12 (MDM2) and 22 (CHEK2). These genomic regions and oncogenes are consistent with previous microarray CGH studies on TNBC patients15. In addition to the frequent CNAs, we also identified many unique high-level focal amplifications (< 10 mb) that occurred exclusively in individual patients (Supplementary Fig. 8). These focal amplifications are consistent with previous reports in TNBC patients15,24. We further investigated inter-patient tumor heterogeneity by integrating single cell data from all of the TNBC patients. Dimensionality reduction was performed using t-SNE25, which shows that single cells cluster according to the patient from which they were isolated (Fig. 8b). Similarly, hierarchical clustering grouped single cells according to patients (Fig. 8c). These data show that single cells from each TNBC patient are genetically more related to each other than to other tumors, suggesting that they share a common ancestral lineage and evolved from a single normal cell in the breast tissue.
Figure 8. Inter-tumor heterogeneity and focal amplifications in TNBCs.
(a) Frequency plot of CNAs across 12 TNBC patients with amplifications in red and deletions in blue. (b) t-SNE plot was calculated using all single aneuploid tumor cells from the 12 TNBC patients. Single cells were colored by individual patients. (c) Hierarchical clustering tree using ward linkage was constructed using pairwise Euclidean distances of all aneuploid tumor cells from the 12 TNBC patients.
Discussion
Collectively, our data support a punctuated model of copy number evolution, in which a large number of CNAs are acquired early in tumor evolution, in a short period of crisis, and remain highly stable as the tumor mass clonally expands (clonal stasis). Despite profiling hundreds of single cells from many spatial regions, we did not detect any gradual intermediate copy number profiles, as the tumor cells evolved from diploid to aneuploid genomes. These data challenge the dogma of gradual tumor evolution4,5 by showing that cancer cells with gradual intermediate copy number profiles are not common during tumor growth. These findings also challenge reports of extensive intratumor genomic heterogeneity in breast cancer16,17,19,26 by showing that CNAs are remarkably stable throughout the tumor mass. However, previous studies focused mainly on point mutations, which may represent different molecular clocks during tumorigenesis18. PCNE is consistent with a ‘big bang’ model for tumor growth27,28,29 in which clonal diversification occurs at the earliest stages of tumor progression, leading to the stable expansion of one or more clones.
An analogous model called ‘Punctuated Equilibrium’ was originally proposed by Gould and Eldredge in 1972 to explain species evolution30,31. This model was mainly supported by evidence in the fossil record and challenged Darwinian gradualism. Several interesting parallels can be drawn between these models: 1) stasis, 2) the lack of gradual intermediates and 3) short bursts of rapid evolution. However, it is important to note that the mechanisms underlying Punctuated Equilibrium in species evolution (e.g. allopatric speciation) are likely to be very different from PCNE in human tumors.
Punctuated copy number evolution and clonal stasis have important implications for tumor evolution, diagnostics and therapy. These data suggest that individual tumor cells may be hard-wired at the earliest stages of tumor growth and intrinsically pre-programmed to become invasive, metastatic or resistant to chemotherapy27,32. This deterministic characteristic may allow oncologists to profile CNAs in early-stage breast cancers (eg. DCIS) to predict whether the tumors should be treated aggressively, or alternatively not at all (‘watchful waiting’). Our single cell data also have important implications for clinical diagnostics, by showing that multi-region sampling may not be necessary for assessing CNAs as biomarkers in TNBC patients, since they are highly stable throughout the tumor mass.
Although most copy number profiles in the TNBC patients were found to be highly clonal, we also identified a minor (< 10%) fraction of cells with non-clonal copy number profiles. These cells were not intermediates in the tumor lineage, but instead showed random chromosome gains or losses. To determine if these cells were due to a tumor-specific field effect, we also profiled cells from normal breast tissue, which showed similar percentages of non-clonal cells (5.9%) to tumors. These data are consistent with recent single cell genomic data on tissue mosaicism, which have reported 1–5% non-clonal aneuploid cells in different normal tissues, including liver, brain and skin33. Because the majority of the non-clonal events involve a single chromosome gain or loss, we speculate that they are due to lagging chromosomes that occur during asymmetric mitoses34. While such events are unlikely to lead to further proliferation in normal tissues, it may provide tumors with additional ‘fuel for evolution’, occasionally leading to the emergence of new tumor subpopulations in the later stages of tumor evolution, as we observed in several polyclonal tumors.
