Abstract
It is becoming clear that Structural Maintenance of Chromosomes (SMC) complexes, such as condensin and cohesin, are involved in the three-dimensional genome organization, yet the exact roles of these complexes in the functional organization remain unclear. This study employs the ChIA-PET approach to comprehensively identify genome-wide associations mediated by condensin and cohesin in fission yeast. We find that although cohesin and condensin often bind to the same loci, they direct different association networks and generate small and larger chromatin domains, respectively. Cohesin mediates local associations between loci positioned within 100 kb; condensin can drive longer-range associations. Moreover, condensin, but not cohesin, connects cell cycle-regulated genes bound by mitotic transcription factors. This study describes the different functions of condensin and cohesin in genome organization and how specific transcription factors function in condensin loading, cell cycle-dependent genome organization, and mitotic chromosome organization to support faithful chromosome segregation.
INTRODUCTION
The three-dimensional genome structure is coupled to important nuclear processes such as transcription, DNA replication and repair1. To understand global genome organization, next-generation sequencing combined with chromosome conformation capture (3C), referred to as Hi-C, has been applied to a variety of organisms from bacteria to humans2–14. These studies collectively reveal that genomes are highly ordered by a hierarchy of organizing events, ranging from gene associations to topologically associating domain (TAD) formation. TADs have recently been identified as functional genome-organizing units related to transcriptional co-regulation and replication timing6–8,15,16. Condensin and cohesin are implicated in TAD organization14,17–19, although their roles in domain formation remain enigmatic.
Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) is a genomic approach that combines chromatin immunoprecipitation (ChIP), proximity ligation, and next-generation sequencing to identify the genome-wide associations mediated by particular proteins20,21. This approach has been used to identify genomic associations involving estrogen receptor, CTCF (CCCTC-binding factor), cohesin, RNA polymerase II, and histone H3 Lys4 methylation22–30. Mechanistically, cohesin is recruited to CTCF binding sites through interaction with CTCF, and mediates associations among genes and their regulatory elements31–34. Another important SMC complex, condensin, is also involved in higher-order genome organization in yeast and other organisms14,17,35,36. Condensin binds to RNA polymerase III-transcribed (Pol III) genes such as tRNA and 5S rRNA, to Pol II-transcribed housekeeping genes, and to long terminal repeat (LTR) retrotransposons, and it clusters these dispersed genetic elements, often associating them with centromeres in fission yeast37,38. It has been shown that cohesin and condensin frequently co-localize to several different genetic elements including Pol III genes, gene promoters/enhancers, and chromatin domain boundaries, in a variety of organisms17,36,39–41. However, it remains largely unclear how these two complexes cooperate to organize a functional genome architecture. Using the ChIA-PET genomic approach, here we show how condensin and cohesin mediate domain organizations and genome-wide associations in the fission yeast model organism.
RESULTS
Specific cell cycle-regulated genes are bound by condensin
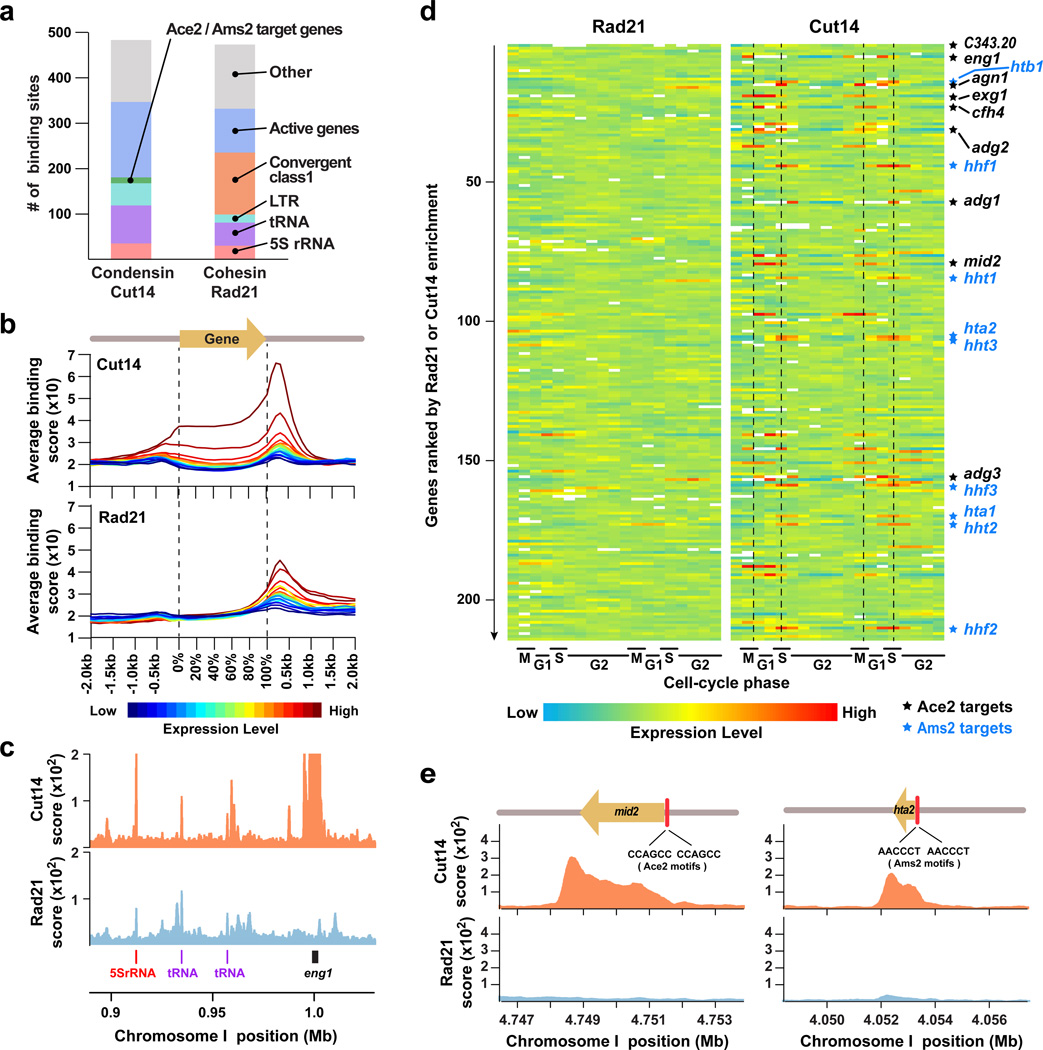
Based on ChIA-PET self-ligation reads, Cut14-Pk and Rad21-Myc proteins were mapped to 485 and 475 significant binding sites, respectively, across the genome (Supplementary Fig. 1a; Supplementary Note). Condensin and cohesin were often co-localized and detected at dispersed repetitive elements including tRNA and 5S rRNA genes, LTR retrotransposons, and centromeres (Fig. 1a; Supplementary Fig 1b,c). They were also localized at highly expressed RNA polymerase II-transcribed (Pol II) genes (Fig. 1b). Transcription levels of Pol II genes appeared to be positively correlated with Cut14-Pk and Rad21-Myc enrichment. The peak of cohesin enrichment was located at the 3’-end of genes, but not within gene bodies. The peak of condensin enrichment was also at the 3’-end, although condensin was also detected at gene bodies to some extent. Previous studies had predicted that condensin and cohesin slide along genes42,43. This result suggests that condensin and cohesin are recruited to highly expressed genes and translocated along gene bodies.
Figure 1. Condensin binds to cell cycle-regulated Pol II genes.
(a) Summary of Cut14-Pk (condensin) and Rad21-Myc (cohesin) binding sites. Active genes are among the top 10% highest transcribed Pol II genes.
(b) Highly transcribed Pol II genes are bound by condensin and cohesin. Pol II genes were classified into 20 groups based on their transcription levels (colors). Each group contains 162 Pol II genes. Average binding patterns of Cut14-Pk (top) and Rad21-Myc (bottom) for the respective gene groups were plotted. Gene sizes from transcriptional initiation to termination sites were normalized to the same length for all the genes investigated.
