Key Points
Hits in driver genes and bi-allelic events affecting tumor suppressors increase apoptosis resistance and proliferation rate–driving relapse.
Excessive biallelic inactivation of tumor suppressors in high-risk cases highlights the need for TP53-independent therapeutic approaches.
Abstract
To elucidate the mechanisms underlying relapse from chemotherapy in multiple myeloma, we performed a longitudinal study of 33 patients entered into Total Therapy protocols investigating them using gene expression profiling, high-resolution copy number arrays, and whole-exome sequencing. The study illustrates the mechanistic importance of acquired mutations in known myeloma driver genes and the critical nature of biallelic inactivation events affecting tumor suppressor genes, especially TP53, the end result being resistance to apoptosis and increased proliferation rates, which drive relapse by Darwinian-type clonal evolution. The number of copy number aberration changes and biallelic inactivation of tumor suppressor genes was increased in GEP70 high risk, consistent with genomic instability being a key feature of high risk. In conclusion, the study highlights the impact of acquired genetic events, which enhance the evolutionary fitness level of myeloma-propagating cells to survive multiagent chemotherapy and to result in relapse.
Introduction
Treatments introduced over the last decade have significantly improved the outcome for patients with multiple myeloma (MM).1 The Total Therapy (TT) program has relied upon the concept of induction, autologous stem cell transplantation (ASCT), consolidation, and maintenance therapy to overcome intra-clonal heterogeneity, leading to cure in a significant proportion of cases.2 Nevertheless, patients continue to relapse, and understanding the underlying molecular mechanisms that mediate this could open new avenues of therapy aimed at overcoming these mechanisms.
Little is known about the mechanisms of disease progression of MM, but some information has come from the study of the transition of smoldering MM to MM using paired samples.3 This analysis highlighted the importance of changes in clonal substructure, rather than defining specific mutational drivers. Other mechanistic information about relapse in MM has been provided by longitudinal copy number aberration (CNA) analyses.4 This approach identified 3 distinct evolution patterns at relapse. In the first pattern, no changes were detectable. The second showed gains of CNAs in a linear fashion. The third was associated with losses and gains of CNAs, indicating branching evolution. More recent longitudinal studies using next-generation sequencing have shown changes consistent with those seen with CNAs but did not address the impact of specific treatments.5,6
In a “Darwinian model” of progression/relapse, treatment can be considered as a selective pressure. Therefore, to understand the molecular mechanisms underlying relapse, the analysis has to be done within the context of a standard treatment. In this work, we aimed to elucidate the molecular mechanisms underlying disease progression during or after ASCT. We included 33 patients enrolled in Total Therapy (TT) protocols including multiagent induction, tandem ASCT, and maintenance with MM plasma cells (PCs) collected at enrollment and at first relapse/disease progression. Methodologically, we used detection of somatic single-nucleotide variants (SNVs) and CNAs to allow more accurate detection of driver aberrations and estimation of the subclonal structure underlying relapse. In this context, next-generation sequencing allows the assessment of both SNVs and CNAs that can be augmented by high-density copy number arrays, and although it has been used to establish the mutational landscape of MM at presentation, there are few data generated longitudinally.5-9
Methods
Patients
We performed a longitudinal whole-exome sequencing (WES) analysis of 33 patients with MM enrolled in TTs, with the first sample collected at presentation and the second at relapse/progression (supplemental Table 1, available on the Blood Web site). The median interval between samples was 2.45 years (range, 0.33-7.45). All patients received multiagent induction therapy including novel agents in the majority of patients (80%) and all except two proceeded with at least one ASCT. Informed consent for treatment and sample procurement in accordance with the Declaration of Helsinki was obtained for all cases included in this study, which was approved by the local institutional review board (#02815).
Gene-expression profiling
Gene-expression profiling (GEP) of CD138-positive PCs was performed as recently described.10 The derivation of the classical GEP70 risk signature,10 the 3-group GEP70 classifier,11 the University of Arkansas for Medical Sciences (UAMS) molecular subgroup classification,12 and the expression-based proliferation index12 have been reported previously. The gene set enrichment analysis was performed using the gauge package in R.
Whole-exome sequencing and high-resolution copy number profiling
DNA was isolated from CD138-positive PCs. Control DNA originated from peripheral blood leukapheresis products collected after induction therapy. WES libraries were prepared using the SureSelectQXT sample prep kit and the SureSelect Clinical Research Exome kit (Agilent), with additional baits covering the Ig and MYC loci.13 Paired-end sequencing was performed to an average sequencing depth of 118× on a HiSeq2500 (Illumina). High-resolution HumanOmni 2.5 SNP array (Illumina) data were available for 30 of 33 patients. The data were preprocessed using GenomeStudio V2011.1 (http://www.Illumina.com/applications/microarrays/microarray-software/genomestudio.html). Details regarding the analysis of sequencing and array data are provided in the supplemental Methods.
