Abstract
Endocrine therapy using estrogen receptor-α (ER-α) antagonists for attenuating horm2one-driven cell proliferation is a major treatment modality for breast cancers. To exploit any DNA repair deficiencies associated with endocrine therapy, we investigated the functional and physical interactions of ER-α with O6-methylguanine DNA methyltransferase (MGMT), a unique DNA repair protein that confers tumor resistance to various anticancer alkylating agents. The ER-α -positive breast cancer cell lines (MCF-7, T47D) and ER- negative cell lines (MDAMB-468, MDAMB-231), and established inhibitors of ER-α and MGMT, namely, ICI-182,780 (Faslodex) and O6-benzylguanine, respectively, were used to study MGMT- ER interactions. The MGMT gene promoter was found to harbor one full and two half estrogen-responsive elements (EREs) and two antioxidant-responsive elements (AREs). MGMT expression was upregulated by estrogen, downregulated by tamoxifen in Western blot and promoter-linked reporter assays. Similarly, both transient and stable transfections of Nrf-2 (nuclear factor-erythroid 2-related factor-2) increased the levels of MGMT protein and activity 3 to 4-fold reflecting novel regulatory nodes for this drug-resistance determinant. Of the different ER-α antagonists tested, the pure anti-estrogen fulvestrant was most potent in inhibiting the MGMT activity in a dose, time and ER-α dependent manner, similar to O6-benzylguanine. Interestingly, fulvestrant exposure led to a degradation of both ER-α and MGMT proteins and O6-benzylguanine also induced a specific loss of ER-α and MGMT proteins in MCF-7 and T47D breast cancer cells with similar kinetics. Immunoprecipitation revealed a specific association of ER-α and MGMT proteins in breast cancer cells. Furthermore, silencing of MGMT gene expression triggered a decrease in the levels of both MGMT and ER-α proteins. The involvement of proteasome in the drug-induced degradation of both proteins was also demonstrated. Fulvestrant enhanced the cytotoxicity of MGMT-targeted alkylating agents, namely, temozolomide and BCNU by 3 to 4-fold in ER-α positive cells, but not in ER–negative cells. We conclude that MGMT and ER-α proteins exist as a complex and are co-targeted for ubiquitin-conjugation and subsequent proteasomal degradation. The findings offer a clear rationale for combining alkylating agents with endocrine therapy.
Keywords: estrogen signaling, MGMT DNA repair, ubiquitin-proteasome pathway, breast cancer, anti-estrogens
Introduction
Estrogens regulate various physiologic processes including cell proliferation, female reproduction, bone stability and cardiovascular functions, all through binding with two cytoplasmic receptors, namely, estrogen receptor alpha (ER-α) and ER-β[1]. These receptors are ligand-activated transcription factors; upon binding with estrogen, they dimerize, recruit coactivators and induce transcription of specific target genes that mediate the estrogenic and cell proliferation functions in breast and other organs[2–3]. ER-α is expressed at low levels, in just < 10% of normal breast epithelia[4]. However, approximately 70% of human breast cancers express significant levels of ER-α[3,5]. Therefore, inhibiting the estrogen signaling pathway with anti-estrogens in premenopausal women or depleting estrogen synthesis in peripheral tissues with aromatase inhibitors in post-menopausal women have long been a standard and effective strategy in breast cancer management[6–7]. The ER-α antagonists dramatically improve long-term disease-free and overall survival for women with ER-positive breast cancer. For example, selective estrogen receptor modulators (SERMs) such as tamoxifen and toremifene, which bind ER-α and inhibit estrogenic signaling in breast tissue, however, possess agonistic properties in the uterus and other tissues, and have been a long-stay in the treatment of breast neoplasms[8]. A long-term use of tamoxifen for 5-10 years is also well-known to reduce the risk for developing breast cancer[9]. In contrast to SERMs, selective estrogen receptor downregulators (SERDs) like fulvestrant (also called ICI-182,780, Faslodex) are pure anti-estrogens and curtail estrogen signaling in all tissues[10]. Fulvestrant disrupts ER dimerization and nuclear localization, completely blocks ER-mediated transcription and facilitates degradation of ER-α through the ubiquitin-proteolytic pathway[11].
Given the multiple physiologic and oncogenic consequences of estrogen signaling, the mechanisms by which these actions control the DNA damage response, DNA repair per se, and the subsequent apoptotic cascades are not well understood. These considerations are highly important because estrogen itself may incur DNA damage, albeit indirectly[12], and ER-signaling will likely affect the processing of other genotoxic insults as well as therapy-induced DNA damage[13]. Furthermore, the proteins regulating the DNA damage responses and DNA repair processing such as the p53 and mismatch repair are frequently mutated or altered in hormone-responsive breast cancers[14–15].
One such important DNA repair protein highly expressed in breast cancers is O6-methylguanine DNA methyltransferase (MGMT)[16]. MGMT maintains the genomic integrity through its ability to remove methyl and alkyl groups introduced at the O6-position of guanine, thereby, effectively protecting the cellular genome and critical oncogenic genes from the mutagenic actions of endogenous and exogenous alkylating agents[17–18]. MGMT functions by a unique stoichiometric and suicidal reaction mechanism in which the alkyl groups bound to the O6-position of guanine are transferred to a cysteine in its active site, resulting in direct restoration of the normal base and self-inactivation of MGMT protein[17]. Because many anticancer alkylating agents induce O6-guanine alkylations in DNA and kill tumor cells through mutations (e.g. temozolomide, dacarbazine, and procarbazine) or producing DNA crosslinks (BCNU, cyclophosphamide), increased expression of MGMT is a major mechanism of tumor resistance[19–20]. Therefore, inhibition of MGMT by powerful pseudosubstrates such as O6-benzylguanine (BG) and O6-(4-bromothenyl)guanine (BTG) has emerged as a prominent strategy to enhance the antitumor activity of alkylating agents[21–22].
