Abstract
Alzheimer's disease (AD) and age-related cognitive decline represent a growing health burden and involve the hippocampus, a vulnerable brain region implicated in learning and memory. To understand the molecular effects of aging on the hippocampus, this study characterized the gene expression changes associated with aging in rodents using RNA-sequencing (RNA-seq). The glutamate modulator, riluzole, which was recently shown to improve memory performance in aged rats, prevented many of the hippocampal age-related gene expression changes. A comparison of the effects of riluzole in rats against human AD datasets revealed that many of the genes changes in AD are reversed by riluzole. Expression changes identified by RNA-Seq were validated by qRT-PCR open arrays. Riluzole is known to increase the glutamate transporter EAAT2's ability to scavenge excess glutamate, regulating synaptic transmission. RNA-seq and immunohistochemistry confirmed an increase in EAAT2 expression in hippocampus, identifying a possible mechanism underlying the improved memory function after riluzole treatment.
INTRODUCTION
Aging is associated with cognitive decline in humans which impairs quality of life and contributes significantly to healthcare costs1. This decline is also observed in rodents and nonhuman primates2. Aging is the primary risk factor for the dementia of Alzheimer's disease (AD) and with significant increases in life expectancy, the prevalence of AD and age-related cognitive disorders is rising3. The neural circuits affected in aging and AD are similar, involving the glutamatergic connections between cortical areas and with the hippocampal formation memory formation, a brain region in the medial temporal lobe that is critical for . memory formation 4,5,6. The glutamatergic pyramidal neurons of the hippocampus are highly vulnerable to damage in both age-related cognitive decline and in AD5, 7. However, the effects of glutamatergic modulation on aging remains unknown.
Riluzole is a glutamate modulator approved for treatment of ALS8. Importantly, riluzole treatment for 4 months prevented age-related cognitive decline in rodents through clustering of dendritic spines 9, which form the postsynaptic component of most excitatory synapses 10. Clustering of synaptic inputs is an important neuroplastic mechanism that increases synaptic strength, empowering neural circuits 11, 12. Riluzole's ability to induce dendritic spines clustering 9, which is dependent on glutamatergic neuronal activity 13, 14 and long-term potentiation (LTP) 15, suggests that it regulates synaptic glutamatergic activity and prevents glutamate overflow to the extra-synaptic space. Synaptic NMDA activity is critical for LTP and memory formation, while extrasynaptic NMDA activation is associated with long-term depression (LTD) and excitotoxicity16-18.
The excitatory amino acid transporter 2 (EAAT2 or GLT-1; Slc1a2) is a high-affinity, Na+-dependent glutamate transporter and the dominant glutamate transporter in the brain 19, 20. Glutamate transporters including EAAT2 are decreased in aging 21, 22 and in AD 23, 24 and associated with neurodegeneration24. Glutamate transporters play a critical role in determining synaptic and extrasynaptic glutamate levels 20, 25, regulating physiological glutamatergic neurotransmission. Riluzole can act to stabilize the inactivated state of the voltage-gated sodium channel and it can increase EAAT2 expression 22, 26-28, potentiating glutamate uptake 28-30.
Understanding the molecular vulnerabilities of glutamatergic neural circuits can point to novel and more effective treatment targets. Additionally, molecular changes resulting from treatments that prevent cognitive decline remain largely unexplored. This study uses the combination of RNA-seq and open arrays to detect and validate specific molecular pathways that are changed by aging and with riluzole. Importantly, gene expression changes associated with a rescue of cognitive decline under a therapeutic intervention (riluzole) are identified. Additionally, genes modulated by riluzole in the rat hippocampus are enriched in many of the same pathways, and in the opposite direction, as those altered in human AD gene expression datasets. These molecular transcriptional profiles in the aging hippocampus and with glutamatergic modulation by riluzole provide mechanistic insights into age-related cognitive decline and provide support for future studies on the role of glutamate transporters as potential therapeutic targets in the aging brain.
MATERIALS AND METHODS
Animals
Young (3 month old) and aged male Sprague-Dawley rats (retired breeders, 10 months old, Harlan Laboratories) were housed at Rockefeller University for the duration of the experiments. All rats were pair-housed in climate controlled conditions (30-50% humidity, 21 ± 2 °C, 12-h light/dark cycle). Separate cohorts of animals were used for RNA extraction and immunohistochemical experiments. For RNA-seq experiments, each group had n=6: 3 month-old (young rats), 10 month-old (middle age rats), 14 month-old riluzole treated rats and 14 month-old riluzole untreated rats. For immunohistochemistry, 3 month-old rats (n=10), 14 month-old untreated rats (n=9) and 14 month-old riluzole treated rats (n=10) were used. All procedures were in agreement with the National Institutes of Health and The Rockefeller University Institutional Animal Care and Use Committee guidelines. Sample sizes were chosen to minimize the number of animals used given previously published reports using these methodologies 31,32, 33.
Riluzole treatment
Treated rats had ad libitum access to riluzole solution from 10 months to 14 months old (17 weeks), and aged-control and young-control rats had ad libitum access to tap water. All rats had ad libitum access to food. The riluzole compound (Sigma–Aldrich, Inc.) was dissolved in tap water at a concentration of 110 μg/mL, translating to
~4.0 mg·kg−1·d−1 p.o. To make the solution, riluzole was stirred in room temperature tap water for 6 hours. All containers with riluzole were covered in foil to prevent light exposure. Fresh solutions were made every 2–3 d for the duration of treatment.
Tissue processing and Immunohistochemistry
One week after the end of riluzole treatment, rats were deeply anesthetized with 100 mg/kg sodium pentobarbital and transcardially perfused with 1.0% paraformaldehyde in 0.1 M phosphate buffer (1 minute) followed by 4.0% paraformaldehyde + 0.125% Glutaraldehyde in 0.1 M PB (12 minutes). Brains were removed and post-fixed for 6 hours in 4.0% paraformaldehyde + 0.125% Glutaraldehyde in 0.1 M PB (4°C) and transferred to 0.1% sodium azide in PB (4°C) until cutting the following day. Brains were cut on a vibratome (Leica, VT1000S) into 40 μm coronal for immunohistochemistry. Sections were stored in 0.1% sodium azide in PB (4°C). Sections from each animal were washed with phosphate buffered saline (PBS), blocked with 1% bovine serum albumin (BSA), and incubated in primary antibody for GLT-1a (1:1000 dilution in PBS; gift from J. Rothstein Lab) overnight at 4°C. The tissue was then washed in PBS, and incubated with fluorescent secondary antibody (AlexaFluor488) for one hour. The intensity of the labeling was quantified using Nikon Imaging software in 50 um intervals from the cellular layer in each region of hippocampus. Electronic images were coded to blind the rater.
RNA Extraction, Sequencing and Analysis
Wet dissected hippocampus from rapidly decapitated rats were flash frozen on dry ice and stored at −80. RNA was extracted using the RNAeasy Lipid Kit (Qiagen) and Qiacube per the manufacturer's instructions. Samples were pooled for sequencing and final RNA integrity was checked using the Bioanalyzer (Agilent) prior to library preparation. All samples had RNA Integrity Numbers >8. Sequencing libraries were prepared by the Rockefeller University Genomics Core Facility using the TruSeq RNA Library Preparation Kit v2 (Illumina) and bar-coded for multiplexing so that all groups could be run in the same flow cell. 100bp single-stranded reads were collected on a HiSeq 2500 (Illumina) at a sequencing depth of approximately 60 million reads per sample.
