Abstract
Two major complexes form structural bridges that connect the erythrocyte membrane to its underlying spectrin-based cytoskeleton. Although the band 3-ankyrin bridge may account for most of the membrane-to-cytoskeleton interactions, the linkage between the cytoplasmic domain of band 3 (cdb3) and adducin has also been shown to be critical to membrane integrity. In this paper, we demonstrate that adducin, a major component of the spectrin-actin junctional complex, binds primarily to residues 246 through 264 of cdb3, and mutation of two exposed glutamic acid residues within this sequence completely abrogates both α- and β-adducin binding. Because these residues are located next to the ankyrin binding site on cdb3, it seems unlikely that band 3 can bind ankyrin and adducin concurrently, reducing the chances of an association between the ankyrin and junctional complexes that would significantly compromise erythrocyte membrane integrity. We also demonstrate the adducin binds the kidney isoform of cdb3, a spliceoform that lacks the first 65 amino acids of erythrocyte cdb3, including the central strand of a large beta-pleated sheet. Because kidney cdb3 is not known to bind any of the common peripheral protein partners of erythrocyte cdb3, including ankyrin, protein 4.1, glyceraldehyde-3-phosphate dehydrogenase, aldolase, and phosphofructokinase, retention of this affinity for adducin was unexpected.
Keywords: Adducin, cytoplasmic domain of band 3, erythrocyte, membrane structure, junctional complex, anion exchanger 1
INTRODUCTION
The erythrocyte membrane is stabilized by an underlying spectrin-based cytoskeleton connected to the lipid bilayer by at least two major protein bridges. The most prominent of these bridges is the band 3-ankyrin-spectrin linkage that attaches the anion transporter (also known as band 3, SLC4A1 or AE1) to the β subunit of spectrin near the center of the spectrin tetramer [1–4]. Mutations that weaken or reduce the number of these bridges commonly cause membrane instability, leading to a pathology known as hereditary spherocytosis [5–11]. A second set of anchors that connects the lipid bilayer to the spectrin-based cortical cytoskeleton is located at the junctional complex, where spectrin tetramers extend like the spokes of a wheel from a central actin protofilament [12]. The stability of this attachment to the erythrocyte membrane is likely established by multiple interactions, including the association of 1) protein 4.1 with glycophorin C [13–15], 2) dematin and adducin with the glucose transporter [16], 3) adducin with actin [17], and 4) adducin with the cytoplasmic domain of band 3 [17].
The importance of the adducin-cytoplasmic domain of band 3 interaction to erythrocyte membrane structure is emphasized by several observations. First, rupture of the adducin-band 3 bridge causes a profound destabilization of the membrane, leading to membrane fragmentation and vesiculation [17]. Second, erythrocytes from band 3-null mice have reduced adducin content, suggesting either compromised trafficking of the polypeptide to the membrane and/or reduced stability of adducin in the absence of band 3 [17]. Third, disruption of the endogenous adducin-band 3 bridge by addition of competing adducin tail domains results in diminished band 3 retention in detergent-extracted membrane skeletons, even though other membrane structural proteins, such as ankyrin, remain unaffected [17]. Taken together, these data suggest that the adducin-band 3 interaction constitutes an important contributor to the structure of the erythrocyte membrane.
Adducin is thought to exist as a tetramer comprised of either α/β or α/γ heterodimers. Although the α, β, and γ subunits are produced from distinct genes, their sequences are highly homologous, and their domain structures are also similar (i.e. each is comprised of a sequence-related head, neck and tail domain) [18]. Adducin heterodimers are found in virtually all tissues of the body, with the α/γ dimer being ubiquitously expressed and the α/β dimer being primarily restricted to the brain and hematopoietic tissues [19]. In all cases, adducin is found to display a distinct membrane localization where it is likely involved in anchoring of a cortical cytoskeleton to the membrane. Polymorphisms in adducin have been associated with abnormalities in blood pressure [20–22] and deficiencies in α adducin have been found to cause erythrocyte fragility/reduce deformability, while deficiencies in β adducin have been linked to loss of erythrocyte membrane surface area and partial red cell dehydration [23].
Although the ankyrin binding sites on the cytoplasmic domain of band 3 (cdb3) have been well characterized [24–29], little information is available on the band 3-adducin interaction site. In this paper, we map the binding site of adducin to residues 246–264 of cdb3. We further show that mutation of two surface glutamic acid residues within the above sequence generates a cdb3 with no affinity for adducin despite retaining full affinity for other cdb3 binding partners. These data now allow assignment of the adducin docking site on band 3 and permit evaluation of the proximity of this site to the other previously characterized docking sites on band 3 incuding: ankyrin [24–29], carbonic anhydrase 2 [30, 31], protein 4.2 [32, 33], protein 4.1 [34, 35], glycophorin A [36], glyceraldehyde-3-phosphate dehydrogenase [37–39], aldolase [38–40], phosphofructokinase [38, 39, 41], and deoxyhemoglobin [42].
EXPERIMENTAL
Expression and isolation of recombinant protein domains/fragments
An expression plasmid for a glutathione S-transferase (GST) fusion construct of β-adducin tail (amino acids 335–726) was a kind gift from Dr. Vann Bennett (Duke University). The plasmid was expressed in E. Coli and purified on a GST column (GE Healthcare) as previously described [17]. The cDNA of α-adducin tail (amino acids 430–737, also a kind gift from Dr. Van Bennett, Duke University) was cloned into the expression plasmid, pT7-7, attached to a (His)6-tag at its C-terminus, expressed in E. Coli, and purified by Ni2+-affinity chromatography (Qiagen).
Similarly, both erythrocyte and kidney cdb3 expression vectors were constructed to contain a (His)6-tag at the COOH-terminus and were expressed in the pT7-7 expression system using isopropylthiogalactoside induction for 4 hrs at 28°C. After expression, the protein was purified by Ni2+-affinity chromatography (Qiagen).
Label transfer experiments and mass spectrometry
In order to identify the adducin binding site on cdb3, a photoactivated biotin transfer method was applied with several modifications (19). Briefly, recombinant GST-β-adducin tail (150 – 500 µg/mL) was labeled with sulfo-N-hydroxysuccinimidyl-2-(6-[biotinamido]-2-(p-azido benzamido)-hexanoamido) ethyl-1,3′-dithiopropionate (sulfo-SBED) in 137 mM NaCl, 2.7 mM KCl, 8.1 mM NaH2PO4, 1.5 mM KH2PO4, 4 mM MgCl2, pH 7.0 (PBS buffer) as described by the manufacturer (Pierce Biotechnology, Inc.). Purified (His)6-tagged cdb3 (210 µg protein) was then gently mixed for 1 h in the dark at 4°C with the sulfo-SBED-adducin to allow the two proteins to associate. Photoactivated cross-linking of the biotinylated adducin to cdb3 was then performed by illumination for 10 min with a 302 nm light source (18.4 W) placed 5 cm from the sample. Dithriothreitol (10 mM) was added to 100 µL of the cross-linked sample to reduce the disulfide bond within the cross-linking reagent, and 2 M urea (pH 8.0) was then added to dissociate the band 3-adducin complex. One of several different proteases (trypsin, chymotrypsin, proteinase K, or endoproteinase Lys-C + trypsin) was then incubated with the sample to promote digestion of the biotinylated cdb3 into peptide fragments, as outlined in the manufacturer’s guidelines (Sigma-Aldrich). Biotinylated fragments were captured on an avidin column, washed with buffer, and eluted with a mixture of 50% acetonitrile/0.4% TFA. Isolated biotinylated peptides were then characterized by matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (Proteomics Center-Purdue University).
Site-directed mutagenesis and purification of mutated cdb3
Site-directed mutagenesis of cdb3 was performed using a QuickChange mutagenesis kit (Stratagene) according to manufacturer’s instructions. For substitution of the two glutamic acid residues (position 252 and 254 on cdb3) with lysines, the following oligonucleotide was synthesized by Integrated DNA Technologies and used for site-directed mutagenesis: 5’-aggaggcagcga agctgaaggcggtggag-3’. A (His)6 tag was introduced at the COOH terminus of cdb3 in a pT7-7 plasmid and used as the template. The resultant mutant cDNA was sequenced to verify integration of the desired mutations, and the plasmid was transformed into BL21 pLysS cells and expressed and purified as described above.
