Abstract
In honeybee (Apis mellifera) colonies, queens and workers are alternative forms of the adult female honeybee that develop from genetically identical zygotes but that depend on differential nourishment. Queens and workers display distinct morphologies, anatomies and behavior, better known as caste differentiation. Despite some basic insights, the exact mechanism responsible for this phenomenon, especially at the molecular level, remains unclear although some progress has been achieved. In this study, we examined mRNA levels of the TOR (target of rapamycin) and Dnmt3 (DNA methyltransferase 3) genes, closely related to caste differentiation in honeybees. We also investigated mRNA expression of the S6K (similar to RPS6-p70-protein kinase) gene linked closely to organismal growth and development in queen and worker larvae (1-day and 3-day old). Last, we investigated the methylation status of these three genes in corresponding castes. We found no difference in mRNA expression for the three genes between 1st instar queen and worker larvae; however, 3rd instar queen larvae had a higher level of TOR mRNA than worker larvae. Methylation levels of all three genes were lower in queen larvae than worker larvae but the differences were not statistically significant. These findings provide basic data for broadening our understanding of caste differentiation in female honeybees.
Keywords: Caste differentiation, DNA methylation, DNA methyltransferase 3, Honeybee, Target of rapamycin
The honeybee (Apis mellifera) is one of the most socially advanced and hitherto best-studied species in Hymenopterans (Amdam et al, 2004; Omholt & Amdam, 2004). A honeybee colony is typically composed of a single queen, none to a few thousand drones (male) depending on reproductive requirements, and 20−40 thousand workers (Winston, 1987). Queens and workers develop from genetically identical fertilized eggs, however, queens are twice as big as workers, have a much longer lifespan, are equipped with specialized anatomy and develop faster from egg to adult. Queens are fertile but workers are sterile. Workers perform various tasks that change with their age such as nest construction and cleaning, larva nursing, food processing and foraging and guarding, while the queens’ sole duty is laying eggs for the reproduction of their colonies (Camazine, 1991; Hoover et al, 2005; Kucharski et al, 2008; Page & Peng, 2001; Rueppell et al, 2005, 2007).
Caste differentiation in social insects has been investigated over several decades and the honeybee has been used as a model animal since the eighteenth century (Corona et al, 1999; Cristino et al, 2006; Page & Peng, 2001; Patel et al, 2007; Rembold et al, 1974; Wheeler et al, 2006). Consequently, the association between nourishment and developmental trajectory has been recognized for more than 100 years (Corona et al, 1999; Patel et al, 2007). It is clear that queen-destined larvae are fed royal jelly, whereas other female larvae are fed worker jelly. Royal jelly contains approximately 12% sugar (wet mass) while worker jelly has only 4% sugar (Corona et al, 1999; Ishay et al, 1976). In addition, protein type and content are different between royal and worker jelly. In a recent study, Kamakura (2011) showed that royalactin in royal jelly induces the differentiation of honeybee larvae into queens through an epidermal growth factor receptor-mediated signaling pathway. However, whether other mechanisms regulate caste differentiation remains unknown.
Many aspects of honeybee biology have benefited from new molecular biological technologies and the accomplishment of whole genome sequencing of Apis mellifera (Honeybee Genome Sequencing Consortium, 2006). The third to fourth day in larval development is a critical period of gene expression/regulation during diphenic caste development in female honeybees (Barchuk et al, 2007; Corona et al, 1999; Evans & Wheeler, 1999; Wheeler et al, 2006). TOR (target of rapamycin) is a nutrient- and energy-sensing kinase and a key player in the caste determination pathway in Apis mellifera; 3rd instar queen larvae have about two-fold higher TOR mRNA levels than 3rd instar worker larvae (Patel et al, 2007). Using rapamycin/FK506 pharmacology and RNA interference (RNAi) gene knockdown methods, Patel et al (2007) proved that in queen-destined larvae, rapamycin can specifically inhibit the activity of TOR and induce the development of worker characteristics, and that TOR gene knockdown shifts larval developmental fate from queen-destined to worker (Patel et al, 2007).Another important discovery is that fruit flies and mice deficient in S6K, another kinase governing cell growth, exhibit severe abnormalities such as developmental delay and smaller body sizes (Montagne & Steward, 1999; Shima et al, 1998).
