Abstract
Background
Personal exposure to fungal bioaerosols derived from contaminated building materials or agricultural commodities may induce or exacerbate a variety of adverse health effects. The genomic mechanisms that underlie pulmonary immune responses to fungal bioaerosols have remained unclear.
Objective
The impact of fungal viability on the pulmonary microRNA and messenger RNA profiles that regulate murine immune responses was evaluated following subchronic inhalation exposure to Aspergillus fumigatus conidia.
Methods
Three groups of naïve B6C3F1/N mice were exposed via nose-only inhalation to A. fumigatus viable conidia, heat-inactivated conidia, or HEPA-filtered air twice a week for 13 weeks. Total RNA was isolated from whole lung 24 and 48 hours post final exposure and was further processed for gene expression and microRNA array analysis. The molecular network pathways between viable and heat-inactivated conidia groups were evaluated.
Results
Comparison of datasets revealed increased Il4, Il13, and Il33 expression in mice exposed to viable versus heat-inactivated conidia. Of 415 microRNAs detected, approximately 50% were altered in mice exposed to viable versus heat-inactivated conidia 48 hours post exposure. Significantly downregulated (P ≤ 0.05) miR-29a-3p was predicted to regulate TGF-β3 and Clec7a, genes involved in innate responses to viable A. fumigatus. Also significantly downregulated (P ≤ 0.05), miR-23b-3p regulates genes involved in pulmonary IL-13 and IL-33 responses and SMAD2, downstream of TGF-β signaling. Using Ingenuity Pathway Analysis, a novel interaction was identified between viable conidia and SMAD2/3.
Conclusion and Clinical Relevance
Examination of the pulmonary genetic profiles revealed differentially expressed genes and microRNAs following subchronic inhalation exposure to A. fumigatus. MicroRNAs regulating genes involved in the pulmonary immune responses were those with the greatest fold change. Specifically, germinating A. fumigatus conidia were associated with Clec7a and were predicted to interact with Il13 and Il33. Furthermore, altered microRNAs may serve as potential biomarkers to evaluate fungal exposure.
INTRODUCTION
Exposure to fungal bioaerosols in indoor environments has been associated with adverse respiratory health effects [1, 2]. Interest in understanding the mechanisms associated with fungal exposures has increased following recent natural disasters, including hurricanes Katrina, Rita, and more recently Sandy, that have resulted in the prolific contamination of flooded indoor building materials [3]. Aspergillus fumigatus is a saprophytic mold naturally found in the environment that produces respirable asexual conidia [4, 5]. In some damp indoor environments, A. fumigatus can be found growing on common building materials such as wood, gypsum board, and chipboard [6] and disturbance of these contaminated building materials can result in elevated personal exposure [7]. To date, the influence of conidia viability on the microRNA (miRNA) and messenger RNA (mRNA) profiles underlying pulmonary immune and toxicological responses to A. fumigatus exposure are not fully characterized.
Our laboratory has developed a murine inhalation model of fungal conidia exposure to reproduce exposures encountered in fungal contaminated indoor, outdoor and occupational environments [8]. Dry homogenous aerosolized fungal conidia are delivered to mice housed in a multi-animal nose-only inhalation chamber. Initially, our laboratory reported an IL-17-secreting CD8a+ cytotoxic T cell population (Tc17) dominant in the lungs of mice after 4 weeks of inhalation exposure to A. fumigatus that correlated to the onset of in vivo conidia germination [8]. By 13 weeks, inhalation of A. fumigatus was found to elicit a mixed Type 1 T helper (Th1) and Type 2 T helper (Th2) response in mice exposed to viable A. fumigatus conidia [9]. Subchronic exposure to viable conidia significantly increased CD4+ T cells and associated cytokines, such as interleukin 13 (IL-13) and interferon gamma (IFNγ) compared to heat-inactivated conidia (HIC). This study corroborated previous observations demonstrating that conidia germination was a crucial parameter in the induction of allergic inflammation in the lungs.
Compared to viable conidia, heat-inactivated conidia were produced through heat and pressure treatment (autoclaving) that appears to degrade the proteins without changing the morphology of the conidia. The degraded proteins render conidia inactivated, ultimately inhibiting germination. Previously, we reported an accumulation of eosinophils in the airways of mice exposed to repeated instillations of heat inactivated conidia over a short period of time [10]. Another study demonstrated that a Th1 dominant response follows exposure to heat inactivated conidia, compared to a mixed Th1 and Th2 response in murine airways following exposure to viable A. fumigatus conidia [11]. A mixed response was also observed in mice exposed to viable A. fumigatus conidia for 13 weeks [12]. Heat inactivation and protein degradation may inhibit the secretion of critical allergens influencing pulmonary immune responses [13]. Inhalation of A. fumigatus conidia in mice is known to elicit an allergic inflammatory phenotype, with the hallmarks of allergic asthma such as the recruitment of eosinophils and lymphocytes [14]. In humans, A. fumigatus is known to cause allergic bronchopulmonary aspergillosis that results from a hypersensitization to Aspergillus antigen exposure characterized by a dominant Th2 immune response and a lower presence of Th1 cytokines, as well as eosinophilic inflammation [15, 16]. Despite extensive characterization of A. fumigatus induced pulmonary immune responses, few studies have evaluated the molecular processes associated with subchronic fungal exposure.
MiRNAs are noncoding RNAs located in the 3′ untranslated region that are 19 – 25 nucleotides in length and capable of influencing gene expression through a variety of mechanisms [17–21]. Early studies suggested that miRNAs could only degrade the target messenger RNA or inhibit translation; however, more recent data suggests that miRNA also activate the translation of certain target mRNA [19, 20]. MiRNA regulation of gene expression is complex because not only can hundreds of genes be targeted by one miRNA, but also many miRNAs can target one individual gene. These small RNAs play a critical regulatory role in numerous diseases and cellular processes such as cancer, heart disease, cellular development, apoptosis, and immune responses [18, 21, 22].
MiR-21 is a miRNA that has been extensively studied as part of the response to inflammatory stimuli and in a variety of disease models including, cancer [23–25], allergic airway inflammation [26], psoriasis [27], as well as in viral [28, 29], bacterial and protozoan infection models [30, 31]. MiR-21, miR-22, miR-27b, miR-31, miR-126, miR-155, miR-210, and miR-301a expression was increased after dermal exposure to toluene 2,4-diisocyanate [32]. MiR-126 and miR-145 have been shown to be upregulated in the airways following exposure to house dust mite allergen [33, 34]. Inhibition of miR-126 in a house dust mite-induced asthma model resulted in decreased allergic inflammation [33]. Airborne pollutants are also known to alter the miRNA environment in the murine lung; for example cigarette smoke downregulates let-7c, let-7f, miR-34b, miR-34c and miR-222 [35–38]; diesel exhaust upregulates miR-135b [39]; and volatile organic compounds have been shown to upregulate miR-125a and miR-466, while downregulating miR-125b [36, 40, 41]. Previous studies have reported the dysregulation of miRNAs following Staphylococcal enterotoxin B [42] and nicotine exposure [43].
Although other studies have examined miRNA and mRNA environments in other models and to different stimuli, this study is among the first to evaluate the pulmonary miRNA and mRNA environment following subchronic exposure to aerosolized fungal conidia. To our knowledge, this is the first study to examine the influence of viable A. fumigatus conidia on the miRNA and mRNA profiles and pathways involved in pulmonary immune responses.
