Abstract
Metazoan linker histones are essential for development and play crucial roles in organization of chromatin, modification of epigenetic states and regulation of genetic activity. Vertebrates express multiple linker histone H1 isoforms, which may function redundantly. In contrast, H1 isoforms are not present in Dipterans, including D. melanogaster, except for an embryo-specific, distantly related dBigH1. Here we show that Drosophila BEN domain protein Elba2, which is expressed in early embryos and was hypothesized to have insulator-specific functions, can compensate for the loss of H1 in vivo. Although the Elba2 gene is not essential, its mutation causes a disruption of normal internucleosomal spacing of chromatin and reduced nuclear compaction in syncytial embryos. Elba2 protein is distributed ubiquitously in polytene chromosomes and strongly colocalizes with H1. In H1-depleted animals, ectopic expression of Elba2 rescues the increased lethality and ameliorates abnormalities of chromosome architecture and heterochromatin functions. We also demonstrate that ectopic expression of BigH1 similarly complements the deficiency of H1 protein. Thus, in organisms that do not express redundant H1 isoforms, the structural and biological functions performed by canonical linker histones in later development, may be shared in early embryos by weakly homologous proteins, such as BigH1, or even unrelated, non-homologous proteins, such as Elba2.
DNA in the nuclei of all eukaryotic cells is packaged into a compact nucleoprotein complex called chromatin1,2. Chromatin is organized into repeating units of nucleosomes that constitute the fundamental structural unit of chromatin. Each nucleosome consists of an octamer of two molecules of each of the four core histones H2A, H3B, H3 and H4 around which is wrapped ~145 bp of DNA. Chromatin also contains a fifth type of histone usually referred to as the linker histone H1. H1 binds to nucleosomes as well as the DNA between nucleosomes (linker DNA) and protects an additional ~20 bp of the linker DNA. In vitro studies indicate that binding of H1 to oligonucleosomal arrays stabilizes the association of DNA with the core histone octamer and facilitates the folding of the arrays into more compact structures3,4. Binding of H1 also increases the spacing between nucleosomes and restricts their mobility. Thus, in vitro studies indicate that H1 plays key roles in the structure of the chromatin fiber. This view is supported by a limited number of studies in vivo. For example, studies of Drosophila larvae that have been depleted of H1 by RNAi show marked changes in polytene chromosome structure including misalignment of sister chromatids, as well as changes in the structural integrity of heterochromatin, including the deposition of its characteristic histone marks (H3K9me2 and H4K20me2)5. Additional support for H1 as a key structural component of chromatin comes from the fact that the stoichiometry of H1 in chromatin ranges from 0.5 to nearly 1 in a wide variety organisms and cell types6. However, recent evidence indicates that H1 functions in chromatin involve more than its structural contributions. H1 interacts with a large number of chromatin-associated proteins7. Although in most instances the functional significance of such interactions have not yet been defined, in a few cases interactions of H1 with other proteins have been shown to be required for H1-mediated processes in chromatin. For example, H1 interacts directly with the H3K9-specific histone methyltransferase Su(var)3–9 and recruits it to chromatin to promote H3K9 methylation of pericentric heterochromatin and repression of transposable element transcription8,9.
Although histones are highly conserved proteins, most multicellular organisms express several variants of each type of histone, except H4. Among the histone classes, the H1 linker histones are the most divergent group. For example, mammals express 11 H1 variants10, some of which appear to have overlapping or redundant functions11. Some of the mammalian H1 variants exhibit tissue-restricted expression. For example, the murine oocyte-specific linker histone (H1oo) is present exclusively in oocytes and very early embryos12. Considering that many organisms express multiple H1 variants, D. melanogaster is quite unique, since it expresses only a single H1 protein during most of its development. Recently, an H1-like protein called dBigH1 was identified in Drosophila13. BigH1 is expressed prior to the cellular blastoderm stage of embryogenesis, a period when Drosophila H1 is not detectable and appears to confer on chromatin some of the structural features associated with H1. However, whether it is able to substitute for H1 has not been tested. The discovery of BigH1 also raises the question whether other proteins with H1-like properties remain to be discovered in Drosophila.
The BEN domain (BEND) is an ~90-amino-acid α-helical module conserved in diverse polydnavirus and cellular metazoan proteins, such as human BANP/SMAR1, NAC1 and Drosophila Mod(mdg4)14. A family of “BEN-solo” factors is characterized by the presence of BEND as a single conserved module of the proteins15. However, BEN domains frequently appear in tandem copies of two to four or are linked to other evolutionary conserved motifs (BTB/POZ, coiled-coiled regions, C4DM, C2H2 fingers, etc). Based on contextual conservation of BEND-containing proteins, it was predicted that they function as DNA-binding factors or adaptor molecules that recruit chromatin-modifying complexes14.
Drosophila BEN solo protein Insv promotes peripheral nervous system development, acts as a nuclear corepressor for Su(H), a Notch transcription factor16, and is recruited to chromatin via binding to the CSL-type transcription factor, a primary effector of Notch signaling17. Mammalian proteins BEND5 and BEND6 are highly homologous to Insv. Murine BEND5 is strongly expressed in the brain cortex, and human BEND5 can substitute for Insv in transient transfection assays15. On the other hand, BEND6 exhibits key attributes of a true functional ortholog of Insv: it binds CSL, associates with and represses Notch targets and restricts Notch signaling in neural stem cells18. Consistent with these properties, a BEND6 transgene is able to rescue the insv phenotypes in vivo19. Genome-wide (ChIP-seq) analyses of Insv distribution and reporter assays suggested that it binds with a high specificity to a novel recognition motif (TCYAATHRGAA)15. The crystal structure of an Insv BEND-DNA target complex revealed homodimeric association of the BEN domain and specific DNA binding through extensive nucleotide contacts with its α helices and C-terminal loop15. Thus, BEND was postulated to function in a DNA sequence-specific manner.
Recently, we performed a mis-expression genetic screen for Drosophila modifiers of His120. One of the strongest suppressors of lethality due to H1 depletion was a UAS allele of Elba2, which encodes a BEN solo protein. Elba2 was originally characterized in studies that described sequence-specific DNA affinity purification of Drosophila embryonic proteins that bind to the Fab-7 chromatin boundary element of the Bithorax complex21. Two BEN solo proteins, Elba1/Bsg25A and Elba2, together with another polypeptide Elba3 exhibited high-affinity, cooperative binding to the Fab-7 element in vitro and were proposed to form a tripartite complex (Elba) that functions as an insulator factor in early embryogenesis21. Among other outcomes of our genetic screen, we expected to identify factors that could, when ectopically overexpressed, replace multiple functions of H1 and thus relieve the stringent requirement for its high-level constitutive expression. Elba2 is a small (43 kDa) basic (pI ~9.8) protein. It is an abundant chromatin component in Drosophila embryos but it is not strongly expressed in larvae21. Therefore, we decided to check whether ectopic ubiquitous expression of Elba2 could rescue cellular/chromosome defect phenotypes associated with H1 depletion in L35. In this paper, we demonstrate that ectopic overexpression of Elba2 in larvae strongly complements all known phenotypes of “hypomorphic” His1 RNAi alleles, and thus, Elba2 can partially substitute for multiple functions of H1 in vivo.