DNA methyltransferase Dnmt3, which is in charge of de novo methylation of cytosines in CpG dinucleotides of genomic DNA, is a possible key driver of epigenetic global reprogramming in honeybees (Kucharski et al, 2008). When treated with Dnmt3 siRNA in newly hatched honeybee larvae, most worker-destined individuals emerge as queens with fully developed ovaries (Kucharski et al, 2008). Further, Kucharski et al (2008) have shown that silencing Dnmt3 can cause widespread gene expression changes and shifts the larvae developmental trajectory.
Based on these findings, we speculated that the TOR, S6K and Dnmt3 genes are differentially expressed in 3rd instar queen and worker larvae and were eager to know if the activities of TOR and S6K are regulated through DNA methylation by Dnmt3. Lower methylation levels of DNA can promote mRNA expression (Jones & Gonzalgo, 1997; Razin, 1998; Weiss & Cedar, 1997; Wolffe et al, 1999; Yamada et al, 2008) in vertebrates. Here, we measured mRNA expression levels of TOR, S6K and Dnmt3 and examined CpG methylation levels for these genes in queen and worker 3-day old larvae (using gene expression patterns of 1st instar larvae as the control). Our results indicate the relative quantity of TOR mRNA level in 3rd instar queen larvae is higher than in worker larvae, but there is no difference in S6K or Dnmt3 mRNA levels between queens and workers. The CpG methylation quantities of the three genes in queen larvae are lower (~20%) than in worker larvae, but the differences are not statistically significant.
MATERIALS AND METHODS
Honeybee larvae
Honeybee larvae were raised in a strong double 8 frame hive using a queen excluder. The colony was fed 1:1 w/v sugar water at the rate of 500 mL every second day. The queen was confined to lay eggs in a square (20 cm×20 cm) of empty comb using wire gauze. Newly emerged larvae were grafted into queen cups using a transferring tool and remaining larvae in the comb were raised as worker larvae. Queen cups were carefully placed in the centre of the top box. After 24 and 72 h larvae were collected for experimentation (Kucharski et al, 2008).
DNA extraction
Nine larvae (queen or worker) from the hive were pooled together for DNA extraction using the TIANamp Genomic DNA kit (DP304). The procedure was performed as described by the manufacturer.
RNA isolation and reverse transcription
Live larvae were grafted into 1.5 mL Eppendorf centrifuge tubes and frozen in liquid nitrogen immediately. RNA extraction was performed using Trizol according to the manufacturer’s instructions (Invitrogen, Cat.No.15596026). We collected larvae from two bee colonies (A and B). In colony A, a total of 48 1st instar larvae (queen, n=24; worker, n=24) and 52 3rd instar larvae (queen, n=26; worker, n=26) were used for total RNA isolation; for colony B, 46 1st instar larvae (queen, n=23; worker, n=23) and 47 3rd instar larvae (queen, n=24; worker, n=23) were used. Reverse transcription was performed using reverse transcriptase (Promega, Catalog# M1701) following the manufacturer’s instructions.
Quantification of TOR, S6K and Dnmt3 mRNA levels in queen and worker larvae
TOR, S6K andDnmt3mRNA levels were determined by using real-time PCR following previously described methods (Amdam et al, 2004; Patel et al, 2007) (Table 1). In brief, real-time PCR was performed in a total volume of 20 μL reaction solution, containing SYBR mix 10 μL, cDNA 1 μL, Primer 1 (1 μmol/L)1 μL, Primer 2 (1 μmol/L)1 μL and ddH2O 7 μL. Target fragments were amplified under the following thermal cycle conditions: a denaturation cycle for 3 min at 95 ℃, followed by 40 cycles of 10 s at 94 ℃, 10 s at 58 ℃ and 10 s at 72 ℃. The signal was collected at 72 ℃ per cycle.