METHODS
Animals
Female B6C3F1/N mice, aged 5–6 weeks old were acquired from the National Toxicology Program and mouse colony housed at Taconic (Germantown, NY). Mice were acclimated for approximately one week prior to exposures and housed in filtered, ventilated polycarbonate cages in groups of 5 on autoclaved hardwood chip bedding. Mice were provided with NIH-31 modified 6% irradiated rodent chow (Harlan Teklad) and tap water ad libitum. The National Institute for Occupational Safety and Health (NIOSH) animal facility is an environmentally controlled barrier facility that is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. All animal procedures were performed under a NIOSH Animal Care and Use Committee approved protocol.
Fungal cultures
Aspergillus fumigatus strain B5233/ATCC 13073 (ATCC, VA) conidia were inoculated on malt extract agar (MEA) plates for 7–10 days at 25°C and harvested in 2 milliliters (mL) of filter sterilized, endotoxin-free water (Sigma Aldrich, St. Louis, MO). Autoclaved Mahatma brown rice (10g) (Riviana Foods Inc., Houston, TX) was inoculated with 3 mL of 2.5 × 106 conidia/mL and incubated for 10 to 14 days. Heat inactivated conidia were prepared by autoclaving the A. fumigatus laden rice cultures for 15 minutes at 121°C. This high temperature and pressure treatment did not alter the morphology of the conidia, but did reduce the viability by at least 97%. It is assumed that the potential germination of the remaining 3% or less viable conidia did not have an appreciable effect on the pulmonary immune response. Viable and heat inactivated conidia were transferred to a desiccator for 3 days prior to inhalation exposures.
Aspergillus fumigatus conidia exposures
Mice were acclimated to the nose-only exposure chamber for 1 week prior to exposures. Eighteen mice (3/exposure group) were separated into 3 exposure groups: 1) 1 × 105 A. fumigatus viable conidia; 2) 1 × 105 heat inactivated A. fumigatus conidia (HIC); or 3) HEPA-filtered air only control. The conidia numbers represented the estimated conidia deposited in the lungs of animals for each exposure as described previously [8]. Mice were exposed in the nose-only chamber of the acoustical generator system (AGS) twice a week for a total of 13 weeks (26 exposures) in a manner similar to Buskirk et. al. [8]. The AGS automatically shut off conidia aerosolization once the desired estimated lung deposition had been reached for each exposure. At 24 and 48 hours post-final exposure, mice were sacrificed via intraperitoneal injection of 200 mg/kg sodium pentobarbital solution (Fatal-Plus, Vortech Pharmaceuticals, LTD., Dearborn, MI) and once determined to be unconscious and unresponsive, the mice were exsanguinated via cardiac puncture.
Harvesting of lung tissue
The lungs were harvested from mice at 24 and 48 hours post final exposure. Total RNA was isolated from whole lung homogenate using Exiqon’s miRCURY RNA Isolation Kit for tissue following the manufacturer’s protocol (Exiqon, MA) or by using Qiagen’s RNeasy Plus Universal RNA Isolation Kit following the manufacturer’s protocol (Qiagen, CA).
MicroRNA Array Profiling
All miRNA experiments were conducted at Exiqon Services, Denmark. Total RNA (500 ng) from both sample and reference was labeled using the miRCURY LNA™ microRNA Hi-Power Labeling Kit, Hy3™/Hy5™ (Exiqon Inc., Denmark) following the manufacturer’s instructions. The Hy3™-labeled samples and a Hy5™-labeled reference RNA sample were mixed pair-wise and hybridized to the miRCURY LNA™ microRNA Array 7th Gen (Exiqon, Denmark) according to the miRCURY LNA™ microRNA Array instruction manual, using a Tecan HS4800™ hybridization station (Tecan, Austria). The miRCURY LNA™ microRNA array slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., USA), and the image analysis was completed using the ImaGeneR 9 (miRCURY LNA™ microRNA Array Analysis Software, Exiqon Inc., Denmark). The quantified signals were then corrected for background noise and normalized using the global Lowess regression algorithm. The array analysis examined all miRNAs for human, mouse or rat registered in miRBASE 18.0.
Gene Expression Arrays
Total RNA (5 μg) was converted to cDNA by the RT2 First Strand Kit (Qiagen, CA). Equal amounts of cDNA were then subjected to real-time PCR according to manufacturer’s instructions (Qiagen, CA). The genes examined were grouped into Th1–Th2 response and Th17 response pathways. The primers were included in prefabricated PCR plates (RT2 Profiler™ PCR Array Mouse Th1 & Th2 Responses and RT2 Profiler™ PCR Array Mouse Th17 Response). RT2 SYBR Green ROX qPCR Mastermix was used for quantification of cDNA (Qiagen, CA). Data were normalized by gene expression relative to the levels of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) for the Th1–Th2 response pathway and beta-glucuronidase (Gusb) for the Th17 response pathway. If specific genes included in the response pathways resulted in undetermined expression levels, the genes were removed from the heat maps (generated using R software) that are reported in Figures 1A and 1B.
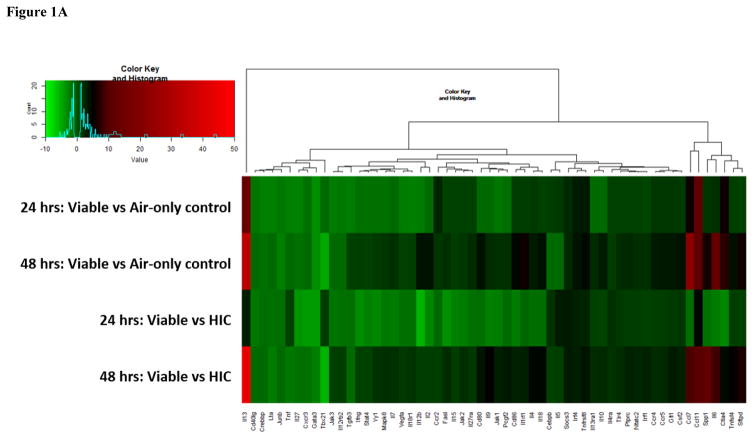
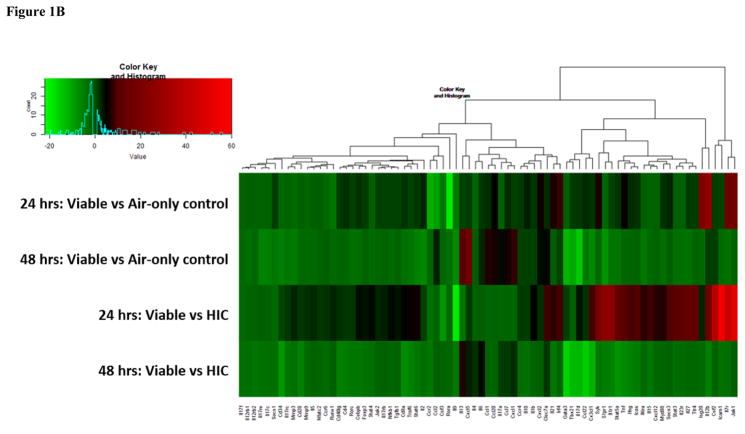
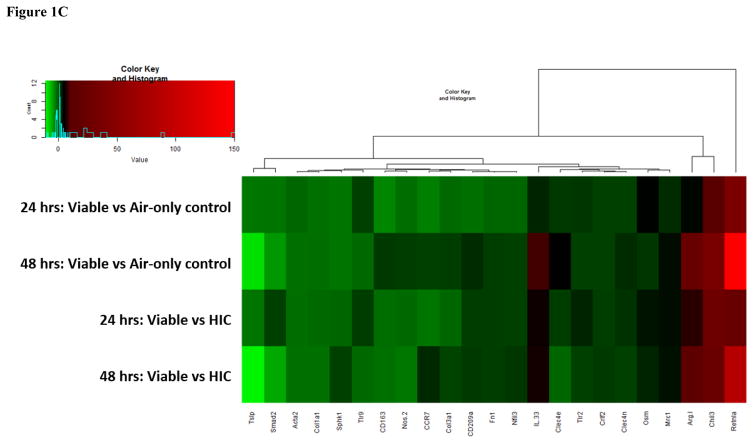
Figure 1.