Results
Increased GEP70-defined risk and proliferation index at relapse
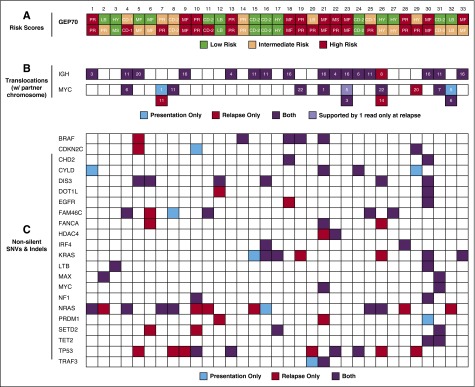
In the majority of cases, GEP70 risk scores increased from presentation to relapse. To illustrate this increase, we used a GEP70 classifier distinguishing 3 risk groups.11 At presentation, we detected 14 low-risk (LR), 7 intermediate risk (IntR), and 12 high-risk (HR) patients (Figure 1A). At relapse, more than one third of patients (n = 13) shifted to a more unfavorable risk group. Only one patient had a downgraded risk status from HR to IntR.
Figure 1.
GEP70 risk, IgH, and MYC translocations and nonsilent mutations in selected genes. (A) The risk status according to the recently published 3-group GEP70 classifier11 is shown for the 33 patients. Intermediate-risk cases would be classified as low risk according to the classical GEP70.10 UAMS GEP groups are shown within the boxes, consisting of CD-1, CD-2, HY, LB, MS, MF, and PR. The upper and lower rows present the status at presentation and relapse, respectively. (B) IgH and MYC translocations were called from whole-exome sequencing data using MANTA16 and are shown with the corresponding translocation partner chromosome. C: Non-silent SNVs and indels in a set of genes previously implicated in the pathogenesis of MM.17,18 CoLRs indicate whether an abnormality was detected at presentation, relapse, or both.
A gene set enrichment analysis of paired presentation/relapse GEP data showed significant upregulation of the cell cycle, DNA replication, and pyrimidine metabolism (P < .001), consistent with increased cell division. To further investigate changes in proliferation, we used a GEP-based proliferation index (PI).12 We detected a significantly higher PI at relapse (median presentation 3.57, median relapse 10.33, P < .001). This effect was seen mainly in the 13 patients with a higher-risk classification at relapse according to the 3-group GEP70 classifier (P < .001 vs P = .08; supplemental Figure 1). This was also reflected by a higher number of cases assigned to the UAMS proliferation (PR) molecular subgroup at relapse (7 at presentation vs 12 at relapse), a group characterized by upregulation of proliferation-related genes12 (Figure 1A).
Accumulation of deletion 17p, ongoing processes at 1q, and parallel evolution involving the MYC locus
We analyzed chromosomal aberrations with known prognostic value and observed a twofold increase in deletions involving 17p13 from presentation to relapse (n = 5 at presentation and n = 10 at relapse; supplemental Table 2). Four patients acquired new CNAs on chromosome 1 at relapse—3 patients with gain of 1q (+1q) and 1 patient with deletion at 1p. Furthermore, 4 patients with preexisting CNAs at chromosome 1 showed additional CNAs at relapse, and in 2 patients a subclone containing +1q became dominant. These results indicate ongoing processes at this chromosome, which occur during progression. Additional acquired CNAs at relapse, frequently seen in this data set, but which lack prognostic significance, were deletions or loss of heterozygosity (LOH) at 6q and 16q found in 5 patients each (supplemental Table 3).
Important molecular events with a prognostic role are the primary IgH translocation t(4;14) and secondary translocations involving the MYC locus.14,15 We did not detect losses or gains of t(4;14) or any of the other recurrent primary etiologic IgH translocations. However, in 3 patients a new secondary MYC translocation was detected at relapse (Figure 1B).16-18 In one of these patients, a clone with a distinct and different MYC translocation disappeared, indicating events at the MYC locus can be subclonal. In line with this observation, 2 other patients had >1 MYC translocation at presentation. In one of these cases, the dominant translocation at presentation disappeared, whereas a minor clone containing the second translocation expanded. In the second patient, one clone declined below the detection level at relapse.
Increased mutational rate and enrichment of driver gene mutations at relapse
We identified a median of 87 (16-898) SNVs at presentation, with 43 (7-419) being nonsilent (missense, nonsense, and splice site). At relapse, the number of total (median 116 [27-745]) and nonsilent (median 60 [15-328]) SNVs was significantly higher (P < .001 for both; supplemental Figure 2). The same observation holds true for indels with a median of 13 (3-23) at presentation and 15 (4-31) at relapse (P = .004). Nonsilent SNVs in NRAS (n = 7), DIS3 (n = 6), KRAS (n = 5), CYLD (n = 4), and BRAF (n = 3) were the most frequent mutations at presentation. In addition, we detected indels in FAM46C (n = 3) and TP53 (n = 2). The distribution of SNVs and indels in these and other genes implicated in the pathogenesis of MM17,18 is shown in Figure 1C. Overall, NRAS and TP53 were the genes most often acquiring SNVs at relapse, with both being mutated in 6 additional patients.