Table 1.
Primers for quantifying mRNA levels and DNA methylation levels of the TOR, S6K and Dnmt3 genes
| Gene | Primer sequence (5'-3') | Length of fragment (bp) |
| All primers were designed in this study. | ||
| For real-time PCR | ||
| TOR | F: CTGCCACATTACCAAAGAAAGG R: AACTTGACGTTGAACACTCAATG | 170 |
| S6K | F: TAAATGCTAGAAGTCCACGTAGAG R: TTAGCCTATTTCGATCATTTCTG | 150 |
| Dnmt3 | F: ACTCGAATGTGGAACACCTGG R: GTCTTGGTCTATCTCGCTCGC | 155 |
| Actin | F: GTATGCCAACACTGTCCTTTCTG R: AAGAATTGACCCACCAATCCA | 160 |
| For examination of DNA methylation | ||
| TOR | Distal primer pair F: GATGGTTTTAGGTTATGATTA R: ACACTACATAATACTCCTTTC | 615 |
| Proximal primer pair F: TTATGATTATTTTATGTTTATG R: ATAACAATAAAAAAATCTAATAC | 573 | |
| S6K | Distal primer pair F: AAGTGAATTTGTTAATAGAG R: CGATCATTTCTATATCTTCTAC | 481 |
| Proximal primer pair F: GTTAATAGAGTATTTTAAGTGTG R: ATCCAACTCCATTATATCTAC | 441 | |
| Dnmt3 | Distal primer pair F: TTTTTGGGATGGTGATTATTTG R: ACAAACCTAAAAACATCTATCCTCT | 1205 |
| Proximal primer pair F: GATAAAGAAGATTGTTTAAGGT R: AATTTCTAAAAACATCTATCTTC | 476 | |
Examination of DNA methylation
Total DNA Bisulfite conversion was carried out using the QIAGEN EpitectR Bisulfite Kit (Code No. 59104) as recommended by the manufacturer. Converted DNA was amplified using gene-specific primers (Table 1) in a volume of 20 μL containing 10×PCR Buffer 2 μL, dNTP 1.6 μL, LA Taq DNA polymerase 0.2 μL, Primer 1 (10 μmol/L)0.4 μL, Primer 2 (10 μmol/L)0.4 μL, DNA template 1 μL, ddH2O 14.4 μL. PCR amplification was performed with an initial denaturation cycle for 5 min at 95 ℃, followed by 40 cycles of 1 min at 95 ℃, 1 min at 50 ℃ and 1 min at 72 ℃. We used 0.5 μL of the first PCR product as template for the second round of PCR amplification under the same thermal cycle conditions. Purified PCR products were cloned into PGM-T vector (TIANGEN, VT202-02). Recombinant clones were selected by plating the transformed DH5α cells (TIANGEN, CB101) onto Amp+ LB plates. Fourteen to 20 clones were chosen for each fragment for sequencing.
Data analysis
Analyses were performed with Prism 4.0. t-testwas performed on differential expression in queen and worker larvae. A P value < 0.05 was considered statistically significant.