(c) Distributions of Cut14-Pk (red) and Rad21-Myc (green) at the genomic region including the Ace2 target gene (eng1) and Pol III genes (tRNA and 5S rRNA).
(d) Expression of Pol II genes bound by cohesin (Rad21-Myc, left) or condensin (Cut14-Pk, right) during the cell cycle. Expression data were downloaded from the Bähler lab homepage45. Pol II genes were ordered from top to bottom based on enrichment of Rad21-Myc or Cut14-Pk. Dotted lines indicate M and S phases, when Ace2 (black) and Ams2 (blue) target genes are up-regulated, respectively.
(e) Binding patterns of Cut14-Pk and Rad21-Myc at the Ace2 target gene (mid2) and Ams2 target histone gene (hta2). Red bars indicate Ace2- and Ams2-binding motifs.
It has been shown that cohesin often localizes at intergenic regions between convergent genes39,44. We observed that cohesin was enriched at convergent gene loci, and that cohesin was more enriched at convergent gene loci in the class 1 context than in class 2, while condensin was only slightly enriched at convergent gene loci (Supplementary Fig. 1d).
Although condensin and cohesin frequently shared the same binding sites across the genome, we found that binding of condensin and cohesin showed a clear difference at some loci; e.g. the eng1 locus in Fig. 1c. Such condensin-specific sites often coincided with histone genes and a group of genes highly expressed during S and M-G1 phases (Fig. 1d). It is known that the expression of histone genes and of M-G1 phase genes is induced by the cell cycle-regulated transcription factors, Ams2 and Ace2, respectively45,46. Ams2 is a GATA-type transcription factor and responsible for cell cycle-dependent transcription of histone genes46. Ace2 is known to control the cell cycle-dependent expression of genes required for septin ring formation and cell separation47,48; two potential Ace2 target genes, exg1 and SPAC343.20, abbreviated as C343.20, were identified (Supplementary Fig. 1e,f). Condensin, but not cohesin, was most enriched at the 3’-end of the Ace2 and Ams2 target genes, implying that condensin is recruited to gene promoters through its interaction with Ace2 and Ams2 and slides along gene bodies (Fig. 1e). Among the 18 Ace2/Ams2 target genes, 18 (100%) are bound by condensin and 6 (33%) are bound by cohesin. We observed prominent condensin peaks at the Ace2/Ams2 target genes, but negligible or no cohesin peaks were present (Fig. 1c,e).
Different-sized domains formed by condensin and cohesin
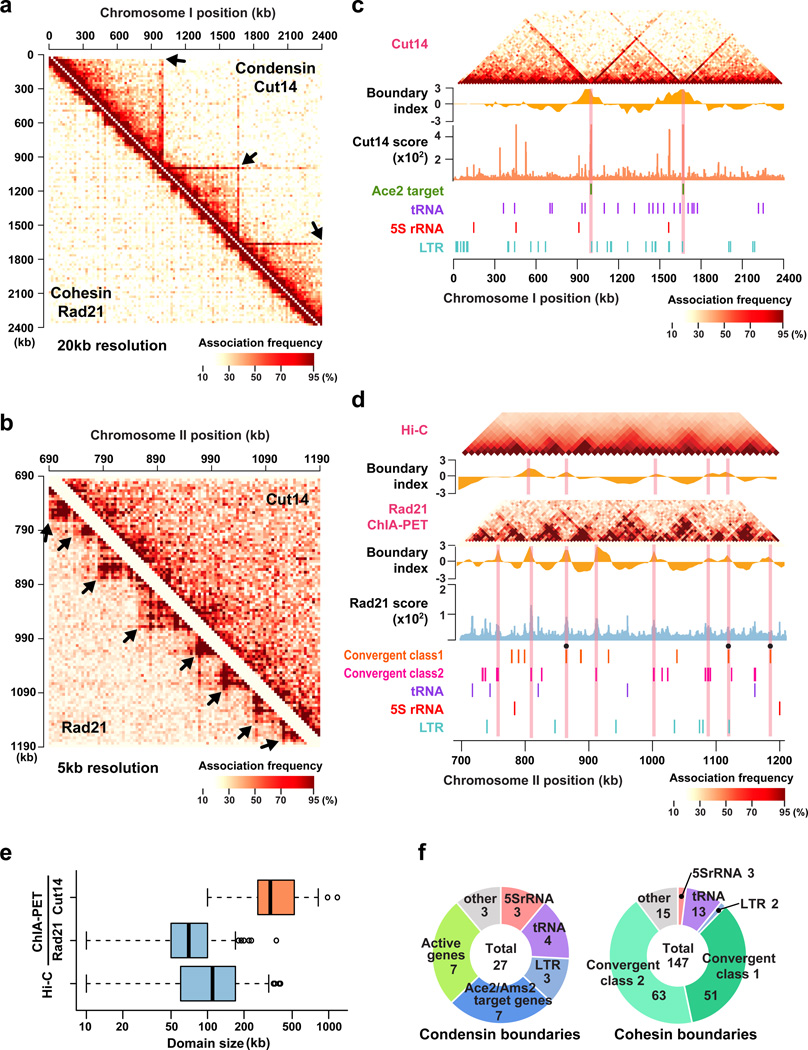
To understand the general features of condensin- and cohesin-mediated genomic associations, ChIA-PET reads were mapped against the fission yeast genome divided into 20 kb sections, and we found that both condensin- and cohesin-mediated associations were most enriched along the diagonal, indicating that both condensin and cohesin drive local associations to the highest degree (Fig. 2a). At a 20 kb resolution, we observed chromatin domains with associations mediated by condensin, but not cohesin (Fig. 2a; Supplementary Fig. 2a). In contrast, at a resolution of 5 kb, smaller chromatin domains with associations mediated by cohesin, but not condensin, were detected (Fig. 2b). Cohesin-mediated associations were observed between flanking gene loci positioned less than 100 kb apart (Supplementary Fig. 2b). The frequency of condensin-mediated associations was reduced as distances between two loci increased, but the decay curve was not as acute as for cohesin-dependent associations, and condensin-mediated associations were frequent between loci separated by as far as 1 Mb (Supplementary Fig. 2b). Note that condensin-mediated associations also occur beyond 1 Mb, as further analyzed in the following sections. These associations are non-randomly distributed across the genome, thereby showing the domain organizations mediated by condensin and cohesin.
Figure 2. Different sizes of chromatin domains are formed by condensin and cohesin.
(a) Heat maps of condensin- and cohesin-mediated associations at a 20 kb resolution. Association frequencies between 20 kb genomic sections were counted as number of reads assigned to respective combinations. ChIA-PET data from biological and technical replicas were all combined. Arrows indicate chromatin domains.
(b) Condensin- and cohesin-mediated genomic associations at a 5 kb resolution.
(c) Condensin-mediated associations between 20 kb genomic sections. Boundary indexes were calculated as described in Supplementary Note. Boundary index, Cut14-Pk binding score, and gene annotations are shown below the association map. Pink lines indicate positions of predicted domain boundaries.
(d) Comparison between Rad21 ChIA-PET and Hi-C data10. Association frequencies between genomic sections and boundary indexes were calculated for Hi-C and cohesin ChIA-PET data at 10 kb and 5 kb resolutions, respectively, and shown with Rad21-Myc binding score and gene annotations. Filled circles represent potential boundaries formed at the class I convergent gene loci. The class II convergent loci at significant cohesin binding sites are also shown.
(e) Distributions of domain sizes predicted from condensin and cohesin ChIA-PET data and Hi-C data. Boxes show center quartiles, and whiskers extend to the data point which is no more than 1.5x the interquartile range from the box.
(f) Breakdown charts of condensin and cohesin boundaries.
