Summary
In this study, we characterized the role of an apple cytosolic malate dehydrogenase gene (MdcyMDH) in the tolerance to salt and cold stresses and investigated its regulation mechanism in stress tolerance. The MdcyMDH transcript was induced by mild cold and salt treatments, and MdcyMDH‐overexpressing apple plants possessed improved cold and salt tolerance compared to wild‐type (WT) plants. A digital gene expression tag profiling analysis revealed that MdcyMDH overexpression largely altered some biological processes, including hormone signal transduction, photosynthesis, citrate cycle and oxidation–reduction. Further experiments verified that MdcyMDH overexpression modified the mitochondrial and chloroplast metabolisms and elevated the level of reducing power, primarily caused by increased ascorbate and glutathione, as well as the increased ratios of ascorbate/dehydroascorbate and glutathione/glutathione disulphide, under normal and especially stress conditions. Concurrently, the transgenic plants produced a high H2O2 content, but a low production rate was observed compared to the WT plants. On the other hand, the transgenic plants accumulated more free and total salicylic acid (SA) than the WT plants under normal and stress conditions. Taken together, MdcyMDH conferred the transgenic apple plants a higher stress tolerance by producing more reductive redox states and increasing the SA level; MdcyMDH could serve as a target gene to genetically engineer salt‐ and cold‐tolerant trees.
Keywords: apple (Malus domestica B.), cytosolic malate dehydrogenase gene, overexpression, salt and cold tolerance, redox state, salicylic acid
Introduction
Plant tissues possess multiple isoforms of malate dehydrogenase (MDH, EC 1.1.1.37), which belongs to the group of oxidoreductases and catalyses the interconversion of malate and oxaloacetate (OAA) coupled to the reduction or oxidation of the NAD(H) or NADP(H) pool. NAD‐dependent MDHs are located in the cytosol and in organelles, including plastids, mitochondria, peroxisomes and microbodies; additionally, chloroplasts contain an NADP‐dependent MDH (Gietl, 1992; Scheibe, 2004). In the Arabidopsis genome, eight putative NAD‐MDH isoforms have been identified, where two are mitochondrial MDH (mMDH), two are peroxisomal MDH, one is a plastidial MDH, and three have no detectable target sequences and are thought to be cytosolic MDH (cyMDH) (Beeler et al., 2014).
The redox state of the cells is especially critical during exposure to abiotic stresses, which are known to induce oxidative stress at the cellular level; under these conditions, regulation of multiple redox and reactive oxygen species (ROS) signals in plants requires a high degree of coordination and balance between the signalling and metabolic pathways in different cellular compartments (Suzuki et al., 2012). In green tissues, an important aspect is the chloroplast/cytosol/mitochondrion cooperation under stress to modulate cell redox homeostasis (Raghavendra and Padmasree, 2003). MDH reaction is involved in central metabolism and redox homeostasis between organelle compartments (Tomaz et al., 2010). MDH isozymes in the chloroplast inner envelope membrane, stroma, mitochondrial membrane and cytosol are the components of malate/OAA and malate/aspartate (Asp) shuttles (malate valves) (Gietl, 1992; Taniguchi and Miyake, 2012). When plants are subjected to abiotic stresses, changes in the activities of the enzymes of the malate valves and expression levels of the MDH isoforms can be observed (Scheibe, 2004), and malate valves play major roles in reductant export under stress conditions (Taniguchi and Miyake, 2012). Chloroplast NADP‐MDH makes the link between the redox states of the chloroplast and the cytosol and other cell compartments, such as peroxisomes (Heyno et al., 2014). In Arabidopsis, malate/OAA transporter knockout impairs the malate valve function of chloroplasts, and the knockout plants show enhanced photoinhibition and ROS accumulation in response to a high‐intensity light, due to a greater accumulation of reducing equivalents in the stroma (Lemaire et al., 1996). Regarding mMDH, its repression modifies the ascorbate‐mediated link between the energy‐generating processes of respiration and photosynthesis (Nunes‐Nesi et al., 2005). Additionally, mMDH lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis (Tomaz et al., 2010). In contrast, cyMDH is thought to function as a key player in the transfer of reducing equivalents from the chloroplast or mitochondria to other destinations in plant cells (Gietl, 1992; Hara et al., 2006; Scheibe, 2004).
On the other hand, plant hormones play central roles in the ability of plants to adapt to abiotic stresses by mediating a wide range of adaptive responses (Peleg and Blumwald, 2011). For example, evidence of the role of salicylic acid (SA) in the oxidative damage generated by NaCl and osmotic stress was reported, and the NPR1‐dependent SA signalling pathway was found to play pivotal roles in enhancing salt and oxidative stress tolerance in Arabidopsis (Borsani et al., 2001; Jayakannan et al., 2015); in contrast, ABA‐dependent and ABA‐independent signalling pathways in response to osmotic stress were widely investigated (Yoshida et al., 2014). Additionally, interactions between hormone and redox signalling pathways can control plant growth and cross‐tolerance to stress (Bartoli et al., 2013). Up to now, evidence that cyMDH and other MDHs can enhance plant stress tolerance by regulating redox and hormone levels, as well as from their interactions, is still lacking.
Because most of the arable lands are used for growing grain crops, fruit trees are increasingly grown on marginal land. Because fruit trees cannot be rotated to avoid stresses, in contrast to annual crops, they often encounter multiple environmental stresses simultaneously. It has become an emerging challenge for scientists to clarify how fruit trees respond to and grow against adverse environments and to improve the stress tolerance of fruit trees by genetic modification. Presently, it has been reported that cyMDH confers superior manganese tolerance by mediating malate synthesis and secretion and inhibiting ROS generation in Stylosanthes guianensis (Chen et al., 2015). Our previous study also showed that MdcyMDH conferred transgenic tomato plants high tolerance to cold and salt stresses alongside with decreased ROS levels (Yao et al., 2011a). However, more research is needed to unravel the mechanism of MdcyMDH in regulating abiotic stresses. Therefore, this study is aimed to assess the potential application of MdcyMDH in engineering salt‐ and cold‐tolerant apple trees and to better understand the mechanism of MdcyMDH‐mediated abiotic stress tolerance by modifying redox homeostasis and hormone levels.
Results
The identification of cyMDH genes and their expression in different tissues and under cold and salt treatments in apple
In our previous study (Yao et al., 2008), the MdcyMDH (DQ221207) ORF was shown to encode a 332‐aa polypeptide which shared over 90% similarity with cyMDHs from other species, but we failed to locate this gene in the present apple genome database. In contrast, the other four cyMDH isoforms were identified from the entire apple genome, and their deduced proteins possessed the highly conserved domains of cytosolic malate dehydrogenase (Figure S1). Highly similar protein tertiary structures were observed between MdcyMDH and MDP0000174740, as well as between MDP0000197620 and MDP0000926135 (Figure 1a). Transcripts of the five cyMDH genes could be detected in all tissues, but they exhibited varying expression levels in different tissues; MdcyMDH, MDP0000174740 and MDP0000197620 were expressed primarily in the stems and roots (Figure 1b,c). MdcyMDH, MDP0000174740 and MDP0000170418 expression were induced by 50 mm NaCl and temperature of 8 °C (Figure 1c). Therefore, MdcyMDH and MDP0000174740 most likely possessed similar functions in the light of their highly similar tertiary structures, spatial expressions and expression responses to salt and cold in apple.
Figure 1.
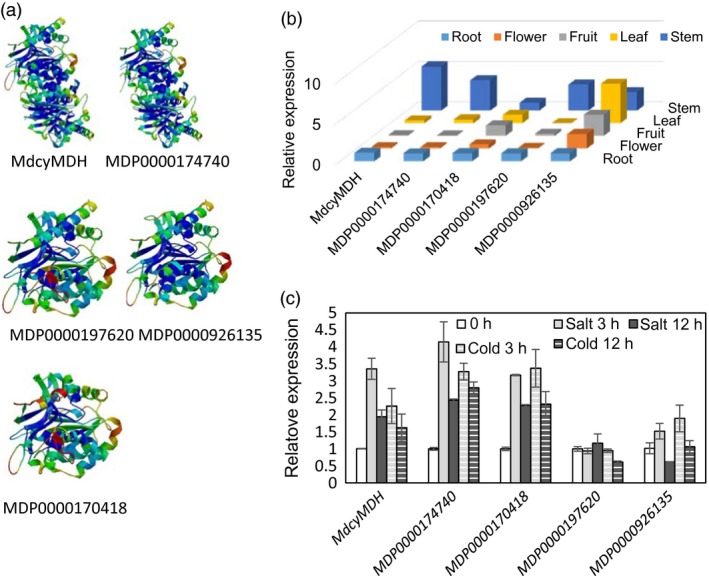
Predicted protein tertiary structures of the different cyMDH isoforms (a) and expression patterns of the cyMDH genes in different tissues (b) and in response to 8 °C temperature and 50‐mm salt in the leaves of apple in vitro shoot cultures (c). Protein tertiary structures were predicted using the SWISS‐MODEL (http://swissmodel.expasy.org/). Values represent the means ± SD of three replicates.