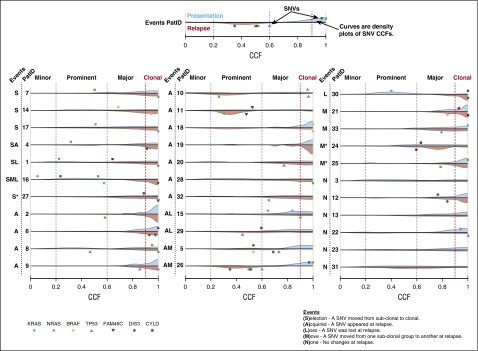
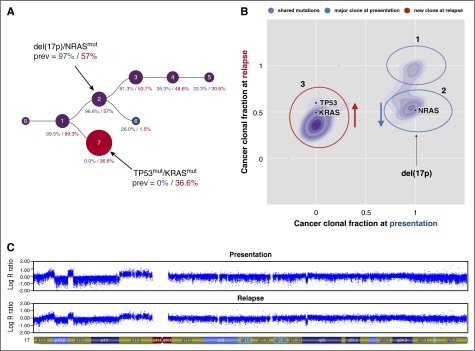
There were several examples of frequently mutated genes being present as subclonal events at presentation, mediating parallel evolution and/or selection of subclones, which increased in size at relapse (Figure 2). Mutations in NRAS were selected in 3 patients, and KRAS-, BRAF-, DIS3-, or FAM46C-mutated subclones expanded in one patient each at relapse. In 2 patients, CYLD mutations were no longer detectable at relapse. Competition among NRAS-, KRAS-, and BRAF-mutated subclones was seen in 4 patients, where one mutation was lost and another was either gained or maintained at relapse. One of these patients also showed TP53 aberrations in different subclones. A major clone with a del(17p) declined in frequency but was still detectable at relapse. At the same time, a new dominant clone with a TP53 mutation appeared at relapse (Figure 3).19
Figure 2.
Cancer clonal fraction. The cancer clonal fraction (CCF) for somatic single-nucleotide variants in 7 genes that were mutated in at least 4 patients at presentation (top) and/or relapse (bottom) is depicted. In addition, a density plot for the cancer clonal fraction for all other nonsilent mutations (presentation: blue, relapse: red) is presented for each patient. Variant allele frequencies and copy numbers used for the calculation of CCFs were derived from whole-exome sequencing data. To describe changes in more detail, we classified clones containing these mutations as minor (<20% CCF), prominent (<60% CCF), or major (≥60% CCF) and called mutations with a CCF ≥90% clonal. Patients are ordered according to events: (S)election of subclonal mutation(s), (A)cquisition of a new SNV at relapse, (L)oss of a SNV at relapse, (M)ovement from one clonal classification to another, and (N)o changes.
Figure 3.
Independent events involving the TP53 locus. Copy number profiles and mutation data were derived from high-resolution copy number arrays and whole-exome sequencing data, respectively. The clonal evolution in one patient from presentation to relapse as predicted by PhyloWGS19 and as further interpreted after inclusion of copy number data are shown in (A). The cancer clonal fractions of mutations detected in this patient are presented in (B). Mutations were manually assigned to clusters/subclones. In both plots, blue denotes aberrations/clones detectable only at presentation, red indicates aberrations/clones found only at relapse, and aberrations/clones present at both time points are shown in purple. Mutations in (B) that were not assigned to a cluster are depicted in gray. The LogR ratio for chromosome 17 at presentation and relapse is shown in (C). The major clone at presentation contained a NRAS mutation and a deletion at 17p involving the TP53 locus. This clone declined, whereas a new clone appeared with a KRAS and a TP53 mutation at relapse.
At relapse, biallelic inactivation of tumor-suppressor genes is frequently seen
TP53 molecular abnormalities, including nonsilent mutations, indels, and deletions, were seen in 15 patients (45%) at relapse. In 4 patients, the clonal double-hit events del(17p)/TP53mut or del(17p)/TP53del were present. Plasma cell leukemia and/or extramedullary disease rapidly developed in all 4 patients. In a fifth patient, two subclonal hits appeared, del(17p) and TP53mut, probably a double-hit event within a single subclone. The 5 patients died within one year after relapse.
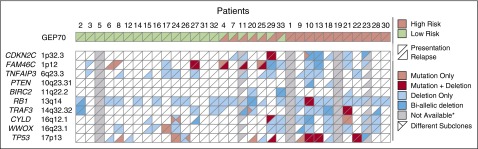
Expanding on this observation, we selected 9 other tumor-suppressor genes with a potential impact on MM pathogenesis17,20 and investigated them at presentation and relapse in 30 cases (Figure 4). Across all 10 genes, at presentation, there were a total of 19 biallelic events in 11 patients, whereas at relapse the number increased to 26 events in 15 patients. Besides TP53, FAM46C (n = 6) was the most common gene showing biallelic events, indicating its biological relevance. Of the 10 genes, only TNFAIP3 lacked biallelic events, but newly acquired deletions involving it were detected in 4 patients at relapse.
Figure 4.
Aberrations affecting tumor-suppressor genes. Deletions and nonsilent mutations for 10 selected tumor-suppressor genes with a potential impact on MM pathogenesis.17,20 Mutations and copy number profiles were derived from whole-exome sequencing and high-resolution copy number arrays, respectively. Samples without high-resolution copy number data were excluded from the analysis but are shown as not available in the plot. The risk status was determined using the classical GEP70 classifier.