Raw data files were uploaded to Galaxy 34, 35 and checked for integrity by FastQC. To remove sequencing artifacts and residual adaptor sequences, reads were trimmed by 5-10bp at the 5’ and 3’ ends and then filtered to remove reads with quality scores <20. Reads were aligned to the rat genome (rn5) using TopHat2 36 and then loaded into Strand (Agilent) for quantification of read density by DESeq. Differential expression analyses were conducted in Strand using Z-Tests that were Benjamin-Hochberg corrected for false discovery rates. Venn diagrams and scatter plots based on significant gene lists were generated using Strand and Microsoft Excel.
Open Array Analysis
RNA was extracted from whole hippocampus as described above. cDNA was synthesized using the VILO kit (Life Technologies) using 2 micrograms of RNA for each reaction. Open array plates were loaded from a 384 well plate as described in the standard Open Array protocol (Life Technologies) and run on a Quantstudio 12k Flex thermocycler. Counts were exported to Microsoft Excel and used to calculate fold change using the ΔΔCt method 37. All values were normalized to pgk1 expression. Open array fold change values were plotted against RNA-seq results to generate scatter plots and calculate R^2 values (Prism Software).
Pathway Analysis
Gene lists from RNA-Seq results were uploaded to the DAVID bioinformatics database (http://david.abcc.ncifcrf.gov/home.jsp). The functional clusters, enrichment scores, and gene ontology (GO) terms for these categories were obtained from the functional annotation clustering tool. In all of the analysis, enrichment scores above 1.3 were considered significant (refelects p<0.05). (http://www.nature.com/nprot/journal/v4/n1/full/nprot.2008.211.html).
The enrichment scores from clusters with similar GO terms were used to compare pathways that were altered in both riluzole and control conditions. Histograms were generated in Excel (Microsoft).
Comparisons with previous human AD expression studies
AD expression data were analyzed from the GEO and AMP-AD databases and lists of significantly changed genes (p<0.05) were generated for each study. Some studies examined whole hippocampus 38,39,40-42,88, whereas others used laser capture microdissection to examine specific subregions of hippocampus, such CA1 and CA3 regions 43 or CA1 alone44, or dentate gyrus and entorhinal cortex45. Therefore, to control for gene differences that arise from using different regions of the hippocampus and identify the most robust findings that were replicated across studies, only genes identified as significant in at least two studies were used for analysis. There were 2,024 genes that were up regulated and 1,870 genes that were down regulated with AD that met these criteria. These gene lists were analyzed with the DAVID functional annotation clustering tool as described above. The enrichment scores from these clusters were used to compare genes altered with AD to genes altered with riluzole treatment and the pathways with the highest combined enrichment scores are presented.
Genes significantly changes in aging studies in rats (p<0.05) were also analyzed using the GEO database. One study examined gene expression in the CA1 subregion at different points across the lifespan 46. Gene lists showing differential expression between 3 month and 23 month old rats, and between 3 month and 12 month old rats were obtained. A second study compared the genes from the whole hippocampus of 4-6 month40 and 24-26 month old rats 47. To control for the differences between regions of the hippocampus, only genes that were significant in both studies were used for analysis. There were 508 genes upregulated and 270 genes that were down-regulated in aged rats. These gene lists were analyzed with DAVID functional annotation clustering tool as described above. The enrichment scores from these clusters were compared to the enrichment scores of similar clusters obtained from the 443 genes that were down regulated and the 674 genes that were upregulated with age in our RNA-seq data.
RESULTS
Riluzole rescues age-related expression changes in rats
Hippocampal transcriptional profiles change markedly across the life span. In this study, rats between 3 months (young) and 10 months of age (middle-aged) showed 268 genes increased and 254 genes decreased. There were nearly twice as many changes from the 10 month to 14 month old (aged) rats, with 674 genes increased and 443 genes decreased, demonstrating that transcriptional changes are stable across adulthood, but accelerate from middle age onwards (Figure 1a; Supp.Table1).
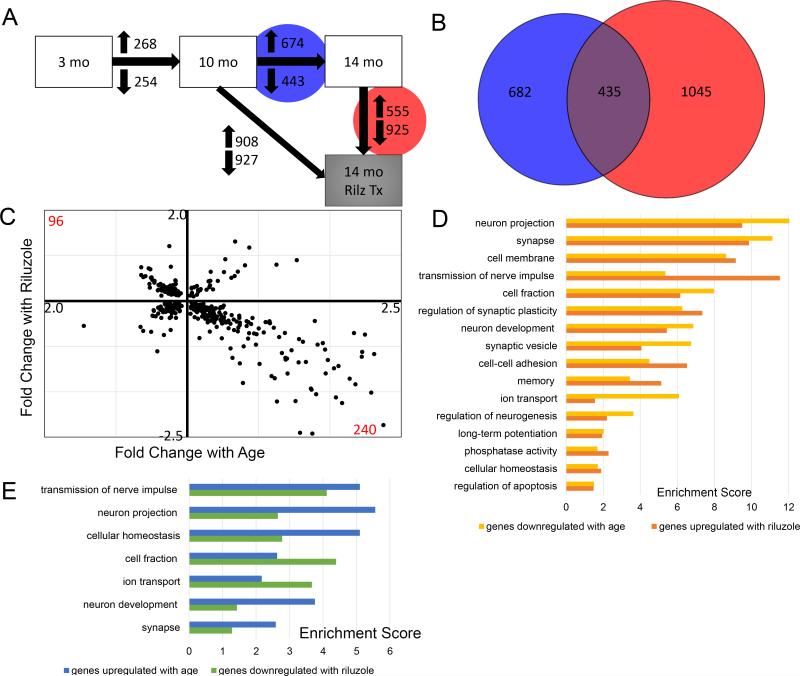
Figure 1. Riluzole treatment of aged rats rescues age-related gene expression changes in the hippocampus.
A. Differential expression analysis revealed gene expression changes across age and with riluzole treatment. Between 10 month old and 14 month old rats (Blue Circle), 674 genes were upregulated and 443 genes downregulated. 1,480 genes were changed between age-matched riluzole treated rats and controls (red Circle), with 555 genes increased and 925 genes decreased. B. Venn diagram illustrating the overlap of 435 genes that were changed by both aging (blue; 10 to 14 months) and by riluzole treatment (Red). C. Scatter plot illustrating the 435 overlapping genes showing fold change by age (x-axis; 10-14 month) against fold change with riluzole (y-axis). The upper left quadrant represent 96 genes that had decreased expression with age and increased expression after riluzole treatment. Conversely, the lower right quadrant illustrates 240 genes that were increased with age and decreased by riluzole. D. Histograms illustrating significantly enriched pathways based on genes differentially expressed by either aging or riluzole treatment (Enrichment score >1.3 reflects p<0.05). Similar pathways and enrichment scores were observed when comparing genes decreased by aging (yellow bars) and increased by riluzole (orange bars), as well as for genes increased with age (blue bars) and decreased by riluzole (green bars).