Results
Elba2 expression is up-regulated in the UAS/GAL4-dependent mis-expression allele P{EP}Elba2 G17999
Abrogation of H1 expression by RNAi in vivo leads to a significant reduction of adult fly eclosion rates due to lethality at the larval-pupal transition5. Depending on the strength of the RNAi allele and temperature, adult viability is affected over a wide range, from a moderate decrease to complete lethality. We have recently identified a number of Drosophila His1 modifiers in a lethality-based genetic screen of EP misexpression alleles20. Among them, P{EP}Elba2G17999 strongly suppresses lethality caused by moderate depletion of H1 by RNAi (to ~50% of wild-type levels). The allele harbors a P{EP} insertion22 on the second chromosome in the intergenic region between Insv and Elba2 genes. Suppression of H1 depletion phenotypes by P{EP}Elba2G17999 could be caused by up-regulation of Insv and/or Elba2. To assign the suppressor function to a specific gene, we analyzed the expression of insv and Elba2 transcripts in whole L3 larvae by quantitative RT-PCR (Fig. 1a,b). The combination of the ubiquitous Act5C-GAL4 driver with P{EP}Elba2G17999 results in strong (>50-fold) activation of Elba2, relative to wild-type controls. In contrast, insv expression is not appreciably increased (<2-fold). Thus, P{EP}Elba2G17999 is a specific mis-expression allele of Elba2. Additionally, H1 depletion by RNAi does not substantially affect the expression of Elba2 in control or mis-expression backgrounds.
Figure 1. Regulation of Elba2, insv and His1 by the UAS-Elba2 mis-expression allele.
(a) Elba2 is strongly up-regulated in the Act5C-GAL4/P{EP}Elba2G17999 background. Elba2 mRNA expression level (relative to that of rp49) in whole L3 larvae of various genotypes was measured by quantitative RT-PCR. Error bars, standard deviation. (b) insv expression is not substantially affected in the Act5C-GAL4/P{EP}Elba2G17999 background. insv mRNA expression was analyzed as in (a). Error bars, standard deviation. (c) Elba2 expression is only moderately increased in Act5C-GAL4/P{EP} Elba2G17999 ovaries. Elba2 mRNA expression in was measured in adult ovaries as in (a). Error bars, standard deviation. (d) Depletion of H1 protein by RNAi is not rescued in the Act5C-GAL4/P{EP}Elba2G17999 background. H1 protein (green) expression was measured by quantitative western blot of lysates prepared from whole larvae of the indicated genotypes. Tubulin (red) served as loading control.
According to MODENCODE RNA-seq data, endogenous Elba2 is only weakly expressed in whole L3 larvae. On the other hand, it is strongly expressed in adult ovaries and early (0–4 hr) embryos, where it is presumably maternally loaded. We decided to compare the Elba2 expression level in Act5C-GAL4/ P{EP}Elba2G17999 larvae to the physiologically relevant level in wild-type ovaries (Fig. 1c). Interestingly, the maximal ovarian Act5C-GAL4-driven mRNA expression in the UAS allele was close to that in whole larvae and only 2–3 times higher than wild-type control levels. Therefore, ectopic overexpression of Elba2 under the control of the ubiquitous Act5C-GAL4 driver in whole larvae, although much higher than in wild type, is comparable to normal levels of endogenous Elba2 expression in ovaries.
Elba2 overexpression does not reverse depletion of H1 protein by RNAi
One explanation for suppression of H1 depletion-related phenotypes by P{EP}Elba2G17999 is an effect on H1 expression. Since Elba2 has been hypothesized to act as a DNA-binding transcription and/or insulator factor21, it is possible that its overexpression results in activation of His1 transcription, causing a reversal of RNAi-dependent abrogation of His1 expression. To test this hypothesis, we examined H1 protein levels in H1 RNAi-depleted larvae with and without Elba2 overexpression (Fig. 1d). RNAi decreases H1 expression >3-fold, relative to wild-type control, however P{EP}Elba2G17999 had no detectable effect on H1 expression. We conclude that Elba2 function is non-epistatic with that of His1, and the genetic interaction between them is due to similar but independent effects of their encoded proteins on downstream cell function(s) in vivo.
Reduced viability of H1-depleted flies is rescued by ectopic expression of Elba2
To examine the effect of Elba2 overexpression on adult viability, we crossed recombinant Act5C-GAL4, pINT-H14M/SM5 females to homozygous P{EP}Elba2G17999 males at 27 °C and examined the relative numbers of Cy+ (H1-depleted) and Cy (H1-normal) adults in the offspring (Table 1). In these conditions (and in all other experiments), H1 was depleted to 30–50% of wild-type levels5,8. w1118 males or Act5C-GAL4/SM5 females were substituted in control crosses. Indeed, we observed strong and statistically significant Elba2-dependent suppression of lethality caused by H1 depletion. The complementation effect can be recapitulated by Elba2 overexpression produced by an independent UAS-Elba2 transgenic insertion introduced on the second chromosome19.
Table 1. Ectopic expression of Elba2 or BigH1 partially restores viability in H1-depleted animals.
| Cross | Act5C-GAL4/SM5 | Act5C-GAL4, pINT-H14M/SM5 | p-value |
|---|---|---|---|
| w1118 (wt control) | 119/256 (128), 93% | 24/187 (94), 26% | N/A |
| P{EP}Elba2G17999 | 125/246 (123), 102% | 69/193 (97), 72% | 2.1 · 10-7 |
| UAS-Elba2 (II) | 153/295 (148), 104% | 84/232 (116), 72% | 5.4 · 10-8 |
| UAS-hBEND6 (II) | 82/174 (87), 94% | 33/195 (98), 34% | 0.26 |
| UAS-insv (II) | 142/346 (173), 82% | 2/234 (117), 2% | 3.9 · 10-7 |
| P{EP}BigH1G18579 | 93/219 (110), 102% | 53/170 (102%), 102% | 2.6 · 10-5 |
All crosses were performed at 27 °C. Males homozygous for UAS mis-expression EP alleles or UAS-driven transgenes on the II chromosome were mated to heterozygous Act5C-GAL4/SM5, Cy or Act5C-GAL4, pINT-1-H14M/SM5, Cy females. Viability was scored as the number of eclosed Cy+ adults relative to the total number of offspring scored (columns 2 and 3). The expected numbers of Cy+ flies (calculated from the Mendelian distribution) are shown in parentheses; percent viability relative to the expected numbers is also shown. Low percentage numbers that indicate increased lethality are highlighted in red. Probability values are calculated by the chi-square two-way test (column 4). Statistically significant results (p < 0.05) are highlighted in bold typeface. N/A, not applicable.
Next, we investigated whether suppression of the H1 RNAi-induced lethality by Elba2 is specific. We observed that similar UAS-driven overexpression transgenes for Insv or its mammalian functional counterpart BEND619 did not lead to suppression of lethality (Table 1). In fact, ectopic overexpression of Insv strongly enhanced the adult lethality. Thus, the functional interaction between H1 and Elba2 is specific and not shared by other BEND proteins. Accentuation of the H1 RNAi-induced lethality by the UAS-insv transgene may arise in part from a “dominant negative” effect of Insv overexpression, which may lead to interference with endogenous Elba2 function in attenuating the H1 depletion phenotypes.