Various lines of evidence provide a strong link between MGMT, estrogen and ER-signaling. Although there was no change in liver MGMT during the estrous cycle, the level of mammary epithelial cell MGMT activity on estrus and proestrus was significantly higher than on metestrus in rat[23–24]. Consequently, the methylnitrosourea (MNU, a carcinogen which generates O6-methylguanine) injections given on different days of the estrous cycle resulted in differential carcinogenesis[23,25]. A study by Teo et al.[26] has proposed that inactivated MGMT may suppress ER-α signaling. MGMT could also play a significant role in altering the breast cancer risk associated with estrogen signaling[26]. Several studies have reported up to four times higher MGMT levels in breast tumors relative to normal breast tissue[27–30]. Additionally, MGMT has been implicated in resistance to phosphoramide mustards used in breast cancer therapy and the ability of acrolein (a byproduct of cyclophosphamide decomposition) to inhibit MGMT activity[30–31]. Work in our laboratory has suggested that MGMT may have other functions besides DNA repair; in this context, the human MGMT protein was shown to interact specifically with proteins involved in DNA replication, cell cycle, RNA metabolism, heat shock stress, ubiquitin and cellular metabolism[32]. Most notably, we showed that MGMT interacts with MCM2, PCNA, ORC1, DNA polymerase δ, MSH2, CDK1 and p21cip1. Building on these data, we suggest that MGMT plays a role in sensing and integrating the DNA damage/repair related signals with replication, cell cycle progression and genomic stability[32]. Therefore, we screened the MGMT gene promoter for the presence of cis-acting regulatory elements responsive to estrogen, and investigated the physical and functional interactions between ER-α and human MGMT using fulvestrant and BG, which curtail their functional activities, respectively. Our results showed a tight protein association and mutual dependence on steady-state protein levels as well as the elimination of inactivated proteins for these partners.
Materials and methods
Cell lines and cell culture
Human breast epithelial adenocarcinoma cell lines MCF7, MDAMB-231, HCC1937 and MDAMB-468 and human breast epithelial ductal carcinoma cell line T47D were purchased from American Type Culture Collection (ATCC). The cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS), and antibiotics. Estrogen was added to culture medium as specified. The MGMT-proficient HT29 colon cancer cells were also used and grown without estrogen under the same conditions.
Chemicals, antibodies and nucleic acids
General chemicals and reagents were obtained from Sigma-Aldrich Company. Tamoxifen citrate, toremifene citrate and fulvestrant were the products of Selleck Chemicals (Houston, TX, USA). Monoclonal antibodies to MGMT and actin were purchased from Millipore Corporation whereas anti-ER-α antibody was from Cell Signaling Technology. Antibodies to PCNA, DNA Polδ, topoisomerase I, CDK1, and APE1 were procured from the Millipore Company (Boston, MA, USA). The proteasome inhibitor Velcade (PS-341, Bortezomib), BCNU and temozolomide were obtained from a local oncology pharmacy. siRNA and shRNA specific for human MGMT were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and Qiagen (Valencia, CA, USA), respectively. The 1 kb promoter of MGMT linked with luciferase gene was a kind gift from Dr. Sankar Mitra (University of Texas Medical Branch, Galveston, TX, USA). The NRF2 expression vector was provided by Dr. Anil K. Jaiswal, University of Maryland, Baltimore, MD, USA.
Assay for DNA repair activity of MGMT
MGMT activity was measured by the transfer of [3H]-labeled methyl groups from the O6-position of guanine in the DNA substrate to the MGMT protein as described previously[33]. The DNA substrate enriched in O6-methylguanine was prepared by reacting [3H]-methylnitrosourea (GE Healthcare, 60 Ci/mmol)[33]. Briefly, the cell pellets were washed with cold Tris-buffered saline (TBS), disrupted by sonication in the assay buffer (30 mmol/L Tris-HCl pH 7.5, 0.5 mmol/L DTT, 0.5 mmol/L EDTA, 5% glycerol, and 20 mmol/L spermidine) and centrifuged. The extracts (50-150 μg protein) were supplemented with the [3H]-DNA (1 μg; 10,000 cpm) and incubated at 37°C for 30 minutes. The reactions were terminated with 20% trichloroacetic acid, the DNA substrate was hydrolyzed at 80°C, and following filtration on glass fiber discs (GF/C), the radioactivity present in protein precipitates was solubilized and quantitated[33].
MGMT promoter reporter assays
FAST CAT (deoxy) chloramphenicol acetyltransferase assay kits which use the green fluorescent substrate (BODIPY FL 1-deoxychloramphenicol) and yield a single product were purchased from Thermo Fisher Scientific Company. Briefly, extracts from the cells transfected with the CAT-linked MGMT promoter were prepared in 250 mmol/L Tris-HCl (pH 7.5) by two freeze–thaw cycles. Extracts with 50-100 μg protein containing 1.1 mmol/L acetyl-CoA and 1μg substrate (100 μL,) were incubated for 40 minutes at 37°C. The reactions were stopped by adding 1 mL of ethyl acetate followed by centrifugation. The upper organic solvent layer containing the CAT substrate and product was removed and dried. The contents were dissolved in 30 μL ethyl acetate followed by thin-layer chromatography (TLC) on silica gel. The plates were developed with chloroform: methanol (85:15 V:V), dried and photographed under UV light. For further quantification, the single fluorescent spots corresponding to the product (acetylated chloramphenicol) were scraped into a microfuge tube, dissolved in 250 μL methanol; the contents were centrifuged, and the supernatants were read using a fluorometer at 540 nm excitation and 570 nm emission.
Electrophoretic mobility shift assay (EMSA) for ER-α binding with DNA
Binding of ER-α to its consensus recognition sequence was examined in fulvestrant and BG-treated MCF-7 cells using EMSA. A double-stranded 30-mer oligonucleotide containing two copies of the ER-α recognition sequence 5′GGTCACABTGACC3′ was labeled with biotin at 5′ end on one strand (Integrated DNA Technologies, Coralville, IA, USA)[34]. Nuclear extracts were prepared from cells as described previously[35] and 5 μg protein samples were incubated in a binding buffer (10% glycerol, 1 mmol/L MgCl2, 0.2 mmol/L EDTA, 1 mmol/L dithiothreitol, 75 mmol/L NaCl, 10 mmol/L Tris-HCl, 0.1 mg/mL calf thymus DNA), and 2 µg of poly(dI-dC) for 30 minutes at room temperature. The protein/DNA complexes were separated on a non-denaturing 5% polyacrylamide gel. The gel was transferred to a nylon membrane, and the biotin-labeled oligonucleotides were detected using strepatavidin-HRP and enhanced chemiluminescence.
