Abstract
Sustained expression of the oestrogen receptor alpha (ESR1) drives two-thirds of breast cancer and defines the ESR1-positive subtype. ESR1 engages enhancers upon oestrogen stimulation to establish an oncogenic expression program1. Somatic copy number alterations involving the ESR1 gene occur in approximately 1% of ESR1-positive breast cancers2–5, implying that other mechanisms underlie the persistent expression of ESR1. We report the significant enrichment of somatic mutations within the set of regulatory elements (SRE) regulating ESR1 in 7% of ESR1-positive breast cancers. These mutations regulate ESR1 expression by modulating transcription factor binding to the DNA. The SRE includes a recurrently mutated enhancer whose activity is also affected by a functional inherited single nucleotide variant (SNV) rs9383590 that accounts for several breast cancer risk-loci. Our work highlights the importance of considering the combinatorial activity of regulatory elements as a single unit to delineate the impact of noncoding genetic alterations on single genes in cancer.
Noncoding regulatory elements are the primary target of inherited risk variants6–8 and their functional relevance to cancer is supported by the mutational constraint observed within these elements across tumours9,10. Functional noncoding SNVs can underlie “single gene” diseases11 confirming their ability to exert large phenotypic effects commonly associated with coding variants. This is highlighted in sporadic and familial melanoma, where somatic and germline genetic alterations in the promoter of the telomerase (TERT) gene favour oncogenesis through an increase in TERT expression12,13.
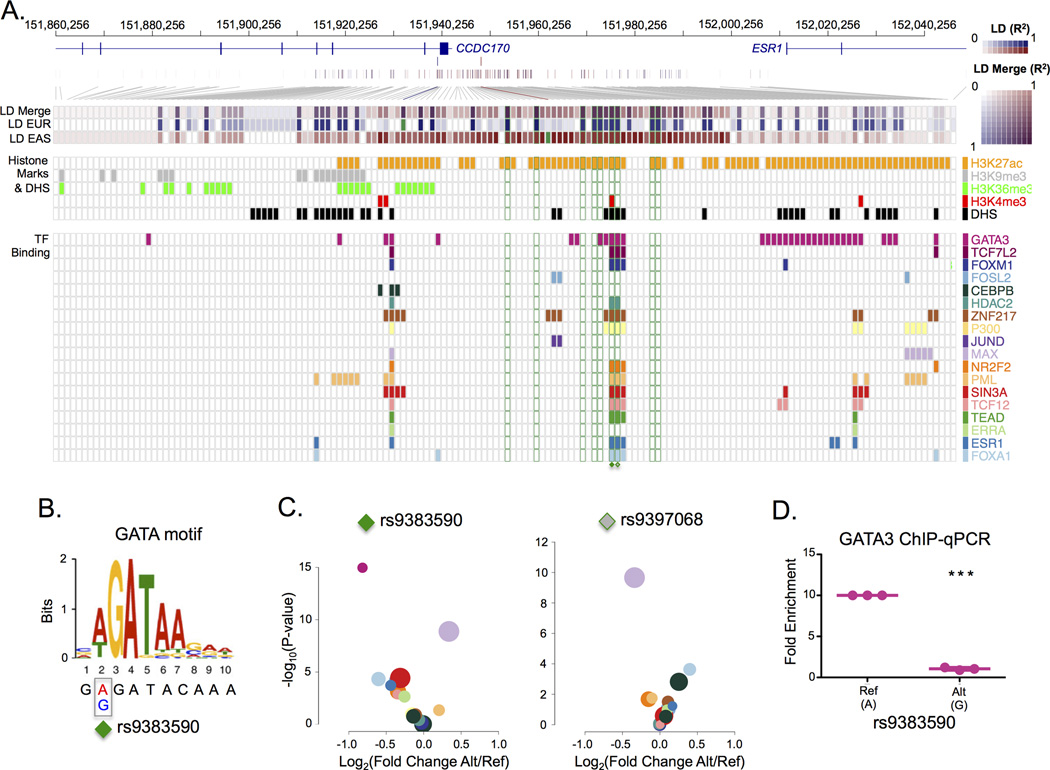
GWAS studies identified several breast cancer risk-associated SNVs at the ESR1 locus among individuals of European and East Asian ancestry14–18. The population-specific patterns of linkage disequilibrium (LD) between the different lead SNVs are consistent with a single underlying causal SNV. GWAS risk-loci are enriched in regulatory elements and function by altering gene expression6–8. To identify the functional SNV(s), we first intersected all SNVs within a five megabase window of the original ESR1 locus lead SNVs with functional annotations generated by the Encyclopaedia of DNA Elements (ENCODE) project19 in MCF-7 and T-47D ESR1-positive breast cancer cells. We then calculated the population-specific LD between the European and the East Asian lead SNVs (rs3734805 and rs2046210, respectively) and the neighbouring SNVs using the genotype data from the 1,000 genomes project20. We identified nine SNVs common to both Europeans and East Asians that share LD with the original population-specific lead SNVs (r2 ≥ 0.8 in both populations). Two SNVs, rs9383590 and rs9397068, in perfect LD with one another and located 95 base pairs (bp) apart within the same DNaseI hypersensitivity site (DHS) coincided with multiple functional genomic annotations generated by the ENCODE project19 (Figure 1A, Supplementary Figure 1 & 2). These SNVs are also in strong LD (r2=0.81) with another European breast cancer rs9383938 lead SNV17. The rs9383590 SNV maps to the second position of a GATA DNA recognition motif (Figure 1B). The intra-genomic replicates (IGR) tool6 (Online Methods) predicts a decrease in the chromatin binding intensity of GATA3 for the variant allele (Figure 1C) confirmed by allele-specific ChIP-qPCR in the heterozygous HCC1419 breast cancer cells (Figure 1D).
Figure 1. Identification of a functional risk-associated SNV shared between Europeans and East Asians.
A) The shared linkage disequilibrium (LD) between the European and East Asian lead SNVs. The composite strength of the LD (LD Merge) for the European and East Asian lead SNVs is shown (purple). The European LD (LD EUR) pattern for the rs3734805 SNV (blue) and the East Asian LD (LD EAS) pattern for rs2046210 SNV (red) are shown. The squares corresponding to the population-specific lead SNVs (rs3734805 and rs2046210) are filled in green. The 9 LD SNVs with an r2 ≥ 0.8 with both the European lead SNV and the East Asian lead SNV are outlined in green boxes. The overlapping functional annotations (DHS, histones modifications and transcription factor binding) observed in breast cancer cells (MCF-7 or T-47D) profiled by the ENCODE project19 are represented as boxes coloured according to the legend (right). B) Location of the rs9383590 SNV within the GATA3 DNA recognition motif. C) A volcano plot of the IGR results for all transcription factors overlapping rs9383590 and rs9397068 in (A). Transcription factors are coloured according to the legend in (A). The area of the circle is proportional to the maximum average signal intensity of the two alleles. D) Allele-specific ChIP-qPCR for GATA3 produced by the rs9383590 SNV. Statistical significance was determined with a one-sample t-test. Reported p-values are two-sided. The mean and standard error of the mean are shown.