Improved tolerance to salt and cold in transgenic apple plants via MdcyMDH overexpression
To further characterize the biological function of the MdcyMDH gene, the MdcyMDH‐overexpressing lines were obtained by agrobacterium‐mediated transformation and confirmed by PCR detection of the target gene (Figure 2a,b). Lines 5 and 7, with high MdcyMDH expression level compared to the wild‐type (WT) plants, were selected to be further assayed (Figure 2b). When subjected to 50 mm salt and 8 °C stresses, no obvious phenotype differences were observed between the transgenic and WT apple in vitro shoot cultures at 10 days after the treatments; after nine more days of 100 mm NaCl and seven more days of 0 °C treatment, both the transgenic and WT apple cultures showed the necrosis phenotypes, but the transgenic plants exhibited less severe phenotypes (Figure 2d,e). In addition, after 3 weeks of NaCl treatment, the leaves of the 3‐year‐old WT plants exhibited symptom of etiolation and marginal necrosis but the leaves of the transgenic plants exhibited near‐to‐normal colour (Figure 2g). Moreover, the leaves of the transgenic apple cultures and trees possessed a much higher chlorophyll content than the leaves of the WT plants after the stress treatments although the chlorophyll contents were reduced in the WT and transgenic plants when subjected to the stress treatments (Figure 2h,i). Therefore, MdcyMDH overexpression enhanced the tolerance of transgenic apple plants to salt and cold stresses.
Figure 2.
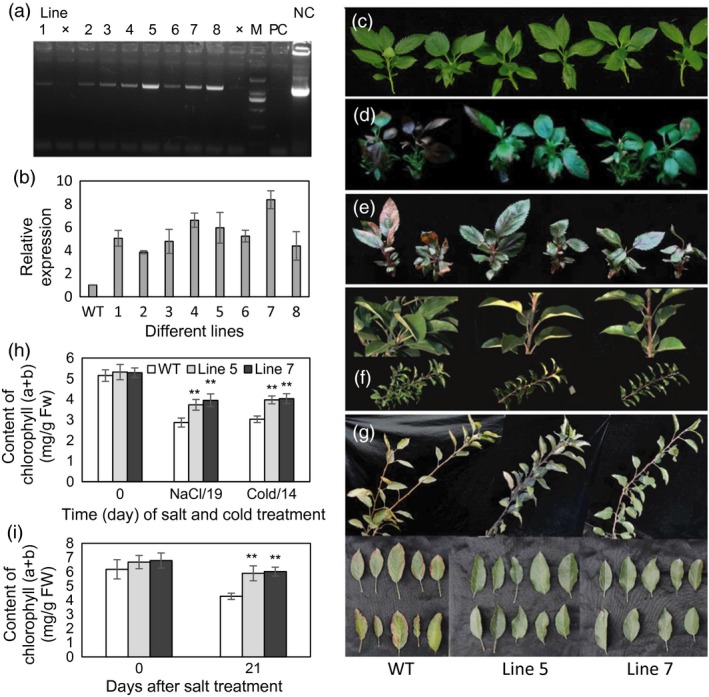
PCR identification of MdcyMDH‐overexpressing apple plants and the evaluation of salt and cold tolerance of transgenic plants. (a) PCR identification using DNA templates and the specific primers from 35S and MdcyMDH. (b) MdcyMDH expression in the leaves of wild‐type (WT) and transgenic lines. (c–e) WT and transgenic apple in vitro shoot cultures at 20 days after a subculture were taken as the control (c); the cultures with the same subculture conditions as the control were subjected to stress treatments, that is at 50‐mm NaCl for 10 days and subsequent 100‐mm NaCl for 9 days (d); at 8 °C for 7 days and subsequent 0 °C for 7 days in a 14‐h light photoperiod (e). (f, g) Shoots and leaves of the 3‐year‐old pot‐cultured WT and transgenic apple trees prior to stress treatments (f) and after 100‐mm NaCl for 14 days and subsequent 150‐mm NaCl for 7 days (g). (h, i) Contents of chlorophyll (a + b) of the leaves from the transgenic apple cultures (h) and trees (i) under normal and stress conditions. Values represent the means ± SD of three replicates. M, DNA marker; NC, negative control (H2O); PC, positive control (plasmid DNA of 35S::MdcyMDH). **Highly significant difference, P < 0.01.
Comparison of expression profiles between the WT and MdcyMDH‐overexpressing plants
To explore the regulation mechanism of MdcyMDH in abiotic stresses, a digital gene expression (DGE) tag profiling analysis between the WT and mixed transgenic plants at 3 h after the cold treatment was conducted to quantify gene changes. It was found that 961 and 958 genes were up‐ and down‐regulated by at least onefold in the transgenic lines, respectively, demonstrating that a massive transcriptional reprogramming took place in the MdcyMDH‐overexpressing plants. According to the putative homology to sequences present in public databases, the differentially expressed genes were classified into 12 different cellular component categories; the cytosol contained the most differentially expressed genes (40.1%), followed by the membrane (33.3%), chloroplast (6.4%), nucleus (2.8%) and mitochondria (2.7%) (Figure 3a). All of the DGE genes are associated with 22 biological processes. The two processes of plant–pathogen interaction and plant hormone signal transduction contain the most DGE genes; additionally, photosynthesis, citrate cycle (TCA cycle) and the oxidation–reduction process were also the clearly changed biological processes (Figure 3b). Moreover, the expression levels of the 28 most differentially expressed annotated genes in the cold‐treated shoot cultures were detected by qRT‐PCR, which contained genes related to SA biosynthesis and signal transduction, chloroplast and mitochondrial metabolism, redox process and abiotic stresses; similar expression changes to DGE tag profiles were found, validating the reliability of the DGE genes (Figure 3c). Additionally, the 28 genes exhibited similar expression changes under salt treatment as that under cold treatment (Figure 3c). Besides, most of the above genes were induced by cold and/or salt in the WT and transgenic plants (Figure S2). Therefore, it is suggested that MdcyMDH improved the stress tolerance of the transgenic apple plants at least partly via modifying hormone signal transduction, photosynthesis, TCA cycle and the oxidation–reduction process.
Figure 3.
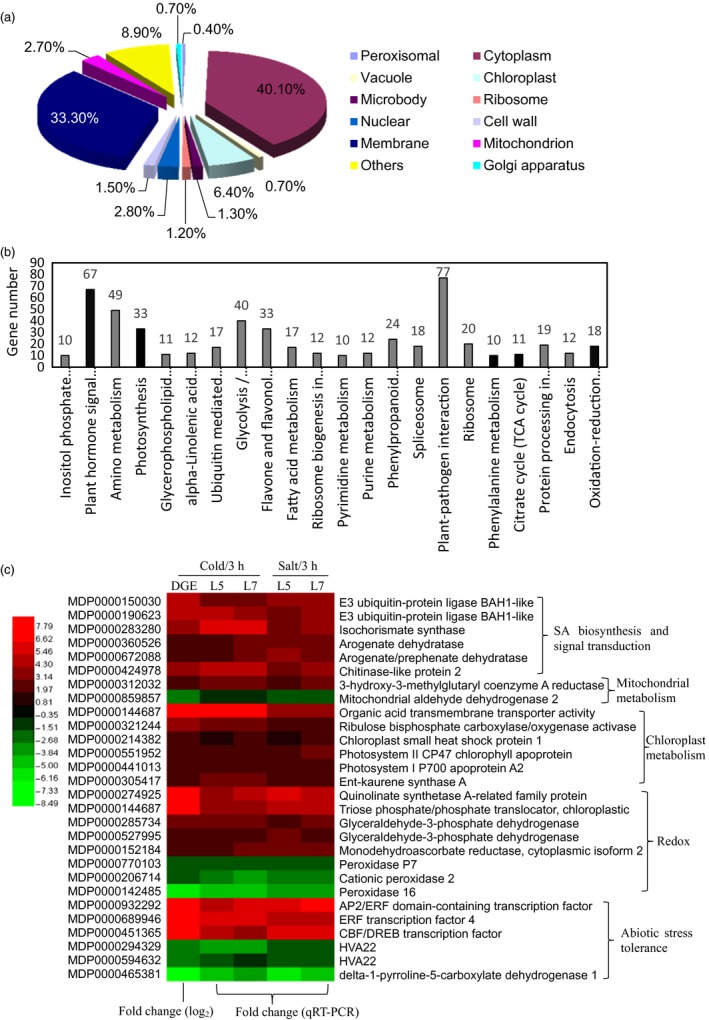
Cell component (a) and biological process (b) of the digital gene expression (DGE) genes classified by Gene Ontology and the most differentially expressed genes related to SA biosynthesis and signalling, mitochondrial and chloroplast metabolism, and redox and abiotic stress tolerance (c). In the heat map of panel (c), the ID and annotation of each gene are provided. Fold changes from qRT‐PCR at 3 h after cold and salt treatments were used to validate the DGE genes and to determine the expression changes of the DGE genes induced by MdcyMDH overexpression under stress treatments. Fold change from qRT‐PCR is calculated by comparing the relative expression values of the selected genes in the transgenic and wild‐type (WT) plants. Data are presented as the means of three replicates.