We next estimated the positions of chromatin boundaries formed by condensin, based on a boundary index (Supplementary Note). Condensin-mediated association domains, referred to as condensin domains, were demarcated by boundaries (condensin boundaries), where condensin was generally highly enriched (Fig. 2c; Supplementary Fig. 2a). Similarly, cohesin domains were separated by boundaries where cohesin was enriched (Fig. 2d). The average sizes of condensin and cohesin domains were 84 kb (median 70 kb) and 349 kb (median 300 kb), respectively (Fig. 2e).
A previous Hi-C study indicated that cohesin mediates the formation of 100 kb chromatin domains across the fission yeast genome10. Consistently, we observed cohesin domains of similar size, and the positions of the boundaries estimated from the cohesin ChIA-PET heavily overlapped those determined from Hi-C data (Fig. 2e; Supplementary Fig. 2c), although the cohesin ChIA-PET data showed clearer domains, probably due to the purity of the data only reflecting cohesin-mediated associations. Cohesin boundaries were in general formed at cohesin binding sites, most typically at convergent gene loci (Fig. 2f; Supplementary Fig. 2d,e).
Quite differently, condensin boundaries (where condensin was highly enriched) coincided with positions of Ace2/Ams2-target genes (eng1, C343.20, adg2, adg1, cfh4, exg1, hta1/htb1 genes), tRNA genes, LTR retrotransposons, and highly transcribed Pol II genes (Fig. 2f). The 7 condensin boundaries, out of 27 total, were formed at the Ace2/Ams2 target gene loci, and only 11 loci consist of all 18 Ace2/Ams2 target genes. That boundaries are so often formed at the Ace2/Ams2 targets suggests that these transcription factors are involved in the boundary organization, as further examined below. Together, these results demonstrate that the condensin and cohesin complexes establish distinct sizes of chromatin domains demarcated by boundaries bound by these complexes. It is important to note that the distance decay in the Rad21 ChIA-PET data was clearly different from the decay in the Hi-C data, indicating that associations detected by ChIA-PET are not derived from randomly sampled in vivo associations (Supplementary Fig. 2f).
Condensin associates Ace2 and Ams2 target genes
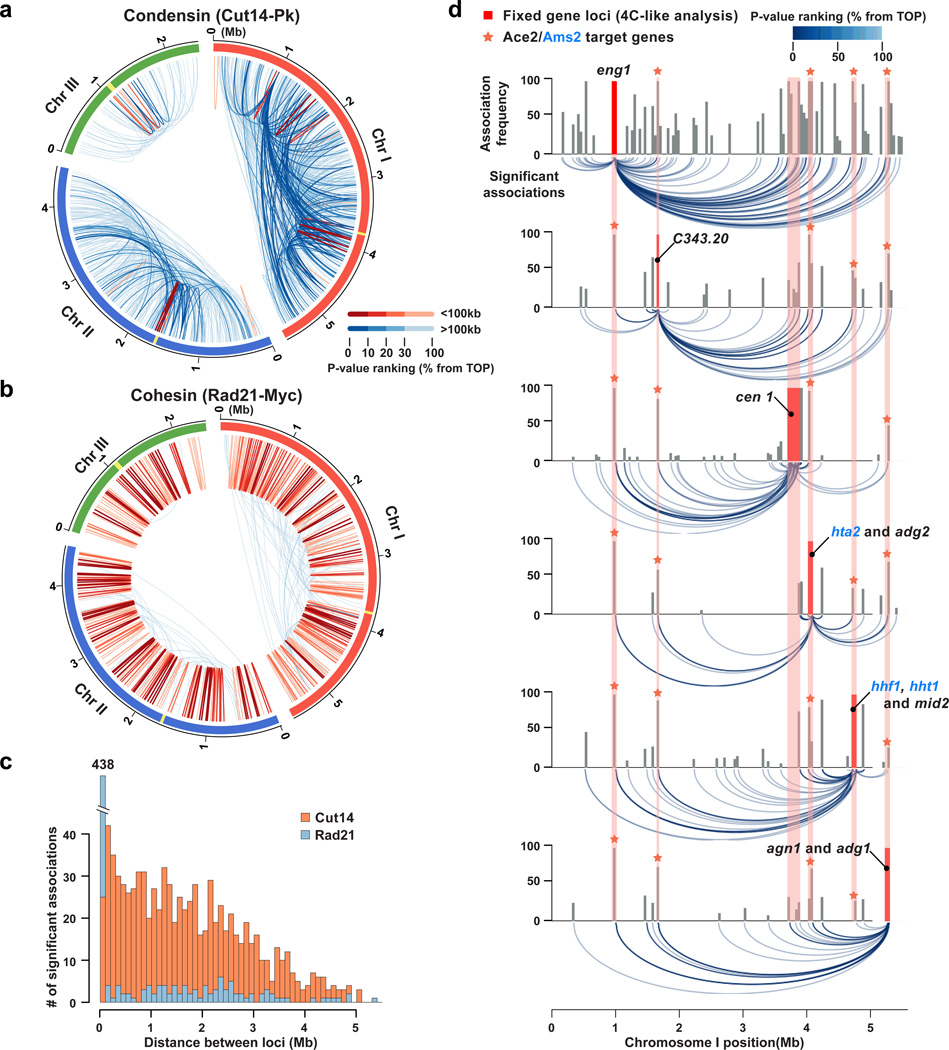
To elucidate how condensin and cohesin build association networks across the genome, we focused on significant associations among association spots. The recently developed algorithm, Mango, predicts association spots and significance of the observed associations by considering the distance-dependent background association frequency49,50. Note that the association spots predicted by the Mango algorithm and the condensin binding sites estimated based on self-ligation reads (Fig. 1a; Supplementary Fig. 1a) overlapped heavily, but association spots generally included relatively weak binding sites: every condensin and cohesin binding site, except for one condensin site, was included in association spots. Condensin and cohesin association spots were 1554 and 1340, and 905 and 805 significant associations between association spots were mediated by condensin and cohesin, respectively (Supplementary Table 1). Using this algorithm, we found that condensin mainly associated two loci positioned more than 100 kb apart (Fig. 3a). In clear contrast, cohesin-dependent associations were restricted to loci separated by less than 100 kb (Fig. 3b). Condensin- and cohesin-mediated associations appeared to depend upon distances between two loci (Fig. 3c).
Figure 3. Condensin and cohesin mediate different gene associations.
(a,b) Significant associations mediated by condensin (Cut14-PK; a) and cohesin (Rad21-Myc; b). Significant associations were defined by Mango software. Associations were categorized into two groups, < 100 kb and > 100 kb, according to the distance between two loci. Color intensities correlate with P-values of respective associations. The graphs were drawn with circos software. Yellow indicates centromeres.
(c) Frequencies of condensin- and cohesin-mediated significant associations were plotted against distance. Associations were divided into 100 kb bins.
(d) 4C-like plots for Cut14-Pk ChIA-PET data. The Ace2/Ams2 target gene loci (eng1, C343.20, hta2/adg2, hhf1/hht1/mid2, and agn1/adg1) and cen1 were used as baits. The entire chromosome I was divided into non-overlapping 20 kb windows, and numbers of significant associations derived from the bait regions were plotted (top). Significant associations from the bait regions are also shown (bottom).
Regarding the association context, we found that condensin connected loci carrying LTR retrotransposons, Pol III genes (tRNA and 5S rRNA), or highly transcribed Pol II genes to a certain degree (Supplementary Fig. 3a). Condensin-mediated associations involving Ace2/Ams2 target genes consistently occurred with much greater frequencies than associations estimated by the randomized simulation, suggesting that Ace2/Ams2 targets serve as association hot spots (Supplementary Fig. 3a). Among cohesin binding sites, convergent loci preferentially associated together (Supplementary Fig. 3b).
We observed condensin-mediated long-range associations among boundaries and centromeres (Supplementary Fig. 3c). Relatively strong associations were often derived from condensin boundaries (Supplementary Fig. 3d). We previously showed that condensin binding sites are preferentially targeted to centromeres36–38. The condensin ChIA-PET data also demonstrated that many condensin bindings sites dispersed across the genome were connected to centromeres and centromere-proximal regions (Supplementary Fig. 3c). In sharp contrast, cohesin solely mediated short-range associations between cohesin boundaries present at 50–70 kb intervals (Supplementary Fig. 3e).