Western blotting assay
After trypsinization, the cell pellets were washed with cold TBS, and subjected to sonication in 50 mmol/L Tris–HCl (pH 8.0) containing 1% glycerol, 1 mmol/L EDTA, 0.5 mmol/L PMSF and 2 mmol/L benzamidine and centrifuged. Equal protein amounts from different treatments were electrophoresed on 12% SDS–polyacrylamide gels. Proteins were electro-transferred to Immobilon-P membranes. The membranes were blocked with 5% non-fat dry milk in TBS (pH 8.0) containing 0.1% Tween 20 for 3 hours, and subsequently incubated with appropriate primary antibodies. The antigen–antibody complexes were visualized by enhanced chemiluminescence (Pierce Company, Woburn, MA, USA). Band intensities were quantified using a Bio-Rad VersaDoc Imaging system.
Co-immunoprecipitation
Cell pellets were solubilized in TBS containing 1 mmol/L PMSF, 2 mmol/L benzamidine, 0.5 mmol/L EDTA, 5% glycerol and 0.5% NP-40. The resulting cell extracts (∼300 µg) were pre-cleaned with 10 mL of protein A/G-agarose then incubated overnight at 4°C with either the anti-MGMT monoclonal antibody (1 µg) or the rabbit monoclonal anti-ER-α antibody (1 µg). Subsequently, 30 mL of protein A/G agarose were added, and the incubation was continued for an additional 3 hours at 4°C. The agarose pellets were washed, SDS sample buffer was added and immunoblotting was performed as described above.
RNAi studies
MCF7 cells were transiently transfected with shRNA plasmid DNA for human MGMT or using Fugene 6 reagent (Roche Co.) pcDNA 3.1 vector DNA was used as control. Similarly, the MCF-7 cells were transfected with MGMT-specific siRNA using Lipofectamine (Invitrogen China). Cells were trypsinized and processed for western blot analyses.
RNA isolation and reverse transcription- polymerase chain reaction (RT-PCR) analysis
MCF7 cells were treated with 1 μmol/L fulvestrant for 48 hours or 50 μmol/L BG for 20 hours. Total RNA was isolated from control and drug-treated cells using the TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. The integrity of RNA was verified by electrophoresis on 1% non-formaldehyde agarose gels at pH 8.5. For RT-PCR, 1 µg RNA was reverse-transcribed using Qiagen One Step RT PCR kit, again following the manufacturer's instructions. Briefly, there was an initial HotStarTaq DNA polymerase activation step for 15 minutes at 95°C. Three-step cycling involved 1 minute of denaturation at 94°C, followed by 35 cycles at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 2 minutes, and a final extension at 72°C for 10 minutes using a Biometra thermal cycler. The amplified DNA sizes were 492 bp for TSP1, 767 bp for IGFBP4, 479 bp for AMD1, 481 bp for OAZ1, 650 bp for ER-α, 500 bp for MGMT, and 650 bp for actin. The Primers used were: TSP1[36], sense 5′-ACCGCATTCCAGAGTCTGGC-3′ and antisense 5′-ATGGGGACGTCCAACTCAGC-3′; IGFB-P4[37], sense 5′-TTCATCCCCATCCCCAACTGC-3′ and antisense 5′-CTGCTACCCCACGCTTCCTTA-3′; AMD1[38], sense 5′-GATGGAACTTATTGGACTATTCACATCAC-3′ and antisense 5′-CTGTGCGACATTTAGAACTCTGATTAAC-3′; OAZ1[38], sense 5′-GA-CAGCTTTGCAGTTCTCCTGG-3′ and antisense 5′-T-TCGGAGCAAGGCGGCTC-3′; ER-α[39], sense 5′-TA-CTGCATCAGATCCAAGGG-3′ and antisense 5′-AT-CAATGGTGCACTGGTTGG-3′; MGMT[40], sense 5′-CCTTGGTACTTAAGCTTATGGACAAG-3′ and antisense 5′-CTACTGCACGAATTCAG-3′; actin, sense 5′- TGACGGGGTCACCCACACCCACACTGTGCCCATCTA and antisense 5′-CTAGAAGCATTTGCGGT-GGACGATGGAGGG. The PCR products were analyzed by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining under UV light. The bands were quantitated by densitometry.
Assay for BCNU-induced interstrand DNA crosslinking in tumor cells
MCF7 and MDAMB-468 cells were first exposed to 1μmol/L fulvestrant for 48 hours and subsequently to BCNU at 100 μmol/L for 1 hour. BCNU-treated cells were next suspended in fresh medium without any drugs to allow DNA crosslinking and its repair up to 72 hours. DNA from cells was isolated by lysis in TE buffer (10 mmol/L Tris, 1 mmol/L EDTA, pH 8.0) containing 0.5% SDS, and 100 μg/mL RNase A and proteinase K (1 mg/mL) digestion. DNA was precipitated with ethanol and dissolved in TE buffer. The drug-induced DNA crosslinking was measured by the ethidium bromide fluorescence assay as described previously[41]. Briefly, 5-10 μg DNA was dissolved in the assay buffer (20 mmol/L potassium phosphate and 2 mmol/L EDTA, pH 11.8) in duplicate. One set of tubes was heated at 100°C for 10 minutes and cooled to room temperature. Ethidium bromide was added to 1μg/mL, and the fluorescence was measured (305 nm excitation and 585 nm emission) using an LS-50 variable wavelength spectrofluorometer (Perkin Elmer). The fluorescence readings were used to compute the crosslink index, CLI, as described earlier[41].
Cell survival assays
Cell survival following various drug treatments was determined by using the yellow tetrazolium dye (3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyl tetrazolium bromide) (MTT) as described previously[33]. Tumor cells (15,000/well) in 24-well plates were treated with 1 μmol/L fulvestrant for 72 hours before exposure to the alkylating agents. The purple color developed due to formazan was measured at 570 nm using a SPECTRAFluor Plus plate reader (Tecan, Crailsheim, Germany)
Statistical analysis
Most experiments were performed in triplicate three times independent of each other. Results were assessed by Student's t-test. When more than two samples were involved, the one-way analysis of variance was used to determine whether there were significant differences between the means of experimental groups. Significance was defined as P equal to or < 0.05.