Our study has addressed several key questions regarding copy number evolution in TNBC patients, but it has also raised several new lines of inquiry. How can genome instability be turned on and off at the earliest stages of tumor evolution in a reversible manner? One possibility for a reversible switch is telomere inactivation and reactivation, which could lead to complex aneuploid rearrangements in just a few cell divisions, in manner that can be reversed by telomerase reactivation. This mechanism has previously been described as ‘episodic telomere crisis’ and was demonstrated using experimental systems35–37. However we speculate that telomerase inactivation alone is insufficient to cause punctuated evolution, since TP53 inactivation and genome duplication are also requirements for PCNE. Indeed previous work using in vivo systems has shown that TP53 and telomere loss can cooperate to drive tumorigenesis35. Another important question is how tumor cells with complex aneuploid rearrangements can undergo symmetric cell divisions and stable clonal expansions (clonal stasis). For tumor cells to undergo symmetric cell divisions with supernumerary chromosomes, we speculate that aneuploid cells must cluster multiple centrioles together to align chromosomes equally along the metaphase plate38,39. Addressing these interesting questions will require future work and should be performed using in vitro and in vivo systems.
We also investigated whether we sequenced a sufficiently large number of cells in each TNBC patient to detect the major tumor subpopulations. To answer this question, we calculated posterior saturation curves with multinomial distributions (Supplementary Fig. 9). The resulting data suggest that 20–40 single cells were necessary to detect the major subpopulations with 95% power, suggesting that our sample size was sufficient (mean = 83). Another question we considered is whether an alternative model to PCNE could explain the observed single cell data, in which evolution is gradual until a clone with high fitness emerges at the later stages of progression, leading to a ‘selective sweep’. Despite extensive testing with mathematical modeling, we found that selective sweeps were highly uncommon during gradual evolution even when clones with high fitness emerged. Furthermore, a clonal sweep is inconsistent with studies that support early clonal diversification and selection27–29.
In summary, our single cell copy number data and mathematical modeling suggest that clonal stasis and PCNE are common in TNBC patients. This process leads to complex aneuploidy copy number profiles that are remarkably stable during tumor growth and ubiquitous throughout the tumor mass. Our preliminary data in other tumors (colon, prostate, liver, lung) suggest that PCNE may not be restricted to breast cancer, and is also likely to be operating in other human cancers. This model has important implications for our evolutionary understanding of cancer dynamics and for the clinical treatment of TNBC patients.
Online Methods
Triple Negative Breast Cancer Samples
Frozen tumors from 12 triple-negative breast cancer patients were selected with poorly differentiated and high grade (III) invasive ductal carcinomas as determined by the Bloom-Richardson score. The triple-negative status of the tumor samples was determined by IHC for estrogen receptor (<1%) and progesterone receptor (<1%), and FISH analysis of the Her2 amplification using the CEP-17 centromere control probe (ratio of Her2/CEP17 < 2.2). The frozen tumor samples and matched normal breast tissues were obtained from the UT MD Anderson Cancer Center Breast Tissue Bank. Two frozen tumor samples (T11 and T12) were obtained from the cooperative human tissue network (CHTN). This study was approved by the Internal Review Board (IRB) at the University of Texas MD Anderson Cancer Center.