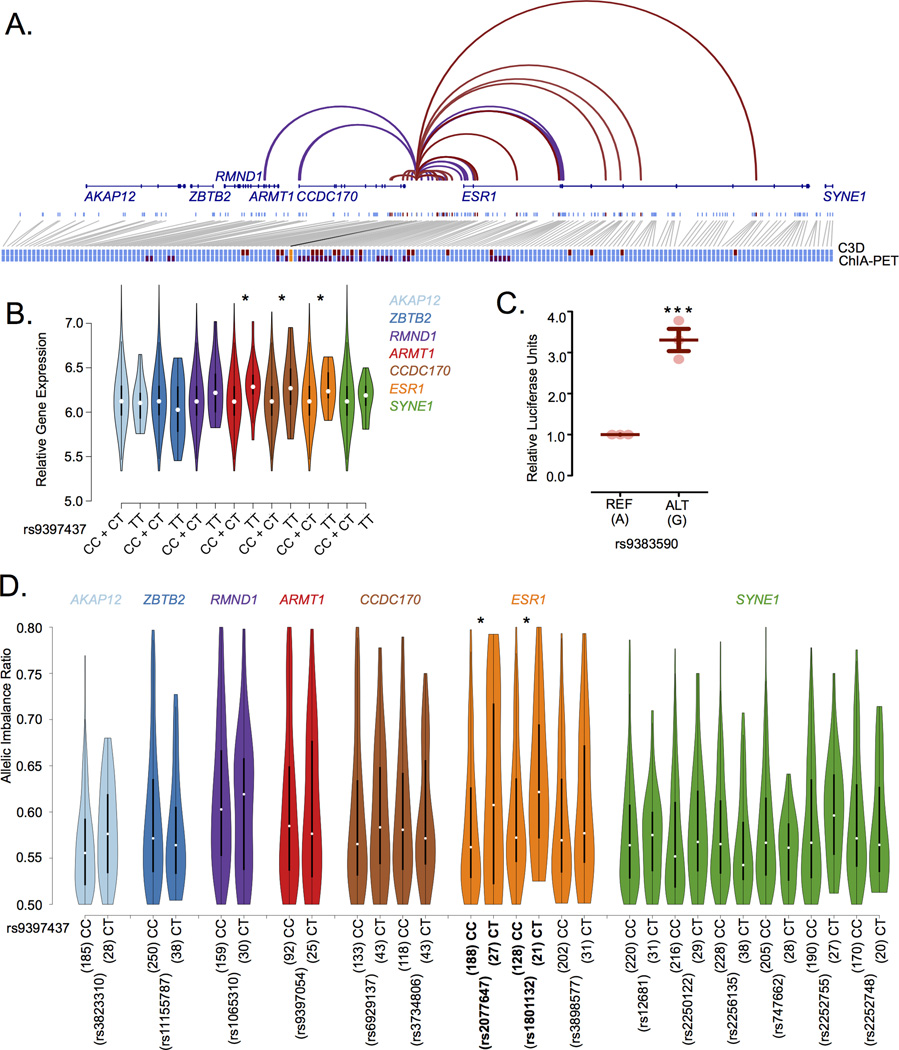
Enhancers regulate gene expression through physical interaction with their target gene(s) promoter. The Cross-Cell type Correlation in DNaseI hypersensitivity (C3D) method21,22 (r≥0.7)(Online Methods) identifies the enhancer harbouring the rs9383590 SNV as potentially interacting with the ESR1 promoter (Figure 2A). This interaction is confirmed in the RNA polymerase 2 (POL2) ChIA-PET dataset from MCF-7 cells produced by the ENCODE project19 (Figure 2A). To determine whether the rs9383590 SNV affects ESR1 gene expression we performed an expression quantitative trait locus (eQTL) analysis. We did not observe an additive association between the variant allele of the rs9383590 SNV and ESR1 expression in ESR1-positive breast tumours profiled by The Cancer Genome Atlas (TCGA) or the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC)23. However, a linked SNV, rs9397435 (r2=0.97 and r2=1 with rs9383590 in Europeans and East Asians, respectively), was previously reported as a recessive eQTL associated with ESR1 expression in breast tumour samples18. Consistently, we observe a weak recessive eQTL among the luminal breast tumours in the larger METABRIC sample, using the rs9397437 SNV as proxy for the rs9383590 SNV (r2=1 among Europeans and East Asians) (n=970, p=0.039)(Figure 2B)(Online Methods). This eQTL should be interpreted with caution, since it was not observed within the TCGA samples. However, Li, et al.24 observed a significant allelic imbalance among TCGA breast tumours heterozygous for the East Asian lead SNV, rs2046210. Using the rs9397437 SNV as a proxy, we observe a consistent allelic imbalance in ESR1 expression among heterozygous breast tumours measured with two independent coding marker SNVs (rs2077647 & rs1801132)(p≤0.05)(Figure 2D)(Online Methods) and within the heterozygous HCC1419 breast cancer cells (p=1.13×10−4)(Supplementary Figure 3)(Online Methods). A similar result for the rs9397437 SNV was reported by Dunning et al.25. In addition, a luciferase reporter assay reveals increased enhancer activity for the rs9383590 SNV variant allele (Figure 2C). The variant allele of the rs9397068 SNV also increased enhancer activity (Supplementary Figure 4). However, the effect of both SNVs does not appear to be additive (Supplementary Figure 4). Together with the reference allele-biased binding of GATA3 these results suggests that GATA3 acts as repressor, which has been previously reported by others26.
Figure 2. The rs9383590 SNV interacts with the ESR1 promoter altering gene expression.
A) Cross-Cell Type Correlation in DNaseI Hypersensitivity (C3D) predicted (red) and POL2 ChIA-PET determined (purple) chromatin interactions between the breast cancer risk-locus enhancer DHS and neighbouring DHS sites are shown. Single DHS resolution is presented for the C3D approach. All DHSs within a paired-end tag are considered as interacting in ChIA-PET data. DHS sites with no evidence of a chromatin interaction are also shown (light blue). The position of nearby genes (dark blue) is also shown. All DHSs interacting with the breast cancer risk-locus enhancer (orange) either predicted (red) or experimentally determine (purple) are enlarge to reveal the overlap at the bottom of the figure. B) Violin plots of the gene expression values for the genes at the ESR1 locus by rs9397437, a proxy of rs9383590, genotypes. Statistical significance was determined using linear regression under a recessive model C) Reporter assay results for the rs9383590 SNV. Statistical significance was determined with a one-sample t-test. D) Allelic imbalance of the genes at the ESR1 locus among TCGA breast tumours profiled by RNA-Seq. The allelic imbalance ratio represents the frequency of the most abundant allele within the RNA-Seq reads. Statistical significance was determined with an approximate Fisher-Pitman test using 10,000 permutations. All reported p-values are two-sided. Lines indicate the mean and the standard error of the mean. *,**,*** denotes the level of significance, a p-value less than 0.05, 0.01 and 0.005, respectively.