Results
Evidence for estrogen regulation of MGMT expression in human breast cancer cells
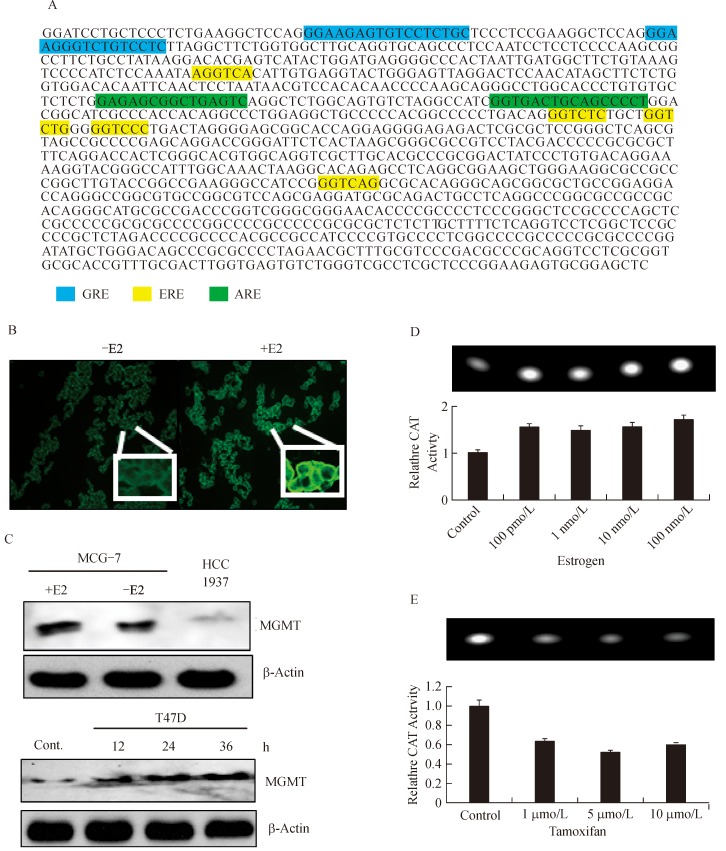
Guanine is the most preferred base for alkylation by endogenous metabolites such as the S-adenosylmethionine, nitroso compounds, polycyclic aromatic hydrocarbons, environmental carcinogens and the adducts at the O6-guanine are particularly critical, because the O6-alkylguanines pair aberrantly with thymine, resulting in GC to AT transitions. Human MGMT is a primary defense against these O6-alkylations. However, because of its stoichiometric reaction mechanism, the physiologic regulation of MGMT by endocrine hormones has not received much attention. Nevertheless, it is essential to recognize that MGMT is an important player in both in breast carcinogenesis and breast cancer treatment. Previously, a report described the downregulation of MGMT through ubiquitin-proteolysis in tamoxifen treated HT29 cells[42]. Recently, Bobustuc et al. reported a significant upregulation of MGMT in tamoxifen resistant breast cancer cells[28]. With this background, we sought to determine the estrogenic regulation of MGMT by searching the human MGMT promoter (GenBank: X61657.1) for estrogen response elements and related cis-acting sequences. Indeed, our search using the TRANSFAC 7.0 and JASPAR databases revealed the presence of one full and two half EREs in MGMT promoter (Fig. 1A). In addition, we also found two antioxidant response elements (AREs) that bind the Nrf2 for the first time and glucocorticoid response elements (GREs) in the MGMT promoter (Fig. 1A). The role of GREs in MGMT expression has been analyzed previously[44].
Fig. 1. Evidence for estrogenic regulation of human MGMT.
A: Presence of estrogen-responsive elements (EREs) in human MGMT promoter. The 1 kb minutesimal promoter[43] shows the presence of one full ERE and two half ERE sequences, highlighted in yellow. Two antioxidant-responsive elements (ARE), besides the previously identified glucocorticoid elements were also found and shown. B: Increased expression of MGMT protein in MCF-7 cells grown in the presence of 20 nmol/L estrogen (E2). Permeabilized cells were stained with a monoclonal antibody for MGMT followed by FITC-conjugated secondary antibodies. C: Western blot analysis of MGMT in ER-α-positive MCF-7 and T47D cells. MCF-7 cells were cultured with and without 20 nmol/L E2 for 24 hours. The HCC1937 breast cancer cells which harbour a BRCA1 mutation were used as a control. The T47D cells grown in the presence of 50 nmol/L of E2 showed a time-dependent upregulation of MGMT protein. D: Increased activity of MGMT- promoter linked with the CAT gene induced by estrogen in MCF-7 cells. The cells were transfected for 12 hours and then exposed to E2 at concentrations shown. The upper panel shows the TLC pattern of the fluorescent acetylated chloramphenicol spots and the quantitation of fluorescence associated with the spots is shown in the lower panel. The bar graph represents the mean of 3 separate experiments. ANOVA showed that the CAT activity between control and 100 pmol/L estrogen was significant (P = 0.05) and not significant with increasing hormone concentrations. E: Attenuation of MGMT promoter-CAT reporter activity in MCF-7 cells exposed to tamoxifen (1-10 μmol/L) for 24 hours. Results from three independent experiments were assessed by ANOVA; they were significant between the control and 1 μmol/L tamoxifen, but not so with increasing tamoxifen concentrations. MGMT: O6-methylguanine DNA-methyltransferase; h: hour.
The next series of experiments characterized the functional aspects of ERE in MGMT expression using two ER-α positive breast cancer cell lines (MCF-7 and T47D, both are MGMT-proficient). The ER-α negative MDAMB-231 (MGMT-deficient) and MDAMB-468 (MGMT- proficient)[45] were used as controls in some assays. First, immunofluorescence staining of MGMT in MCF-7 cells cultured in the presence of 20 nM estrogen for 24 hours clearly showed increased expression of the repair protein (Fig. 1B). Western blot analysis in the same setting confirmed the moderate upregulation of MGMT in MCF-7 cells; the BRCA1 mutant and MGMT-deficient H1937 breast cancer cells served as a control in this blot (Fig. 1C, upper panel). The ER-α expressing T47D cells also showed a time-dependent increase of MGMT levels in the presence of 50 nM estrogen (Fig. 1C, lower panel).
Further, direct evidence for the enhancement of MGMT transcription by estrogen was obtained by transient expression of the chloramphenicol acetyltransferase (CAT) reporter gene driven by a human MGMT promoter fragment[44]. The CAT gene in this construct was placed under the control of 5′-flanking region of 0.7 kb MGMT promoter containing all the EREs. The reporter construct was transfected into the MCF-7 cells using lipofectamine. Twelve hours after transfection, MCF-7 cells were cultured without or with estrogen (100 pmol/L-100 nmol/L) for 20 hours. CAT assays were performed in cell extracts using a fluorescent substrate that underwent a single acetylation reaction and yielded a single spot of the product. Fig. 1D shows the generation of the fluorescent product in relation to estrogen treatment; a 1.6 to 1.7 – fold increase in CAT activity was observed and suggests that the ERE mediates the transcriptional upregulation of the MGMT gene. The MGMT promoter-CAT reporter assays were also used to determine the effect of tamoxifen on MGMT transcription. For this, the MCF-7 cells were transiently transfected with the reporter and then exposed to tamoxifen in the range of 1-10 μmol/L. Fig. 1E shows that 1 μmol/L tamoxifen reduced the promoter activity by 40%, which did not change with increasing concentrations of tamoxifen. Taken together, the results presented in this section provide strong evidence that estrogen can upregulate MGMT expression in ER-α positive breast cancers.