Elba2 is not essential but exhibits genetic interactions with His1
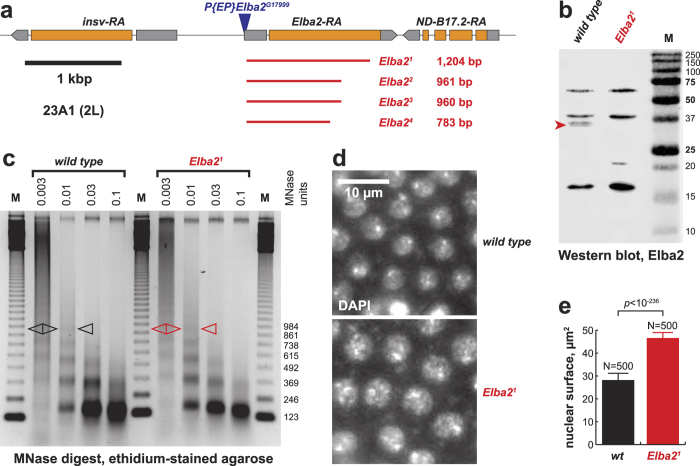
To analyze the biological functions of Elba2 in vivo, we generated mutant alleles of Elba2 by imprecise excision of P{EP}Elba2G17999. We recovered four deficiency alleles that contain deletions of 783 to 1,204 bp downstream of the P-element insertion site (Fig. 2a). In the largest deficiency, Df(2L)Elba21, 1,204 bp (−108 to +1,086 relative to the start codon of Elba2) are excised and replaced with a 26-bp fragment of the P-element. Thus, almost the entire coding sequence, with the exception of the last C-terminal 19 codons, is eliminated. Therefore, Elba21 is a null allele.
Figure 2. The function(s) of Elba2 in global compaction of embryonic chromatin.
(a) Schematic of the Drosophila Elba2 locus. A 6-kbp genomic interval in D. melanogaster 23A1 cytological region (left arm of chromosome 2) encompassing insv, Elba2 and ND-B17.2 genes is shown. Blue triangle, P-element insertion; red lines, genomic deficiencies in corresponding mutant Elba2 alleles; thick black line, scale bar, 1 kbp; thin black lines, introns and intergenic regions; gray boxes, non-coding parts of exons; orange boxes, coding regions of exons; exon-intron boundaries are depicted as identified for major gene transcripts (named in the top line). (b) Elba21 is a null mutant allele. Lysates of nuclei from 0–4 h embryos (homozygous Elba21 and isogenic wild-type control) were analyzed by immunoblotting. A polypeptide with an apparent molecular mass of ~36 kDa (red arrowhead) is recognized by mouse anti-Elba2 antibody in wild-type but is absent in Elba21 embryos. M, molecular mass marker; marker sizes (kDa) are shown on the right. (c) Nucleosome repeat length (NRL) is slightly reduced in chromatin of homozygous Elba2 null mutant embryos. Samples of nuclei from wild-type control and Elba21 embryos were subjected to partial digestion with the indicated number of units of micrococcal nuclease (MNase), and the DNA was analyzed by agarose gel electrophoresis and EtBr staining. Pentanucleosome bands are calculated to be 947 bp long in wild type (black open triangles) and 910 bp long in Elba21 (red open triangles). M, 123-bp DNA ladder; marker sizes are shown on the right. (d) Elba2 mutation leads to a decrease of nuclear sizes in syncytial embryos. Wild-type control and homozygous Elba21 null embryos were stained with DAPI. DAPI-stained nuclei of an embryo in the interphase of nuclear division cycle 12 are shown. Scale bar (white), 10 μm. (e) The sizes of interphase nuclei in Elba2 null syncytial embryos are dramatically decreased. Surface areas of interphase nuclei (division cycles 11–13) were measured in homozygous Elba21 (red bar) and isogenic wild-type control (black bar) embryos based on DAPI staining (d). Mean values (μm2) are plotted. Error bars, standard deviations; N = 500 (10 nuclei each in 50 randomly selected embryos); probability value was calculated by two-tailed paired Student T-test.
First, we checked whether Elba2 mutations alone affect fly viability. To this end, we crossed heterozygous Elba2/CyO flies inter se and examined the distribution of Cy+ and Cy genotypes in the F1 progeny (Table S1). It appears that Elba2 is not essential. Significantly, there was no appreciable effect of Elba2 mutations on fly development or adult eclosion, since the relative proportion of Elba2/Elba2 homozygotes in the offspring of the crosses was very close to the expected Mendelian ratio. Also, the homozygous mutant flies (both males and females) produce normal progeny in crosses with wild-type counterparts or inter se and are thus, fertile. Since homozygous Elba21 animals are viable and fertile, we sought to confirm that they do not express Elba2 protein. To this end, we raised polyclonal antibodies to full-length recombinant Elba2 and used them for immunochemical analyzes of 0–4 h embryos, where endogenous Elba2 RNA is highly expressed21. We observed a strong expression of a 36 kDa protein specifically recognized by Elba2 antibody in nuclei prepared from wild-type control embryos. However, its expression was not detected in Elba21 embryos (Fig. 2b). Thus, as expected, Elba21 embryos do not express Elba2.
The genetic interaction between Elba2 and His1 described above (Table 1) relies on artificial ectopic overexpression of Elba2 in larvae, where the endogenous gene has limited transcriptional activity (Fig. 1a). Therefore, we decided to test whether endogenous Elba2 and His1 also exhibit genetic interactions. To this end, we examined how Elba2 mutation affects viability of H1-depleted flies. We crossed Tub-GAL4/TM3,Sb and pINT-H11M/TM3, Sb flies in homozygous Elba21 and wild-type (isogenic, precise excision) backgrounds and scored the distributions of H1-depleted (Sb+) and control (Sb) eclosed adults in the progeny. We discovered that the Elba2 null mutation significantly reduced viability of H1-depleted flies (Table 2). Thus, as expected, Elba2 null and hypomorphic His1 mutations exhibit a strong synthetic lethal interaction. Abrogation of H1 expression results in more severe lethality when performed in the Elba2 null background. Therefore, endogenous Elba2 partially compensates for H1 depletion in vivo.
Table 2. Endogenous Elba2 and His1 exhibit synthetic lethal genetic interactions.
| Cross | Viability | p-value |
|---|---|---|
| +; Tub-GAL4/TM3,Sb × +; pINT-H11M/TM3, Sb | 17/135 (68), 25% | N/A |
| Elba21; Tub-GAL4/TM3,Sb × Elba21; pINT-H11M/TM3, Sb | 8/235 (118), 7% | 7.0 · 10-4 |
All crosses were performed at 26 °C. In a control cross, heterozygous males that carry the Tub-GAL4 driver on the III chromosome balanced with TM3, Sb were mated to females that carry pINT-1-H11M His1 RNAi allele balanced with TM3, Sb. In the experimental cross, the parents additionally carried a homozygous Elba21 allele on the II chromosome. Viability of the progeny was scored as the number of eclosed Sb+ adults relative to the total number of offspring (column 2). The expected numbers of Sb+ flies (calculated from the Mendelian distribution) are shown in parentheses; percent viability relative to the expected numbers is also shown. Probability value is calculated by the chi-square two-way test (column 3). Elba2 mutation enhances the H1 depletion-dependent semilethal effect in a statistically significant fashion. N/A, not applicable.