Animals treated with the glutamate modulator, riluzole, from 10 until 14 months had 908 genes increased and 927 genes decreased (Fig. 1A). Importantly, there is a large overlap of genes (435) that were changed with aging and were also altered by riluzole treatment (Fig. 1B). The overlapping genes were plotted to show fold change by age against fold change by riluzole treatment (Fig 1C). The lower right quadrant reflects 240 genes that increased with age and were decreased with riluzole treatment. In the upper left quadrant, 96 genes that were decreased with age were increased with riluzole treatment. This profile suggests that riluzole treatment rescues many age-related gene expression changes in the hippocampus.
Differentially expressed genes were organized into functional pathways using the DAVID pathway tools. The pathway classes that were reversed by riluzole are ranked by significance and divided into those that were upregulated with age and downregulated by riluzole (Fig. 1D) and pathways that were downregulated with age and upregulated by riluzole (Fig. 1E). Many pathways reversed by riluzole treatment were related to synaptic transmission and plasticity. Examples of genes altered by aging that were reversed by riluzole are provided in Table 1. The NMDA receptor subunit NR2b (GRIN2b), a voltage-gated sodium channel subunit (Scn2a1), a calcium/calmodulin protein kinase II alpha (CAMK2A), the microtubule-associated protein 1B (MAP1B), the synaptic scaffolding protein enriched in the postsynaptic density of excitatory synapses SHANK3, the matrix metalloproteinase 9 (MMP9), each decrease with aging and are increased by riluzole treatment, and have been implicated in learning and neuroplasticity 48, 49,50-54. In contrast, isoforms of the GABA receptor (GABRA6) are found to increase with aging and are decreased by riluzole treatment and their blockage may improve memory consolidation 50,55.
TABLE 1.
Pathway groups of genes that are changed with age and reversed by Riluzole
| Pathways | Genes that are downregulated with age and upregulated with riluzole | Pathways | Genes that are upregulated with age and downregulated with riluzole |
|---|---|---|---|
| transmission of nerve impulse | ank2, Clstn3, EIF2B2, GPI, GRIN2B, Nsf, PRKCG Scn2a1, SV2B, Syn2, SYNJ1, Syt1, Uchl1, vamp2 |
transmission of nerve impulse | abca4, ALS2, CACNB4, camk4, Gabra6, GJC3, Grid2, MBP PDE4D, PLP1, scd, Scn1a, SLC12A2, Syt2, thbs2 TRPV4, Unc13c, wfs1 |
| synapse | Ap2a2, BSN, CACNA1E, CADM3, CaIY, Camk2a, cdk5r1 Clstn3, Gabra2, GNG2, GRIN2B, MAP1B, mras, Nsf Scn2a1, SHANK3, SNAP91, SV2B, Syn2, Syt1, vamp2 |
neuron projection | ALS2, aqp1, CALD1, Canx, Car2, Cst3, Gabra6, MBP MCAM, PEX5L, Plcb4, Pvalb, Scn1a, TPH1, wfs1 |
| neuron projection | Camk2a, Cdk5r1, GRIN2B, HTR1A, MAP1B, Nsf PRKCG, Scn2a1, Syt1, Uchl1 |
cellular homeostasis | ADIPOQ, CACNB4, Car2, GJC3, Grid2, HFE, ID2, ITPR1 PLP1, PTPN11, SCARA5, scd, Scn1a, SLC12A2, SLC4A5 MBP, PEX5L, srprb, TEX15, TF, tgm2, TRPV4, VEGF4, wfs1 |
| regulation of synaptic plasticity | Camk2a, GRIN2B, MAP1B, Mmp9, nisch, Syn2, YWHAG | cell fraction | abcc9, ACE, ADAM10, ALS2, BCAS1, CALD1, CTSB, CTSD, GFAP Grid2, ITPR1, PDE4D, PEX5L, Plcb4, PON1, PTPN11, scd Scn1a, SLC12A2, Slco1a5, Steap2, stk39, Syt2, sytl3 |
| neuron development | ank2, Cdk5r1, CELSR2, Dgkg, MAP1B Nnat, slit1, Uchl1, Unc5a. WNT7B |
||
| synaptic vesicle | Ap2a2, atp6v1b2, calY, Camk2a, Clstn3, COX8A, CPE GRIN2B, mdh2, PI4KA, SNAP91, SV2B, Syn2, Syt1 trh, Uqcrh, vamp2, YWHAB |
ion transport | abcc9, aqp1, ATP2A3, CACNB4, Clic6, Gabra6, GABRB2 Grid2, GULP1, ITPR1, KCNE2, Kcnj13, ptgds, RAB11FIP1 SCARA5, Scn1a, SCN4B, SFT2D2, SLC12A2, Slc12a4, Slc13a4 SLC31A1, SLC4A2, SLC4A5, Slc5a5, Slco1a5, srprb, TF, TRPV4, TTR |
| memory | GPI, GRIN2B, PRKCG, SYNJ1, trh, Uchl1 | neuron development | ALS2, BARHL2, bmp7, Chn2, clu, DAB2, En2, etv1, EZR LMX1A, Mmp2, Nrep, OLFM3, Otx2, PTPN11, VEGFA |
| cell membrane | ank2, Ap2a2, atp6v1b2, BSN, CACNA1E, CADM3 calY, Camk2a, cbx6, Cdk5r1, CELSR2, Clstn3, COX8A CTXN1, Gabra2, GNG2, GRIN2B, HTR1A, LPPR2, Isamp MAP1B, mras, nisch, nptxr, PTPRN, Ptprs, Scn2a1 SHANK3, SNAP91, SV2B, Syn2, Syt1, trh, Unc5a, Uqcrh vamp2, WNT7B |
synapse | |
| ACE, ADAM10, ALS2, CACNB4, cadps2, CALD1, cbln1 CDH3, cgnl1, CLDN2, Clic6, CTSC, Gabra6, GABRB2, GJC3 Grid2, htr2c, ITPR1, Kcnj13, Ocln, Plcb4, PRLR, Scn1a sdc1, SLC12A2, Slc12a4, SLC4A2, SLC4A5, Slc5a5, Slco1a5 Syt2, Unc13c | |||
| cell-cell adhesion | CADM3, Cdk5r1, CELSR2, Clstn3, Isamp, PCDH1 PCDHGA10, Ptprs, WNT7B |
||
| cell fraction | ank2, BSN, CPE, GPI, GRIN2B, MAP1B, nisch, PCDHGA10 PRKCG, PTPRN, Scn2a1, SYNJ1, vamp2 |
||
| ion transport | Ap2a2, atp6v1b2, CACNA1E, Camk2a, Gabra2, GRIN2B Nsf, Scn2a1, SV2B, Uqcrh |
||
| cellular homeostasis | ank2, atp6v1b2, CACNA1E, EIF2B2, GRIN2B, Scn2a1 | ||
| phosphatase activity | LPPR2, PTPRG, PTPRN, PTPRO, Ptprs, SYNJ1 | ||
| regulation of apoptosis | Cdk5r1, GPI, Mmp9, UBB, UBC, YWHAB | ||
| long-term potentiation | Camk2a, GRIN2B, PRKCG, tcf3, WNT7B | ||
| regulation of neurogenesis | calY, MAP1B, SYNJ1, WNT7B, YWHAG |
Notably, several neuroprotective genes were increased with riluzole treatment, including tropomyosin receptor kinase B (TrkB; NTRK2), which is a receptor for brain-derived neurotrophic factor (BDNF) 56(SuppTable2). An example of a gene that was significantly decreased with treatment but we did not find to be changed with aging is the cysteine/glutamate antiporter (xCT;Slc7a11) that exchanges extracellular cysteine for intracellular glutamate 57. It is a non-vesicular glutamate release system that may contribute to the regulation of extra-synaptic glutamate levels 57 (Supp Table 2).