Figure 1A: Heat map of genes involved in the Th1 and Th2 immune response pathways. Genes are color-coded (red or green for up- and downregulation, respectively).
Figure 1B: Heat map of genes involved in the Th17 immune response pathways. Genes are color-coded (red or green for up- and downregulation, respectively).
Figure 1C: Heat map of additional genes involved in the immune response. Genes are color-coded (red or green for up- and downregulation, respectively).
Real Time Polymerase Chain Reaction: Validation and Additional Genes
Total RNA (1 μg) was isolated and converted to cDNA by the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Life Technologies, MA). Equal amounts of cDNA were subjected to real-time PCR. Murine genes and associated assay identifiers (Life Technologies, MA) that were validated are listed in Supplemental Table 3, while the additional genes and associated assay identifiers (Life Technologies, MA) are reported in Supplemental Table 4. TaqMan® Fast Universal PCR Master Mix (2x), no AmpErase® UNG (Life Technologies, MA) was used for quantification of cDNA. Data were normalized by gene expression relative to the levels of Gapdh. Heat maps were generated using R software [44] and Bimax Biclustering (‘BCBimax’ clustering algorithm from Package ‘biclust’ version 1.0.2) as the clustering algorithm.
Ingenuity Pathway Analysis
After miRNA identification and mRNA and miRNA quantification was assessed, the fold changes and false discovery rates of the differentially expressed genes were imported into Ingenuity Pathway Analysis (IPA) software (IPA, Qiagen Redwood City, CA) for core, comparative and miRNA target filter analysis. The software was utilized for the construction of interacting mRNA and miRNA networks identified within the viable exposure group and compared to HIC and HEPA-filtered air control groups.
Statistics
All miRNA analytical calculations were performed by Exiqon Inc. using R/Bioconductor software, which included moderated t-tests, accounting for the variance of the entire dataset. MiRNAs with fold changes ≤ −2 (downregulated) or ≥ 2 (upregulated) were considered altered. The adjusted p-values ≤ 0.05 were considered significant.
For the RT2 Profiler PCR array results, which included genes grouped into Th1, Th2, and Th17 response pathways, the Ct values and subsequent statistical analysis was processed using Qiagen’s GeneGlobe Data Analysis Center. For both the RT2 Profiler PCR arrays, the additional genes measured and the validation of genes measured by qRT-PCR, statistics were completed using the same methods described above. Briefly, the fold change was the normalized gene expression in the test sample divided by the normalized gene expression in the control sample. Genes with fold changes ≤ −2 (downregulated) or ≥ 2 (upregulated) were considered altered. The p-values were calculated based on a Student’s t-test of the replicate values for each gene in the control group and treatment groups, and p-values ≤ 0.05 were considered significant.
RESULTS
MicroRNA Analysis
After threshold filtering, 415 murine miRNAs were detected in the Exiqon analysis (Supplemental Table 1). Comparing the viable exposure group to HIC (viable versus HIC) and air-only control (viable versus air-only control) groups 24 hours post exposure, 117 and 115 miRNAs, respectively, were shown to be differentially expressed (Supplemental Table 2). At 48 hours post exposure, 217 and 210 miRNAs were altered, respectively (Supplemental Table 2). Interestingly, all but 4 of the miRNAs that were differentially expressed at 24 hours, were further dysregulated at 48 hours. There were no significantly dysregulated miRNAs when comparing HIC to air-only control for either time point.
The miRNAs with the largest fold change at 24 and 48 hours following exposure were the same miRNAs when comparing viable versus air-only control and viable versus HIC (Table 1). These miRNAs included miR-23b-3p, miR-29a-3p, and miR-30c-5p, as well as miR-2137, miR-677-3p, and miR-1947-3p. Although the miRNAs were not significantly altered at 24 hours post-exposure, the fold changes of these miRNAs reached statistical significance at the 48 hour time point. IPA identified that these dysregulated miRNAs targeted specific genes that participate in the immune response and were associated with different diseases and functions. IPA revealed the involvement of miR-2137 and miR-1947-3p in chemokine- and T-cell receptor signaling, respectively, while miR-23b-3p and miR-29a-3p regulated different genes involved in the inflammatory response.
Table 1. Top 5 largest dysregulated miRNAs.
| 24 hours | 24 hours | ||||
|---|---|---|---|---|---|
| Viable/Air-only control | Viable/HIC | ||||
| Fold change | FDR | Fold change | FDR | ||
| mmu-miR-23b-3p | −16.47 | 0.11 | mmu-miR-23b-3p | −12.08 | 0.16 |
| mmu-miR-30c-5p | −10.58 | 0.11 | mmu-miR-24-3p | −9.51 | 0.16 |
| mmu-miR-24-3p | −10.44 | 0.12 | mmu-miR-30c-5p | −9.06 | 0.16 |
| mmu-miR-30b-5p | −7.65 | 0.11 | mmu-miR-30b-5p | −6.83 | 0.16 |
| mmu-miR-23a-3p | −6.95 | 0.11 | mmu-miR-29a-3p | −6.60 | 0.21 |
| mmu-miR-2137 | 10.23 | 0.11 | mmu-miR-2137 | 7.11 | 0.16 |
| mmu-miR-592-3p | 9.11 | 0.02* | mmu-miR-677-3p | 6.43 | 0.16 |
| mmu-miR-677-3p | 8.84 | 0.11 | mmu-miR-1843b-3p | 6.23 | 0.16 |
| mmu-miR-1947-3p | 6.44 | 0.11 | mmu-miR-1947-3p | 5.91 | 0.16 |
| mmu-miR-1843b-3p | 6.40 | 0.11 | mmu-miR-592-3p | 5.67 | 0.12 |
| 48 hours | 48 hours | ||||
|---|---|---|---|---|---|
| Viable/Air-only control | Viable/HIC | ||||
| Fold change | FDR | Fold change | FDR | ||
| mmu-miR-23b-3p | −62.23 | 0.00* | mmu-miR-23b-3p | −52.84 | 0.00* |
| mmu-miR-24-3p | −34.34 | 0.00* | mmu-miR-24-3p | −32.66 | 0.00* |
| mmu-miR-23a-3p | −25.30 | 0.01* | mmu-miR-29a-3p | −22.03 | 0.01* |
| mmu-miR-30c-5p | −24.90 | 0.00* | mmu-miR-23a-3p | −21.05 | 0.01* |
| mmu-miR-29a-3p | −20.97 | 0.01* | mmu-miR-30c-5p | −21.05 | 0.00* |
| mmu-miR-677-3p | 15.98 | 0.00* | mmu-miR-2137 | 13.14 | 0.00* |
| mmu-miR-2137 | 15.35 | 0.00* | mmu-miR-677-3p | 12.42 | 0.00* |
| mmu-miR-1947-3p | 13.52 | 0.01* | mmu-miR-1947-3p | 11.39 | 0.01* |
| mmu-miR-1843b-3p | 13.30 | 0.00* | mmu-miR-1843b-3p | 10.64 | 0.00* |
| mmu-miR-3103-3p | 12.69 | 0.00* | mmu-miR-3103-3p | 10.22 | 0.00* |
p-value ≤ 0.05 is considered significant.
