Summary
Objective
Obesity is a metabolic disease. However, the underlying molecular mechanisms linking metabolic profiles and weight gain are largely unknown.
Methods
Here, we used semi‐targeted metabolomics to assay 156 metabolites selected from 25 key metabolic pathways in plasma samples from 300 non‐smoking healthy women identified from Mano‐A‐Mano, the Mexican American Cohort study. The study subjects were randomly divided into two cohorts: training (N = 200) and testing (N = 100) cohorts. Linear regression and Cox proportional hazard regression were used to assess the effect of body mass index (BMI) at baseline on metabolite levels and the effects of metabolites on significant weight gain during a 5‐year follow‐up.
Results
At baseline, we observed 7 metabolites significantly associated with BMI in both training and testing cohorts. They were Methyl succinate, Asparagine, Urate, Kynurenic acid, Glycine, Glutamic acid, and Serine. In further analysis, we identified 6 metabolites whose levels at baseline predicted significant weight gain during 5‐year follow‐up in both cohorts. They were Acetylcholine, Leucine, Hippuric acid, Acetylglycine, Urate, and Xanthine.
Conclusions
The findings establish the baseline metabolic profiles for BMI, and suggest new metabolic targets for researchers attempting to understand the molecular mechanisms of weight gain and obesity.
Keywords: Body mass index (BMI), metabolites, Mexican Americans, weight gain
Introduction
According to the CDC National Health Survey conducted from 2009 to 1012, 40.2% of Mexican origin adult men and 45.2% Mexican origin adult women are obese 1. More alarmingly, the rate of obesity has been steadily increasing. In the early 1980s, only 21% of Mexican Americans were obese. By the middle 2000s, the rate was close to 35%. Obesity has also been linked to a variety of chronic diseases, including diabetes, cardiovascular disease, and cancers 2, 3, 4, 5, 6. Clearly, obesity and related chronic diseases have posed enormous threats to the health of Mexican Americans.
To reduce such huge burden, understanding the underlying molecular mechanisms of how body weight is gained and obesity is developed and consequently developing interventions and therapeutic agents that can facilitate weight loss and conquest obesity are critically needed. In the past several years, metabolomics has been introduced in the obesity research 7. Metabolomic platforms quantify small‐molecule metabolites in biospecimens and can be used to evaluate the role of metabolic alterations in chronic disease. Because it takes into account genetic regulation, altered kinetic activity of enzymes, genomics and proteomics, metabolomics reflects changes in phenotype, and thereby function 8, 9. Thus, the application of metabolomics in obesity research can help elucidate the molecular mechanisms of obesity and weight gain. Using blood samples from 947 participants identified from three cohort studies, Moore et al. identified a total of 37 metabolites associated with body mass index (BMI) in a cross‐sectional analysis 10. In a few other studies, several amino acids (e.g. branched‐chain and aromatic amino acids) were associated with an increased BMI and levels of these amino acids were later found to predict future diabetes risk 11, 12, 13, 14, 15. Weight loss upon behavioral intervention was associated with changes in the blood metabolome 16, 17. In addition, Wahl et al. found that long‐term weight change was globally associated with serum metabolite concentrations 18. However, none of those studies include Mexican American study participants. More importantly, none of those studies have investigated whether metabolites might predict future weight gain prospectively.
To fill the gap, we performed metabolic profiling in plasma samples from 300 healthy Mexican American women from Mano‐A‐Mano, the Mexican American Cohort study. The study subjects were randomly divided into two cohorts: training (N = 200) and testing (N = 100) cohorts. The training cohort had 200 and the testing cohort had 100 study subjects. The objectives were two‐folded. First, we examined cross‐sectional associations between metabolite levels with BMI at baseline; second, we investigated whether baseline metabolite levels predicted significant weight gain during a 5‐year follow‐up.
Materials and methods
Study population
The samples for the current study were drawn from participants in a large population‐based cohort of Mexican origin households recruited from the Houston‐area. This ongoing prospective cohort of 1st and 2nd generation Mexican origin immigrant households in Houston, Texas, was initiated in July 2001 and maintained by the Department of Epidemiology at the University of Texas MD Anderson Cancer Center. A detailed description of the sampling and recruitment strategy has been published previously 19. Briefly, participants have been recruited through block walking in predominantly Mexican American neighborhoods, from community centers and local health clinics, and networking through currently enrolled participants. Of the identified eligible households, ~88% agreed to participate in the study. After written informed consent was obtained, trained bilingual research interviewers conducted a structured face‐to‐face interview lasting ~45 minutes, using a standardized and validated questionnaire in the participant's preferred language, either Spanish or English. The questionnaire elicited information on birthplace and residential history, social‐demographic characteristics, lifestyle behaviors, levels of physical activity, personal medical history, family history of chronic disease, acculturation, and occupational exposure. Participants have been actively followed up via annual telephone re‐contact to update body weight, selected exposures and new diagnosis of selected chronic diseases, including cancer, diabetes, and hypertension. Current study included 300 healthy female non‐smokers who have no major chronic diseases reported at baseline (e.g. hypertension, cardiovascular disease, diabetes, and cancer). BMI at baseline was calculated using measured height and weight data during the in‐person interview according to a standard protocol by trained interviewers. BMI during follow‐up was calculated using self‐reported weight and height at baseline. The study protocol was approved by the Institutional Review Board of The University of Texas M. D. Anderson Cancer Center.