Enrichment of mutations in TP53 and PI(3)K/RAS signaling at relapse
To identify pathways with a potential role in disease progression during treatment, we investigated the frequency of mutations in networks recently identified by a pan-cancer analysis.21 At relapse, at least 3 patients acquired SNVs (silent and nonsilent) for each of the networks “NOTCH”, “TP53”, “RTK” (receptor tyrosine kinase), and “PI(3)K/RAS” (supplemental Figure 3). The latter showed mutations in a significantly higher number of patients at relapse (n = 15 vs n = 24, P = .04), in line with results of the single-gene analysis for NRAS, KRAS, and BRAF. The “Condensin complex” also showed mutations in at least 3 additional patients at relapse using only nonsilent SNVs (supplemental Figure 4). When considering patients with preexisting mutations in these networks, an enrichment of mutated genes in “PI(3)K/KRAS” and “TP53” networks was seen in 12 and 14 patients, respectively (9 and 11 for nonsilent SNVs only).
Resistance to immunomodulatory drugs (IMiD) and proteasome inhibitors was recently associated with mutations in IMiD response genes IRF4, CRBN, DDB1, CUL4A, CUL4B, IKZF1, IKZF2, and IKZF3 or in the proteasome inhibitor response genes PSMB5 and PSMG2, respectively.22,23 These genes showed no difference in mutation status in this analysis, but in 2 cases with an IRF4 mutation, the respective clone expanded.
Recent studies identified frequent mutations in genes coding for ribosomal proteins (RP) in tumors of the B-cell and T-cell lineage.24-26 In our set, 3, 4, and 2 patients showed SNVs (silent + nonsilent) in genes coding for mitochondrial (M) RPs, RPs or members of the ribosomal protein S6 kinase (RPS6K) family at presentation, respectively (supplemental Figures 3 and 4). The total number of patients increased from 8 to 14 at relapse, with 6 patients having a mutation in MRP, 7 in RP, and 4 in RPS6K family genes. Nonsilent SNVs in RPL3L and silent SNVs in RPL10 were detected in 2 patients each.
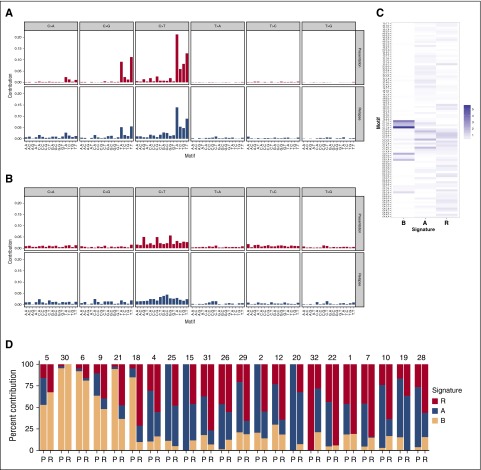
Non-negative matrix factorization identified enrichment of a novel mutation signature at relapse
Mutational signatures can identify the processes mediating initiation and progression of cancer. To identify signatures associated with relapse, we applied non-negative matrix factorization. Based on visual inspection of somatic spectra, we set the number of signatures to be obtained to 3. Recently, an enrichment of C>G and C>T mutations in the context of TpCpA and TpCpT was found in cases with translocations involving MAF or MAFB that are associated with deregulated activity of APOBECs.13 These enzymes are involved in somatic hypermutation during antibody affinity maturation and, when abnormally expressed, may cause collateral genomic damage.27 Our data set contained 8 MF cases, which were characterized by upregulation of MAF or MAFB.12 To account for the association with the “APOBEC signature” in downstream analyses of signatures, we differentiated between MF and non-MF cases. Only paired samples with at least 30 acquired mutations at relapse were included (6 MF, 16 non-MF cases).
At presentation, signature B that corresponds to the “APOBEC signature” was enriched in MF-cases (P < .001), whereas signature A showing a weak enrichment of C>T mutations at CpG dinucleotides mainly contributed to the mutation spectrum in non-MF cases (P = .002, Figure 5).28 Of note, whereas in 4 MF cases the contribution of signature B was similar at presentation and relapse, in 2 MF cases its contribution dropped from 94% to 37% and from 85% to 10% for acquired mutations, respectively (Figure 5D).
Figure 5.
Enrichment of a novel mutational signature. Mutation signatures associated with relapse were identified with the SomaticSignatures R package28 using non-negative matrix factorization. To account for the association of MAF translocations with the “APOBEC signature,” we differentiated between cases assigned to the MF molecular subgroups (MF) and other cases (noMF). The mutational spectra at presentation and relapse are shown in (A) and (B) for all MF and noMF cases, respectively. For the identification of signatures, only paired samples with at least 30 acquired mutations at relapse were included (6 MF, 16 non-MF cases). (C) The 3 identified signatures are presented. (D) Shown are the contribution of these signatures to the mutation load at presentation (P) and acquired mutations at relapse (R) in the 22 analyzed cases.
