Abstract
Left ventricular non-compaction (LVNC) is the third most prevalent cardiomyopathy in children and its pathogenesis has been associated with the developmental defect of the embryonic myocardium. We show that patient-specific induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) generated from LVNC patients carrying a mutation in the cardiac transcription factor TBX20 recapitulate a key aspect of the pathological phenotype at the single-cell level and was associated with perturbed transforming growth factor beta (TGFβ) signaling. LVNC iPSC-CMs have decreased proliferative capacity due to abnormal activation of TGFβ signaling. TBX20 regulates the expression of TGFβ signaling modifiers including a known genetic cause of LVNC, PRDM16, and genome editing of PRDM16 caused proliferation defects in iPSC-CMs. Inhibition of TGFβ signaling and genome correction of the TBX20 mutation were sufficient to reverse the disease phenotype. Our study demonstrates that iPSC-CMs are a useful tool for the exploration of pathological mechanisms underlying poorly understood cardiomyopathies including LVNC.
Introduction
Left ventricular non-compaction (LVNC) is increasingly recognized as a cause of cardiomyopathy1, 2, especially in children. In a recent study, LVNC accounted for 9.2% of all children with primary cardiomyopathies, and was the third most prevalent form of cardiomyopathy, after dilated cardiomyopathy (DCM) and hypertrophic cardiomyopathy (HCM)2. LVNC is characterized by deep and extensive hypertrabeculation of the left ventricle, and causes heart failure, arrhythmias, and thromboembolism.
LVNC has been theorized to result from the arrest of compaction of the developing LV myocardium, as it passes through several distinct evolutionally conserved steps. Trabeculations in the human embryo emerge after looping of the primitive heart tube at the end of the fourth week of gestation3. Trabecular remodeling begins at 8 weeks with an increase in LV volume compressing the trabeculations, leading to an increase in thickness of the compacted myocardium. Serial pathologic studies suggest that LVNC arises from impaired/arrested compaction of the myocardium, abnormalities of vascularization, or in development of the multilayered spiral system3, 4. Among these steps, emergence of trabeculations and trabecular remodeling are thought to be the key steps to understanding LVNC. The trabeculation patterns are ventricle-specific, which are generally thicker and the corresponding intertrabecular spaces are larger in the LV than in the right ventricle. When this embryonic pattern persists postnatally, the morphologic appearance strongly resembles the embryonic “spongiform” myocardium, which was the original nomenclature for this cardiomyopathy.
Like many congenital cardiomyopathies, the genetics of LVNC is complex and the full spectrum of the disorder is still undefined. The mechanisms that lead to LVNC are not well understood, although animal models of LVNC have suggested that abnormal regulation of growth signals, including the transforming growth factor beta (TGFβ)5–9, NOTCH, and NRG1/ERBB210, 11, may be causative factors. Since most of these animal models harboring non-compaction-like myocardium showed alterations in cell cycle regulation in developing cardiomyocytes, it is thought that the abnormal proliferation of embryonic cardiomyocytes may be associated with the pathogenesis of LVNC. However, studies have differed on whether this proliferation is increased or decreased5–7, 9. Furthermore, recent human studies have identified mutations in genes which are associated with regulation of cardiomyocyte proliferation11, 12. However, it is still unclear which phenotypes in developing cardiomyocytes are actually associated with the pathogenesis seen in humans and investigation of this disease has been challenging due to its complex genetic basis.
To overcome the problems for the investigation of human cardiac cell development with pathological background of LVNC, we used patient-specific induced pluripotent stem cells (iPSCs). Here we demonstrated the use of human iPSC-derived cardiomyocytes (iPSC-CMs) from patients carrying the TBX20 mutation affected by LVNC as a model to define cell-specific phenotypes and elucidate potential mechanisms of this disease.
Results
TBX20 mutation is a candidate genetic cause of LVNC
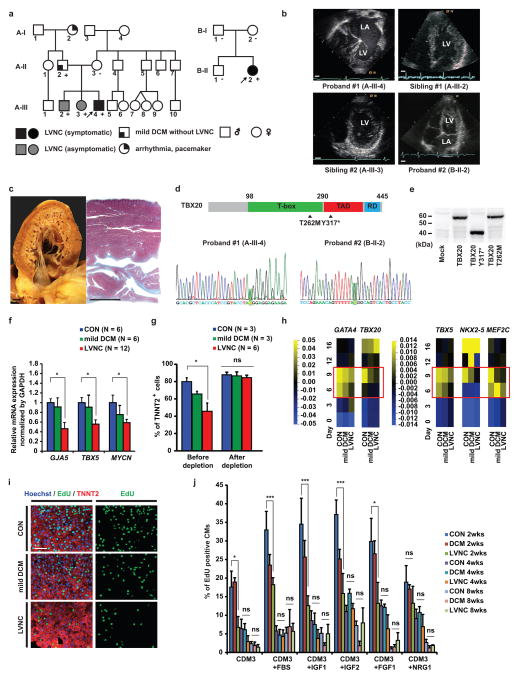
To identify potential genetic causes of LVNC, we recruited a family with LVNC including the proband #1 (A-III-4), who had undergone heart transplantation for restrictive physiology, two siblings (A-III-2 and A-III-3) with significantly deeper and more extensive trabeculation of the left ventricle (a form-fruste of LVNC referred to clinically as “hypertrabeculation”) but with normal systolic function, and the father (A-II-2) with asymptomatic dilated cardiomyopathy (DCM) without LVNC (Figs. 1a–c and Supplementary Table 1). Genetic testing by genome-wide exome sequencing revealed a stop-gain mutation in the TBX20 gene (Y317*) in the proband #1, two siblings, and father (Fig. 1d). No mutations in maternally-transmitted and de novo modifiers known to contribute to cardiomyopathies were detected (Supplementary Table 2). To investigate whether TBX20 mutations are seen in other LVNC patients, we performed genetic testing in an additional 77 LVNC patients and detected another de novo mutation (T262M) from one additional isolated LVNC patient (proband #2: B-II-2) (Figs. 1a, 1b, 1d and Supplementary Table 1).
Figure 1. Characterization of patient-specific LVNC iPSC-CMs carrying TBX20 mutation.
a, Schematic pedigree of two families with LVNC. The probands are indicated by arrow (A-III-4 and B-II-2). “+” and “−” signs indicate presence and absence of the TBX20 Y317* mutation in the family A and T262M mutation in the family B, respectively. b, LVNC phenotype of the proband #1 (A-III-4), two siblings (A-III-2 and A-III-3), and an isolated proband #2 (B-II-2) are assessed by echocardiography. LA, left atrium; LV, left ventricle. Scale bars, 1 cm. c, The proband #1’s explanted heart (left) and Masson’s trichrome staining of the left ventricle (right). Scale bars, 1 cm. d, Schema of TBX20 and position of Y317* and T262M mutations (Upper). Confirmation of the Y317* (c. 951C>A) and T262M (c. 785C>T) mutation on the TBX20 gene (highlighted in green) (Lower). TAD, transactivation domain; RD repression domain. e, Western blot of FLAG tagged wild-type, Y317* and T262M TBX20 mutant protein overexpressed in HEK293 cells. f, Significant downregulation of TBX20 downstream target gene mRNA expression in LVNC iPSC-CMs. g, The efficiency of cardiac differentiation of patient-specific iPSC lines validated by FACS sorting. h, Heat map showed mRNA expression of cardiac transcription factors in iPSCs from day 0 to day 16 after induction of cardiac differentiation. The LVNC iPSCs showed significant decrease of cardiac transcription factors in day 6 and day 9 (red boxes). n=6 independent experiments. Mean=0. i, Immunostaining of nuclear (blue), TNNT2 (red), and EdU (green) in iPSC-CMs at 2 weeks. Scale bars, 100 μm. j, Percentage of EdU+ cardiomyocytes in control, mild DCM, and LVNC iPSC-CMs with or without serum (n=10, 10 and 30 for CON, mild DCM and LVNC independent experiments respectively at 2 weeks; n=4 independent experiments per each group at 4 and 8 weeks) or with growth factors (n=4 independent experiments per each group at 2, 4, and 8 weeks). CON, unrelated controls. *p < 0.05, ***p < 0.005; ns, not significant in one-way ANOVA followed by Tukey post hoc test. The bar graphs show the mean and error bars represent s.e.m. Statistics source data can be found in Supplementary Table 12. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
TBX20 is an essential cardiac transcription factor that regulates cardiomyocyte differentiation and proliferation13, 14. TBX20 has also been described in conjunction with congenital heart disease15–17 and DCM18, 19. The TBX20 Y317* mutation resulted in a truncated protein (Fig. 1e). TBX20 Y317* and T262M mutant proteins showed disturbed synergistic activity with other cardiac transcription factors, including NKX2-5, GATA4, and TBX5 (Supplementary Figs. 1a and 1b). Furthermore, both TBX20 Y317* and T262M mutations were found to impair the negative transcriptional regulatory function of TBX20 compared to wild-type (Supplementary Fig. 1c). These results suggest that LVNC-associated mutations caused disturbance of transcriptional regulation by TBX20.
LVNC iPSC-CMs possess a defect of proliferative capacity
To investigate the pathological mechanism of LVNC, we generated iPSCs using Sendai virus reprogramming from a family with LVNC history along with three unrelated control volunteers20, 21; two clones per patient were established (Supplementary Figs. 1d–f and Supplementary Table 3). We classed all lines into three subgroups: unrelated control, mild DCM (A-II-2), and LVNC (A-III-2, 3, 4). iPSCs were differentiated into cardiomyocytes (iPSC-CMs)22, followed by glucose deprivation to enrich for iPSC-CMs (Supplementary Fig. 2a)23. Although LVNC iPSC-CMs were similar to control iPSC-CMs in terms of structural and electrophysiological phenotype as well as sarcomeric gene expression at 2–4 weeks, LVNC iPSC-CMs showed approximately 50% reduction of the expression of TBX20 downstream target genes compared with control iPSC-CMs (Fig. 1f, Supplementary Figs. 2b–j and Supplementary Table 4). Furthermore, the differentiation efficiency was also halved in LVNC iPSC lines compared to the control prior to glucose deprivation (Fig. 1g). On the other hand, mild DCM iPSC-CMs showed intermediate defects in TBX20 target gene expressions and differentiation efficiency between LVNC and control iPSC-CMs. Despite comparable expression of mesodermal markers including MESP1 and brachyury (T) across all lines during cardiac differentiation (Supplementary Fig. 2k), LVNC iPSCs had significantly lower expression of cardiac transcription factors compared to control cell lines especially between day 6 and 9 (Fig. 1h and Supplementary Fig. 3a). These results suggest an impaired induction of cardiac lineage from mesodermal progenitors in LVNC iPSCs.
