Abstract
Background & Aims
The Lgr family of transmembrane proteins (Lgr4, 5, 6) act as functional receptors for R-spondin proteins (Rspo 1, 2, 3, 4), and potentiate Wnt signaling in different contexts. Lgr5 is arguably the best characterized of the Lgr family members in a number of adult and embryonic contexts in mice. However, the function of LGR family members in early embryonic development is unclear, and has not been explored during human development or tissue differentiation in detail.
Methods
We interrogated the function and expression of LGR family members using human pluripotent stem cell–derived tissues including definitive endoderm, mid/hindgut, and intestinal organoids. We performed embryonic lineage tracing in Lgr5-GFP-IRES-CreERT2 mice.
Results
We show that LGR5 is part of the human definitive endoderm (DE) gene signature, and LGR5 transcripts are induced robustly when human pluripotent stem cells are differentiated into DE. Our results show that LGR4 and 5 are functionally required for efficient human endoderm induction. Consistent with data in human DE, we observe Lgr5 reporter (eGFP) activity in the embryonic day 8.5 mouse endoderm, and show the ability to lineage trace these cells into the adult intestine. However, gene expression data also suggest that there are human–mouse species-specific differences at later time points of embryonic development.
Conclusions
Our results show that LGR5 is induced during DE differentiation, LGR receptors are functionally required for DE induction, and that they function to potentiate WNT signaling during this process.
Keywords: Pluripotent Stem Cells, Endoderm, LGR5, WNT, Organoid, Intestine
Abbreviations used in this paper: CDX2, caudal type homeobox2; ChIPseq, chromatin immunoprecipitation sequencing; creER, cre recombinase protein fused to estrogen receptor; Ct, cycle threshold; DE, definitive endoderm; E, embryonic day; GFP, green fluorescent protein; hESC, human embryonic stem cell; mRNA, messenger RNA; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; Rspo, R-spondin protein; shRNA, short hairpin RNA
Summary.
Our results show that LGR5 is induced during definitive endoderm differentiation, LGR receptors are functionally required for definitive endoderm induction, and that they function to potentiate WNT signaling during this process.
Wnt signaling is a critical signaling pathway in a broad number of developmental, homeostatic, and disease contexts.1, 2, 3, 4 Recent work has shown the importance of secreted R-spondin proteins (Rspo 1–4) and their Lgr receptors (Lgr 4, 5, and 6) as important modulators of the Wnt signaling pathway that act to potentiate Wnt signaling.5, 6, 7, 8, 9, 10, 11 For example, R-spondin ligands and Lgr receptors are all required for tight regulation of the crypt base columnar intestinal stem cells of the adult mouse epithelium.2, 7, 12, 13, 14, 15, 16 In adult intestinal stem cells, Lgr4 and Lgr5 are redundantly required for stem cell maintenance,7 whereas in the fetal murine intestine, it appears that Lgr5 is dispensable, although Lgr4 is essential for growth.17, 18, 19 However, despite our increasing understanding of the importance of Rspo/Lgr signaling in the intestine during mouse development and adult homeostasis, nothing is known about the functional role for LGR genes during human endoderm differentiation and little is known about expression in human tissues.
In this study, we used several methods to show that LGR5 is induced robustly in human embryonic stem cell–derived definitive endoderm (DE) and is one of the most highly up-regulated genes in the DE gene signature. Analysis of chromatin immunoprecipitation sequencing (ChIPseq) data show that LGR5 is bound directly by β-catenin upon WNT stimulation in human embryonic stem cells (hESCs) and that H3K27ac is increased at the LGR5 locus during DE differentiation. Genetic lineage tracing in the embryonic day (E)8.5 mouse embryo supports human expression data that Lgr5 is expressed in the early endoderm. We also examined the expression of all 3 LGR family members, LGR4, LGR5, and LGR6, during endoderm lineage specification, hindgut induction, and differentiation into intestinal organoids, and we compared expression in the human fetal intestine and mouse embryonic intestine, where we found notable species-specific differences. In human tissues, LGR5 was expressed most robustly in endoderm, intestinal lineages, and in the human fetal intestine, whereas LGR4 was expressed at lower levels, and LGR6 was undetectable. In contrast, in mice, Lgr4 and Lgr6 were more abundant than Lgr5. Functionally, our data show that RSPO/LGR signaling synergizes with WNT signaling during human endoderm induction, and short hairpin RNA (shRNA) knockdown of either LGR4 or LGR5 significantly impairs the ability of hESCs to differentiate into DE. Taken together, our work details a previously uncharacterized functional role for LGR4 and LGR5 during human DE differentiation, and highlights species-specific gene expression differences between humans and mice during intestine development.
Materials and Methods
hESC Cell Lines, Human Tissue, and Mice
hESCs
All work with hESCs was reviewed and approved by the University of Michigan human pluripotent stem cell research oversight committee. The hESC cell line H9 (WA09; National Institutes of Health stem registry 0062) was obtained from the WiCell Research Institute (Madison, WI). Karyotypically normal cell lines were used for all experiments.
Human tissue
De-identified human intestinal tissue was obtained from the University of Washington Laboratory of Developmental Biology, and was approved by the University of Michigan institutional review board.
Animal use
All mouse work was reviewed and approved by the University Committee on Use and Care of Animals (PRO00005809). All mouse strains used have been published previously and were obtained from Jackson Laboratories (Bar Harbor, ME).12, 20, 21
shRNA Knockdown Cell Lines
Mission pLKO.1-puromycin–resistant lentiviral plasmids were obtained from Sigma-Aldrich (St. Louis, MO) for generating shRNA knockdown lines (see Supplementary Table 1 for The RNAi Consortium Number clone numbers). Unconcentrated lentiviral supernatants were generated by the University of Michigan Viral Vector Core (available from: http://medicine.umich.edu/medschool/research/office-research/biomedical-research-core-facilities/vector). To generate knockdown cell lines, hESCs were dissociated into single cells using Accutase (Sigma-Aldrich). Cells were spun down briefly and resuspended in mTesR1 plus 10 umol/L Y-27632 (both from Stem Cell Technologies). Cells (1 × 106) were infected with pooled lentivirus by mixing 3 different shRNA viral supernatants for the same target gene. Y-27632 was included for the first 24 hours after dissociation, and media was changed daily. After 72 hours, cells were selected by puromycin (3 ug/mL) to generate LGR4 and/or LGR5 knockdown cell lines. For double-knockdown lines, the LGR5 knockdown line was infected sequentially by pooled LGR4 shRNA.
Lineage Tracing
Lgr5-GFP-IRES-CreERT2;Rosa26LacZ and Lgr5-GFP-IRES-CreERT2;Rosa26tdTomato embryos were generated for lineage tracing from embryonic stages. Pregnant mice were given tamoxifen by oral gavage (100 mg/kg) at embryonic stages of E8.5, E10.5, E12.5, and E14.5. For embryonic lineage tracing, whole gastrointestinal tracts were dissected at E16.5 and tissues were fixed in 4% paraformaldehyde in phosphate-buffered saline overnight. X-gal staining was performed the next day as previously described.22 Whole-mount tissues were cleared using Murray’s clear before imaging. For lineage tracing induced in embryos and analyzed at adult stages, whole gastrointestinal tracts were dissected from 6-week-old mice and were fixed in 4% paraformaldehyde in phosphate-buffered saline for 2 hours followed by optimum cutting temperature embedding for histologic analysis.
Differentiation of hESCs
Differentiation of hESCs was performed as previously published.23, 24, 25, 26, 27 In brief, hESCs were grown in feeder-free conditions on Matrigel (Corning, NY) with mTesrR1 medium (Stem Cell Technologies, Vancouver, BC, Canada), passaged into 24-well plates, and grown to approximately 70% confluency. Media then was changed to RPMI1640 media plus Activin A (ACTA) (100 ng/mL; R&D Systems, Minneapolis, MN) for 1 day. The same media was used on days 2 and 3 of differentiation, except that 0.2% and 2% fetal bovine serum (HyClone, GE Life Sciences, Logan, UT) was added on the respective days. For induction of caudal type homeobox2 (CDX2), endoderm was treated for 4 days with RPMI1640 with 2% fetal bovine serum (HyClone) and supplemented with fibroblast growth factor 4 (500 ng/mL), generated as previously described,28 plus 2 umol/L Chir99021 (Tocris Bioscience, Bristol, United Kingdom). Media was changed daily. For organoid experiments, spheroids that had formed on day 4 of CDX2 induction were transferred to a droplet of Matrigel and grown in advanced Dulbecco's modified Eagle medium/F12 supplemented with 1× B27 (Thermo Fisher, Waltham, MA), epidermal growth factor (EGF) (R&D Systems; 100 ng/mL), Noggin (5% conditioned medium),29 and R-Spondin2 (5% conditioned medium).30 Media was changed every 3–4 days.
Histology and Immunofluorescence
Immunofluorescence was performed as previously published22, 25, 31 using antibodies outlined in Supplementary Table 2. Immunofluorescent images were optimized for contrast and brightness in a uniform manner, consistent with the policies of Cellular and Molecular Gastroenterology and Hepatology.