Binding of GST-β-adducin tail to His-tagged cdb3 immobilized on nickel beads
His-tagged wild-type or mutated cdb3 were immobilized on nickel beads and incubated with purified GST-β-adducin tail overnight at 4°C in PBS containing 4 mM MgCl2 and 1 mg/mL BSA. Beads were washed with PBS and eluted with 250 mM imidazole. The amount of bound fusion protein was evaluated by measuring the GST activity on the beads or by performing quantitative dot blot assays using an anti-GST polyclonal antibody (Santa Cruz Biotechnology).
Binding of (His)6-α-adducin tail to cdb3 immobilized on Affi-Gel 15
(His)6-α-adducin tail was immobilized on Affi-Gel 15 according to manufacturer’s instructions (Bio-Rad) by mixing 20 nmol of (His)6-α-adducin tail domain with 500 uL packed Affi-Gel 15 in a total volume of 2 mL overnight at 4°C. Unreacted Affi-Gel sites were blocked by incubation with 1 M ethanolamine, pH 8.0 for 1 hour. Ethanolamine-blocked empty beads were used as a control. Derivatized beads were washed with ice-cold double-distilled H2O and equilibrated in PBS, pH 7.4. Different concentrations of kidney, wild-type, or mutated cdb3 were then incubated with (His)6-α-adducin tail and ethanolamine-derivatized beads for 3 h at 4°C. Beads were pelleted, washed 3 times with PBS to remove unbound proteins, and analyzed by SDS-PAGE followed by immunoblotting. Cdb3 was detected and quantified with an anti-cdb3 polyclonal antibody (generated in our lab).
Binding of GST-D3D4-ankyrin domain to His-tagged cdb3 immobilized on nickel beads
His-tagged wild-type or mutated cdb3 was immobilized on nickel beads and incubated with purified GST-D3D4-ankyrin domain overnight at 4°C in PBS containing 1 mg/mL BSA. Beads were washed with PBS and eluted with 250 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4, pH 8.0. The amount of bound fusion protein was evaluated by measuring the GST activity on the beads or by performing quantitative dot blots using anti-GST polyclonal antibody (Santa Cruz Biotechnology).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity assay
Wild-type or mutant cdb3 was mixed in a cuvette with 25 nM GAPDH in a total volume of 100 µl containing 10 mM imidazole acetate, 0.1 mM EDTA, 0.5 mM sodium arsenate and 1 mM sodium phosphate (pH 7.0). After 5 min incubation, 900 µl of the same buffer containing 250 µg NAD+ and 5 µg glyceraldehyde-3-phosphate was added, and the absorbance at 340 nm was monitored continuously for 3 minutes. GAPDH activity was calculated from the absorbance difference between 0 and 50 second time points.
Intrinsic fluorescence
The cytoplasmic domain of band 3 undergoes a large pH dependent conformational change over the physiological pH range, and this structural change can be monitored by measuring the intrinsic fluorescence of cdb3 as a function of pH [43, 44]. For this purpose, the intrinsic fluorescence of both wild-type and mutant cdb3, equilibrated at 0.5 pH intervals from pH 6.0 to pH 11.0 in 50 mM sodium phosphate, 50 mM sodium borate, 70 mM NaCl (final concentration, 37 µg/mL), was monitored in a Bowman Series 2 Luminescence Spectrophotometer [43]. The excitation and emission monochrometers were set at 290 and 335 nm, respectively, with a bandwidth of 4 nm.
Circular dichroism
Circular dichroism (CD) measurements were conducted at 24°C on a JASCO CD spectropolarimeter Model J-810 as described by Wang and colleagues [45]. Briefly, wild-type and mutant cdb3 were dialyzed against 50 mM sodium phosphate, 50 mM sodium borate, 70 mM NaCl, pH 7.4. Subsequently 21 µg/mL of wild-type or mutant cdb3 was added to the above buffer and a CD spectrum was monitored in a 1 cm length cell.
RESULTS
The NH2-terminus of band 3 is not involved in β-adducin binding
Despite the large size and multidomain structure of band 3, most peripheral protein ligands (e.g. glyceraldehyde-3-phosphate dehydrogenase, aldolase, phosphofructokinase, deoxyhemoglobin, protein 4.1) bind to a short sequence within the first 65 amino acids of band 3 [28, 39, 46, 47]. To determine whether adducin might similarly require the NH2-terminus of band 3 for its association, we expressed the kidney spliceoform of the cytoplasmic domain of band 3 that lacks the extreme NH2-terminus (i.e. residues 1–65) but is otherwise identical to erythrocyte band 3 [46–51]. For the β-adducin ligand, we expressed the highly elongated 33 kDa COOH-terminal tail domain of β-adducin, because it has been shown to contain the residues responsible for band 3 binding [17].
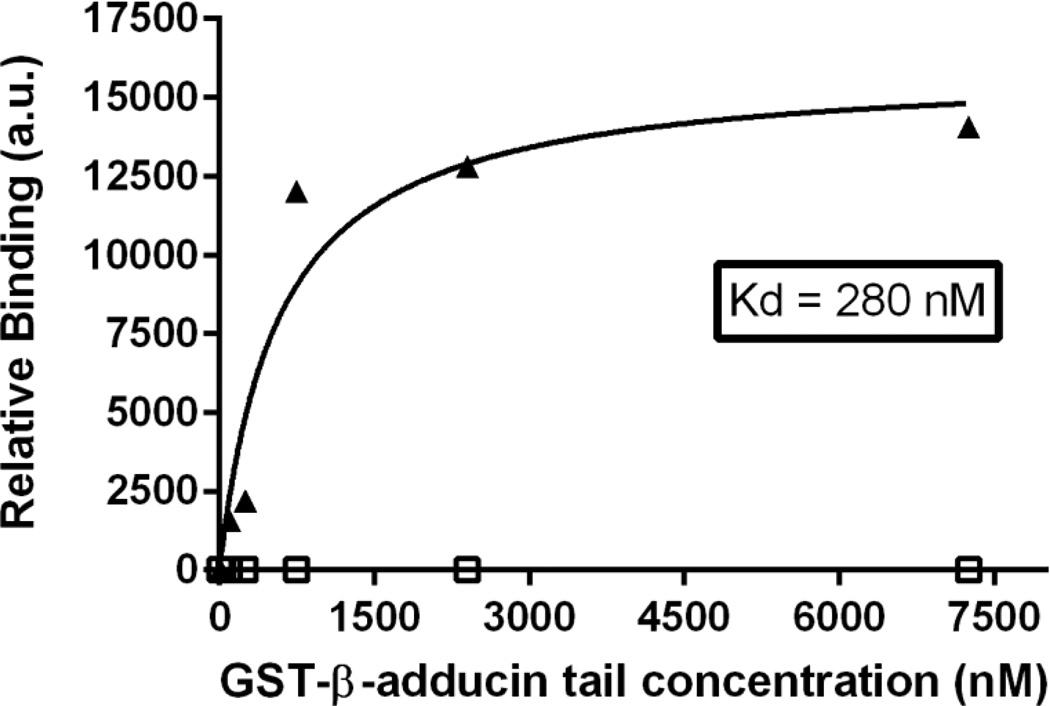
To evaluate whether the truncated kidney cdb3 might bind the tail domain of β adducin, a GST-tagged β-adducin tail construct was incubated with purified (His)6-tagged kidney cdb3 and then pelleted in a standard nickel bead pull-down assay. Immobilized kidney cdb3 was found to associate with the tail domain of β-adducin with a Kd of 280 nM (Figure 1). Because this dissociation constant is similar to that found for erythrocyte cdb3 (i.e. Kd= 260 nM; see Fig. 3), these data imply that the first 65 amino acids of cdb3 are not required for adducin binding and that the conformational rearrangements resulting from the absence of the central beta strand in the major beta sheet in kidney cdb3 (i.e. residues 57–66 of erythrocyte band 3 are absent in kidney band 3) do not perturb the adducin binding interface.
Figure 1.