FDR = False discovery rate.
mRNA Array Analysis
mRNA array analysis demonstrated the expression of 96 genes associated with either the Th1 and Th2 or Th17 response pathways. Figure 1A depicts the heat map that shows the fold changes of genes involved in the Th1 and Th2 response pathways. A greater number of genes were downregulated (indicated by green bars in Figure 1A) in both exposure groups at 24 hours, compared to 48 hour time points. After 48 hours, only 3 genes were downregulated, whereas the majority of genes were upregulated in both the viable versus air-only control, and in the viable versus HIC group (indicated by black and red bars in Figure 1A). While as many as 46 genes involved in the Th17 response pathway were upregulated in both groups 24 hours post-exposure, no more than 12 were upregulated after 48 hours following exposure, whereas the majority of genes were downregulated (Figure 1B). Analysis of the genes involved in the Th1 and Th2 response pathway revealed no significantly dysregulated genes at either time point. In contrast, of the 96 measured genes involved in the Th17 response pathway, 10 genes were dysregulated in the HIC compared to air-only control at 24 hours post-final exposure, whereas only 1 gene was dysregulated at 48 hours following the final exposure.
In both response pathways, GATA binding factor 3 (Gata3) and T-box transcription factor T-Bet (Tbx21) were the two genes with the greatest downregulation in both the viable versus HIC group and the viable versus air-only control at 48 hours. The decrease in Gata3 expression was uncharacteristic of an allergy model; therefore, it was verified by a separate qRT-PCR analysis using TaqMan® and Gata3 primers from a different vendor (data not shown). In contrast, interleukin 13 (Il13) had the greatest degree of upregulation in both groups at 48 hours following fungal exposure in both response pathways.
Previous analyses reported from our laboratory indicated the importance of fungal conidia germination on the resulting pulmonary immune response following subchronic A. fumigatus exposure [8]. C-type lectin domain family 7 member A (Clec7a), also known as Dectin-1, is a surface receptor that recognizes β-glucan on the cell wall of germinating conidia and fungal hyphae. At the 24 hour time point, the expression of Clec7a, measured by both the Th17 response pathway and through validation, showed an 8.9 fold increase in the viable versus HIC group (Figure 1B). Although Clec7a did not show increased expression in the viable versus HIC group in the Th17 response pathway 48 hours post-final exposure, it was significantly increased by 4.5 fold when validated (P ≤ 0.05; data not shown). This difference could be explained by the different technologies used to obtain the measurement of Clec7a expression. Overall, the increase in Clec7a further demonstrates the importance of A. fumigatus conidia germination on ensuing pulmonary immunological responses.
Molecular Network Analysis
Utilizing the microRNA target filter in Qiagen’s IPA software, the interactions between the miRNA data and the mRNA data were further examined. One of the greatest downregulated miRNAs at both time points and in both exposure groups was miR-23b-3p. IPA identified miR-23b-3p as a regulator of genes involved in IL-13 and interleukin 33 (IL-33) responses, such as an upregulation of arginase 1 (Arg1) and mannose receptor c, type 1 (Mrc1). Along with validating several genes in the Th1 and Th2, and Th17, response pathways, a third smaller set of genes involved in a combination of Th1, Th2 and Th17 immune responses were also measured (Figure 1C) and included the expression of Il33, which was also increased at both time points in the viable compared to HIC group.
MiR-29a-3p was also decreased in the viable versus HIC group at 24 hours and was significantly decreased (P ≤ 0.05) at 48 hours. In IPA, the miRNA target filter identified an interaction between Clec7a and miR-29a-3p. As previously mentioned, Clec7a expression was increased in the viable versus HIC group at both time points and was associated with in vivo A. fumigatus conidia germination. IPA also identified miR-29a-3p to regulate transforming growth factor beta 3 (TGF-β3). TGF-β3 was decreased in the viable versus air-only control at 24 hours post-exposure (Figure 1A), while TGF-β1 was increased in the viable versus HIC group (Figure 1B), but neither were altered at 48 hours.
The TGF-β signaling pathway is mediated by SMAD proteins that transduce the signal produced by TGF-β to the nucleus for downstream gene transcription regulation. As one of the greatest downregulated miRNAs, miR-30c-5p was identified as a predicted regulator of SMAD2. To date, SMAD signaling has not been identified as having a mechanistic role in the immune response to subchronic fungal exposures. Generated in IPA, comparative analyses between the Th1 and Th2 pathway mRNA dataset with miRNA dataset at 48 hours in the viable versus HIC group, and filtering for inflammatory disease and either cell-mediated or humoral immune response, not only identified SMAD2/3, but also illustrated that most of the miRNAs regulated by SMAD2/3 further regulate a number of different genes that are involved in the immune response (Figures 2A and 2B). This comparison also highlighted the uncharacterized involvement of interleukin 1 receptor-like 1 protein (IL1RL1) on the immune response elicited by A. fumigatus exposure (Figure 2A). Comparative analyses between the Th17 pathway mRNA dataset with miRNA dataset at 48 hours in the viable versus HIC group, and filtering for inflammatory disease, illustrated associations between Clec7a and genes involved in the inflammatory response, such as IL-13 (Figure 3). Overall, IPA highlighted the importance of viability as a mechanism underlying the elicited immune response and revealed novel associations between genes and miRNAs involved, following subchronic viable A. fumigatus fungal exposure.