Metabolic profiling
Metabolomics profiles were obtained using Liquid chromatography triple quadrupole mass spectrometry (LC‐QQQ‐MS) at Northwest Metabolomics Research Center, University of Washington. Semi‐targeted metabolomics analysis was performed. Briefly, LC‐MS/MS was performed using an electrospray ionization source and the multiple‐reaction‐monitoring mode. A Sciex 5500 QTRAP triple quad MS system equipped with an Agilent 1200 ultra‐high‐pressure liquid chromatography system was utilized. The MS acquisition for each plasma sample (0.050 mL each) target a list of 156 metabolites. In addition, 2 isotope‐labeled standards were included in each sample run for quality control and comparisons with other studies or additional samples run at a different time. The average CV of relative intensities for instrument QC samples was 5.3% for the detected metabolites and the average CV of quantified metabolites using isotope‐labeled internal standards was 3%, indicating good reproducibility. The targeted 156 metabolites are selected from 25 key metabolic pathways, including Alanine, Aspartate, and Glutamate metabolism; Arginine and Proline metabolism; Butanoate metabolism; the Citrate cycle (TCA cycle); Cysteine and Methionine metabolism; Fatty acid metabolism; Glutathione metabolism; Glycine, Serine, and Threonine metabolism; Glycolysis/Gluconeogenesis; Histidine metabolism; Lysine biosynthesis; Lysine degradation; Nitrogen metabolism; Oxidative phosphorylation; Pentose phosphate pathway; Phenylalanine metabolism; Phenylalanine, Tyrosine, and Tryptophan biosynthesis; Purine metabolism; Pyrimidine metabolism; Pyruvate metabolism; Synthesis and degradation of ketone bodies; Tryptophan metabolism; Tyrosine metabolism; Valine, Leucine, and Isoleucine biosynthesis; and Valine, Leucine, and Isoleucine degradation. Certain metabolites are involved in multiple metabolic pathways. The raw data were processed using MultiQuant software (AB SCIEX) to integrate chromatographic peaks, and the data were visually inspected to ensure the quality of signal integration. The peak area was calculated for each individual metabolite.
Statistical analysis
Log‐transformed metabolite data were used in the analysis. Missing values were assumed to be below the limits of detection, and these values were imputed with the compound minimum (minimum value imputation). Training and testing cohorts were analyzed separately, but using the same approach. First, we examined the effect of baseline BMI as continuous variables in relation to metabolite levels using linear aggression, adjusting for age, HbA1c, acculturation score, nativity, years of living in U.S., sedentary lifestyle, sitting time, and biospecimen storage time. The study participants were further grouped into 5 categories based on their BMI, including normal weight (<25), overweight (25‐29.90), class I obesity (30‐34.90), class II obesity (35‐39.90), and class III obesity (40 and above). ANOVA analysis was performed to determine the relationship between baseline BMI category and metabolite levels. To assess whether associations were independent, we evaluated the pairwise correlations between all metabolites significantly associated with BMI. Metabolites whose pairwise correlations greater than 0.5 were considered highly correlated and to have possible redundancy.
During the 5‐year follow‐up, each study participant had at least 3 self‐reported body weights. Because the focus of this study was on weight gain, 15 study subjects who lost weight during follow‐up were excluded from further analysis. Among them, 10 were from the training cohort and 5 were from the testing cohort. To assess the levels of weight gain during the follow‐up, we created a variable, termed “significant weight gain”, which was defined as increase in BMI by at least one category between the baseline and the follow‐up. The creation was based on several reasons. First, it was considered major weight gain in relation to the time horizon of the prediction comprising 5 years. Second, it seems high enough to exclude random variation in body weight while simultaneously allowing for some weight gain as natural part of the aging process. Last, self‐reported BMI was used during the follow‐up. Self‐reported BMI correlates reasonably well with measured BMI values in adults 20, but misclassification is more common among Mexican‐American women than men 21. Cox regression analysis was performed to assess the association of metabolite levels with significant weight gain, adjusting for co‐variates as appropriate. The results from two cohorts were compared. Significant Metabolites in both cohorts were noted. All analyses were performed with STATA software version 10.1 (STATA, College Station, TX) and the R statistical language version 3.2.2.
Results
At baseline, 47.5 and 57.0% of study subjects in the training and testing cohorts were obese (Table 1). The obesity prevalence was further increased to 58.0 and 63.0% during the follow‐up. The mean age was 40 for both training and testing cohorts. The two cohorts were comparable with regard to baseline HbA1c levels, nativity, years living in U.S., acculturation score, and sitting time. However, study participants in the training cohort were less likely to have a sedentary lifestyle than those in the testing cohort (81.5 vs 88.0%).
Table 1.