The association of GST-β-adducin tail with band 3 is not affected by the absence of the first 65 amino acids of cdb3. His-tagged kidney-cdb3 (triangles) or BSA (open squares) was immobilized on nickel beads and incubated with increasing concentrations of GST-β-adducin tail. Beads were pelleted, washed 5 times, and eluted with 250 mM imidazole in PBS. GST activity was then quantified as a measure of the amount of bound adducin. (a.u. represents arbitrary units).
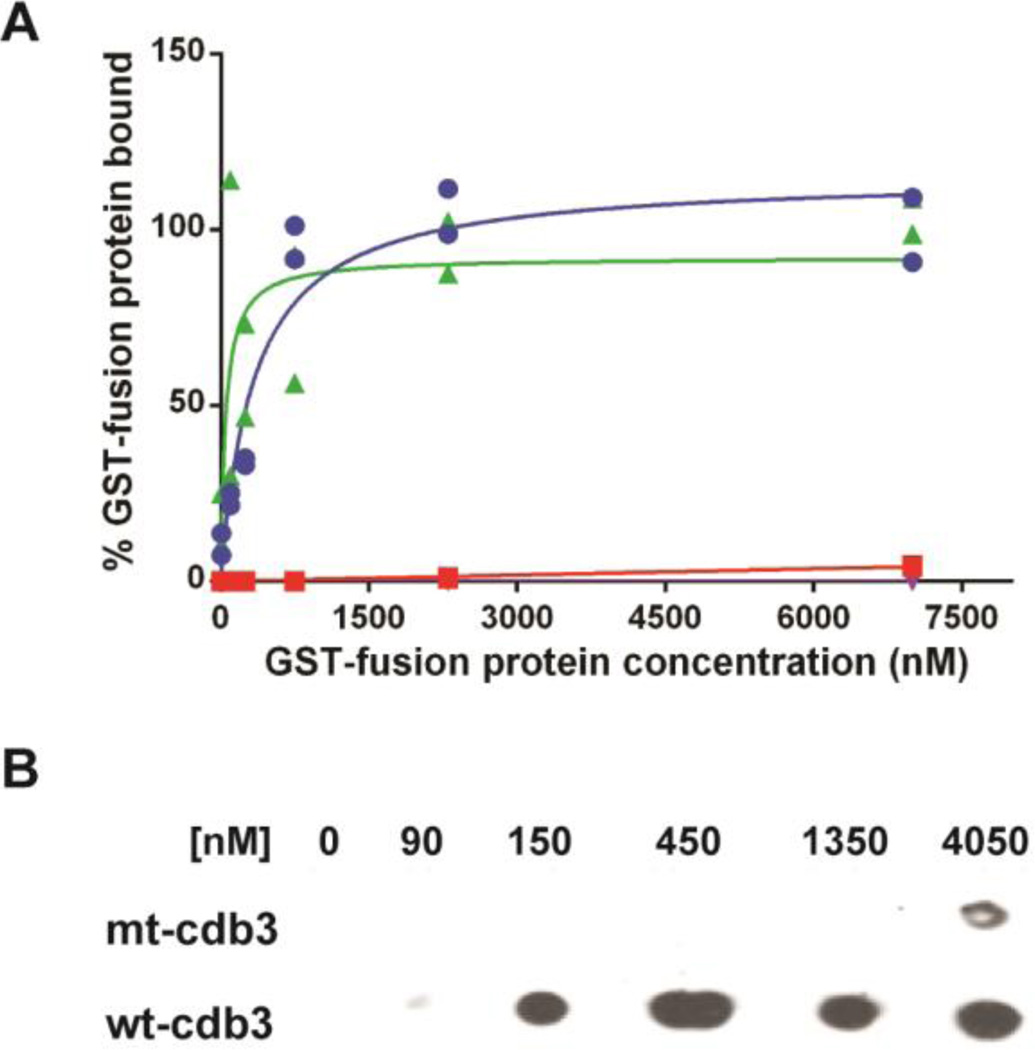
Figure 3.
Binding of β-adducin tail and ankyrin to wild-type and mutant cdb3. Increasing concentrations of GST-β-adducin tail or GST-D3D4-ankyrin were incubated with immobilized wild-type or mutated cdb3 in which glutamates 252 and 254 were mutated to lysines. Beads were washed with PBS and eluted with 250 mM imidazole, and GST activity was quantified as a measure of bound adducin or ankyrin. (A) Binding of β-adducin tail domain to wild type cdb3 (blue, Kd = 260 nM), mutated cdb3 (red), or BSA (purple) is shown as a function of GST-β-adducin tail domain concentration. Binding of D3D4-ankyrin (green) to mutated cdb3 is shown as a function of GST-D3D4-ankyrin concentration. Individual data points (n=2) are plotted with estimated binding curves. (B) Dot blot analysis of β-adducin tail binding to purified wild-type and mutant cdb3. Wild-type or mutated band 3 was immobilized on nickel beads and incubated with increasing concentrations of GST-β-adducin tail domain. Beads were pelleted, washed 4×, and eluted with 250 mM imidazole in PBS. Eluted proteins were transferred to a nitrocellulose membranes and probed for GST-β-adducin tail with an anti-GST polyclonal antibody.
Further localization of the adducin binding site on band 3
To define the docking site of adducin on band 3 with greater accuracy, we labeled both α- and β-adducin tail domains with sulfo-SBED, a photoactivatable cross-linking reagent that transfers a biotin to its nearest neighbor upon illumination with UV light. In the present application, labeled α- and β-adducins were allowed to bind cdb3, and after photo-crosslinking, the protein complexes were digested with a variety of different proteases. Biotin-labeled peptides were then isolated on an avidin column and analyzed by tandem mass spectrometry. As seen in Table 1, regardless of the digestive enzyme used or the isoform of adducin employed as the photolabel donor (i.e. α- or β-adducin), the predominant peptides isolated all contained residues 246-RLQEAAELEAVELPVPIRF-264 of cdb3. These data strongly suggest that both α- and β-adducin bind to the same site on cdb3 and that this site encompasses the region spanning residues 246–264.
Table 1.
Band 3 sequence labeled by photo-activated transfer of biotin from sulfo-SBED-adducin to cdb3.1
| Digestion enzyme |
Sulfo-SBED α-adducin tail |
Sulfo-SBED β-adducin tail |
|---|---|---|
| Trypsin | 247LQEAAELEAVELPVPIR263 | 247LQEAAELEAVELPVPIR263 |
| Chymotrypsin | 251AELEAVELPVPIRF264 | 254EAVELPVPIRF264 |
| Proteinase K | 245VRLQEAAELEAVELPVPIRF264 | 245VRLQEAAELEAVELPVPIRF264 |
| Endo Lys-C + Trypsin |
248QEAAELEAVEL258 | 248QEAAELEAVEL258 |
Sulfo-SBED α- or β-adducin tail domain was allowed to bind cdb3, after which the biotin label in sulfo-SBED was transferred to cdb3 by illumination with UV light. Following proteolysis as indicated below, biotinylated peptides were isolated on an avidin column and analyzed by MALDI-TOF mass spectrometry.
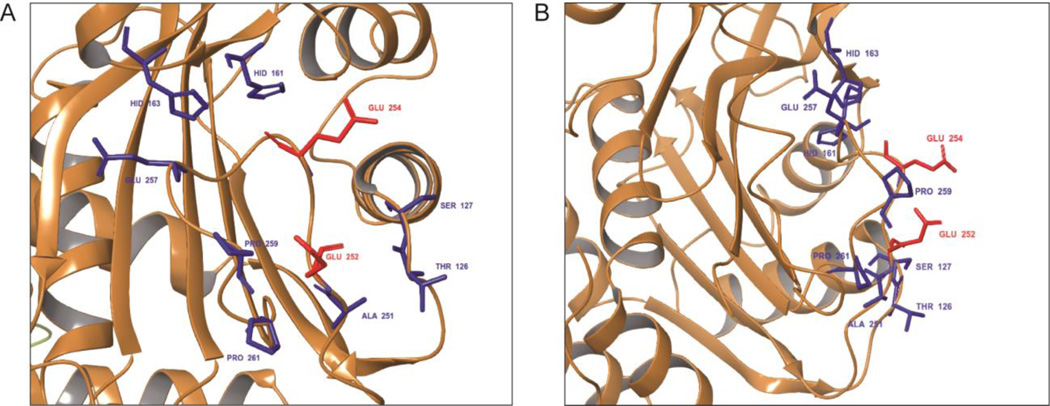
Visualization of residues 246–264 in the crystal structure of cdb3 revealed four important conclusions: 1) residues 246–264 lie near the center of a surface-exposed region located between β strands 9 and 10 of cdb3; 2) although the entire sequence is largely hydrophobic, the prominently protruding residues are negatively charged; 3) the adducin binding sequence is located on the opposite side of cdb3 from its dimerization arm, suggesting that adducin binding cannot interfere with band 3 dimerization; and 4) the docking region is immediately adjacent to the binding site of ankyrin, suggesting that concurrent binding of adducin and ankyrin will not be possible.