MdcyMDH overexpression modulated mitochondrial and chloroplast metabolisms
First, MDH activities in the cytosol, mitochondria and chloroplast were detected in the MdcyMDH‐overexpressing apple in vitro shoot cultures (Figure 4a–c). The MdcyMDH overexpression increased the reductive activities of cyMDH and chMDH and the oxidative activity of mMDH compared to the WT plants; in contrast, the difference of activities was enlarged and reached a significant level under cold and salt stresses. In contrast, the oxidative activities of cyMDH and chMDH and the reductive activity of mMDH were generally not significantly changed in the transgenic plants compared to those in the WT plants. Moreover, the reductive activities of cyMDH and chMDH and the oxidative activity of mMDH were clearly induced by cold and salt treatment in the WT and transgenic plants (Figure 4a–c). Therefore, it can be suggested that the increases in the cyMDH and chMDH reductive activities and the mMDH oxidative activity contributed favourably to the stress tolerance of apple plants in the transgenic plants. Moreover, the significantly increased malate content indicated that the reductive activity of MDH was strengthened against its oxidative activity in the transgenic plants under normal and stress conditions (Figure 5a).
Figure 4.
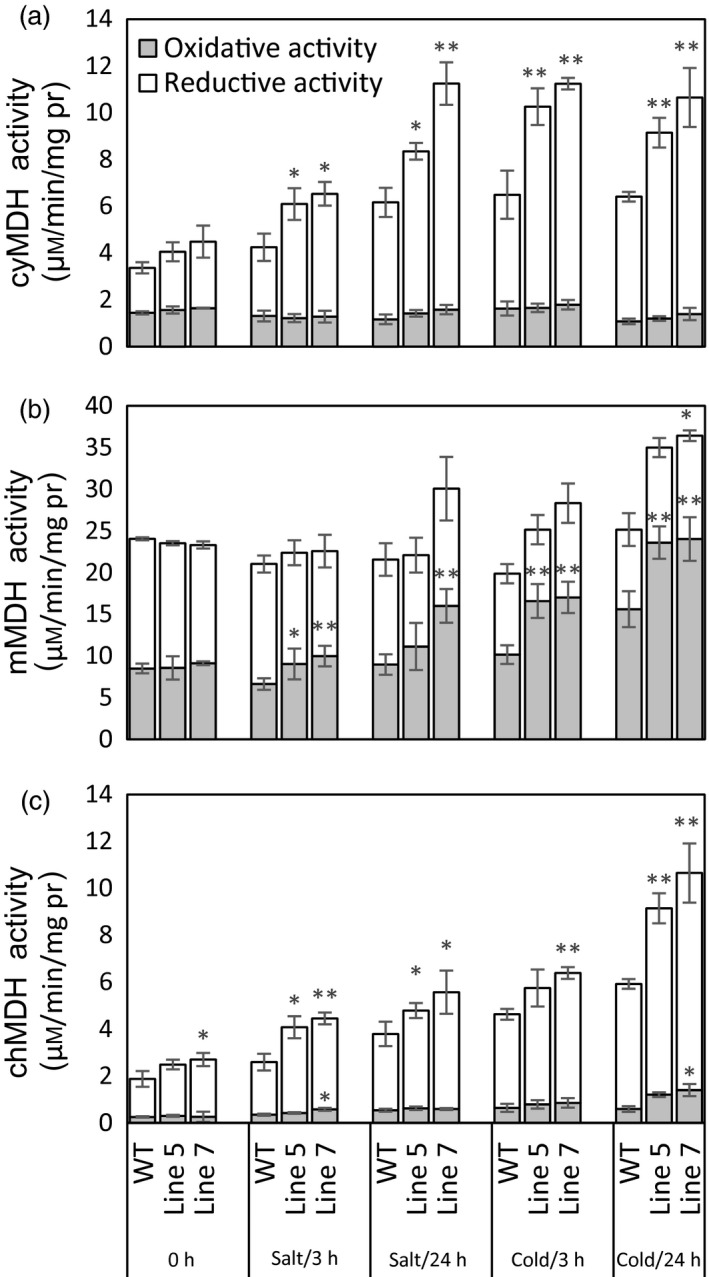
Modifications of reductive and oxidative activities of malate dehydrogenase (MDH) in the cytosol (a), mitochondria (b) and chloroplast (c) in the wild‐type (WT) and transgenic plants in response 50 mm salt and 8 °C stresses. Data are presented as means ± SD (n = 3). The difference level was compared between the transgenic lines and WT at the same treatment time. *Significant difference, P < 0.05; **highly significant difference, P < 0.01.
Figure 5.
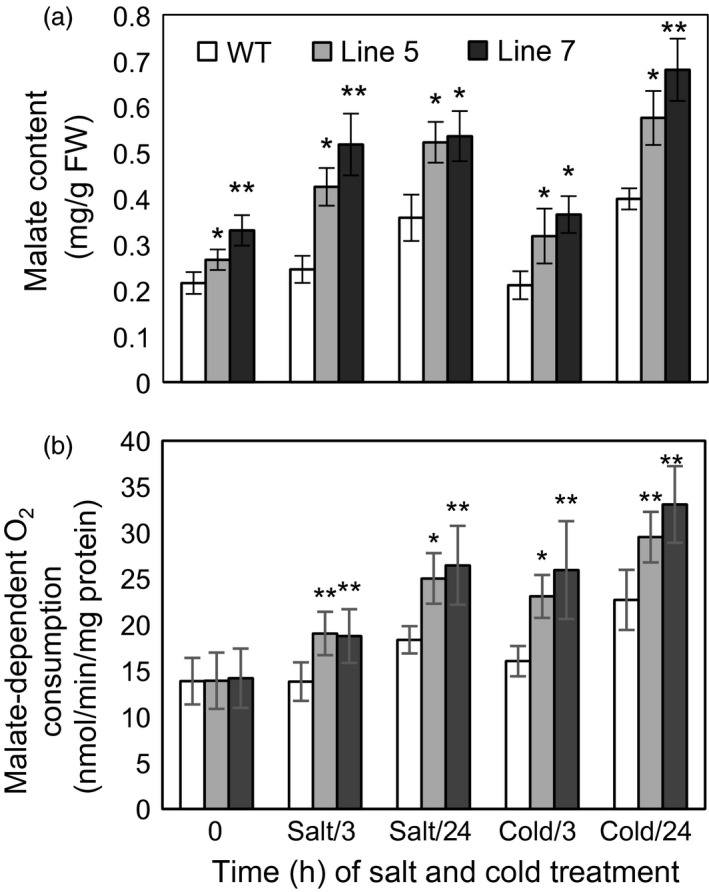
Malate content and malate‐dependent O2 consumption in the wild‐type (WT) and transgenic plants under cold and salt treatments. Data are presented as means ± SD (n = 3). The difference level was compared between transgenic lines and WT at the same treatment time. *Significant difference, P < 0.05; **highly significant difference, P < 0.01.
Because the MDH activities in the cytosol, mitochondria and chloroplast were altered in the transgenic plants and MDH is one of the components of the malate/OAA shuttle between the cytosol and other organelles, it was speculated that the mitochondrial and chloroplast metabolisms would be changed. To verify this speculation, mitochondrial and chloroplast metabolisms were detected in the WT and transgenic plants under normal and stress conditions. On the one hand, malate‐dependent oxygen consumption by isolated mitochondria was measured to assess the change in the mitochondrial activity. The results showed that the oxidation capacity of malate was significantly increased in the transgenic plants under cold and salt treatments (Figure 5b), which was consistent with the increased oxidative activity of mMDH (Figure 4b). On the other hand, parameters related to photosynthesis and chlorophyll fluorescence were determined to assess the changes in chloroplast metabolism in the transgenic plants. The results showed that the NaCl treatment induced a decrease in the photosynthetic rate in the WT and transgenic plants; nevertheless, the MdcyMDH overexpression produced an increase in the photosynthetic rate and a concomitant increase in the stomatal conductance (Figure 6a,b), suggesting the contribution of stomatal factors to the increased photosynthetic rate in the transgenic plants. However, similar intercellular CO2 concentration in the leaves of the WT and transgenic plants also indicated altered activities of the mesophyll by the MdcyMDH overexpression (Figure 6c). Compared to the WT plants, MdcyMDH overexpression did not significantly influence the photoinhibition of PSII in the light of similar ratios of F v/F m under normal and salt treatment (Figure 6d; Björkman and Demmig, 1987); in contrast, MdcyMDH overexpression significantly increased the electron transport rate (ETR), which was used as a quantitative indicator of the electron transport beyond PSII (Maxwell and Johnson, 2000) (Figure 6e). Additionally, the transgenic plants significantly modified the redox level and excitation pressure of PSII, indicated by the altered values of 1−qP (Figure 6f; Maxwell and Johnson, 2000). Taken together, the mitochondrial and chloroplast metabolisms were modified in the MdcyMDH‐overexpressing plants.
Figure 6.