Since past human and mouse studies have shown possible association between cell-cycle defects in developing cardiomyocytes and pathogenesis of LVNC5–7, 12, 24, we next assessed the proliferative potential in iPSC-CMs. We found that iPSC-CMs were responsive to growth factors and had increased numbers within 3 weeks after induction of differentiation although their growth was temporary (Supplementary Fig. 3b). In order to assess the proliferation potential in developing iPSC-CMs, distribution of S-phase cardiomyocytes was validated by EdU incorporation with or without stimulation of serum or growth factors. LVNC iPSC-CMs showed reduced baseline proliferative capacity by ~50% without any stimulation as well as in the presence of serum and growth factors compared to control cells at an earlier time point (2 weeks), whereas mild DCM iPSC-CMs showed a milder reduction of ~30% compared to control (Figs. 1i and 1j). On the other hand, undifferentiated iPSCs showed no significant difference in their growth speed (Supplementary Fig. 3c), suggesting the proliferation defect may be characteristic in differentiated cardiac cells.
Abnormal activation of TGFβ signaling is associated with proliferation defect in LVNC iPSC-CMs
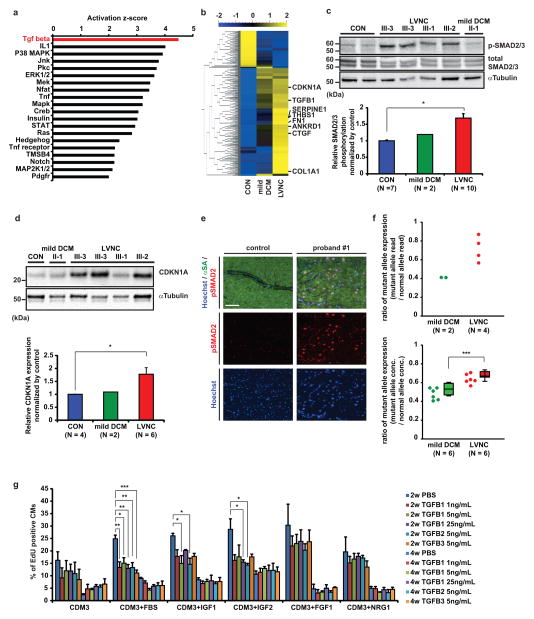
To clarify potential signaling pathways associated with the proliferation defect in LVNC iPSC-CMs, we next performed RNA-sequencing using control, mild DCM, and LVNC iPSC-CMs at 2 weeks. Upstream regulator analysis predicted that activation of TGFβ signaling may be potentially responsible for this differential gene expression between control and LVNC iPSC-CMs (Fig. 2a, Supplementary Fig. 4a and Supplementary Table 5). Most of the TGFβ signaling pathway, especially TGFβ1 associated genes, were upregulated in LVNC and mild DCM iPSC-CMs compared to control iPSC-CMs (Fig. 2b and Supplementary Figs. 4b and 4c). Previous studies have shown that aberrant TGFβ signaling causes incomplete compaction of myocardium in mice6, 9, and that TGFβ signaling inhibits proliferation of embryonic cardiomyocytes25, 26 via upregulation of cyclin-dependent kinase inhibitors (CKI) including CDKN1A27–29. Consistent with these studies, LVNC iPSC-CMs showed significant upregulation of TGFβ signaling-related genes and 1.7-fold higher phosphorylation of SMAD2/3 (Fig. 2c and Supplementary Figs. 4d and 4e), whereas mild DCM iPSC-CMs had less phosphorylation (1.2-fold vs control). LVNC iPSC-CMs also showed a 1.8-fold increased CDKN1A expression compared to control iPSC-CMs (Fig. 2d and Supplementary Figs. 4f and 4g), suggesting that activation of the TGFβ1-CDKN1A regulatory axis may be responsible for the early cell cycle exit seen in LVNC iPSC-CMs. Histological analysis of proband’s explanted LV myocardium revealed higher phosphorylation of SMAD2 in comparison to control LV tissue, providing further evidence of activated TGFβ signaling as a pathogenesis of LVNC (Fig. 2e). Interestingly, LVNC iPSC-CMs showed significantly higher expression of TGFβ signaling pathway associated genes compared to mild DCM iPSC-CMs that were associated with milder phenotypic severity (Supplementary Figs. 4b–g).
Figure 2. Upregulation of TGFβ signaling in LVNC phenotype.
a, Upstream regulator analysis of signaling pathway comparing LVNC and control iPSC-CMs at 2 weeks after induction of cardiac differentiation. The p-value measures whether there is a statistically significant overlap between the dataset genes and the genes that are regulated by a transcription regulator. b, Heat map showing upregulation of TGFβ signaling pathway in LVNC (III-2, 3, 4; mean of four samples) and mild DCM (II-2; mean of two samples) compared to control iPSC-CMs (unrelated controls; mean of two samples). Mean=0, variance=1. c, Western blot of total and phospho-SMAD2/3 in control and patient-specific iPSC-CMs (upper) and densitometry analysis, normalized against α-tubulin (lower). d, Western blot of CDKN1A protein in control and patient-specific iPSC-CMs (upper) and densitometry analysis, normalized against α-tubulin (lower). e, Immunostaining of nuclear (blue), alpha-sarcomeric actin (green), and phospho-SMAD2 (red) in LV of control donor heart tissue vs. explanted heart of proband #1. f, Allele-specific mRNA expression analysis by mRNA-sequencing (upper panel) and digital droplet PCR (lower panel) showed a higher ratio of TBX20 mutant allele expression in LVNC iPSC-CMs compared to mild DCM iPSC-CMs. n=6 independent experiments. g, The effect of TGFβ isoform treatments on the percentage of EdU+ cardiomyocytes in control iPSC-CMs with or without growth factors. PBS treated control; n = 4 independent experiments, TGFβ treated samples; n = 3 independent experiments. CON, unrelated controls. *p < 0.05, **p < 0.01, ***p < 0.005 in unpaired two-tailed t-test or one-way ANOVA followed by Tukey post hoc test. The bar graphs show the mean and error bars represent s.e.m. Scale bars, 100 μm. Statistics source data can be found in Supplementary Table 12. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
In order to understand the difference in disease severity between the father (mild DCM phenotype) and his children (LVNC phenotype), despite carrying the same TBX20 mutation, we next assessed expression of the mutant TBX20 allele by RNA-sequencing and digital droplet PCR. We found that the ratio of mutant allele against wild-type allele expression was higher in LVNC iPSC-CMs than in mild DCM iPSC-CMs (Fig. 2f), suggesting a dosage effect of the TBX20 mutant allele on the ectopic activation level of the TGFβ signaling pathway in patient-specific iPSC-CMs and perhaps explaining for the differences in phenotypic severity among affected family members. In order to confirm that stimulation with TGFβ isoforms could affect the proliferation potential in iPSC-CMs, we treated control iPSC-CMs with TGFβ isoforms with or without serum or growth factors. We found that, like LVNC iPSC-CMs, TGFβ-treated control iPSC-CMs showed a significant decrease of proliferative response to serum, IGF-1, and IGF-2 stimulation at 2 weeks compared to TGFβ-untreated control iPSC-CMs (Fig. 2g). These results suggest that activation of TGFβ signaling has a negative effect on the proliferative potential of developing cardiomyocytes.
Ectopic activation of TGFβ signaling causes a cardiomyocyte proliferation defect in vivo
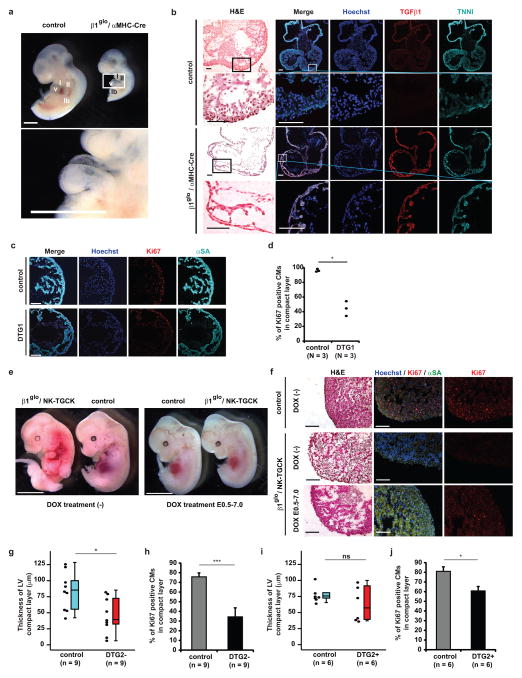
To assess how abnormal TGFβ1 signaling affects myocardial development in vivo, we next generated cardiac-specific TGFβ1 overexpression transgenic mice (β1glo/αMHC-Cre; Supplementary Figs. 5a and 5b). These double-transgenic mice exhibit embryonic lethality by embryonic day (E) 11.5 due to decreased proliferation in developing cardiomyocytes (Figs. 3a–d, and Supplementary Figs. 5c and 5d, and Supplementary Table 6), providing further proof implicating aberrant TGFβ signaling in the embryonic heart with developmental arrest of the compact layer.