Since condensin favorably mediates associations of Ace2/Ams2 target genes, we extracted only the associations involving those genes. This 4C–like analysis again revealed associations among the Ace2/Ams2 targets and centromeres (Fig. 3d). These condensin-dependent associations were detected between two loci separated by 3–4 Mb, indicating that condensin can mediate long-range associations.
Mitotic associations of Ace2/Ams2 targets via condensin
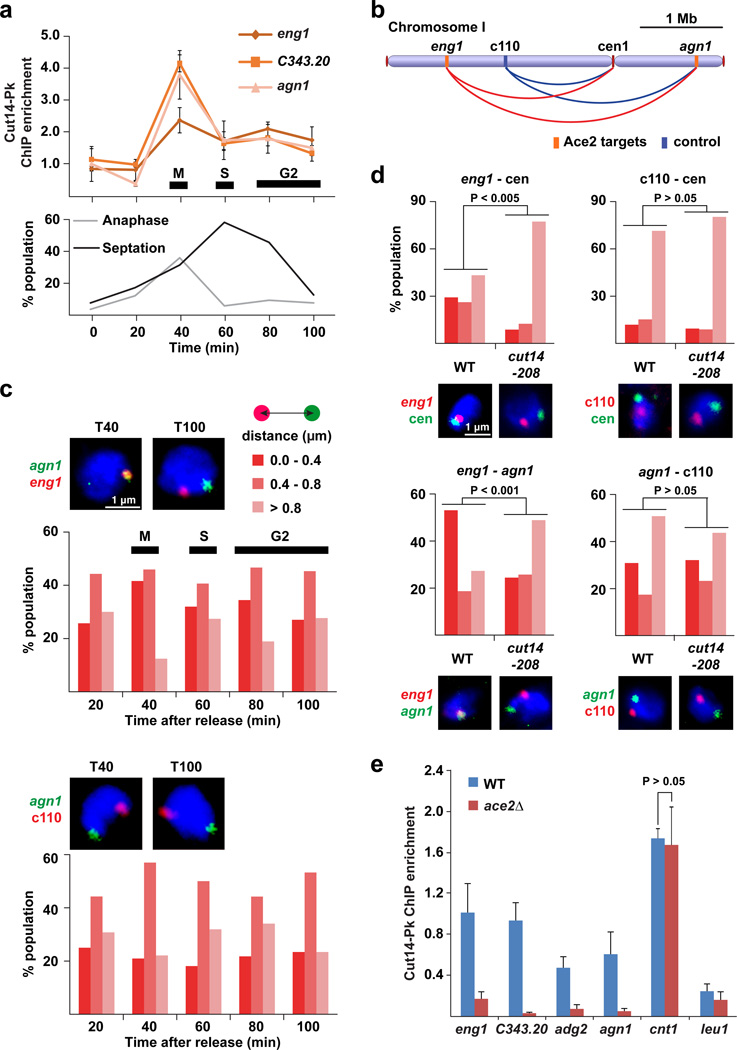
The transcription factors Ace2 and Ams2 are highly expressed during mitosis and up-regulate their target genes during M-G1 and S phases, respectively45. Therefore, we speculated that associations among Ace2/Ams2 target genes and centromeres might also be cell cycle-dependent. We first investigated Cut14 condensin enrichment at Ace2 and Ams2 targets during the cell cycle, and observed that condensin was most enriched at their target genes during mitosis, and that the enrichment decreased as the cell cycle proceeds (Fig. 4a; Supplementary Fig. 4a). We next analyzed associations of Ace2 targets distributed across chromosome I (Fig. 4b) and found that Ace2 targets were best associated with each other during mitosis and to a lesser extent during S and early G2 phases, whereas the association between the Ace2 target (agn1) and the c110 negative control locus was continuously at background level during the cell cycle (Fig. 4c). Likewise, the Ams2 target histone gene (hta1) associated with centromeres to the highest degree during mitosis (Supplementary Fig. 4b). Since condensin-mediated association is a transient event, the association is scored as a frequency of nearby localization of gene loci51.
Figure 4. Condensin-dependent associations of Ace2 target genes.
(a) ChIP result showing Cut14-Pk condensin enrichment at the Ace2 target genes (eng1, C343.20, and agn1) during the cell cycle. The cell cycle was synchronized using the temperature-sensitive cdc25-22 mutation, and monitored by two indexes (binucleated anaphase cells and septation). ChIP enrichment was normalized against the leu1 control locus.
(b) Positions of FISH probes along chromosome I.
(c) FISH analysis co-visualizing the agn1 and eng1 gene loci (top), or the agn1 and c110 loci (bottom). The c110 negative control locus is not bound by condensin. The cell cycle was synchronized as described in panel a. Representative FISH images of mitotic/M (T40) and G2 (T100) cells are shown above the graphs. The distance between two FISH foci was measured in more than 100 cells at the indicated time points and binned into one of the three categories (top right).
(d) FISH analysis visualizing the indicated paired loci in WT and cut14-208 condensin mutant cells during mitosis. Cells were treated by HU for cell-cycle synchronization as explained in Supplementary Fig. 4c. The distance between the paired loci was summarized as described in panel c. Typical FISH images are shown below the graphs.
(e) ChIP result showing enrichment of Cut14-Pk at the Ace2 targets (eng1, C343.20, adg2, and agn1), centromere 1 (cnt1), and leu1 gene (negative control) in WT and ace2Δ cells.
In panels a and e, experiments were independently repeated three times, and data are represented as mean ± SD.
In addition, we observed that the associations of the Ace2 target (eng1) and Ams2 target genes (hht2 and hta1) with centromeres during mitosis were significantly impaired by the cut14-208 condensin mutation (p < 0.005, Mann-Whitney U test), while relative positioning between the c110 negative control locus and centromeres was not affected (Fig. 4d and Supplementary Fig. 4c). Similarly, the mitotic association between the two Ace2 targets (eng1 and agn1) was significantly disrupted by the condensin mutation (p < 0.001, Mann-Whitney U test), while positioning between the agn1 gene and c110 locus was not affected (Fig. 4d). Negative control data indicate that the disruption of gene associations by the condensin mutation did not result from indirect global effects.
Moreover, Cut14 condensin enrichment was reduced in the ace2Δ and ams2Δ mutants at the Ace2 and Ams2 targets, but not at the cnt1 centromeric region of the chromosome I (Fig. 4e and Supplementary Fig. 4d). It has been shown that condensin loading to fission yeast centromeres is compromised by the pcs1Δ monopolin-kinetochore mutation52. We observed that the associations of Ace2 and Ams2 targets with centromeres were significantly impaired by the psc1Δ, which did not affect relative positioning of the c110 control locus and centromeres, suggesting that centromeric condensin loaded by the monopolin is involved in the associations of Ace2 and Ams2 target genes with centromeres (Supplementary Fig. 4e). Together, these results suggest that Ace2 and Ams2 recruit condensin to their target genes most efficiently during mitosis when these transcription factors are highly expressed, and that condensin in turn mediates associations among Ace2/Ams2 targets and centromeres.
Condensin ChIA-PET using mitotic cells
We used asynchronous culture to identify condensin-mediated gene associations (Fig. 3), and our microscopic investigation suggested that those associations are promoted during mitosis (Fig. 4c; Supplementary Fig. 4b). Therefore, we hypothesized that the asynchronous data mainly reflect mitotic associations. To test this hypothesis, we decided to conduct the condensin ChIA-PET analysis with mitotic and G2 cells. We observed that condensin localization patterns in mitosis and G2 cells were in general similar to that in asynchronous cells (Supplementary Fig. 5a), e.g., condensin localization was consistently observed at tRNA and 5S rRNA genes (Supplementary Fig. 5b). Condensin localization at the Ace2/Ams2 targets (mid2 and hta2) was observed in asynchronous and mitotic cells, but not in G2 cells (Supplementary Fig. 5c). In this regard, the nuclear localization of condensin was much enhanced during mitosis compared to interphase, whereas cohesin localized in the nucleus throughout the cell cycle (Supplementary Fig. 5d)53,54.