RESULTS
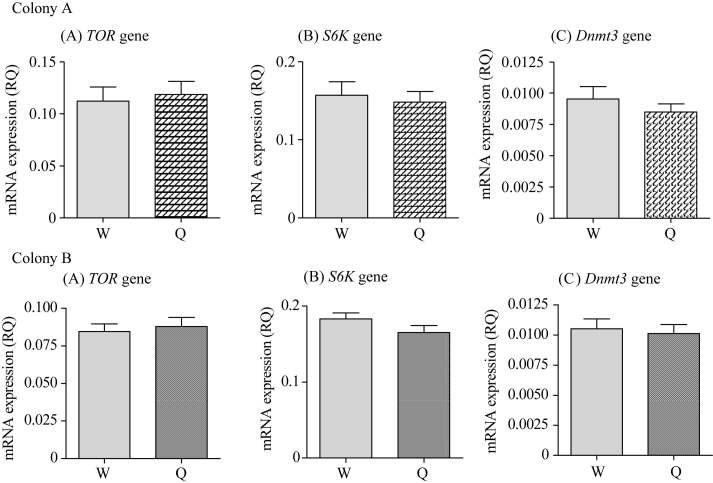
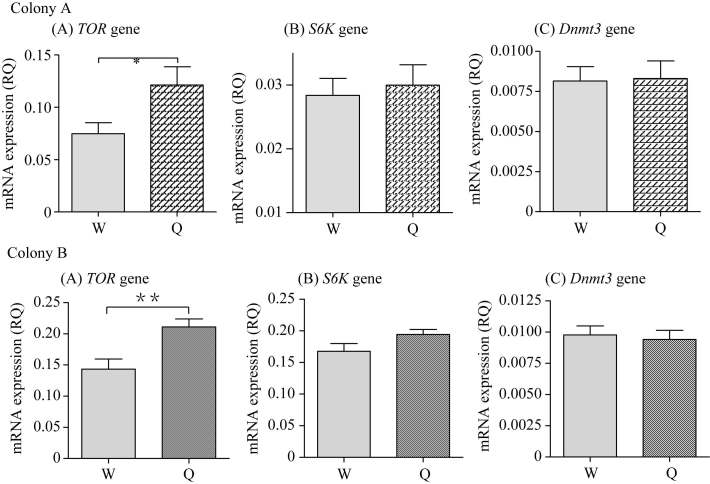
mRNA levels for TOR are different between queen and worker 3rd instar but not 1st instar larvae
Using the 1st and 3rd instar queen and worker larvae reared in colony, we measured the relative mRNA levels of the TOR, S6K and Dnmt3 genes. We found no difference in expression between 1st instar queen and worker larvae (Figure 1). However, TOR mRNA expression levels in 3rd instar queen larvae were higher than in worker larvae, whereas there were no differences for S6K and Dnmt3 mRNA levels between 3rd instar queen and worker larvae (Figure 2). To confirm our results, we examined another colony (Colony B) and obtained similar results (Figure 1, Figure 2).
1.
Comparison of relative quantity (RQ) of TOR gene (A) , S6K gene (B) and Dnmt3 gene (C) mRNA levels in 1st instar larvae
2.
Comparison of relative quantity (RQ) of TOR gene (A) , S6K gene (B) and Dnmt3 gene (C) mRNA levels in 3rd instar larvae
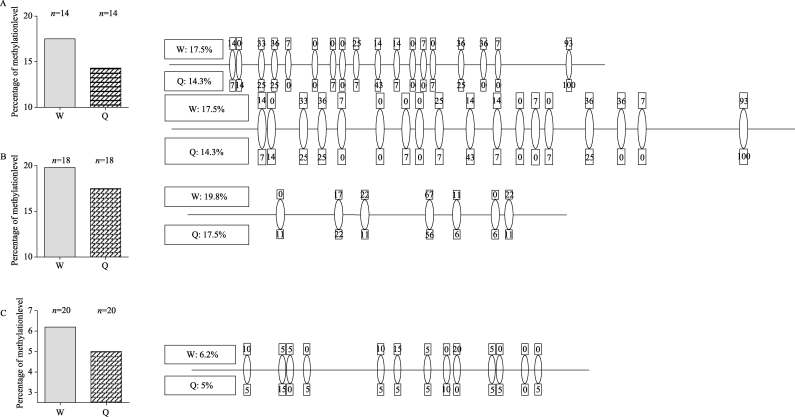
In 3rd instar larvae, DNA methylation level is lower in queen larvae than in worker larvae but the differences are not significant
To ascertain whether the mRNA expression of TOR, S6K and Dnmt3 are regulated by DNA methylation we examined methylation levels for these genes in 3rd instar larvae (1st instar larvae were not analyzed as we did not see any change in mRNA expression for these three genes). Previous studies found that CpG methylation in Apis targets most coding exons (Kucharski et al, 2008; Wang et al, 2006). Therefore, for the TOR gene, we chose exon 4 which contains 18 CpG dinucleotides as the target region for examination. The overall amount of TOR methylation is 14.3% in queen larvae versus 17.5% in worker larvae (Figure 3A). For the S6K gene, we screened exon 10, which contains 7 CpG dinucleotides, and we found that the overall amount of S6K methylation in the queen larvae (17.5%) was lower than that of worker larvae (19.8%) (Figure 3B). For the Dnmt3 gene, we also observed a lower level of methylation in queen larvae (5%) than in worker larvae (6.2%) when we focused on exon 9 of this gene, which harbors 13 CpG dinucleotides (Figure 3C). Overall, the CpG methylation quantities of all three genes in queen larvae were around 20% lower when compared with worker larvae (Figure 3). However, these data were not statistically different, probably due to the limited number of individuals analyzed in this study.