Figure 3. Developmental arrest in cardiomyocyte-specific TGFβ1 overexpression mouse embryo heart.
a, Lateral view of double transgenic embryo (β1glo/αMHC-Cre) at embryonic day (E) 10.5 compared to wild-type littermate (control). The lower panel shows higher magnification view of the double transgenic embryo heart (white box in upper panel). Scale bars, 1 mm. b, Hematoxylin and eosin (H&E) staining and immunostaining of nuclear (blue), Ki67 (red), and αSA (cyan) in control and β1glo/αMHC-Cre double transgenic (DTG1) embryo hearts at E10.5. Scale bars, 100 μm. c, Immunostaining for nuclear (blue), TGFβ1 (red), and TNNI (cyan) in coronal sections of control and DTG1 hearts at E10.5. Higher magnification pictures of compact layer (indicated by white box in upper panels) are shown in lower lane. Scale bars, 100 μm. d, Percentage of Ki67+ cardiomyocytes in compact layer of control and DTG1 embryo hearts at E10.5. e, The systemic phenotypes of partially TGFβ1-overexpressed double transgenic embryo (β1glo/NE-TGCK) at E12.5 compared to wild-type littermate (control) with or without doxycycline (DOX) treatment. Scale bars, 5 mm. f, H&E and immunostaining of nuclear (blue), Ki67 (red), and αSA (cyan) in wild-type (control) and β1glo/NE-TGCK double transgenic embryo hearts at E12.5. Scale bars, 100 μm. g, Dot and box plot of thickness of left ventricle (LV) compact layer in control and β1glo/NE-TGCK double transgenic embryo (DTG2−) hearts without DOX treatment at E12.5. h, DTG2− embryo hearts showed significant decrease of percentage of Ki67+ cardiomyocytes in compact layer at E12.5. i, Dot and box plot of thickness of left ventricle (LV) compact layer in control and DOX-treated β1glo/NE-TGCK double transgenic embryo (DTG2+) hearts at E12.5. j, DTG2+ embryo hearts showed significant decrease of percentage of Ki67+ cardiomyocytes in compact layer at E12.5. v, ventricle; I, 1st pharyngeal arch; II, 2nd pharyngeal arch; lb, limb bud. *p < 0.05, ***p < 0.005; ns, not significant in unpaired two-tailed t-test. The bar graphs show the mean and error bars represent s.e.m. The box plot shows the median, with upper and lower percentiles, and the bars show maxima and minima values. Statistics source data can be found in Supplementary Table 12.
Next, in order to assess the later stages of compact layer development, we crossed β1glo transgenic mice with NK-TGCK transgenic mice that express GFP-Cre fusion protein under the control of the Nkx2-5 cardiac-specific enhancer/promoter and Tet-Off system to negatively regulate GFP-Cre expression upon doxycycline (DOX) exposure (Supplementary Fig. 5e). The αMHC-Cre transgene labeled >90% of cardiomyocytes at E12.5 and the NK-TGCK transgene activated Cre in ~40% of cardiomyocytes without DOX and ~25% of cardiomyocytes with DOX treatment and showed significantly lesser TGFB1 mRNA expression compared to αMHC-Cre/β1glo double transgenic mouse embryos at E10.5 (Supplementary Figs. 5f–h). The double transgenic embryos carrying both NK-TGCK and β1glo (β1glo/NK-TGCK) without DOX treatment showed significant increase of TGFβ downstream target genes whereas the double transgenic embryos with DOX treatment showed only mild increase of these genes at E10.5 (Supplementary Fig. 5i). The β1glo/NK-TGCK double transgenic embryos showed embryonic lethality around E12.5 to E13.5. In contrast, when mice were treated with DOX, the embryos could survive normally at E12.5 (Fig. 3e and Supplementary Table 7). Histological analysis showed a significantly thinner compact layer and reduced Ki67 or phospho-histone H3 (PHH3)-positive cardiomyocytes in the compact layer in β1glo/NK-TGCK double transgenic embryos without DOX compared to wild-type litter mates, although the trabecular layer was well developed in these double transgenic embryos (Figs. 3f–h and Supplementary Figs. 5j and 5k). Half of DOX-treated β1glo/NK-TGCK double transgenic embryos showed reduced thickness of the LV compact layer, and the proportion of Ki67+ or PHH3+ cardiomyocytes in the compact layer in β1glo/NK-TGCK embryos was mildly decreased compared to wild-type littermates (Figs. 3f, 3i and 3j, and Supplementary Figs. 5j and 5k).
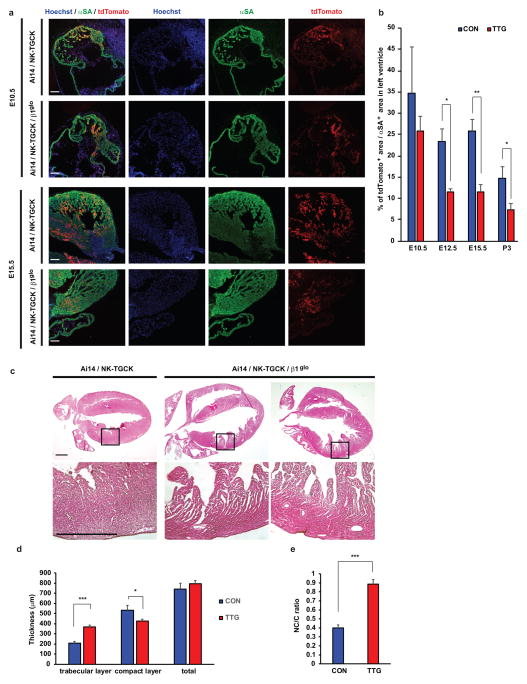
To assess the fate of TGFβ1-activated CMs in DOX-treated transgenic mice, we next generated triple transgenic embryos carrying NK-TGCK/β1glo transgenes with Cre-dependent tdTomato overexpression system (Ai14)30. We found that the proportion of TGFβ1-activated CMs in LV was significantly decreased in triple transgenic embryos and neonates during development compared to control (NK-TGCK/Ai14) mice, suggesting reduced proliferation by TGFβ-expressing CMs (Figs. 4a and 4b). Furthermore, the hearts of triple transgenic neonates at postnatal day 3 showed thicker trabecular layer and thinner compact layer resulting in significantly higher non-compaction/compaction (NC/C) ratio in LV myocardium compared to control neonates (Figs. 4c–e). Taken together, these results support the role of abnormal activation of TGFβ signaling to cause developmental arrest of cardiomyocytes in a temporal and dose-dependent manner in vivo.
Figure 4. Disturbed expansion of embryonic cardiomyocytes and trabecular/compact layer ratio in left ventricle of TGFβ1-overexpression mouse.
a, Immunostaining of nuclear (blue), tdTomato (red), and αSA (green) in coronal sections of control (Ai14/NK-TGCK) and Ai14/NK-TGCK/β1glo triple transgenic embryo hearts with doxycycline treatment at E10.5 and E15.5. Scale bars, 100 μm. b, Percentage of tdTomato positive area per αSA positive area in compact layer of control and Ai14/NK-TGCK/β1glo triple double transgenic mouse hearts with doxycycline treatment at E10.5 (control; n=5 hearts, triple transgenic; n=5 hearts), E12.5 (control; n=6 hearts, triple transgenic; n=5 hearts), E15.5 (control; n=6 hearts, triple transgenic; n=5 hearts), and postnatal day 3 (P3) (control; n=18 hearts, triple transgenic; n=8 hearts). c, Hematoxylin and eosin staining in coronal sections of control (CON: Ai14/NK-TGCK) and Ai14/NK-TGCK/β1glo triple double transgenic mouse (TTG) hearts with doxycycline treatment at postnatal day (P) 3. Lower panels show hyper-magnified views of areas indicated by the black box in the upper panels. d, Thickness of trabecular and compact layer and total myocardium in control (n=8 hearts) and Ai14/NK-TGCK/β1glo triple transgenic mouse (n=11 hearts) hearts with doxycycline treatment at P3. e, Trabecular layer/compact layer (NC/C) ratio in control (n=8 hearts) and Ai14/NK-TGCK/β1glo triple transgenic mouse hearts (n=11 hearts) with doxycycline treatment at P3. Scale bars, 0.5 mm. *p < 0.05, **p < 0.01, ***p < 0.005 in unpaired two-tailed t-test. The bar graphs show the mean and error bars represent s.e.m. Statistics source data can be found in Supplementary Table 12.
Functional disturbance of TBX20 causes abnormal activation of TGFβ signaling
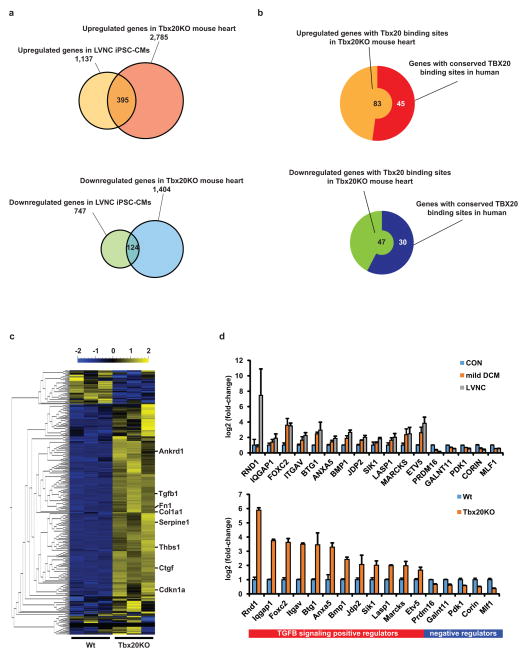
Next, to assess whether the defective TBX20 transcriptional cascade causes abnormal activation of TGFβ signaling, we analyzed the gene expression profile of the Tbx20 knockout mouse31. Interestingly, mRNA expression analysis of Tbx20 knockout mouse heart revealed a similar gene expression profile to that of LVNC iPSC-CMs, including upregulation of TGFβ signaling (Fig. 5a and Supplementary Table 8)31. Furthermore, ChIP-sequencing data of Tbx2031 showed conserved TBX20 binding sites in the genes that are associated with TGFβ signaling regulation and modification between human and mouse (Fig. 5b and Supplementary Table 9). This gene expression profile was significantly disturbed in LVNC iPSC-CMs compared to control iPSC-CMs, as well as in the Tbx20 knockout mouse heart compared to wild-type (Fig. 5c and 5d). We further employed shRNA against TBX20 in H7 human embryonic stem cell (ESC) line (TBX20KD-H7) (Figs. 6a and 6b), and found that TBX20KD-H7-CMs showed less proliferative capacity and increased expression of TGFβ–related genes compared with scramble control lines (Figs. 6c–g). These results confirm that the dysfunction of TBX20 is associated with the pathological proliferation phenotype of LVNC iPSC-CMs through disturbance of the TGFβ signaling.