Basic characteristics of study participants in the training and testing cohorts
| Characteristics | Training (n = 200) | Testing (n = 100) |
|---|---|---|
| BMI at baseline, mean (SD) | 30.6 (5.9) | 31.5 (6.0) |
| BMI category at baseline (kg/m2) | ||
| <25.0 | 32 (16.0%) | 8 (8.0%) |
| 25.0‐29.9 | 73 (36.5%) | 35 (35.0%) |
| 30.0‐34.9 | 54 (27.0%) | 33 (33.0%) |
| 35.0‐39.9 | 24 (12.0%) | 14 (14.0%) |
| 40.0 and above | 17 (8.5%) | 10 (10.0%) |
| BMI at follow‐up, mean (SD) | 31.8 (6.0) | 32.6 (5.6) |
| BMI category at last follow‐up | ||
| <25.0 | 16 (8.0%) | 5 (5.0%) |
| 25.0‐29.9 | 68 (34.0%) | 32 (32.0%) |
| 30.0‐34.9 | 67 (33.5%) | 33 (33.0%) |
| 35.0‐39.9 | 30 (15.0%) | 22 (22.0%) |
| 40.0 and above | 19 (9.5%) | 8 (8.0%) |
| HbA1C, mean (SD) | 4.9 (0.83) | 4.9 (0.44) |
| Follow‐up time (days) | ||
| mean (SD) | 990 (450) | 1459 (326) |
| median (range) | 957 (229‐1825) | 1481 (932‐1825) |
| Age at baseline | ||
| mean (SD) | 40 (10.7) | 40 (10.7) |
| median (range) | 38 (20‐72) | 38 (21‐69) |
| Nativity | ||
| U.S. | 35 (17.5%) | 13 (13.0%) |
| Mexico | 165 (82.5%) | 87 (87.0%) |
| Years living in U.S., mean (SD) | 19.6 (13.9) | 19.9 (14.0) |
| Acculturation score, mean (SD) | 2.25 (0.92) | 2.10 (0.90) |
| Sedentary lifestyle | ||
| no | 38 (19.0%) | 12 (12.0%) |
| yes | 162 (81.0%) | 88 (88.0%) |
| Sitting hours/day, mean (SD) | 2.1 (1.0) | 2.0 (1.0) |
In the training and testing cohorts, we identified a total of 20 and 18 metabolites significantly associated with BMI at baseline after adjusting for age, HbA1c, acculturation score, nativity, years living in U.S., sedentary lifestyle, sitting time, and biospecimen storage time (Table 2). Among them, 7 were observed in both training and testing cohorts, with effect sizes in the same direction and of similar magnitudes between the two cohorts. The specific metabolites, their effect size calculated per 1 unit BMI increase, and P value are shown in Table 3. They were Methyl succinate, Asparagine, Urate, Kynurenic acid, Glycine, Glutamic acid, and Serine. Methyl succinate was the most significant metabolite in the training cohort (P = 0.001). Each 1 unit elevation in BMI was associated with a 4.75% and 3.82% decrease in standard deviation in Methyl succinate levels in the training and testing cohorts. In the pairwise analysis of 7 significant metabolites in the training cohort, the only highly correlated associations were observed between Methyl succinate and Asparagine (ρ = 0.965, P < 0.001), Asparagine and Serine (ρ = 0.543, P < 0.001), and Glycine and Serine (ρ = 0.513, P < 0.001). Neither Glutamic acid, Kynurenic acid, nor Urate was highly correlated with any other metabolites. Similar associations were observed in the testing cohort.
Table 2.
Association of metabolites with BMI at baseline in the training and testing cohorts*
| Training | Testing | ||
|---|---|---|---|
| Metabolite | P value | Metanolite | P value |
| Acetylglycine | 0.004 | Allantoin | 0.037 |
| Asparagine | 0.002 | Asparagine | 0.027 |
| Biotine | 0.003 | Carnitine | 0.027 |
| Choline | 0.015 | Glycocholine | 0.042 |
| Dimethylglycine | 0.048 | Cystathionine | 1.0E‐3 |
| Glutamic acid | 0.034 | Glutamic acid | 0.005 |
| Glycerate | 0.031 | Glycine | 0.037 |
| Glycine | 0.026 | Guanidinoacetate | 0.021 |
| Homocysteine | 0.030 | Histidine | 0.002 |
| Hydrooxyguanosine | 0.011 | Hydroxyguanosine | 2.4E‐05 |
| Isoleucine | 0.025 | Kynurenic acid | 0.024 |
| Kynurenic acid | 0.006 | kynurenine | 0.043 |
| Leucine | 0.025 | Methyl succinate | 0.006 |
| Methyl succinate | 0.001 | Phenylalanine | 0.034 |
| Ornithine | 0.007 | Serine | 0.005 |
| Serine | 0.036 | TMA | 0.041 |
| Sorbitol | 0.036 | Urate | 0.041 |
| Tyrosine | 0.038 | Xanthine | 0.033 |
| Urate | 0.003 | ||
| Valine | 0.022 | ||
Linear regression model includes age, acculturation score, nativity, years living in U.S., BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
Table 3.