The ARE-NRF2 regulatory axis for MGMT expression in human cancers
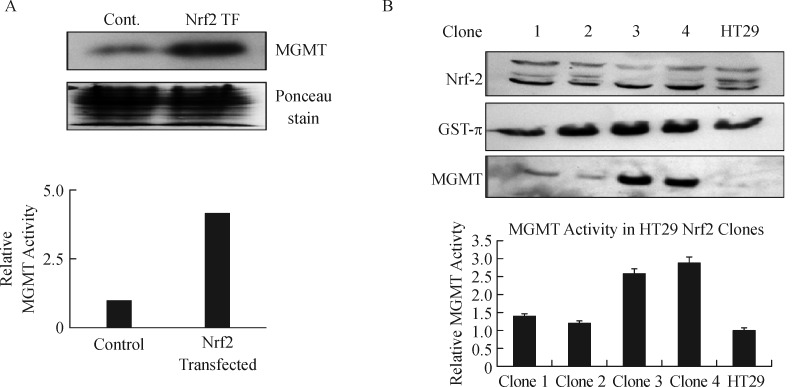
In two previous studies, we showed that human MGMT can be moderately upregulateld by antioxidants such as curcumin, procysteine, neem extract and other natural products[40, 45]. However, the cellular mechanisms mediating the antioxidant regulation of MGMT have remained unclear. The discovery of two AREs in the MGMT promoter (Fig. 1A) encouraged us to probe the ARE-NRF2 interaction as the underlying regulatory mechanism of MGMT expression in response to oxidative stress. The Nrf-2 (nuclear factor erythroid 2-related factor) is a cytoprotective transcription factor activated during redox imbalance and functions to restore the redox homeostasis[46]. Nrf2 controls the basal and induced expression of an array of ARE–dependent genes such as the γ-glutamylcysteine synthase, glutathione S-transferase-π, and NAD(P)H oxidoreductase 1 to regulate the physiologic and pathophysiological outcomes of oxidant exposure[47]. Estrogen signaling has a close relationship with NRF2, because inhibition of estrogenic functions has been shown to activate the NRF2 pathway in breast cancers[48]. Therefore, the control of MGMT by ARE was first probed by transient transfection of a full-length NRF2 expression vector in MCF-7 cells. Fig. 2A shows that the MGMT protein and activity were increased 4-fold 24 hours after transfection. To gain more insight, we also engineered HT29 cells to stably express the NRF2. Four clones arising from the G418 selection were screened for the expression of NRF2, GST-π, and MGMT protein levels relative to the parent HT29 cells (Fig. 2B). The clones showed variable expression of NRF2. GST-π, an established target of the ARE-NRF2 pathway was expressed to a higher extent in clones 2, 3 and 4. MGMT protein was upregulated by 3-fold in clones 3 and 4. MGMT activity levels were equivalent to its protein levels in these clones (lower panel of Fig. 2B). Overall, these experiments firmly establish the NRF2 as a regulator of MGMT gene expression in breast cancer and other human cancers.
Fig. 2. Nrf2 upregulation of MGMT expression in MCF-7 and HT29 cells.
A: Increased MGMT protein levels 24 hours following transient transfection of Nrf2 in MCF-7 cells. The middle panel shows equivalent protein staining on the membrane used for Western blotting. The third panel shows increased DNA repair activity catalyzed by MGMT in Nrf2 transfected cells. B: Increased expression of GST-π and MGMT proteins in HT29 cell clones after stable transfection of Nrf2. Four clones were isolated by G418 selection. The first panel shows the Nrf2 protein expression in these clones and HT29 parent cells. The middle panel displays the enhanced GST-π expression in the clones. The third panel represents the increased expression of MGMT protein in the same background. The bar graph underneath represents the MGMT activity levels in HT29 and its Nrf2 expressing clones. ANOVA revealed that the 2-fold increased DNA repair activity in clones 3 and 4 was significant at P = 0.05. MGMT: O6-methylguanine DNA-methyltransferase; Cont: control; Nrdf2: nuclear factor erythroid 2-related factor.