Elba2 contributes to genome-wide chromatin compaction
Although Elba2 is not essential for viability, we investigated its effects on chromatin structure in early embryos, where it is strongly expressed21 (and where the loading of canonical H1 into chromatin is low13,23). We collected nuclei from 0–4 h Elba21 and Elba2+ embryos and examined their nucleosome structure by partial micrococcal nuclease (MNase) digestion assay (Fig. 2c). We observed a small but reproducible decrease of nucleosome repeat length (NRL) in embryos that do not express Elba2 (from ~189 bp in wild type to ~182 bp), comparable to that observed in animals with reduced expression of prototypical linker histones H1 and BigH15,13. Therefore, similar to linker histone H1, Elba2 is required to maintain a normal NRL of bulk native chromatin and, similar to BigH1, controls global chromatin structure in early embryos.
We also noticed that nuclear pellets from Elba21 and Elba2+ embryos containing equivalent amounts of nucleic acid differed in their volumes by nearly two-fold. Nuclei prepared from Elba2 mutants appeared consistently less dense than wild-type nuclei. To determine whether mutant Elba2 embryos have larger nuclei in vivo, we stained Elba21 and Elba2+ embryos (0–4 h AED) with DAPI (Fig. 2d) and measured the diameters of interphase syncytial nuclei (division cycles 11–13) based on DAPI staining (Fig. 2e). Whereas, the sizes of nuclei were relatively uniform within each genotype, nuclei in Elba21 embryos were on average ~29% larger than nuclei in wild-type embryos (linear size, equivalent to an ~65% increase of the measured surface). We conclude that Elba2 plays an important role in global compaction of chromatin in nuclei of Drosophila embryos.
Elba2 distribution in chromatin is nearly ubiquitous and overlaps with that of H1
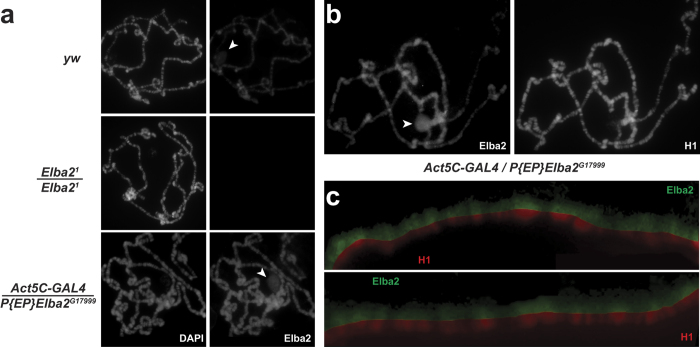
Elba2 and other BEND proteins were proposed to function as sequence-specific, DNA-binding factors based on structural considerations15. Also, Elba2 was originally purified as a component of a putative sequence-specific insulator factor Elba21. These findings suggest that Elba2 may exhibit a restricted distribution pattern in the genome, limited to loci that contain its recognition sequence(s). To examine Elba2 distribution in chromatin in vivo, we used Elba2 antibodies to analyze the distribution of the protein in larval polytene chromosomes by indirect immunofluorescence (IF) microscopy. Endogenous Elba2 can be readily detected by IF staining in wild-type salivary gland cells (Fig. 3a, top). Unexpectedly and contrary to the predicted restricted distribution of Elba2, the observed pattern of Elba2 occupancy is nearly ubiquitous, and the protein localizes mostly to the bands of polytene chromosomes. In contrast, a prototypical insulator protein Su(Hw)24 exhibits a substantially more discrete distribution pattern, consistent with its known locus-specific functions (Fig. S1). Thus, Elba2 does not associate with chromatin in the locus-specific manner expected of a transcription and/or insulator factor. Alternatively, its binding site recognition specificity may be very relaxed.
Figure 3. Elba2 distribution in polytene chromosomes.
(a) Endogenous and ectopically expressed Elba2 is nearly ubiquitously distributed in polytene chromosome arms. Polytene chromosomes of salivary glands from control (yw), Elba2 null and Elba2-ectopically expressing larvae were stained with DAPI and affinity-purified Guinea pig anti-Elba2 antibodies. No Elba2 signal above background is detected in the Elba2 null. Arrowheads, nucleoli. (b) Elba2 distribution in polytene chromosomes is restricted to polytene bands and resembles that of H1. Elba2 and H1 distribution in polytene chromosomes from Elba2-ectopically expressing larval salivary glands were examined by IF as in (a). Arrowhead, nucleolus. (c) Elba2 and H1 exhibit strong co-localization in polytene chromosomes. Split-images of fragments of polytene chromosomes analyzed by indirect immunofluorescence with Elba2 (green) and H1 (red) antibodies.
The observed IF staining of Elba2 is specific, since no signal can be detected in homozygous Elba21 null mutant (Fig. 3a, middle). The intensity of the staining is also moderately increased in polytene chromosomes from L3 larvae that ectopically overexpress Elba2 (in Act5C-GAL4/P{EP}Elba2G17999 animals, Fig. 3a, bottom). Importantly, Elba2 mutation or overexpression does not affect the overall morphology of polytene chromosomes (compare DAPI images in left panels in different genetic backgrounds).
Since endogenous and ectopically expressed Elba2 localizes predominantly to polytene chromosome bands, which contain condensed, repressed chromatin, it is possible that Elba2 loading into chromosomes overlaps with that of H1. Thus, we performed co-staining of polytene chromosomes with Elba2 and H1 antibodies (Fig. 3b). We observed a substantial correlation of Elba2 and H1 IF signals. In high-resolution split images (Fig. 3c), it is clear that Elba2 and H1 patterns are very similar: the majority of H1-negative loci show greatly reduced levels of Elba2 staining and vice versa. In contrast, high-abundance loci for both proteins strongly correlate. The co-localization of Elba2 and H1 in polytene chromosomes suggests a model of direct replacement of H1 by Elba2 upon simultaneous H1 depletion and Elba2 overexpression. When Elba2 is ectopically expressed in H1-depleted animals, it is deposited in loci formerly occupied by H1 and provides repressive and/or chromatin effector recruitment function(s) to partially compensate for the loss of H1.
Ectopic expression of Elba2 ameliorates defects of chromosome structure that are associated with H1 depletion
Depletion of H1 in salivary gland cells results in profound defects of polytene chromosome architecture: the regular band-interband pattern of aligned chromatin fibers is compromised, and the normal singular heterochromatic chromocenter is dissociated into multiple diffuse foci containing HP15. Furthermore, abrogation of H1 expression almost completely eliminates IF staining of polytene chromosomes with H3K9me2-specific antibodies. The latter effect is attributed to the reduced/abolished tethering of the H3K9 methyltransferase Su(var)3–98. Together, these molecular defects likely contribute to the larval/pupal lethality of H1-depleted animals9.