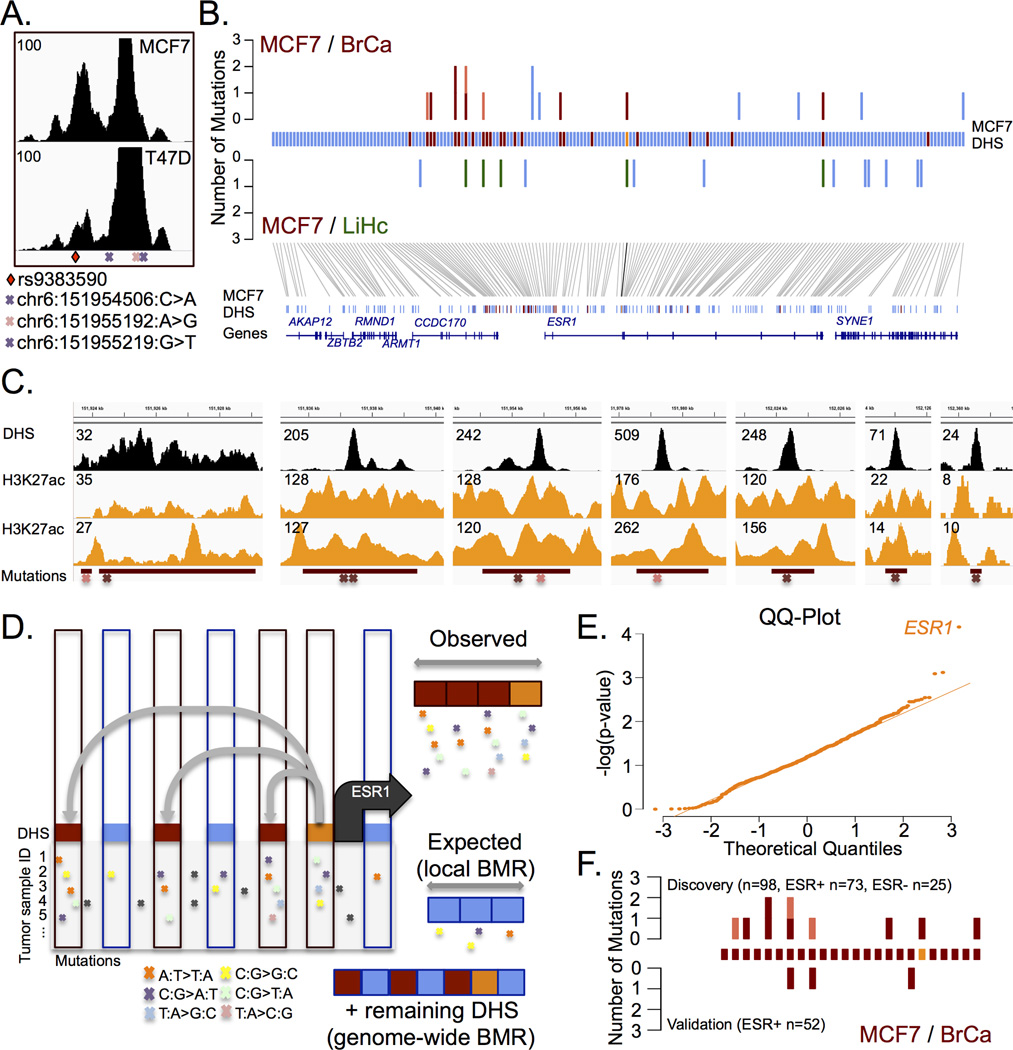
Convergence of inherited risk variants and acquired somatic mutations on regulatory elements occurs at the TERT promoter in melanoma12. Using a set of 98 breast cancer samples profiled by whole genome sequencing (WGS)27, we found two samples harbouring a somatic mutation in the enhancer modulated by the rs9383590 SNV (Figure 3A). Since the SRE of a gene tightly regulates its expression28, we hypothesized that mutations within the SRE of ESR1 could account for its persistent expression in breast cancer. We first delineated the SRE of ESR1 using the C3D method. This predicted the physical interaction of 24 regulatory elements with the ESR1 promoter within a 1 MB window of its transcription start site (r≥0.7)(Supplementary Table 1). 18 of these predicted interactions were validated by first or second order interactions identified in the POL2 ChIA-PET datasets19 (Supplementary Figure 5). We then identified mutations in the ESR1 SRE in approximately 10% of the 98 WGS breast cancer samples (10/98). Nine of these mutations are found in seven enhancers and one localizes in the ESR1 promoter (Figure 3B). We validated the interaction between all mutated enhancers and ESR1 promoter by Chromatin Conformation Capture-based assays in MCF-7 cells (Supplementary Figure 5). Of note, each mutated enhancer is flanked by nucleosomes containing histone H3 acetylated on lysine residue 27 (H3K27ac) in breast cancer cells, a feature of active enhancer elements29 (Figure 3C).
Figure 3. The set of regulatory elements (SRE) of ESR1 is targeted by acquired somatic mutations in breast cancer.
A) DNAse-seq signal across the enhancer harbouring the rs9383590 SNV and three somatic mutations. Two identified in the discovery set of breast tumours cohort and one in the validation set of breast tumours. B) The top panel reveals the enrichment of mutations with DHS sites that interact (red) or not (blue) with the ESR1 promoter (orange) in breast tumours (BrCa). The number of mutations identified in ESR1-positive (red) and ESR1-negative samples (pink) are shown. The lack of enrichment within the SRE for mutations from liver tumours (LiHc) is also shown (green). C) The DNAse-Seq and H3K27ac ChIP-Seq signal profiles (from ENCODE19 and Taberlay et al.36) for each of the regulatory elements harbouring a somatic mutation in breast tumours. D) Schematic representation of the mutational significance within ESR1’s SRE (MuSE) approach. C3D predicted (grey lines) DHS interacting (red rectangle) with the ESR1 gene promoter (orange rectangle) and non-interacting DHS (blue rectangle) are shown. The mutational load in the interacting versus non-interacting DHS define the observed versus expected mutational rate in ESR1’s SRE. E) A QQ-Plot of the observed −log(p-values) for the mutational significance of all SREs defined using MCF-7 cells (r≥0.9). F). The mutational burden within the ESR1 (±250kb) SRE for the discovery (top) and validation (bottom) samples, the enhancers are rank according to panel (C). Red indicates mutations found in ESR1-positive tumours and pink indicates mutations found in ESR1-negative tumours.