Cell Sorting and Cell Counting
For cell counting data presented in Figure 2B and C, H9-LGR5-eGFP hESCs were differentiated into endoderm or CDX2+ mid/hindgut.32, 33 Monolayers were digested to single cells using Accutase, spun down, resuspended, and sorted for GFP+ cells. These cells then were plated for 24 hours and immunostaining was performed. After staining, the total cell number (4′,6-diamidino-2-phenylindole–positive) and FOXA2+/SOX17+ (FOXA2: Forkhead Box A2; SOX17: SRY Box 17) or FOXA2+/CDX2+ cells were counted using MetaMorph Software (Molecular Devices, Sunnyvale, CA), as described previously.34
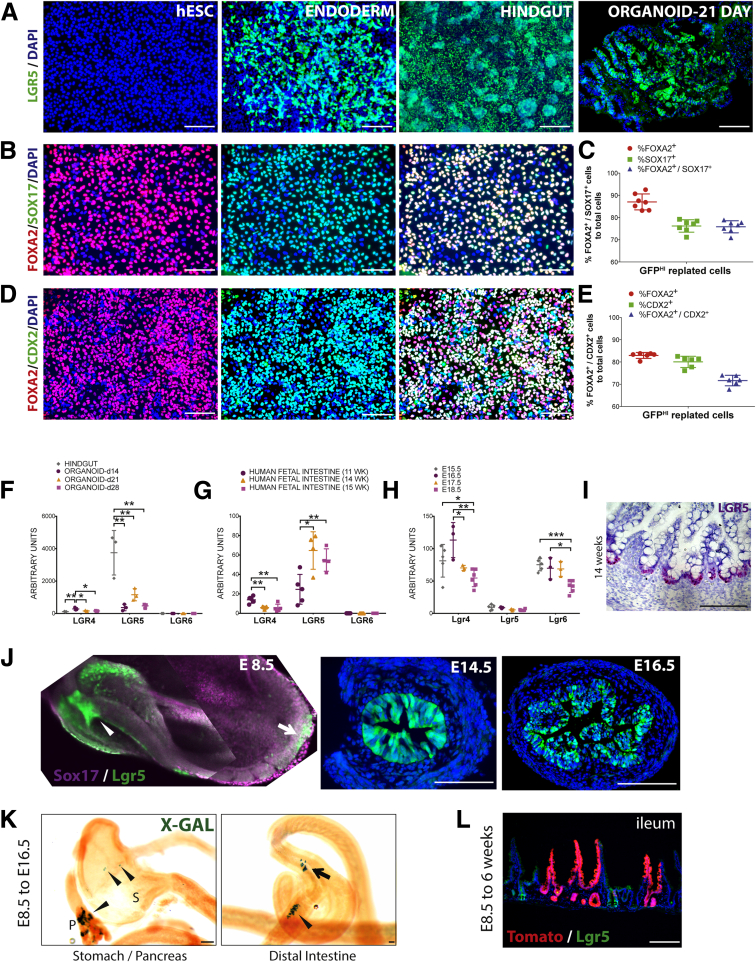
Figure 2.
LGR5 expression in the human and mouse endoderm and intestinal lineages. (A) h9–LGR5–eGFP reporter hESC line showing GFP expression in endoderm, mid/hindgut, and in human intestinal organoids grown for 21 days in vitro, but not in undifferentiated hESCs. (B) GFPHI cells were sorted from endoderm, replated for 24 hours, and stained for FOXA2 (red), SOX17 (green), and DAPI (blue). (C) The percentage of GFPHI cells expressing FOXA2, SOX17, or co-expressing both markers after 24 hours of replating were calculated. (D) GFPHI cells were sorted from mid/hindgut endoderm, replated for 24 hours, and stained for FOXA2 (red), CDX2 (green), and DAPI (blue). (E) The percentage of GFPHI cells expressing FOXA2, CDX2, or co-expressing both markers after 24 hours of replating were calculated. (F) qRT-PCR showing mRNA abundance of LGR genes in mid/hindgut and organoids grown in vitro for different lengths of time (14 days, 21 days, 28 days). (G) qRT-PCR showing mRNA abundance of LGR genes in the human fetal intestine at different weeks of gestation (n = 1 replicate per time point). (H) qRT-PCR showing mRNA abundance of Lgr genes in the mouse fetal intestine at different weeks of gestation (n ≥ 3 replicates per time point). (F–H) One-way analysis of variance was used for comparisons. (I) LGR5 in situ hybridization (using the RNAscope hybridization system, Advanced Cell Diagnostics, Newark, CA) in the human fetal duodenum at 14 weeks of gestation showing strong localization to the intervillus domain. Tissue was counterstained with hematoxylin. (J) GFP expression from Lgr5–eGFP–ires–creER knock-in mouse at E8.5 (whole mount stain, 2 images stitched together) showing strong anterior and posterior endoderm expression (arrowhead and arrow, respectively), and in sections through the E14.5 and E16.5 distal small intestine. (K) Lineage tracing from Lgr5–eGFP–IRES–creER;Rosa26–LacZ embryos, with tamoxifen given at E8.5, and embryos harvested at E16.5 showing lineage labeling (X-gal staining, blue) in the pancreas (p) and stomach (s) (arrowheads) in the left panel, and labeling in the ileum (arrow) and colon (arrowhead) in the right panel. (L) Lineage tracing from Lgr5–eGFP–ires–creER;Rosa26–tdTomato embryos, with tamoxifen given at E8.5, and tissues analyzed at 6 weeks old, showing lineage labeling in the ileum. Scale bars: 200 μm. DAPI, 4',6-diamidino-2-phenylindole. *P ≤ .05, **P ≤ .01, and ***P ≤ .001.
Quantitative Reverse-Transcription Polymerase Chain Reaction
Briefly, RNA isolation was performed using the MagMAX-96 total RNA isolation kit (AM1830; Ambion, Waltham, MA). SuperScript VILO complementary DNA synthesis kit (11754250; ThermoFisher) was used to make complementary DNA from 200 ng RNA. Complementary DNA levels were detected using QuantiTect SYBR Green (608056; Qiagen). Relative gene expression was plotted as arbitrary units, using the following formula: [2^(housekeeping gene Ct - gene Ct)] × 10,000. All primer sequences are listed in Supplementary Table 3. The Ct values for all of the quantitative reverse-transcription polymerase chain reaction (qRT-PCR) experiments are provided in Supplementary Table 4.
Cell Quantification and Statistical Analysis
Cell quantification was conducted as previously described in detail.23, 34 For statistical analysis, data are expressed as the median of each sample set, with each data point represented in the plots. As noted in the figure legends, unpaired t tests and 1-way and 2-way analysis of variance comparisons were performed with GraphPad Prism 5.0 software (GraphPad Software, LaJolla, CA) and data are presented as the means ± SEM. All animal experiments were repeated on at least 3 different embryos or adult animals with representative images shown, and all in vitro experiments were conducted on at least 3 independent biological replicates, and each experiment was repeated on at least 2 separate occasions (independent experiments). The only exception to this was the experiments using human tissues. For these experiments, each stage was represented by only 1 (n = 1) biological sample. For qRT-PCR, the intestine (n = 1 biological sample per time point) was divided into 1-cm sections and RNA was isolated from at least 3 different segments (n = 3). Each segment was counted as an independent sample in an analysis shown in Figure 2G.
RNA and ChIP Sequencing
Processing of RNAseq data was conducted as described.23 The complete sequence alignment and expression analysis scripts can be found at https://github.com/hilldr/Tsai_LGR5_endoderm_2015. For re-analysis of β-catenin and LEF1 and H3k27ac ChIPseq data, public Fastq sequences35 were aligned (hg18) using Bowtie (v1.1.1). Bam files were reads per kilobase of transcript per million mapped reads normalized using BamCoverage to generate BigWig files. Bigwig files were visualized using integrative genomics viewer.
Accession Numbers
RNAseq data have been published previously,23, 34 and FastQ files can be found at the European Molecular Biology Laboratory-European Bioinformatics Institute ArrayExpress: E-MTAB-3158. ChIPseq data have been published previously35, 36 and data can be found at Gene Expression Omnibus: GSE64758 and GSE54471.
Results
LGR5 Is Highly Induced in Definitive Endoderm Derived From hESCs
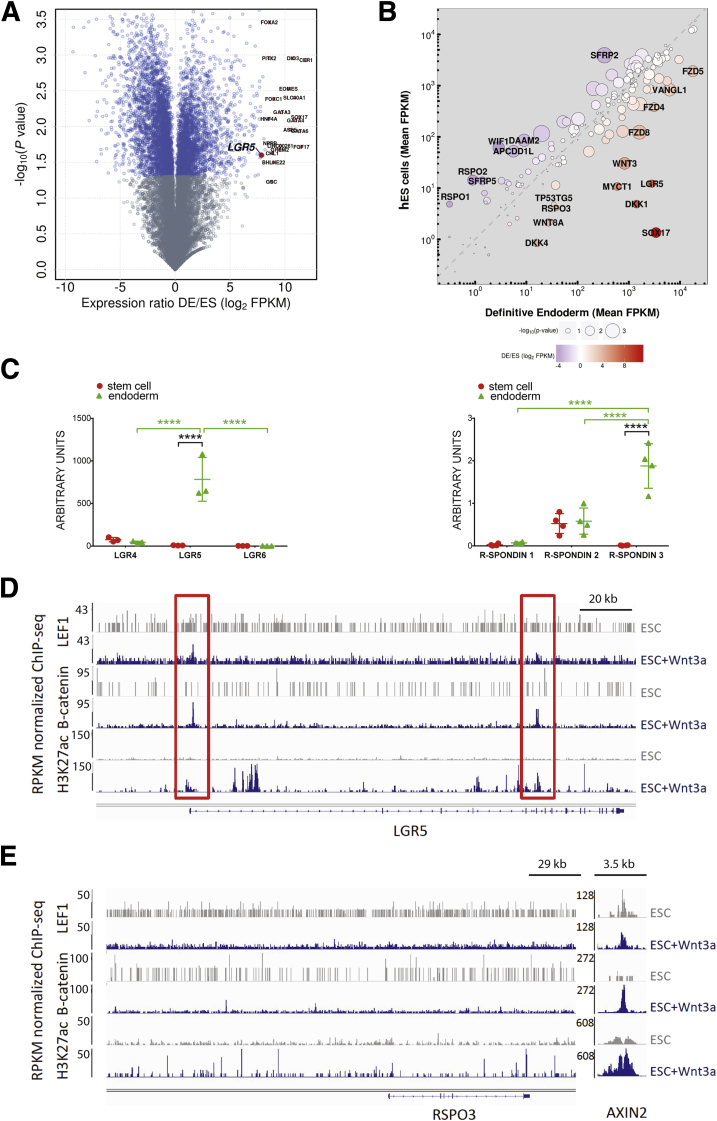
By using publically available RNAseq data generated from the H9 hESC line and H9 DE (EMBL-EBI ArrayExpress accession E-MTAB-3158),23, 34 we used differential expression analysis to identify the most highly up-regulated genes in DE. We observed that LGR5 was among the most highly up-regulated genes in the DE gene signature (Figure 1A and Supplementary Table 5). In addition, because LGR receptors are known to bind RSPO proteins and potentiate WNT signaling, we examined other differentially expressed members of the WNT signaling pathway, including ligands, receptors, modifiers, and inhibitors. We found that among a curated list of Wnt signaling components, LGR5 and RSPO3 were highly induced in DE (Figure 1B and Supplementary Table 6).