At relapse, a distinct previously unreported signature R (relapse), characterized by small peaks at “C>T GCA” and “C>T GCC” motifs (Figure 5C), was significantly enriched in acquired mutations (P < .001, Figure 5D).
Branching as the main evolution pattern during treatment
To more accurately identify evolution patterns leading to relapse, we combined pattern predictions based on CNA (5-MB threshold) and SNV data (supplemental Table 3). The most common pattern leading to relapse was branching evolution (n = 22). The second most frequent pattern was linear evolution (n = 5), followed by differential response in 3 cases, in which the relative proportion of clones changed. Two cases showed a combination of drift and linear evolution, where one clone declined and another clone acquired new mutations. A stable pattern was observed only once. Examples for patterns are shown in supplemental Figure 5.
Based on the interpretation of SNV data, major and prominent clones disappeared in 9 and 15 patients, respectively (supplemental Table 4). In 2 additional patients, major clones significantly declined but were still detectable. In 6 of 7 patients with a partial response, a prominent clone disappeared at the same time as another prominent clone survived, consistent with differential response between clones. For one patient, we collected an additional sample after 2 more years of treatment including carfilzomib, pomalidomide, and trametinib (supplemental Figure 6). During this time, the patient shifted to GEP70 HR. At “relapse 1” the tumor had 6 acquired CNAs, including subclonal del(12p), LOH at 16q, and bi-allelic del(16q12.1–12.2). At a subsequent relapse, LOH at 16q disappeared, the bi-allelic del(16q12.1-12.2) was clonal, and instead of a whole-arm del(12p), we detected a del(12p13.2) only. Importantly, del(17p) as well as an indel in TP53 appeared, a further bi-allelic event.
Association of GEP70 risk with copy-number aberration differences and biallelic events
Taking account of all CNA changes (lost and acquired), patients assigned to GEP70 HR at presentation had a higher number of CNAs between presentation and relapse (HR median 5 [0-20], LR median 2 [0-9]; P = .038; supplemental Figure 7). Using the GEP70 score determined at relapse, the difference seen was similar (HR median 4 [0-20], LR median 2 [0-9]; P = .008). In line with an increased number of CNA differences in HR cases, we also observed a significantly higher number of patients with biallelic events in our set of 10 preselected tumor-suppressor genes at presentation (LR n = 4/19 vs HR n = 7/10; P = .02). At relapse, the difference was even more pronounced (LR n = 3/15 vs HR n = 12/14; P < .001). These observations are consistent, with genomic instability at a chromosomal level being a key feature of HR.
In contrast to these observations, the mutational load at presentation was independent of the GEP70 risk status at presentation as well as at relapse after adjustment for molecular subgroups (supplemental Figure 8). The MF molecular subgroup had a higher mutational load at presentation (median 164.5 vs 81; P = .04) and relapse (median 343 vs 111; P = .01), when compared with non-MF cases.
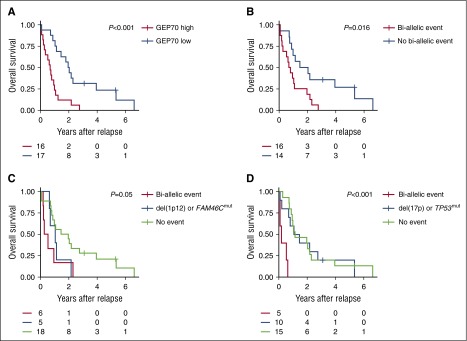
Double-hit events involving tumor-suppressor genes are prognostic at relapse
The GEP70 risk classifier allowed the discrimination of 2 groups, with significantly different overall survival after relapse (P < .001, Figure 6A). Patients with biallelic events involving at least one of our preselected 10 tumor-suppressor genes had a significantly worse outcome (P = .01, Figure 6B). To investigate this association in more detail, we performed an extended analysis with FAM46C, TP53, and TRAF3, the genes showing most of the biallelic events at relapse. Events affecting both alleles of FAM46C (P = .04, Figure 6C) or TP53 (P < .001, Figure 6D) were significantly associated with poor outcome, but not the ones involving TRAF3 (P = .45). Of note, the single events TP53mut or del(17p) were not associated with inferior overall survival after relapse. In contrast, the biallelic events del(17p)/TP53mut or del(17p)/TP53del defined a subgroup with a terrible outcome after relapse (Figure 6D).
Figure 6.
Impact of GEP70 risk and bi-allelic events on overall survival after relapse. Nonsilent mutations and deletions affecting 10 selected tumor-suppressor genes with a potential impact on MM pathogenesis17,20 were identified using whole-exome sequencing and high-resolution arrays, respectively. (A) The overall survival after relapse according to the classical GEP70 risk status determined at relapse is shown. (B) Impact of bi-allelic events in at least one of the 10 selected tumor suppressor genes on outcome. (C-D) Overall survival after relapse according to the type of aberration affecting FAM46C and TP53, respectively. We assumed that the bi-allelic deletion affecting TP53 in patient 23 was still present at relapse (no array data available) and included this patient. Survival rates after relapse were estimated using the Kaplan-Meier method. The log-rank test was used to perform group comparisons.