3.
Comparison of methylation level of cytosines in CpG dinucleotides in exon 4 of the TOR gene (A) , exon 10 of the S6K gene (B) and exon 9 of the Dnmt3 gene (C)
DISCUSSION
The involvement of nutrition in diphenic development in female honeybees has long been recognized (Maeterlinck 1901), but insufficient knowledge has been accumulated regarding the molecular mechanisms regulating diverse developmental fates. Recently, studies have demonstrated that two proteins, TOR and DNA methyltransferase Dnmt3, play pivotal roles in caste differentiation in Apis mellifera (Kucharski et al, 2008; Patel et al, 2007). TOR is known to be a serine/threonine kinase that controls growth in response to nutrition. Patel et al (2007) found that high expression of the TOR gene is essential for a larva to become a queen and Kucharski et al (2008) indicated that knockdown of the Dnmt3 gene in worker-destined larvae results in queen development. Some reports demonstrate that DNA methyltransferase can inactivate or suppress a gene by DNA methylation (Jones & Gonzalgo, 1997; Razin, 1998; Weiss & Cedar, 1997; Wolffe et al, 1999; Yamada et al, 2008). Consistent with the report by Patel et al (2007), the mRNA expression levels of TOR measured here were higher in 3rd instar queen larvae than worker larvae (Figure 2). However, there was no difference for TOR mRNA levels in 1st instar queen and worker larvae (Figure 1). This result suggests that TOR plays its regulatory role in a critical period of development, providing new molecular evidence to confirm the previous finding that the 3rd instar is a critical point for honeybee caste division (de Wilde & Beetsma, 1982; Handre & Lees, 1985). No significant difference in DNA methylation levels of the TOR gene was found between queen and worker larvae, although worker larvae had a relatively lower level of DNA methylation (Figure 3). In addition, there was no difference in mRNA expression of Dnmt3 in both larvae (Figure 2) and this observation implies that TOR mRNA expression may not be mediated by Dnmt3 via DNA methylation.
S6K is another kinase that controls cell growth. The fact that knockout of the S6K gene in fruit flies and mice results in extreme developmental delay and smaller bodies (Montagne & Steward, 1999; Shima et al, 1998) led us to hypothesize that S6K may be involved in caste differentiation in honeybees. We showed that mRNA expression of the S6K gene in queen larvae was slightly higher than worker larvae at the 3rd instar (a critical developmental stage for caste determination), although this did not reach statistical significance (Figure 2). Consistent with mRNA expression patterns, we found a slightly lower level of DNA methylation of the S6K gene in queen larvae than in worker larvae. Combined with the report by Kamakura (2011) that suppression of S6K with RNAi inhibited the increase to adult size induced by royal jelly, we speculate that S6K may play a role in honeybee caste differentiation. Further study regarding the role of S6K in honeybee caste diphenism is clearly essential.