Gene pathways implicated in AD are altered by riluzole
Hippocampal data from nine studies characterizing mortem hippocampal tissue from AD patients 38, 39, 43, 44,40-42, 45,88 were obtained from the GEO (Gene Expression Omnibus, NCBI) and AMP-AD (Accelerating Medicines Partnership-Alzheimer's Disease) databases. Differentially expressed genes identified in at least two of the nine studies were analyzed in DAVID for comparison with the present pathway results (Fig. 2A; Supp.Tables 3,4). Many of the pathways altered across studies in AD are also changed with riluzole treatment, including ones related to transmission of nerve impulse and synaptic plasticity (Fig. 2B). Examples of genes altered in both AD tissue and by riluzole treatment in rats can be found in Table 2. Importantly, the glutamate transporter EAAT2 is significantly decreased in AD, as well as in aging rats, and was rescued by riluzole treatment. Several genes previously implicated in neural transmission and plasticity are diminished in AD and recovered by riluzole treatment, including: ANK3, an integral membrane protein to the underlying spectrin-actin cytoskeleton that mediates synaptic morphology and transmission 58; CAMK2, a calcium/calmodulin kinase protein and major component of the postsynaptic density that is critically involved in induction of synaptic potentiation and memory 51, 59 and Rab3A, a vesicular trafficking protein that is crucial for synaptic plasticity, learning and memory 60, 61.
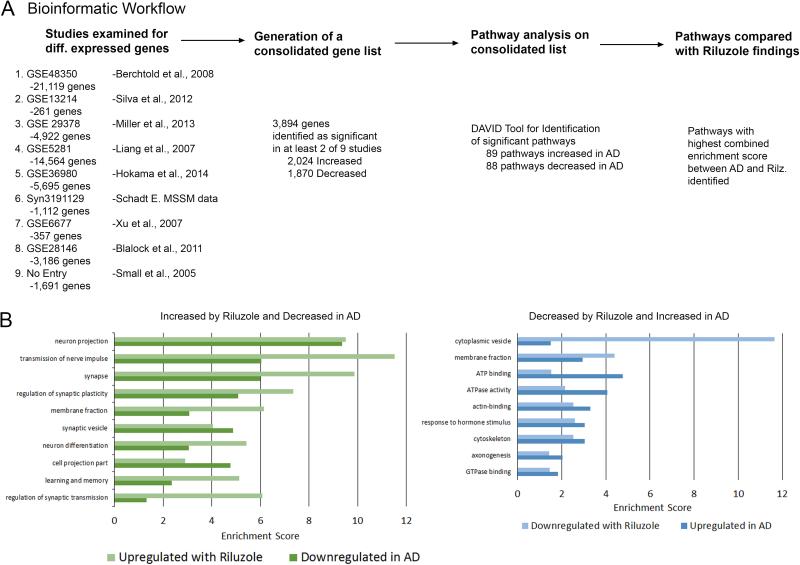
Figure 2. Gene pathways changed by riluzole in aged rats are similar and in the opposite direction to those changed in post-mortem AD brains.
A. Schematic representation of the bioinformatics strategy used to generate pathway lists. Only genes significantly differentially expressed in at least two studies were included. B. Significantly enriched pathways derived from genes upregulated by riluzole (light green bars) were similar to gene pathways downregulated in AD brains (dark green bars). Gene pathways downregulated by riluzole (light blue bars) also showed similarity to pathways that are increased in AD (dark blue bars). Differentially expressed gene lists from AD brains were derived from publically available data posted in GEO and AMP-AD and subjected to pathway analysis using DAVID. (Enrichment score >1.3 reflects p<0.05)
Table 2.
| Selected Genes | |||
|---|---|---|---|
| Pathways | Downregulated with Alzheimer's Upregulated with Riluzole |
Pathways | Upregulated with Alzheimer's Downregulated with Riluzole |
| neuron projection | ANK3, APC, Cdk5r1, CLSTN1, DLGAP2, DLGAP3, EVL Gabbr2, Gad1, GAD2, gas7, GRIN2B, Nsf, PARK7 ptprf, Slc1a2, SLC6A1, Syt1, Uchl1 |
cytoplasmic vesicle |
A2M, abcc4, ADAM10, AP1AR, APP, bmp7 clu, CTSB, CTSD, DAB2 |
| transmission of nerve impulse | atp1a3, CHST10, DLGAP2, DLGAP3, DLGAP4, Egr1, Epas1, Gad1, GAD2, GRIN2B, kcnip3, NCAN, PARK7 | membrane fraction | abcc4, ALS2, APP, ASAM, CALD1, MPDZ |
| ptprf, RAB3A, SLC6A1, Syt1, Uchl1 | ATP binding | abcc4, abcc9, ACSM5, ATAD1, ATAD2, Atp11a, Atrx DDX17, EIF4A1, Kif1c, KIF27, Rragd, SYNCRIP |
|
| synapse | ANK3, APC, BSN, CADM3, Camk2a, Cdk5r1, CLSTN1 Dlg2, DLGAP2, DLGAP3, DLGAP4, EVL, Gabbr2, Gad1 |
ATPase activity | abcc4, abcc9, Atp11a, BHMT2, EIF4A1 |
| GAD2, GRIN2B, ptprf, RAB3A, Slc1a2, SNAP91, SV2B, Syt1 | actin-binding | baiap2l1, CALD1, CALM1, CALM2, CALM3, CDK5RAP2 cgnl1, nrcam |
|
| regulation of synaptic plasticity | Camk2a, Egr1, Epas1, GNAO1, GRIN2B, RAB3A SLC6A1, YWHAG |
response to hormone stimulus |
A2M, ADAM10, aqp1, bmp7, CDKN1A, CTSC, IGF2 |
| membrane fraction | Amfr, ANK3, BSN, DLGAP3, DLGAP4, GNAO1 GRIN2B, ptprf, PTPRN, RAB3A, Slc1a2, SLC6A1 |
cytoskeleton | ADAM10, Adarb1, AKAP12, ALS2, APP, Atrx, baz1b CALD1, CALM1, CALM2, CALM3, CDK5RAP2, CDYL cgnl1, EML5, GFAP, Kif1c, KIF27, MPDZ, PDS5A, STAG2 |
| synaptic vesicle | Ap2a2, atp6v1b2, calY, Camk2a, CAMK2D, Gad1 GAD2, GRIN2B, pam, RAB3A, SV2B, Syt1 |
axonogenesis |
ALS2, APP, baiap2l1, bmp7, clu, DAB2, nrcam |
| neuron differentiation | ANK3, APC, Cdk5r1, CELSR2, DCLK1, dlx1, gas7 GNAO1, RAB3A, Uchl1 |
GTPase binding | ALS2 |
| cell projection part | APC, Cdk5r1, DLGAP3, GRIN2B, Nsf, Slc1a2 | ||
| learning and memory | atp1a3, CHST10, Cx3cl1, Egr1, Epas1, GNAO1 GRIN2B, kcnip3, PARK7, RAB3A, SLC1A2, SLC6A1, Uchl1 |
||
| regulation of synaptic plasticity | GRIN2B | ||
Validation of RNA-Sequencing results
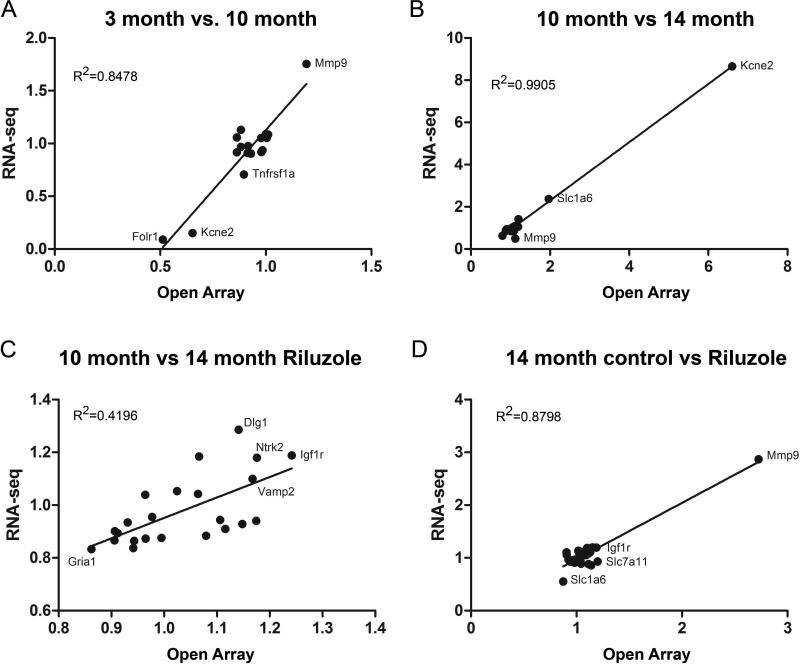
Custom open array technology allows for high through qRT-PCR analysis of gene expression. Fifty-three genes of interest were selected to assay given their known roles in neuroplasticity, glutamate signaling, learning and memory, 39 of which were significant between at least one RNA-seq conditions (Supp Table 5). A strong correlation in fold change was observed between genes identified as significant by RNA-seq and the open array measurements, with three of the comparisons exhibiting R^2 values >0.80 (Fig. 3). Conversely, no significant differences in gene expression were observed in the open array analysis that were not already identified as significant by RNA-seq. This concordance, of both the positive and negative data for these genes, demonstrates that our sequencing analysis reflects reliable changes in gene expression.