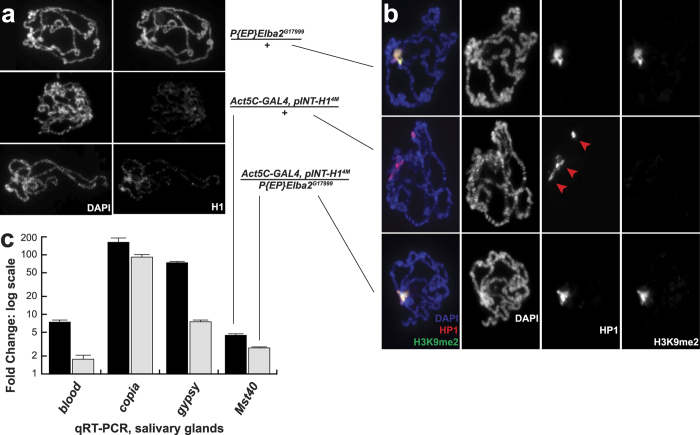
We investigated whether these abnormalities are rescued by ectopic expression of Elba2. First, we compared the gross morphology of polytene chromosomes in wild-type control and H1-depleted salivary gland cells, as well as in cells where H1 depletion was accompanied by Elba2 overexpression. To this end, we performed DAPI staining of polytene spreads from various progeny of crosses between Act5C-GAL4, pINT-H14M/T(2;3)B3, CyO: TM6B, Tb and P{EP}Elba2G17999/P{EP}Elba2G17999 or w1118 parents (Fig. 4a). In addition, to verify H1 depletion, we stained these preparations with an H1-specific antibody. Whereas control polytene spreads (exemplified by P{EP}Elba2G17999/T(2;3)B3, CyO: TM6B or w1118 animals) exhibited regular banded structure (Fig. 4a, top), the banding pattern was severely disrupted in Act5C-GAL4, pINT-H14M/+ salivary glands (Fig. 4a, middle). However, the band-interband structure was restored to normal when the H1 depletion was combined with Elba2 overexpression in Act5C-GAL4, pINT-H14M/ P{EP}Elba2G17999 animals (Fig. 4a, bottom). Similarly, the singular chromocenter that is easily identified in DAPI-stained control polytene spreads is not apparent in H1-depleted polytene chromosomes but restored upon Elba2 overexpression. Consistent with immunoblot analyses (Fig. 1d), H1 expression or deposition into chromatin is not substantially elevated in P{EP}Elba2G17999-rescued chromosomes in comparison to H1-depeletd chromosomes (Fig. 4a, right panels).
Figure 4. Rescue of H1 depletion phenotypes in vivo by ectopic expression of Elba2.
(a) Elba2 mis-expression restores regular polytene band-interband structure in salivary glands depleted of H1 by RNAi. Polytene chromosomes of salivary glands from control, H1-depleted larvae and larvae that are simultaneously depleted of H1 and ectopically expressing Elba2 were stained with DAPI and H1 antibodies. (b) Polytene chromosome defects due to H1 depletion in vivo are ameliorated by ectopic mis-expression of Elba2. Polytene chromosomes prepared from animals of the indicated genotypes as in (a) were stained with DAPI (blue) and antibodies against HP1 (red) and H3K9me2 (green). Red arrowheads, multiple HP1-rich foci that do not coalesce into a singular chromocenter in H1-depleted salivary glands. (c) Derepression of transposable elements upon H1 depletion is partially reversed by ectopic expression of Elba2. Transcripts of the indicated TE’s in salivary glands were measured by quantitative RT-PCR in control (P{EP}Elba2G17999/+), Act5C-GAL4, pINT-H14M/+ and Act5C-GAL4, pINT-H14M/P{EP}Elba2G17999 backgrounds. Transcript levels were normalized to that of rp49, and the fold change brought about by H1 depletion (black bars) and H1 depletion with simultaneous Elba2 overexpression (gray bars) was calculated relative to the control allele. All RT-PCR reactions were performed on three independent biological samples in duplicate. Significance testing was performed by applying two-tailed paired Student T-test to normalized ∆Ct values (with and without ectopically expressed Elba2); p-values are <0.02 for all transcripts. Error bars, standard deviation.
We also examined these polytene spreads for deposition of the heterochromatin-specific histone mark H3K9me2 and distribution of HP1 (Fig. 4b). As described previously5, H1 depletion disrupts the singular chromocenter and produces multiple HP1-rich foci in polytene spreads; it also severely reduces polytene chromosome staining with anti-H3K9me2 antibodies (compare top and middle panels of Fig. 3b). In contrast, simultaneous depletion of H1 and ectopic expression of Elba2 reverses the two phenotypes to that of the control (Fig. 4b, bottom). Thus, ectopically overexpressed Elba2 partially compensates for the loss of H1 and ameliorates all of the described microscopic defects of polytene chromosome structure that are caused by H1 depletion in larvae.
H1 RNAi-dependent derepression of transposable elements is reversed upon ectopic expression of Elba2
Normal levels of H1 in chromatin are essential for the proper regulation of genetic activity in flies. Reducing the abundance of H1 leads to positive and negative effects on expression of multiple Drosophila genes in salivary glands and Kc cells8. In particular, H1 depletion causes an extremely strong (up to 600-fold) derepression of transposable elements (TE’s) and other repetitive sequences. This up-regulation is partially reversed by overexpression of Su(var)3–98. Since H3K9 methylation is restored in H1-depleted animals also expressing ectopic Elba2 (Fig. 4b), we determined whether Elba2 overexpression in salivary glands also reverses derepression of TE’s in H1 RNAi animals. We observed a significant (3- to 20-fold) negative effect of Elba2 ectopic expression on TE transcript levels in H1-depleted salivary glands (Fig. 4c). Thus, ectopic Elba2, like H1 itself, promotes repression of repetitive sequences and counteracts the derepression of these elements occurring upon H1 depletion.
Ectopic BigH1 or Elba2 have similar restorative activity in reversing lethality and defects in chromosome structure in H1-depleted larvae
We showed previously that ectopic expression of His1 cDNA transgenes and duplications of the histone gene cluster can partially rescue lethality due to the RNAi-mediated abrogation of H1 expression5,9. The preceding series of experiments demonstrate that ectopic expression of Elba2 also compensates for RNAi-mediated loss of endogenous H1, resulting in rescue of lethality and chromosomal defects (Table 1 and Fig. 4). Therefore, ectopic Elba2 can assume several of the biological functions of H1. Recently, an H1-like protein dBigH1 was identified in Drosophila13. Its expression, similar to that of Elba2, is largely limited to ovaries and early embryos. We sought to determine whether ectopic ubiquitous expression of BigH1 in larvae counteracts the effects of H1 depletion. To this end, we used the existing P{EP}BigH1G18579 UAS misexpression allele in which BigH1 transcription can be stimulated by GAL4. We observed that ectopic expression of BigH1 under the control of Act5C-GAL4 driver rescues viability of H1-depleted adults (larvae) (Table 1). Therefore, the P{EP}Elba2G17999 and P{EP}BigH1G18579 alleles phenocopy each other, and mis-expression of either Elba2 or BigH1 complements a deficiency of H1 protein.