Association of metabolites with BMI at baseline in the training and testing cohorts at baseline*
| Training | Testing | |||
|---|---|---|---|---|
| Metabolite | Effect size# | P value | Effect size# | P value |
| Methyl succinate | ‐4.35% | 0.001 | ‐3.82% | 0.006 |
| Asparagine | ‐4.11% | 0.002 | ‐4.14% | 0.027 |
| Urate | 3.92% | 0.003 | 3.60% | 0.041 |
| Kynurenic acid | 3.34% | 0.006 | 4.06% | 0.024 |
| Glycine | ‐2.91% | 0.026 | ‐3.79% | 0.037 |
| Glutamic acid | 2.54% | 0.034 | 5.06% | 0.005 |
| Serine | ‐2.29% | 0.036 | ‐4.88% | 0.005 |
. Linear regression model includes age, acculturation score, nativity, years living in U.S., BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
. Effect size indicates the percentage change in standard deviation in metabolite levels per one BMI unit increase for BMI.
We further investigated whether levels of metabolites differed by BMI category. In the training cohort, 20 metabolites were significantly different by BMI category (Table 4). Among them, 8 remained significant in the testing cohort. They were Urate (P = 0.001 and 0.043), Methyl succinate (P = 0.004 and 0.015), Glutamic acid (P = 0.007 and 0.003), Asparagine (P = 0.008 and 0.047), Glycine (P = 0.010 and 0.047), and Acetyl glycine (P = 0.027 and 0.008), Serine (P = 0.028 and 0.006), and Kynurenic acid (P = 0.045 and 0.025). The levels of the top 4 validated significant metabolites by BMI category in the training cohort were shown in Figure 1.
Table 4.
Association of metabolites with BMI category at baseline in the training and testing cohorts*
| Training | Testing | ||
|---|---|---|---|
| Metabolite | P value | Metabolite | P value |
| Acetoacetate | 0.045 | Acetyl glycine | 0.008 |
| Acetyl glycine | 0.027 | Asparagine | 0.047 |
| Asparagine | 0.008 | Carnitine | 0.043 |
| Carnitine | 0.020 | Cystathionine | 0.031 |
| Glucose | 0.031 | Glutamic acid | 0.003 |
| Glutamic acid | 0.007 | Glycine | 0.047 |
| Glycine | 0.010 | Guanidinoacetate | 0.036 |
| Guanidinoacetate | 0.011 | Hydroxyguanosine | 0.023 |
| Histidine | 0.044 | Kynurenic acid | 0.025 |
| Homocysteine | 0.011 | kynurenine | 0.043 |
| Isoleucine | 0.028 | Methyl succinate | 0.015 |
| Kynurenic acid | 0.045 | Phenylalanine | 0.024 |
| Leucine | 0.028 | Serine | 0.006 |
| Methyl succinate | 0.004 | TMA | 0.043 |
| Nitrotyrosine | 0.033 | Urate | 0.043 |
| Ornithine | 0.012 | Xanthine | 0.027 |
| Serine | 0.028 | ||
| Tyrosine | 0.001 | ||
| Urate | 0.001 | ||
| Valine | 0.019 | ||
Linear regression model includes age, acculturation score, nativity, years living in U.S., BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
Figure 1.
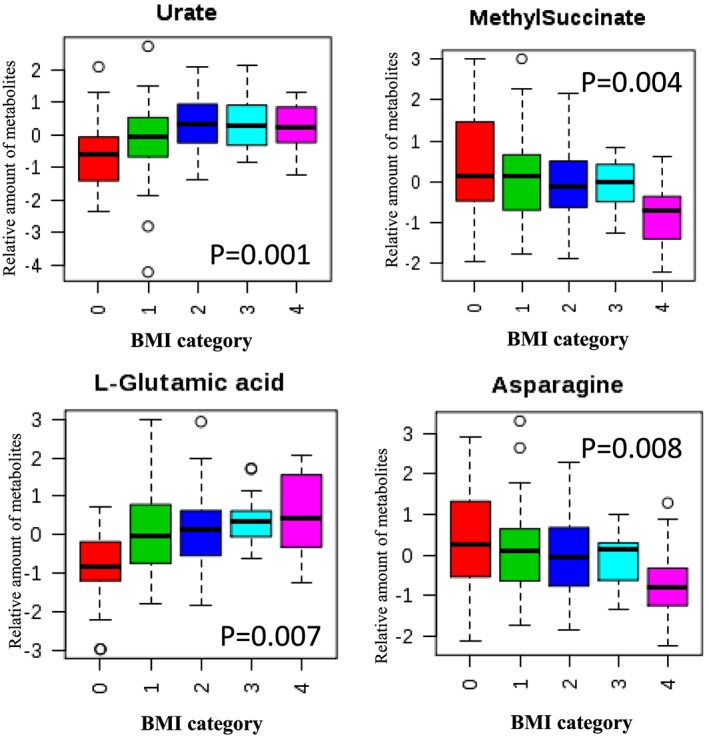
Box‐plot of top 4 metabolites differed by BMI category in the discovery cohort. BMI category is defined as 0: normal weight (<25); 1: overweight (25‐29.90); 2: class I obesity (30‐34.90); 3: class II obesity (35‐39.90); 4: class III obesity (40 and above).