Figure 5. TBX20 regulates the expression of TGFβ signaling modifier genes in developing cardiomyocytes.
a, Venn diagram to show the overlap between genes upregulated (upper) or downregulated (lower) in LVNC iPSC-CMs compared to control iPSC-CMs (q<0.05), and upregulated (upper) or downregulated (lower) in Tbx20 knockout mouse heart compared to wild type (q<0.05). The mouse data were obtained from NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo)31. b, Predicted number of upregulated (left) or downregulated (right) genes with Tbx20 binding sites in Tbx20KO mouse heart and number of genes with conserved Tbx20 binding site between mouse and human. The ChIP-sequencing data were obtained from NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM734426). c, The heat map of TGFβ signaling pathway showing upregulation of TGFβ signaling pathway in Tbx20 knockout mouse heart compared to wild-type mouse heart. Mean=0, variance=1. d, Significant mRNA expression changes of TBX20 downstream target genes that are involved in TGFB signaling pathway as validated by RNA-sequencing. These genes were found to have conserved binding sites in both human (LVNC vs. control iPSC-CMS: upper) and mouse (Tbx20 knockout vs. wild type mouse heart: lower). The bar graphs show the mean and error bars represent s.e.m.
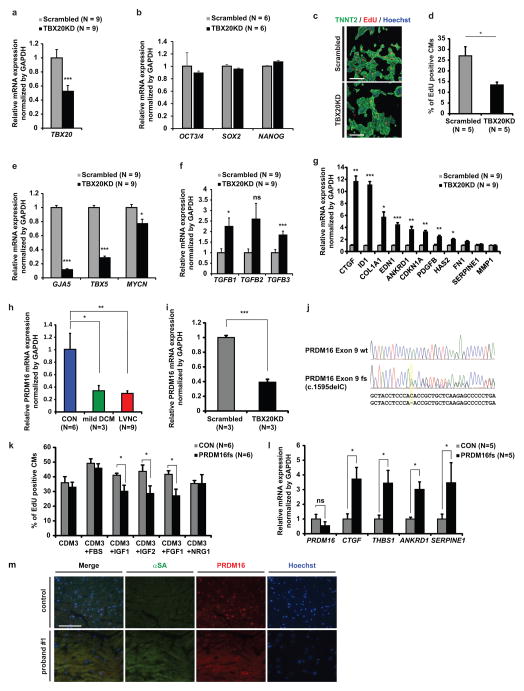
Figure 6. PRDM16 is a candidate target gene of TBX20 in cardiomyocytes.
a, Validation of TBX20 shRNA knockdown efficiency in lentiviral TBX20 shRNA-transduced H7 ESC-CMs (TBX20KD) compared to scrambled shRNA-transduced H7 ESC- CMs (Scrambled) at 2 weeks. n=9 independent experiments per each group. b, Validation of pluripotent gene expression profile in TBX20KD and scrambled ESCs by qRT-PCR. c, Immunostaining of nuclear (blue), TNNT2 (green), and EdU (red) in scrambled and TBX20KD ESC-CMs at 2 weeks. d, Percentage of EdU+ ESC-CMs in scrambled and TBX20KD ESC-CMs at 2 weeks. e, Significant decrease of TBX20 downstream target gene (GJA5, TBX5, and MYCN) expressions in TBX20KD ESC-CMs compared to scrambled ESC-CMs at 2 weeks. f–g Significant increase of TGFβ isoforms (f) and TGFβ downstream target gene (g) expression in TBX20KD ESC-CMs compared to scrambled ESC-CMs at 2 weeks. h–i, Significant decrease of PRDM16 mRNA expression in LVNC iPSC-CMs compared to control iPSC-CMs (h) and in TBX20KD-H7-CMs compared to scrambled H7-CMs (i). j, CRISPR/Cas9-based frame shift mutation in exon 9 of PRDM16 gene in control iPSC lines (PRDM16 p.T532fs*8, c.1595delC: highlighted in yellow). k, Percentage of EdU+ cardiomyocytes in control and PRDM16 frame shift mutation-created iPSC-CMs (PRDM16fs) with or without growth factors. l, Significant increase of TGFβ downstream target gene expressions in PRDM16fs iPSC-CMs. m, Immunostaining of nuclear (blue), alpha-sarcomeric actin (green), and PRDM16 (red) in LV of donor’s control heart tissue and explanted heart of proband #1. CON, unrelated controls. *p < 0.05. **p < 0.01. ***p < 0.001; ns, not significant in unpaired two-tailed t-test or one-way ANOVA followed by Tukey post hoc test. The bar graphs show the mean and error bars represent s.e.m. Scale bars, 100 μm. Statistics source data can be found in Supplementary Table 12.
TBX20-PRDM16 axis regulates TGFβ signaling and contributes to cardiomyocyte proliferation
To better understand the regulatory mechanism of TGFβ signaling by TBX20, we next studied TGFβ modifiers with conserved TBX20 binding sites31 between human and mouse (Supplementary Table 9), and selected PRDM16 gene as one of the potential downstream targets of TBX20. PRDM16, a repressor of TGFβ signaling and known genetic cause of LVNC and DCM, was significantly downregulated in all LVNC iPSC-CMs, TBX20KD-H7-CMs, and the Tbx20 knockout mouse heart (Figs. 5d, 6h and 6i). A previous genetic study of human LVNC with the PRDM16 mutation also revealed that truncation in exon 9 of PRDM16 is associated with LVNC phenotype12. To confirm whether LVNC iPSC-CMs with the PRDM16 mutation showed abnormal activation of TGFβ signaling and a proliferation defect, as seen in LVNC iPSC-CMs with the TBX20 mutation, we next generated genome-edited iPSC line carrying the frameshift mutation in exon 9 of the PRDM16 gene (PRDM16fs) and induced cardiac differentiation (Fig. 6j). Importantly, PRDM16fs iPSC-CMs showed significantly decreased proliferative response when exposed to growth factors and upregulation of TGFβ downstream target genes (Figs. 6k and 6l). Furthermore, the explanted heart tissue of proband #1 showed a significant decrease of PRDM16 expression compared to control donor heart tissue (Fig. 6m), suggesting the TBX20-PRDM16-TGFβ axis as one of the mechanisms causing LVNC.
Modification of TGFβ signaling or genetic correction of the TBX20 mutation exacerbate the proliferation defect in LVNC iPSC-CMs
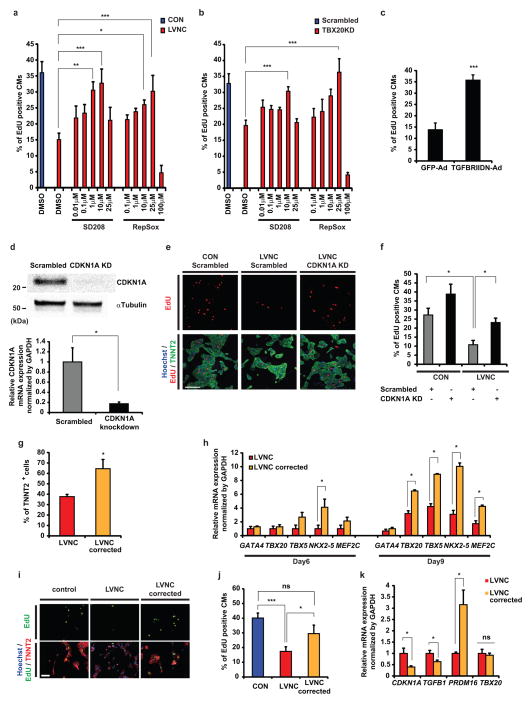
Finally, we evaluated whether modification of aberrant TGFβ signaling could rescue the proliferation defect in LVNC iPSC-CMs. Inhibition of TGFβ signaling with TGFβ receptor-1 inhibitors or overexpression of dominant negative form of TGFβ receptor-2 increased S-phase cells in both LVNC iPSC-CMs and TBX20KD-H7-CMs compared to untreated cells (Figs. 7a-c and Supplementary Figure 6). Likewise, knockdown of the downstream effector, CDKN1A, also improved the distribution of S-phase LVNC iPSC-CMs (Figs. 7d–f). To validate the effects of the TBX20 Y317* mutation on the phenotype of LVNC iPSC-CMs, we generated TBX20 Y317* mutation-corrected (LVNC-corrected) iPSCs from the proband’s iPSCs (Supplementary Figs. 7a–e). LVNC-corrected iPSC-CMs showed increased cardiac differentiation efficiency, expression of cardiac transcription factors, and restored proliferative capacity compared to non-corrected LVNC iPSC-CMs along with decreased TGFβ signaling activity (Figs. 7g–k). These results suggest that the TBX20 mutation contributes to the pathological phenotype of LVNC iPSC-CMs via disturbance of TGFβ signaling, which is associated with developmental defects of the compact layer during embryogenesis (Supplementary Fig. 7f).
Figure 7. Rescue of pathological features of LVNC iPSC-CMs.
a–b, Percentage of EdU+ control and LVNC iPSC-CMs (a) or scramble control and TBX20 knock-down ESC-CMs (b) at 2 weeks after induction of cardiac differentiation with or without treatment of TGFβ receptor-1 inhibitors (SD208 or RepSox) for 2 continuous days. n=7 independent experiments. c, Adenoviral mediated overexpression of dominant negative form of TGFβ receptor-2 (TGFBRIIDN) significantly restored proliferative potential in LVNC iPSC-CMs compared to control (adenoviral mediated GFP overexpression; GFP-Ad). n=8 independent experiments. d, Knockdown efficiency of CDKN1A protein (upper) and mRNA (lower) in iPSC-CMs by siRNA. n=6 independent experiments. e, Immunostaining for nuclear (blue), TNNT2 (green), and EdU (red) in control and LVNC iPSC-CMs with CDKN1A or scramble siRNA knockdown. f, Percentage of EdU+ iPSC-CMs at 2 weeks after induction of cardiac differentiation with CDKN1A or scramble siRNA knockdown. n=6 independent experiments. g, The efficiency of cardiac differentiation of LVNC and mutation corrected LVNC iPSC lines before glucose deprivation as validated by FACS for TNNT2. n=5 independent experiments per each group. h, mRNA expression of cardiac transcription factors in differentiating LVNC and mutation corrected LVNC iPSCs at day 6 and day 9 after induction of cardiac differentiation. LVNC n=6 independent experiments; LVNC corrected n=5 independent experiments. i, Immunostaining for nuclear (blue), TNNT2 (green), and EdU (red) in control, LVNC, and TBX20 mutation corrected (LVNC corrected) iPSC-CMs. j, Percentage of EdU+ iPSC-CMs at 2 weeks in control, LVNC, and LVNC corrected group. n=6 independent experiments. k, Reversible CDKN1A, TGFB1, and PRDM16 mRNA expression abnormality in LVNC corrected iPSC-CMs compared to LVNC iPSC-CMs. n=6 independent experiments. CON, unrelated controls. Scale bars, 100 μm. *p < 0.05, **p < 0.01, ***p < 0.005; ns, not significant in unpaired two-tailed t-test or one-way ANOVA followed by Tukey post hoc test. The bar graphs show the mean and error bars represent s.e.m. Statistics source data can be found in Supplementary Table 12. Unprocessed original scans of blots are shown in Supplementary Fig. 8.