Highly Multiplexed Single Nucleus Sequencing
Nuclei from frozen tumors were isolated using a NST/DAPI buffer (800 mL of NST [146 mM NaCl, 10 mM Tris base at pH 7.8, 1 mM CaCl2, 21 mM MgCl2, 0.05% BSA, 0.2% Nonidet P-40]), 200 mL of 106 mM MgCl2, 10 mg of DAPI, and 5mM EDTA. The frozen tumors were dissociated into nuclear suspensions by mincing with no.11 surgical scalpels in 1ml of NST-DAPI cytoplasmic lysis buffer at 4°C using ice blocks in a plastic Petri dish. Nuclear suspensions were filtered through 37-μm plastic mesh prior to flow-sorting into a 5-ml polystyrene tube (Falcon). Single nuclei were flow-sorted into 96-well plates by FACS using the Aria II flow cytometer (BD Biosciences). Ploidy distributions were gated by differences in their total genomic DNA content as determined by DAPI intensity. To establish the fluorescence DAPI intensity corresponding to diploid (2N) a lymphoblast control cell line (REFM) was first flow-sorted to establish gates. Prior to flow-sorting single nuclei, a few thousand cells were first sorted to establish the DNA content distributions for gating by Ploidy. Single nuclei were collected from both diploid and aneuploid gated fractions. Additionally, nuclei were collected from each tumor by gating broadly across all ploidy distributions. Single nuclei were deposited into individual wells on a 96-well plate with 10ul of lysis solution in each well from the Sigma-Aldrich GenomePlex© WGA4 kit, along with negative control reactions, in which no nuclei were deposited.
Whole-Genome-Amplification & Barcoded Library Construction
Whole genome amplification (WGA) was performed on single flow-sorted nuclei using degenerative-oligonucleotide-PCR (DOP-PCR) as described in the Sigma- Aldrich GenomePlex WGA4 kit (cat # WGA4-50RXN) protocol. For QC of WGA performance the DNA concentration was measured (ThermoFisher Scientific, Qubit 2.0 Fluorometer) and reactions were run out using gel electrophoresis to determine size distributions. To prepare sequencing libraries by TA ligation cloning, 500ng of DNA were acoustically sonicated to 200bp using the Sonicator S220 (Covaris). Fragmented WGA products underwent end repair (New England Biolabs (NEB), #E6050L) and were purified with the DNA Clean & Concentrator-5 Kit (Genesee, #11-303 or 11-306). Libraries were constructed using NEBNext® DNA library Prep enzymes (NEB, #E6050L, E6053L, E6056L/M0202L, and M0541L) for end-repair, 3′ adenylation, ligation and PCR amplification according to manufacturer’s instructions, but using different P7 adapters to barcode each single cell library with a unique 8bp identifier and common P5 adapters for sample multiplexing. The 96 unique P7 indexes are NEXTflex-96 barcodes that were purchased from Bio scientific. Following the ligation, DNA underwent a negative and positive selection with Ampure XP beads (Beckman Coulter, #A63881), 0.7× and 0.15× respectively, prior to PCR amplification. Final library concentrations were measured using the Qubit 2.0 Fluorometer and 48–96 single cell libraries were pooled together in equimolar concentrations. Final concentration of the pooled libraries was measured by quantitative PCR using KAPA Library Quantification Kit (KAPA Biosystems, KK4835) and ABI PRISM real-time machine (Applied Biosystem 7900HT), as well as 2100 Bioanalyzer (Agilent).
Multiplexed Illumina Next-Generation Sequencing
Pooled libraries containing 48–96 barcoded single cell libraries were sequenced at 76 single-end cycles on the HiSeq2000 system (Illumina) in Sequencing Core Facility of Genetics Department at MD Anderson Cancer Center to obtain a target coverage depth of 0.1× per single cell library. Data was processed using the CASAVA 1.8.1 pipeline (Illumina Inc.) and sequence reads were converted to a master FASTQ files. Sequencing reads from each single cell were demultiplexed using an in-house perl script (demultiplex.pl) into 48–96 independent FASTQ files, where each file represented the sequencing reads from one cell.
Sequence Alignment and Data Processing
After barcodes and sequencing adaptors were trimmed, sequence reads in FASTQ format were mapped to the human assembly US National Center for Biotechnology Information (NCBI) build 37 (HG19/NCBI37) using Bowtie2 alignment software40 with default parameters to generate SAM files. Samtools (0.1.19) was used to convert SAM files to compressed BAM files and sort the BAM files by chromosome coordinates41. To eliminate PCR duplicates, Samtools was used to remove sequence reads with identical start coordinates. Sequence reads with low mapping quality (MQ<40) were also filtered using Samtools.