Further, we explored whether metabolite levels at baseline might prospectively predict significant weight gain during 5‐year follow‐up. In the training and testing cohorts, 55 (28.2%) and 22 (22.0%) study subjects, respectively, were observed to have an increase in BMI category during the 5‐year follow‐up. In the training cohort, a total of 22 metabolites were observed to predict significant weight gain during follow‐up after adjusting for age, acculturation, nativity, years living in U.S., baseline BMI, HbA1c, sedentary lifestyle, sitting time, and biospecimen storage time (Table 5). The most significant metabolite based on P value was Acetylcholine (P < 1.0E‐05). With one standard deviation in levels of Acetylcholine increase, the risk of having significant weight gain during follow‐up was doubled (HR = 2.07, 95% CI: 1.44, 2.99). In the testing cohort, 6 of the 22 significant metabolites from the training cohort were validated. They were Acetylcholine, Leucine, Hippuric acid, Acetylglycine, Urate, and Xanthine. An elevated risk of significant weight gain during follow‐up was associated with High levels of Acetylcholine, Leucine, Acetylglycine, and Urate, but with reduced levels of Hippuric acid and Xanthine. In the testing cohort, when all 6 metabolites were included in the model together, the risk of having significant weight gain during follow‐up remained significant for Leucine (HR = 1.49, 95%CI: 1.05, 3.56), Hippuric acid (HR = 0.78, 95%CI: 0.45, 0.89), and Urate (HR = 1.46, 95%CI: 1.08, 2.43) (Table 6). For all 6 metabolites, same analysis was performed using BMI as a continuous variable. Similar associations were observed (data not shown).
Table 5.
Individual metabolites predicting significant weight gain during follow‐up*
| Metabolites | HR (95%CI)# | P value | HR (95%CI)# | P value |
|---|---|---|---|---|
| Training | Testing | |||
| Acetylcholine | 2.07 (1.44, 2.99) | <1.0E‐05 | 1.46 (1.07, 4.55) | 0.032 |
| Transaconitic acid | 2.11 (1.36, 3.04) | 1.80E‐05 | 1.43 (0.85, 4.69) | 0.142 |
| Leucine | 2.23 (1.31, 3.80) | 5.10E‐05 | 1.54 (1.09, 5.23) | 0.025 |
| Aminoadipic acid | 1.36 (1.13, 1.64) | 2.24E‐04 | 1.73 (0.78, 4.32) | 0.265 |
| Leucic acid | 1.50 (1.22, 1.83) | 4.48E‐04 | 1.32 (0.56, 1.86) | 0.548 |
| Methyladenosine | 1.48 (1.17, 1.72) | 4.65E‐04 | 1.24 (0.73, 2.57) | 0.436 |
| Pantothenic acid | 1.25 (1.19, 1.57) | 0.001 | 1.09 (0.54, 1.73) | 0.642 |
| Betaine | 1.72 (1.21, 2.45) | 0.002 | 1.44 (0.73, 2.32) | 0.473 |
| Hippuric acid | 0.80 (0.29, 0.90) | 0.003 | 0.77 (0.52, 0.95) | 0.029 |
| Hydroxyguanosine | 1.96 (1.21, 3.15) | 0.005 | 1.35 (0.81, 3.06) | 0.352 |
| Cysteamine | 1.29 (1.14, 1.67) | 0.006 | 1.05 (0.73, 1.89) | 0.436 |
| Acetylglycine | 1.44 (1.13, 1.63) | 0.008 | 1.56 (1.01, 2.78) | 0.040 |
| Urate | 1.54 (1.18, 3.07) | 0.008 | 1.53 (1.09, 2.87) | 0.025 |
| Alphadglucuronic acid | 1.75 (1.25, 2.18) | 0.009 | 1.23 (0.73, 2.79) | 0.658 |
| TMA | 1.39 (1.11, 1.92) | 0.018 | 1.09 (0.64, 4.52) | 0.821 |
| Fructose | 1.27 (1.08, 1.95) | 0.018 | 1.31 (0.88, 2.54) | 0.169 |
| Glucose | 1.26 (1.06, 2.02) | 0.02 | 1.29 (0.84, 3.02) | 0.243 |
| Guanidinoacetate | 0.60 (0.38, 0.92) | 0.021 | 0.69 (0.35, 1.04) | 0.077 |
| (s)‐b‐Aminoisobutyric acid | 1.41 (1.05, 1.89) | 0.024 | 1.03 90.56, 2.46) | 0.872 |
| Carnitine | 1.43 (1.03, 2.01) | 0.032 | 0.89 (0.41, 2.09) | 0.764 |
| Xanthine | 0.62 (0.39, 0.79) | 0.035 | 0.55 (0.34, 0.97) | 0.031 |
| Glycocholate | 1.24 (1.04, 1.53) | 0.038 | 1.05 (0.82, 1.25) | 0.762 |
. Cox hazard regression was performing adjusting age, acculturation score, nativity, years living in U.S., baseline BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
. The hazard ratio and 95% confidence interval were calculated as one standard deviation in levels of the individual metabolite increase.
Table 6.