Comparing condensin domains between asynchronous and mitotic cells, we observed that positions of condensin boundaries in asynchronous and mitotic cells were very similar across the genome (Supplementary Fig. 5e,f). As in asynchronous cells, the distance decay in the condensin ChIA-PET data from mitotic cells was clearly different from that in the cohesin data (Supplementary Fig. 5g); e.g., cohesin-mediated associations were enhanced between flanking gene loci positioned less than 100 kb apart. Condensin ChIA-PET data were in general highly similar between asynchronous and mitotic cells (r=0.962; Supplementary Fig. 5h). Moreover, condensin-mediated significant associations were detected mainly between two loci positioned more than 100 kb apart in both asynchronous and mitotic cells, whereas cohesin-dependent associations were predominantly restricted to loci separated by less than 100 kb (Supplementary Fig. 5i,j). Together, these results strongly suggest that condensin ChIA-PET data from asynchronous cells mainly reflect domain organization and gene associations during mitosis.
Ace2 binding sites form domain boundaries
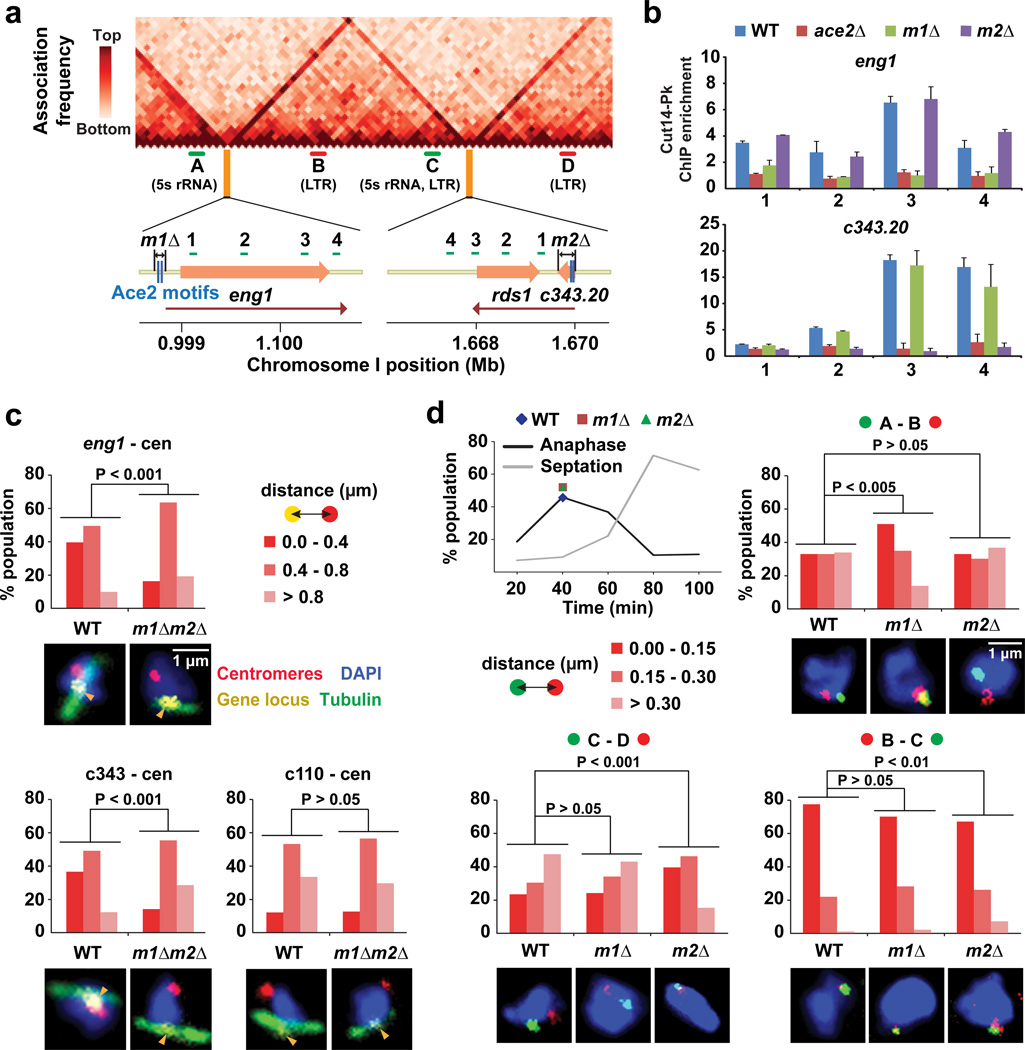
We posited that the two Ace2 target genes, eng1 and C343.20, form domain boundaries (Fig. 5a). To test this hypothesis, we deleted the two Ace2 binding motifs from the eng1 gene and the entire C343.20 gene containing the two motifs; these deletions we refer to as m1Δ and m2Δ (Fig. 5a). We observed that condensin enrichment around the eng1 and C343.20 genes were clearly diminished in m1Δ and m2Δ cells, respectively, as well as in ace2Δ cells, suggesting that Ace2 recruits condensin to its target genes (Fig. 5b). Consistently, the condensin-mediated associations of the eng1 and C343.20 genes with centromeres were significantly disrupted by the m1Δm2Δ double deletions (p < 0.001, Mann-Whitney U test; Fig. 5c). Note that relative positioning of the c110 control locus and centromeres was not affected by the deletions (Fig. 5c).
Figure 5. Ace2 target sites form domain boundaries.
(a) Deletions of the Ace2 binding sites. The m1Δ removes the two Ace2 binding motifs from the eng1 gene promoter, while the m2Δ deletes the C343.20 gene. Red lines indicate transcribed regions.
(b) ChIP results showing enrichment of condensin (Cut14-Pk) around the eng1 and C343.20 genes in the WT, ace2Δ, m1Δ, and m2Δ strains. Primer positions are indicated in panel a. ChIP enrichment was normalized against the leu1 control locus. Experiments were independently repeated three times, and data are represented as mean ± SD.
(c) FISH-IF data summarizing distances between the indicated gene loci and centromeres in the WT and m1Δm2Δ double-deletion strains. Cells were treated by HU for the cell-cycle synchronization. Mitotic cells were defined by spindle staining (tubulin), and the distance between two FISH foci was measured in more than 50 mitotic cells and binned into one of the three categories (top right). Typical FISH images are shown below the graphs.
(d) FISH analysis was performed to visualize the indicated loci (A, B, C, and D in panel a) during mitosis in the WT, m1Δ, and m2Δ strains. The regions between these loci are consistently ~350 kb. Mitotic cells were prepared as described in Fig. 4a (40 minutes after cell-cycle release; top left chart). The distance between two FISH foci was measured in more than 100 cells and binned into one of the three categories (middle left). Typical FISH images are shown below the graphs.
We next asked whether the eng1 and C343.20 genes function as domain boundaries. For this purpose, we investigated how the m1Δ and m2Δ impact intra- and inter-domain associations. The m1Δ, but not the m2Δ, significantly enhanced the inter-domain associations between the A and B loci bound by condensin (p < 0.001, Mann-Whitney U test; Fig. 5d). Differently, the m2Δ specifically increased the inter-domain associations between the C and D loci. Both the m1Δ and m2Δ slightly reduced the intra-domain association between the B and C loci. These microscopic observations together with the condensin ChIA-PET data strongly suggest that the two Ace2 binding sites serve as domain boundaries to prevent inter-domain associations.