Integer Copy Number Calculation from Single Cell Data
The sequencing data was counted in 11,927 genomic bins with variable start and stop coordinates, using the ‘variable binning’ method as previously described11,12. The median genomic length spanned by each bin is 220kb. This variable binning approach reduces mappability errors and false deletion events when compared to scaffolds using uniform length-fixed bins. A blacklist of ‘aberrant bins’ was filtered to remove false-positive amplifications in the centromeric and telomeric regions. The ‘aberrant bins’ are defined as bins where 5% to 95% percentile of ratios are distant from the ground states (|difference of ratio| > 0.5) in at least 2/3 normal single cell populations or the bins with systematic artifacts where the ratios are extremely high (ratio >10) or low (ratio < 0.001) across all single cells. Only single cells with median reads/bin greater than or equal to 50 were included for downstream copy number analysis. We then applied Loess normalization to correct for GC bias11. The copy number profiles were segmented using the Circular Binary Segmentation (CBS)42 followed by MergeLevels43 to join adjacent segments with non-significant differences in segmented ratios. The parameters used for CBS segmentation is alpha=0.0001 and undo.prune=0.05 respectively. Default parameters were used for performing MergeLevels, which successfully joined false positive detections of erroneous breakpoints. The exp.mad was calculated as the median distance between the log2 transformed ratio and segmented values. Only segmentations having at least 1.48 times exp.mad deviation were retained as copy number aberrations. Finally, the integer copy number was calculated by scaling segmented ratios with average DNA ploidy determined by flow-sorting indexes and rounding to the closest integer values. When DNA ploidy information was unavailable for universally gated single cells, the least-square rounding method was applied to obtain the optimum scaling factor that has least sum of deviations from the closet integers after rounding44. Lastly, we filtered diploid single cells with variant coefficients of bin counts larger than 0.4 or having ‘mean_resid’ values bigger than 0.03. We calculated the ‘mean_resid’ as the average deviation of scaled ratios from the true ground state integer values, i.e. 2, since they are 2N diploid cells. We filtered aneuploid single cells with MAD of genome-wide ratios bigger than 0.62 or autocorrelation values of neighboring data points less than 0.53. Autocorrelation values were calculated as moving windows using a 10-bin interval size across the 11,927 bins in the human genome scaffold. This window results in low correlation values in regions where adjacent data points have random values. These steps remove single cells with poor WGA performance for the subsequent multivariate data analysis.
Cancer Gene Annotations
Amplifications and deletions identified in the single cell copy number profiles were annotated for known cancer genes, which was consisted of 413 genes that were compiled from multiple databases, including the Cancer Gene Census45, The Cancer Gene Atlas Project (TCGA) and the NCI cancer gene index (Sophic Systems Alliance Inc., Biomax Informatics A.G). BEDtools46 was used with the IntersectBED function to find the intersection of the known cancer gene BED file and regions of chromosome amplifications and deletions that were detected in the single cell sequencing datasets.
Clustering for Single Cell Copy Number Profiles
To construct the clustering heatmaps, the Euclidean distances were calculated from copy number data matrix where each column represents one single cell and each row contains the log2(ratio+0.1) transformed data of each segment. The one-dimensional hierarchical clustering was performed in R using the heatmap.2 function from the ‘gplots’ package available on CRAN47. Each column representing one single cell are hierarchically clustered using ‘ward’ linkage based on pairwise Euclidean distances, and the X-axis is ordered by genome positions. To estimate optimal numbers of clustering for each patient, we performed partition around medoids clustering with optimum Calinski-Harabasz index48 or average silhouette width using the pamk49 function from the ‘fpc’ package. Clusters with singleton cells were collapsed and penalized on pamk criteria to minimize technical artifacts. The K-medoids clustering was performed on a range from K=1 to K=20 clusters.
High-Dimensional Data Analysis Methods
For each individual tumor, the numeric matrix containing integer copy numbers was used to perform principle component analysis (PCA) using ‘prcomp’ function in R47. The columns in the numeric matrix are segmented bins and each row is an individual single cell. The first two principle components were plotted in x and y-axis respectively. Each dot on the PCA plots represented single cell copy number profiles and are colored according to cells that clustered together into subpopulations that were identified by the hierarchical clustering analysis. To determine the genomic relationship of all aneuploid tumor cells from the 12 TNBC patients, the t-distribution Stochastic Neighbor Embedding (t-SNE)50 method was applied based on the pairwise Euclidean distances of the ratio data. The t-SNE method is an improved nonlinear dimensionality reduction and visualization method, with which both local and global structures in high-dimensions can be visualized in low-dimensional plots, while avoiding dramatic masking of very similar data points seen in PCA plots.