Combined analysis to identify metabolites predicting weight gain during follow‐up*
| metabolite | HR (95% CI)# | P value |
|---|---|---|
| acetylcholine | 1,64 (1.06, 3.42) | 0.022 |
| Leucine | 1.49 (0.90, 3.56) | 0.073 |
| hippuric acid | 0.78 (0.45, 0.89) | 0.004 |
| acetylglycine | 1.15 (0.73, 1.98) | 0.436 |
| Urate | 1.46 (1.08, 2.43) | 0.032 |
| xanthine | 0.87 (0.41, 1.80) | 0.621 |
. Cox hazard regression was performing adjusting age, acculturation score, nativity, years living in U.S., baseline BMI, HbA1C, sedentary lifestyle, sitting time, and biospecimen storage time.
. The hazard ratio and 95% confidence interval were calculated as one standard deviation in levels of the individual metabolite increase.
Discussion
In the current study with 300 Mexican American non‐smoking healthy women, we identified 7 metabolites associated with baseline BMI. Furthermore, we identified 6 metabolites, namely, Acetylcholine, Leucine, Hippuric acid, Acetylglycine, Urate, and Xanthine that predicted significant weight gain during 5‐year follow‐up. In further combined analysis with all 6 metabolites in the model, Acetylcholine, Hippuric acid, and Urate remained significant.
Overall, our findings of BMI associated metabolites are consistent with previous findings in non‐Hispanic populations 10, 22, 23. Of 37 metabolites significantly associated with BMI in a meta‐analysis by Moore et al. 10, 13 were analyzed in the current study. Among them, 9 and 7 were significantly associated with BMI in the training and testing cohorts, respectively. And 4 were significant in both cohorts. In addition to those 4 metabolites, we observed 3 more significant metabolites, namely Urate, Serine, and Methylsuccinate. The linear positive relationship between serum Urate and BMI is previously reported in several population‐based studies 22, 24. In a study among youth with obesity and type‐2 diabetes, Mihalik et al. reported that Serine levels in fasting plasma were inversely associated with BMI 23. The only metabolite whose association with BMI has not been reported previously is Methylsuccinate. Methylsuccinate is involved in Isoleucine catabolism 25. Previous work also demonstrated that methyl esters of succinate are potent insulin secretagogues in pancreatic islets 26. On the other hand, Newgard et al. compared urinary levels of Methylsuccinate between obese and lean individuals and found no difference 11. Clearly, further assessment of the relationship between Methylsuccinate and BMI is needed.
In the individual metabolite analysis, we identified a total of 6 metabolites that could predict significant weight gain during follow‐up. And 3 of them remained significant when we included all 6 metabolites in multivariate analysis. Among them, Urate was the only metabolite that was significantly associated with BMI and also predicted significant weight gain. This may suggest the metabolic profile associated with BMI may not overlap with the one predicting weight gain. Given how quickly the individuals can gain significant amount of weight, research particularly focusing on metabolites predicting weigh gain is urgently needed.
Of the highest significance is the positive association between Acetylcholine levels and risk of significant weight gain. Our observation is consistent with the role of Acetylcholine in metabolic regulation. Acetylcholine is a neurotransmitter derived from an ester of acetic acid and choline. The central and peripheral Acetylcholine system plays a very important role in glucose and energy homeostasis 27. In animal models of obesity and hyperinsulinemia, obesity could lead to enhanced Acetylcholine release from neurons 27, 28, 29. More interestingly, in feeding studies, a possible link of choline, Acetylcholine, and obesity/diabetes has been suggested 30, 31, 32. Wu et al. found that choline deficiency attenuates body weight gain and improves glucose tolerance in obese mice 32. Acetylcholine exerts its functions by binding to two cell‐surface receptors: the nicotinic and muscarinic acetylcholine receptors 27. In American Indians, Zhu et al. reported that multiple variants in the nicotinic acetylcholine receptor gene family were jointly associated with abdominal obesity, independent of general obesity and cigarette smoking per se. Interestingly, In recent studies, the potential of muscarinic acetylcholine receptors as a therapeutic target of obesity has been proposed 33, 34.
Our observation of an inverse association between Hippuric acid and risk of significant weight gain is consistent with a previous report that urinary levels of Hippuric acid were significantly lower in obese individuals than normal weight controls 35. Hippuric acid is a gut flora‐derived metabolite from low‐molecular‐weight aromatic compounds and polyphenols generated by a range of gut microbes 36. Urinary levels of Hippuric acid have been shown to correlate with the obese phenotype in different animal models and to associate with the fecal counting of several bacterial species in a rat model of obesity 37, 38, 39. Thus, the observed relationship in the current study may suggest a link between gut microflura metabolism and obesity phenotype. Recent works have shown that obesity is associated with phylum‐level changes in the microbiota, reduced bacterial diversity, and altered representation of bacterial genes and metabolic pathways 40, 41. On the other hand, gut microbiome may modulate metabolic pathways, lead to weight loss and eventually reduce the obese burden 42.
Both Xanthine and Urate were associated with significant weight gain, but at different directions. This is consistent with the biological function of these two metabolites since Xanthine is converted to Urate by the action of the xanthine oxidase enzyme. In our study, Xanthine is highly correlated with Urate (ρ = 0.672, P < 0.001). Recent studies show that fructose‐induced Urate generation causes mitochondrial oxidative stress that stimulates fat accumulation independent of excessive caloric intake 43. Interestingly, we also observed a significant positive relationship between Fructose and significant weight gain in the training cohort. Fructose may increase the risk for weight gain by altering satiety, resulting in increased food intake. Thus, the results from this study support the roles of Urate and Fructose in obesogenic development.