To determine if the burden of mutations found in the SRE of ESR1 is significantly more than expected by chance. We designed a conservative analytical approach, termed MuSE (Figure 3D and Online Methods). Briefly, we consider all regulatory elements, or DHSs, predicted to interact with the ESR1 promoter as a single unit, analogous to splicing together the exons of a gene. We then test for an excess of mutations within the ESR1 SRE using a binomial probability test given a genome-wide and local background mutation rate (gBMR and lBMR, respectively). The gBMR is calculated from all DHSs including the ESR1 SRE. The lBMR is calculated from the DHSs surrounding the ESR1 gene that are not connected to its promoter based on C3D (Figure 3D). Each type of mutation is tested separately and the p-values are combined using Fisher’s method (Online Methods). This approach reveals the significant enrichment of noncoding somatic mutations within the ESR1 SRE (SRE r≥0.7, size=20,744bp, n=10, p=8.06×10−3)(Figure 3B and Supplementary Table 2). The number of nucleotides considered exceeds what is typical of coding sequences, which hinders the statistical significance. For example, the median length of a human protein is 375 amino acids30, which translates into 1,125 nucleotides. In comparison, 20,744 nucleotides encompass the SRE of ESR1. Increasing the stringency of the C3D predicted promoter-enhancer interactions improved the significant enrichment of mutations in the ESR1 SRE, despite including fewer mutations (C3D r≥0.9, size=7,746, n=6 p=2.57×10−4). The statistical enrichment is also improved by restricting the analysis to ESR1-positive tumours (C3D r≥0.9, size=7,746, n=5, p=7.02×10−5)(~7% (5/73))(Supplementary Table 2). The mutational significance appears specific to breast cancer mutations, since we do not detect an enrichment of somatic mutations within the ESR1 SRE defined in breast cancer cells using mutations called in WGS of 88 liver hepatocellular carcinomas27 (Figure 3B). To determine whether the observed enrichment is greater than expected by chance, we performed a genome-wide MuSE analysis restricted to mutations called in ESR1-positive breast cancer. Focusing on all RefSeq annotated genes with a promoter DHS in MCF-7 cells connecting to at least one regulatory element (C3D r≥0.9), we found a significant enrichment of mutations in only the SRE of ESR1 (Figure 3E)(FDR q-value=0.045).
To independently confirm that the ESR1 SRE is recurrently altered in breast cancer we sequenced the ESR1 SRE in an independent set of 52 primary ESR1-positive breast tumours from the Princess Margaret IMPACT and COMPACT trials. We identified three (~6%) somatic point mutations, chr6:151955219:G>T, chr6:151979547:A>G and chr6:152075097:G>C within enhancers interacting with the ESR1 promoter (C3D; r≥0.7)(Figure 3F). These mutations had a tumour fraction of 0.42, 0.32 and 0.03, respectively (Supplementary Table 3). The chr6:151955219:G>T falls within the enhancer altered by the rs9383590 SNV (Figure 3A) and is located 27bp away from the previously characterized chr6:151955192:A>G mutation.
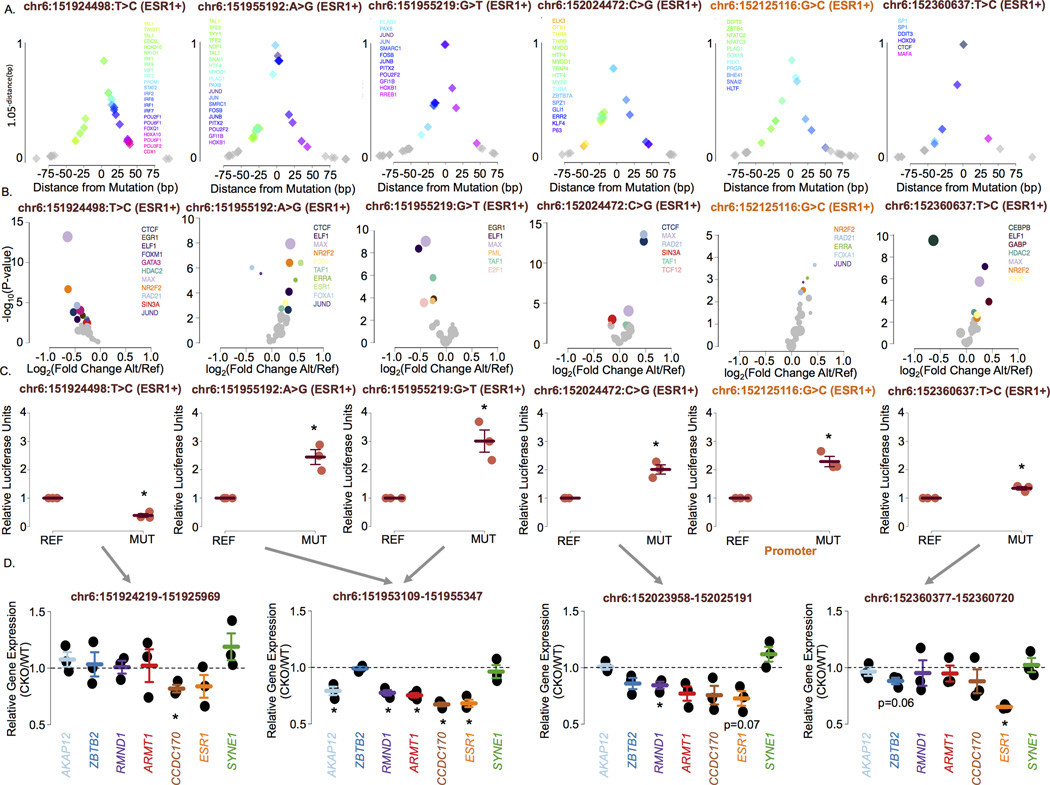
Similar to inherited risk variants, noncoding somatic mutations can impact transcription factor activity31. All somatic mutations found in the ESR1 SRE fall within or map nearby relevant transcription factor DNA recognition motifs (Figure 4A & Supplementary Figure 6). In addition, all mutations were predicted by IGR6 to modulate the chromatin binding intensity of known regulators of ESR1 expression, including GATA3, Cohesin, SIN3A and ESR1 (Figure 4B & Supplementary Figure 7). 10 of 11 tested mutations significantly altered the transactivation potential of their regulatory elements (Figure 4C & Supplementary Figure 8). Next, we focused on the mutations within the four enhancers with the strongest predicted interaction with the ESR1 promoter (r ≥ 0.9) and the promoter itself. These elements correspond to the MuSE analysis that passed multiple testing correction (FDR < 0.05). All six mutations affecting these regulatory elements identified in ESR1-positive tumours significantly impact their transactivation potential, including the chr6:151955219:G>T mutation from the validation set (Figure 4C). Except for the chr6:151924498:T>C mutant allele, which decreased enhancer activity compared to the wild-type sequence, the remaining five (83%) mutant alleles significantly increased reporter gene expression compared to the wild-type sequence. We confirmed the regulatory role of these enhancers on ESR1 gene expression by deleting each of the affected enhancers using the CRISPR-Cas9 system in T-47D cells stably expressing the Cas9 (Online Methods). The deletion of two of the enhancers significantly decreased ESR1 expression and a trend was observed for the remaining enhancers (Figure 4D). While each deletion is relatively large in size, they correspond to a small fragment of the SRE arguing in favour of the significant contribution of single elements to ESR1 expression.
Figure 4. Noncoding somatic mutations targeting ESR1 increase gene expression.