To more precisely identify amino acids that might be critical in adducin binding, we evaluated the accessible surface area of all amino acids within 12Å of the geometric center of this sequence. As shown in Figure 2, two Glu residues (Glu 252 and 254) distinguish themselves as being prominently exposed and available for formation of docking interactions with the highly basic tail domain of both adducins. To learn whether these two Glu residues might be important for adducin binding, we mutated them to lysines and re-evaluated adducin’s binding affinity. As seen in Figure 3A, mutation of the Glu 252 and 254 to lysines abrogates β-adducin’s interaction with cdb3. Thus, the binding curve for association of GST-β-adducin tail with wild-type cdb3 yielded a Kd of ~260 nM (i.e. similar to kidney cdb3), whereas the binding isotherm for the substitution mutant was barely distinguishable from a BSA control. Moreover, when the same binding analysis was performed by dot blot assay, the same result was qualitatively confirmed (Figure 3B).
Figure 2.
Constellation of surface protruding residues located in the binding region comprising residues 246–264 of the lower pH conformation of cdb3. Accessible surface amino acids residing within 12Å of the geometric center of the above sequence were determined from the crystal structure of cdb3, and are shown with an (A) en face view and (B) right-rotated view. The two most exposed residues, Glu 252 and Glu 254, are highlighted in red. Less prominently exposed residues are highlighted in blue.
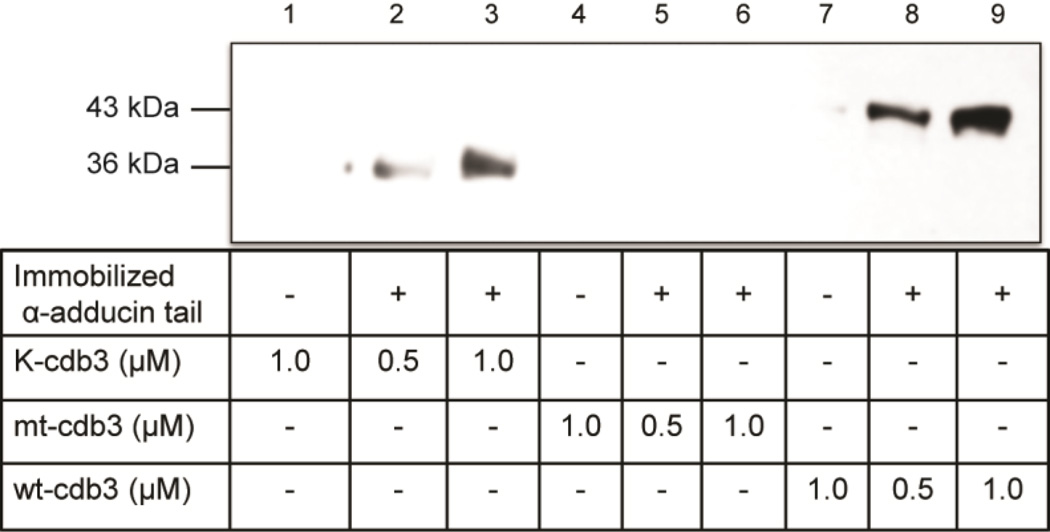
Because the adducin tetramer is comprised of α and β subunit heterodimers [52–54], we also wished to determine whether α-adducin might experience a similar loss of affinity for band 3 when residues 252 and 254 are mutated. For this purpose, (His)6-α-adducin tail was immobilized on Affi-gel 15 beads (Bio-Rad) and different concentrations of either kidney, erythrocyte, or the above mutated cdb3 were added prior to binding analysis in a co-pelleting assay. As seen in Figure 4, both kidney and erythrocyte cdb3 co-pellet with α-adducin, whereas the mutated cdb3 does not.
Figure 4.
α-adducin tail binds kidney and erythrocyte cdb3, but not mutant cdb3, in a dose-dependent manner. α-adducin tail domain was reacted with Affi-Gel 15 beads. Different concentrations of His-tagged kidney (K-cdb3), erythrocyte (wt-cdb3), and mutant cdb3 (mt-cdb3) were incubated with the derivatized beads, and beads were pelleted, washed 4×, eluted and separated by SDS-PAGE. Cdb3 was then crudely quantitated by western blotting using anti-cdb3 antibody.
Mutation of cdb3 residues 252 and 254 does not alter cdb3 function or structure
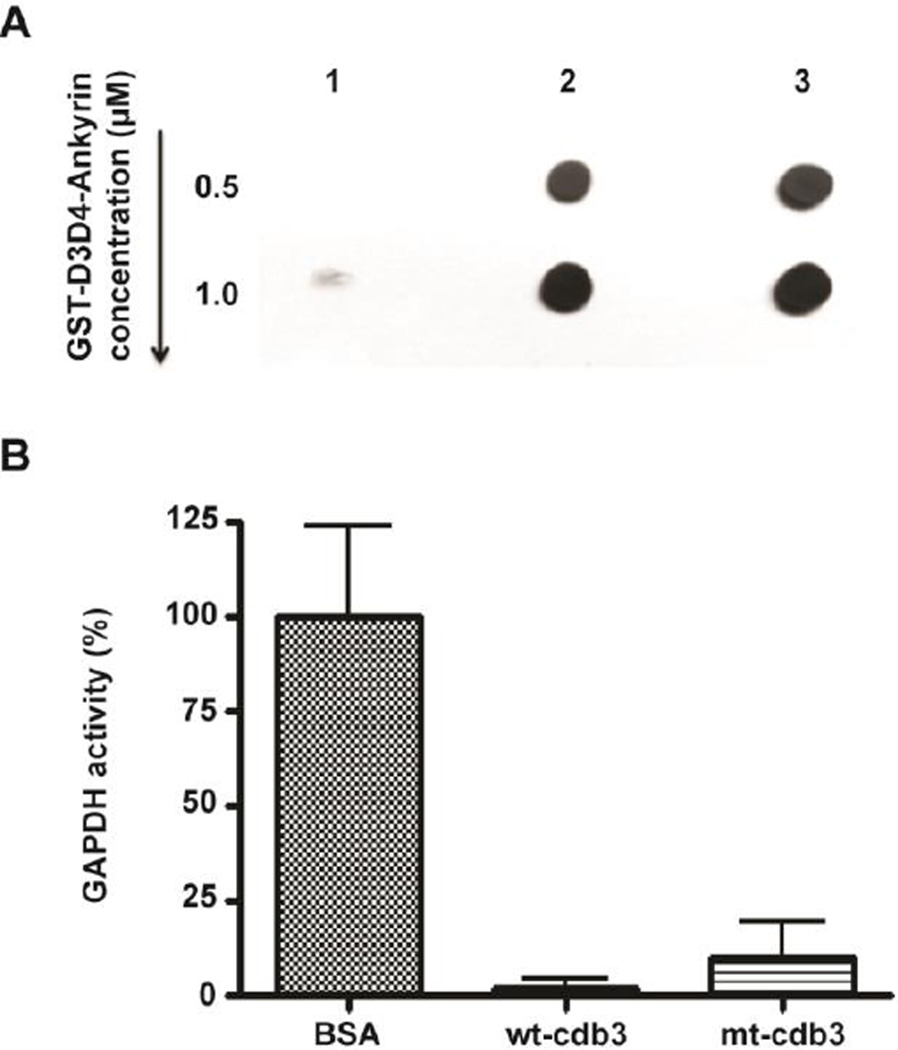
To establish that substitution of the two glutamic acids with lysines does not prevent adducin binding by causing a global perturbation of cdb3 structure, the interaction of the mutated cdb3 with ankyrin was examined. For this purpose, a recombinant band-3-binding domain of ankyrin fused to glutathione-S-transferase (GST-D3D4-ankyrin; [28, 29]) was incubated with (His)6-tagged wild-type or mutated cdb3 immobilized on standard nickel beads. After washing to remove unbound ankyrin domain, bound D3D4-ankyrin was quantitated by measuring either the GST activity retained on the nickel beads (Fig. 3A) or by assaying the amount of fused GST using an anti-GST polyclonal antibody (Figure 5A). As shown in Fig. 3A, D3D4-ankyrin binds to mutant cdb3 in a saturable manner, and qualitative analysis of the co-pelleting of D3D4-ankyrin with mutant cdb3 reveals approximately the same amount of co-pelleting ankyrin as that seen with native cdb3 (Figure 5A). These data confirm that substitution of glutamic acids 252 and 254 with lysine residues does not change the affinity of cdb3 for ankyrin.