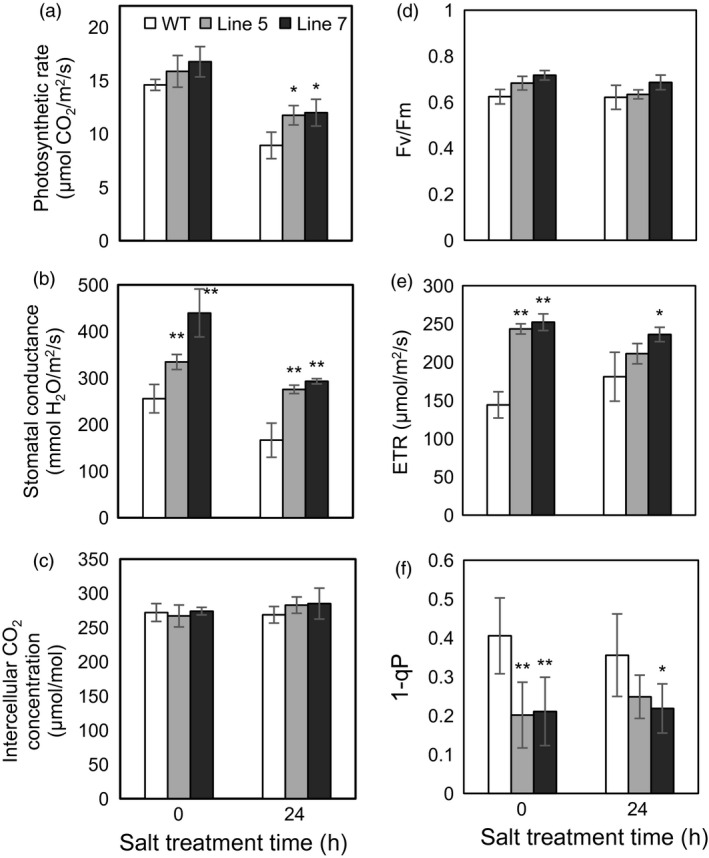
Changes of the photosynthetic rate (a), stomatal conductance (b), intercellular CO 2 concentration (c), maximum quantum yield of PSII (F v/F m) (d), photosynthetic electron transport rate (ETR) (e), and PSII excitation pressure (1−qP) (f) in the leaves of the wild‐type (WT) and transgenic plants at 0 and 24 h after 50‐mm NaCl treatment. Data are presented as means ± SD (n = 3). *Significant difference, P < 0.05; **highly significant difference, P < 0.01.
Overexpression of MdcyMDH increases the reducing power and modifies the ROS level under cold and salt stresses
The three most abundant antioxidants in plant cells, that is NAD(P)H, ascorbate and glutathione (Queval and Noctor, 2007), and their reductive and oxidized forms were determined in the WT and transgenic plants under normal and stress conditions (Figure 7). MdcyMDH overexpression increased the NADH levels and the ratios of NADH/NAD+ although generally not in a statistically significant manner under normal and stress conditions (Figure 7a). In contrast, the NADPH content was slightly enhanced but the ratios of NADPH/NADP+ were almost unchanged (Figure 7b,e). Moreover, accumulation of both reduced ascorbate (AsA) and oxidized ascorbate (DHA) was induced by cold and salt stresses (Figure 7c). The contents of both AsA and DHA increased in the transgenic plants compared to the WT plants; additionally, the increments in AsA and AsA/DHA reached statistical significance at most of the treatment points (Figure 7c,e). Similarly, salt and cold induced the accumulation of oxidized glutathione (GSSG) and especially reduced glutathione (GSH); MdcyMDH overexpression significantly promoted the GSH content and GSH/GSSG under stress conditions (Figure 7d,e). Taken together, MdcyMDH overexpression increased the reducing power predominantly via enhancing the levels of AsA and GSH, as well as the ratios of AsA/DHA and GSH/GSSG.
Figure 7.
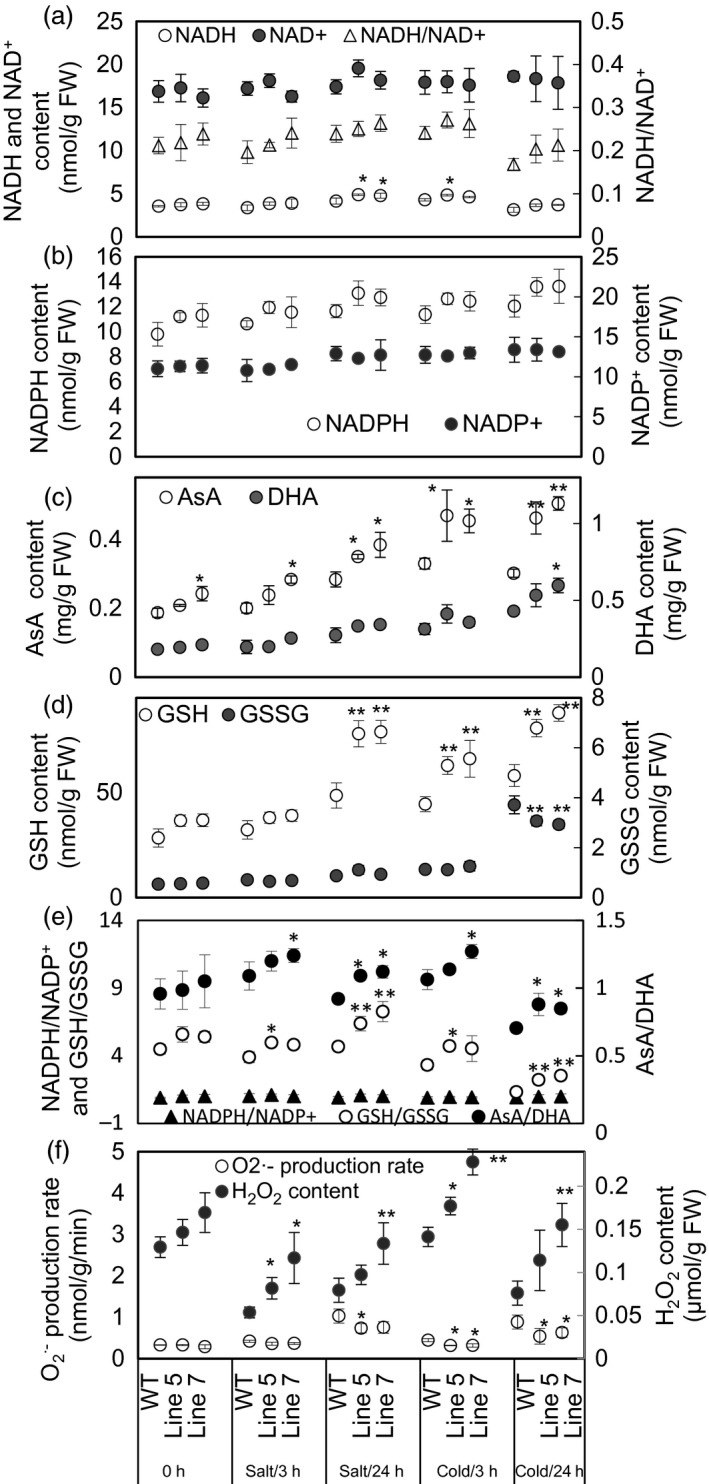
Assays of redox couples and reactive oxygen species (ROS) in the wild‐type (WT) and transgenic plants under 50 mm NaCl and 8 °C treatments. (a) The contents of the reduced form of NADH and the oxidized form of NAD +, as well as the ratio of NADH/NAD +. (b–d) The contents of the reduced form of redox couples (NADPH, AsA and GSH) and the oxidized form of redox couples (NADP +, DHA and GSSG). (e) The ratios of the reduced and oxidative forms of redox couples. (f) The contents of and H2O2. Data are presented as means ± SD (n = 3). *Significant difference, P < 0.05; **highly significant difference, P < 0.01. The analysis of the significant difference was performed between the transgenic lines and WT plants at the same treatment point.
On the other hand, the transgenic lines generated a lower production rate but a higher H2O2 content than the WT plants under normal and stress conditions (Figure 7f).
The level of SA is elevated in the transgenic plants
To examine whether MdcyMDH overexpression impacted hormone levels, the contents of ABA, IAA, GA3 and SA were determined. It was found that IAA and GA3 were not significantly altered in the transgenic plants. In contrast, the ABA level was slightly decreased, while the SA level was largely increased in the transgenic plants (Figure 8a). Free and total SA contents were further evaluated in the WT and transgenic plants under stress conditions. Compared to the WT controls, MdcyMDH overexpression elevated the free and total SA contents, and the increments reached a significant level at some time points under the stress treatments (Figure 8b,c). Free SA and total SA showed a fast decline induced by salt and cold temperature, but the decline was not intensified by the extension of the salt and cold treatments in the WT and transgenic plants. Moreover, MdcyMDH overexpression did not impact the ratio of free SA in the total SA (Figure 8b,c).
Figure 8.