A) Distance of the transcription factor DNA recognition motifs to the identified mutations. The y-axis is a function (1.05−distance) of the distance to each mutation to emphasize the closest motifs. This function has a range of 0 – 1 within 100bp of the mutation. Each diamond represents the location of a transcription factor DNA recognition motif. B) Volcano plots presenting the p-value versus the fold change in chromatin binding intensity predicted by the intra-genomic replicates (IGR) analysis for transcription factors for each mutation in the ESR1’s SRE. All transcription factors profiled by ChIP-seq in MCF-7 or T-47D by ENCODE19 were tested for each mutation. Only those whose binding intensity for the chromatin is predicted to be modulated by the mutations (p<0.005) are coloured. Others are grey. C) Reporter assays revealing the impact of six mutations targeting the ESR1 SRE in ESR1-positive breast tumours on gene expression. Error bars indicate the standard error of the mean. D) Gene expression levels assessed by RT-qPCR in wild-type T-47D (WT) and T-47D cells with a CRISPR/Cas9-based deletion of the respective enhancer (CKO) region. All reported p-values are two-sided. * denotes significant (p<0.05).
The deletion of the enhancer harbouring the rs9383590 SNV, chr6:151953109–151955347, led to a significant decrease in ESR1 expression (Figure 4D) reinforcing the direct impact of this SNV on ESR1 expression. This enhancer also harbours three mutations. The chr6:151955192:A>G and chr6:151955219:G>T mutations were discovered in ESR1-positive tumours. However, the chr6:151954506:C>T mutation was found within an ESR1-negative tumour. Interestingly, this recapitulates the observed association between the GWAS lead SNVs at the ESR1 locus and both ESR1-positive and ESR1-negative breast cancer and suggest the presence of an additional oncogene(s) co-regulated with ESR1 at this locus. Of note, deletion of this enhancer in the T47D cells stably expressing Cas9 also led a significant reduction in the expression of ARMT1, CCDC170 and RMND1 (Figure 4D). The co-expression between ESR1 and the CCDC170 and RMND1 genes has been reported32 and their allele-specific expression may account for the association between variants at the ESR1 locus and ESR1-negative breast cancer33. In fact, silencing RMND1 significantly reduced the proliferation of ESR1-positive and ESR1-negative breast cancer cells (MCF7 and MDAMB436, respectively)(Supplementary Figure 9).
By demonstrating that the inherited risk variant and somatic point mutations that populate the SRE of ESR1 behave as gain-of-function genetic alterations, our results provide a mechanism to explain the sustained expression of ESR1 in approximately 7% of ESR1-positive breast cancer patients. This finding contrasts with gain-of-function coding mutations that typically present as mutations recurrently targeting a single codon34. Hence, noncoding mutational hotspots may be rare. Instead, mutations affecting distinct regulatory elements converging on the same gene, such as those reported here, may represent the mutational pattern of noncoding driver mutations. These do not need to directly target DNA recognition motifs. Indeed, recent work revealed that noncoding mutations still influence transcription factor activity, despite falling outside DNA recognition motifs35. Taken together, our work supports the idea that noncoding mutations relevant to cancer development and the genes they target can be identified using an approach focused on SREs that is inclusive of mutations outside of DNA recognition motifs.
Online Methods
Genotype Calling, Linkage Disequilibrium & Multidimensional Scaling
The raw genotype data of the METABRIC samples23 were downloaded from the European Genome-Phenome Archive. The raw genotype data of the TCGA samples were downloaded from the TCGA data portal. The genotypes of the METABRIC and TCGA samples were called using Birdseed37. The phase 3 genotype data for the 1,000 genomes project samples20 were downloaded from the 1,000 genomes website. Linkage disequilibrium (LD) was calculated using VCFtools38 within the continental European (CEU, TSI, FIN, GBR & IBS; n=503) and East Asian (CHB, JPT, CHS, CDX & KHV; n=504) groups. The SNVs in LD with GWAS lead SNVs are presented in Supplementary Table 4. Ethnicity was determined by merging the genotype data with the 1,000 genomes samples and performing multidimensional scaling (MDS) of the genotype data using PLINK39 (Supplementary Figure 10 & 11).
Intra-genomic replicates (IGR)
The functional impact of SNVs on transcription factor binding was predicted using the IGR tool as previously described6. Briefly, we compare the average ChIP-Seq signal intensity across genomic loci that contain short DNA sequences seven nucleotides in length (7mers) that match the reference allele and its surrounding DNA sequence against the average signal intensity at genomic loci that contain 7mers that differ only by the variant allele of each SNV. All 7mers from a sliding window surrounding each SNV are considered. The 7mer with the highest average intensity matching the reference allele is tested against the 7mer with the highest average intensity that matches the variant allele. The genomic locations of all 7mers are filtered to include only sites within open chromation. The wgEncodeUwDnaseMcf7PkRep1.narrowPeak and wgEncodeUwDnaseT47dPkRep1.narrowPeak called DHS sites produced as part of the ENCODE project19 were used as filters for the MCF-7 and T-47D cells, respectively. The aligned ChIP-Seq files were downloaded from the ENCODE website. The complete list of files used in the analysis is available in Supplementary Table 5. Signal files were generated using MACS40. All analyses were performed using hg19.
eQTL analysis
We used the sample of breast tumours profiled by METABRIC23 and TCGA. The expression data for the METABRIC samples were downloaded from the European Genome-Phenome Archive The RNA-Seq data for the TCGA breast cancer samples were downloaded from Cancer Genomics Hub
The reads were aligned to human reference GRCh37 with Gencode version 15 human transcript annotation using STAR41 in two-pass mode. Gene level expression values for each sample were quantified using Cufflinks42. The expression of ESR1 is bimodal in METABRIC and TCGA and is explained by ESR1-positive and ESR1-negative tumours (Supplementary Figure 12). Consistent with the METABRIC23 analysis, we determined the expression status of TCGA tumours for ESR1, PGR, and ERBB2 among TCGA samples using MClust in R. We fitted a gaussian finite mixture model with two components. We restricted the analysis to luminal-type tumours, those that express both ESR1 and PGR, but do not overexpress ERBB2. TCGA tumour samples with low expression of ERBB2 were also identified as belonging to a separate distribution by MClust and were removed. We merged the identified luminal METABRIC discovery and validation samples and performed a quantile normalization of the merged samples using the preprocessCore library43 in R. Statistical significance was determined using linear regression under an additive and recessive model. The reported p-values are two-sided. To control for potential population stratification we included the first three components of the MDS analysis as covariates. The rs9397437 SNV was used as a proxy for the rs9383590 (r2=1.0 & r2=1.0) among Europeans and East Asians, respectively. The gene expression values stratified by SNV genotype are presented in Supplementary Figure 13.
Allelic Imbalance
We analyzed the TCGA breast tumours profiled by RNA-Seq. Duplicate reads were removed using Picard. The number of aligned reads containing either the reference or variant alleles of coding marker SNVs was determined using the ABC tool44. The default settings of ABC were used. Marker SNVs were identified by intersecting the common SNV database with refSeq exon annotations using bedTools45. We calculated the allelic imbalance (AI) ratio as the number of reads containing either the reference or variant allele, whichever was larger, divided by the total number of reads. We removed samples with an AI ratio greater than 0.8, since they could represent sequencing error within homozygous individuals24. Individuals heterozygous for the rs9397437 SNV, a proxy of the rs9383590 SNV, were compared to individuals homozygous for the common allele using an approximate Fisher-Pitman test with 10,000 permutations implemented in the coin library in R. We included markers SNVs with at least 20 samples heterozygous for the rs9397437 SNV.