Discussion
This is the first longitudinal study of relapse in MM using copy number profiles, gene expression, and mutation data to systematically analyze the pattern of evolution and drivers of relapse in patients with MM treated with combination high-dose chemotherapy. Furthermore, the availability of temporally separated paired samples enhances the ability to describe evolutionary patterns and drivers. In Total Therapies, multiagent chemotherapy and ASCT aim to eradicate all subclones to maximize response and clinical outcomes. Notably, this approach has been reported to lead to cure in a significant number of cases.2
In evolutionary terms, treatment can be considered as selective pressure, interacting with acquired events within subclones best thought of as “units of selection” present within the overall clonal population. We show that in 75% of cases, subclones were reduced to below the detection threshold of our sequencing approach, suggesting subclonal “units of selection” can be eradicated in the majority of cases. In contrast to the current study, significantly lower rates of clonal eradication of 27% to 43% have been seen in other longitudinal studies, probably reflecting the lower treatment intensity used.4,5 In addition, >30% of cases in these studies showed a “stable” evolution pattern at relapse. In contrast we found this pattern in only one patient who did not respond to therapy, and as such should be considered primary refractory. There are other methodologic differences between the studies. In our study, the first sample was taken at presentation, whereas in the other it was collected after first relapse, a time when clonal selection may already have occurred.5 The depth of sequencing also differed. Despite differences, there is a consistent message of branching evolutionary pathways frequently leading to relapse.
One of the key therapeutic strategies of the TT approach is to enhance DNA damage by maximizing chemotherapy exposure and, consequently, increase apoptosis. Elucidating the molecular mechanisms during progression from this aggressive therapeutic strategy is important and could indicate ways to prevent relapse. Within this context, variants leading to escape from apoptosis and increasing proliferation are expected because they lead to both drug resistance and tumor expansion. Therefore, it is not surprising that we observed increased proliferation and clones with inactivating events involving TP53 and other tumor-suppressor genes at relapse. A proportion of increased proliferation may be explained by the appearance of mutations in NRAS, KRAS, and BRAF, as well as MYC deregulation as a result of secondary translocations to the Ig and other loci. The importance of these variants is further supported by our description of parallel evolution of subclones containing them. Impaired apoptosis and enhanced proliferation is similar to what is seen in double-hit lymphomas, where the ability to resist apoptosis at the same time as proliferating has very poor prognostic implications,29 and the same clinical course is also seen in MM with this combination of variants.
Surprisingly, single inactivating events at the TP53 locus, an established prognostic marker for patients newly diagnosed with MM, was not associated with impaired outcome after relapse. In contrast, bi-allelic events likely leading to complete inactivation of TP53 defined the group with the worst outcome after relapse. These results have important implications for use of del(17p13) as an adverse prognostic factor at presentation. We hypothesize the negative prognosis of TP53 aberrations at presentation is a result of the dramatic impact of TP53 bi-allelic events on survival. Intuitively, over time patients with mono-allelic events involving 17p13 will have a higher risk of acquiring a second hit, leading to impaired survival. This hypothesis is supported by the short progression-free survival (<1 year), from presentation, of 4 cases with TP53 bi-allelic events in our set and the results of a recent study showing an accumulation of TP53 mutations in del(17p13)-positive cases at presentation.30 A further important consideration is whether the effect is mediated by TP53 alone, because mouse models of 17p deletions support the potential effects of other genes located in the same region.31
In addition to TP53, we detected a high frequency of bi-allelic alterations affecting other tumor-suppressor genes, potentially further contributing to reduced apoptosis and drug resistance. The tumor-suppressor genes analyzed are most often located in regions of frequent loss of copy number in MM including 1p, 6q, and 16q, which showed newly acquired alterations at relapse. Further support for the critical importance of copy number loss in MM and the inactivation of tumor-suppressor genes located within them is the poor prognosis associated with a hypodiploid karyotype, which is characterized by multiple whole-chromosome deletions.32
The biological basis for poor outcome in MM remains unclear. In this work, we found an excess of bi-allelic inactivation events and more copy numbers changes from presentation to relapse in HR cases, consistent with increased genomic instability being a feature of HR. In contrast to these findings, we did not find a correlation between the GEP70 and mutational load or the number of lost or acquired SNVs, indicating that processes leading to CNAs or SNVs may act independently, and highlighting the critical role of CNAs in HR biology and relapse.
The mutagenic effects of chemotherapy are well described, with distinct mutation signatures being associated with DNA alkylating and intercalating agents.33 In the scenario of relapse after Total Therapy, multiagent chemotherapy could potentially cause harmful DNA damage, increasing the likelihood of driver abnormalities within resistant “units of selection,” which could therefore enhance relapse risk. Indeed, we found significantly more mutations and an enrichment for a novel signature, characterized by peaks at “C>T GCA” and “C>T GCC” motifs at relapse. To our knowledge, this signature has not been described previously and it certainly does not have the classic features of alkylating agent or cisplatin exposure. However, mutagenesis leading to relapse could also be driven by intrinsic mechanisms (eg, aberrant APOBEC activity). Although at presentation our data confirmed the association of MAF translocations with increased mutational load and an “APOBEC signature,”13 data generated at relapse, in 2 MF cases, showed the contribution of signature B significantly dropped being consistent with the role of APOBECs being less pronounced after evolution under the selective pressure of treatment.