In our DNA methylation experiment we chose one exon as the target to estimate the overall methylation quantity of each gene. Although CpG methylations may be heterogeneous throughout the gene, at least 7 CpGs in our target fragment are capable of representing the overall methylation level by and large, as demonstrated in previous studies (Kucharski et al, 2008; Wang et al, 2006). In spite of the fact that some special genes have alternative exons, DNA methylation or other epigenetic modifications are correlated with alternative splicing patterns (Chittka & Chittka, 2010; Luco et al, 2011; Lyko et al, 2010) and methylation promotes expression of some neurogenic genes by functionally antagonizing Polycomb repression (Wu et al, 2010). Reducing the DNA methylation quantity of a gene can facilitate its expression for the overwhelming majority of genes. Nevertheless, it is difficult to determine the direct relationship between DNA methylation and mRNA expression of certain genes because widespread gene expression differences can be caused by changes to a small number of upstream regulators (Carone et al, 2010). Differences in protein expression between 3-day old worker and queen larvae are known, indicating that the fate of queen and worker larvae have already been decided before the 3rd instar (Li et al, 2010).
In conclusion, we investigated mRNA expression and DNA methylation of Dnmt3, TOR and S6K in queen and worker-destined larvae in order to determine if mRNA expression of TOR and S6K is controlled by Dnmt3 through DNA methylation. Our results showed no difference in gene expression (except for TOR) between 3rd instar queen and worker larvae. In addition, DNA methylation levels of these three genes in both larvae did not differ, although queen larvae had a relatively lower level of methylation. These results suggest that mRNA expression of TOR might not be regulated by Dnmt3; whether S6K is regulated by Dnmt3 remains obscure. Thus, the significance of CpG methylation in exons in honeybees needs to be reevaluated.
Acknowledgements
We would like thank members of the Yao Lab at the Kunming Institute of Zoology, Chinese Academy of Sciences (KIZ, CAS) for their helpful discussion.
Funding Statement
This work was supported by the Yunnan Government (2009CI119) and Modern Agricultural Industry Technology System (Honeybee) (CARS-45-kxj14)
References
- 1. Amdam GV, Norberg K, Fondrk MK, Page RE Jr. 2004. Reproductive ground plan may mediate colony-level selection effects on individual foraging behavior in honey bees. Proceedings of the National Academy of Sciences of the United States of America, 101 (31): 11350- 11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP, Maleszka R. 2007. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Developmental Biology, 7 1- 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camazine S. 1991. Self-organizing pattern formation on the combs of honey bee colonies. Behavioral Ecology and Sociobiology, 28 (1): 61- 76. [Google Scholar]
- 4. Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li RW, Bock C, Li CJ, Gu HC, Zamore PD, Meissner A, Weng ZP, Hofmann HA, Friedman N, Rando OJ. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell, 143 (7): 1084- 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chittka A, Chittka L. 2010. Epigenetics of royalty. PLoS Biology, 8 (11): e1000532- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corona M, Estrada E, Zurita M. 1999. Differential expression of mitochondrial genes between queens and workers during caste determination in the honeybee Apis mellifera. Journal of Experimental Biology, 202 (8): 929- 938. [DOI] [PubMed] [Google Scholar]
- 7. Cristino AS, Nunes FMF, Lobo CH, Bitondi MMG, Simões ZLP, Costa Lda F, Lattorff HMG, Moritz RFA, Evans JD, Hartfelder K. 2006. Caste development and reproduction: a genome-wide analysis of hallmarks of insect eusociality. Insect Molecular Biology, 15 (5): 703- 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Wilde J, Beetsma J. 1982. The physiology of caste development in social insects. Advances in Insect Physiology, 16 167- 246. [Google Scholar]
- 9. Evans JD, Wheeler DE. 1999. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proceedings of the National Academy of Sciences of the United States of America, 96 (10): 5575- 5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Handre J, Lees AD. 1985. In: Kerkut GA, Gilbert LI. Comprehensive insect physiology, Biochemistry and Pharmacology Vol 8. England: Pergamon Press, 473- 480. [Google Scholar]