Figure 1.
LGR5 is up-regulated in hPSC-derived definitive endoderm. (A) The log2-transformed ratio of expression in DE relative to undifferentiated hESC is plotted against the -log10-transformed P value of a 2-tailed Student t test evaluating the difference in expression between the 2 groups (volcano plot). Genes that differ significantly (P < .05) are plotted in blue. The 20 most highly up-regulated genes in DE are highlighted in red and labeled. (B) The mean fragments per kilobase of transcript per million mapped reads of a subset of genes involved in WNT signaling is plotted for the DE (X-axis) against the mean FPKM in hES cells (Y-axis). A dashed grey line represents equivalent expression, with deviation to the right or left indicating relatively greater expression in DE or hES, respectively. The log2-transformed expression ratio and -log10-transformed P value are given as the color and size of the points, respectively, as indicated in the legend. The top 20 differentially regulated genes are labeled. (C) qRT-PCR showing expression (arbitrary units) of LGR and RSPO mRNA in undifferentiated stem cells (red circles) or in DE (green triangles). Statistical tests used were as follows: 1-way analysis of variance (green bars) and unpaired t test (black bar). (D and E) Re-analysis of published LEF1 and β-catenin ChIPseq analysis at the (D) LGR5 and (E) RSPO3 loci in undifferentiated hESCs and hESCs treated with WNT3A35 and H3K27ac ChIPseq analysis at the same loci in ESCs and DE.36 ****P ≤ .0001.
To corroborate the RNAseq data, we used qRT-PCR to examine the expression of LGR4–6 and RSPO1–3 in undifferentiated hESCs, as well as in hESC-derived DE. We found that LGR4 was expressed in both hESCs and DE, whereas LGR6 was almost undetectable. We observed that only LGR5 was highly induced in endoderm (Figure 1C). Furthermore, RSPO1 was expressed at very low levels, RSPO2 was expressed in both hESCs and DE, and RSPO3 was almost undetectable in hESCs, but was highly induced in DE (Figure 1C).
Because Lgr5 is a known Wnt signaling target gene in different contexts,12, 37 we examined publicly available β-catenin, LEF1, and H3K27ac ChIPseq data during hESC differentiation to determine transcription factor binding and enhancer activity at the LGR5 and RSPO3 loci.35, 36 Consistent with our RNAseq data, H3K27ac levels were increased within the 180-kb region surrounding the LGR5 locus after DE induction, supporting the notion that LGR5 transcriptional activity increases after DE induction. hESCs treated with Wnt3A for 6 hours showed rapid binding of β-catenin and LEF1 to the LGR5 locus as well, indicating that LGR5 is a direct target of these transcription factors (Figure 1D). Interestingly, β-catenin and LEF1 were not similarly recruited to the RSPO3 locus when comparing undifferentiated hESCs with DE, whereas H3K27ac was increased only modestly. In contrast, AXIN2, a well-characterized WNT signaling target gene (used here as a positive control), showed increased binding of β-catenin and enhanced H3K27ac in response to Wnt3a (Figure 1E). Together, the results presented in Figure 1 show that LGR5 and RSPO3 are increased during human DE differentiation, and that these genes are bound directly by β-catenin and LEF1 upon WNT stimulation.
LGR5 Is Expressed Across Development in Human and Mouse Endoderm Lineages
Our findings show that LGR5 is expressed early during endoderm differentiation from human ES cells. To determine if LGR5 is expressed at other stages of intestinal development/differentiation in human tissue, we took advantage of a previously described bacterial artifical chromosome transgenic H9 LGR5-eGFP hESC reporter line,24, 38 as well as a human intestinal organoid culture system that recapitulates intestine development in vitro.25, 39 Our results show that eGFP was undetectable in hESCs, whereas eGFP was abundant in endoderm, hindgut, and organoids (Figure 2A). To verify that LGR5-eGFP–positive cells indeed were enriched for endoderm lineages, we used fluorescence-activated cell sorting to purify GFPHI cells from DE cultures (Figure 2B) or mid/hindgut cultures (Figure 2D), replated them for 12 hours, and used immunofluorescence to co-stain endoderm for SOX17 and FOXA2 (Figure 2B) or mid/hindgut for FOXA2 and CDX2 (Figure 2D). Quantification of co-stained cells showed that approximately 85% of GFPHI endoderm stained positive for FOXA2 and approximately 75% stained positive for SOX17 or co-stained for both FOXA2 and SOX17 (Figure 2C). Similarly, approximately 80% of GFPHI cells were positive for FOXA2 or CDX2 and approximately 72% were positive for both markers (Figure 2E).
To corroborate our findings with the bacterial artifical chromosome transgenic H9 LGR5-eGFP hESC reporter line, we conducted qRT-PCR at different stages of hESC-differentiated intestinal tissue, including the mid/hindgut stage, and in organoids after days 14, 21, and 28 in vitro. We found that LGR4 was expressed at similar levels across these stages, that LGR6 expression was undetectable, and that LGR5 expression was highest at the hindgut stage, but was maintained at all organoid stages examined (Figure 2F). Similarly, we examined human fetal intestinal tissue by qRT-PCR at 11, 14, and 15 weeks of gestation (n = 1 biological replicate per time point), and we found that LGR4 messenger RNA (mRNA) was expressed at low levels at all stages, that LGR5 was the most highly expressed LGR family member, and that LGR6 was undetectable (Figure 2G). The expression of LGR5 was confirmed by in situ hybridization on sections at 14 weeks of development (Figure 2I). Interestingly, when we examined expression of Lgr4, 5, and 6 in the developing mouse intestine at 15.5, 16.5, 17.5, and 18.5 days of gestation, qRT-PCR results consistently showed that Lgr4 and Lgr6 had more abundant levels of transcript relative to Lgr5, highlighting gene expression differences between human beings and mice (Figure 2H).