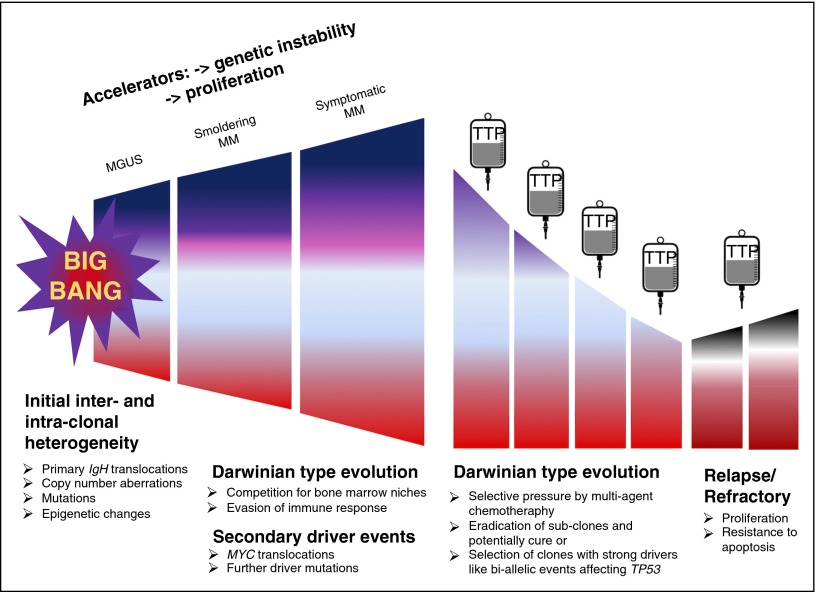
Alternatively, molecular aberrations detectable at relapse may reflect subclonal heterogeneity at diagnosis with the selection of rare preexisting events during treatment. Recently a “Big Bang model” has been put forward to explain the development of colorectal cancer.34 This model postulates that most of the detectable intratumor heterogeneity develops early after the transition to an invasive tumor. According to our and other recent data,3,5,8,9 the majority of driver events are subclonal at presentation, consistent with “Big Bang dynamics” in MM development. We postulate a model combining “Big Bang” and Darwinian type of evolution (Figure 7). In this model, “Big Bang” dynamics lead to the early establishment of intratumor heterogeneity, followed by Darwinian-type evolution in which treatment can be thought of as a significant population bottleneck, after which residual “units of selection” compete with each other and normal hematopoiesis for access to an appropriate bone marrow niche.
Figure 7.
Model of multiple myeloma development and relapse. We postulate a model combining the recently introduced “Big Bang model”34 and Darwinian type of evolution. According to this model, “Big Bang” dynamics lead to the early establishment of intra-tumor heterogeneity, followed by a Darwinian type of evolution in which different subclones acquire additional aberrations and compete with each other and normal hematopoiesis for access to an appropriate bone marrow niche. Treatment can be thought of as generating a significant population bottleneck, which eradicates some subclones but may simultaneously select for clones with strong driver events that increase proliferation and resistance to apoptosis.
In conclusion, our study stresses the biological relevance of del(1p), +1q and del(17p), MYC translocations, and mutated driver genes such as BRAF, NRAS, KRAS, and TP53 on the fitness level of MM cells to survive multiagent chemotherapy. Mechanistically, bi-allelic loss of tumor-suppressor genes is a crucial mechanism, allowing units of selection to evade treatment-induced apoptosis with the acquisition of subsequent proliferative advantage leading to their outgrowth. Using alternate therapies with different mechanisms of action during the postinduction phase, especially treatments that induce cell death via tumor-suppressor independent pathways, would be predicted to overcome these mechanisms and prevent relapse.
Acknowledgments
The authors thank the patients and staff of the Myeloma Institute, UAMS.
This work was supported in part by the National Cancer Institute, National Institutes of Health (P01 CA 55819), and the Deutsche Forschungsgemeinschaft (L.R.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.J.M. and N.W. conceived and designed the study; G.J.M., C.J.H., B.B., F.v.R., M.Z., S.T., C. Schinke, F.E.D., E.T., and J.E. provided the study material and patients: O.W.S., R.T., and S.D. performed sample management and processing; P.H.P. performed gene-expression profiling; O.W.S. and R.T. performed whole-exome sequencing; C.A. and N.W. performed bioinformatic and statistical analyses; S.S.C., M.A.B., C. Stein, T.M., T.B., G.M., and E.A.P. performed additional analyses; N.W., G.J.M., S.C., C.A., C. Stein, L.R., T.M., and B.A.W. interpreted data; N.W., L.R., C.J.H., C.A., and G.J.M. wrote the paper; and all authors reviewed and approved the paper.