Figure 3. Expression differences determined by qRT-PCR are highly correlated with RNA-seq results.
Scatter plots illustrating fold change levels determined by qRT-PCR open arrays (x-axis) plotted against fold change level calculated from RNA-seq analysis (y-axis) for each comparison group. Only genes that reached significance between each condition in the RNA-seq analyses are represented in the scatter plots (Benjamin-Hochberg corrected p<0.05). Several genes of interest and R^2 values for each comparison are highlighted.
To further validate the RNA-seq findings, results were compared against previous reports using microarray technology to study gene expression changes with aging in rat hippocampus 46, 47. Differentially expressed gene lists from rat studies were obtained using the GEO database and analyzed for pathway enrichment using DAVID. Many of the same gene pathways were changed in both the RNA-seq data and previously published reports of aged rats (Supp.Fig. 1). This is despite differences in the exact age of the animals, tissue collection techniques and the different array technologies used across these studies. Together, the bioinformatics and open array results suggest the RNA-sequencing data are representative of age and riluzole induced gene expression changes.
Riluzole rescues EAAT2 levels after aging
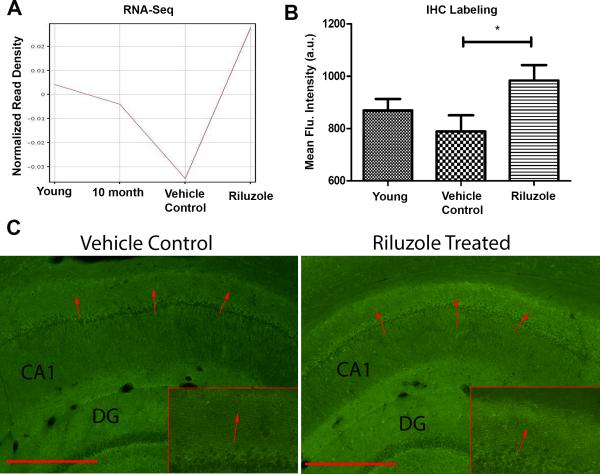
Riluzole is known to increase EAAT2 expression22, 26-28, which helps maintain the correct amount of glutamate in the synaptic cleft 20. Failure of EAAT2 leads to glutamate spillover to the extrasynaptic space, which can cause decreased synaptic efficiency, long-term depression and excitotoxicity17, 62. EAAT2 is expressed in neurons, axon terminals and glial cells 63-67. Previous studies have shown EAAT2 is decreased with aging and AD 21, 24, 23. RNA-seq results from our rodent experiments confirm EAAT2 is downregulated with age, but importantly, levels of this gene are rescued by Riluzole treatment (Fig. 4A). Further, we identified increased immunohistochemical labeling for EAAT2 in the distal portion of CA1 (Fig. 4B,C), which confirms the RNA-seq findings, and suggests a potential mechanism by which riluzole may rescue cognitive function to be further validated in future studies. Importantly, this region corresponds to the area in which increased dendritic spine clustering occurred in response to riluzole treatment 9.
Figure 4. Riluzole increases EAAT2 expression.
A. Normalized expression values from RNA-seq data for EAAT2 (y-axis) show that gene expression decreases with age (x-axis) but is restored by riluzole treatment. B. Quantification of fluorescent intensity (y-axis) of CA1 hippocampal sections labeled for EAAT2. Riluzole significantly increased labeling in the region 150-200 um from the pyramidal cell bodies in aged rats (p<0.05). C. Example images from 14 month control and 14 month old rats treated with riluzole. Red arrows indicate regions of difference.
DISCUSSION
This study reveals gene expression changes that occur with aging and glutamatergic modulation by riluzole in the rat hippocampus. The majority of transcriptional changes identified occurred from middle age to aged rats, rather than from young to middle aged animals, suggesting that a loss of the transcriptional stability during adulthood occurs with aging (Fig. 1A). Importantly, riluzole treatment reversed many of the age-related expression changes in the rat hippocampus (Fig. 1B,C), which primarily occurred in pathways associated with synaptic function (Fig. 1D,E). A similar inverse comparison of the pathways changed by riluzole with those identified in the hippocampus of AD patients demonstrated extensive commonality of affected genes (Fig. 2/Table 2), establishing riluzole's potential as a therapeutic agent for AD. The expression changes identified by sequencing were highly correlated with qRT-PCR results using open array technology, which validated over 50 genes of interest (Fig. 3). Finally, changes in the levels of EAAT2, a gene known to be increased by riluzole treatment 22, 26, 27, were validated by immunohistochemical labeling in hippocampus (Fig. 4). This finding raises the possibility that modulation of glutamate transporters is one mechanism by which riluzole can improve cognitive performance in aging. Additionally, these high-throughput studies offer an essential library of new targets that warrant further investigation into their role in glutamatergic transmission in the hippocampus and age-related cognitive decline.