Lgr5 Is Expressed in Definitive Endoderm in Mice
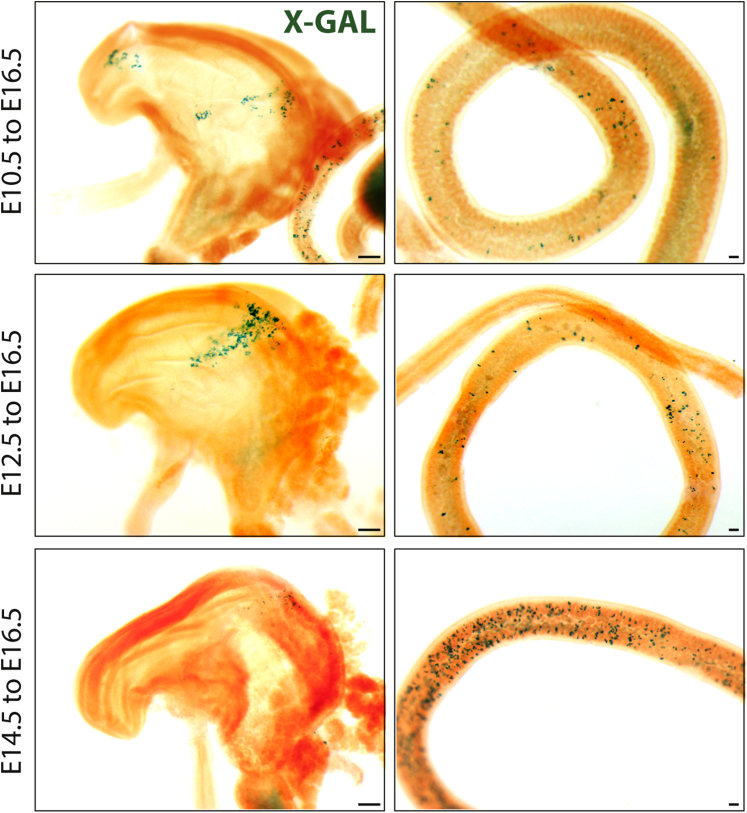
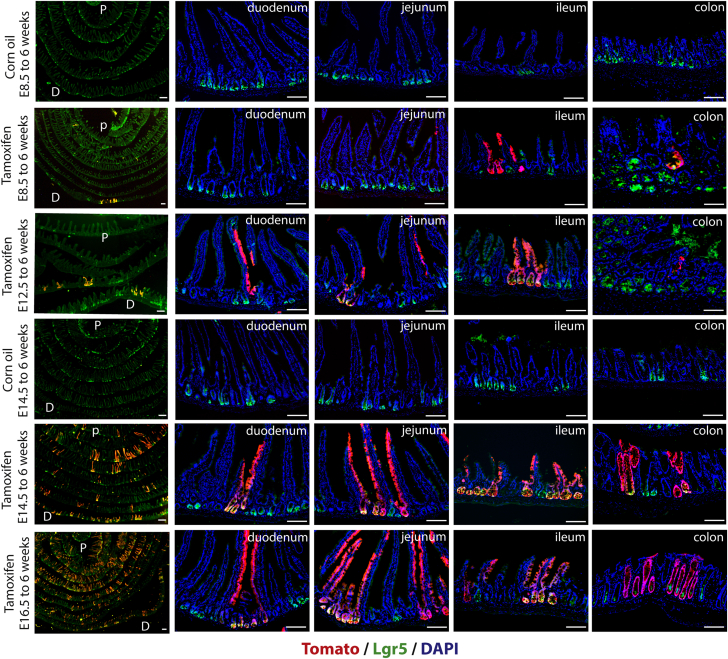
To determine if earlier endodermal Lgr5 expression is conserved between humans and mice, and to show that endodermal expression is detectable in an in vivo animal model, we used Lgr5-GFP-IRES-CreERT2 mice, in which GFP faithfully reports Lgr5 expression in many different contexts.12, 40, 41 By using whole-mount immunofluorescence in E8.5 embryos co-stained for Sox17 and GFP, we observed staining for GFP in the anterior foregut endoderm (Figure 2J, arrowhead), as well as in Sox17+ hindgut endoderm (Figure 2J, arrow) (note that 2 images were stitched together to generate the E8.5 image in Figure 2J). Examining Lgr5-driven GFP expression across time in the developing mouse intestine, we also observed GFP at E14.5 and E16.5. Consistent with our data, Lgr5-GFP recently was reported in the developing mouse intestine at E12.5.42 We conducted lineage tracing at several different developmental times to determine if early embryonic Lgr5+ cells contribute to adult tissues (Figure 2K and L, and Supplementary Figures 1 and 2). Lineage labeling was induced in timed pregnant Lgr5-GFP-IRES-CreERT2 female mice crossed to Rosa26–LSL–LacZ or Rosa–LSL–tdTomato mice at 8.5 days of gestation. Lineage-labeled embryos were harvested at E16.5 (Figure 2K) or at 6 weeks old (Figure 2L). We observed small patches of lineage labeling in the pancreas, stomach, and intestine at E16.5, and labeling that persisted into adulthood (Figure 2K). It is likely that recombination efficiencies in our experiments were poor given that lower doses of tamoxifen were administered by oral gavage to avoid miscarriage at early stages. Similarly, embryos given tamoxifen at later time points (E10.5, E12.5, E14.5) showed embryonic and adult labeling, however, the contribution of Lgr5 lineage-labeled cells in the pancreas and stomach decreased over time, whereas intestinal labeling increased over time (Supplementary Figures 1 and 2). Similarly, the contribution of Lgr5 lineage-labeled cells to the adult intestinal lineage increased over time (Supplementary Figure 2). Together, these results show that E8.5 endoderm expresses Lgr5, that cells expressing Lgr5 are found across development, and that Lgr5+ progenitor cells labeled at all embryonic times examined (E8.5–E16.5) contribute to the adult intestinal epithelium.
Supplementary Figure 1.
Lgr5–eGFP–ires–creER–mediated lineage tracing across time. Timed pregnant females were given tamoxifen when embryos were E10.5 (top row), E12.5 (middle row), or E14.5 (bottom row), and all embryos were collected at E16.5. E10.5 and E12.5 lineage tracing was evident in the stomach and intestine, whereas less lineage tracing was seen in the stomach and more was seen in the intestine when tamoxifen was given at E14.5. Lineage tracing was performed with Rosa26–LacZ and X-gal staining is shown in blue. Scale bar: 200 μm.
Supplementary Figure 2.
Lgr5–eGFP–ires–creER–mediated lineage tracing from embryo to adult. Timed pregnant females were given tamoxifen when embryos were E8.5, E12.5, E14.5, or E16.5, and whole intestines were examined using Swiss rolls in 6-week-old adults so that the proximal (P) and distal (D) intestine could be visualized in one section. Lineage tracing was performed with Rosa26-tdTomato and lineage tracing is shown in red. Lgr5-eGFP is visualized in green. Increased lineage labeling can be seen as developmental time progresses. For controls, corn oil was administered to timed pregnant females (Lgr5–eGFP–ires–creER;Rosa–LSL–Tomato) when embryos were E8.5 or E14.5, and whole intestines were examined in 6 week old adults via Swiss roll. Corn oil controls showed no lineage labeling. DAPI, 4',6-diamidino-2-phenylindole; Scale bars: 200 μm.
R-Spondin Proteins Potentiate WNT Signaling During DE Differentiation
Our studies used well-described methods to generate DE using ACTA, but not WNT ligands.25, 26 However, Wnt signaling plays an important role in the commitment to the endoderm lineage both in vitro and in vivo.43, 44, 45, 46, 47, 48, 49 Therefore, we wanted to determine the following: (1) if WNT signaling was required during our ACTA differentiation protocol; (2) if inhibition of WNT signaling led to decreased LGR5 expression; and (3) if RSPO/LGR signaling potentiated WNT signaling during endoderm differentiation. To determine if DE induced by ACTA requires WNT signaling, we added ACTA to hESC cultures with and without inhibitor of WNT processing 2 (IWP2), a small molecule that inhibits Porcupine, leading to disrupted processing and inhibition of endogenous WNT ligand secretion from cells.50 We found that inclusion of IWP2 during ACTA-induced DE differentiation almost completely abolished the induction of FOXA2/SOX17+ cells (Figure 3A). Similarly, ACTA-stimulated induction of FOXA2, SOX17, and LGR5 mRNA was blocked completely by addition of IWP2 during differentiation (Figure 3B). These experiments (Figure 3A and B) inhibited WNT signaling at the onset of differentiation. To determine if WNT signaling is required for LGR5 expression after DE induction, we treated hESCs with ACTA for 3 days to induce DE, and then cultured cells for an additional 2 days with ACTA ± IWP2 (Figure 3C). The results showed that DE treated with IWP2 had reduced LGR5 levels, suggesting that sustained WNT signaling is required for expression of these genes (Figure 3C), and supporting β-catenin and LEF1 ChIPseq data (Figure 1D and E) showing that LGR5 is a WNT target gene.
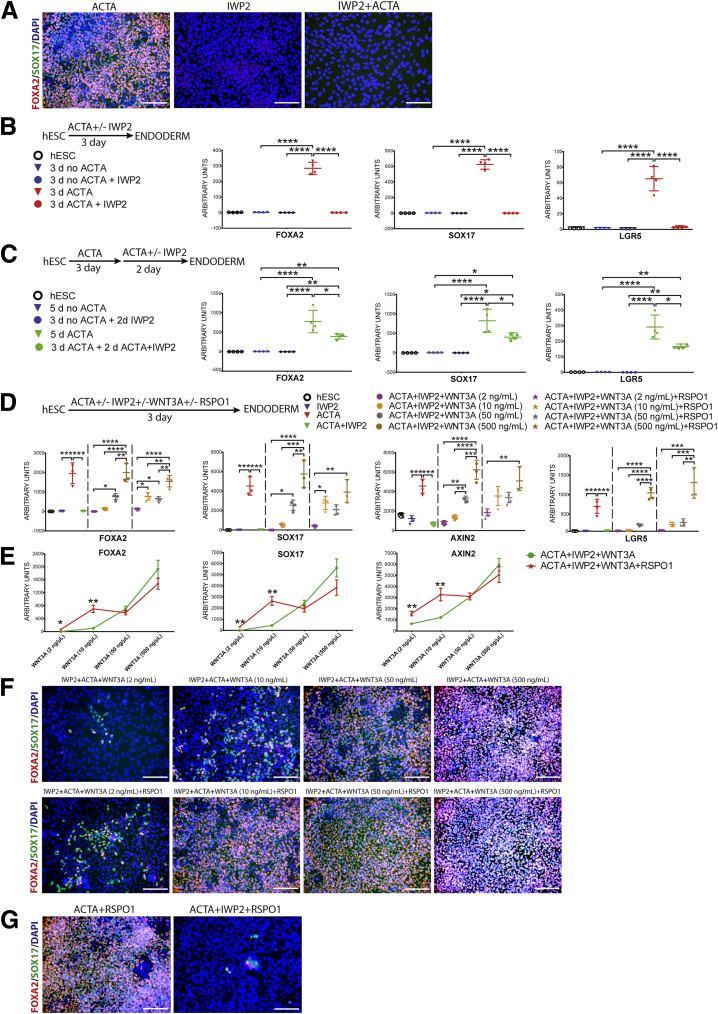
Figure 3.