Mitotic transcription factors recruit condensin
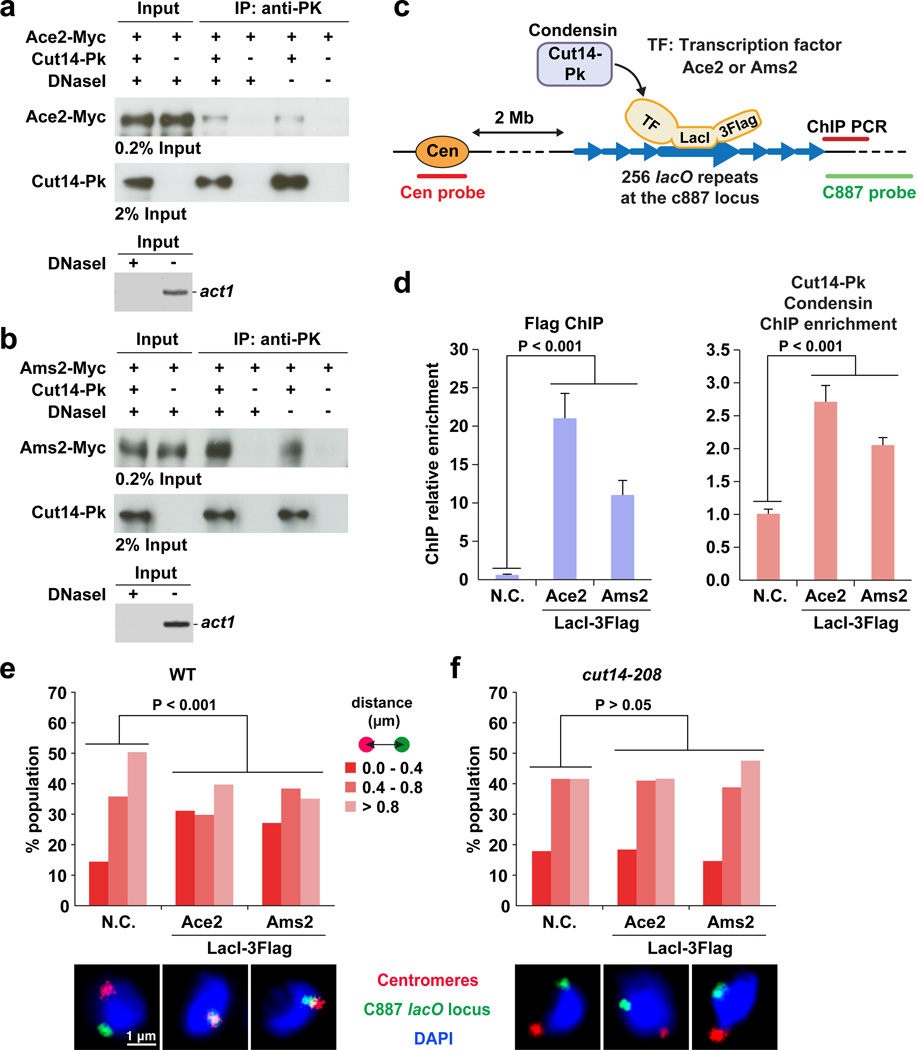
Since it was still not clear whether Ace2 and Ams2 play a direct role in recruiting of condensin to their targets, we performed co-immunoprecipitation (co-IP) experiments and found that condensin (Cut14-Pk) interacts with Ace2 and Ams2, and that these interactions remained even after DNase I treatment (Fig. 6a,b).
Figure 6. Condensin recruited by Ace2 and Ams2 associates a gene locus with centromeres.
(a) Co-IP result showing the protein interaction between the Cut14-Pk and Ace2-Myc. Epitope-tagged proteins were expressed from the endogenous gene loci with their own promoters. Co-IP assay was performed with and without DNase I treatment. DNA digestion was verified by a PCR-based assay to detect act1 gene region as a control (bottom panel).
(b) Co-IP result showing the interaction between the Cut14-Pk and Ams2-Myc.
(c) A schematic explanation of the tethering assay.
(d) Tethering of Ace2- or Ams2-LacI-3Flag is sufficient to recruit condensin to the lacO locus. Cells expressing Ace2 or Ams2 fused to LacI-3Flag were subjected to ChIP experiments to detect LacI-3Flag fusion proteins (left) and the Cut14-Pk condensin subunit (right) at the lacO repeats-flanking region. As a negative control (N.C.), cells carrying an empty vector were used for the same analysis. Experiments were independently repeated three times, and data are represented as mean ± SD.
(e) The c887 lacO locus frequently localizes near centromeres in cells expressing Ace2- or Ams2-LacI-3Flag. The distance between the lacO locus and centromeres was measured in more than 100 cells and binned into one of the three categories. Representative FISH images are shown below the graph.
(f) The same FISH analysis as described in panel e was performed with the cut14-208 condensin mutant. The cut14-208 mutant was cultured at the restrictive temperature (36°C) for 1 hour.
We employed a system to test the hypothesis that tethering of Ace2 and Ams2 could result in the recruitment of condensin to a chromatin locus (Fig. 6c)55. Specifically, lacO repeats were integrated to the c887 locus, where no significant condensin binding peaks were observed in our previous ChIP-seq data37,51. We also made strains expressing either Ace2- or Ams2-LacI-3Flag. It had previously been shown that the condensin loading process could be validated by the same approach38. After expressing Ace2- or Ams2-LacI-3Flag, we first observed, using ChIP, that Ace2- and Ams2-LacI-3Flag were enriched at the lacO locus, indicating that these LacI-tagged proteins were successfully tethered to the lacO repeats (Fig. 6d). We then found that tethering of Ace2- and Ams2-LacI-3Flag resulted in Cut14 condensin becoming enriched at the lacO locus, firmly establishing that Ace2 and Ams2 play an important role in recruiting of condensin to a gene locus (Fig. 6d).
Using the tethering system we also examined the association of the lacO locus with centromeres by FISH analysis (Fig. 6e). We found that when Ace2- or Ams2-LacI was expressed, the lacO locus was frequently located near centromeres, and that the localization patterns in cells with and without expression of the LacI-fused proteins were significantly different (p < 0.001, Mann-Whitney U test; Fig. 6e). This co-localization of the lacO locus with centromeres by expression of the Ace2/Ams2-LacI was not observed in the cut14-208 condensin mutant, indicating that the Ace2/Ams2-mediated associations of the locus with centromeres are dependent upon condensin (Fig. 6f).