Defining Sets of Regulatory Elements (SREs)
We took advantage of the known relationship between the Cross-Cell type Correlation in DNaseI hypersensitivity signals (C3D) and chromatin interactions21 to predict connections between regulatory elements. We used the uniformly processed DNaseI hypersensitivity sequencing signal files for 79 cell lines available from the ENCODE project19. We performed the correlation of DNaseI signal intensities in a cell type-specific manner by interrogating only DHS identified in the MCF-7 cell line22. The DHS used in our study were identified by the Hotspots algorithm46 and produced as part of the ENCODE project19. We validated the predicted interactions called for breast cancer with a POL2 ChIA-PET data, profiled in MCF-7 cells, created by the ENCODE Project19. We combined all four replicates for our analyses.
Calculating Mutational Significance within the Regulatory Element Set (MuSE)
DHSs predicted to interact with the gene promoter at a given correlation threshold (r ≥ 0.7 – 0.9) are combined to create the test region or SRE. We use the binomial test implement in R to assess whether the observed number of mutations within the test region is greater than expected given both a genome-wide and local background mutation rate (lBMR and gBMR, respectively). The approach is comparable to that employed by MutSig47 and MuSiC48, but applied to non-coding regions and mutations. We calculate the lBMR using the remaining DHSs that are below the correlation threshold but within the specified window surrounding the anchor DHS. This approach is thought to be conservative, since it is possible that mutations included in the lBMR are functional. It is important that the lBMR and gBMR be calculated from DHSs and that these DHSs be cell type-specific, because somatic mutations have been shown to preferentially fall in heterochromatic noncoding regions in a cell type-specific manner9,10. To control for different rates of mutations, a separate binomial test is performed for each of the six mutation types (n) and a final combined p-value is calculated using Fisher’s method from a χ2 distribution with 2n degrees of freedom in R. Only one mutation within the test region is counted per tumour. However, all mutations contribute to the BMR calculation, which again should be conservative. If we are unable to calculate the lBMR for a given mutation type, because we do not observe a mutation within the window, we use the gBMR for that mutation type. We excluded tumours profiled by whole exome sequencing, because they are typically sequenced to a greater depth and regulatory elements co-occur with coding sequencings49.
Mutation Data (Discovery)
The breast and liver cancer mutations used in the MuSE calculations were reported by Alexandrov et al27. We used the cleaned mutation dataset in all analyses. We included only those samples with known ESR1 status (n=98) in our analysis50. The identifiers of the ESR1-positivity and TNBC tumours are listed in Supplementary Table 6).
Targeted Sequencing of the ESR1 Set of Regulatory Elements (SRE) (Validation)
We validated the enrichment of mutations within the ESR1 SRE in an independent set of 52 primary ESR1-positive breast tumours and matched normal blood samples from the Integrated Molecular Profiling in Advanced Cancer Trial (IMPACT) and the Community Oncology Molecular Profiling in Advanced Cancer Trial (COMPACT) trials conducted at the Princess Margaret Cancer Centre (PMCC) The research ethics board of the University Health Network (UHN) approved the retrospective analysis of the breast cancer samples. Informed consent was obtained from all study participants. We used hybrid capture to isolate the ESR1 SRE elements using a custom panel of xGen Lockdown Probes (Integrated DNA Technology Inc). The 120bp probes were spaced 60bp apart. The probe sequences and the targeted regions are available in Supplementary Table 1 and Supplementary Table 7. Captured fragments were sequenced using 150 bp paired-end reads from a NextSeq 500 sequencer (Illumina) at the Princess Margaret Genomics Centre. Tumours and normal samples were sequenced to median >600× coverage.
Calling Somatic Point Mutations (Validation)
Reads were aligned to the human reference genome, hg19, using BWA51. Base recalibration and realignment around insertions and deletions (indels) was performed with GATK52. Duplicate reads were marked with Picard. Somatic point mutations were called from tumour/normal pairs using muTect53. The identified mutations are available in Supplementary Table 3 and Supplementary Table 8.
Localization of Transcription Factor Motifs Surrounding Mutations
We searched the flanking sequence (±100bp) surrounding each somatic mutation for human transcription factor DNA recognition motifs using position weight matrices compiled in the Homo Sapiens Comprehensive Model Collection (HOCOMOCO)54 with FIMO55.
Annotation of LD SNVs and Identification of Active Enhancers
We downloaded all available called peaks and signal files for the two breast cancer model cell lines, MCF-7 & T-47D, that were produced as part of the ENCODE project19. To verify the identified enhancers we downloaded an additional H3K27ac ChIP-Seq data profiled by Taberlay, et al.36 from the Gene Expression Omnibus (GEO)(GSM1383859). The reads were aligned to the reference genome using BWA51 and signal files were generated using MACS240.
Cell Culture
HCC1419 cells (ATCC) were grown to 95% confluence in RPMI 1640 medium supplemented with 10% foetal bovine serum. MCF-7 (in house), T-47D (in house) and MDAMB436 (in house) cell lines were grown in DME medium supplemented with 10% foetal bovine serum. The cell lines used in this study have not been listed as cross-contaminated or commonly misidentified by the international cell line authentication committee (ICLAC). All cell lines were determined to be mycoplasma free using the EZ-PCR mycoplasma test kit (Biological Industries).
Western Blot
We verified the ESR1 protein expression status of the HCC1419 and MDAMB436 by western blot. Cells were lysed for 5 minutes on ice in lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris-HCl pH=8.1) supplemented with a protease inhibitor cocktail (Roche), followed by sonication. The protein concentration of the lysates was determined using a Pierce Micro BCA Protein Assay Kit (Thermo Scientific). Equal amounts of protein (25µg) were electrophoresed on TGX Protein Gels (Bio-Rad), and then transferred to a PVDF membrane (Bio-Rad). Membranes were blocked with 5% BSA (AMRESCO) in Tris-buffered saline (TBS) with 0.1% Tween-20 (Fisher Scientific) for 2 hours at room temperature. Primary antibodies were incubated overnight at 4°C. Western blots for ESR1 were performed with the rabbit anti-ESR1 antibody (sc-543X, Santa Cruz) and blots for β-Actin were performed using the rabbit anti-β-Actin (4970, Cell Signalling). Membranes were washed then probed with anti-rabbit IgG, HRP-linked antibody (7074, Cell Signalling) at room temperature for 1 hour. Bands were visualized with ECL Western Blotting Detection Reagent (GE Healthcare) and scanned using the FluorChemQ (Alpha Innotech).