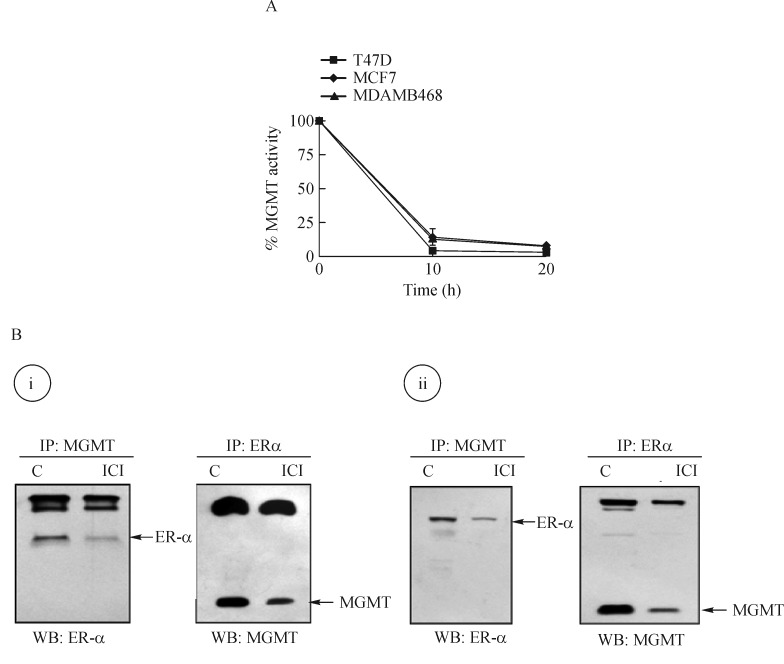
BG-induced inhibition of MGMT, physical association of MGMT with ER-α protein and evidence suggesting their co-degradation in breast cancer cells
BG is a free-base pseudosubstrate for MGMT. BG irreversibly forms an S-benzylcysteine moiety at the cysteine acceptor site (Cys145), thereby inactivating the DNA repair protein. Since this linkage is covalent, the inactivated protein is not recycled and becomes a substrate for ubiquitin-conjugation and subsequent proteasomal degradation[21]. The resulting depletion of MGMT facilitates greater DNA alkylation and in turn an increased tumor cell killing. In the context of current study, we tested the extent of MGMT inhibition by BG in T47D, MCF-7 (ER-α positive) and MDAMB-468 (ER-α negative) cells. The kinetics and duration of MGMT inactivation were similar irrespective of the ER-status (Fig. 3A). Based on the previous observations[26], we hypothesized that human MGMT and ER-α proteins associate physically with each other. To verify this interaction, the lysates from MCF-7 and T47D cells were immunoprecipitated with antibodies to MGMT or ER-α proteins, the complexes resolved by Western blotted using reciprocal antibodies as the probes. We found that in both ER-α positive cell lines, MGMT protein was present in the immunocomplexes of ER-α and similarly, ER-α was associated MGMT protein (Fig. 3B). These observations confirm a protein–protein association of MGMT with the ER in hormone-responsive breast cancer cells. Fulvestrant (Faslodex, ICI 182,780) is a pure anti-estrogen used in endocrine therapy of breast cancers. Unlike the SERMs such as tamoxifen, fulvestrant functions as an ER-α antagonist in all human tissues and has no agonistic activities[49]. Mechanistically, fulvestrant inhibits ER- dimerization and induces a dramatic and rapid degradation of the ER-α[49] to eliminate the estrogenic signaling and halts malignant proliferation. The cells treated with fulvestrant reproducibly showed reduced levels of ER-α protein in MGMT immunocomplexes and decreased MGMT levels in ER-α immunoprecipitates compared with their respective untreated controls (Fig. 3B). These unexpected data suggested a mutual co-degradation of the partner proteins(ER-α and MGMT) in response to fulvestrant.
Fig. 3. Inhibition of MGMT activity by O6-benzylguanine (BG) and physical association of ER-α and MGMT proteins breast cancer cells.
A: Inhibition of the DNA repair activity of MGMT by BG (75 μmol/L over 20 hours in MCF7, T47D and MDA-MB-468 cells. Equivalent inhibition of MGMT at all time points was found irrespective of the ER status. B: Direct interaction between ER-α and MGMT proteins was detected following immunoprecipitation (IP) and Western blot analyses in breast cancer cells. Endogenous ER-α and MGMT proteins were physically associated in (i) MCF7 and (ii) T47D cells. Cell lysates were immunoprecipitated using either MGMT or ER-α antibodies. The resulting Western blots (WB) were probed with reciprocal antibodies to show the association. Similar results were obtained in two independent experiments. MGMT: O6-methylguanine DNA-methyltransferase. ER: estrogen receptor; C: control.
BG-induced co-elimination of MGMT and ER-α proteins in MCF-7 and T47D cells
In further exploring the possible co-degradation of the partner proteins observed in Fig. 3, we surmised that either inhibition of MGMT or that of ER-α by their known inhibitors will lead to a degradation of both proteins. Therefore, we exposed the ER-positive and MGMT-proficient MCF-7 and T47D cells to BG and performed Western blot analyses. Fig. 4A and B show that as expected, the MGMT protein levels deceased by approximately. 50% in both cell lines reflective of its degradation through the ubiquitin-proteolytic pathway[21]. Consistent with our postulate, the ER-α protein levels were also diminished with similar kinetics as MGMT (Fig. 4AB). Interestingly, the same extracts showed a time-dependent disappearance of the CDK inhibitor p21cip1 (which we have shown to associate with MGMT[32]. The co-elimination of proteins, however, appeared to be a specific phenomenon, because the steady-state levels of other proteins such as the DNA polδ, PCNA, topo I, CDK1 and APE1 which also bind with MGMT[32] were not altered in BG-treated MCF-7 cells (Fig. 4C).
Fig. 4. Specific degradation of estrogen receptor-α (ER-α) and MGMT proteins by BG in breast cancer cells.
A: Elimination of ER-α, MGMT and p21cip1 proteins with similar kinetics in BG-treated MCF-7 cells. B: Degradation of ER-α and MGMT proteins in BG-treated T47D cells. C: Specificity in protein degradation induced by BG. The same MCF7 extracts used in Fig. 4A were probed with antibodies to DNA pol δ, PCNA, topoisomerase 1 (Topo I), CDK1, and APE1. Please note the lack of alterations in their steady-state levels after BG treatment. MGMT: O6-methylguanine DNA-methyltransferase; BG: O6-benzylguanine; C: control.
ER-dependent inhibition of MGMT activity by fulvestrant in breast cancer cells
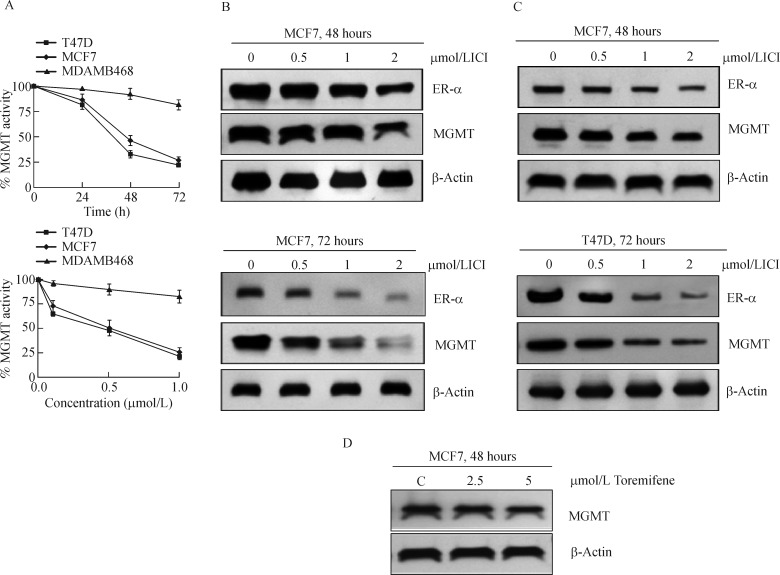
The attenuation of MGMT protein observed in fulvestrant-treated cells (Fig. 3B) implied a downregulation of the associated DNA repair activity. Therefore, the catalytic activity of MGMT was assayed in fulvestrant-treated MCF-7 and T47D cells. In both of these ER-α positive cells, the inhibition was 50%-65% at 48 hours and about 75% at 72 hours after 1 μmol/L fulvestrant treatment; in contrast, the anti-estrogen did not inhibit MGMT in the ER-negative MDAMB-468 cells (Fig. 5A, upper panel). The inhibition of MGMT activity was also proportionate with the fulvestrant concentration (0-1 μmol/L) during 72 hours incubation (Fig. 5A, lower panel). These results demonstrate that fulvestrant curtails the MGMT activity only in ER-α expressing cells.