Calculation of the Subclonal Diversity Index
To calculate the subpopulation diversity index for each tumor, we performed hierarchical clustering of copy number data to cluster the aneuploid tumor cells into 1–3 major groups (‘species’) based on Euclidean distances. Cells within each subpopulations were defined as highly correlated with mean R2 >0.8. We then calculated the proportion (p) of cells that belong to each distinct group. The subpopulation diversity index is then calculated as Shannon Index: Dc = −Σi(pi×lnpi), where larger values representing higher subclonal diversity within the tumor.
Clonal Frequency of Subpopulations
To calculate the clonal frequencies of each clonal subpopulation, we first identified clusters of genotypes by hierarchical clustering and the optimal clustering results were selected based on Calinski-Harabasz index48 or average silhouette width. We then counted the number of cells that were classified into each sub-clusters. The relative clonal frequencies were calculated as the number cells that fell into each specific sub-clusters divided by the total number of clonal aneuploid cells. The singleton cells that formed the only one member of a sub-cluster were defined as non-clonal and were extruded from this calculation.
Copy Number Aberration Frequency Calculation and Plots
Consensus copy number integer profiles for each tumor were calculated using the median integer copy number segment values of all aneuploid single cells from each tumor. To calculate the frequency plot of the 12 TNBC samples, the mean copy number values across the genomic bins of each cell were treated as the ground state copy number and 1.5 times the standard deviation (SD) across the genome as deviation cutoff values. If a copy number was higher than mean+1.5×SD, then a significant amplification was designated, while for the copy number lower than mean−1.5×SD, a significant deletion was designated. The amplification and deletion frequencies across all tumors were calculated by first counting the total number of consensus tumor profiles that having significant amplifications or deletions in each of the 11,927 bins across the genome and then dividing the counts by the total number of consensus profiles.
Multi-Cell Segmentation and Event Matrix Construction
To detect common chromosome breakpoints and segments that are shared between single cell samples, we applied a multi-sample population segmentation algorithm using bioconductor R package (“copynumber”)21, with regularization parameter γ=40 (default). Segments smaller than 20 bins were removed, and their flanking segments joined, or separated at the center of the removed segment if they differed significantly (Wilcoxon test, Hommel-adjusted p-value < 0.05 in at least 2 cells)51. The ground state of each cell was calculated by rounding its expected ploidy to the nearest integer44. For each tumor, a median matrix M was constructed, in which M is the median of the ith segment in the jth cell. From this median matrix, an event matrix E was calculated as follows: Let gi be the ground state of the jth cell. Eij = 1 (amplification) if Mij − gi > 0.6, Eij = −1 (deletion) if Mij − gi < − 0.6, and Eij = 0 (neutral) if |Mij − gi | < 0.4. If 0.4 ≤ |Mij − gi |≤ 0.6, Eij was treated as missing with systematic artifact. Segments that have missing values systematically across all cells were removed if they satisfied the following criterion:
where u is the probability density function (p.d.f.) of a uniform distribution on (0, 1), C0.2 is the cardioid p.d.f. 52 with concentration parameter ρ=0.2, and {Mij} is the fractional part of Mij. This formula is a Bayes factor for comparing a model in which ploidy-scaled segment medians do not cluster around integer values to one in which they do, and the chosen cutoff represents 99% certainty of the first model assuming equal prior probabilities.
Phylogenetic Tree Construction using Maximum Parsimony
Maximum parsimony trees were calculated from event matrices using the parsimony ratchet algorithm with R package “phangorn”53. Amplification, neutral and deletion were treated as characters, and missing values as ambiguous sites. Events occurring on sex chromosomes were ignored. Metastable cells were removed from each tumor for phylogenetic analyses, since they do not share CNAs in the main tumor lineages. Cells were also removed if at least one third of events were missing values. Branch lengths and ancestral character probability distributions were inferred using the Acctran algorithm53. Altered sites on each edge were estimated as the sites such that:
where A and B are the ancestral sequences estimated at each node and c is a character.