RSPO potentiates WNT signaling during endoderm differentiation. (A) hESC-derived DE after 3 days of ACTA (left panel) shows robust FOXA2 (red) and SOX17 (green) staining. In contrast, IWP2- or ACTA + IWP2–treated cells show no FOXA2 or SOX17 (middle and right panels, respectively). Nuclei are marked by DAPI (blue) in all panels. (B) Schematic of experimental set-up is shown on the left, along with a sample legend. qRT-PCR showing mRNA abundance in all samples for FOXA2, SOX17, and LGR5. All 3 of these genes are induced robustly in DE (3d ACTA), and expression is inhibited by IWP2 (3d ACTA + IWP2). One-way analysis of variance was used for statistical analysis. (C) Schematic of experimental set-up is shown on the left, along with a sample legend. mRNA abundance in all samples for FOXA2, SOX17, and LGR5 shows that all 3 of these genes are induced robustly in DE (5d ACTA), and that endoderm expression is reduced significantly by IWP2 (3d ACTA + 2d ACTA + IWP2). One-way analysis of variance was used for statistical analysis. (D) Schematic of experimental set-up is shown on top, along with a sample legend. qRT-PCR showing mRNA abundance in all samples for FOXA2, SOX17, AXIN2, and LGR5. Data show that IWP2 blocks ACTA induction of all 4 transcripts (compare red triangles with green triangles), and that addition of exogenous WNT3A ligand rescues the IWP2 effect in a dose-dependent manner at 10, 50, and 500 ng/mL. Addition of exogenous RSPO1 (500 ng/mL) potentiates WNT3A rescue of the IWP2 effect such that gene expression for FOXA2, SOX17, and AXIN2 is enhanced significantly in RSPO + WNT3A at 2 ng/mL along with LGR5 at 10 ng/mL compared with low doses of WNT3A alone (compare purple circle with purple star, and yellow circle with yellow star). One-way analysis of variance was used for statistical analysis. (E) Data shown in panel D plotted with mRNA abundance (y-axis) at different doses of WNT3A (x-axis) for ACTA + IWP2 + WNT3A (green line) and ACTA + IWP2 + WNT3A + RSPO1 (red line). An unpaired t test was used for statistical analysis. (F) Immunostaining for FOXA2 (red) and SOX17 (green) on ACTA + IWP2–treated hESCs treated with increasing doses of exogenous WNT3A (top row) or increasing doses of WNT3A, and a constant dose of RSPO1 (500 ng/mL) (bottom row). (G) Controls to show that RSPO1 treatment alone does not overcome the effect of IWP2 on ACTA-treated cells. DAPI, 4',6-diamidino-2-phenylindole. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001. All scale bars represent 200 μm.
Finally, we asked if RSPO/LGR signaling synergized with WNT signaling during endoderm induction. For this experiment, we took advantage of the fact that IWP2 abolished the ability of ACTA to induce DE (Figure 3A and B), and we rescued WNT signaling by adding increasing doses (2, 10, 50, 500 ng/mL) of exogenous WNT3A ± RSPO1 (500 ng/mL) (Figure 3D–F). Importantly, high doses of RSPO1 (500 ng/mL) alone did not rescue IWP2-inhibited DE induction, indicating that in the absence of WNT ligand, RSPO1 does not have a role in this process (Figure 3G). We hypothesized that if RSPO/LGR potentiates WNT signaling during DE differentiation, that low doses of WNT3A plus RSPO1 should have more efficient DE induction compared with WNT3A alone. Indeed, we observed that ACTA + IWP2 plus low doses of WNT3A (2 or 10 ng/mL) resulted in minimal DE induction as measured by FOXA2 and SOX17 mRNA (Figure 3D and E) and immunofluorescence (Figure 3F), whereas low doses of WNT3A (2, 10 ng/mL) plus RSPO1 resulted in significantly higher levels of FOXA2 and SOX17 mRNA and noticeably more positively stained cells (Figure 3D–F). Moreover, at low doses of WNT3A, expression of the WNT signaling target gene, AXIN2, was increased when comparing ACTA + IWP2 + WNT3A vs ACTA + IWP2 + WNT3A + RSPO1 (Figure 3D and E). The synergistic effect of WNT3A + RSPO1 was apparent only at low doses and was lost when exogenous WNT3A exceeded 50 ng/mL (Figure 3E). Based on this data, we conclude that ACTA-stimulated DE differentiation requires WNT signaling, and that RSPO proteins act to potentiate WNT signaling during this process.
LGR4 and LGR5 Are Functionally Required for Human Endoderm Differentiation
Because LGR4 and LGR5 are expressed in hESC-derived endoderm, hindgut, and intestinal organoids, as well as in the human fetal intestine (Figure 1, Figure 2, Figure 3), we wanted to determine if LGR4 and 5 are functionally required for differentiation. To do this, we used lentiviral shRNAs to generate a control (scrambled shRNA) H9 hES cell line and to generate LGR4–shRNA, LGR5–shRNA, and LGR4/5–shRNA knockdown lines. To determine the efficiency of knockdown, we differentiated all cell lines into DE, and performed qRT-PCR for LGR4 and LGR5 (Figure 4A). Interestingly, in LGR4–shRNA and LGR5–shRNA knockdown lines, we observed a significant reduction in both receptors for each line (Figure 4A). This observation likely is explained by the reduced efficiency of endoderm induction in any of the knockdown lines (Figure 4A and B). Double knockdown showed a significant reduction in both transcripts (Figure 4A).
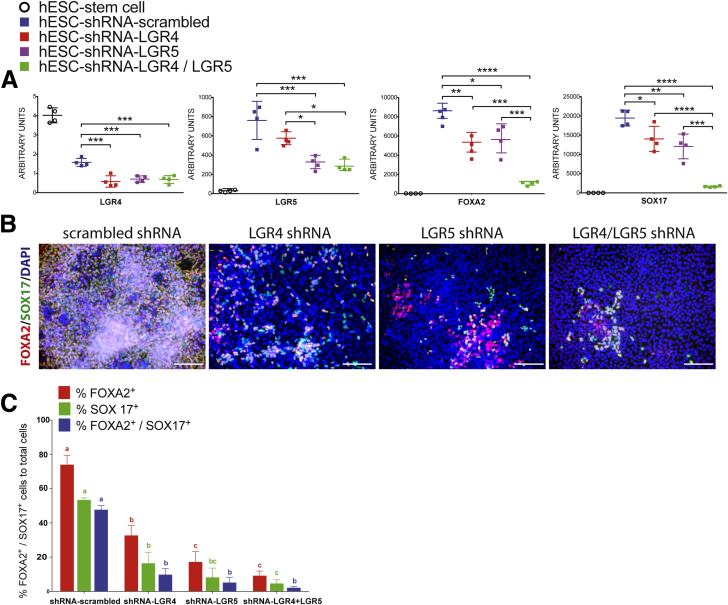
Figure 4.
LGR4 and LGR5 are required for endoderm differentiation. (A) qRT-PCR showing mRNA abundance in all samples for LGR4, LGR5, FOXA2, and SOX17. h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation. One-way analysis of variance was used for statistical analysis. (B) h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation, and immunostained for FOXA2 (red) and SOX17 (green). Nuclei were stained with DAPI (blue). Scale bars represent 200 μm. (C) Cell staining shown in panel B was quantitated. Each color group, corresponding to the percentage of FOXA2+ cells (red), the percentage of SOX17+ cells (green), and the percentage of double-positive cells (blue) was compared using 1-way analysis of variance. Dissimilar letters in a specific color group indicate significantly different values (P ≤ .05), whereas similar letters in a color group indicate no difference. DAPI, 4',6-diamidino-2-phenylindole. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001.
To assess the consequences of LGR knockdown, we generated DE from scrambled control and knockdown lines, and performed qRT-PCR (Figure 4A) and immunofluorescence for FOXA2/SOX17 (Figure 4B and C). LGR4–shRNA and LGR5–shRNA knockdown lines showed significantly reduced FOXA2 and SOX17 expression compared with control, and LGR4/5–shRNA double knockdown showed an even larger reduction in these transcripts (Figure 4A). qRT-PCR results were supported by immunofluorescence staining for FOXA2 and SOX17 (Figure 4B and C), which showed a robust and significant reduction in co-staining, indicating that DE differentiation was impaired in knockdown lines. The reduction in the number of cells stained for FOXA2, SOX17, or for both proteins was statistically reduced in all 3 knockdown lines compared with controls (Figure 4C).
Differentiation Potential of LGR4 and/or LGR5 Knockdown Cell Lines
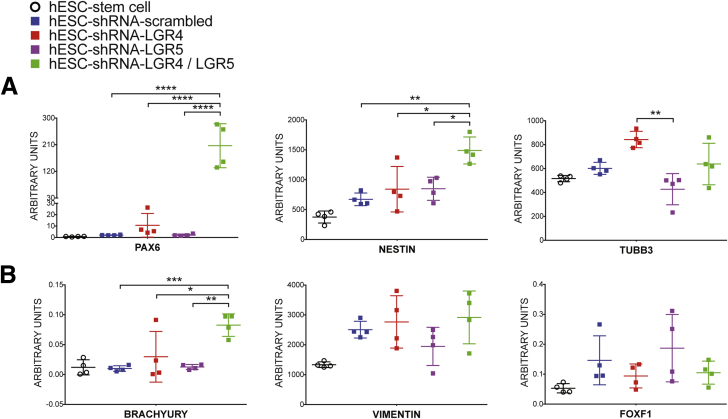
Given that knockdown of LGR4, LGR5, and LGR4/5 resulted in poor endoderm formation compared with the scrambled control, we wanted to determine the fate of these cells after 3 days of exposure to ACTA (Supplementary Figure 3A). LGR4/5 knockdown cells showed significantly higher levels of neural markers PAX6 and NESTIN and the mesodermal lineage marker T (encoding the protein BRACHYURY), indicating that these cultures are enriched with neural and mesenchymal lineages (Supplementary Figure 3A and B).
Supplementary Figure 3.
Assessment of lineage markers in control, LGR4, LGR5, and LGR4/5 knockdown hESC lines. h9–shRNA–scrambled, h9–shRNA–LGR4, h9–shRNA–LGR5, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation. qRT-PCR showing mRNA expression above hESCs for (A) neural markers PAX6 and NESTIN, and (B) mesenchymal lineage markers BRACHYURY, VIMENTIN, and FOXF1. One-way analysis of variance was used for statistical analysis. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001.