Discussion
Previously, both accelerative9, 11 and decelerative proliferation5–7, 12 in embryonic cardiomyocytes have been reported in animal models. In our transgenic mouse model, high overexpression of TGFB1 in the developing myocardium led to a severe developmental arrest in the compact layer and the severity of this non-compaction phenotype was directly correlated with the levels of TGFB1 overexpression. These observations suggest that at the early developmental stage of LV compact layer remodeling, proper activation of TGFβ signaling in the embryonic heart is required to ensure normal ventricular development.
Human iPSC-CMs have been used to model familial dilated cardiomyopathy23, 32, 33, familial hypertrophic cardiomyopathy34, 35, long QT syndrome36–38, among others39. In this study, we demonstrate that iPSC-CM technology is useful not only for delineating the detailed molecular pathogenesis of congenital developmental defects, but also to clarify genetic causes in such diseases. We successfully found an association between functional disturbance of TBX20 and ectopic activation of TGFβ signaling in patient-specific iPSC-CMs as well as perturbed regulation of putative downstream targets of TBX20. We showed that PRDM16 is a possible downstream target gene of TBX20. PRDM16 is a known cause of LVNC in humans and morpholino knockdown of PRDM16 causes a proliferation defect in the developing zebrafish heart12. PRDM16 is a repressor of TGFβ signaling40–42 via binding of SMAD2/3, and our results revealed ectopic activation of TGFβ signaling in LVNC iPSC-CMs consistent with these previous studies. Although the comprehensive RNA expression data and ChIP-sequencing data showed that TBX20 has a large number of potential downstream target genes associated with TGFβ signaling regulation, our data suggests that the TBX20/PRDM16/TGFβ signaling pathway may be one of the key regulatory cascades for proper development of the compact/trabecular layer.
It is known that patients with LVNC generally show a wide spectrum of clinical manifestations. Some are asymptomatic, and the age at presentation can vary considerably, from as early as the newborn period to as late as older adulthood43, 44. This may be explained by varying expression levels of mutant allele-specific mRNA expression between mild DCM and LVNC as shown here. TBX20 is known to interact with other cardiac transcription factors including NKX2-5, GATA4, and TBX5 that are essential for embryonic cardiomyocyte proliferation and ventricular development. As shown here, the disturbance of interaction between TBX20 and these cardiac transcription factors may negatively impact the expansion of embryonic cardiomyocytes and impair the formation of the multilayered compact myocardium (Supplementary Fig. 7f). The differences of allele-specific TBX20 expression and the background expression of these cardiac transcription factors which may also be affected by heterogeneous factors could then impact the various cardiac phenotypes in patients with TBX20 mutations.
In conclusion, iPSC-CMs recapitulate the proliferative defects associated with LVNC5–7, 12 at the single cell level. Importantly, this study presents the supporting evidence of a proliferation defect, a consequence of abnormal activation of TGFβ signaling, as a pathological feature of human LVNC. Whether this defect is common to all cases of LVNC or just a genetic subset remains to be determined. Our data suggest that iPSC-CMs may be useful in therapeutic screening to identify potential interventions for this cardiomyopathy in the future.
Methods
Derivation of human induced pluripotent stem cells (iPSCs)
Fibroblasts or peripheral blood mononuclear cells (PBMCs) were obtained from family members with LVNC with informed consent under protocols approved by the Stanford University Human Subjects Research Institutional Review Board. Human skin-punch biopsies were digested with Collagenase II and transferred into 6-well culture dish (BD Biosciences). At passage 2 or later, primary fibroblasts were used for reprogramming to iPSCs using the non-integrating Sendai virus-based CytoTune™-iPS Reprogramming Kit (Life Technologies). After 24 h and 72 h, the medium was replaced. At day 4, cells were detached using TrypleE (Life Technologies) and replated onto mouse embryonic fibroblasts (MEF)-coated 6-well plates in DMEM/Glutamax with 10% FBS and Rho-associated, coiled-coil containing protein kinase (ROCK) inhibitor, Y27632 (Selleckchem). From day 5 onwards, cells were cultured in mTeSR medium (STEMCELL Technologies). Colonies were picked after day 20 and transferred to a Matrigel-coated culture dish (BD Biosciences), and subsequently passaged using Accutase (Global Cell Solutions). After passage 10, culture medium was changed to Essential 8 (Life Technologies)20. PBMC reprogramming was performed as described previously21. Briefly, PBMCs were isolated using a Ficoll-Paque Premium (GE Healthcare) from 20 ml of whole blood and plated at 1 million cells per milliliter in 2 ml of blood medium21. After day 9, 1×106 cells were plated in blood medium with Sendai virus. Cells were transferred to mTeSR medium in a MEF-coated 6-well plate at day 3. Colonies were picked after day 20 and transferred to a new Matrigel-coated culture dish and cultured as described above.
Culture and maintenance of iPSCs and ESCs
Human ESCs and iPSCs were grown on Matrigel-coated plates using chemically-defined E8 medium as described before20. The medium was changed daily and cells were passaged every 4 days using EDTA.
Cardiac differentiation
iPSCs were grown to 90% confluence and differentiated subsequently into beating cardiomyocytes, using a small molecule-based monolayer method described in detail previously22. Ten days after cardiac differentiation, iPSC-CM monolayers were purified using RPMI-1640 without glucose (Life Technologies) with B27 supplement (Life Technologies). Non-glucose culture medium was changed every 2 days. After 5 days, iPSC-CMs were reseeded onto Matrigel-coated plates in culture medium with glucose.
Pluripotency marker analysis
Human ESC and iPSC colonies grown in Matrigel-coated 8-well chamber glasses (Thermo Scientific) were fixed using 4% paraformaldehyde (PFA) and permeabilized with 0.5% Triton X-100. After blocking samples with 5% goat serum in PBST (PBS with 0.1% Tween20), cells were stained with mouse anti-SSEA4 (R&D systems), rabbit anti-OCT3/4 (Santa Cruz Biotechnology), rabbit anti-NANOG (Santa Cruz Biotechnology), and mouse anti-TRA1-60 (EMD Millipore) antibodies. Cells were then incubated with AlexaFluor-conjugated secondary antibodies (Life Technologies) and Hoechst 33342 (Life Technologies) to visualize the specific stains. Image acquisition was performed on an Eclipse 80i fluorescent microscope (Nikon Instruments).
Teratoma formation assay
1×106 iPSCs were suspended in 25 μL PBS, mixed with equal volumes of Matrigel, and injected into the subcutaneous regions of the anterior thigh of immunodeficient female SCID mice (n=2 spots per group of mice) (Charles River Laboratories). Fifty days after transplantation, teratomas were explanted, fixed with 4% paraformaldehyde, set in paraffin, sectioned, and stained with hematoxylin & eosin (H&E).
Plasmid construction and site-directed mutagenesis
The Flag-tagged Human TBX20 expression vector (TBX20-pCMV tag2B), GATA4 expression vector (GATA4-pcDNA3.1), NKX2.5 expression vector (NKX2.5-pcDNA3.1), TBX5 expression vector (TBX5-pcDNA3.1), and the NPPA promoter luciferase reporter vector (NPPA-pGL3 basic) were kindly provided by Hiroyuki Yamagishi. Site-directed mutagenesis was performed using a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s instructions. The MSX2 promoter sequence containing a 3.9-kb fragment of the MSX2 5′ untranslated and franking region was subcloned into pGL3-Basic (Promega), as described previously45. All constructed vectors were verified by sequencing.
Luciferase assay
The HeLa cells were purchased from ATCC. The cells were not authenticated or tested for mycoplasma contamination recently. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. HeLa cells were transfected using Lipofectamine 2000 (Life Technologies) with 50 ng reporter vector, 50 ng expression vectors, and 0.2 ng pRL-SV40 internal control vector (Promega). Luciferase activity was measured 48 h after transient transfection by using Dual-Glo Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Two duplicate, six independent assays were performed.
Immunocytochemistry
Cells grown on cover slips were fixed using 4% PFA, permeabilized with 0.5% Triton X-100, incubated with primary antibodies and Hoechst 33342, and detected using Alexa Fluor conjugated secondary antibodies. Primary antibodies used include mouse anti-FLAG M2 (Sigma Aldrich), rabbit anti-cardiac troponin T (Abcam), mouse anti-cardiac troponin T (Thermo Scientific), mouse anti-sarcomeric alpha-actinin (Sigma Aldrich), or rabbit anti-TBX20 (Abcam), as published previously45. Image acquisition was performed on an Eclipse 80i fluorescent microscope and a confocal microscope (Carl Zeiss, LSM 510 Meta) and ZEN software (Carl Zeiss). Measurement of cell surface area and length was performed using NIS-Elements Basic Research 3.0 software (Nikon). A detailed list for antibodies used is shown in Supplementary Table 10.
EdU-based proliferation analysis
iPSC-CMs were cultured in RPMI with B27 supplement and 2% FBS 2 days prior to EdU staining to accelerate the proliferation. Staining was performed using the Click-it EdU Imaging Kit (Life technologies) according to the manufacturer’s instructions. EdU was incorporated for 24 h prior to experiment. Cells were fixed with 4% PFA, permeabilized with 0.5% TritonX-100 in PBS, and stained with Alexa Fluor-conjugated azide. After washing and blocking with 5% goat serum in PBST, cells were incubated with a rabbit anti-TNNT2 antibody (Abcam) and visualized with anti-rabbit Alexa Fluor-conjugated secondary antibody and Hoechst 33342. At least more than one hundred cells were counted per sample and more than three independent studies per group were performed. Measurement of the fluorescent signals was performed using NIS elements BR analysis 4.13.04 64-bit software (Nikon).