Phylogenic tree visualization
Phylogenic trees were exported in Newick format from R-studio and plotted as square trees using Matlab (MathWorks Inc). The trees were re-rooted by the top node of diploid cells. Each individual single cell was represented as tips of the tree and the nodes were colored based on the sub-clonal populations. Single cells from the same subpopulations were flipped to physically nearby with each other to favor visualization.
Mathematical Modeling of Gradual and Punctuated Evolution
Please see ‘Supplementary Notes’ for details on mathematical modeling of gradual and punctuated tumor growth.
Statistical Fitting of Copy Number Aberrations
We first counted the total number of CNA events within each single cell from the collapsed trinary event matrix. To minimize technical noise, singleton CNAs that existed in only one cell or CNA events with missing values in > 50% cells, or cells with > 40% missing values were excluded from analysis. Subsequently, single cells were sorted based on the total number of CNA events within each cell. For the gradual linear model, we assume tumor cells evolved through intermediate genomes and therefore the total CNAs increased gradually over time. We therefore fit a linear model of total segments with a slope: CNAs=Time + Error. For the punctuated model, we assumed tumor cells lack gradual intermediate species and therefore the total CNAs changed over one or more critical evolutionary steps reflected in the jumping of events. The punctual model is CNAs=Step + Error, with no slope, where the fitted values are simply the average numbers of CNAs per cell within each step. Models were fit using the lm function in R47. The Bayesian Information Criteria (BIC), Akaike Information Criterion (AIC) and adjusted R2 values were calculated to measure the best fit of each model for the tumor datasets.
Saturation Analysis to Estimate Required Sample Sizes
A post hoc saturation analysis was performed to determine whether we sequenced sufficient cells for the purpose of this study. We first obtained the total number of subpopulations and the fractions of each subpopulation within each tumor by hierarchical clustering single cell with copy number data as described above. We then calculated the accumulative probability of observing at least 3 single cells in each subpopulation given numbers of sequenced cells, by assuming the number of observed cells following a binomial distribution for bi-clonal tumors and multinomial distribution for tri-clonal tumors. The two monogenomic tumors were excluded from this analysis. The cumulative probabilities were calculated in R47.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Lefkofsky Family Foundation. N.N. is a Nadia’s Gift Foundation Damon Runyon-Rachleff Innovator (DRR-25-13). This work is also supported by a grant to N.N. from NCI (1RO1CA169244-01) and from the American Cancer Society (129098-RSG-16-092-01-TBG). N.N. is a T.C. Hsu Endowed Scholar, AAAS Wachtel Scholar and an Andrew Sabin Family Fellow. The study is also supported by the Moonshot Knowledge Gap Award and the Center for Genetics & Genomics. This study was supported by the MD Anderson Sequencing Core Facility Grant (no. CA016672) and the Flow Cytometry Facility grant from NIH (no. CA016672). Additional funding support includes the Rosalie B. Hite Fellowship (A.C.); Center for Genetics & Genomics Postdoctoral Fellowship (R.G.); NIH UL1 TR000371 (F.M.B.); Nellie B Connally Breast Cancer Research Endowment (F.M.B), Susan Komen SAC10006 (F.M.B.), CPRIT RP110584 (F.M.B), and the MD Anderson Cancer Center Support grant (NIH/NCI P30 CA016672). F.M. gratefully acknowledges support from the Dana-Farber Cancer Institute Physical Science Oncology Center (U54CA193461-01). We thank Mary Edgerton, Jude Kendall, Michael Wigler and James Hicks for their support and discussions. We are also very grateful to the breast cancer patients at MD Anderson for generously donating their tumor tissues to our research studies.
Footnotes
Accession Codes
All data from this study has been deposited to the Sequence Read Archive (SRA) under accession number: SRP064210 for tumors T1–T10, SRA018951 for tumors T11 and T12.