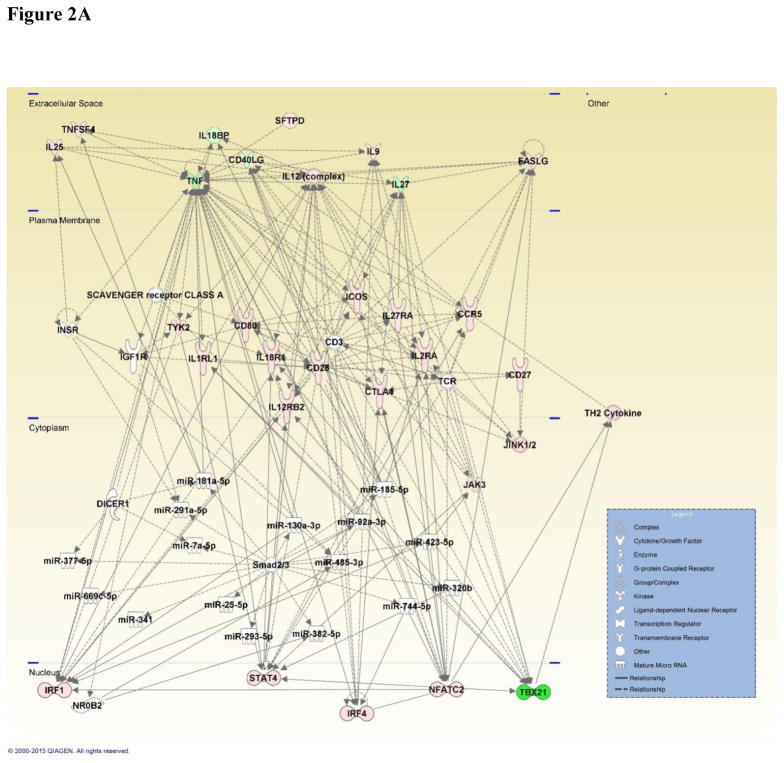
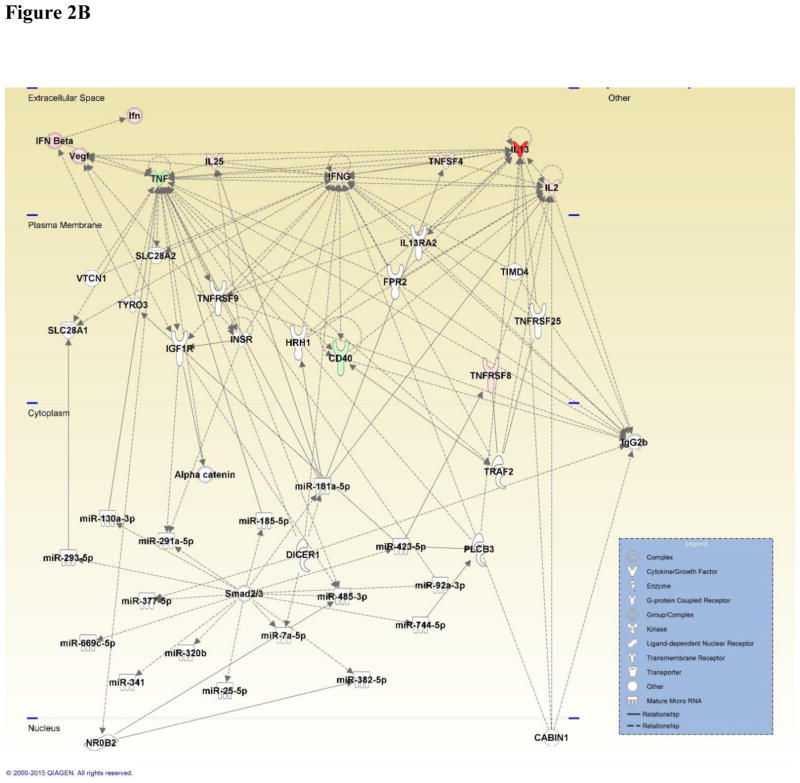
Figure 2.
Figure 2A: Network map generated by IPA depicting genes involved in the Th1 and Th2 immune response and associated miRNA involved in inflammatory and cell-mediated immune response, shaped by gene function and positioned by subcellular location. Genes are color-coded (red or green for up- and downregulation, respectively) for the expression of Th1 and Th2 genes in the viable versus HIC group 48 hours post-exposure.
Figure 2B: Network map generated by IPA depicting genes involved in the Th1 and Th2 immune response and associated miRNA involved in inflammatory and humoral immune response, shaped by gene function and positioned by subcellular location. Genes are color-coded (red or green for up- and downregulation, respectively) for the expression of Th1 and Th2 genes in the viable versus HIC group 48 hours post-exposure.
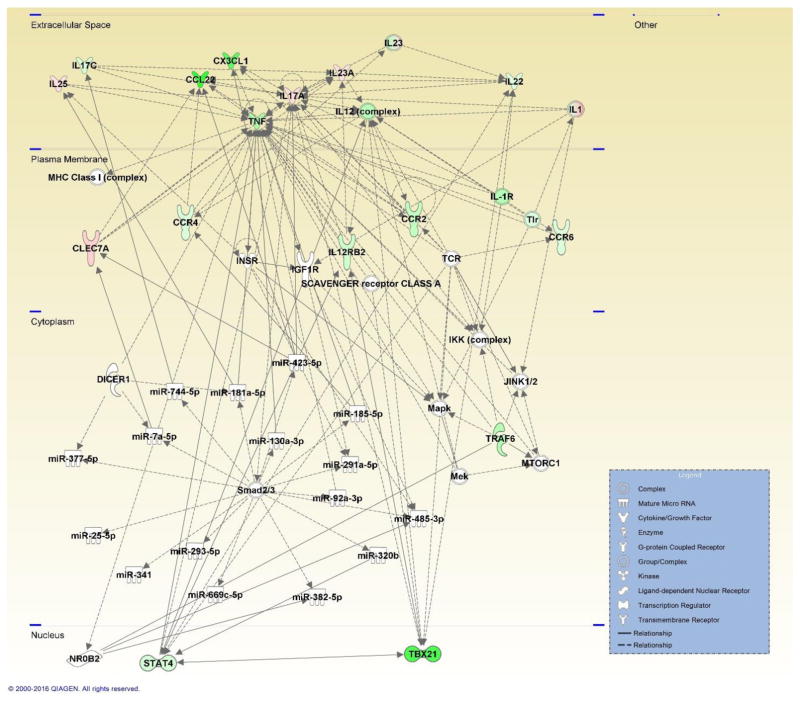
Figure 3. Network map generated by IPA depicting genes involved in the Th17 immune response and associated miRNA involved in the inflammatory response, shaped by gene function and positioned by subcellular location.
Genes are color-coded (red or green for up- and downregulation, respectively) for the expression of Th17 genes in the viable versus HIC group 48 hours post-exposure.
DISCUSSION
The goal of this study was to evaluate the role of fungal viability on the murine pulmonary miRNA and mRNA profiles that regulate immune responses following a subchronic A. fumigatus exposure. Using a previously described AGS, mice were exposed via inhalation to either viable or HIC A. fumigatus conidia, which allowed for the influence of viability to be assessed. Utilizing genomic microarray and RT-PCR technologies, the effects of fungal exposure on the expression of specific miRNAs and genes involved in the Th1, Th2, and Th17 response pathways were evaluated. Qiagen’s IPA software was then used to identify associations between these miRNAs and targeted genes that play a role in the immune response following subchronic fungal exposure.
After examining miRNAs that had demonstrated the most significant changes following subchronic A. fumigatus exposure, IPA identified miR-29a-3p which targets two critical genes in the inflammatory response, Clec7a and TGF-β. Clec7A is important relative to conidia germination and align with previous studies that have reported A. fumigatus germination to elicit an inflammatory response due to the recognition of β-glucan on the hyphal cell wall [45–50]. Previous studies from our laboratory showed germination of A. fumigatus conidia in the lungs of mice exposed to viable A. fumigatus conidia [8, 12]. These results are further supported by increased Clec7A expression in the viable versus HIC group at both time points. The decrease in miR-29a-3p is one potential mechanism that could contribute to the increase in Clec7A.