The identification of pathways associated with the maintenance of synaptic health as changed by riluzole (Fig. 1D,E; Table 1) is consistent with previous work that demonstrated riluzole prevented age-related cognitive decline through clustering of dendritic spines 9, an important neuroplastic mechanism that has been shown by electrophysiological studies and computational models to allow non-linear summation of synaptic inputs 11, 68. Some examples of genes implicated in learning and plasticity that are reduced in aging and increased by riluzole treatment include: MMP9, which induces structural spine modifications54; NR2B, which is important for LTP31; MAP1B, which helps maintainaince of structural plasticity in the adult brain52 and SHANK3, which plays an important role in synaptic regulation53.
Many of riluzole's effects on the aging rat hippocampus were opposite the changes observed in human hippocampus from AD patients (Fig 2B). This indicates that many of the key pathways altered by glutamate modulation with riluzole are implicated in the development of AD pathology, suggesting riluzole may have therapeutic potential. Notably, as in aging, the top AD pathways identified also involved synaptic transmission and plasticity and were reversed by riluzole. Some examples of genes recovered by riluzole treatment include: ANK3, which mediates synaptic morphology and transmission 58; CAMK2, involved in induction of synaptic potentiation and memory 51, 59 and Rab3A, which is crucial for synaptic plasticity, learning and memory 60, 61. Genes that are consistently altered in AD brains and are reversed by riluzole provide potential future therapeutic targets.
Synaptic dysfunction is a critical pathophysiological mechanism in AD that highly correlates with cognitive decline 69, 70. Amyloid-β (Aβ) and phosphorylated tau toxicities, the hallmarks of the neuropathology of AD, are intimately related to glutamatergic dysregulation: oligomers of Aβ disrupt glutamate uptake, inhibiting LTP through excessive activation of extrasynaptic NMDA receptors 71, 72. Oligomers of Aβ also facilitate LTD71, 73 and decrease surface expression of synaptic NMDA receptors74. Additionally, dysregulated glutamate increases release of Aβ75 and tau 76, 77 and enhances tau phosphorylation 78 and expression 79, forming a vicious cycle of neurotoxicity. Importantly, previous work has shown that EAAT2 haploinsufficiency accelerates cognitive deficits in AD mouse model (AβPPswe/PS1ΔE9) 80 and EAAT2 overexpression improves cognitive and pathological markers in APPSw,Ind AD mouse model 81. These studies support the hypothesis that improved regulation of glutamatergic signaling via enhanced EAAT2 uptake could potentially mitigate toxicities in AD brains.
Finally, increased immunoreactivity for EAAT2 was observed in the same region as increased spine clustering was previously identified in riluzole-treated rats,9 suggesting a potential mechanism by which riluzole can increase cognitive performance. Glutamate transporters play the key role of regulating synaptic transmission, and thereby learning and memory20, 25. They prevent glutamate spillover to the extrasynaptic space and minimize cross-talk between neighboring synapses20, 62. Importantly, they also control the time course of synaptic glutamate82, 83. More recent work suggests that EAAT2 surface trafficking also shapes synaptic transmission84. Previous studies have suggested that riluzole increases glutamate uptake through both increased EAAT2 expression and stabilization of the inactivated state of voltage-gated sodium channels 22, 29, 30,26, 28. A recent in vivo study using microelectrode arrays coupled with amperometry has shown that riluzole reduces extrasynaptic glutamate levels and enhances cognitive performance which correlated with the increased glutamate uptake measures 27. These mechanisms of riluzole have been hypothesized to facilitate synaptic glutamatergic activity and to increase glutamate-glutamine cycling while preventing glutamate overflow to the extra-synaptic space 26, 85, 86. Activation of extra-synaptic NMDA receptors has been associated with LTD and excitotoxicity, and it is a likely important mechanism in many neurodegenerative diseases, including AD17, 18.
In conclusion
these findings identify molecular pathways implicated in aging that are rescued by administration of a known glutamate modulator, riluzole. Modeling the expression differences in response to riluzole establishes a framework of changes associated with improved learning and memory, against which other treatments can be compared. Further, many of the pathways changed by riluzole have been implicated across multiple studies in the pathophysiology of AD, suggesting glutamate modulators may represent novel treatments for both age-related cognitive decline and AD. Future studies will seek to conclusively demonstrate whether increased expression and activity of glutamate transporters are the essential mechanism underlying riluzole's ability to improve cognitive performance.
Supplementary Material
Acknowledgments
This work was supported by DANA Foundation, the Rockefeller University Women & Science Initiative and Alzheimer's Drug Discovery Foundation to ACP, NIH grant F32 MH102065 to JDG, NIA grant R37 AG06647 to JHM and partial support by grant # 8 UL1 TR000043 from the National Center for Research Resources and the National Center for Advancing Translational Sciences (NCATS).
Footnotes
Conflict of Interest: The authors have no conflict to declare.
Supplementary Information is linked to the online version of the paper.
Reference List
- 1.Buckner RL. Memory and executive function in aging and AD: multiple factors that cause decline and reserve factors that compensate. Neuron. 2004;44(1):195–208. doi: 10.1016/j.neuron.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nat Rev Neurosci. 2006;7(1):30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- 3.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimers Dement. 2007;3(3):186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 4.Morrison JH, Hof PR. Life and death of neurons in the aging brain. Science. 1997;278(5337):412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- 5.Morrison JH, Hof PR. Selective vulnerability of corticocortical and hippocampal circuits in aging and Alzheimer's disease. Prog Brain Res. 2002;136:467–486. doi: 10.1016/s0079-6123(02)36039-4. [DOI] [PubMed] [Google Scholar]
- 6.Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9(1):65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 7.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 8.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330(9):585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 9.Pereira AC, Lambert HK, Grossman YS, Dumitriu D, Waldman R, Jannetty SK, et al. Glutamatergic regulation prevents hippocampal-dependent age-related cognitive decline through dendritic spine clustering. Proc Natl Acad Sci U S A. 2014;111(52):18733–18738. doi: 10.1073/pnas.1421285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420(6917):812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- 11.Larkum ME, Nevian T. Synaptic clustering by dendritic signalling mechanisms. Curr Opin Neurobiol. 2008;18(3):321–331. doi: 10.1016/j.conb.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Polsky A, Mel BW, Schiller J. Computational subunits in thin dendrites of pyramidal cells. Nat Neurosci. 2004;7(6):621–627. doi: 10.1038/nn1253. [DOI] [PubMed] [Google Scholar]