We then examined the ability of ectopically expressed BigH1 to rescue chromosome defects in H1-depleted salivary glands. We stained Act5C-GAL4, pINT-H14M/+; P{EP}BigH1G18579/+ polytene chromosomes with DAPI, HP1 and H3K9me2 antibodies and observed a normal chromosome architecture, including regular band-interband structure, single chromocenter and strong dimethylation of H3K9, identical to that seen in wild type polytene chromosomes (Fig. S2). Therefore, Elba2 and BigH1 have very similar biological activities such that ectopic expression of either protein can substitute for H1 when it is depleted by RNAi in vivo.
Discussion
Although BEN domain proteins were proposed to function as sequence-specific factors15, the relationship between their DNA binding activity and distribution in the genome in vivo remains enigmatic. For instance, reporter assays indicate that Drosophila Bsg25A and Elba2 polypeptides can individually recognize the palindromic Insv DNA binding motif with high affinity. Furthermore, the crystal structure of the Bsg25A BEND-DNA complex suggests that Bsg25A shares key aspects of DNA binding in vitro with Insv19. On the other hand, the Fab-7 Elba motif, the presumed in vivo target of Bsg25A and Elba2, is quite distinct from the Insv site19,21. Also, Insv exhibits extensive co-binding with class I insulator elements that generally do not conform to its binding consensus19. Of interest, mammalian BEND5-VP16 fusion protein can specifically activate 4x-Insv reporters in transient transfection assays, and GST-BEND5 can bind EMSA probes that bear an intact Insv motif15. On the other hand, similar fusions of mammalian BEND6 fail to perform in these assays19, yet a BEND6 transgene strongly complements homozygous insv mutation in flies in vivo19. Therefore, BEND proteins may utilize alternative mechanisms for tethering to their functional sites in the genome, such as recruitment by adapter proteins or deposition by specialized chaperones.
In this work, we show that Drosophila BEND protein Elba2 exhibits a distribution pattern in salivary gland polytene chromosomes that is inconsistent with its putative sequence-specific binding. Unambiguously, instead of restricted binding to a number of discrete bands that would be expected of a sequence-specific factor, Elba2 is broadly distributed to virtually all polytene bands, which mostly represent compact, silent chromatin also occupied by the linker histone H1 (Fig. 3). It is possible that sequence recognition by Elba2 is very relaxed, and thus, it can tolerate substantial degeneracy among its binding sites. Alternatively, deposition of Elba2 in chromosomes may rely on completely sequence-independent mechanisms.
The postulated global, genome-wide function of Elba2 in vivo is further supported by the effect of the endogenous protein on the chromatin NRL and nuclear compaction in early embryos (Fig. 2). Although the overall oligonucleosome structure and periodicity is not affected in the bulk native chromatin of Elba2 mutant embryos, the nucleosomes are more closely spaced, like those in H1-depleted larvae5 and BigH1 mutant embryos13. The decrease of NRL in BigH1 mutant embryos was reported to be more substantial than that in Elba2 embryos (17 bp versus 7 bp) and even that in larvae depleted of about 95% of H15. However, differences in the methods used for quantifying NRL changes in the study of BigH1 and the other studies (including the current work) may account for the discrepancy. Interestingly, we also observe that Elba2 null mutation leads to a significant increase of the volume of syncytial nuclei, which further supports a global role for Elba2 in chromatin condensation. Although at the present time, we cannot exclude an indirect effect of Elba2 on the compaction of nuclear DNA, other evidence indicates that Elba2 functions via mechanisms similar to those of linker histone H1.
The expression of endogenous Elba2 protein is limited to the early embryo21; it is only weakly expressed in larvae (e.g., Fig. 1). On the other hand, H1 is expressed ubiquitously throughout development, and abrogation of its expression in L3, results in a dramatically reduced rate of adult eclosion. Upon moderate to strong depletion of H1, lethality occurs during the larval to pupal transition. We demonstrate here that the Elba2 polypeptide, when ectopically expressed in larvae under the control of a ubiquitous driver, can compensate for the loss of H1 in vivo (Table 1). Importantly, ubiquitous mis-expression of Elba2 rescues all known phenotypes observed in H1-depleted animals5,8, including polytene chromosome structure abnormalities as well as changes in the composition and genetic activity of heterochromatin (Fig. 4). Thus, Elba2 is expected to share multiple biochemical activities with H1: it may compact an oligonucleosome fiber in vitro and may also physically interact with natural H1 partner proteins, such as Su(var)3–9 and STAT92E8,25. Unfortunately, we were unable to express and purify recombinant Elba2 polypeptide in a soluble form. Both E. coli and S. frugiperda (baculovirus) expression systems produced highly insoluble Elba2 under several tested conditions (not shown), which hampered further biochemical experimentation. It is possible that native Elba2 in solution exists as a part of a heteromeric complex and thus, critically depends on the presence of additional subunits for solubility. In fact, Elba2 has been proposed to form a heterotrimeric complex Elba21. It has been demonstrated that Elba subunits synergistically bind to the Fab-7 insulator element in vitro. However, (i) the existence of a stable Elba complex was not further confirmed, (ii) its complete composition is unknown, and (iii) Elba (or an Elba-like complex) has not been demonstrated to be the major complex of Elba2. In the future, it will be interesting to characterize the major native form of Elba2, using an unbiased approach that does not rely on the use of a sequence-specific DNA affinity resin. A complex of Elba2 (Elba or an alternative complex) may modify the structure of the chromatin fiber in vitro and/or physically interact with H1 partner proteins. In addition, it remains to be tested whether the nearly ubiquitous deposition of Elba2 in chromatin (Fig. 3) takes place in the context of a putative complex or as an individual polypeptide.
Most multi-cellular eukaryotes, including animals and plants, express several variants of linker histone H1, as many as 11 in mammals10. Although their variant-specific biological/biochemical functions are largely unknown, H1 variant genes exhibit heterogeneity in tissue-, cell cycle- and developmental stage-specific expression patterns. For example, murine H1oo protein is expressed only in the oocyte and very early embryo10,12. In contrast, Drosophilids typically express a single H1 isoform throughout their life cycle. The only known exception is D. virilis, which expresses three H1 variants. However, the differences amongst their sequences are more characteristic of polymorphic variants and therefore, it is likely that they are not true functional protein isoforms. Intriguingly, analysis of chromatin from very early Drosophila embryos (0–2 hours after egg deposition) indicates that H1 is not loaded into chromosomes at this stage13,23, when Elba2 is highly expressed. Thus, Elba2 may function as a “replacement linker histone” that carries out some of the normal biological activities of H1 in early embryonic chromatin, when canonical H1 is not present. Likewise, when ectopically expressed in larval cells with limiting H1 (depleted by RNAi), Elba2 is able to assume H1’s functions.