TGF-β plays a critical role in immunity by maintaining tolerance to self- or non-infectious antigens and by exerting both stimulatory and inhibitory effects during an immune response [51]. TGF-β1 and TGF-β3 are two isoforms of TGF-β that had opposing altered expression levels at 24 hours following A. fumigatus exposure. This result could be explained by the differing functions between the two isoforms, as well as the environment of cytokines present at each time point. One cytokine that counters the activity of TGF-β is IFNγ, which was increased in both groups at 24 hours, then decreased at 48 hours. Identified by the miRNA target filter in IPA, IFNγ is also regulated by miR-29a-3p. Similar to the reported increased IFNγ expression following subchronic fungal exposure in our studies, Ma et. al. demonstrated that decreased miR-29 participated in Th1 responses to intracellular pathogens by upregulating IFNγ expression [52]. Specifically, T cells from Mycobacterium bovis Bacillus Calmette-Guérin-infected or Listeria monocytogenes-infected mice presented lower amounts of miR-29 that in turn, assisted in the production of IFNγ. In the current study, a possible mechanism underlying the lack of change in TGF-β1 and TGF-β3 expression 48 hours post fungal exposure could be the increase of IFNγ at 24 hours.
Downstream of the TGF-β signaling pathway are transcription factors known as SMADs. Analysis of the 48 hour miRNA dataset revealed a network that contained a large number of the miRNAs, along with other prominent genes associated with the inflammatory disease and response pathway. This specific network reported a predicted association with SMAD2/3 and to our knowledge, this is the first time SMADs have been implicated in a fungal exposure-elicited immune response. There are several isoforms of SMAD and the two identified included SMAD2 and SMAD3. Phosphorylation of these receptor-activated SMADs result in a complex formation that consists of both SMAD2 and SMAD3, as well as SMAD4. Together, these act to transduce the signal from TGF-β in the cytoplasm into the nucleus for further gene transcription regulation. The expression levels of SMAD2 revealed a decrease 48 hours post-exposure in the viable versus HIC group and the viable versus air-only control. Upon further examination, the miRNA target filter identified several miRNAs that targeted SMAD2. Although miR-30c-5p was one of the greatest downregulated miRNAs and was predicted by IPA to interact with SMAD2, the expression of both the miRNA and the gene were both decreased. MiR-302a-3p was the only miRNA increased at 48 hours in the viable versus HIC group and viable versus air-only control and was also predicted to interact with SMAD2. This suggests that miR-302a-3p may serve as the more likely regulator of SMAD2 during fungal exposures. Taken together, this study demonstrates the potential for a functional role of SMAD2/3 in the inflammatory response and miR-302a-3p as a potential therapeutic target to reduce inflammation elicited from subchronic A. fumigatus exposure.
IL-13 is a hallmark cytokine of allergic response. Mice exposed to viable A. fumigatus conidia (1 × 106) intratracheally for 10 days showed that β–glucan recognition of Dectin-1 resulted in the production of IL-4, IL-13, IFNγ, and IL-33 [53]. In the present study, increased expression of Il13 and Il33 in the viable versus HIC group was observed at 48 hours. Increased Clec7a in the viable versus HIC group was also observed at both time points, as well as conidia germination in the lungs of mice exposed to viable A. fumigatus conidia[9]. These results further highlight the influence of fungal conidia germination on allergy-induced immune responses that have been previously shown using in vitro approaches [13].
An unexpected finding of this study was the decrease in Gata3 expression. Gata3 is a transcription factor expressed during the Th2 immune response to allergens [54]. This gene is also known to have a critical role in the expression of interleukin 4 (IL-4), interleukin 5 (IL-5) and IL-13; all cytokines that are characteristic of a Th2 response. Although Il5 was increased in the viable versus HIC at 24 hours and not 48 hours post exposure, both Il4 and Il13 were increased at 48 hours in viable versus HIC group. Gata3 expression has been shown to be regulated by Stat6, whose expression was significantly downregulated in the viable versus HIC group at 48 hours post exposure [55]. Also significantly downregulated in the viable versus HIC group at 48 hours post exposure was the expression of Stat3, which has been shown to regulate the binding of STAT6 with GATA3 [56]. Corn et al. showed that Bcl-3-deficient cells exhibited decreased Gata3, consistent with evidence that Bcl-3 can transactivate a Gata3 promoter [57]; however, Bcl3 expression was not measured in the present study. In vitro, TGF-β has been reported to inhibit Th2 mediated immune responses through the inhibition of Gata3 expression [58]. Although possible mechanisms exist that may negatively impact Gata3 expression, the increase in the pro-inflammatory cytokines regulated by Gata3, along with previously reported Gata3 induction during inflammation, warrants Gata3 expression to be further examined in future fungal exposure studies.
MiRNA profiles have also been examined in cell lines and activated cell populations following exposure to different pathogens. Along with targeting IFNγ and subsequently regulating the immune response following a fungal or bacterial exposure, miR-29 has also been reported to regulate the production and infectivity of human immunodeficiency virus type 1 [59]. MiR-29a is upregulated in the serum and sputum of Mycobacterium tuberculosis-infected humans [60], but in contrast, miR-29 is downregulated in murine T-cells infected with M. bovis [52], which is in agreement with the current study. Presently, miR-155-5p decreased 48 hours post fungal exposure; however, this miRNA was upregulated in different cell types following infections from bacterial pathogens, including Salmonella enterica, Helicobacter pylori, L. monocytogenes, M. avium and M. smegmatis [30, 61–69]. In contrast to the present study, miR-23b, miR-30b, miR-30c, miR-125b, miR-15b, miR-16, miR-27b, miR-24 and miR-21 were up-regulated in Cryptospordium parvum-infected human host biliary epithelial cells [31]. In that same study, both miR-98 and miR-214 were decreased, a finding that is in agreement with the currently reported miRNAs following A. fumigatus exposure. Another study reported decreased miR-16 and miR-451 in the plasma of human patients infected with Plasmodium vivax [70], which was also observed in the murine lung homogenates derived from the group of mice exposed to viable A. fumigatus conidia. Taken together, the results from the present study suggest that subchronic A. fumigatus exposures result in similar miRNA profiles that include the same highly dysregulated miRNAs that have also been observed in bacterial and protozoan pathogen models.
This study offers preliminary insight into the miRNA environment and identified miRNA-mRNA interactions following subchronic exposure to aerosolized A. fumigatus conidia; however, there are a number of limitations associated with this study that need to be taken into consideration. In this study, the pulmonary exposure consisted of 26 exposures over a 13 week interval and data was collected only at 24 and 48 hour time points following the final exposure. As a result, the miRNA environment was not evaluated at shorter time points, nor after time points beyond 48 hours, post final exposure. It is important to note that IPA predicts interactions between miRNAs and mRNA, even if the interactions have not been scientifically observed. In the current study, all of the interactions between the described miRNAs in the present study and SMAD2/3 were predicted based on interactions that have only been observed in human cell culture and not in a murine inhalation exposure study. Lastly, the genetic environment was assessed using whole lung homogenate and not a specific cell population or cell line; however, specific cell populations from the same study were examined by flow cytometry and reported in a separate manuscript [9]. The current manuscript is focused on the miRNA and mRNA environments following a subchronic fungal exposure, but further characterization of the immune response by flow cytometric analyses, gene and protein expression from the same study are reported separately [9]. Critical genes identified in the current manuscript to be involved in the immune responses were also confirmed through proteomic analyses reported in the separate manuscript [9]. Although not all fungal species with comparable sized conidia may behave in a similar manner [71], the findings in the current study provide unique insight into the molecular environment within the murine lung following a subchronic A. fumigatus exposure.