Figure 5.
Mutation of cdb3 does not affect other protein functions. (A) Dot blot analysis of GST-D3D4-ankyrin interaction with wild-type and mutant cdb3. 1) Empty bead control, 2) wild-type cdb3, or 3) mutant cdb3 were immobilized on nickel beads and incubated with increasing concentrations of GST-D3D4-ankyrin. After washing and elution of proteins with 250 mM imidazole, eluted proteins were dot blotted on to nitrocellulose and probed with anti-GST antibody. (B) Effect of wild type and mutant cdb3 on GAPDH activity in solution. Wild-type erythrocyte band 3 has already been established to bind and inhibit GAPDH [39], whereas BSA has no effect on GAPDH activity. Data points represent mean ± S.D., n = 4.
To further demonstrate that this site-directed mutagenesis has no major impact on global cdb3 structure, we also examined the ability of cdb3 to inhibit the activity of the glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase, i.e. a well-established property of wild-type erythrocyte band 3 [39]. As demonstrated in Figure 5B, both wild-type and mutant cdb3 inhibit the enzyme’s catalytic activity with approximately equal potency.
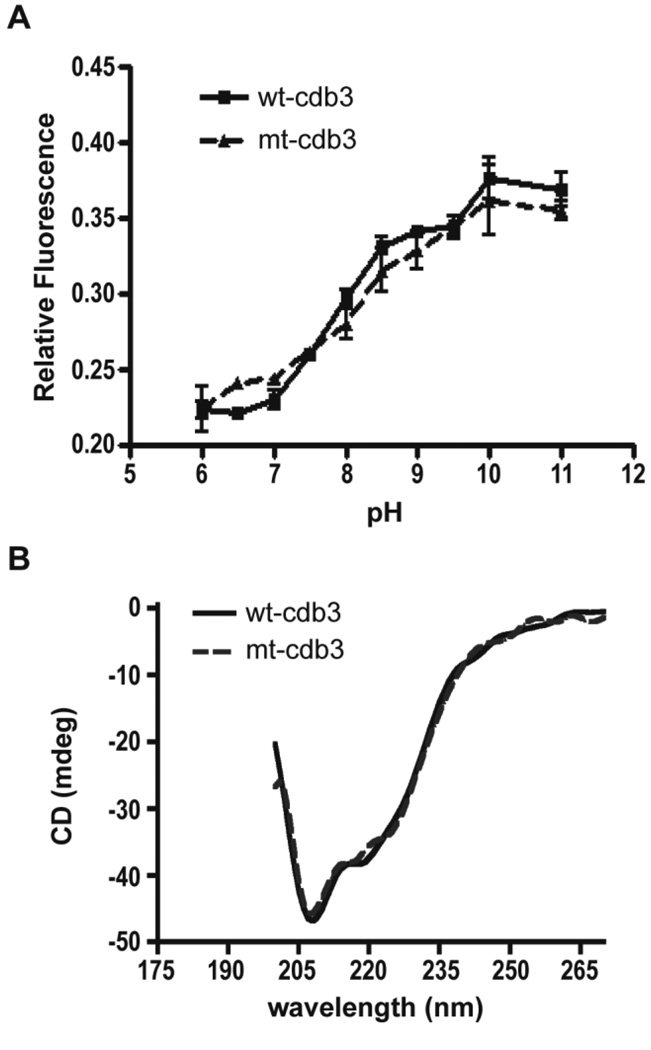
Finally, to unequivocally establish that the above two amino acid substitutions have no global effect on cdb3 conformation, the reversible pH-dependent conformational change characteristic of native cdb3 was examined (Figure 6). In this conformational change, a protonated His residue quenches the fluorescence of a proximal Trp at low pH. However, as pH is raised from pH 6.0 to 11.0, the two residues move apart, leading to dequenching of the Trp fluorescence and a doubling of the protein’s intrinsic fluorescence [55, 56]. As seen in Figure 6A, titration of the mutated cdb3 from pH 6.0 to 11.0 yields the fluorescence increase characteristic of wild-type cdb3, suggesting the dramatic conformational change in cdb3 is unaffected by the Glu for Lys substitutions. Furthermore, comparison of the circular dichroism spectra of wild-type and mutant cdb3 reveals no major differences (Figure 6B). Taken together, these data demonstrate that the loss of cdb3’s binding affinity for adducin associated with replacement of the two negatively charged glutamic acids for lysines arises from a localized change in electrostatic interactions and not from a global perturbation of cdb3 structure. We therefore conclude that residues 252 and 254 of band 3 comprise a critical interaction site for association of band 3 with adducin.
Figure 6.
Cdb3 mutants that cannot bind adducin retain normal structural properties. (A) Comparison of the pH-dependent conformational changes of wild-type and mutant cdb3. The intrinsic fluorescence (emission at 335 nm) of purified recombinant wild-type and mutant cdb3 was measured following excitation 290 nm and plotted as a function of pH. Data points represent mean ± S.D., n = 2. (B) Comparison of the circular dichroism spectra of wild-type and mutant cdb3. The CD spectrum of wild-type and mutant cdb3 was monitored in 50 mM sodium phosphate, 50 mM sodium borate, 70 mM NaCl, pH 7.4 at 24°C using a Jasco Model J810 CD spectropolarimeter.
DISCUSSION
We have presented three lines of evidence that the major binding site of adducin on cdb3 resides between residues 246 and 264, and that Glu 252 and 254 within this sequence constitute critical amino acids in mediating adducin binding. First, the first 65 amino acids of erythrocyte cdb3, which comprise the NH2-terminus, are not required for adducin binding. Second, biotin label transfer studies reveal that essentially the same cdb3 region (i.e. residues 246–264) is biotinylated by sulfo-SBED-labeled adducin, regardless of whether α- or β-adducin is used as the biotin donor, and independent of whether trypsin, chymotrypsin, proteinase K, or a mixture of endoproteinase Lys-C plus trypsin is used for digestion of cdb3. Finally, mutation of two highly exposed Glu residues within this labeled region totally abrogates cdb3 affinity for both α- and β-adducin without affecting the binding of cdb3 to other peripheral proteins. Taken together, these data suggest that the most prominent α- and β-adducin binding site on band 3 lies between residues 246 and 264. However, because our mutagenesis studies did not explore the possible involvement of adjacent regions on the cdb3 surface, we cannot exclude participation of additional amino acids near this site. Indeed, the second most highly photolabeled peptide on cdb3 resides directly adjacent to the mutated peptide in the crystal structure of the polypeptide (data not shown), but further experiments will be needed to determine whether it is directly involved in binding adducin.