Mitotic defects caused by ace2 Δ and boundary deletions
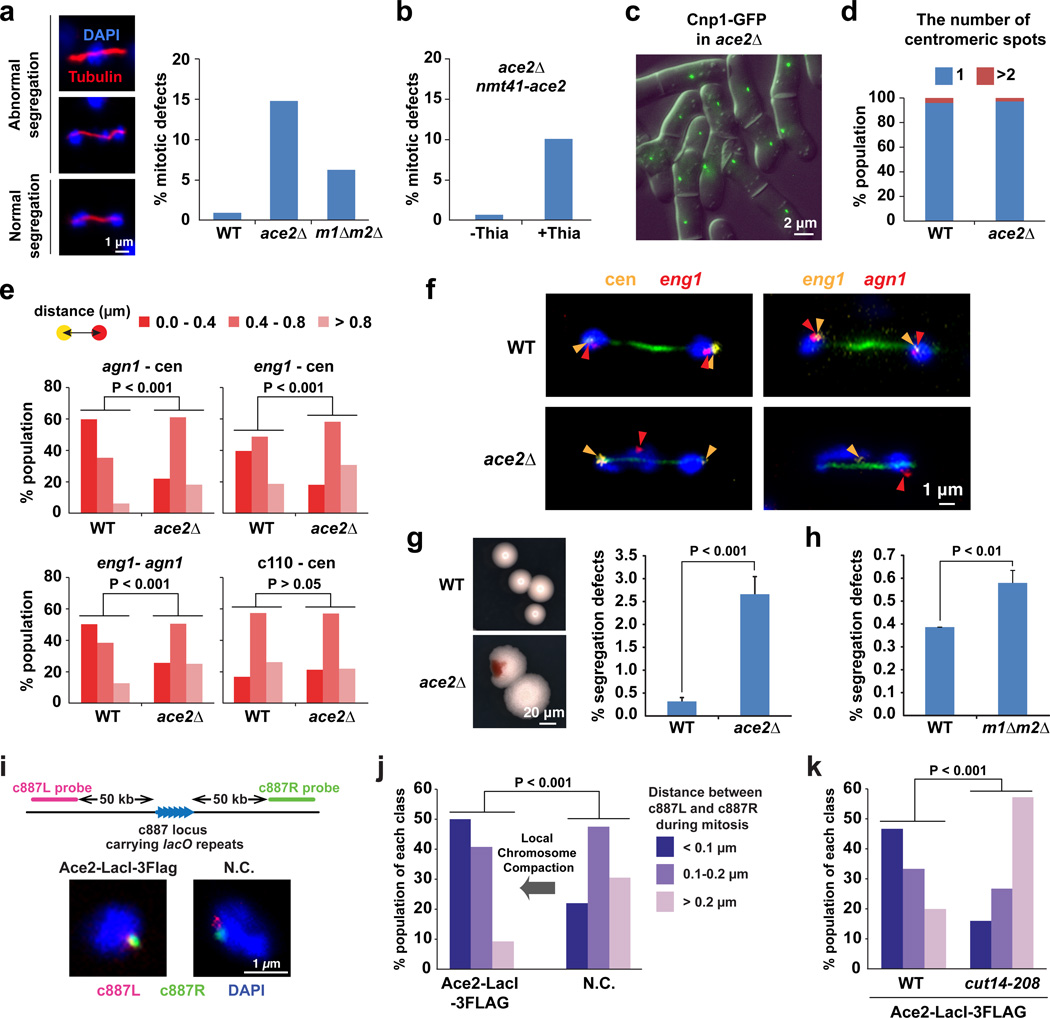
We next investigated functions of Ace2-dependent gene associations in mitotic chromosome organization and segregation. Mitotic defects, including lagging chromosomes, were observed in ace2Δ and m1Δm2Δ cells, but not in WT cells (Fig. 7a). Note that the m1Δm2Δ double deletions remove the two Ace2 binding sites (eng1 and C343.20), which serve as strong association hot spots and as domain boundaries (Fig. 3d; Fig. 5). The mitotic defects in ace2Δ cells were suppressed by exogenous expression of Ace2 proteins (Fig. 7b). Since Ams2 is involved in loading of CENP-A (centromeric histone H3 variant) to centromeres and thereby chromosome segregation, it is difficult to assess the mitotic roles of Ams2-dependent gene associations56. In contrast, we observed that CENP-A loading and centromeric clustering were not affected by the ace2Δ mutation (Fig. 7c,d). During mitosis, in ace2Δ cells, the centromeric associations of the Ace2 target genes (ang1 and eng1) and the association between the Ace2 target genes were significantly disrupted (p < 0.001, Mann-Whitney U test), whereas positioning between the c110 negative control locus and centromeres was not affected (Fig. 7e). In WT anaphase cells, the Ace2 target genes associated together, and centromeres associating with the Ace2 target gene locus were pulled by the spindle microtubules and the mitotic chromosomes were properly segregated (Fig. 7f). In contrast, in the ace2Δ mutant, the associations among the Ace2 target genes and centromeres were disrupted, while centromeres were still pulled by the microtubules, causing a defect in mitotic chromosome segregation (Fig. 7f). Re-examining the fidelity of chromosome segregation in the ace2Δ mutant using the Chr16 mini-chromosome system57, we found that the frequency of mini-chromosome loss was roughly 10-times more elevated in the ace2Δ mutant compared to WT (Fig. 7g). Since the mini-chromosome does not consist of Ace2 target genes, the observed mini-chromosome loss in ace2Δ cells likely accompanies segregation defects of other chromosomes. Moreover, we observed modest, but significant, defects in mitotic chromosome segregation when the two Ace2 target sites were removed by the m1Δm2Δ double deletions (Fig. 7h). These results collectively suggest that associations among Ace2 target genes and centromeres, and potentially domain organization, contribute to the faithful segregation of mitotic chromosomes.
Figure 7. Mitotic defects caused by the ace2Δ and the boundary deletions.
(a) Rates of mitotic defects in WT, ace2Δ, and m1Δm2Δ double-deletion cells (right). IF images show abnormal chromosome segregation in ace2Δ cells (left).
(b) Mitotic defects in ace2Δ cells are suppressed by the exogenous expression of the ace2 gene. The ace2 gene is under control of the nmt41 promoter, which is activated by the absence of thiamine (-Thia) in culture medium.
(c) Centromeric localization of CENP-A (Cnp1 in fission yeast) was not disrupted by ace2Δ.
(d) FISH was performed to visualize the three centromeres.
(e) FISH-IF data summarizing distances between the indicated gene loci in WT and ace2Δ mitotic cells.
(f) Representative FISH-IF images showing the indicated gene loci (yellow and red) and tubulin (green) in WT and ace2Δ cells during mitosis.
(g,h) Effect of ace2Δ and the boundary deletions (m1Δm2Δ) on Chr16 mini-chromosome stability. This experiment was performed as described in Supplementary Note. Experiments were independently repeated three times: n > 500 for each time. Data are represented as mean ± SD.
(i) FISH data showing the c887L (red) and c887R (green) regions in mitotic cells expressing Ace2-LacI-3Flag or carrying an empty vector (N.C.).
(j) The distance between the c887L and c887R regions was measured in more than 50 mitotic cells, and binned into one of the three categories (right).
(k) The same analysis as described in panel j was performed with the cut14-208 condensin mutant.
Using the tethering system, we examined local chromosomal compaction around the c887 lacO locus in mitotic cells (Fig. 7i). We observed that the two foci representing the c887L and c887R regions completely overlapped in approximately 50% of mitotic cells when Ace2-LacI was expressed (Fig. 7i,j). In contrast, we primarily observed two separate foci when the LacI-fused proteins were not expressed, indicating that the local chromosome compaction around the lacO locus was significantly facilitated by the expression of Ace2-LacI proteins (p < 0.001, Mann-Whitney U test; Fig. 7j). The local compaction mediated by Ace2-LacI was diminished in the cut14-208 condensin mutant, indicating that this compaction process is mediated by condensin (Fig. 7k). Since Ace2-LacI recruits condensin to the lacO locus (Fig. 6d) and facilitates local chromosomal compaction during mitosis, local compaction is mediated by condensin molecules recruited by Ace2 and participates in mitotic chromosome assembly (mechanisms detailed in discussion).
DISCUSSION
In this study, we find that both the condensin and the cohesin complexes frequently bind to the same loci including tRNA and 5S rRNA genes, LTR retrotransposons, and highly expressed Pol II genes. Only condensin – but not cohesin – additionally localizes at the cell cycle-regulated genes bound by the transcription factors Ace2 and Ams2. It has been shown by a ChIP-seq approach that condensin is enriched at mitotically activated genes58,59. However, our present study indicates that condensin is preferentially targeted to specific genes bound by these mitotic transcription factors. Moreover, we now show that Ace2 and Ams2 interact with condensin and recruit it to chromatin (Fig. 6). Since Ace2 and Ams2 bind only to their target genes, these transcription factors allow condensin to bind to specific genes, although other factors may interact with Ace2 and Ams2 to help recruit condensin to their targets.
Even though condensin and cohesin frequently bind to the same loci, they drive distinct genome-wide association networks. Cohesin-dependent associations are mostly restricted to loci positioned within 100 kb, while condensin can associate gene loci separated by more than 100 kb (Fig. 3a,b). The condensin-mediated associations among Ace2/Ams2 target genes and centromeres occur at loci positioned beyond 3–4 Mb, indicating that condensin can mediate long-range associations. Cohesin and condensin form on average 50–100 and 300–500 kb chromatin domains, respectively, and domain boundaries are occupied by these complexes (Fig. 2).