Fig. 5. Concentration and time-dependent inhibition/elimination of MGMT and ER-α proteins in response to fulvestrant in breast cancer cells.
A: Time dependent loss of MGMT activity in ERα-positive MCF7 and T47D tumor cells but not in ERα-negative MDAMB-468 cells after 1 μmol/L Fulvestrant treatment (upper panel). Concentration dependent inhibition of MGMT activity in MCF7 and T47D cells but not in MDA-MB-468 cells after a 72 hours exposure to fulvestrant (lower panel). The data represents the mean of two independent experiments in duplicate and the last time point (72 hours) as assessed by student's t-test was significant at P < 0.05. B: Decreased MGMT and ER-α protein levels in MCF7 cells after 48 hours and 72 hours fulvestrant exposure. ICI is fulvestrant, also called ICI-182,780. C: Decreased MGMT and ER-α protein levels in T47D cells after 48 hours and 72 hours fulvestrant exposure. D: Marginal degradation of MGMT in MCF7 cells treated with toremifene, a SERM. MGMT: O6-methylguanine DNA-methyltransferase; C: control.
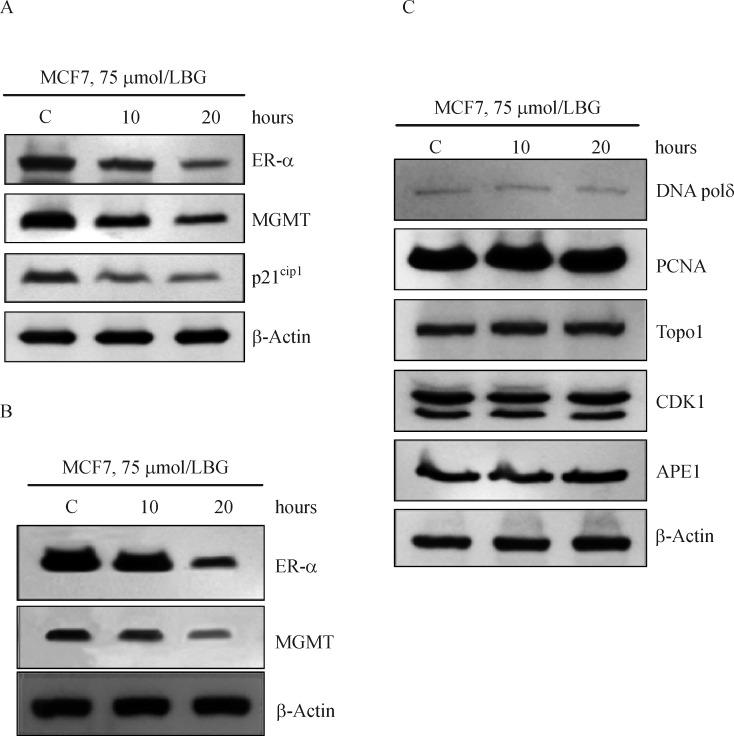
Time-dependent co-elimination of ER-α and MGMT proteins by fulvestrant in MCF-7 and T47D cells
Since MGMT performs the de-alkylation from O6-position of guanine in a stoichiometric reaction, the fulvestrant-induced inhibition (Fig. 5A) is likely to result from decreased MGMT protein levels. Furthermore, fulvestrant destabilizes the ER-α and triggers its degradation through the ubiquitin-proteasome pathway[11, 49–50]. Therefore, direct Western blot analysis was performed to determine the protein degradation patterns in fulvestrant-treated cells (Fig. 5B& C). It is clear that fulvestrant induced a simultaneous decrease of ER-α and MGMT proteins, apparently with similar kinetics; protein disappearance was more evident at 72 hours in MCF-7 cells and well discernible at both 48 and 72 hours in T47D cells. Collectively, the results point to fulvestrant-mediated co-destruction of the partner proteins presented in a complex (Fig. 5B, 5C).
Toremifene induces only a weak elimination of MGMT protein
Estrogen receptor binding with its ligands (irrespective of the type, natural ligand, SERM or SERD) is known to induce the degradation of the receptor protein, and this elimination occurs through the ubiquitin (ub)-proteasomal pathway[51-52]. The stability of the receptor-ligand complex, rate of receptor degradation, however, varies depending on the ligand, its affinity and the consequent structural perturbation of the receptor[51]. Thus, the binding of the natural ligand, estrogen, also leads to ubiquitination-dependent elimination of the ER, however, providing adequate time for the signaling to occur[51-52]. Similarly, the SERM, tamoxifen appears to trigger the degradation of ER through the ubiquitin proteolysis[51], and fulvestrant follows the same route as well[11,49-50]. Consistent with this notion and supportive of our observations of co-elimination of ER and MGMT is a weak degradation of MGMT seen in tamoxifen treated HT29 colon cancer cells[42]. Toremifene, a SERM and an analog of tamoxifen induced only < 10% decrease of MGMT protein in MCF-7 cells (Fig. 5D), thereby, supporting the concept that cellular catabolism of ER and MGMT is linked and driven by the proteolytic rate of these partner proteins.