Cell viability assays following silencing of RMND1
Two sets of siRNA against RMND1 were designed through Thermo Fisher Scientific (RMND1 Silencer Select #s29968 and s29969, catalogue #4392420). A scrambled siRNA was used as a negative control. RNA was isolated from MCF-7 and MDAMB436 cells using the RNeasy Mini kit (Qiagen) according to the manufacturer's protocol. Reverse transcription (RT) was performed to convert RNA into cDNA (iScript cDNA Synthesis Kit, BioRad). The resulting cDNA was subjected to quantitative PCR (RT-qPCR) using SensiFAST SYBR (Bioline) to confirm the silencing effect of the siRNAs against RMND1. Expression levels were quantified using the ΔΔCt method with Actin as the calibrator56 and then normalized to RMND1 RNA levels in cells that were treated with negative control siRNA. The primers are as listed in Supplementary Table 9.
The relative proportion of viable MCF-7 and MDAMB436 breast cancer cells were measured using AlamarBlue reagent (Thermo Fisher Scientific) 72 hours after silencing of RMND1. AlamarBlue reagent was added to the wells and the cells were incubated at 37°C for four hours. Fluorescence was read at excitation and emission wavelengths of 550nm and 590nm respectively.
RNA-Seq and Allele-specific Expression within HCC1419 cells
Total RNA was extracted from HCC1419 cells using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. Two RNA-seq libraries were prepared using the Truseq Stranded mRNA kit (Illumina). RNA was sequenced using 75bp paired end reads. Reads were mapped to the reference genome, hg19, using TopHat42. Allele-biased expression was called using a binomial test with the ABC tool44. The sequencing data is available through the NCBI (National Center for Biotechnical Information) gene expression omnibus (GEO) accession number GSE74718.
Allele-Biased Chromatin-immunoprecipitation
HCC1419 cells were cross-linked with 1% formaldehyde at 37°C for 10 minutes. Cells were rinsed with ice-cold PBS plus 5% BSA followed by PBS and harvested with PBS plus 1× protease inhibitor cocktail (Roche Molecular Biochemicals, IN). Harvested cells were centrifuged for 2 min at 3,000 rpm. Cells were lysed in 0.35 mL of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, 1× protease inhibitor cocktail) by sonication (Diagenode Biorupter 300). The lysed cells were subjected to centrifugation at maximum speed for 15 minutes. Supernatants were collected and diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1).
12.5ug of GATA3 antibody (Santa Cruz, sc268x) was prebound for 6 hours to protein A and protein G Dynal magnetic beads (Dynal Biotech, Norway) and washed three times with ice-cold PBS plus 5% BSA and then added to the diluted chromatin for overnight immunoprecipitation. The magnetic bead-chromatin complexes were collected and washed six times in RIPA buffer (50 mM HEPES [pH 7.6], 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCl) and then washed twice with TE buffer. Cross-linking was reversed with decrosslinking buffer (1% SDS, 0.1 M NaHCO3) overnight at 65°C. DNA fragments were purified with a QIAquick Spin Kit (Qiagen, CA).
Allele-biased binding was assessed using MAMA primers based qPCR57 and verified by Sanger sequencing (Supplementary Figure 14). Fold enrichment was calculated over input. Significance of the differential enrichment was calculated using the unpaired t-test. A complete list of primers is available in Supplementary Table 9.
Luciferase Reporter Assays
Each enhancer was PCR-amplified using PfuUltraII fusion polymerase from Human DNA (Roche Molecular Biochemicals, IN) and then cloned at the BamHI (BamHI-HF, NEB) restriction site into the pGL3 and pGL4.23 promoter vector (Promega, WI) in the antisense and/or sense direction. Site-directed mutagenesis was performed using QuickChange XLII kit (Agilent) according to manufacturer’s instructions to generate the mutant alleles. The results of luciferase assay in the sense orientation are presented in Supplementary Figure 15. All sequences were verified by Sanger sequencing (Supplementary Figure 16. Wild-type and mutant enhancer constructs were independently transfected in T-47D cells grown in oestrogen depleted media together with a Renilla reporter plasmid at a 1:100 ratio. 48 hours after transfection, the cells were stimulated with full media and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Final readings are reported as firefly luciferase normalised to renilla luciferase activity per well. Fold change in luciferase activity of the mutant allele compared to the reference allele was calculated. The values were log transformed and significance was tested using a one-sample t-test. Two-sided p-values are reported.
Chromatin Conformation Capture
Chromosome conformation capture (3C) coupled with qPCR was performed according to a published protocol58. Briefly, 7.5 million MCF-7 cells were cross-linked using formaldehyde treatment (1%, 10 min at room temperature), followed by HindIII-HF digestion (400 units, overnight at 37°C) and ligation (T4 DNA ligase 4000 units, 4h at 16°C). A phenol:chloroform extraction was performed on the DNA fragments, followed by ethanol precipitation. The ligated fragments were quantified by qPCR. Genomic DNA amplicons of 60 primer pairs spanning the ESR1 SRE region (Supplementary Table 9) were mixed in equimolar amounts, digested and ligated to generate a randomly ligated control template. This was used to verify primer efficiency and to normalize the 3C interaction frequency. To normalize our 3C data analysis, we generated the Ct value standard curve using the control template for each tested ligation. Then we quantified the ligation products between the ESR1 promoter and each of the tested 3C sites using the parameters from each corresponding standard curve (b: intercept, a: slope): value = 10(Ct − b)/a58.
CRISPR-Cas9 Enhancer deletion in Stable Cas9 expressing T-47D cells
Lentivirus carrying the human codon-optimized S. pyogenes Cas9 gene was generated using 293FT cells transfected with Lipofectamine 3000, the viral plasmids VSVG and PAX2, and a lentiviral-Cas9 vector (Addgene plasmid #52962). T-47D cells were infected with the Cas9 lentivirus and selected for 14 days with blasticidin. Cas9 protein expression was verified by western blotting using an antibody from Diagenode (#C15200203)(Supplementary Figure 17A).
Stable Cas9 expressing T-47D cells were transfected with a pool of four enhancer targeting crRNA/tracrRNA complexes (Integrated DNA technologies) or two non-targeting crRNA/tracrRNA complexes. Briefly, 125 pmol of each of four different enhancer targeting crRNA/tracrRNA complexes (or 250 pmol of two non-targeting complexes) were combined with 6 µL Lipofectamine RNAiMAX and 200 µL OptiMEM, incubated for 20 minutes at room temperature, and added to each well prior to plating cells. 150,000 cells were then plated per well for each crRNA/tracrRNA pool targeting different enhancers or non-targeting controls. Transfected cells were incubated for 72 hours followed by extraction of RNA and genomic DNA using the Qiagen AllPrep DNA/RNA mini kit according the manufacturer’s recommended protocol. RNA was reverse transcribed into cDNA using the Bioline SensiFAST cDNA synthesis kit and RT-qPCR performed using the Bioline SensiFAST SYBR mix (No-Rox) on a BioRad CFX96 Real-Time PCR instrument. Threshold qPCR values for each gene were first normalized to Actin mRNA and were then normalized to the non-targeting crRNA/tracrRNA treated cell values for each of three independent replicates. The fold changes were log transformed and significance was tested using a one-sample t-test. Two-sided p-values are reported. Deletions were verified through gel electrophoresis (Supplementary Figure 17B).