Author Contributions
RG analyzed the data and wrote the manuscript. AD analyzed the data. TM and FM performed mathematical modeling and wrote the manuscript. ES, XS, PT and JL performed experiments. AC and YW analyzed the data. HZ and FMB provided tumor samples and interpreted the data. NN analyzed the data, led the project and wrote the manuscript.
Competing Interests
The authors have no competing interests to declare.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Hicks J, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–79. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Hoglund M, Gisselsson D, Hansen GB, Sall T, Mitelman F. Multivariate analysis of chromosomal imbalances in breast cancer delineates cytogenetic pathways and reveals complex relationships among imbalances. Cancer Res. 2002;62:2675–80. [PubMed] [Google Scholar]
- 6.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicks J, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navin N, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navin NE, Hicks J. Tracing the tumor lineage. Mol Oncol. 2010;4:267–83. doi: 10.1016/j.molonc.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baslan T, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–41. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 14.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 15.Turner N, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–23. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almendro V, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–27. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21:751–9. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds APRG, Iglesia B, Rayward-Smith Clustering Rules: Comparison of Partitioning and Hierarchical Clustering Algorithms. Journal of Mathmatical Modeling and Algorithms. 2006;5:475–504. [Google Scholar]
- 21.Nilsen G, et al. Copynumber: Efficient algorithms for single- and multi-track copy number segmentation. BMC Genomics. 2012;13:591. doi: 10.1186/1471-2164-13-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foo J, et al. An Evolutionary Approach for Identifying Driver Mutations in Colorectal Cancer. PLoS Comput Biol. 2015;11:e1004350. doi: 10.1371/journal.pcbi.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–91. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lips EH, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015;17:134. doi: 10.1186/s13058-015-0642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bushati N, Smith J, Briscoe J, Watkins C. An intuitive graphical visualization technique for the interrogation of transcriptome data. Nucleic Acids Res. 2011;39:7380–9. doi: 10.1093/nar/gkr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–44. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–16. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling S, et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc Natl Acad Sci U S A. 2015;112:E6496–505. doi: 10.1073/pnas.1519556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–44. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould SJ, Eldredge N. Punctuated equilibrium comes of age. Nature. 1993;366:223–7. doi: 10.1038/366223a0. [DOI] [PubMed] [Google Scholar]
- 31.Eldredge N, Gould SJ. Punctuated equilibria: an alternative to phyletic gradualism. Models in Paleobiology. 1972:82–115. [Google Scholar]
- 32.DePinho RA, Polyak K. Cancer chromosomes in crisis. Nature Genetics. 2004;36:932–934. doi: 10.1038/ng0904-932. [DOI] [PubMed] [Google Scholar]
- 33.Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A. 2014;111:13409–14. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–38. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 36.Artandi SE, DePinho RA. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr Opin Genet Dev. 2000;10:39–46. doi: 10.1016/s0959-437x(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 37.Chin K, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–8. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 38.Leber B, et al. Proteins required for centrosome clustering in cancer cells. Sci Transl Med. 2010;2:33ra38. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- 39.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 43.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array CGH data for downstream analyses. Bioinformatics. 2005;21:4084–91. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 44.Garvin T, et al. Interactive analysis and assessment of single-cell copy-number variations. Nat Methods. 2015 doi: 10.1038/nmeth.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Team, R.C. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 48.Calinski RB, Harabasz J. A Dendrite Method for Cluster Analysis. Communications in Statistics. 1974;3:27. [Google Scholar]
- 49.Kaufman L, Rousseeuw PJ. Finding groups in data: an introduction to cluster analysis. xiv. Wiley; Hoboken, N.J: 2005. p. 342. [Google Scholar]
- 50.Maaten LJPvd, Hinton GE. Visualizing High-Dimensional Data Using t-SNE. Journal of Machine Learning Research. 2008;9:27. [Google Scholar]
- 51.Hommel G. A Stagewise Rejective Multiple Test Procedure Based on a Modified Bonferroni Test. Biometrika. 1988;75:383–386. [Google Scholar]
- 52.Pewsey A, Neuhäuser M, Ruxton GD. Circular statistics in R. xiv. Oxford University Press; Oxford; New York: 2013. p. 183. [Google Scholar]
- 53.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.