RNA sequencing
Eight paired-end cDNA libraries (two biological replicates of proband iPSC-CMs, two biological replicates of father iPSC-CMs, one biological replicate of two sibling iPSC-CMs, and one biological replicate of two control iPSC-CMs) were prepared and sequenced. Total RNA was extracted and quantified using miRNeasy Kit (Qiagen) according to the manufacturer’s protocol. 10 μg of total RNA was used to generate index-tagged paired-end cDNA libraries. Briefly, mRNAs were purified by polyA enrichment procedure using Dynal Oligo(dT) beads (Life technologies). mRNA fragmentation was performed using RNA Fragmentation Reagents (Life technologies) to obtain 200–300 bp fragments. cDNA was generated using SuperScript Double-Stranded cDNA Synthesis Kit (Life technologies). Illumina sequencing adapters were ligated to cDNA using LigaFast (Promega) and PE Adapter Oligo Mix (Illumina, San Diego, CA). PCR was performed on the adapter-ligated cDNA with 2X Phusion DNA polymerase Master Mix (New England Biolabs, Ipswich, MA). Sequencing was performed with Illumina’s HiSeq2000 or 2500 platform using paired end reads at an average length of 100 bps (2×100).
RNA-seq data processing and differential expression analysis
Paired-end fastq sequence reads from each sample were assembled against hg19 using the Tophat v2.0.6 (http://tophat.cbcb.umd.edu) with the Illumina-supplied hg19 gene-model annotation file (gtf annotation). The expression level (FPKM, Fragments Per Kilobase of exon per Million fragments mapped) was estimated by Cufflinks (http://cufflinks.cbcb.umd.edu). Q-value is same as false discovery rate. Cuffdiff was used to call differentially expressed genes with a false discovery rate less than 0.05. Cuffdiff was run against the UCSC iGenomes GTF file from Illumina (http://cufflinks.cbcb.umd.edu/igenomes.html). Subsequent to determining which genes were differentially expressed, FPKM was used for filtering and visualization purposes. Only genes with expression values > 1 FPKM in at least one sample were considered for subsequent analysis. Samtools (http://samtools.sourceforge.net/) was used to calculate allele-specific expression of TBX20 gene. We used this software to analyze the differentially expressed genes between controls and LVNC patients. Using IPA Upstream Regulator analysis, the cascade of upstream transcriptional regulators was predicted, and this explained the observed gene expression changes. This upstream regulator analysis is based on prior knowledge of expected effects between transcriptional regulators and their target genes stored in the Ingenuity® Knowledge Base. Activation z-score was validated to infer the activation states of predicted transcriptional regulators by the IPA Upstream Regulator analysis. The p-value measures whether there is a statistically significant overlap between the dataset genes and the genes that are regulated by a transcription regulator. The p-value calculated using Fisher’s Exact Test, and significance is generally attributed to p < 0.01. RNA and ChIP-sequencing data of wild-type and Tbx20 knockout mouse heart were obtained from NCBI GEO database (RNA-sequencing: http://www.ncbi.nlm.nih.gov/geo, ChIP-sequencing: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM734426)31. All of the RNA sequencing data can be accessed as the GEO reference GSE63161 (URL: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=kdizeyiyzlilpcn&acc=GSE63161).
Quantifying allele specific expression using RNA-seq data
Allele specific expression of TGFB gene was measured using RNA-Seq data. Alignment was performed using tophat v2.0.6 (http://tophat.cbcb.umd.edu) and processed using SAMtools v0.1.2, which produces site-specific allele frequencies using overlapping reads (read pileup). Allele specific expression was calculated by determining whether or not each overlapping read at mutant site fitted the reference or the mutant allele. These summed counts represented our measures of relative allelic abundance at that site. Any deviation from equal allelic abundance was reflected as allelic imbalance46.
Quantitative RT-PCR
Total RNA was prepared using RNAeasy plus kit (Qiagen). cDNA was synthesized by the High Capacity cDNA Reverse Transcription Kit (Life Technologies). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on a StepOne Real-Time PCR System (Life Technologies) using the TaqMan Universal PCR Master Mix (Life technologies) according to the manufacturer’s protocol. Relative quantification was normalized against GAPDH. More than three respective sets of experiments were performed.
Digital droplet PCR for the detection and quantification of TBX20 wild-type and mutant allele-specific mRNA
Total RNA of 2 week old LVNC and mild DCM iPSC-CMs were extracted and prepared using RNAeasy plus kit (Qiagen). cDNA was synthesized by the High Capacity cDNA Reverse Transcription Kit (Life Technologies). The quantification of allele specific mRNA expression was analyzed using the QX100 Droplet Digital PCR system (Bio-Rad, Hercules, CA, USA). Y317* mutation was detected with an FAM probe: 5′ FAM-ZCCATCCGTACCTACGGAGGAGAX- Black Hole Quenche 3′, and the wild-type allele with another HEX probe: 5′ HEX-ZCCATCCGTACCTAAGGAGGAGAX- Black Hole Quencher 3′. Each reaction mix consisted of the following: 2x ddPCR Supermix, 20x specific target primers including the forward primer (5′ GGAAAGTGTGGAGAGCCTGA 3′) and the reverse primer (5′ TGACTCTCATCCCCCAAGAC 3′), 20x mutant-specific FAM probes, 20x wild-type-specific HEX-probe, 5 ng cDNA template, and the mixture was adjusted with PCR grade water to a final volume of 20 μl. The thermal cycling conditions were as follows: enzyme activation, 95°C for 10 min (1 cycle); denaturation, 94°C for 30 s (40 cycles); annealing/extension, 61°C for 1 min (40 cycles); and hold 98°C for 10 min (1 cycle). The ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad), and the quantification of either the deletion or the insertion allele was presented as the number of copies per microliter of PCR mixture.
Western blotting
Following SDS-PAGE, proteins were transferred to 0.45 μm nitrocellulose membranes (Bio-Rad) using a mini Bio-Rad Mini PROTEAN 3 Cell system in NuPAGE transfer buffer (Life technologies). The membrane was then blocked in Membrane Blocking Solution (Life technologies) and incubated with primary antibody overnight at 4 °C. Blots were incubated with the appropriate secondary antibodies for 1 h at room temperature and visualized using ECL Western Blotting Analysis System (GE Healthcare). Primary antibodies used were biotinylated SMAD2/3 (Cell Signaling), phospho-SMAD2/3 (Cell Signaling), CDKN1A (Cell Signaling), TBX20 (Sigma Aldrich), and HRP conjugated α–Tubulin (Cell Signaling). A detailed list of antibodies used is shown in Supplementary Table 10.
Ca2+ imaging
Briefly, 30 day old iPSC-CMs were dissociated with TrypLE and reseeded on Matrigel-coated 22-mm round coverslips. The Fluo-4 Direct Calcium Assay kit was used (Life Technologies) as per the manufacturer’s instructions. Fluo-4 loading solution was incubated with the cells at 37 °C for 30 min and fluorescence was measured at 495±20 nm excitation and 515±20 nm emission. Videos were taken of iPSC-CMs spontaneously beating or electrically field-stimulated at 1 and 2 Hz, 10 V/cm, and 10 ms biphasic pulse width. Measurements were taken on an AxioObserver Z1 inverted microscope (Carl Zeiss) equipped with a Lambda DG-4 300 W Xenon light source (Sutter Instruments), an ORCA-ER CCD camera (Hamamatsu), and AxioVison 4.7 software (Zeiss). All experiments were conducted at 37 °C with normal culture medium (RPMI-1640 with B27 supplement). The mean Ca2+ signals of ten paced beats were analyzed with ImageJ software. More than three sets of experiments were performed.
Patch clamp
Whole cell action potentials were recorded with the use of standard patch-clamp technique, as previously described22. Briefly, cultured iPSC-CMs were plated on No. 18 mm glass cover slips (Warner Instruments) coated with Matrigel, placed in a RC-26C recording chamber (Warner Instruments), and mounted onto the stage of an inverted microscope (Nikon, Tokyo, Japan). The chamber was continuously perfused with warm (35–37 °C) extracellular solution (pH 7.4) of following composition: (mM) 150 NaC1, 5.4 KCl, 1.8 CaCl2, 1.0 MgCl2, 1.0 Na pyruvate, 15 HEPES, and 15 glucose. Glass micropipettes were fabricated from standard wall borosilicate glass capillary tubes (Sutter BF 100-50-10, Sutter Instruments) using a programmable puller (P-97; Sutter Instruments) and filled with the following intracellular solution (pH 7.2): 120 KCl, 1.0 MgCl2, 10 HEPES, 10 EGTA, and 3 Mg-ATP. A single beating cardiomyocyte was selected and action potentials (APs) were recorded in whole cell current clamp mode using an EPC-10 patch-clamp amplifier (HEKA). Data were acquired using PatchMaster software (HEKA) and digitized at 1.0 kHz. The following criteria are used for classifying observed APs into ventricular-, atrial-, and nodal-like iPSC-CMs. For ventricular-like, the criteria were a negative maximum diastolic membrane potential (<−50 mV), a rapid AP upstroke, a long plateau phase, AP amplitude > 90 mV, and AP duration at 90% repolarization/AP duration at 50% repolarization (APD90/APD50) < 1.4. For atrial-like, the criteria were an absence of a prominent plateau phase, a negative diastolic membrane potential (<−50 mV), and APD90/APD50 > 1.7. For nodal-like, the criteria were a more positive MDP, a slower AP upstroke, a prominent phase 4 depolarization, and APD90/APD50 between 1.4 and 1.722.
Transgenic mice
αMHC-Cre mice47, mTmG mice48, and NK-TGCK mice49 were previously described. The tdTomato reporter mice (Ai14)30 and transgenic mice that conditionally express active TGF-β1 upon genomic recombination by Cre recombinase (β1glo)50 were purchased from the Jackson Laboratory. All transgenic mice were backcrossed with C57BL6 background more than five times. αMHC-Cre mice and NK-TGCK mice were crossed with mTmG reporter transgenic mice to observe the Cre activity in embryonic heart. αMHC-Cre mice and NK-TGCK mice were also crossed with β1glo mice to obtain cardiac-specific TGFβ1 overexpression (αMHC-Cre/β1glo or NK-TGCK/β1glo) double-transgenic embryos. Doxycycline (2 mg/ml plus 50 mg/ml sucrose) was administrated orally in the drinking water and replaced every 4 days. Pregnant female mice were treated with doxycycline from embryonic day 0.5 to 7.0. For the generation of the NK-TGCK/β1glo double-transgenic adult female mice, pregnant female mice were treated with doxycycline from embryonic day 0.5 to 12.5 to inactivate the NK-TGCK transgene during pregnancy. NK-TGCK/β1glo double-transgenic female mice were crossed with Ai14 male mice to obtain triple transgenic embryos and neonates. All experimental procedures were approved by the ethics committee of Stanford University and were in accordance with the Guide for the Care and Use of Laboratory Animals (U.S. National Institutes of Health).