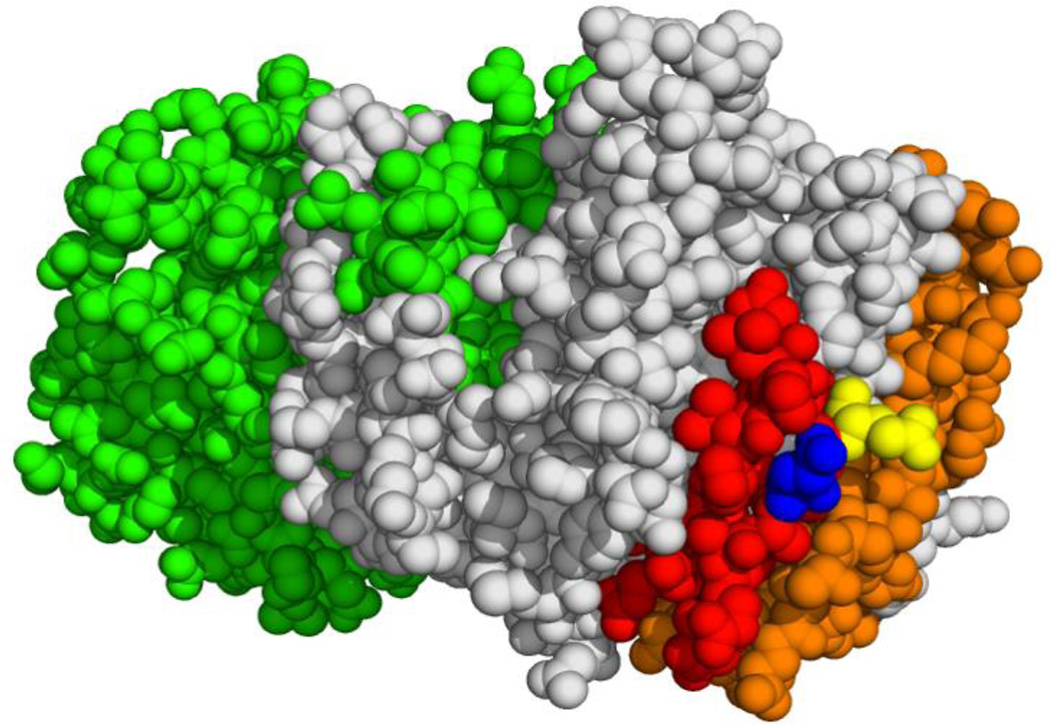
Visualization of the binding sites of adducin and ankyrin on the crystal structure of cdb3 [57] suggests that simultaneous binding of the two proteins is highly unlikely (see Fig. 7). The established docking site of ankyrin on band 3 involves residues 63–73, 118–141, 155–162, and 174–195 [25–29, 47], which encompass a significant fraction of cdb3’s surface area. Given that adducin has an approximate diameter of 89 Å [54] and that the band 3 binding domain of ankyrin has a diameter of ~74 Å [58], the probability of significant steric overlap is very high, considering the distances from the critical adducin binding residues (i.e. E252 and E254) to the major ankyrin binding residues are only ~21, 22, 9, and 20 Å, respectively. Indeed, preliminary studies demonstrate that the binding of D3D4-ankyrin to cdb3 is inhibited by addition of the α-adducin tail domain [59]. This mutual exclusivity in cytoskeletal bridge formation is likely important, since it should help preserve the separation between the ankyrin and junctional complexes that is necessary for maintenance of erythrocyte membrane structure.
Figure 7.
Structure of the cdb3 dimer with the adducin (red, blue, and yellow) and ankyrin (orange) binding sites shown on the white subunit. Glutamates 252 and 254 which are critical for adducin binding are colored in blue and yellow, respectively. A structurally identical unlabeled subunit of cdb3 is shown in green.
The unexpected retention of binding affinity between adducin and kidney cdb3 may have implications for the role of adducin in the kidney. We originally examined the binding of adducin to kidney cdb3 in order to determine whether the first 65 amino acids of erythrocyte cdb3, which comprise the NH2-terminus and are deleted in kidney cdb3, were necessary for the association. However, the observation that the truncated kidney cdb3 binds adducin was surprising in view of the fact that kidney cdb3 displays no known affinity for any other protein with which erythrocyte cdb3 normally interacts, including ankyrin, protein 4.1, glyceraldehyde-3-phosphate dehydrogenase, aldolase, phosphofructokinase, and deoxyhemoglobin [28, 39, 46, 47]. Although the interactions of kidney band 3 have received limited study, established protein ligands of kidney cdb3 include integrin-linked kinase, nephrin, and the β1 subunit of the Na,K-ATPase [60–62]. While we were not able to find any literature demonstrating a direct band 3-adducin interaction in the kidney, several studies suggest that mutations in adducin can lead to alterations in levels of nephrin, Na,K-ATPase activity, renal sodium retention, and blood pressure [20–22, 63–65]. Taken together, these data raise a question whether the unexpected association of kidney band 3 and adducin could potentially contribute to maintenance of a membrane complex involving nephrin and the Na+,K+-ATPase that could in turn be involved in the proper control of sodium homeostasis in the kidney. While it is an intriguing possibility, the extent of band 3’s involvement in these renal processes remains unknown at this time.
SUMMARY STATEMENT.
The linkage between band 3 and adducin is critical to the integrity of the erythrocyte membrane. In this paper, we identify residues on the cytoplasmic domain of band 3 that are necessary for binding of adducin.
Acknowledgments
The expression plasmid for GST fusion construct of β-adducin tail (amino acids 335–726) and the α-adducin tail fragment (amino acids 430–737) were kindly donated by Dr. Vann Bennett, Duke University School of Medicine. We thank Ann G. Liu, PhD for assistance in drafting and editing the manuscript. We also thank Karson Putt and Som Dutt for assistance with figure preparation.
FUNDING INFORMATION
This work was supported by National Institutes of Health Grant GM024417-36. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- cdb3
cytoplasmic domain of band 3
- GST
glutathione-S-transferase
- sulfo-SBED
sulfo-N-hydroxysuccinimidyl-2-(6-[biotinamido]-2-(p-azido benzamido)-hexanoamido) ethyl-1, 3′-dithiopropionate
- MALDI-TOF
matrix assisted laser desorption ionization time-of-flight
- CD
circular dichroism
Footnotes
AUTHOR CONTRIBUTIONS
TF and PSL conceived and designed the studies. TF and HC performed the experiments and analyzed and interpreted the data. TF and PSL drafted the manuscript. All authors read and approved the final manuscript.