In conclusion, mice exposed to viable A. fumigatus conidia highlighted the influence of conidia germination on gene expression controlling the ensuing pulmonary immune responses. This was specifically supported by data that showed increased Clec7a expression and its association with IL-13 and IL-33. Downregulated miRNAs predicted to target Il13, Il33, and Clec7a (Figure 4) were identified, suggesting a possible mechanism that contributes in part to an immune response following subchronic exposures to A. fumigatus. Furthermore using IPA, novel interactions between SMAD2/3 and other genes and miRNAs involved in the inflammatory response were able to be resolved for the first time. These altered miRNAs may serve as potential biomarkers to evaluate fungal exposure.
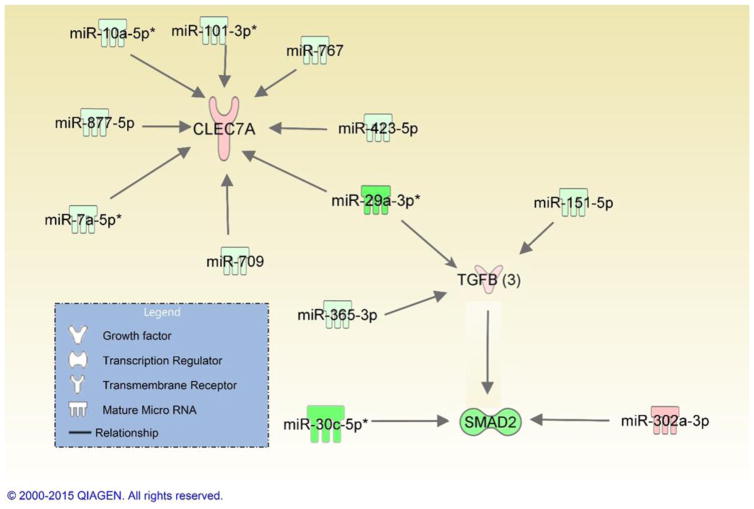
Figure 4. Summary of network map depicting the three differentially expressed genes of interest and their associated miRNAs involved in the inflammatory immune responses.
Genes and miRNAs are color-coded (red or green for up- and downregulation, respectively) for the expression of Clec7a, TGF-β3, SMAD2/3 and miRNAs in the viable versus HIC group 48 hours post-exposure.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Michael Kashon in the Biostatistics and Epidemiology Branch, National Institute for Occupational Safety and Health (NIOSH) for assistance with statistical analysis and representation, as well as Brandon F. Law in the Allergy and Clinical Immunology Branch, NIOSH for assistance in harvesting and processing samples. The authors would also thank Dr. Paivi Salo from the National Institute of Environmental Health Sciences (NIEHS) and Dr. John Noti, NIOSH for participating in the internal NIEHS and NIOSH review process. This study was supported in part by an interagency agreement between NIOSH and NIEHS (AES12007001-1-0-6) as a collaborative National Toxicology Program research activity. This study was also funded in part by Centers for Disease Control and Prevention-NIOSH intramural funds (927ZLCT). The findings and conclusions in this study are those of the authors and do not necessarily represent the views of National Institute for Occupational Safety and Health. The authors declare no conflict of interest.
References
- 1.Damp Indoor Spaces and Health. Washington (DC): National Academies Press (US); 2004. [PubMed] [Google Scholar]
- 2.WHO Guidelines for Indoor Air Quality: Dampness and Mould. Geneva: World Health Organization; 2009. [PubMed] [Google Scholar]
- 3.Benedict K, Park BJ. Invasive fungal infections after natural disasters. Emerging infectious diseases. 2014;20:349–55. doi: 10.3201/eid2003.131230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balloy V, Chignard M. The innate immune response to Aspergillus fumigatus. Microbes and infection/Institut Pasteur. 2009;11:919–27. doi: 10.1016/j.micinf.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Abad A, Fernandez-Molina JV, Bikandi J, Ramirez A, Margareto J, Sendino J, Hernando FL, Ponton J, Garaizar J, Rementeria A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Revista iberoamericana de micologia. 2010;27:155–82. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Nieminen SM, Kärki R, Auriola S, Toivola M, Laatsch H, Laatikainen R, Hyvärinen A, von Wright A. Isolation and Identification of Aspergillus fumigatus Mycotoxins on Growth Medium and Some Building Materials. Applied and Environmental Microbiology. 2002;68:4871–75. doi: 10.1128/AEM.68.10.4871-4875.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauni R, Uitti J, Jauhiainen M, Kreiss K, Sigsgaard T, Verbeek JH. Remediating buildings damaged by dampness and mould for preventing or reducing respiratory tract symptoms, infections and asthma (Review) Evidence-Based Child Health: A Cochrane Review Journal. 2013;8:944–1000. doi: 10.1002/ebch.1914. [DOI] [PubMed] [Google Scholar]
- 8.Buskirk AD, Green BJ, Lemons AR, Nayak AP, Goldsmith WT, Kashon ML, Anderson SE, Hettick JM, Templeton SP, Germolec DR, Beezhold DH. A murine inhalation model to characterize pulmonary exposure to dry Aspergillus fumigatus conidia. PloS one. 2014;9:e109855. doi: 10.1371/journal.pone.0109855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nayak AP, Croston TL, Lemons AR, Goldsmith WT, Marshall NB, Kashon ML, Germolec DR, Beezhold DH, Green BJ. Fungal spore viability drives allergic responses to inhaled conidia. 2016 doi: 10.1016/j.anai.2018.04.008. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Templeton SP, Buskirk AD, Law B, Green BJ, Beezhold DH. Role of germination in murine airway CD8+ T-cell responses to Aspergillus conidia. PloS one. 2011;6:e18777. doi: 10.1371/journal.pone.0018777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera A, Van Epps HL, Hohl TM, Rizzuto G, Pamer EG. Distinct CD4(+)-T-Cell Responses to Live and Heat-Inactivated Aspergillus fumigatus Conidia. Infection and immunity. 2005;73:7170–79. doi: 10.1128/IAI.73.11.7170-7179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nayak AP, Green BJ, Lemons AR, Marshall NB, Goldsmith WT, Kashon ML, Anderson SE, Germolec DR, Beezhold DH. Subchronic exposures to fungal bioaerosols promotes allergic pulmonary inflammation in naive mice. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2016 doi: 10.1111/cea.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green BJ, Mitakakis TZ, Tovey ER. Allergen detection from 11 fungal species before and after germination. The Journal of allergy and clinical immunology. 2003;111:285–9. doi: 10.1067/mai.2003.57. [DOI] [PubMed] [Google Scholar]
- 14.Samarasinghe AE, Hoselton SA, Schuh JM. A comparison between intratracheal and inhalation delivery of Aspergillus fumigatus conidia in the development of fungal allergic asthma in C57BL/6 mice. Fungal Biol. 2011;115:21–9. doi: 10.1016/j.funbio.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kousha M, Tadi R, Soubani AO. Pulmonary aspergillosis: a clinical review. Eur Respir Rev. 2011;20:156–74. doi: 10.1183/09059180.00001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss RB. Allergic bronchopulmonary aspergillosis. Clin Rev Allergy Immunol. 2002;23:87–104. doi: 10.1385/CRIAI:23:1:087. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 20.Li L-C, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proceedings of the National Academy of Sciences. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. The Journal of allergy and clinical immunology. 2013;132:3–13. doi: 10.1016/j.jaci.2013.04.039. quiz 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–76. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer research. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 24.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragine N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foa R, Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–51. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 26.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Stahle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PloS one. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology. 2008;382:257–66. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS pathogens. 2013;9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eulalio A, Schulte L, Vogel J. The mammalian microRNA response to bacterial infections. RNA biology. 2012;9:742–50. doi: 10.4161/rna.20018. [DOI] [PubMed] [Google Scholar]
- 31.Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS pathogens. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson SE, Beezhold K, Lukomska E, Richardson J, Long C, Anderson K, Franko J, Meade BJ, Beezhold DH. Expression kinetics of miRNA involved in dermal toluene 2,4-diisocyanate sensitization. Journal of immunotoxicology. 2014;11:250–9. doi: 10.3109/1547691X.2013.835891. [DOI] [PubMed] [Google Scholar]