The positive association between circulating levels of Leucine and BMI has been reported previously 10, 11 as well as in the discovery cohort. Leucine has also been reported as a predictor for type‐2 diabetes 11, 14. However, no study has reported Leucine as a positive predictor for weight gain. Acetylglycine levels were inversely associated with baseline BMI (P = 0.061 and 0.022 in the discovery and validation cohorts, respectively). However, we found high Acetylglycine levels were associated with increased risk of significant weight gain. Among functionally‐limited adults, Lustgarten et al. reported Acetylglycine was positively associated with dietary fiber intake, muscle strength, and mobility. However, dietary fiber has been frequently linked to aid in energy intake control and reduced risk for development of obesity. Unfortunately, we don't have dietary intake data on those study participants.
Several potential limitations should be considered. The study sample size of 300 total participants is relatively small. Considering that the effect size of many metabolites may be very small 44, some of the associations may not be detected in the current study. Our metabolomics analysis was limited to metabolites that could be detected by the platforms used and our data were measured in terms of relative concentrations rather than actual concentrations. In addition, the metabolite levels in our study were based on blood samples from a single point in time that may not reflect the true associations over time. Nevertheless, this is the first metabolomics study to identify metabolites associated with BMI and to further discover metabolites prospectively predicting weight gain in Mexican Americans. Our results provide strong rationales for future large prospective studies to further clarify the associations among obesity/weight gain, metabolites, and chronic disease risks.
Conflict of Interest Statement
Authors have declared there are no any competing financial interests in relation to the work described.
Funding
The Mexican American Cohort receives funds collected pursuant to the Comprehensive Tobacco Settlement of 1998 and appropriated by the 76th legislature to The University of Texas MD Anderson Cancer Center and from the Caroline W. Law Fund for Cancer Prevention and the Duncan Family Institute for Risk Assessment and Cancer Prevention.
Acknowledgements
We thank the field staff for their ongoing work with participant recruitment and follow‐up. Most importantly, we thank our study participants and their parents for their cooperation and participation, without which this research would not be possible.
Zhao, H. , Shen, J. , Djukovic, D. , Daniel‐MacDougall, C. , Gu, H. , Wu, X. , and Chow, W.‐H. (2016) Metabolomics‐identified metabolites associated with body mass index and prospective weight gain among Mexican American women. Obesity Science & Practice, 2: 309–317. doi: 10.1002/osp4.63.
References
- 1. Fryar CD, Wright JD, Eberhardt MS, Dye BA. Trends in nutrient intakes and chronic health conditions among Mexican‐American adults, a 25‐year profile: United States, 1982‐2006. Natl Health Stat Report. 2012(50):1‐20. [PubMed]
- 2. Thow AM, Jan S, Leeder S, Swinburn B. The effect of fiscal policy on diet, obesity and chronic disease: a systematic review. Bull World Health Organ 2010; 88: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Renzaho AM, Halliday JA, Nowson C. Vitamin D, obesity, and obesity‐related chronic disease among ethnic minorities: a systematic review. Nutrition 2011; 27: 868–879. [DOI] [PubMed] [Google Scholar]
- 4. Kushner RF. Weight loss strategies for treatment of obesity. Prog Cardiovasc Dis 2014; 56: 465–472. [DOI] [PubMed] [Google Scholar]
- 5. Egger G, Dixon J. Beyond obesity and lifestyle: a review of 21st century chronic disease determinants. Biomed Res Int 2014; 2014: 731685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chaffee BW, Weston SJ. Association between chronic periodontal disease and obesity: a systematic review and meta‐analysis. J Periodontol 2010; 81: 1708–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abu Bakar MH, Sarmidi MR, Cheng KK, et al. Metabolomics ‐ the complementary field in systems biology: a review on obesity and type 2 diabetes. Mol Biosyst 2015; 11: 1742–1774. [DOI] [PubMed] [Google Scholar]
- 8. Griffin JL, Atherton H, Shockcor J, Atzori L. Metabolomics as a tool for cardiac research. Nat Rev Cardiol 2011; 8: 630–643. [DOI] [PubMed] [Google Scholar]
- 9. Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res: an official journal of the American Association for Cancer Research 2009; 15: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moore SC, Matthews CE, Sampson JN, et al. Human metabolic correlates of body mass index. Metabolomics 2014; 10: 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newgard CB, An J, Bain JR, et al. A branched‐chain amino acid‐related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009; 9: 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 2012; 125: 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaudet MM, Falk RT, Stevens RD, et al. Analysis of serum metabolic profiles in women with endometrial cancer and controls in a population‐based case‐control study. J Clin Endocrinol Metab 2012; 97: 3216–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011; 17: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Floegel A, Stefan N, Yu Z, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013; 62: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oberbach A, von Bergen M, Bluher S, Lehmann S, Till H. Combined serum proteomic and metabonomic profiling after laparoscopic sleeve gastrectomy in children and adolescents. J Laparoendosc Adv Surg Tech A 2012; 22: 184–188. [DOI] [PubMed] [Google Scholar]