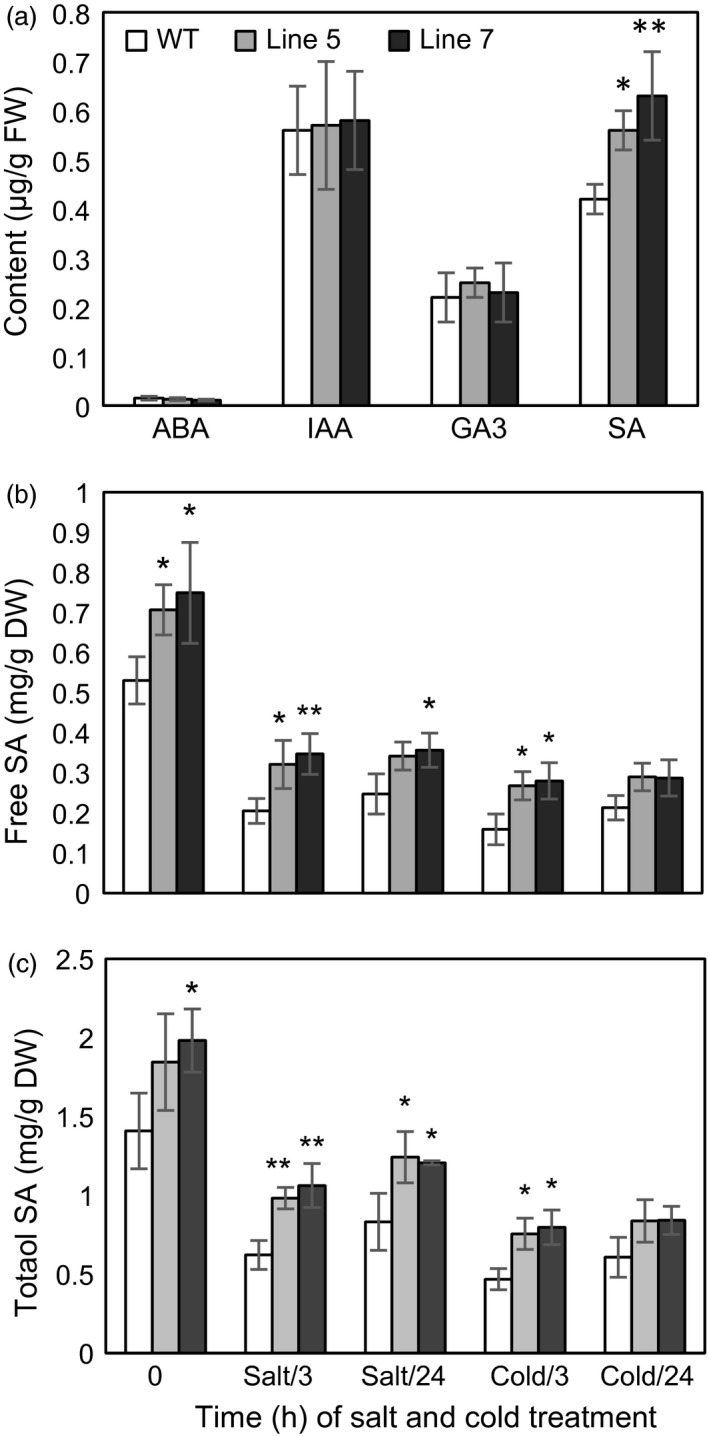
Hormone content in the leaves of the transgenic and wild‐type (WT) plants. (a) Contents of ABA, IAA, GA 3 and SA in the leaves under normal growth conditions. (b) Total and free SA contents in the leaves under salt and cold treatments. Data are presented as means ± SD (n = 3). *Significant difference, P < 0.05; **highly significant difference, P < 0.01.
Discussion
cyMDH reaction favours malate synthesis under normal conditions and especially under abiotic stresses
The kinetic properties of recombinant cyMDH proteins showed that the cyMDH reaction favoured malate production in vitro in apple (Yao et al., 2011b), wheat (Ding and Ma, 2004) and pineapple (Cuevas and Podestá, 2000). Additionally, it was demonstrated that MdcyMDH promoted malate accumulation in MdcyMDH‐overexpressing apple callus (Yao et al., 2011b) and leaves (Figure 5a). Similarly, malate synthesis was promoted through the overexpression of cyMDH in tobacco and Stylosanthes guianensis (Chen et al., 2015; Wang et al., 2010). Therefore, regeneration of NAD+ and malate inside the cytosol was the most primary role of cyMDH, although the enzyme kinetics of the MDH reaction in vivo were affected by substrate/product ratios and the NAD+ redox state (Tomaz et al., 2010). Moreover, cyMDH reductive activity increased in response to cold and salt stresses in the WT and transgenic plants (Figure 4a), and cyMDH conferred superior manganese tolerance by increasing malate synthesis in Stylosanthes guianensis (Chen et al., 2015). Therefore, cyMDH primarily catalysed malate synthesis under stresses, and the reaction positively contributed to the tolerance of abiotic stresses.
Because cyMDH primarily catalysed the reductive reaction, more NADH was supposed to be converted into NAD+. However, a high ratio of NADH/NAD+ was found in the transgenic plants (Figure 7a), which can be explained, at least partly, by the following facts. First, NADH and NAD+ were used to generate other antioxidants and provide electron cofactors for other oxidoreductases and hence were tied to every other redox pair and large amounts of oxidoreductases (Hashida et al., 2009); second, large redox differences in the NAD(H) pools existed between cell compartments (Igamberdiev and Gardeström, 2003); third, MdcyMDH overexpression affected the expression of genes involved in NAD(H) metabolism, such as quinolinate synthetase A and dihydrolipoamide dehydrogenase (Figure 3c; Ceciliani et al., 2000). Therefore, the sole reaction catalysed by MdcyMDH could not determine the NAD(H) level of the cell, and an enhanced NADH/NAD+ might result from the wide metabolism modification mediated by MdcyMDH overexpression.
MdcyMDH enhances the tolerance of the transgenic plants to cold and salt by improving the cell reducing power
Abiotic stresses, such as drought, salt and low temperature, can damage cells and lead to oxidative stress by producing certain deleterious chemical entities called ROS, which include H2O2, , hydroxyl radical (OH−), etc. Enzymatic and nonenzymatic antioxidants can be employed to directly scavenge ROS and free radicals. In our previous study, MdcyMDH overexpression increased SOD and CAT activities in the transgenic apple callus and tomato plants (Yao et al., 2011b). In this study, the significantly increased nonenzymatic antioxidants (AsA and GSH) in the transgenic plants favoured the direct scavenging of ROS (Figure 7c,d; Noctor and Foyer, 1998). In contrast, AsA/DHA, GSH/GSSH, NADH/NAD+ and NADPH/NADP+ couples were placed at the heart of the cell's redox metabolism (Potters et al., 2010). Presently, the regulatory roles of cellular redox couples have predominantly come from studies using rather severe stress treatments that induced large amounts of ROS or programmed cell death (Potters et al., 2010). For example, severe salt stress caused a 10‐fold decrease in the GSH/GSSH ratio in Arabidopsis (Borsani et al., 2001). In contrast, the redox state and responses to mild stress remain a conundrum (Potters et al., 2010). Here, mild cold and salt stresses generated increased ratios of AsA/DHA, GSH/GSSG and NAD(P)H/NAD(P)+ (Figure 7a,e). Similarly, CO2‐induced heat stress alleviation can be associated with the reduction of oxidative stress through increasing AsA/DHA and GSH/GSSG ratios in tomato plants (Li et al., 2015). Therefore, it is suggested that the increased ratio of AsA/DHA, GSH/GSSG and NAD(P)H/NAD(P)+ contributed favourably to the mild oxidative stress tolerances.
Redox compounds are primarily derived from the mitochondrial and chloroplast metabolisms (Chew et al., 2003; Green and Fry, 2005). The modified chloroplast and mitochondrial metabolisms (Figures 5 and 6) might be one of the factors affecting the level of redox compounds in the transgenic plants. Similarly, it was reported that mMDH can alter photosynthetic, photorespiration and leaf respiration and affect ascorbate content in tomato and Arabidopsis (Nunes‐Nesi et al., 2005; Tomaz et al., 2010); plastid NAD‐MDH can alter glutathione levels and the respiration rate in Arabidopsis (Beeler et al., 2014). Additionally, modifications in the mMDH and chMDH activities were found in the MdcyMDH transgenic plants (Figure 4b,c). Therefore, it is speculated that MDHs from different cell compartments might cooperate to modify the chloroplast and mitochondrial metabolisms via malate valves. Moreover, another shuttle system (triose‐phosphate/3‐phosphoglycerate shuttle) between the chloroplast and the cytosol is suggested to be activated by MdcyMDH overexpression, based on the large expression modification of the key genes in this shuttle system, that is triose‐phosphate/phosphate translocator and glyceraldehyde‐3‐phosphate dehydrogenase (Figure 3c; Guo et al., 2012; Taniguchi and Miyake, 2012). Therefore, MdcyMDH might modify chloroplast and mitochondrial metabolisms and hence the redox state via different shuttle systems.