DECLARATIONS OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- 1.Bennett V. Proteins involved in membrane--cytoskeleton association in human erythrocytes: spectrin, ankyrin, and band 3. Methods in enzymology. 1983;96:313–324. doi: 10.1016/s0076-6879(83)96029-9. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy SP, Warren SL, Forget BG, Morrow JS. Ankyrin binds to the 15th repetitive unit of erythroid and nonerythroid beta-spectrin. J Cell Biol. 1991;115:267–277. doi: 10.1083/jcb.115.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis L, Abdi K, Machius M, Brautigam C, Tomchick DR, Bennett V, Michaely P. Localization and structure of the ankyrin-binding site on beta2-spectrin. J Biol Chem. 2009;284:6982–6987. doi: 10.1074/jbc.M809245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ipsaro JJ, Huang L, Mondragon A. Structures of the spectrin-ankyrin interaction binding domains. Blood. 2009;113:5385–5393. doi: 10.1182/blood-2008-10-184358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lux SE, Tse WT, Menninger JC, John KM, Harris P, Shalev O, Chilcote RR, Marchesi SL, Watkins PC, Bennett V, et al. Hereditary spherocytosis associated with deletion of human erythrocyte ankyrin gene on chromosome 8. Nature. 1990;345:736–739. doi: 10.1038/345736a0. [DOI] [PubMed] [Google Scholar]
- 6.Eber S, Lux SE. Hereditary spherocytosis--defects in proteins that connect the membrane skeleton to the lipid bilayer. Seminars in hematology. 2004;41:118–141. doi: 10.1053/j.seminhematol.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Jarolim P, Murray JL, Rubin HL, Taylor WM, Prchal JT, Ballas SK, Snyder LM, Chrobak L, Melrose WD, Brabec V, Palek J. Characterization of 13 novel band 3 gene defects in hereditary spherocytosis with band 3 deficiency. Blood. 1996;88:4366–4374. [PubMed] [Google Scholar]
- 8.Palek J, Lux SE. Red cell membrane skeletal defects in hereditary and acquired hemolytic anemias. Seminars in hematology. 1983;20:189–224. [PubMed] [Google Scholar]
- 9.Agre P, Casella JF, Zinkham WH, McMillan C, Bennett V. Partial deficiency of erythrocyte spectrin in hereditary spherocytosis. Nature. 1985;314:380–383. doi: 10.1038/314380a0. [DOI] [PubMed] [Google Scholar]
- 10.Eber SW, Gonzalez JM, Lux ML, Scarpa AL, Tse WT, Dornwell M, Herbers J, Kugler W, Ozcan R, Pekrun A, Gallagher PG, Schroter W, Forget BG, Lux SE. Ankyrin-1 mutations are a major cause of dominant and recessive hereditary spherocytosis. Nature genetics. 1996;13:214–218. doi: 10.1038/ng0696-214. [DOI] [PubMed] [Google Scholar]
- 11.Hassoun H, Vassiliadis JN, Murray J, Njolstad PR, Rogus JJ, Ballas SK, Schaffer F, Jarolim P, Brabec V, Palek J. Characterization of the underlying molecular defect in hereditary spherocytosis associated with spectrin deficiency. Blood. 1997;90:398–406. [PubMed] [Google Scholar]
- 12.Bennett V, Gilligan DM. The spectrin-based membrane skeleton and micron-scale organization of the plasma membrane. Annu Rev Cell Biol. 1993;9:27–66. doi: 10.1146/annurev.cb.09.110193.000331. [DOI] [PubMed] [Google Scholar]
- 13.Reid ME, Takakuwa Y, Conboy J, Tchernia G, Mohandas N. Glycophorin C content of human erythrocyte membrane is regulated by protein 4.1. Blood. 1990;75:2229–2234. [PubMed] [Google Scholar]
- 14.Marfatia SM, Lue RA, Branton D, Chishti AH. In vitro binding studies suggest a membrane-associated complex between erythroid p55, protein 4.1, and glycophorin C. J Biol Chem. 1994;269:8631–8634. [PubMed] [Google Scholar]
- 15.Chang SH, Low PS. Regulation of the glycophorin C-protein 4.1 membrane-to-skeleton bridge and evaluation of its contribution to erythrocyte membrane stability. J Biol Chem. 2001;276:22223–22230. doi: 10.1074/jbc.M100604200. [DOI] [PubMed] [Google Scholar]
- 16.Khan AA, Hanada T, Mohseni M, Jeong JJ, Zeng L, Gaetani M, Li D, Reed BC, Speicher DW, Chishti AH. Dematin and adducin provide a novel link between the spectrin cytoskeleton and human erythrocyte membrane by directly interacting with glucose transporter-1. J Biol Chem. 2008;283:14600–14609. doi: 10.1074/jbc.M707818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anong WA, Franco T, Chu H, Weis TL, Devlin EE, Bodine DM, An X, Mohandas N, Low PS. Adducin forms a bridge between the erythrocyte membrane and its cytoskeleton and regulates membrane cohesion. Blood. 2009;114:1904–1912. doi: 10.1182/blood-2009-02-203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuoka Y, Li X, Bennett V. Adducin: structure, function and regulation. Cell Mol Life Sci. 2000;57:884–895. doi: 10.1007/PL00000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilligan DM, Lozovatsky L, Gwynn B, Brugnara C, Mohandas N, Peters LL. Targeted disruption of the beta adducin gene (Add2) causes red blood cell spherocytosis in mice. Proc Natl Acad Sci U S A. 1999;96:10717–10722. doi: 10.1073/pnas.96.19.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bianchi G, Tripodi G, Casari G, Salardi S, Barber BR, Garcia R, Leoni P, Torielli L, Cusi D, Ferrandi M, et al. Two point mutations within the adducin genes are involved in blood pressure variation. Proc Natl Acad Sci U S A. 1994;91:3999–4003. doi: 10.1073/pnas.91.9.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casari G, Barlassina C, Cusi D, Zagato L, Muirhead R, Righetti M, Nembri P, Amar K, Gatti M, Macciardi F, et al. Association of the alpha-adducin locus with essential hypertension. Hypertension. 1995;25:320–326. doi: 10.1161/01.hyp.25.3.320. [DOI] [PubMed] [Google Scholar]
- 22.Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M, Glorioso N, Lanzani C, Manunta P, Righetti M, Rivera R, Stella P, Troffa C, Zagato L, Bianchi G. Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349:1353–1357. doi: 10.1016/S0140-6736(97)01029-5. [DOI] [PubMed] [Google Scholar]
- 23.Franco T, Low PS. Erythrocyte adducin: a structural regulator of the red blood cell membrane. Transfus Clin Biol. 2010;17:87–94. doi: 10.1016/j.tracli.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thevenin BJ, Willardson BM, Low PS. The redox state of cysteines 201 and 317 of the erythrocyte anion exchanger is critical for ankyrin binding. J Biol Chem. 1989;264:15886–15892. [PubMed] [Google Scholar]
- 25.Willardson BM, Thevenin BJ, Harrison ML, Kuster WM, Benson MD, Low PS. Localization of the ankyrin-binding site on erythrocyte membrane protein, band 3. J Biol Chem. 1989;264:15893–15899. [PubMed] [Google Scholar]
- 26.Davis L, Lux SE, Bennett V. Mapping the ankyrin-binding site of the human erythrocyte anion exchanger. J Biol Chem. 1989;264:9665–9672. [PubMed] [Google Scholar]
- 27.Ding Y, Kobayashi S, Kopito R. Mapping of ankyrin binding determinants on the erythroid anion exchanger, AE1. J Biol Chem. 1996;271:22494–22498. doi: 10.1074/jbc.271.37.22494. [DOI] [PubMed] [Google Scholar]
- 28.Chang SH, Low PS. Identification of a critical ankyrin-binding loop on the cytoplasmic domain of erythrocyte membrane band 3 by crystal structure analysis and site-directed mutagenesis. J Biol Chem. 2003;278:6879–6884. doi: 10.1074/jbc.M211137200. [DOI] [PubMed] [Google Scholar]
- 29.Grey JL, Kodippili GC, Simon K, Low PS. Identification of contact sites between ankyrin and band 3 in the human erythrocyte membrane. Biochemistry. 2012;51:6838–6846. doi: 10.1021/bi300693k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vince JW, Reithmeier RA. Carbonic anhydrase II binds to the carboxyl terminus of human band 3, the erythrocyte C1−/HCO3− exchanger. J Biol Chem. 1998;273:28430–28437. doi: 10.1074/jbc.273.43.28430. [DOI] [PubMed] [Google Scholar]
- 31.Vince JW, Reithmeier RA. Identification of the carbonic anhydrase II binding site in the Cl(−)/HCO(3)(−) anion exchanger AE1. Biochemistry. 2000;39:5527–5533. doi: 10.1021/bi992564p. [DOI] [PubMed] [Google Scholar]
- 32.Korsgren C, Cohen CM. Associations of human erythrocyte band 4.2. Binding to ankyrin and to the cytoplasmic domain of band 3. J Biol Chem. 1988;263:10212–10218. [PubMed] [Google Scholar]