It has been proposed that a weakly homologous dBigH1 protein may function as an embryonic replacement H1 in Drosophila13. Importantly, the reversal of phenotypes associated with H1 depletion by ectopic overexpression of Elba2 is indistinguishable from that produced by ectopic overexpression of BigH1 (Table 1, Figs 4 and S2). Thus, at least in the context of reduced H1, ectopic Elba2 and BigH1 play nearly identical biological roles. The functional similarity in vivo between Elba2 and H1-homologous BigH1 further supports our idea that Drosophila Elba2 has features of an H1 replacement protein.
Surprisingly, both BigH1 and Elba2 are expressed in Drosophilids but lack true orthologs in other organisms. We surveyed several sequenced eukaryotic genomes and found a negative correlation between the conservation of BigH1 and Elba2 orthologs in a particular species and the existence of multiple linker histone H1 isoforms. Furthermore, BigH1 and Elba2 orthologs, when present in eukaryotic genomes, are always found together. It is possible that in ancient metazoan ancestor organisms, some of the functions of linker histone H1 may have been shared by proteins that are not structurally similar (Elba2) or only distantly related (BigH1) to H1. In early embryos, these proteins may perform biological functions (chromatin compaction and tethering of effector enzymes) that are characteristic of the canonical H1 during later stages of development. The emergence of H1 isoforms that are expressed early in development, such as H1oo in mammals, may have obviated the selective pressure to maintain H1 replacement proteins similar to Drosophila BigH1 and Elba2, which resulted in their evolutionary loss.
Methods
Fly strains and genetics
Flies were maintained on standard corn meal, sugar and yeast medium with Tegosept at 18–27 °C as indicated. His1 RNAi and P{EP}Elba2G17999 alleles are described elsewhere5,20. UAS-Elba2, UAS-insv and UAS-hBEND6 transgenes are a generous gift of Eric Lai (Memorial Sloan Kettering Cancer Center). P{EP}BigH1G18579, various balancer and GAL4 driver alleles were obtained from the Bloomington Stock Center. Genetic rescue experiments and adult fly viability analyses were performed at 27 °C as described previously20. See legends to Tables 1, 2 and S1 for details. To genotype L3 larvae that contain the Act5C-GAL4, pINT-H14M transgene combination, the allele was balanced with T(2;3)B3, CyO: TM6B, Tb translocation and used in crosses. Tb+ F1 progeny contain the recombinant second chromosome with the GAL4 driver and RNAi transgene.
Elba2 null allele(s) were generated by imprecise excision of P{EP}Elba2G17999 insertion as described26. 107 w– excision alleles were analyzed by genomic DNA PCR with the following primers: AGGTGGCATGAATCTGGGATAGCA and CAAGTACAAGTGGATAGCGGACCA. The size of wild-type PCR product is 3,198 bp. Four alleles with large deletions (>500 bp, see Fig. 2a) and several precise excision alleles (isogenic controls) were balanced with CyO; their genomic DNA flanking Elba2 was amplified and sequenced. For viability tests, 10 heterozygous males and females each were crossed inter se, allowed to mate for 4 days and discarded; the distribution of balancer markers was scored in the adult progeny. For fertility tests, 10 homozygous males or females were crossed with 10 yw counterparts of the opposite sex, allowed to mate for 4 days and discarded. The appearance of advanced development stage/adult progeny of the crosses indicated fertility of Elba2 males and females, accordingly.
To examine genetic interactions of Elba2 and His1, synthetic alleles that harbor homozygous Elba21 second chromosome and heterozygous balanced GAL4 driver or UAS-H1-RNAi transgenes on the third chromosome (Elba21; Tub-GAL4/TM3,Sb and Elba21; pINT-H11M/TM3, Sb) were generated in a series of crosses. The two alleles were mated at 26 °C and the distribution of Sb+ (H1-depleted) and Sb (H1-normal) progeny was scored to assess the effect of H1 depletion in the Elba21 background. For comparison, similar synthetic alleles with a precise excision allele (“+”) were generated and used in crosses.
Real-time RT-PCR
Expression of transcripts for Elba2, insv and transposable elements was measured by quantitative RT-PCR on an ABI Prizm 770 sequence detection system (Applied Biosystems) as described previously5. Total RNA was prepared from 2–5 whole L3 larvae, 20 pairs of salivary glands or 20 adult ovaries dissected from animals of specific genotypes. The expression levels were quantitated relative to an endogenous reference gene, rp49. The following primers were used for Elba2: GATCAGGACTCGTGTCCTAACC and CGCTGGGCAGGATAGCAGTC; and insv: CGGACCCGCAAGTGGAGGTC and CATGAATCTGGGATAGCAGATCC, the primers for rp49 and transposable elements blood, copia, gypsy and Mst40 are described elsewhere8. All experiments were performed on three independent biological samples in duplicate, along with no-template controls.
Partial micrococcal nuclease (MNase) digestion assay
Homozygous Elba21 and Elba2+ embryos were collected 0–4 h after egg deposition (AED) and dechorionated. Nuclei were isolated and treated with MNase as described5. Briefly, ~200 mg of freshly prepared embryos were resuspended in 500 μl ice-cold Buffer A (15 mM Tris-HCl, pH 7.5, 15 mM NaCl, 60 mM KCl, 0.34 M sucrose, 0.5 mM spermidine, 0.15 mM spermine, 0.1% β-mercaptoethanol, 0.25 mM PMSF) additionally containing 2 mM EDTA and 0.5 mM EGTA and homogenized on ice using Roto-Dounce (Fisher Scientific). The nuclei were pelleted in a cold microcentrifuge (5 min at 13,000 rpm), washed twice with Buffer A (no EDTA/EGTA) and resuspended in 200 μl Buffer A. Nucleic acid concentrations were estimated by A260 measurement, and the volumes of two samples were adjusted with Buffer A to achieve equal concentrations. Aliquots of nuclei equivalent to 0.3 optical units (A260) were digested in four separate reactions with 0.003–0.1 units of MNase (Sigma) in a buffer containing 2 mM CaCl2 in a total volume of 50 μl for 10 min at room temperature. The digestions were stopped by adding EDTA to 25 mM, and nuclear RNA was degraded with 25 μg RNase A (Sigma) for 20 min at 37 °C. The samples were deproteinized by treating with Proteinase K and phenol-chloroform extraction as described27. DNA was precipitated with ethanol, and one-third of each sample was loaded on a 1.3% agarose gel in 1x TBE. The gel was stained with EtBr after electrophoresis.
DAPI staining of Drosophila syncytial embryos
Homozygous Elba21 and Elba2+ embryos were collected 0–4 h AED, dechorionated and fixed with methanol as described28. The embryos were mounted in Vectashield (Vector Laboratories) with 1 μg/ml DAPI and observed under a Zeiss Axiovert 200M. Fixed embryos were staged by the number of surface nuclei per unit area29. For each genotype, 50 embryos in the interphase of division cycles 11–13 were randomly selected, and a random area from the middle of each embryo was used to score the sizes of 10 blastoderm nuclei using AxioVision digital image processing software. Mean, standard deviation and two-tailed paired Student T-test values were calculated using Microsoft Excel.