- 13.Kavalali ET, Klingauf J, Tsien RW. Activity-dependent regulation of synaptic clustering in a hippocampal culture system. Proc Natl Acad Sci U S A. 1999;96(22):12893–12900. doi: 10.1073/pnas.96.22.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleindienst T, Winnubst J, Roth-Alpermann C, Bonhoeffer T, Lohmann C. Activitydependent clustering of functional synaptic inputs on developing hippocampal dendrites. Neuron. 2011;72(6):1012–1024. doi: 10.1016/j.neuron.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 15.De Roo M, Klauser P, Muller D. LTP promotes a selective long-term stabilization and clustering of dendritic spines. PLoS Biol. 2008;6(9):e219. doi: 10.1371/journal.pbio.0060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardingham GE. Pro-survival signalling from the NMDA receptor. Biochem Soc Trans. 2006;34(Pt 5):936–938. doi: 10.1042/BST0340936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11(10):682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusakov DA, Kullmann DM. Extrasynaptic glutamate diffusion in the hippocampus: ultrastructural constraints, uptake, and receptor activation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18(9):3158–3170. doi: 10.1523/JNEUROSCI.18-09-03158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuta A, Rothstein JD, Martin LJ. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17(21):8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tzingounis AV, Wadiche JI. Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nat Rev Neurosci. 2007;8(12):935–947. doi: 10.1038/nrn2274. [DOI] [PubMed] [Google Scholar]
- 21.Potier B, Billard JM, Riviere S, Sinet PM, Denis I, Champeil-Potokar G, et al. Reduction in glutamate uptake is associated with extrasynaptic NMDA and metabotropic glutamate receptor activation at the hippocampal CA1 synapse of aged rats. Aging Cell. 2010;9(5):722–735. doi: 10.1111/j.1474-9726.2010.00593.x. [DOI] [PubMed] [Google Scholar]
- 22.Brothers HM, Bardou I, Hopp SC, Kaercher RM, Corona AW, Fenn AM, et al. Riluzole partially rescues age-associated, but not LPS-induced, loss of glutamate transporters and spatial memory. J Neuroimmune Pharmacol. 2013;8(5):1098–1105. doi: 10.1007/s11481-013-9476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob CP, Koutsilieri E, Bartl J, Neuen-Jacob E, Arzberger T, Zander N, et al. Alterations in expression of glutamatergic transporters and receptors in sporadic Alzheimer's disease. J Alzheimers Dis. 2007;11(1):97–116. doi: 10.3233/jad-2007-11113. [DOI] [PubMed] [Google Scholar]
- 24.Masliah E, Alford M, DeTeresa R, Mallory M, Hansen L. Deficient glutamate transport is associated with neurodegeneration in Alzheimer's disease. Annals of neurology. 1996;40(5):759–766. doi: 10.1002/ana.410400512. [DOI] [PubMed] [Google Scholar]
- 25.Huang YH, Bergles DE. Glutamate transporters bring competition to the synapse. Curr Opin Neurobiol. 2004;14(3):346–352. doi: 10.1016/j.conb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Banasr M, Chowdhury GM, Terwilliger R, Newton SS, Duman RS, Behar KL, et al. Glial pathology in an animal model of depression: reversal of stress-induced cellular, metabolic and behavioral deficits by the glutamate-modulating drug riluzole. Mol Psychiatry. 2010;15(5):501–511. doi: 10.1038/mp.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunsberger HC, Weitzner DS, Rudy CC, Hickman JE, Libell EM, Speer RR, et al. Riluzole rescues glutamate alterations, cognitive deficits, and tau pathology associated with P301L tau expression. J Neurochem. 2015 doi: 10.1111/jnc.13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gourley SL, Espitia JW, Sanacora G, Taylor JR. Antidepressant-like properties of oral riluzole and utility of incentive disengagement models of depression in mice. Psychopharmacology (Berl) 2012;219(3):805–814. doi: 10.1007/s00213-011-2403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frizzo ME, Dall'Onder LP, Dalcin KB, Souza DO. Riluzole enhances glutamate uptake in rat astrocyte cultures. Cell Mol Neurobiol. 2004;24(1):123–128. doi: 10.1023/B:CEMN.0000012717.37839.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fumagalli E, Funicello M, Rauen T, Gobbi M, Mennini T. Riluzole enhances the activity of glutamate transporters GLAST, GLT1 and EAAC1. European journal of pharmacology. 2008;578(2-3):171–176. doi: 10.1016/j.ejphar.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Faherty SL, Campbell CR, Larsen PA, Yoder AD. Evaluating whole transcriptome amplification for gene profiling experiments using RNA-Seq. BMC Biotechnol. 2015;15:65. doi: 10.1186/s12896-015-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maag JL, Panja D, Sporild I, Patil S, Kaczorowski DC, Bramham CR, et al. Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front Neurosci. 2015;9:351. doi: 10.3389/fnins.2015.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray JD, Rubin TG, Hunter RG, McEwen BS. Hippocampal gene expression changes underlying stress sensitization and recovery. Mol Psychiatry. 2014;19(11):1171–1178. doi: 10.1038/mp.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19: Unit 19 10 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Hokama M, Oka S, Leon J, Ninomiya T, Honda H, Sasaki K, et al. Altered expression of diabetes-related genes in Alzheimer's disease brains: the Hisayama study. Cereb Cortex. 2014;24(9):2476–2488. doi: 10.1093/cercor/bht101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28(3):311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berchtold NC, Coleman PD, Cribbs DH, Rogers J, Gillen DL, Cotman CW. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer's disease. Neurobiol Aging. 2013;34(6):1653–1661. doi: 10.1016/j.neurobiolaging.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva AR, Grinberg LT, Farfel JM, Diniz BS, Lima LA, Silva PJ, et al. Transcriptional alterations related to neuropathology and clinical manifestation of Alzheimer's disease. PLoS One. 2012;7(11):e48751. doi: 10.1371/journal.pone.0048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu PT, Li YJ, Qin XJ, Kroner C, Green-Odlum A, Xu H, et al. A SAGE study of apolipoprotein E3/3, E3/4 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Mol Cell Neurosci. 2007;36(3):313–331. doi: 10.1016/j.mcn.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JA, Woltjer RL, Goodenbour JM, Horvath S, Geschwind DH. Genes and pathways underlying regional and cell type changes in Alzheimer's disease. Genome Med. 2013;5(5):48. doi: 10.1186/gm452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer's disease. J Chem Neuroanat. 2011;42(2):118–126. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, et al. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Annals of neurology. 2005;58(6):909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- 46.Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM, et al. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29(6):1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowe WB, Blalock EM, Chen KC, Kadish I, Wang D, Barrett JE, et al. Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(12):3098–3110. doi: 10.1523/JNEUROSCI.4163-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao X, Cui Z, Feng R, Tang YP, Qin Z, Mei B, et al. Maintenance of superior learning and memory function in NR2B transgenic mice during ageing. Eur J Neurosci. 2007;25(6):1815–1822. doi: 10.1111/j.1460-9568.2007.05431.x. [DOI] [PubMed] [Google Scholar]
- 49.Nayak TK, Sikdar SK. Time-dependent molecular memory in single voltage-gated sodium channel. J Membr Biol. 2007;219(1-3):19–36. doi: 10.1007/s00232-007-9058-4. [DOI] [PubMed] [Google Scholar]
- 50.Shen K, Teruel MN, Connor JH, Shenolikar S, Meyer T. Molecular memory by reversible translocation of calcium/calmodulin-dependent protein kinase II. Nat Neurosci. 2000;3(9):881–886. doi: 10.1038/78783. [DOI] [PubMed] [Google Scholar]
- 51.Glazewski S, Giese KP, Silva A, Fox K. The role of alpha-CaMKII autophosphorylation in neocortical experience-dependent plasticity. Nat Neurosci. 2000;3(9):911–918. doi: 10.1038/78820. [DOI] [PubMed] [Google Scholar]