MdcyMDH increases cold and salt tolerance by modifying the redox signal
The redox pairs within the cellular stress response network are more complex and do not simply act as a scavenger; redox metabolism and its associated signalling are important machinery during abiotic stress (Munné‐Bosch et al., 2013). It is suggested that ascorbate and glutathione pools have precise and distinct roles in redox signalling (Munné‐Bosch et al., 2013) and participate in the signal transduction pathways (Ramel et al., 2012). Changes in the glutathione content and in the ratio of GSH/GSSG clearly modify redox‐ and H2O2‐dependent gene regulation (Han et al., 2013). In this study, the increased ratios of AsA/DHA and GSH/GSSG (Figure 7e) may affect the redox signalling in the transgenic plants. On the other hand, ROS, such as H2O2 and , are proposed to function in signalling between different organelles and the nucleus; the cytosol should be considered as a hub where cross‐communication between divergent ROS signals occurs (Choudhury et al., 2013). However, ROS accumulation at high levels causes oxidative stress leading to cell death (Choudhury et al., 2013). The dual function of ROS implies that its cellular concentration has to be tightly controlled (Apel and Hirt, 2004). In this study, the H2O2 level was increased, but was decreased by MdcyMDH overexpression (Figure 7f); this modification possibly altered the ROS signal, which was also suggested by the two largely up‐regulated glyceraldehyde 3‐phosphate dehydrogenase genes, related to H2O2 signal transduction (Figure 3c; Guo et al., 2012); the altered redox signalling might positively contribute to the increased cold and salt tolerance in the transgenic plants.
It is proposed that malate valves play an essential role in the regulation of catalase activity and the accumulation of a H2O2 signal by transmitting the redox state of the chloroplast to other cell compartments (Heyno et al., 2014). Therefore, the increase in H2O2 content in the transgenic plants may be influenced by the altered MdcyMDH‐mediated regulation of malate valves. Additionally, the expression of three peroxidases was largely down‐regulated (Figure 3c), suggesting a possible decrease in H2O2 scavenging, which led to the increase of H2O2 (Passardi et al., 2004). Moreover, can be rapidly converted to H2O2 by SOD1 or SOD2 (Murphy, 2009); the decreased (Figure 7f) might contribute to the increase of H2O2 in the transgenic plants.
The promoted interaction of redox and SA favours the tolerance of the transgenic plants to cold and salt stresses
It is known that the SA signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis (Jayakannan et al., 2015). MdcyMDH overexpression might affect the stress tolerance of the transgenic plants via the SA signalling pathway based on the increased free and total SA (Figure 8b,c). SA is synthesized by two distinct pathways: The phenylalanine ammonia‐lyase pathway in the cytoplasm and the isochorismate (ICS1) pathway in the chloroplast. The ICS1 pathway is now thought to be the predominant pathway responsible for SA synthesis (Jayakannan et al., 2015). In the transgenic plants, an ICS1 gene, marker gene in the ICS1 pathway and two arogenate dehydratase genes involved in the biosynthesis of phenylalanine were largely transcriptionally up‐regulated (Figure 3c), implying that MdcyMDH increased SA biosynthesis by promoting the above pathways. The defectiveness in the expression of the ICS1 gene is hypersensitive to salt stress in Arabidopsis (Asensi‐Fabado and Munné‐Bosch, 2011), and salt‐ and cold‐induced ICS1 expression was found in apple plants (Figure S2), implying that this pathway was essential for salt and cold tolerance in plants. Noticeably, two E3 ubiquitin‐protein ligase BAH1‐like genes were largely enhanced by MdcyMDH overexpression (Figure 3c), and they were largely induced by cold and salt (Figure S2); this type of gene is involved in the negative regulation of SA accumulation independent of ICS1 (Kim et al., 2010) and might be one the factors decreasing the SA level when the transgenic and WT plants were subjected to stresses (Figure 8b,c). Taken together, MdcyMDH increased the SA level by several SA metabolism pathways including synthesis and degradation, which are likely to maintain the most suitable SA level under stress.
The modification of the redox state might be an important factor affecting the SA level. It has been found that an elevated NAD+ level did influence SA turnover and biosynthesis via downstream ICS1 genes (Pétriacq et al., 2012). Other antioxidants, such as ascorbate and glutathione, are also central redox regulators of the SA signalling pathway (Bartoli et al., 2013). Moreover, ROS can influence the SA metabolism, and the H2O2‐dependent activation of SA‐dependent pathways is well characterized (Munné‐Bosch et al., 2013). On the other hand, the tight correlation between SA, H2O2 and GSH contents was observed in plants, implying an essential role of SA in the acclimation processes and in regulating the redox homeostasis of the cell (Mateo et al., 2006). Taken together, the complex network of cross‐communication between oxidants and antioxidants in the redox signalling hub and the different hormone signalling pathways regulates plant survival upon exposure to stress (Bartoli et al., 2013), which might be the key mechanism of MdcyMDH‐mediated abiotic stress tolerance.
Thus to conclude, MdcyMDH overexpression improved the tolerance of the transgenic apple plants to salt and cold conditions by the following possible mechanisms. First, the reducing power was enhanced, and the level of was decreased; second, changes in the redox couples and the level of H2O2 contributed favourably to stress tolerance as redox signals; third, the stress‐resistant pathway mediated by SA was strengthened; finally, the widely modified gene expression in the transgenic plants implicated that redox compounds and SA participated in the chloroplast‐to‐nucleus and mitochondria‐to‐nucleus retrograde communication, which served to coordinate gene expression in the nuclear and cytoplasmic genomes and played an important role in the stress acclimation of plants (Cela et al., 2011; Ramel et al., 2012).
Experimental procedures
Plant materials and growth conditions
‘Gala’ apple in vitro shoot cultures were used for the gene expression assays, genetic transformation and other analyses. The ‘Gala’ cultures were grown on MS subculture media containing 0.6 mg/L of 6‐BA and 0.2 mg/L of IAA at 25 °C under a 16‐h photoperiod with a light intensity of 600 μmol/m2/s. Three‐year‐old pot‐cultured WT and transgenic apple trees were used for the salt treatment, photosynthesis and chlorophyll fluorescence assays.
RNA extraction and gene expression analysis with real‐time quantitative PCR
Total RNAs were extracted from apple in vitro shoot cultures using TRIzol Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Two micrograms of total RNAs was used to synthesize first‐strand cDNA. For real‐time quantitative PCR, the specific primer pairs are given in Table S1. The reactions were performed using SYBR Green MasterMix (SYBR Premix EX Taq TM, Dalian, China), as described by the manufacturer. Real‐time quantitative PCRs were performed using a BIO‐RAD iQ5 (Hercules, CA) instrument. The expression values obtained were normalized against 18s rRNA by the cycle threshold (C t) method (Software IQ5 2.0) (Bio‐Rad, Hercules, CA).
Agrobacterium‐mediated genetic transformation of MdcyMDH into apple in vitro shoot cultures
The MdcyMDH ORF was obtained by PCR using the forward primer 5′‐GGTCTAGAATGGCGAAAGAACCAGTTC‐3′ and the reverse primer 5′‐CGTCGACAGTCGAAGTGTCCGAATAGAAT‐3′. The forward primer contained a Xba I digestion site, and the reverse primer contained a SalI site (both underlined). Subsequently, the PCR product was cloned into the pMD18‐T vector (TaKaRa, Dalian, China). MdcyMDH was double‐digested with XbaI and SalI, and then ligated into the pBI121 vector, downstream of the CaMV 35S promoter. The resultant constructs were introduced into the Agrobacterium strain LBA4404 and transformed into apple leaves from in vitro shoot cultures, as described by our previous study (Feng et al., 2012). The specific primers from 35s (5′‐GACGCACAATCCCACTATCC‐3′) and MdcyMDH (5′‐GGATCCAGAGAGGCAAGAGTA‐3′) were used in PCR to determine whether MdcyMDH was integrated into the apple plants. A plasmid DNA containing the pBI121::MdcyMDH construct and H2O was used as positive and negative controls.
Enzyme extraction and isolation of mitochondria and chloroplast
Cytosolic enzyme extractions were performed according to our previous study (Yao et al., 2011b). Mitochondria and chloroplast were isolated mechanically from 15 and 10 g of leaves of apple in vitro shoot cultures, respectively, and then purified by Percoll gradient centrifugation as described by Osmond and Makino (1991). Mitochondrial integrity was detected by measuring the cytochrome c oxidase activity with and without 0.05% Triton X‐100. In the tested samples, the membrane integrity exceeded 86%. The intactness of the purified chloroplast fraction was over 84% as judged by the ferricyanide test. The purities of the cytosolic, mitochondria and chloroplast fractions were verified by measuring the activities of maker enzymes located in other organelles (Table S2). Protein amounts were quantified using dye‐binding assay with bovine serum albumin (Bradford, 1976).
MDH activity assays and malate‐dependent respiratory rates
MDH reductive activity was measured in 1 mL of a reaction mixture containing 50 mm Tris–HCl (pH 7.8), 2 mm MgCl2, 0.5 mm EDTA, 0.2 mm NADH, 2 mm OAA and 50 μL of extract at 30 °C. The reaction was initiated by adding OAA. MDH oxidative activity was measured in 1 mL of a reaction mixture containing 50 mm Tris–HCl (pH 8.9), 2 mm MgCl2, 0.5 mm EDTA, 0.2 mm NAD+ and 25 mm malate. The reaction was initiated by adding malate. For each reaction, 5 min of spectrophotometric change at 340 nm was monitored automatically at 40‐s intervals, and the activities were calculated according to the slopes of the obtained lines. The assay conditions were not optimized with respect to the concentration of each component of the reaction mixture. However, linear relationships between the activity and time and amount of the extract were found.