Conflict-of-interest disclosure: B.B. has received research funding from Celgene and Millennium, is a consultant to Celgene and Millennium, and is a coinventor on patents and patent applications related to use of GEP in cancer medicine that have been licensed to Signal Genetics Inc. G.J.M. has received research funding from Celgene and Janssen, honoraria from Celgene, Takeda-Millennium, and Bristol-Myers Squibb, and consultancy fees from Celgene, Takeda Millennium, and Bristol-Myers Squibb. F.E.D. has received honoraria from Celgene, Takeda-Millennium, Amgen, and Onyx, and consultancy fees from Celgene, Takeda-Millennium, and Amgen. The remaining authors declare no competing financial interests.
Correspondence: Niels Weinhold, Myeloma Institute, University of Arkansas for Medical Sciences, 4301 W. Markham St, #816, Little Rock, AR 72205; e-mail: nweinhold@uams.edu.
References
- 1.Röllig C, Knop S, Bornhäuser M. Multiple myeloma. Lancet. 2015;385(9983):2197–2208. doi: 10.1016/S0140-6736(14)60493-1. [DOI] [PubMed] [Google Scholar]
- 2.Barlogie B, Mitchell A, van Rhee F, Epstein J, Morgan GJ, Crowley J. Curing myeloma at last: defining criteria and providing the evidence. Blood. 2014;124(20):3043–3051. doi: 10.1182/blood-2014-07-552059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker BA, Wardell CP, Melchor L, et al. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28(2):384–390. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keats JJ, Chesi M, Egan JB, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120(5):1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kortüm KM, Langer C, Monge J, et al. Longitudinal analysis of 25 sequential sample-pairs using a custom multiple myeloma mutation sequencing panel (M(3)P). Ann Hematol. 2015;94(7):1205–1211. doi: 10.1007/s00277-015-2344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker BA, Boyle EM, Wardell CP, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33(33):3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohr JG, Stojanov P, Carter SL, et al. Multiple Myeloma Research Consortium. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaughnessy JD, Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 11.Weinhold N, Heuck CJ, Rosenthal A, et al. Clinical value of molecular subtyping multiple myeloma using gene expression profiling. Leukemia. 2016;30(2):423–430. doi: 10.1038/leu.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker BA, Wardell CP, Murison A, et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun. 2015;6:6997. doi: 10.1038/ncomms7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinhold N, Kirn D, Seckinger A, et al. Concomitant gain of 1q21 and MYC translocation define a poor prognostic subgroup of hyperdiploid multiple myeloma. Haematologica. 2016;101(3):e116–e119. doi: 10.3324/haematol.2015.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker BA, Wardell CP, Brioli A, et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014;4:e191. doi: 10.1038/bcj.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 17.Prideaux SM, Conway O'Brien E, Chevassut TJ. The genetic architecture of multiple myeloma. Adv Hematol. 2014;2014 doi: 10.1155/2014/864058. 864058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 19.Deshwar AG, Vembu S, Yung CK, Jang GH, Stein L, Morris Q. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015;16:35. doi: 10.1186/s13059-015-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang H, Qi XY, Claudio J, Zhuang L, Patterson B, Stewart AK. Analysis of PTEN deletions and mutations in multiple myeloma. Leuk Res. 2006;30(3):262–265. doi: 10.1016/j.leukres.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Leiserson MD, Vandin F, Wu H-T, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015;47(2):106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan JB, Kortuem KM, Kurdoglu A, et al. Extramedullary myeloma whole genome sequencing reveals novel mutations in Cereblon, proteasome subunit G2 and the glucocorticoid receptor in multi drug resistant disease. Br J Haematol. 2013;161(5):748–751. doi: 10.1111/bjh.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krönke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343(6168):301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljungström V, Cortese D, Young E, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2015 doi: 10.1182/blood-2015-10-674572. blood-2015-2010-674572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Keersmaecker K, Atak ZK, Li N, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45(2):186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao S, Lee SY, Gutierrez A, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120(18):3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narvaiza I, Landry S, Weitzman MD. APOBEC3 proteins and genomic stability: the high cost of a good defense. Cell Cycle. 2012;11(1):33–38. doi: 10.4161/cc.11.1.18706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31(22):3673–3675. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aukema SM, Siebert R, Schuuring E, et al. Double-hit B-cell lymphomas. Blood. 2011;117(8):2319–2331. doi: 10.1182/blood-2010-09-297879. [DOI] [PubMed] [Google Scholar]
- 30.Kortüm KM, Langer C, Monge J, et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M(3) P) in -17p high risk disease. Br J Haematol. 2015;168(4):507–510. doi: 10.1111/bjh.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Chen C, Xu Z, et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature. 2016;531(7595):471–475. doi: 10.1038/nature17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smadja NV, Bastard C, Brigaudeau C, Leroux D, Fruchart C Groupe Français de Cytogénétique Hématologique. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98(7):2229–2238. doi: 10.1182/blood.v98.7.2229. [DOI] [PubMed] [Google Scholar]
- 33.Szikriszt B, Póti Á, Pipek O, et al. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol. 2016;17(1):99. doi: 10.1186/s13059-016-0963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sottoriva A, Kang H, Ma Z, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]