- 52.Nothias F, Fischer I, Murray M, Mirman S, Vincent JD. Expression of a phosphorylated isoform of MAP1B is maintained in adult central nervous system areas that retain capacity for structural plasticity. J Comp Neurol. 1996;368(3):317–334. doi: 10.1002/(SICI)1096-9861(19960506)368:3<317::AID-CNE1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20(15):3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dziembowska M, Wlodarczyk J. MMP9: a novel function in synaptic plasticity. Int J Biochem Cell Biol. 2012;44(5):709–713. doi: 10.1016/j.biocel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 55.Kim DH, Kim JM, Park SJ, Cai M, Liu X, Lee S, et al. GABA(A) receptor blockade enhances memory consolidation by increasing hippocampal BDNF levels. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37(2):422–433. doi: 10.1038/npp.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein R, Nanduri V, Jing SA, Lamballe F, Tapley P, Bryant S, et al. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell. 1991;66(2):395–403. doi: 10.1016/0092-8674(91)90628-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massie A, Boillee S, Hewett S, Knackstedt L, Lewerenz J. Main path and byways: nonvesicular glutamate release by system x as an important modifier of glutamatergic neurotransmission. J Neurochem. 2015 doi: 10.1111/jnc.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith KR, Kopeikina KJ, Fawcett-Patel JM, Leaderbrand K, Gao R, Schurmann B, et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron. 2014;84(2):399–415. doi: 10.1016/j.neuron.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3(3):175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- 60.Thakker-Varia S, Alder J, Crozier RA, Plummer MR, Black IB. Rab3A is required for brain-derived neurotrophic factor-induced synaptic plasticity: transcriptional analysis at the population and single-cell levels. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21(17):6782–6790. doi: 10.1523/JNEUROSCI.21-17-06782.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Castillo PE, Janz R, Sudhof TC, Tzounopoulos T, Malenka RC, Nicoll RA. Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature. 1997;388(6642):590–593. doi: 10.1038/41574. [DOI] [PubMed] [Google Scholar]
- 62.Asztely F, Erdemli G, Kullmann DM. Extrasynaptic glutamate spillover in the hippocampus: dependence on temperature and the role of active glutamate uptake. Neuron. 1997;18(2):281–293. doi: 10.1016/s0896-6273(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 63.Furness DN, Dehnes Y, Akhtar AQ, Rossi DJ, Hamann M, Grutle NJ, et al. A quantitative assessment of glutamate uptake into hippocampal synaptic terminals and astrocytes: new insights into a neuronal role for excitatory amino acid transporter 2 (EAAT2). Neuroscience. 2008;157(1):80–94. doi: 10.1016/j.neuroscience.2008.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petr GT, Sun Y, Frederick NM, Zhou Y, Dhamne SC, Hameed MQ, et al. Conditional deletion of the glutamate transporter GLT-1 reveals that astrocytic GLT-1 protects against fatal epilepsy while neuronal GLT-1 contributes significantly to glutamate uptake into synaptosomes. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35(13):5187–5201. doi: 10.1523/JNEUROSCI.4255-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen W, Mahadomrongkul V, Berger UV, Bassan M, DeSilva T, Tanaka K, et al. The glutamate transporter GLT1a is expressed in excitatory axon terminals of mature hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24(5):1136–1148. doi: 10.1523/JNEUROSCI.1586-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Danbolt NC, Storm-Mathisen J, Kanner BI. An [Na+ + K+]coupled L-glutamate transporter purified from rat brain is located in glial cell processes. Neuroscience. 1992;51(2):295–310. doi: 10.1016/0306-4522(92)90316-t. [DOI] [PubMed] [Google Scholar]
- 67.Lehre KP, Levy LM, Ottersen OP, Storm-Mathisen J, Danbolt NC. Differential expression of two glial glutamate transporters in the rat brain: quantitative and immunocytochemical observations. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15(3 Pt 1):1835–1853. doi: 10.1523/JNEUROSCI.15-03-01835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Govindarajan A, Kelleher RJ, Tonegawa S. A clustered plasticity model of long-term memory engrams. Nat Rev Neurosci. 2006;7(7):575–583. doi: 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- 69.Selkoe DJ. Alzheimer's disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 70.DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann Neurol. 1990;27(5):457–464. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- 71.Li S, Hong S, Shepardson NE, Walsh DM, Shankar GM, Selkoe D. Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron. 2009;62(6):788–801. doi: 10.1016/j.neuron.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li S, Jin M, Koeglsperger T, Shepardson NE, Shankar GM, Selkoe DJ. Soluble Abeta oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(18):6627–6638. doi: 10.1523/JNEUROSCI.0203-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng L, Yin WJ, Zhang JF, Qi JS. Amyloid beta-protein fragments 25-35 and 31-35 potentiate long-term depression in hippocampal CA1 region of rats in vivo. Synapse. 2009;63(3):206–214. doi: 10.1002/syn.20599. [DOI] [PubMed] [Google Scholar]
- 74.Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY, et al. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8(8):1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 75.Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, et al. APP processing and synaptic function. Neuron. 2003;37(6):925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- 76.Yamada K, Holth JK, Liao F, Stewart FR, Mahan TE, Jiang H, et al. Neuronal activity regulates extracellular tau in vivo. J Exp Med. 2014;211(3):387–393. doi: 10.1084/jem.20131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pooler AM, Phillips EC, Lau DH, Noble W, Hanger DP. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep. 2013;14(4):389–394. doi: 10.1038/embor.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sindou P, Lesort M, Couratier P, Yardin C, Esclaire F, Hugon J. Glutamate increases tau phosphorylation in primary neuronal cultures from fetal rat cerebral cortex. Brain research. 1994;646(1):124–128. doi: 10.1016/0006-8993(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 79.Esclaire F, Lesort M, Blanchard C, Hugon J. Glutamate toxicity enhances tau gene expression in neuronal cultures. J Neurosci Res. 1997;49(3):309–318. doi: 10.1002/(sici)1097-4547(19970801)49:3<309::aid-jnr6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 80.Mookherjee P, Green PS, Watson GS, Marques MA, Tanaka K, Meeker KD, et al. GLT-1 loss accelerates cognitive deficit onset in an Alzheimer's disease animal model. J Alzheimers Dis. 2011;26(3):447–455. doi: 10.3233/JAD-2011-110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi K, Kong Q, Lin Y, Stouffer N, Schulte DA, Lai L, et al. Restored glial glutamate transporter EAAT2 function as a potential therapeutic approach for Alzheimer's disease. J Exp Med. 2015;212(3):319–332. doi: 10.1084/jem.20140413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mennerick S, Zorumski CF. Glial contributions to excitatory neurotransmission in cultured hippocampal cells. Nature. 1994;368(6466):59–62. doi: 10.1038/368059a0. [DOI] [PubMed] [Google Scholar]
- 83.Tong G, Jahr CE. Block of glutamate transporters potentiates postsynaptic excitation. Neuron. 1994;13(5):1195–1203. doi: 10.1016/0896-6273(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 84.Murphy-Royal C, Dupuis JP, Varela JA, Panatier A, Pinson B, Baufreton J, et al. Surface diffusion of astrocytic glutamate transporters shapes synaptic transmission. Nat Neurosci. 2015;18(2):219–226. doi: 10.1038/nn.3901. [DOI] [PubMed] [Google Scholar]
- 85.Chowdhury GM, Banasr M, de Graaf RA, Rothman DL, Behar KL, Sanacora G. Chronic riluzole treatment increases glucose metabolism in rat prefrontal cortex and hippocampus. J Cereb Blood Flow Metab. 2008;28(12):1892–1897. doi: 10.1038/jcbfm.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brennan BP, Hudson JI, Jensen JE, McCarthy J, Roberts JL, Prescot AP, et al. Rapid enhancement of glutamatergic neurotransmission in bipolar depression following treatment with riluzole. Neuropsychopharmacology. 2010;35(3):834–846. doi: 10.1038/npp.2009.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.