Malate‐dependent respiratory rates on isolated intact mitochondria were performed according to the method of Tomaz et al. (2010).
Determination of malate content
Malate content was determined using a capillary electrophoresis system (Beckman P/ACE, Palo Alto, CA) as described in our previous study (Yao et al., 2007).
Measurement of photosynthesis and chlorophyll content
Photosynthesis was measured using a portable photosynthesis system (CIRAS‐2; PPS Co. Ltd., Hitchin, UK). Photosynthesis was measured three times for each selected leaf, so that nine measurements were made for each plant. Measurements were made between 8:30 and 11:00 h on a sunny day. During each measurement, CO2 concentration was maintained at 396 ± 21 μL/L by the CIRAS system at an air temperature of approximately 25 °C and a relative humidity of 85 ± 0.9%. Chlorophyll content was determined according to the methods described by Zhu et al. (1990).
Measurement of chlorophyll fluorescence
Chlorophyll fluorescence was measured with a FMS‐2 pulse‐modulated fluorometer (Hansatech, Norfolk, UK). The light‐fluorescence measurement protocol was as follows: The light‐adapted leaves were continuously illuminated by actinic light at 100 μmol/m2/s from the FMS‐2 light source; steady‐state fluorescence (F s) was recorded after 2 min of illumination; and 0.8 s of saturating light of 8000 μmol/m2/s was imposed to obtain a maximum fluorescence in the light‐adapted state (F m). The actinic light was then turned off, and the minimum fluorescence in the light‐adapted state (F 0) was determined by 3 s of illumination with far‐red light. The following parameters were then calculated as follows (Maxwell and Johnson, 2000):
Extractions and assays of redox couples and ROS
Extractions and determinations of the four redox couples, that is NADH/NAD+, NADPH/NADP+, reduced ascorbate (AsA)/oxidized ascorbate (DHA) and reduced glutathione (GSH)/oxidized glutathione (GSSG), were performed as described by Queval and Noctor (2007). Hydrogen peroxide (H2O2) and superoxide radical () were extracted and measured according to the methods described by Zhu et al. (1990).
Hormone extraction and determination
Lyophilized leave (0.5 g) was ground into a powder. The powdered samples were extracted three times in 5 mL of cold 80% methanol (v/v) mixed with 30 μg/mL of sodium diethyldithiocarbamate. The extracts were centrifuged at 8000 g and 4 °C for 10 min. The supernatant was concentrated to dryness under vacuum. The residue was dissolved in 4 mL of 0.4 m phosphate buffer (pH 8.0). The solution was mixed with 4 mL of trichloromethane to remove the pigment. After centrifuging at 8000 g and 4 °C for 10 min, the aqueous phase was collected and supplemented with polyvinylpyrrolidone to remove the phenolics, and then centrifuged at 8000 g and 4 °C for 10 min. The supernatant was extracted with 4 mL of ethyl acetate (pH 3.0) twice. The upper phase was collected and concentrated to dryness under vacuum. The residue was dissolved in 1 mL of 0.5% (v/v) acetic acid/methanol (55 : 45, v/v) and finally filtered through a 0.45 μm filter. The obtained solution was used to determine ABA, GA3, IAA and SA. Extractions of free and conjugated SA were performed as described by Fragnière et al. (2011).
The extracted hormones were quantified using a LC‐ESI‐MS (Thermo, San Jose, CA) instrument. Ten microlitres of the sample was injected into a Thermo Scientific Hypersil Gold column (50 × 2.1 mm, 1.9 μm) in the Thermo Scientific Ultimate 3000 HPLC system (Thermo, San Jose, CA). The HPLC solvents were as follows: A, 0.04% acetate acid in water and B, methanol (0.4 mL/min). The two mobile phases were used in the gradient mode under the following time/concentration (min/%) of B: 0.0/20, 0.5/20, 2.5/90, 3.5/90, 3.6/20 and 5.0/20. Detection and quantification were performed using the TSQ Quantum Access MAX system (Thermo). SA was detected in the ESI negative mode and selected reaction monitoring with the following parameters: parent mass by charge (m/z) of 263.1, daughter mass by charge (m/z) of 153.0 and collision energy of 14 eV. The parameters for the ion source were set as follows: ion spray voltage of 3000 V, temperature of 350 °C, collision gas pressure of 1.5 mTorr, sheath gas of 25 arbitrary units and auxiliary gas of 15 arbitrary units. The amount of free and conjugated SA was calculated in mg/g DW with reference to the amount of internal standard (ortho‐anisic acid).
Sequence‐based DGE
Sample preparation and sequencing were performed with the Illumina Gene Expression Sample Prep kit and Solexa Sequencing Chip (flowcell) according to the manufacturer's introductions on the Illumina Cluster Station and Illumina HiSeq™ 2000 system (Illumina Inc., San Diego, CA). The sequencing lengths of raw reads were 49 bp in each line of the flowcell tunnel. The raw sequences were transformed into 17 bp clean tags, and tag counting was carried out using the Illumina Pipeline.
All tags were annotated using the apple genome database (http://genomics.research.iasma.it/). Briefly, a preprocessed database containing all possible CATG + 17‐base tag sequences was created. All clean tags were mapped to the reference sequence, and only 1 bp mismatch was considered. Clean tags mapped to the reference sequences from multiple genes were filtered. The remainder clean tags were designed as unambiguous clean tags. The number of unambiguous clean tags for each gene was calculated, and then normalized to TPM (number of transcripts per million clean tags).
Finally, a rigorous algorithm developed by the Beijing Genomics Institute (BGI) referring to ‘the significance of digital gene expression profiles’ (false discovery rate < 0.001) (Audic and Claverie, 1997) was used to identify differentially expressed genes between two samples, and absolute value of log2ratio ≥1 (minimum of twofold difference) was used as the threshold to judge the significance of gene expression differences.
Conflict of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1 Conserved domains (a) and sequence alignment (b) of MdcyMDH and the other four cyMDH isoforms.
Figure S2 Expression modifications of the selected DGE genes related to SA biosynthesis and signalling, mitochondrial and chloroplast metabolism, redox and abiotic stress tolerance in response to cold and salt treatments.
Table S1 Primer sequence for real‐time quantitative RT‐PCR.
Table S2 Marker enzyme activities in isolated cytosolic and mitochondrial fractions of the leaves under normal growth conditions.
Acknowledgements
This work was supported by the Special Financial Grant from the China Postdoctoral Science Foundation (2012T50623), the General Financial Grant from the China Postdoctoral Science Foundation (2011M501157), China's Agricultural Research System (CARS‐30‐ZP‐06), and the Changjiang Scholars and Innovative Research Team in University (IRT1155).