Stabilization of β-Catenin Downstream of Ligand-Receptor Interactions Rescues DE Differentiation With Lgr4 and Lgr5 Knockdown
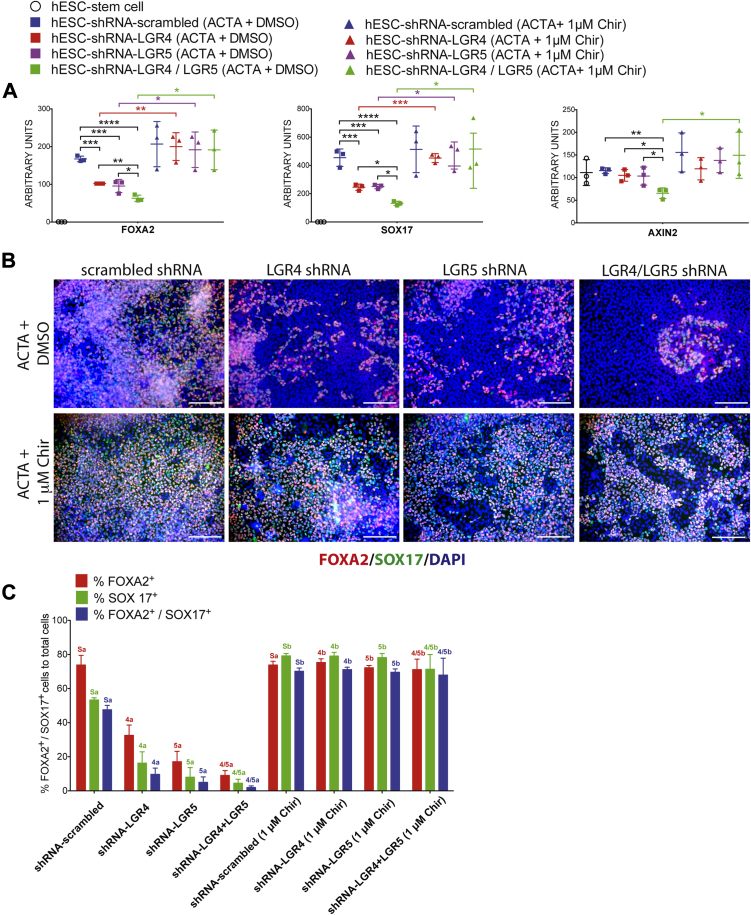
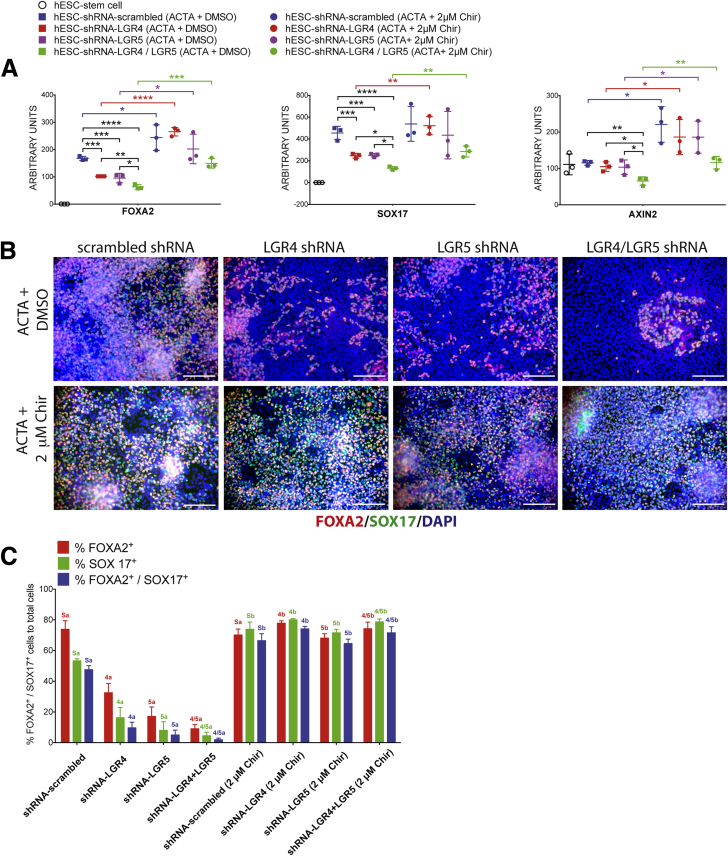
Given that RSPO/LGR interacts with WNT signaling at the cell surface, and knockdown of LGR4 and/or LGR5 leads to reduced DE formation (Figure 4), we hypothesized that stabilization of β-catenin downstream of ligand/receptor complexes should rescue endoderm induction in LGR shRNA knockdown hESC lines. To do this, we used Chir99021, a potent and specific GSK3β inhibitor involved in the β-catenin destruction complex, such that inhibition of GSK3β with Chir99021 allows β-catenin stabilization downstream of ligand/receptors.11, 51, 52 We differentiated all 4 hESC lines (control, knockdown) using a 3-day ACTA protocol along with dimethyl sulfoxide (control) or with Chir99021 (1 umol/L) (Figure 5). qRT-PCR (Figure 5A) and immunofluorescence (Figure 5B and C) showed that Chir99021 was able to rescue the defective endoderm differentiation observed in LGR4, LGR5, and LGR4/5 shRNA knockdown both at the gene expression and cellular level. Our results also showed that addition of Chir99021 led to a rescue in the mRNA expression level of the WNT/β-catenin target gene AXIN2 (Figure 5A). We also conducted rescue experiments with higher doses of Chir99021 (2 umol/L) and observed that this dose showed similar results to the lower dose (Supplementary Figure 4).
Figure 5.
β-catenin stabilization rescues loss of LGR4 or LGR5 knockdown. (A) qRT-PCR showing mRNA abundance in all samples for FOXA2, SOX17, and LGR5. h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA, or with ACTA + 1 umol/L Chir99021 (Chir) to induce endoderm formation. LGR4, 5, and 4/5 knockdown lines treated with Chir showed a statistically significant rescue of expression levels for all genes in all knockdown lines, except for AXIN2 in the LGR4 knockdown line. One-way analysis of variance was used for statistical analysis across groups. (B) h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation or with ACTA + 1 umol/L Chir, and immunostained for FOXA2 (red) and SOX17 (green). Nuclei were stained with DAPI (blue). Scale bars represent 200 μm. (C) Cell staining shown in panel B was quantitated. Each color group, corresponding to the percentage of FOXA2+ cells (red), the percentage of SOX17+ cells (green), and the percentage of double-positive cells (blue) was compared using 1-way analysis of variance. Statistical comparisons were performed between a particular knockdown group and the corresponding Chir-treated group (ie, the percentage of FOXA2-positive in shRNA-LGR5 vs shRNA–LGR5+ Chir, and so forth). Dissimilar labels in a color group denote statistically significant differences (ie, 5a vs 5b) (P ≤ .05), whereas similar labels in a color group indicate no difference (ie, Sa vs Sa). DAPI, 4',6-diamidino-2-phenylindole. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001.
Supplementary Figure 4.
β-catenin stabilization rescues loss of LGR4 or LGR5 knockdown. (A) qRT-PCR showing mRNA expression above hESCs for FOXA2, SOX17, and AXIN2. h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA, or with ACTA + 2 umol/L Chir99021 (Chir) to induce endoderm formation. LGR4, 5, and 4/5 knockdown lines treated with Chir showed a statistically significant rescue of expression levels for all genes in all knockdown lines, except for SOX17 in the LGR5 knockdown line. One-way analysis of variance was used for statistical analysis across groups. (B) h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation or with ACTA + 2 umol/L Chir, and immunostained for FOXA2 (red) and SOX17 (green). Nuclei were stained with DAPI (blue). (C) Cell staining shown in (B) was quantitated. Each color group, corresponding to %FOXA2+ cells (red), %SOX17+ cells (green), % double positive (blue) was compared using one-way ANOVA. Statistical comparisons were carried out between a particular knockdown group and the corresponding RSPO1 treated group (ie, %FOXA2 positive in shRNA-LGR5 vs shRNA-LGR5+RSPO1, etc). Dissimilar labels in a color group denote statistically significant differences (ie, 5a vs 5b) (P ≤ .05), whereas similar labels in a color group indicate no difference (ie, Sa vs Sa). Dissimilar labels in a color group denote statistically significant differences (ie, 5a vs 5b) (P ≤ .05), whereas similar labels in a color group indicate no difference (ie, Sa vs Sa). DAPI, 4',6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide. Scale bars: 200 μm. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001.