- 11. Honeybee Genome Sequencing Consortium. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature, 443 (7114): 931- 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoover SE, Higo HA, Winston ML. 2005. Worker honey bee ovary development: seasonal variation and the influence of larval and adult nutrition. Journal of Comparative Physiology B, 176 (1): 55- 63. [DOI] [PubMed] [Google Scholar]
- 13. Ishay J, Fischl J, Alpern G. 1976. Study of honeybee caste differentiation by glucose level differences during development. Insectes Sociaux, 23 (1): 23- 28. [Google Scholar]
- 14. Jones PA, Gonzalgo ML. 1997. Altered DNA methylation and genome instability: A new pathway to cancer?. Proceedings of the National Academy of Sciences of the United States of America, , 94 (6): 2103- 2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kamakura M. 2011. Royalactin induces queen differentiation in honeybees. Nature, 473 (7348): 478- 483. [DOI] [PubMed] [Google Scholar]
- 16. Kucharski R, Maleszka J, Foret S, Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science, 319 (5871): 1827- 1830. [DOI] [PubMed] [Google Scholar]
- 17. Li JK, Wu J, Begna Rundassa D, Song FF, Zheng AJ, Fang Y, Delprato AM. 2010. Differential protein expression in honeybee (Apis mellifera L). larvae: underlying caste differentiation. PLoS ONE, 5 (10): e13455- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. 2011. Epigenetics in alternative pre-mRNA splicing. Cell, 144 (1): 16- 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biology, 8 (11): e1000506- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maeterlinck M. 1901. The Life of the Bee. [Google Scholar]
- 21. Montagne J, Steward MJ. 1999. Drosophila S6 kinase: a regulator of cell size. Science, 285 (5436): 2126- 2129. [DOI] [PubMed] [Google Scholar]
- 22. Omholt SW, Amdam GV. 2004. Epigenetic regulation of aging in honeybee workers. Science of Aging Knowledge Environment, 2004 (26): pe28- [DOI] [PubMed] [Google Scholar]
- 23. Page RE Jr, Peng CYS. 2001. Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Experimental Gerontology, 36 (4-6): 695- 711. [DOI] [PubMed] [Google Scholar]
- 24. Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K, Amdam GV. 2007. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS ONE, 2 (6): e509- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Razin A. 1998. CpG methylation, chromatin structure and gene silencing-a three-way connection. The EMBO Journal, 17 (17): 4905- 4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rembold H, Czoppelt C, Rao PJ. 1974. Effect of juvenile hormone treatment on caste differentiation in the honeybee, Apis mellifera. Journal of Insect Physiology, 20 (7): 1193- 1202. [DOI] [PubMed] [Google Scholar]
- 27. Rueppell O, Fondrk MK, Page RE. 2005. Biodemographic analysis of male honey bee mortality. Aging Cell, 4 (1): 13- 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rueppell O, Bachelier C, Fondrk MK, Page RE. 2007. Regulation of life history determines lifespan of worker honey bees (Apis mellifera L. ). Experimental Gerontology, 42 (10): 1020- 1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. 1998. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO Journal, 17 (22): 6649- 6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Jorda M, Jones PL, Maleszka R, Ling X, Robertson HM, Mizzen CA, Peinado MA, Robinson GE. 2006. Functional CpG methylation system in a social insect. Science, 314 (5799): 645- 647. [DOI] [PubMed] [Google Scholar]
- 31. Weiss A, Cedar H. 1997. The role of DNA demethylation during development. Genes to Cells, 2 (8): 481- 486. [DOI] [PubMed] [Google Scholar]
- 32. Wheeler DE, Buck N, Evans JD. 2006. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Journal of Insect Physiology, 15 (5): 597- 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winston ML. 1987. The Biology of the Honey Bee. Cambridge Massachusetts: Harvard University Press, 1 [Google Scholar]
- 34. Wolffe AP, Jones PL, Wade PA. 1999. DNA demethylation. Proceedings of the National Academy of Sciences of the United States of America, 96 (11): 5894- 5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu H, Coskun V, Tao JF, Xie W, Ge WH, Yoshikawa K, Li E, Zhang Y, Sun YE. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science, 329 (5990): 444- 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S. 2008. MUC1 Expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Research, 68 (8): 2708- 2716. [DOI] [PubMed] [Google Scholar]