References
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Asensi‐Fabado, M. and Munné‐Bosch, S. (2011) The aba3‐1 mutant of Arabidopsis thaliana withstands moderate doses of salt stress by modulating leaf growth and salicylic acid levels. J. Plant Growth Regul. 30, 456–466. [Google Scholar]
- Audic, S. and Claverie, J.M. (1997) The significance of digital gene expression profiles. Genome Res. 7, 986‐995. [DOI] [PubMed] [Google Scholar]
- Bartoli, C.G. , Casalongué, C.A. , Simontacchi, M. , Marquez‐Garcia, B. and Foyer, C.H. (2013) Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environ. Exp. Bot. 94, 73–88. [Google Scholar]
- Beeler, S. , Liu, H.C. , Stadler, M. , Schreier, T. , Eicke, S. , Lue, W.L. , Truernit, E. et al (2014) Plastidial NAD‐Dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis . Plant Physiol. 164, 1175–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman, O. and Demmig, B. (1987) Photon yield of O2 evolution and chlorophyll fluorescence characteristics at 77K among vascular plants of diverse origins. Planta, 170, 489–504. [DOI] [PubMed] [Google Scholar]
- Borsani, O. , Valpuesta, V. and Botella, M.A. (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 126, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Ceciliani, F. , Caramori, T. , Ronchi, S. , Tedeschi, G. , Mortarino, M. and Galizzi, A. (2000) Cloning, overexpression, and purification of Escherichia coli quinolinate synthetase. Protein Expr. Purif. 18, 64–70. [DOI] [PubMed] [Google Scholar]
- Cela, J. , Chang, C. and Munné‐Bosch, S. (2011) Accumulation of g‐ rather than a‐tocopherol alters ethylene signaling gene expression in the vte4 mutant of Arabidopsis thaliana . Plant Cell Physiol. 52, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J. , Sun, L.L. , Liu, P.D. , Liu, G.D. , Tian, J. and Liao, H. (2015) Malate synthesis and secretion mediated by a manganese‐enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis . Plant Physiol. 167, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, O. , Whelan, J. and Millar, A.H. (2003) Molecular definition of the ascorbate/glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877. [DOI] [PubMed] [Google Scholar]
- Choudhury, S. , Panda, P. , Sahoo, L. and Panda, S.K. (2013) Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 8, 23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas, I.C. and Podestá, F.E. (2000) Purification and physical and kinetic characterization of an NAD+‐dependent malate dehydrogenase from leaves of pineapple (Ananas comosus). Physiol. Plant. 108, 240–248. [Google Scholar]
- Ding, Y. and Ma, Q.H. (2004) Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate reduction in wheat. Biochimie, 86, 509–518. [DOI] [PubMed] [Google Scholar]
- Feng, X.M. , Zhao, Q. , Zhao, L.L. , Qiao, Y. , Xie, X.B. , Li, H.F. , Yao, Y.X. et al (2012) The cold‐induced basic helix‐loop‐helix transcription factor gene MdCIbHLH1 encodes an ICE‐like protein in apple. BMC Plant Biol. 12, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragnière, C. , Serrano, M. , Abou‐Mansour, E. , Métraux, J. and L'Haridon, F. (2011) Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 585, 1847–1852. [DOI] [PubMed] [Google Scholar]
- Gietl, C. (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim. Biophys. Acta, 1100, 217–234. [DOI] [PubMed] [Google Scholar]
- Green, M.A. and Fry, S.C. (2005) Vitamin C degradation in plant cells via enzymatic hydrolysis of 4‐O‐oxalyl‐l‐threonate. Nature, 433, 83–87. [DOI] [PubMed] [Google Scholar]
- Guo, L. , Devaiah, S.P. , Narasimhan, R. , Pan, X.Q. , Zhang, Y.Y. , Zhang, W.H. and Wang, X.M. (2012) Cytosolic glyceraldehyde‐3‐phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell, 24, 2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Chaouch, S. , Mhamdi, A. , Queval, G. , Zechmann, B. and Noctor, G. (2013) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal. 18, 2106–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, S. , Motohashi, K. , Arisaka, F. , Romano, P.G.N. , Hosoya‐Matsuda, N. , Kikuchi, N. , Fusada, N. et al (2006) Thioredoxin‐h1 reduces and reactivates the oxidized cytosolic malate dehydrogenase dimer in higher plants. J. Biol. Chem. 281, 32065–32071. [DOI] [PubMed] [Google Scholar]
- Hashida, S.N. , Takahashi, H. and Uchimiya, H. (2009) The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 103, 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyno, E. , Innocenti, G. , Lemaire, S.D. , Issakidis‐Bourguet, E. and Krieger‐Liszkay, A. (2014) Putative role of the malate valve enzyme NADP–malate dehydrogenase in H2O2 signalling in Arabidopsis . Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev, A.U. and Gardeström, P. (2003) Regulation of NAD‐and NADP‐dependent isocitrate dehydrogenases by reduction levels of pyridine nucleotides in mitochondria and cytosol of pea leaves. Biochim. Biophys. Acta, 1606, 117–125. [DOI] [PubMed] [Google Scholar]
- Jayakannan, M. , Bose, J. , Babourina, O. , Shabala, S. , Massart, A. , Poschenrieder, C. and Rengel, Z. (2015) The NPR1‐dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis . J. Exp. Bot. 66, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.H. , Bae, J.M. and Huh, G.H. (2010) Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweetpotato in response to plant developmental stage and environmental stress. Plant Cell Rep. 29, 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, M. , Issakidis, E. , Ruelland, E. , Decottignies, P. and Miginiac‐Maslow, M. (1996) An active‐site cysteine of sorghum leaf NADP‐malate dehydrogenase studied by site‐directed mutagenesis. FEBS Lett. 382, 137–140. [DOI] [PubMed] [Google Scholar]
- Li, X. , Ahammed, G.J. , Zhang, Y.Q. , Zhang, G.Q. , Sun, Z.H. , Zhou, J. , Zhou, Y.H. et al (2015) Carbon dioxide enrichment alleviates heat stress by improving cellular redox homeostasis through an ABA‐independent process in tomato plants. Plant Biol. 17, 81–89. [DOI] [PubMed] [Google Scholar]
- Mateo, A. , Funck, D. , Mühlenbock, P. , Kular, B. , Mullineaux, P.M. and Karpinski, S. (2006) Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57, 1795–1807. [DOI] [PubMed] [Google Scholar]
- Maxwell, K. and Johnson, G.N. (2000) Chlorophyll fluorescence‐a practical guide. J. Exp. Bot. 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Munné‐Bosch, S. , Queval, G. and Foyer, C.H. (2013) The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol. 161, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, M. (2009) How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor, G. and Foyer, C.H. (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Biol. 49, 249–279. [DOI] [PubMed] [Google Scholar]
- Nunes‐Nesi, A. , Carrari, F. , Lytovchenko, A. , Smith, A.M.O. , Loureiro, M.E. , Ratcliffe, R.G. , Sweetlove, L.J. et al (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond, B. and Makino, A. (1991) Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol. 96, 355–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi, F. , Longet, D. , Penel, C. and Dunand, C. (2004) The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry, 65, 1879–1893. [DOI] [PubMed] [Google Scholar]
- Peleg, Z. and Blumwald, E. (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14, 290–295. [DOI] [PubMed] [Google Scholar]
- Pétriacq, P. , de Bont, L. , Hager, J. , Didierlaurent, L. , Mauve, C. , Guérard, F. , Noctor, G. et al (2012) Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst‐AvrRpm 1 . Plant J. 70, 650–665. [DOI] [PubMed] [Google Scholar]
- Potters, G. , Horemans, N. and Jansen, M.A.K. (2010) The cellular redox state in plant stress biology‐A charging concept. Plant Physiol. Biochem. 48, 1–9. [DOI] [PubMed] [Google Scholar]
- Queval, G. and Noctor, G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 363, 58–69. [DOI] [PubMed] [Google Scholar]
- Raghavendra, A.S. and Padmasree, K. (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8, 546–553. [DOI] [PubMed] [Google Scholar]
- Ramel, F. , Birtic, S. , Ginies, C. , Soubigou‐Taconnat, L. , Triantaphylidès, C. and Havaux, M. (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl Acad. Sci. USA, 109, 5535–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe, R. (2004) Malate valves to balance cellular energy supply. Physiol. Plant. 120, 2126. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Koussevitzky, S. , Mittler, R. and Miller, G. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M. and Miyake, H. (2012) Redox‐shuttling between chloroplast and cytosol: integration of intra‐chloroplast and extra‐chloroplast metabolism. Curr. Opin. Plant Biol. 15, 252–260. [DOI] [PubMed] [Google Scholar]
- Tomaz, T. , Bagard, M. , Pracharoenwattana, I. , Lindén, P. , Lee, C.P. , Carroll, A.J. , Stroher, E. et al (2010) Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis . Plant Physiol. 154, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q.F. , Zhao, Y. , Yi, Q. , Li, K.Z. , Yu, Y.X. and Chen, L.M. (2010) Overexpression of malate dehydrogenase in transgenic tobacco leaves: enhanced malate synthesis and augmented Al‐resistance. Acta Physiol. Plant, 32, 1209–1220. [Google Scholar]
- Yao, Y.X. , Li, M. , Liu, Z. , Hao, Y.J. and Zhai, H. (2007) A novel gene, screened by cDNA‐AFLP approach, contributes to lowering the acidity of fruit in apple. Plant Physiol. Biochem. 45, 139–145. [DOI] [PubMed] [Google Scholar]
- Yao, Y.X. , Hao, Y.J. , Li, M. , Pang, M.L. , Liu, Z. and Zhai, H. (2008) Gene cloning expression and enzyme activity assay of a cytosolic malate dehydrogenase from apple fruits. Acta Horticult. Sin. 35, 181–188 (in Chinese, with English abstract). [Google Scholar]
- Yao, Y.X. , Dong, Q.L. , Zhai, H. , You, C.X. and Hao, Y.J. (2011a) The functions of an apple cytosolic malate dehydrogenase gene in growth and tolerance to cold and salt stresses. Plant Physiol. Biochem. 49, 257–264. [DOI] [PubMed] [Google Scholar]
- Yao, Y.X. , Li, M. , Zhai, H. , You, C.X. and Hao, Y.J. (2011b) Isolation and characterization of an apple cytosolic malate dehydrogenase gene reveal its function in malate synthesis. J. Plant Physiol. 168, 474–480. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Mogami, J. and Yamaguchi‐Shinozaki, K. (2014) ABA‐dependent and ABA‐independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. [DOI] [PubMed] [Google Scholar]
- Zhu, G.L. , Zhang, H.W. and Zhang, A.Q. (1990) Plant Physiological Experiments, 1st edn Beijing: Peking University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Conserved domains (a) and sequence alignment (b) of MdcyMDH and the other four cyMDH isoforms.
Figure S2 Expression modifications of the selected DGE genes related to SA biosynthesis and signalling, mitochondrial and chloroplast metabolism, redox and abiotic stress tolerance in response to cold and salt treatments.
Table S1 Primer sequence for real‐time quantitative RT‐PCR.
Table S2 Marker enzyme activities in isolated cytosolic and mitochondrial fractions of the leaves under normal growth conditions.