Antibodies, immunoblot analyses and immunohistochemistry
Full-length recombinant His-tagged Elba2 was expressed in E. coli and purified in denaturing conditions (8 M urea) on Ni-NTA agarose (Qiagen). Details of expression construct cloning and purification procedures are available upon request. After urea was removed by dialysis in PBS, the protein was precipitated out of solution, and the suspension of insoluble protein was used to raise polyclonal mouse (AECOM Hybridoma Facility) and Guinea pig antibodies (Pocono Rabbit Farm and Lab). Guinea pig immunoglobulins were purified from plasma using Bakerbond ABx (J. T. Baker). They were further affinity-purified on the resin obtained by cross-linking denatured Elba2 polypeptide to NHS-activated Sepharose (Pierce). Polyclonal rabbit anti-Su(Hw) antibody24 was a generous gift of V. Corces; other antibodies were previously described in ref. 5.
Western blotting was performed as described5 using whole larval L3 lysates or lysates of embryonic (0–4 h AED) nuclei in SDS-PAGE loading buffer. Polyclonal rabbit anti-H1 and mouse anti-Elba2 antisera as well as monoclonal mouse anti-tubulin E7 (loading control) and infrared dye secondary antibodies5 were used at 1:50,000, 1:1,000, 1:500 and 1:10,000 dilutions, respectively. Images were obtained and quantitated using a LI-COR Odyssey system.
Indirect immunofluorescence (IF) staining of polytene chromosomes was performed exactly as described5. Briefly, salivary glands of the wandering third instar larvae were dissected in PBS +0.1% Triton X-100. Polytene chromosomes were fixed in 3.7% paraformaldehyde for 30 sec, squashed in 45% acetic acid +3.7% formaldehyde, and frozen in liquid nitrogen. They were incubated overnight in PBS +10% fat-free milk +0.1%Triton X-100 with primary antibodies, washed twice in PBS +400 mM NaCl +0.2% NP-40 for 30 min and stained with secondary antibodies in PBS +0.1% Triton X-100. The final preparations were mounted in Vectashield mounting solution (Vector) and stained with DAPI (0.5 mg/mL). Affinity purified rabbit anti-H1, Guinea pig anti-Elba2 and rabbit anti-H3K9me2 (Abcam) were used at 1:5,000, 1:200 and 1:100 dilutions, respectively. Mouse anti-HP1 C1A9 antibody, rabbit anti-Su(Hw) and appropriate secondary goat Alexa Fluor (Molecular Probes) antibodies were used at 1:50, 1:200 and 1:200 dilutions, respectively. The preparations were examined using epifluorescence on a Zeiss Axiovert 200 microscope, and images were captured using a high-resolution CCD camera.
Additional Information
How to cite this article: Xu, N. et al. BEN domain protein Elba2 can functionally substitute for linker histone H1 in Drosophila in vivo. Sci. Rep. 6, 34354; doi: 10.1038/srep34354 (2016).
Supplementary Material
Acknowledgments
We are grateful to E. Lai for fly stocks and V. Corces for antibodies. We thank S. Buhl and M. Scharff for raising mouse polyclonal antibodies. We also thank K. Beirit and A. Konev for helpful discussions and critical reading of the manuscript. This work was supported by grants from the National Institutes of Health to D.V.F. (GM074233) and A.I.S. (GM093190 and GM116143). N.X. was supported in part by the NIH IRACDA/K12 training grant (K12GM102779). T.J.B. was supported in part by a NIH F32 Fellowship (GM115210).
Footnotes
Author Contributions N.X., X.L., H.K., A.V.E., T.J.B. and E.V. designed and conducted the experiments and analyzed the results; A.I.S. and D.V.F. conceived the experiments, analyzed the results and wrote the manuscript. All authors reviewed the manuscript.
References
- van Holde K. E. Chromatin (ed. Rich A.) (Springer-Verlag, 1989). [Google Scholar]
- Wolffe A. Chromatin: Structure and Function (eds. Picknett T. & Davies S.) (Academic Press, 1998). [Google Scholar]
- Ramakrishnan V. Histone H1 and chromatin higher-order structure. Crit. Rev. Eukaryot. Gene Expr. 7, 215–230 (1997). [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Histone H1. Int. J. Biochem. Cell Biol. 29, 1463–1466 (1997). [DOI] [PubMed] [Google Scholar]
- Lu X. et al. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 23, 452–465 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. L., Skoultchi A. I. & Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chrom. Res. 14, 17–25 (2006). [DOI] [PubMed] [Google Scholar]
- Kalashnikova A. A., Rogge R. A. & Hansen J. C. Linker histone H1 and protein-protein interactions. Biochim. Biophys. Acta 1859, 455–461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science 340, 78–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi H., Emelyanov A. V., Fyodorov D. V. & Skoultchi A. I. Independent biological and biochemical functions for individual structural domains of Drosophila linker histone H1. J. Biol. Chem. 291, 15143–15155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo A., Kamieniarz K. & Schneider R. The histone H1 family: specific members, specific functions? Biol. Chem. 389, 333–343 (2008). [DOI] [PubMed] [Google Scholar]
- Fan Y. et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23, 4559–4572 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G. et al. Mouse oocytes and early embryos express multiple histone H1 subtypes. Biol. Reprod. 68, 1569–1576 (2003). [DOI] [PubMed] [Google Scholar]
- Perez-Montero S., Carbonell A., Moran T., Vaquero A. & Azorin F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell 26, 578–590 (2013). [DOI] [PubMed] [Google Scholar]
- Abhiman S., Iyer L. M. & Aravind L. BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 24, 458–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q. et al. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 27, 602–614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H. et al. Insensitive is a corepressor for Suppressor of Hairless and regulates Notch signalling during neural development. EMBO J. 30, 3120–3133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C. Notch signaling: control of cell communication and cell fate. Development 131, 965–973 (2004). [DOI] [PubMed] [Google Scholar]
- Dai Q. et al. BEND6 is a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Development 140, 1892–1902 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q. et al. Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev. 29, 48–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi H. et al. A genetic screen and transcript profiling reveal a shared regulatory program for Drosophila linker histone H1 and chromatin remodeler CHD1. G3 (Bethesda) 5, 677–687 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T., Sarkeshik A., Yates J. & Schedl P. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. eLife 1, e00171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93, 12418–12422 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner S. S. & Travers A. A. HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 13, 1817–1822 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C. Y., Lei E. P., Ghosh D. & Corces V. G. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16, 737–748 (2004). [DOI] [PubMed] [Google Scholar]
- Xu N., Emelyanov A. V., Fyodorov D. V. & Skoultchi A. I. Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenetics Chromatin 7, 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov D. V., Blower M. D., Karpen G. H. & Kadonaga J. T. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 18, 170–183 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov D. V. & Levenstein M. E. Chromatin assembly in Drosophila systems In Current Protocols in Molecular Biology (ed. Chanda V. B.) 21.27.21–21.27.27 (Wiley & Sons, 2002). [DOI] [PubMed] [Google Scholar]
- Konev A. Y. et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317, 1087–1090 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E. & Alberts B. M. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31–70 (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.