- 33.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18704–9. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. The Journal of allergy and clinical immunology. 2011;128:160–67. e4. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:3243–50. doi: 10.1096/fj.09-135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izzotti A, Bagnasco M, Cartiglia C, Longobardi M, Balansky RM, Merello A, Lubet RA, De Flora S. Chemoprevention of genome, transcriptome, and proteome alterations induced by cigarette smoke in rat lung. European journal of cancer. 2005;41:1864–74. doi: 10.1016/j.ejca.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, Jiang Y, Zhang H, Greenlee AR, Han Z. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life sciences. 2010;86:192–8. doi: 10.1016/j.lfs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Bourdon JA, Saber AT, Halappanavar S, Jackson PA, Wu D, Hougaard KS, Jacobsen NR, Williams A, Vogel U, Wallin H, Yauk CL. Carbon black nanoparticle intratracheal installation results in large and sustained changes in the expression of miR-135b in mouse lung. Environmental and molecular mutagenesis. 2012;53:462–8. doi: 10.1002/em.21706. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Li C, Liu W, Jin Y. Modulation of microRNA expression by volatile organic compounds in mouse lung. Environmental toxicology. 2014;29:679–89. doi: 10.1002/tox.21795. [DOI] [PubMed] [Google Scholar]
- 41.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–9. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 42.Rao R, Nagarkatti P, Nagarkatti M. Role of miRNA in the regulation of inflammatory genes in staphylococcal enterotoxin B-induced acute inflammatory lung injury and mortality. Toxicological sciences : an official journal of the Society of Toxicology. 2015;144:284–97. doi: 10.1093/toxsci/kfu315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taki FA, Pan X, Zhang B. Chronic nicotine exposure systemically alters microRNA expression profiles during post-embryonic stages in Caenorhabditis elegans. Journal of cellular physiology. 2014;229:79–89. doi: 10.1002/jcp.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Team RC. R: A Language and Environment for Statisical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 45.Amarsaikhan N, Templeton SP. Co-recognition of beta-glucan and chitin and programming of adaptive immunity to Aspergillus fumigatus. Frontiers in microbiology. 2015;6:344. doi: 10.3389/fmicb.2015.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Dea EM, Amarsaikhan N, Li H, Downey J, Steele E, Van Dyken SJ, Locksley RM, Templeton SP. Eosinophils Are Recruited in Response to Chitin Exposure and Enhance Th2-Mediated Immune Pathology in Aspergillus fumigatus Infection. Infection and immunity. 2014;82:3199–205. doi: 10.1128/IAI.01990-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latge JP. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–21. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 48.Luther K, Torosantucci A, Brakhage AA, Heesemann J, Ebel F. Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the dectin-1 beta-glucan receptor and Toll-like receptor 2. Cellular microbiology. 2007;9:368–81. doi: 10.1111/j.1462-5822.2006.00796.x. [DOI] [PubMed] [Google Scholar]
- 49.Bozza S, Gaziano R, Spreca A, Bacci A, Montagnoli C, di Francesco P, Romani L. Dendritic cells transport conidia and hyphae of Aspergillus fumigatus from the airways to the draining lymph nodes and initiate disparate Th responses to the fungus. J Immunol. 2002;168:1362–71. doi: 10.4049/jimmunol.168.3.1362. [DOI] [PubMed] [Google Scholar]
- 50.Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, Pamer EG. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS pathogens. 2005;1:e30. doi: 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annual review of immunology. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 52.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nature immunology. 2011;12:861–9. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 53.Lilly LM, Gessner MA, Dunaway CW, Metz AE, Schwiebert L, Weaver CT, Brown GD, Steele C. The beta-glucan receptor dectin-1 promotes lung immunopathology during fungal allergy via IL-22. J Immunol. 2012;189:3653–60. doi: 10.4049/jimmunol.1201797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barnes PJ. Role of GATA-3 in allergic diseases. Current molecular medicine. 2008;8:330–4. doi: 10.2174/156652408785160952. [DOI] [PubMed] [Google Scholar]
- 55.Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunologic research. 2011;50:87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175:2102–10. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- 58.Gorelik L, Fields PE, Flavell RA. Cutting edge: TGF-beta inhibits Th type 2 development through inhibition of GATA-3 expression. J Immunol. 2000;165:4773–7. doi: 10.4049/jimmunol.165.9.4773. [DOI] [PubMed] [Google Scholar]
- 59.Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Molecular cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. Journal of clinical microbiology. 2011;49:4246–51. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. The Journal of infectious diseases. 2009;200:916–25. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 62.Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, Kalali B, Mollenkopf HJ, Beigier-Bompadre M, Karlas A, Schneider T, Churin Y, Gerhard M, Meyer TF. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PloS one. 2010;5:e9500. doi: 10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Muller A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic Gastritis and Colitis. J Immunol. 2011;187:3578–86. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- 64.Matsushima K, Isomoto H, Inoue N, Nakayama T, Hayashi T, Nakayama M, Nakao K, Hirayama T, Kohno S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. International journal of cancer Journal international du cancer. 2011;128:361–70. doi: 10.1002/ijc.25348. [DOI] [PubMed] [Google Scholar]
- 65.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. The EMBO journal. 2011;30:1977–89. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnitger AK, Machova A, Mueller RU, Androulidaki A, Schermer B, Pasparakis M, Kronke M. Papadopoulou N, Listeria monocytogenes infection in macrophages induces vacuolar-dependent host miRNA response. PloS one. 2011;6:e27435. doi: 10.1371/journal.pone.0027435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Izar B, Mannala GK, Mraheil MA, Chakraborty T, Hain T. microRNA response to Listeria monocytogenes infection in epithelial cells. International journal of molecular sciences. 2012;13:1173–85. doi: 10.3390/ijms13011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PloS one. 2011;6:e20258. doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajaram MV, Ni B, Morris JD, Brooks MN, Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB, Schlesinger LS. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17408–13. doi: 10.1073/pnas.1112660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chamnanchanunt S, Kuroki C, Desakorn V, Enomoto M, Thanachartwet V, Sahassananda D, Sattabongkot J, Jenwithisuk R, Fucharoen S, Svasti S, Umemura T. Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Experimental Parasitology. 2015;155:19–25. doi: 10.1016/j.exppara.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 71.Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus--what makes the species a ubiquitous human fungal pathogen? PLoS pathogens. 2013;9:e1003743. doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.