Exogenous RSPO1 Rescues DE Differentiation in LGR4 and LGR5 Knockdown hESC Lines
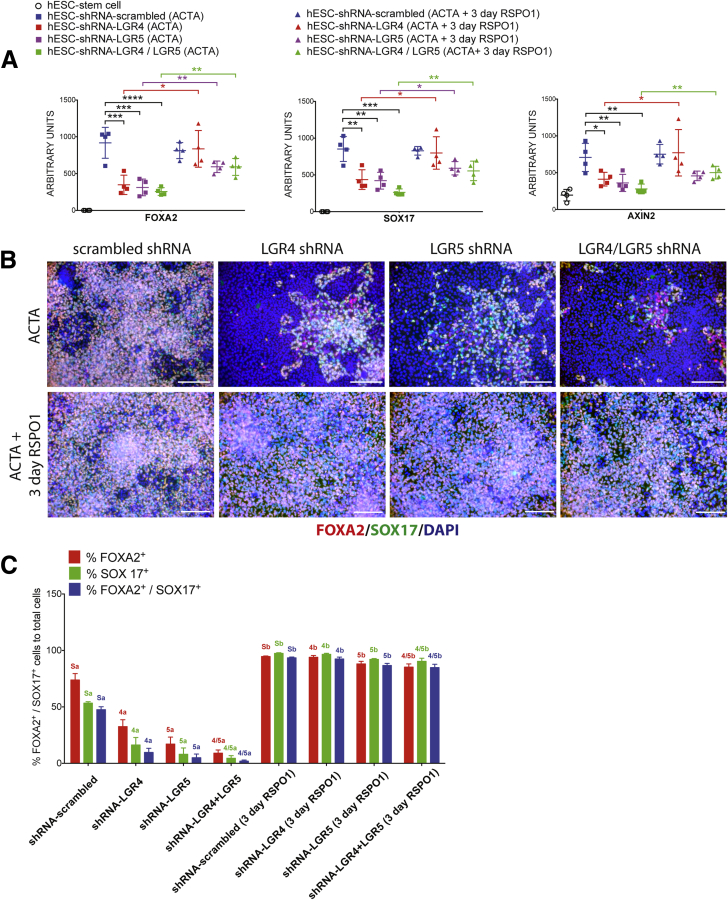
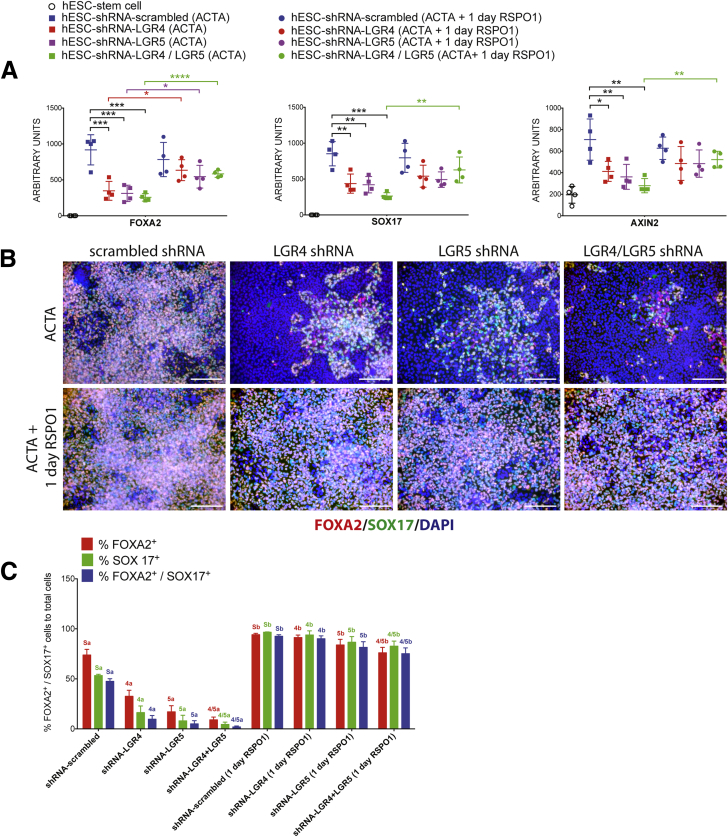
Because shRNAs reduce, but do not completely inhibit, gene expression, and because LGR receptors function redundantly, we reasoned that it also may be possible to rescue DE induction in knockdown lines by adding exogenous RSPO proteins because the current differentiation paradigm relies on endogenous RSPO production. Therefore, we added RSPO1 (500 ng/mL) throughout the differentiation (3 days of ACTA + RSPO1) and assayed DE induction with qRT-PCR and immunofluorescence (Figure 6). qRT-PCR showed that mRNA levels of FOXA2 and SOX17 significantly were restored when RSPO1 was added for 3 days (Figure 6A). In LGR4 and LGR5 knockdown lines, addition of RSPO1 for 3 days led to a statistically significant increase in SOX17 and FOXA2, however, AXIN2 mRNA expression was rescued in the LGR4 knockdown and LGR4/5 double-knockdown line, but not in the LGR5 knockdown group (Figure 6A). Consistent with mRNA data, addition of RSPO1 to cultures during DE induction led to a significant rescue of the number of FOXA2, SOX17, or double-positive DE cells present in the culture (Figure 6B and C). We also repeated the earlier-described experiment, but added RSPO1 only on the first day of ACTA treatment, followed by 2 days of ACTA alone (Supplementary Figure 5). Interestingly, in this experiment, at the mRNA level, FOXA2 was rescued in all knockdown lines whereas SOX17 and AXIN2 were rescued only in the LGR4/5 double-knockdown line. Rescue was seen at the protein level in all knockdown lines treated with 1 day of RSPO1 during endoderm induction (Supplementary Figure 5B and C).
Figure 6.
Exogenous RSPO1 rescues loss of LGR4 or LGR5 knockdown. (A) qRT-PCR mRNA abundance in all samples for FOXA2, SOX17, and AXIN2. h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA, or with ACTA + RSPO1 (500 ng/mL) to induce endoderm formation. LGR4, 5, and 4/5 knockdown lines treated with RSPO1 showed a statistically significant rescue of expression levels for all genes in all knockdown lines, except for AXIN2 in the LGR4 knockdown line. One-way analysis of variance was used for statistical analysis across groups. (B) h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation or with ACTA + RSPO1, and immunostained for FOXA2 (red) and SOX17 (green). Nuclei were stained with DAPI (blue). Scale bars represent 200 μm. (C) Cell staining shown in panel B was quantitated. Each color group, corresponding to the percentage of FOXA2+ cells (red), the percentage of SOX17+ cells (green), and the percentage of double-positive cells (blue) was compared using 1-way analysis of variance. Statistical comparisons were performed between a particular knockdown group and the corresponding RSPO1-treated group (ie, the percentage of FOXA2-positive in shRNA–LGR5 vs shRNA–LGR5 + RSPO1, and so forth). Dissimilar labels in a color group denote statistically significant differences (ie, 5a vs 5b) (P ≤ .05), whereas similar labels in a color group indicate no difference (ie, Sa vs Sa). DAPI, 4',6-diamidino-2-phenylindole. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001.
Supplementary Figure 5.
Exogenous RSPO1 rescues loss of LGR4 or LGR5 knockdown. (A) qRT-PCR showing mRNA expression above hESCs for FOXA2, SOX17, and AXIN2. h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA, or with ACTA+RSPO1 (500 ng/mL) on day 1 only, followed by 2 additional days of ACTA to induce endoderm formation. LGR4, 5, and 4/5 knockdown lines treated with RSPO1 showed a statistically significant rescue of expression levels for FOXA2 in all knockdown lines, whereas only the LGR4/5 knockdown lines showed rescue for SOX17 and AXIN2 gene expression. One-way analysis of variance was used for statistical analysis across groups. (B) h9–shRNA–scrambled, h9–shRNA–LGR4 knockdown, h9–shRNA–LGR5 knockdown, and h9–shRNA–LGR4/5 knockdown lines all were treated for 3 days with ACTA to induce endoderm formation or with ACTA+RSPO1 on day 1, followed by 2 days of ACTA, and immunostained for FOXA2 (red) and SOX17 (green). Nuclei were stained with DAPI (blue). (C) Cell staining shown in panel B was quantitated. Each color group, corresponding to the percentage of FOXA2+ cells (red), the percentage of SOX17+ cells (green), and the percentage of double-positive cells (blue) was compared using 1-way analysis of variance. Statistical comparisons were performed between a particular knockdown group and the corresponding RSPO1-treated group (ie, the percentage of FOXA2 positive in shRNA–LGR5 vs shRNA–LGR5+RSPO1, and so forth). Dissimilar labels in a color group denote statistically significant differences (ie, 5a vs 5b) (P ≤ .05), whereas similar labels in a color group indicate no difference (ie, Sa vs Sa). DAPI, 4',6-diamidino-2-phenylindole. Scale bars: 200 μm. *P ≤ .05, **P ≤ .01, ***P ≤ .001, and ****P ≤ .0001.
Discussion
Wnt signaling is known to have important roles in regulating pluripotency53, 54 and endoderm differentiation both in vitro and in vivo,55, 56, 57 however, it remains unclear how the Wnt signal might be tailored to the specific contexts of pluripotency vs endoderm. Based on our results, it is interesting to speculate that LGR4 and LGR5 might function in stage-dependent contexts to act as a rheostat for WNT signaling during this transition. Our findings show that LGR4 is expressed constitutively in hESCs and their differentiated progeny, whereas LGR5 is induced highly during DE differentiation and is maintained in CDX2+ endoderm derivatives (Figure 1). Thus, for example, it is possible that LGR4 function is most important during the initial stages of DE induction to amplify WNT/β-catenin signaling, whereas LGR5 function is important to maintain high levels of WNT signaling later during differentiation, however, this idea has not yet been tested experimentally. In an analogous manner, Lgr-directed Wnt signaling has been proposed to work in context-specific roles in the crypt base columnar stem cells to direct Achaete-Scute Family BHLH Transcription Factor 2-mediated stem cell control.58
One finding of the current study showed that in both mouse and human systems, LGR5 expression is present at very early stages of development. Although Lgr5 expression has been documented in numerous contexts in mice, much less is known about early developmental contexts, and expression in developing human tissues is particularly lacking. Other studies have reported embryonic expression in the early mouse gut before villus formation,42 and functional studies have supported the notion that in mice Lgr4 plays a more important functional role in the embryonic intestine than Lgr5.17, 18, 19 Our data in the developing mouse intestine are consistent with published studies, indicating that Lgr4 is expressed more highly than Lgr5. Interestingly, using human intestinal organoid differentiation as a surrogate for human development, and by examining human fetal intestines, we found that LGR5 consistently was expressed more highly relative to LGR4 across progressive stages of intestine differentiation, and in the human fetal intestine. Thus, although expression and function of Lgrs in the mouse are better characterized than in human beings, our studies suggest that expression differences between species exist, opening the possibility that functional differences also may exist. Future studies will be required to further clarify functional differences between human beings and mice in light of these findings.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This research was performed as a project of the Intestinal Stem Cell Consortium, a collaborative research project funded by the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Allergy and Infectious Diseases (U01DK103141 to J.R.S. and M.P.V.). This work also was supported, in part, by the University of Michigan Center for Gastrointestinal Research (National Institute of Diabetes and Digestive and Kidney Diseases P30DK034933 and K01DK091415), the University of Michigan M-cubed initiative (J.R.S.), and the National Cancer Institute (R01CA190558 to M.P.V.). The University of Washington Laboratory of Developmental Biology was supported by NIH Award Number 5R24HD000836 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development.