TAD organization is observed among eukaryotic species and linked to transcriptional co-regulation and replicating timing coordination6–8,15,16. Condensin has recently been implicated in TAD organization, and the work here supports this finding by demonstrating that fission yeast condensin is involved in forming chromatin domains that are similar to the mammalian TADs in size, ranging from 100 kb to 1 Mb14,17. We have observed that the mitotic domain organization is disrupted by the deletions of Ace2 binding sites, demonstrating that this mitotic transcription factor participates in the cell cycle-dependent arrangement of chromatin domains.
Using the LacI tethering system, this study indicates that condensin is recruited by Ace2 and mediates both the association of a gene locus with centromeres and local chromosomal compaction during mitosis. Condensin is targeted to centromeres through a mechanism involving kinetochore proteins52,60. Condensin is known to interact with another condensin molecule to mediate associations between chromatin fibers61,62. Therefore, condensin molecules present at centromeres and Ace2 target genes can mediate their associations through condensin-condensin interactions.
It has also been shown that the condensin complex interacts with another condensin in the presence of DNA and introduces positive supercoiling into the chromatin loop61,63. We thus hypothesize that chromatin loops generated by associations among gene loci and centromeres are supercoiled by condensin, leading to local chromosomal compaction. This looped and supercoiled chromosome arrangement contributes to the efficient transmission of physical force at the kinetochore to the chromosomal arms, improving the fidelity of chromosome segregation. Therefore, when gene associations are disrupted by the ace2Δ mutation, Ace2 target genes no longer associate with centromeres, but centromeres still segregate without holding the Ace2 target genes, leading to chromosomal segregation defects (Fig. 7f). We propose that the mitotic transcription factors interact with and recruit condensin to mitotic chromosomes and participate in the mitotic chromosome organization required for the fidelity of chromosome segregation.
ONLINE METHODS
Strains and culture conditions
Cut14, Rad21, Ace2, Ams2, and Sep1 proteins were tagged with Myc, Flag, or Pk at the C-termini of their proteins using a PCR-based module method64,65. Tagged proteins were expressed from their endogenous gene loci with their own promoters. Strain constructions were performed using conventional genetic crosses. The fission yeast Schizosaccharomyces pombe strains were cultured in Yeast-Extract Adenine (YEA) or Edinburgh Minimal Medium (EMM).
ChIA-PET
Chromatin was fixed with 3% paraformaldehyde followed by further cross-linking with 10 mM dimethyl adipimidate (DMA). Crosslinked chromatin was sheared into 300–500 bp DNA fragments by Bioruptor (Diagenode). Cut14-Pk and Rad21-Myc proteins were immunoprecipitated using mouse monoclonal anti-PK (SV5-Pk1, Serotec) or mouse monoclonal anti-Myc (9E10, Clontech), and protein G-coupled Dynabeads (Life Technologies). Immunoprecipitated chromatin was subjected to the ChIA-PET procedure20. ChIP DNA was split into an equal amount and separately ligated to either A or B linkers, and mixed ligation products were further subjected to proximity ligation. The ligation product was digested by EcoP15I and purified using streptavidin-coupled magnetic beads (Invitrogen). Illumina sequencing adaptors were added to DNA fragments, which were then subjected to PCR amplification (Phusion PCR Master Mix; New England Biolabs). This PCR was performed in a total of 32 separate tubes for each ChIA-PET library. Assembled PCR products were sequenced on the Illumina HiSeq 2000 platform to obtain 100 bp single end reads.
Supplementary Material
Acknowledgments
We would like to thank the Wistar Institute Genomics and Bioinformatics Facilities for high-throughput sequencing and genomic data analyses; the Imaging Facility for microscopic analysis; and the Yeast Genetic Resource Center (YGRC) for fission yeast strains. We also thank Drs. Louise Showe, Paul Lieberman, and Rachel Locke for critically reading the manuscript, and Sylvie Shaffer for editorial assistance. This work was supported by the G. Harold & Leila Y. Mathers Charitable Foundation and the NIH Director’s New Innovator Award Program of the National Institutes of Health under award number [DP2-OD004348 to K.N.]. Support for Shared Resources utilized in this study was provided by Cancer Center Support Grant (CCSG) P30CA010815 to the Wistar Institute.
Footnotes
URLs
Bähler lab homepage, http://128.40.79.33/projects/cellcycle/; Mango software, https://github.com/dphansti/mango; circos software, http://circos.ca/software/download/circos/.
ACCESSION NUMBERS
Data described in this paper were deposited into the National Center for Biotechnology Information Sequence Read Archive database with accession number SRP061635.
AUTHOR CONTRIBUTIONS
H.T. performed the bioinformatics analyses. O.I. performed the tethering assays. K-D.K. performed the ChIA-PET and other experiments. K.N. conceived and designed the study. All authors contributed to the data analyses and the writing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodley CD, Bertels F, Jones B, O’Sullivan JM. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet. Biol. 2009;46:879–886. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanizawa H, et al. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le TB, Imakaev MV, Mirny LA, Laub MT. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizuguchi T, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516:432–435. doi: 10.1038/nature13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grob S, Schmid MW, Grossniklaus U. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell. 2014;55:678–693. doi: 10.1016/j.molcel.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Feng S, et al. Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell. 2014;55:694–707. doi: 10.1016/j.molcel.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fullwood MJ, Han Y, Wei CL, Ruan X, Ruan Y. Chromatin interaction analysis using paired-end tag sequencing. Chapter 21,Unit 21 15 1–25. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb2115s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeMare LE, et al. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji X, et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nativio R, et al. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol. Cell Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwasaki O, Tanaka A, Tanizawa H, Grewal SI, Noma K. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell. 2010;21:254–265. doi: 10.1091/mbc.E09-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka A, et al. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol. Cell. 2012;48:532–546. doi: 10.1016/j.molcel.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki O, et al. Interaction between TBP and condensin drives the organization and faithful segregation of mitotic chromosomes. Mol. Cell. 2015;59:755–767. doi: 10.1016/j.molcel.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt CK, Brookes N, Uhlmann F. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 2009;10:R52. doi: 10.1186/gb-2009-10-5-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dowen JM, et al. Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Reports. 2013;1:371–378. doi: 10.1016/j.stemcr.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Bortle K, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lengronne A, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Ambrosio C, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 45.Rustici G, et al. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- 46.Takayama Y, Takahashi K. Differential regulation of repeated histone genes during the fission yeast cell cycle. Nucleic Acids Res. 2007;35:3223–3237. doi: 10.1093/nar/gkm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso-Nunez ML, et al. Ace2p controls the expression of genes required for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell. 2005;16:2003–2017. doi: 10.1091/mbc.E04-06-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petit CS, Mehta S, Roberts RH, Gould KL. Ace2p contributes to fission yeast septin ring assembly by regulating mid2+ expression. J. Cell Sci. 2005;118:5731–5742. doi: 10.1242/jcs.02687. [DOI] [PubMed] [Google Scholar]
- 49.Phanstiel DH, Boyle AP, Heidari N, Snyder MP. Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics. 2015;31:3092–3098. doi: 10.1093/bioinformatics/btv336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heidari N, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim KD, et al. Centromeric motion facilitates the mobility of interphase genomic regions in fission yeast. J. Cell Sci. 2013;126:5271–5283. doi: 10.1242/jcs.133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 53.Sutani T, et al. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamazaki H, Tarumoto Y, Ishikawa F. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012;26:241–246. doi: 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell. 2003;11:175–187. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 57.Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 58.Nakazawa N, et al. RNA pol II transcript abundance controls condensin accumulation at mitotically up-regulated and heat-shock-inducible genes in fission yeast. Genes Cells. 2015;20:481–499. doi: 10.1111/gtc.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sutani T, et al. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat. Commun. 2015;6:7815. doi: 10.1038/ncomms8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakazawa N, et al. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshimura SH, et al. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
- 63.Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- 64.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 65.Gadaleta MC, Iwasaki O, Noguchi C, Noma K, Noguchi E. New vectors for epitope tagging and gene disruption in Schizosaccharomyces pombe. Biotechniques. 2013;55:257–263. doi: 10.2144/000114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.