Supplementary Material
Acknowledgments
We thank Drs. Albiruni Razak, Christine Elser, Dave Cescon, David Warr, Eitan Amir, Lillian Siu, Natasha Leighl and Srikala Sridhar for their involvement in recruiting the IMPACT/COMPACT samples used in this study. We also thank Dr. Mathieu Lemaire for helpful discussions. We thank Drs. Robert Rottapel and Oliver Kent for use of and help with the Glomax Multi-Detection system. We acknowledge the ENCODE consortium and the ENCODE production laboratories that generated the data sets provided by the ENCODE Data Coordination Center used in the manuscript. We also acknowledge the Cancer Genome Project for making all the breast cancer and liver cancer called mutations publicly available and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) for making the genotyping and expression data from primary breast tumours data available. Finally we acknowledge the Princess Margaret Genomics Centre and the Bioinformatics group for providing the infrastructure assisting us with the targeted sequencing and analysis of the ESR1 SRE. The National Cancer Institute (NCI) at the National Institute of Health (NIH) (R01CA155004 to M.L.), the Princess Margaret Cancer Foundation (T.J.P. and M.L.), The Canadian Cancer Society (CCSRI702922 to M.L.), Susan G. Komen Foundation (CCR15332792 to T.J.P), and the Gattuso-Slaight Personalized Cancer Medicine Fund/PMCF (B.H-K.) supported this research. M.L. holds a young investigator award from the Ontario Institute for Cancer Research (OICR), a new investigator salary award from the Canadian Institute of Health Research (CIHR) and a Movember Rising Star award from Prostate Cancer Canada (PCC) (RS2014-04). K.J.K and R.C.P. are Canadian Breast Cancer Foundation (CBCF) postdoctoral fellowship recipients. S.D.B. is a Knudson and CIHR postdoctoral fellowship recipient.
Footnotes
URLs
The Cancer Genome Atlas (TCGA)(http://cancergenome.nih.gov/)
The Princess Margaret Genomics Centre (www.pmgenomics.ca).
The European Genome-Phenome Archive (https://www.ebi.ac.uk).
The TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp).
The 1,000 genomes project samples (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/).
The ChIP-Seq files for the ENCODE (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeHaibTfbs/ and http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeSydhTfbs/).
The Cancer Genomics Hub (https://browser.cghub.ucsc.edu/).
R (www.r-project.org).
Picard (http://picard.sourceforge.net).
The common SNV database (ftp://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/snp142Common.txt.gz)
The uniformly processed DNaseI hypersensitivity sequencing signal files for 79 cell lines (http://ftp.ebi.ac.uk/pub/databases/ensembl/encode/integration_data_jan2011/byDataType/signal/jan2011/bigwig/)
The Hotspots algorithm (https://github.com/rthurman/hotspot)
The MCF7 DHS identified by the Hotspots algorithm (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwDnase/wgEncodeUwDnaseMcf7HotspotsRep1.broadPeak.gz).
The MCF7 POL2 ChIA-PET data created by the ENCODE Project (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeGisChiaPet/).
The breast cancer and liver cancer mutations reported by Alexandrov et al.18 are available here ftp://ftp.sanger.ac.uk/pub/cancer/AlexandrovEtAl/.
The Integrated Molecular Profiling in Advanced Cancer Trial (IMPACT) and the Community Oncology Molecular Profiling in Advanced Cancer Trial (COMPACT) trials conducted at the Princess Margaret Cancer Centre (PMCC)(https://clinicaltrials.gov/ct2/show/NCT01505400).
The Gene Expression Omnibus (GEO)(http://www.ncbi.nlm.nih.gov/geo/).
MACS2 (https://github.com/taoliu/MACS).
Author contributions statement
The concept of interrogating the mutational load in regulatory elements converging on single genes arose through discussions between S.D.B., N.A.S-A., R.C.S. and M.L.. S.D.B. designed and/or implemented all the computational and statistical approaches, except for IGR and analyzed the results under the supervision of M.L. Experimental assessment of the effect of SNVs on enhancer activity, transcription factor binding and gene expression was designed by K.D., S.D.B. and M.L. and conducted by K.D. with assistance from K.J.K., A.T. and X.W. The CRISPR/Cas9 based enhancer deletion was conducted by K.D., K.J.K., K.L.T., J.S. and D.W.C. under the supervision of T.W.M. and M.L. P.M. and N.A.S-A. implemented the IGR approach to predict allele-bias binding of transcription factors on SNVs following improvements to IGR by N.A.S-A., and R.C.S.. R.C.P. and P.L.B. assessed the ESR1, PR and HER2 expression status on primary breast tumours included in our validation cohort. S.Y.C.Y. performed the alignment and gene expression quantification of the TCGA RNA-Seq data. M.D. assisted in the DNA capture sequencing of the primary breast tumour validation cohort under T.J.P.’s supervision. B.H-K. oversaw the expression analysis of the METABRIC dataset. M.L. oversaw the project. Figures were designed and prepared by S.D.B. and K.D. while the manuscript was written by S.D.B., K.D. and M.L. with assistance from all other authors.
Competing financial interests:
The authors declare no competing financial interests.
References
- 1.Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer. 2007;7:713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- 2.Vincent-Salomon A, Raynal V, Lucchesi C, Gruel N, Delattre O. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:809. doi: 10.1038/ng0708-809a. author reply 810-2. [DOI] [PubMed] [Google Scholar]
- 3.Brown LA, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:806–807. doi: 10.1038/ng0708-806. author reply 810-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horlings HM, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:807–808. doi: 10.1038/ng0708-807. author reply 810-2. [DOI] [PubMed] [Google Scholar]
- 5.Reis-Filho JS, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet. 2008;40:809–810. doi: 10.1038/ng0708-809b. author reply 810-2. [DOI] [PubMed] [Google Scholar]
- 6.Cowper-Sallari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nature Genetics. 2012;44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Research. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polak P, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32:71–75. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weedon MN, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2014;46:61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 13.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher O, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 16.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–5384. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacey SN, et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ENCODE. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey SD, et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nature Communications. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunning AM, et al. Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat Genet. 2016 doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frietze S, et al. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012;13:R52. doi: 10.1186/gb-2012-13-9-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature Biotechnology. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33:3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell RJ, et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunbier AK, et al. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto-Ibusuki M, et al. C6ORF97-ESR1 breast cancer susceptibility locus: influence on progression and survival in breast cancer patients. Eur J Hum Genet. 2015;23:949–956. doi: 10.1038/ejhg.2014.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katainen R, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
References
- 36.Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014;24:1421–1432. doi: 10.1101/gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 44.Bailey SD, Virtanen C, Haibe-Kains B, Lupien M. ABC: a tool to identify SNVs causing allele-specific transcription factor binding from ChIP-Seq experiments. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature Genetics. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dees ND, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stergachis AB, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju YS, et al. Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Res. 2015;25:814–824. doi: 10.1101/gr.190470.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulakovskiy IV, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–D202. doi: 10.1093/nar/gks1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Li B, Kadura I, Fu DJ, Watson DE. Genotyping with TaqMAMA. Genomics. 2004;83:311–320. doi: 10.1016/j.ygeno.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Hagege H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.