Supplementary Material
Supplementary Table 1.
shRNA clone identifiers
| Target genes | The RNAi Consortium number |
|---|---|
| shRNA LGR4 | TRCN0000285015 |
| shRNA LGR4 | TRCN0000273590 |
| shRNA LGR4 | TRCN0000004742 |
| shRNA LGR5 | TRCN0000011585 |
| shRNA LGR5 | TRCN0000011586 |
| shRNA LGR5 | TRCN0000011587 |
Supplementary Table 2.
Antibodies
| Antibody | Source | Dilution |
|---|---|---|
| CDX2 | MU392A-UC; Bio-Genex (Fremont, CA) | 1:500 |
| FOXA2 | WRAB 1200; Seven Hill, Cincinnati, OH | 1:500 |
| GFP | Ab13970; Abcam (Cambridge, MA) | 1:500 |
| SOX17 | AF 1924; R&D Systems (Minneapolis, MN) | 1:500 |
Supplementary Table 3.
qRT-PCR primers
| Gene | Sequence |
|---|---|
| h-AXIN2-F | AGTGTGAGGTCCACGGAAAC |
| h-AXIN2-R | CTGGTGCAAAGACATAGCCA |
| h-BRACHYURY-F | GCAAAAGCTTTCCTTGATGC |
| h-BRACHYURY-R | ATGAGGATTTGCAGGTGGAC |
| h-FOXA2-F | CGACTGGAGCAGCTACTATGC |
| h-FOXA2-R | TACGTGTTCATGCCGTTCAT |
| h-FOXF1-F | AGTCCCCAATGCAAAGACAC |
| h-FOXF1-R | TCAGCAGAATTCCTGTGTGG |
| h-LGR4-F | GCCTGAATGGGCTAAATCAA |
| h-LGR4-R | CCTTTCTCCTGTGCCACACT |
| h-LGR5-F | CAGCGTCTTCACCTCCTACC |
| h-LGR5-R | TGGGAATGTATGTCAGAGCG |
| h-LGR6-F | CAAGCCCTGGATCTTAGCTG |
| h-LGR6-R | TTTTGGGAAACTGTCCTTGG |
| h-NESTIN-F | GCCCTGACCACTCCAGTTTA |
| h-NESTIN-R | GGAGTCCTGGATTTCCTTCC |
| h-OCT4-F | GTGGAGGAAGCTGACAACAA |
| h-OCT4-R | GGTTCTCGATACTGGTTCGC |
| h-PAX6-F | GTTGGTATCCGGGGACTTC |
| h-PAX6-R | TCCGTTGGAACTGATGGAGT |
| h-RSPONDIN 1-F | TGATTGGCATGTTACCCAAA |
| h-RSPONDIN 1-R | ACCATCACTGGAAGCCTACG |
| h-RSPONDIN 2-F | GAGCGAATGGGGAACTTGTA |
| h-RSPONDIN 2-R | TCCTCTTCTCCTTCGCCTTT |
| h-RSPONDIN 3-F | GCATCCTTCAGCAAAGGGTA |
| h-RSPONDIN 3-R | TCGTTGCTCTGGGATTTCTT |
| h-SOX17-F | CAGAATCCAGACCTGCACAA |
| h-SOX17-R | TCTGCCTCCTCCACGAAG |
| h-VIMENTIN-F | CTTCAGAGAGAGGAAGCCGA |
| h-VIMENTIN-R | ATTCCACTTTGCGTTCAAGG |
Supplementary Material
References
- 1.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Schepers A., Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol. 2012;4:a007989. doi: 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon M.D., Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan K.M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 5.Glinka A., Dolde C., Kirsch N. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffner H., Sprunger J., Charlat O. R-spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One. 2012;7:e40976. doi: 10.1371/journal.pone.0040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lau W., Barker N., Low T.Y. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 8.Carmon K.S., Gong X., Lin Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao H.-X., Xie Y., Zhang Y. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 10.Koo B.-K., Spit M., Jordens I. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 11.Fearon E.R., Spence J.R. Cancer biology: a new RING to Wnt signaling. Curr Biol. 2012;22:R849–R851. doi: 10.1016/j.cub.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker N., van Es J.H., Kuipers J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 13.Sato T., Vries R.G., Snippert H.J. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 14.Ootani A., Li X., Sangiorgi E. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhnert F., Davis C.R., Wang H.-T. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2003;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto D. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia M.I., Ghiani M., Lefort A. LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the fetal intestine. Dev Biol. 2009;331:58–67. doi: 10.1016/j.ydbio.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Kinzel B., Pikiolek M., Orsini V. Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice. Dev Biol. 2014;390:181–190. doi: 10.1016/j.ydbio.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Mustata R.C., Vasile G., Fernandez-Vallone V. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 2013;5:421–432. doi: 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Madisen L., Zwingman T.A., Sunkin S.M. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2009;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 22.Spence J.R., Lange A.W., Lin S.-C.J. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkbeiner S.R., Hill D.R., Altheim C.H. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep. 2015;4:1140–1155. doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCracken K.W., Catá E.M., Crawford C.M. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spence J.R., Mayhew C.N., Rankin S.A. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Amour K.A., Agulnick A.D., Eliazer S. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 27.Finkbeiner S.R., Freeman J.J., Wieck M.M. Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids. Biol Open. 2015;4:1462–1472. doi: 10.1242/bio.013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie J.L., Huang S., Opp J.S. Persistence and toxin production by Clostridium difficile within human intestinal organoids results in disruption of epithelial paracellular barrier function. Infect Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heijmans J., de Jeude J., Koo B.K. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 2013;3:1128–1139. doi: 10.1016/j.celrep.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 30.Bell S.M., Schreiner C.M., Wert S.E. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135:1049–1058. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- 31.Rockich B.E., Hrycaj S.M., Shih H.-P. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci U S A. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCracken K.W., Howell J.C., Spence J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue X., Ramakrishnan S., Anderson E. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dye B.R., Hill D.R., Ferguson M.A. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estarás C., Benner C., Jones K.A. SMADs and YAP compete to control elongation of β-catenin:LEF-1-recruited RNAPII during hESC differentiation. Mol Cell. 2015;58:780–793. doi: 10.1016/j.molcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang A., Yue F., Li Y. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chai R., Xia A., Wang T. Dynamic expression of Lgr5, a Wnt target gene, in the developing and mature mouse cochlea. J Assoc Res Otolaryngol. 2011;12:455–469. doi: 10.1007/s10162-011-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson C.L., Mahe M.M., Múnera J. An in vivo model of human small intestine using pluripotent stem cells. Nat Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells J.M., Spence J.R. How to make an intestine. Development. 2014;141:752–760. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker N., Huch M., Kujala P. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Barker N., Rookmaaker M.B., Kujala P. Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2012;2:540–552. doi: 10.1016/j.celrep.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 42.Shyer A.E., Huycke T.R., Lee C. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161:569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zorn A.M., Butler K., Gurdon J.B. Anterior endomesoderm specification in Xenopus by Wnt/beta-catenin and TGF-beta signalling pathways. Dev Biol. 1999;209:282–297. doi: 10.1006/dbio.1999.9257. [DOI] [PubMed] [Google Scholar]
- 44.Rankin S.A., Kormish J., Kofron M. A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Dev Biol. 2011;351:297–310. doi: 10.1016/j.ydbio.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lickert H., Kutsch S., Kanzler B. Formation of multiple hearts in mice following deletion of β-catenin in the embryonic endoderm. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 46.Gadue P., Huber T.L., Paddison P.J. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nostro M.C., Sarangi F., Ogawa S. Stage-specific signaling through TGF family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherwood R.I., Chen T.Y., Melton D.A. Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biechele S., Cox B.J., Rossant J. Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev Biol. 2011;355:275–285. doi: 10.1016/j.ydbio.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 50.Chen B., Dodge M.E., Tang W. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett C.N., Ross S.E., Longo K.A. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 52.Ring D.B., Johnson K.W., Henriksen E.J. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- 53.Sato N., Meijer L., Skaltsounis L. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 54.Miyabayashi T., Teo J.-L., Yamamoto M. Wnt/β-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2007;104:5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muñoz-Descalzo S., Hadjantonakis A.-K., Arias A.M. Wnt/ß-catenin signalling and the dynamics of fate decisions in early mouse embryos and embryonic stem (ES) cells. Semin Cell Dev Biol. 2015;47:101–109. doi: 10.1016/j.semcdb.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zernicka-Goetz M., Hadjantonakis A.-K. From pluripotency to differentiation: laying foundations for the body pattern in the mouse embryo. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130535. doi: 10.1098/rstb.2013.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherwood R.I., Maehr R., Mazzoni E.O. Wnt signaling specifies and patterns intestinal endoderm. Mech Dev. 2011;128:14. doi: 10.1016/j.mod.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuijers J., Junker J.P., Mokry M. Ascl2 Acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.