- 33.Jarolim P, Palek J, Rubin HL, Prchal JT, Korsgren C, Cohen CM. Band 3 Tuscaloosa: Pro327----Arg327 substitution in the cytoplasmic domain of erythrocyte band 3 protein associated with spherocytic hemolytic anemia and partial deficiency of protein 4.2. Blood. 1992;80:523–529. [PubMed] [Google Scholar]
- 34.Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. An alternative binding site for an element of the membrane skeleton. J Biol Chem. 1985;260:3676–3683. [PubMed] [Google Scholar]
- 35.Lombardo CR, Willardson BM, Low PS. Localization of the protein 4.1-binding site on the cytoplasmic domain of erythrocyte membrane band 3. J Biol Chem. 1992;267:9540–9546. [PubMed] [Google Scholar]
- 36.Bruce LJ, Ring SM, Anstee DJ, Reid ME, Wilkinson S, Tanner MJ. Changes in the blood group Wright antigens are associated with a mutation at amino acid 658 in human erythrocyte band 3: a site of interaction between band 3 and glycophorin A under certain conditions. Blood. 1995;85:541–547. [PubMed] [Google Scholar]
- 37.Tsai IH, Murthy SN, Steck TL. Effect of red cell membrane binding on the catalytic activity of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1982;257:1438–1442. [PubMed] [Google Scholar]
- 38.Campanella ME, Chu H, Low PS. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc Natl Acad Sci U S A. 2005;102:2402–2407. doi: 10.1073/pnas.0409741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu H, Low PS. Mapping of glycolytic enzyme-binding sites on human erythrocyte band 3. The Biochemical journal. 2006;400:143–151. doi: 10.1042/BJ20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murthy SN, Liu T, Kaul RK, Kohler H, Steck TL. The aldolase-binding site of the human erythrocyte membrane is at the NH2 terminus of band 3. J Biol Chem. 1981;256:11203–11208. [PubMed] [Google Scholar]
- 41.Higashi T, Richards CS, Uyeda K. The interaction of phosphofructokinase with erythrocyte membranes. J Biol Chem. 1979;254:9542–9550. [PubMed] [Google Scholar]
- 42.Chu H, Breite A, Ciraolo P, Franco RS, Low PS. Characterization of the deoxyhemoglobin binding site on human erythrocyte band 3: implications for O2 regulation of erythrocyte properties. Blood. 2008;111:932–938. doi: 10.1182/blood-2007-07-100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Low PS, Westfall MA, Allen DP, Appell KC. Characterization of the reversible conformational equilibrium of the cytoplasmic domain of erythrocyte membrane band 3. J Biol Chem. 1984;259:13070–13076. [PubMed] [Google Scholar]
- 44.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 45.Wang CC, Badylak JA, Lux SE, Moriyama R, Dixon JE, Low PS. Expression, purification, and characterization of the functional dimeric cytoplasmic domain of human erythrocyte band 3 in Escherichia coli. Protein Sci. 1992;1:1206–1214. doi: 10.1002/pro.5560010913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CC, Moriyama R, Lombardo CR, Low PS. Partial characterization of the cytoplasmic domain of human kidney band 3. J Biol Chem. 1995;270:17892–17897. doi: 10.1074/jbc.270.30.17892. [DOI] [PubMed] [Google Scholar]
- 47.Ding Y, Casey JR, Kopito RR. The major kidney AE1 isoform does not bind ankyrin (Ank1) in vitro. An essential role for the 79 NH2-terminal amino acid residues of band 3. J Biol Chem. 1994;269:32201–32208. [PubMed] [Google Scholar]
- 48.Tanner MJ. The structure and function of band 3 (AE1): recent developments (review) Molecular membrane biology. 1997;14:155–165. doi: 10.3109/09687689709048178. [DOI] [PubMed] [Google Scholar]
- 49.Schofield AE, Martin PG, Spillett D, Tanner MJ. The structure of the human red blood cell anion exchanger (EPB3, AE1, band 3) gene. Blood. 1994;84:2000–2012. [PubMed] [Google Scholar]
- 50.Brosius FC, 3rd, Alper SL, Garcia AM, Lodish HF. The major kidney band 3 gene transcript predicts an amino-terminal truncated band 3 polypeptide. J Biol Chem. 1989;264:7784–7787. [PubMed] [Google Scholar]
- 51.Kollert-Jons A, Wagner S, Hubner S, Appelhans H, Drenckhahn D. Anion exchanger 1 in human kidney and oncocytoma differs from erythroid AE1 in its NH2 terminus. Am J Physiol. 1993;265:F813–F821. doi: 10.1152/ajprenal.1993.265.6.F813. [DOI] [PubMed] [Google Scholar]
- 52.Gardner K, Bennett V. A new erythrocyte membrane-associated protein with calmodulin binding activity. Identification and purification. J Biol Chem. 1986;261:1339–1348. [PubMed] [Google Scholar]
- 53.Dong L, Chapline C, Mousseau B, Fowler L, Ramsay K, Stevens JL, Jaken S. 35H, a sequence isolated as a protein kinase C binding protein, is a novel member of the adducin family. J Biol Chem. 1995;270:25534–25540. doi: 10.1074/jbc.270.43.25534. [DOI] [PubMed] [Google Scholar]
- 54.Joshi R, Gilligan DM, Otto E, McLaughlin T, Bennett V. Primary structure and domain organization of human alpha and beta adducin. J Cell Biol. 1991;115:665–675. doi: 10.1083/jcb.115.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Appell KC, Low PS. Partial structural characterization of the cytoplasmic domain of the erythrocyte membrane protein, band 3. J Biol Chem. 1981;256:11104–11111. [PubMed] [Google Scholar]
- 56.Pang AJ, Bustos SP, Reithmeier RA. Structural characterization of the cytosolic domain of kidney chloride/bicarbonate anion exchanger 1 (kAE1) Biochemistry. 2008;47:4510–4517. doi: 10.1021/bi702149b. [DOI] [PubMed] [Google Scholar]
- 57.Zhang D, Kiyatkin A, Bolin JT, Low PS. Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood. 2000;96:2925–2933. [PubMed] [Google Scholar]
- 58.Stefanovic M, Puchulu-Campanella E, Kodippili G, Low PS. Oxygen regulates the band 3-ankyrin bridge in the human erythrocyte membrane. The Biochemical journal. 2013;449:143–150. doi: 10.1042/BJ20120869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Franco T. Characterization and mapping the interaction between the cytoplasmic domain of band 3 and adducin. Purdue University; 2011. ed.)^eds.) [Google Scholar]
- 60.Wu F, Saleem MA, Kampik NB, Satchwell TJ, Williamson RC, Blattner SM, Ni L, Toth T, White G, Young MT, Parker MD, Alper SL, Wagner CA, Toye AM. Anion exchanger 1 interacts with nephrin in podocytes. J Am Soc Nephrol. 2010;21:1456–1467. doi: 10.1681/ASN.2009090921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keskanokwong T, Shandro HJ, Johnson DE, Kittanakom S, Vilas GL, Thorner P, Reithmeier RA, Akkarapatumwong V, Yenchitsomanus PT, Casey JR. Interaction of integrin-linked kinase with the kidney chloride/bicarbonate exchanger, kAE1. J Biol Chem. 2007;282:23205–23218. doi: 10.1074/jbc.M702139200. [DOI] [PubMed] [Google Scholar]
- 62.Su Y, Al-Lamki RS, Blake-Palmer KG, Best A, Golder ZJ, Zhou A, Karet Frankl FE. Physical and functional links between anion exchanger-1 and sodium pump. J Am Soc Nephrol. 2015;26:400–409. doi: 10.1681/ASN.2013101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrandi M, Cusi D, Molinari I, Del Vecchio L, Barlassina C, Rastaldi MP, Schena FP, Macciardi F, Marcantoni C, Roccatello D, Peters LL, Armelloni S, Min L, Giardino L, Mattinzoli D, Camisasca C, Palazzo F, Manunta P, Ferrari P, Bianchi G. alpha- and beta-Adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy. J Mol Med (Berl) 2010;88:203–217. doi: 10.1007/s00109-009-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Efendiev R, Krmar RT, Ogimoto G, Zwiller J, Tripodi G, Katz AI, Bianchi G, Pedemonte CH, Bertorello AM. Hypertension-linked mutation in the adducin alpha-subunit leads to higher AP2-mu2 phosphorylation and impaired Na+,K+-ATPase trafficking in response to GPCR signals and intracellular sodium. Circ Res. 2004;95:1100–1108. doi: 10.1161/01.RES.0000149570.20845.89. [DOI] [PubMed] [Google Scholar]
- 65.Torielli L, Tivodar S, Montella RC, Iacone R, Padoani G, Tarsini P, Russo O, Sarnataro D, Strazzullo P, Ferrari P, Bianchi G, Zurzolo C. alpha-Adducin mutations increase Na/K pump activity in renal cells by affecting constitutive endocytosis: implications for tubular Na reabsorption. Am J Physiol Renal Physiol. 2008;295:F478–F487. doi: 10.1152/ajprenal.90226.2008. [DOI] [PubMed] [Google Scholar]