- 17. Reinehr T, Wolters B, Knop C, et al. Changes in the serum metabolite profile in obese children with weight loss. Eur J Nutr 2015; 54: 173–181. [DOI] [PubMed] [Google Scholar]
- 18. Wahl S, Vogt S, Stuckler F, et al. Multi‐omic signature of body weight change: results from a population‐based cohort study. BMC Med 2015; 13: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chow WH, Chrisman M, Daniel CR, et al. Cohort Profile: The Mexican American Mano a Mano Cohort. Int J Epidemiol 2015. doi: 10.1093/ije/dyv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuczmarski MF, Kuczmarski RJ, Najjar M. Effects of Age on Validity of Self‐Reported Height, Weight, and Body Mass Index: Findings from the Third National Health and Nutrition Examination Survey, 1988–1994. J Am Diet Assoc 2001; 101: 28–34. [DOI] [PubMed] [Google Scholar]
- 21. Villanueva EV. The validity of self‐reported weight in US adults: a population based cross‐sectional study. BMC Public Health 2001; 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huffman JE, Albrecht E, Teumer A, et al. Modulation of genetic associations with serum urate levels by body‐mass‐index in humans. PLoS One 2015; 10 e0119752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mihalik SJ, Michaliszyn SF, de las Heras J, et al. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: evidence for enhanced mitochondrial oxidation. Diabetes Care 2012; 35: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yue JR, Huang CQ, Dong BR. Association of serum uric acid with body mass index among long‐lived Chinese. Exp Gerontol 2012; 47: 595–600. [DOI] [PubMed] [Google Scholar]
- 25. Nowaczyk MJ, Lehotay DC, Platt BA, et al. Ethylmalonic and methylsuccinic aciduria in ethylmalonic encephalopathy arise from abnormal isoleucine metabolism. Metabolism 1998; 47: 836–839. [DOI] [PubMed] [Google Scholar]
- 26. MacDonald MJ. Metabolism of the insulin secretagogue methyl succinate by pancreatic islets. Arch Biochem Biophys 1993; 300: 201–205. [DOI] [PubMed] [Google Scholar]
- 27. Gautam D, Jeon J, Li JH, et al. Metabolic roles of the M3 muscarinic acetylcholine receptor studied with M3 receptor mutant mice: a review. J Recept Signal Transduct Res 2008; 28: 93–108. [DOI] [PubMed] [Google Scholar]
- 28. Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit Rev Neurobiol 1996; 10: 69–99. [DOI] [PubMed] [Google Scholar]
- 29. Caulfield MP, Birdsall NJ. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol Rev 1998; 50: 279–290. [PubMed] [Google Scholar]
- 30. Jacobs RL, Zhao Y, Koonen DP, et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N‐methyltransferase‐deficient mice are protected from diet‐induced obesity. J Biol Chem 2010; 285: 22403–22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubio‐Aliaga I, Roos B, Sailer M, et al. Alterations in hepatic one‐carbon metabolism and related pathways following a high‐fat dietary intervention. Physiol Genomics 2011; 43: 408–416. [DOI] [PubMed] [Google Scholar]
- 32. Wu G, Zhang L, Li T, et al. Choline Deficiency Attenuates Body Weight Gain and Improves Glucose Tolerance in ob/ob Mice. J Obes 2012; 2012: 319172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maresca A, Supuran CT. Muscarinic acetylcholine receptors as therapeutic targets for obesity. Expert Opin Ther Targets 2008; 12: 1167–1175. [DOI] [PubMed] [Google Scholar]
- 34. Marrero MB, Lucas R, Salet C, et al. An alpha 7 Nicotinic Acetylcholine Receptor‐Selective Agonist Reduces Weight Gain and Metabolic Changes in a Mouse Model of Diabetes. J Pharmacol Exp Ther 2010; 332: 173–180. [DOI] [PubMed] [Google Scholar]
- 35. Calvani R, Miccheli A, Capuani G, et al. Gut microbiome‐derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes 2010; 34: 1095–1098. [DOI] [PubMed] [Google Scholar]
- 36. Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol 2005; 3: 431–438. [DOI] [PubMed] [Google Scholar]
- 37. Waldram A, Holmes E, Wang Y, et al. Top‐down systems biology modeling of host metabotype‐microbiome associations in obese rodents. J Proteome Res 2009; 8: 2361–2375. [DOI] [PubMed] [Google Scholar]
- 38. Salek RM, Maguire ML, Bentley E, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics 2007; 29: 99–108. [DOI] [PubMed] [Google Scholar]
- 39. Williams RE, Lenz EM, Evans JA, et al. A combined (1)H NMR and HPLC‐MS‐based metabonomic study of urine from obese (fa/fa) Zucker and normal Wistar‐derived rats. J Pharm Biomed Anal 2005; 38: 465–471. [DOI] [PubMed] [Google Scholar]
- 40. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009; 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol 2009; 587(Pt 17): 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341: 1079, 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson RJ, Nakagawa T, Sanchez‐Lozada LG, et al. Sugar, uric acid, and the etiology of diabetes and obesity. Diabetes 2013; 62: 3307–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sampson JN, Boca SM, Shu XO, et al. Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev 2013; 22: 631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]