Immunohistochemistry
Mouse embryos and neonates were collected, fixed in 4% PFA, embedded in OCT, and frozen in dry iced hexane. Frozen sections were blocked with 5% goat serum, stained with mouse anti-TNNT2 (Thermo Scientific), mouse anti-sarcomeric alpha-actinin (Sigma Aldrich), rabbit anti-TNNI3 (Santa Cruz), rabbit anti-TGFβ1 (Abcam), rabbit anti-Ki67 (Thermo Scientific), rabbit anti-phospho-Histone H3 (Cell Signaling Technology), and Hoechst 33342, and visualized using Alexa Fluor-conjugated secondary antibodies (Life Technologies). Image acquisition was performed on a Zeiss LSM510 Meta inverted confocal microscope (Carl Zeiss) or an Eclipse 80i fluorescent microscope (Nikon). Transgenes were detected by PCR from yolk sac DNA of embryos or tail DNA of neonatal pups. For human heart tissues, formalin-fixed paraffin embedded tissue-sections were rehydrated and autoclaved with Citrate Buffer, pH 6.0 (Sigma-Aldrich) and stained with rabbit anti-phospho-Smad2 (EMD Millipore), rabbit anti-PRDM16 (Abcam), mouse anti-sarcomeric alpha-actinin (Sigma Aldrich) and Hoechst 33342, and visualized using Alexa Fluor-conjugated secondary antibodies (Life Technologies). Image acquisition was performed on an Eclipse 80i fluorescent microscope (Nikon). A detailed list of antibodies used is shown in Supplementary Table 10.
Exome sequencing in case-parent trio
Exome sequencing was performed on patient and both parents using the Agilent SureSelectXT HumanAllExon V4 (50 Mb) (Agilent Technologies). Briefly, 3 μg of genomic DNA was sheared in 130 μl of low TE buffer to a peak size of 150–200 bp using Covaris E220, and then purified with AmpPure XP beads to remove fragments less than 100 bp. The purified DNA fragments were then subjected to Agilent SureSelect Library preparation Kit, ILM, to be end-repaired, A-tailed, and ligated to indexing-specific paired-end adaptor. The adapter-ligated libraries were amplified for five cycles using Herculase II (Agilent Technologies). Amplified Pre-capture libraries (750 ng) were concentrated in 3 μl and hybridized to the target specific baits (SureSelectXT Human All Exon V4; Agilent Technologies) according to the manufacturer’s recommendations. Hybridized material was captured using streptavidin-coated beads (Invitrogen, Paisley, UK) and amplified for 10 cycles. Captured libraries were pooled in pairs and paired-end sequenced on one lane of the Illumina HiSeq 2000 at the Stanford Center for Genomics and Personalized Medicine.
Exome-sequencing data analysis
For each exome, raw reads in FASTQ format were aligned to hg19 using the Burrows-Wheeler aligner (BWA, http://bio-bwa.sourceforge.net/) to produce a BAM (binary alignment/map) file. For case and each parent, approximately 12 Gb high quality mappable data were obtained with an average read coverage of 178X, and 90% of bases in the captured region covered more than 49X. SBgenomics BWA-GATK- and Ingenuity Variant Calling platforms version 2.0.20130604 were used to identify potentially pathogenic variants associated to LVNC. An initial variant dataset (in variant call format, VCF) was generated for each sample using SBgenomics BWA+GATK+Coverage Exome Analysis pipeline. Variants filtering were done based on quality, allele frequency in known populations, pathological potential, genetic analysis, and biological relevance. The initial variant list that contains 2160040 variants, affecting 19706 genes, was filtered for high confidence variants that met the following quality control: (i) SNPs and Indels with overall quality more than 20 and (ii) variants with read depth greater than 10. The “usual suspect” genes were excluded by only keeping variants both outside top 0.2% of most exonically variable 100 base windows and 1% of most exonically variable gene in the 1000 Genomes Project (the database of healthy public genomes; http://www.1000genomes.org/home). To obtain more possible pathogenic variants, low-frequency variants with an allele frequency of more than 0.1% in the databases of 1000 Genomes Project, public Complete Genomics genomes (http://www.completegenomics.com/public-data/) and NHLBI ESP exomes (http://evs.gs.washington.edu/EVS/) were excluded. In addition, suspicious mutations were selected with a focus on suspicious genetic causes that were associated with pathogenic, likely pathogenic, and uncertain significance phenotype. These selected genes have sequence variants associated with 1) non-synonymous mutations including gain of function, loss of function, frame shift, in-frame in-del, start/stop codon change missense which were not tolerated by SIFT or PolyPhen-2, or 2) nucleotide changes in microRNA, or 3) nucleotide changes in likely splice site loss up to 2 bases into intron and structural changes. Finally, 102 variants affecting 108 genes were listed by applying a genetic filtering that selected the genotype occurring in at least 2 out of the 3 case samples and not in control samples, as well as by using a biological filtering that kept only variants known to be implicated in LVNC phenotype within 1 hop upstream. Analyzed data access URL: https://variants.ingenuity.com/Jahanbani2016. Raw data can be accessed as the SRA reference SRP080041.
Genotyping of LVNC patients
The genotype of each family member was determined by isolating genomic DNA from ~5×106 human fibroblasts or PBMCs, using the DNeasy Blood & Tissue kit (Qiagen) according to the manufacturer’s instructions. PCR and direct-sequencing was performed as described previously51. A detailed list of primers used is shown in Supplementary Table 11.
shRNA-mediated knockdown
Knockdown of TBX20 was performed using GIPZ Lentiviral Human TBX20 shRNA (V2LHS_286836) and GIPZ Non-silencing Lentiviral shRNA Control (GE Healthcare Dharmacon) according to the manufacturer’s protocol. Knockdown efficiency was measured by qRT-PCR.
siRNA-mediated knockdown
Gene knockdown experiments were performed using Lipofectamine RNAiMax (Life Technologies) according to manufacturer’s instructions. Cells were transfected with either scrambled or siRNA against p21 (Life Technologies; siRNA s415; Ambion Silencer Select, 25 nM/well) for 36 h before being subjected to subsequent downstream analysis.
Adenoviral mediated dominant negative form of TGFβRII overexpression
The HEK293T cells were purchased from Thermo Fisher Scientific. The cells were not authenticated or tested for mycoplasma contamination recently. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. For adenoviral vector production, ~80% confluent HEK293T cells were transfected for 48 h with an adenovirus expressing a dominant-negative TGFβ type II receptor52 or eGFP adenovirus (Vector biolabs). The collected supernatants were used for the second transduction to concentrate the virus titer. Titers were measured by using Adeno-X™ qPCR Titration Kit (Clontech). 1×104 of LVNC iPSC-CMs were transducted with concentrated viral supernatant containing 2.5×104 transducing particles and incubated for 24 h.
Treatment with TGFβ receptor-1 inhibitors
TGFβ receptor-1 inhibitors (SD208, RepSox: Selleckchem) were dissolved in DMSO. An equal concentration of solvent (DMSO) was used as control. Both iPSC-CMs and ESC-CMs were treated with SD208 or RepSox for 48 h prior to experiment.
CRISPR/Cas9-mediated genome editing
CRISPR/Cas9-mediated genome editing was performed as previously described53. Single-guide DNA oligonucleotides were subcloned into vector PX458 (Addgene), and were delivered into control iPSCs by nucleofection using P3 Primary Cell 4D-Nucleofector X Kit (Lonza). After 48 h following nucleofection, transfected cells were enriched by fluorescence-activated cell sorting, and established clones were genotyped by PCR and bidirectional direct sequencing.
Transcription activator-like effector nuclease (TALEN)-mediated genome editing
TALEN pair vectors were designed and constructed using the rapid TALEN assembly system as previously described54. 500 bp fragments of wild-type TBX20 exon 7 and adjacent intronic sequences were synthesized as GeneArt String DNA fragments (Life Technologies) to make left and right homologous arms, and cloned into PB-MV1Puro-TK vector (Transposagen), as previously described55. Four silent mutations in homologous arms were inserted in order to create an artificial TTAA site and to avoid re-cleavage of the genomic sequence. Both TALEN pair and targeting vectors were delivered into LVNC iPSCs by nucleofection using P3 Primary Cell 4D-Nucleofector X Kit (Lonza). Afterwards, cells with correct targeting vector integration were selected by puromycin (Life Technologies) and genotyped. To excise the selection cassette, the transient expression of piggyBac transposase was performed by nucleofection of excision-only piggyBac transposase plasmid, PBx (Transposagen). After negative selection using ganciclovir (Sigma Aldrich), the established clones were genotyped by PCR and bidirectional direct sequencing.
Statistics and reproducibility
The experiments were not randomized and no statistical method was used to predetermine sample size. No inclusion/exclusion criteria was applied to the animal study. The investigators were not blinded to allocation during experiments and outcome assessment and replicate experiments were performed on the basis of the severity and variability of phenotypes obtained. Data were expressed as mean ± s.e.m. Immunoblots shown are representative of at least two independent experiments. All other experiments are average of at least two independent assays, and for cell number calculation in immunostaining assays, at least 100 cells per sample were counted for each independent experiments. To confirm the reproducibility, in vitro experiments were performed by two independent operators. Statistical analyses were performed using SPSS statistics 21 software (IBM). An unpaired two-tailed Student’s t-test was used to calculate significant differences between two groups. Multiple comparison correction analysis was performed using ANOVA followed by Tukey’s Post Hoc HSD test. A p-value of <0.05 was considered statistically significant. For RNA-sequencing, raw p values were adjusted for multiple testing with the Benjamini-Hochberg procedure. Genes with adjusted p value of 0.05 or less were termed as differentially expressed genes. The investigators were not blinded to allocation during experiments and outcome assessment. Source data is provided in Supplementary Table 12.
Data availability
Primary RNA-seq data-sets have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE63161. The Exome-sequencing processed data can be accessed on https://variants.ingenuity.com/Jahanbani2016 and raw data can be accessed as the SRA reference SRP080041. Source data for Figures 1–7 and Supplementary Figures 1–5 have been provided in Supplementary Table 12. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Ning Sun, Shijun Hu, and Jaecheol Lee for their help with functional assessments; Hiroyuki Yamagishi for providing plasmid; Adam B. Glick for providing dominant negative TGFB1RII overexpression adeno virus; Bruno Huber, Bhagat Patlolla and Gerald Berry for their help with analyzing in vivo study. We are grateful for the support provided by the Neuroscience Microscopy Service (NMS), and FACS Core at the Institute for Stem Cell Biology and Regenerative Medicine, Stanford University. This work is supported by the Uehara Memorial Foundation Research Fellowship, American Heart Association Postdoctoral Fellowship 15POST25160016 (KK), American Heart Association Postdoctoral Fellowship 15POST22940013 and NIH K99HL130416 (SO), The Ministry of Education, Culture, Sports, Science and Technology in Japan (FI), National Institutes of Health P01 GM099130 (MPS), NIH K18 HL11708301, Children’s Cardiomyopathy Foundation (DB), AHA Established Investigator Award, NIH R01 HL113006, NIH R01 HL130020, NIH R01 126527, NIH R01 128170, NIH R01 HL123968, and NIH R24 HL117756 (JCW).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Referenced accessions
RNA-sequencing: GSE63161
Exome-sequencing: Variant Analysis; URL: https://variants.ingenuity.com/Jahanbani2016, SRA reference; SRP080041.
AUTHOR CONTRIBUTIONS
All authors have read and approved the manuscript. K.K., S.-G.O., D.B. and J.C.W. designed research, performed the experiments and wrote the manuscript; F.I., D.B. and J.C.W. recruited the patients; K.K., J.M.C., and S.-G.O. generated and performed experiments on iPSCs/ESCs with the help of A.D.E. and G.J.; K.K., F.J., K.H., K.I. and J.M.C. performed sequencing and bioinformatics analysis; S.M.W. generated NK-TGCK transgenic mouse; K.K., V.T. and I.K. generated genome-corrected iPSC lines; K.K., P.S. and O.J.A. performed and interpreted the patch clamping and calcium imaging data; M.P.S. provided scientific advice; and J.C.W. provided funding and supervised the entire research project.
References
- 1.Kohli SK, et al. Diagnosis of left-ventricular non-compaction in patients with left-ventricular systolic dysfunction: time for a reappraisal of diagnostic criteria? Eur Heart J. 2008;29:89–95. doi: 10.1093/eurheartj/ehm481. [DOI] [PubMed] [Google Scholar]
- 2.Nugent AW, et al. The epidemiology of childhood cardiomyopathy in Australia. The N Engl J Med. 2003;348:1639–1646. doi: 10.1056/NEJMoa021737. [DOI] [PubMed] [Google Scholar]
- 3.Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 4.Chin TK, Perloff JK, Williams RG, Jue K, Mohrmann R. Isolated noncompaction of left ventricular myocardium. A study of eight cases. Circulation. 1990;82:507–513. doi: 10.1161/01.cir.82.2.507. [DOI] [PubMed] [Google Scholar]
- 5.Kosaka Y, et al. 14-3-3epsilon plays a role in cardiac ventricular compaction by regulating the cardiomyocyte cell cycle. Mol Cell Biol. 2012;32:5089–5102. doi: 10.1128/MCB.00829-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, et al. Smad7 is required for the development and function of the heart. J Biol Chem. 2009;284:292–300. doi: 10.1074/jbc.M807233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMichele LA, et al. Transient expression of FRNK reveals stage-specific requirement for focal adhesion kinase activity in cardiac growth. Circ Res. 2009;104:1201–1208. doi: 10.1161/CIRCRESAHA.109.195941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartram U, et al. Double-Outlet Right Ventricle and Overriding Tricuspid Valve Reflect Disturbances of Looping, Myocardialization, Endocardial Cushion Differentiation, and Apoptosis in TGF- 2-Knockout Mice. Circulation. 2001;103:2745–2752. doi: 10.1161/01.cir.103.22.2745. [DOI] [PubMed] [Google Scholar]
- 9.Shou W, et al. Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature. 1998;391:489–492. doi: 10.1038/35146. [DOI] [PubMed] [Google Scholar]
- 10.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luxan G, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- 12.Arndt AK, et al. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am J Hum Genet. 2013;93:67–77. doi: 10.1016/j.ajhg.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty S, Yutzey KE. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev Biol. 2012;363:234–246. doi: 10.1016/j.ydbio.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi JK, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 15.Hammer S, et al. Characterization of TBX20 in human hearts and its regulation by TFAP2. J Cell Biochem. 2008;104:1022–1033. doi: 10.1002/jcb.21686. [DOI] [PubMed] [Google Scholar]
- 16.Posch MG, et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet. 2010;47:230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, et al. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51:580–587. doi: 10.1016/j.ejmg.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Qian L, et al. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc Natl Acad Sci U S A. 2008;105:19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirk EP, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81:280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Churko JM, Burridge PW, Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol Biol. 2013;1036:81–88. doi: 10.1007/978-1-62703-511-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, et al. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised beta-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem Cell. 2015;17:89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Amato G, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelmann GL, Boehm KD, Birchenall-Roberts MC, Ruscetti FW. Transforming growth factor-beta 1 in heart development. Mech Dev. 1992;38:85–97. doi: 10.1016/0925-4773(92)90001-z. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura R, et al. Stage-specific role of endogenous Smad2 activation in cardiomyogenesis of embryonic stem cells. Circ Res. 2007;101:78–87. doi: 10.1161/CIRCRESAHA.106.147264. [DOI] [PubMed] [Google Scholar]
- 27.Hauck L, et al. Critical role for FoxO3a-dependent regulation of p21CIP1/WAF1 in response to statin signaling in cardiac myocytes. Circ Res. 2007;100:50–60. doi: 10.1161/01.RES.0000254704.92532.b9. [DOI] [PubMed] [Google Scholar]
- 28.Akli S, Zhan S, Abdellatif M, Schneider MD. E1A Can Provoke G1 Exit That Is Refractory to p21 and Independent of Activating Cdk2. Circ Res. 1999;85:319–328. doi: 10.1161/01.res.85.4.319. [DOI] [PubMed] [Google Scholar]
- 29.Brooks G, Poolman RA, Li JM. Arresting developments in the cardiac myocyte cell cycle: role of cyclin-dependent kinase inhibitors. Cardiovasc Res. 1998;39:301–311. doi: 10.1016/s0008-6363(98)00125-4. [DOI] [PubMed] [Google Scholar]
- 30.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakabe NJ, et al. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21:2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun N, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra147. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis J, et al. A Tension-Based Model Distinguishes Hypertrophic versus Dilated Cardiomyopathy. Cell. 2016;165:1147–1159. doi: 10.1016/j.cell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan F, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinson JT, et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moretti A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 37.Itzhaki I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64:451–459. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsa E, Ahrens JH, Wu JC. Human Induced Pluripotent Stem Cells as a Platform for Personalized and Precision Cardiovascular Medicine. Physiol Rev. 2016;96:1093–1126. doi: 10.1152/physrev.00036.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahata M, et al. SKI and MEL1 cooperate to inhibit transforming growth factor-beta signal in gastric cancer cells. J Biol Chem. 2009;284:3334–3344. doi: 10.1074/jbc.M808989200. [DOI] [PubMed] [Google Scholar]
- 41.Warner DR, et al. PRDM16/MEL1: a novel Smad binding protein expressed in murine embryonic orofacial tissue. Biochim Biophys Acta. 2007;1773:814–820. doi: 10.1016/j.bbamcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oechslin E, Jenni R. Left ventricular non-compaction revisited: a distinct phenotype with genetic heterogeneity? Eur Heart J. 2011;32:1446–1456. doi: 10.1093/eurheartj/ehq508. [DOI] [PubMed] [Google Scholar]
- 44.Sen-Chowdhry S, McKenna WJ. Left ventricular noncompaction and cardiomyopathy: cause, contributor, or epiphenomenon? Curr Opin Cardiol. 2008;23:171–175. doi: 10.1097/HCO.0b013e3282fdc939. [DOI] [PubMed] [Google Scholar]
- 45.Kodo K, et al. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci U S A. 2009;106:13933–13938. doi: 10.1073/pnas.0904744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson KR, Coolon JD, Wittkopp PJ. Sources of bias in measures of allele-specific expression derived from RNA-sequence data aligned to a single reference genome. BMC Genomics. 2013;14:536. doi: 10.1186/1471-2164-14-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abel ED, et al. Cardiac hypertrophy with preserved contractile function after selective deletion of GLUT4 from the heart. J Clin Invest. 1999;104:1703–1714. doi: 10.1172/JCI7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 49.Chen WP, Liu YH, Ho YJ, Wu SM. Pharmacological inhibition of TGFbeta receptor improves Nkx2.5 cardiomyoblast-mediated regeneration. Cardiovasc Res. 2015;105:44–54. doi: 10.1093/cvr/cvu229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall BE, et al. Conditional overexpression of TGF-beta1 disrupts mouse salivary gland development and function. Lab Invest. 2010;90:543–555. doi: 10.1038/labinvest.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang B, et al. 14-3-3epsilon gene variants in a Japanese patient with left ventricular noncompaction and hypoplasia of the corpus callosum. Gene. 2013;515:173–180. doi: 10.1016/j.gene.2012.12.049. [DOI] [PubMed] [Google Scholar]
- 52.Markell LM, Masiuk KE, Blazanin N, Glick AB. Pharmacologic inhibition of ALK5 causes selective induction of terminal differentiation in mouse keratinocytes expressing oncogenic HRAS. Mol Cancer Res. 2011;9:746–756. doi: 10.1158/1541-7786.MCR-11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding Q, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat Protoc. 2013;8:2061–2078. doi: 10.1038/nprot.2013.126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary RNA-seq data-sets have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE63161. The Exome-sequencing processed data can be accessed on https://variants.ingenuity.com/Jahanbani2016 and raw data can be accessed as the SRA reference SRP080041. Source data for Figures 1–7 and Supplementary Figures 1–5 have been provided in Supplementary Table 12. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.