Evidence for proteasomal involvement in the inhibitor–induced co-degradation of ER-α and MGMT proteins
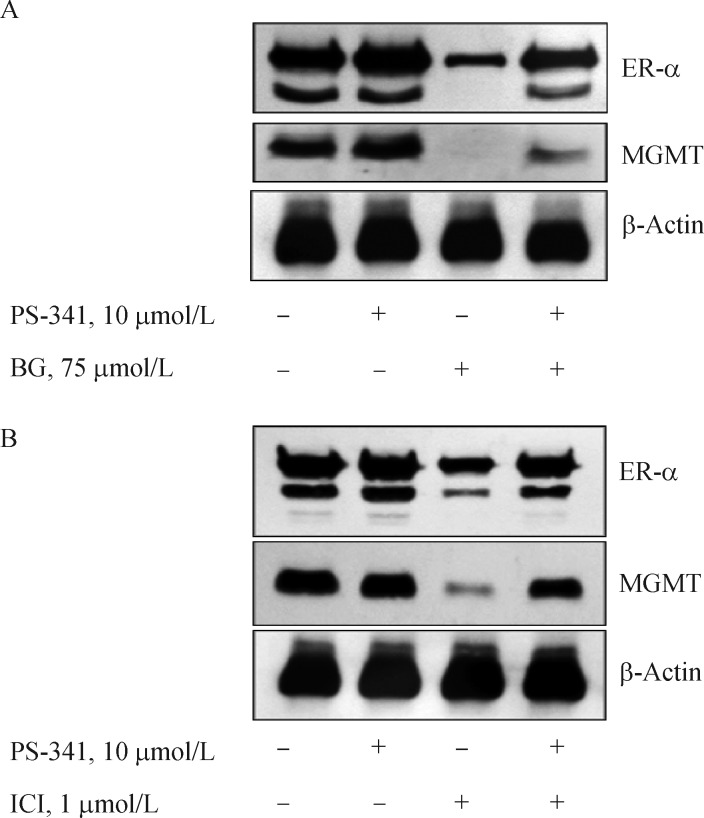
In Figs. 4 and 5, we showed the degradation of both ER-α and MGMT proteins by BG and fulvestrant respectively. Because the inactivation of both proteins is known to trigger ubiquitation and subsequent digestion by the proteasome[21,50], bortezomib (PS-341), a clinically used proteasome inhibitor, was used to verify the co-degradation induced by the reciprocal inhibitors. MCF-7 cells were pretreated with or without PS-341 for 6 h followed by exposure to 75 μmol/L BG for 24 h. Western blots clearly showed that proteasome inhibition stabilized both ER-α and MGMT from BG-induced degradation (Fig. 6A). Similarly, the PS341 pretreatment protected against the fulvestrant-induced elimination of both ER-α and MGMT proteins (Fig. 6B). These results conclusively prove that both proteins are targeted for destruction even one of them is inactivated and confirms proteasome participation in the digestion.
Fig. 6. Proteasome inactivation curtailed the degradation of both ER-α and MGMT proteins in BG or fulvestrant treated MCF-7 cells.
A: BG treated cells, B: Fulvestrant treated cells. MCF7 cells were treated with 10 μmol/L PS341 alone for 6 hours (lane 2), with 75 μmol/L BG (24 hours) or 1 μmol/L Fulvestrant (72 hours) alone (lane 3), pretreated 6 hours with 10 μmol/L PS341 followed by 75 μmol/L BG (24 hours) or 1 μmol/L Fulvestrant (72 hours) for 12 hours (lane 4). Cell lysates were Western blotted for ER-α and MGMT proteins. Similar results were observed in three separate experiments. PS-341 is Bortezomib or Velcade, a clinically used proteasome inhibitor. MGMT: O6-methylguanine DNA-methyltransferase; ER: estrogen receptor; BG: O6-benzylguanine; ICI: fulvestrant; C: control.
BG or fulvestrant did not alter the mRNA levels of their respective targets
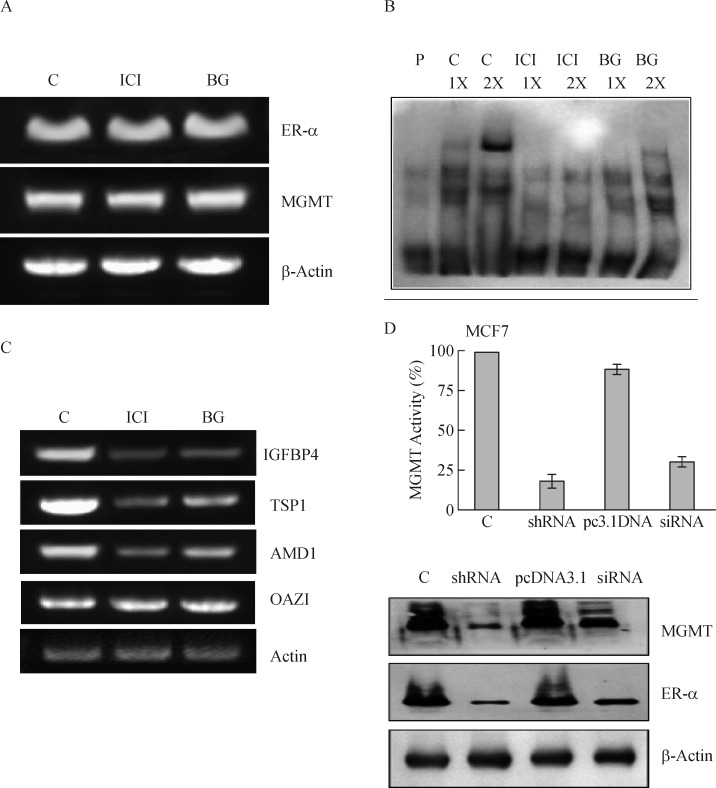
To rule out the possibility of transcriptional or posttranscriptional effects as factors in the attenuated levels of ER-α and MGMT proteins (Fig. 4 and 5), we performed RT-PCR assays in BG or fulvstrant treated MCF-7 cells. As shown in Fig. 7A, the gene transcript levels for MGMT and ER-α remained similar 24 h after inhibitor treatments. The data supports the suggestion that protein decreases observed in response to the reciprocal inhibitors occurred mainly at the level of protein elimination.
Fig. 7. BG-induced abrogation of ER-α binding with DNA, attenuated expression of ER-targeted genes, and mutual dependence in the steady-state levels of ER-α and MGMT proteins in MCF-7 cells.
A: No alterations occur in gene transcripts for ER-α and MGMT after BG or fulvestrant treatments. Total RNA (1 μg) isolated from control and drug-treated cells (1 μmol/L fulvestrant or 50 μmol/L BG for 24 h) followed by RT-PCR. The PCR products were electrophoresed on an agarose gel, stained with ethidium bromide and photographed. B: BG or fulvestrant treatment of MCF-7 cells resulted in the elimination of ER-α binding to its recognition sequence. EMSA was performed as described in Methods. C: Loss of ER-α-protein induced by BG or fulvestrant affects the expression of ER-target genes similarly. Total RNA was extracted from control, 50 μmol/L BG or 1 μmol/L fulvestrant treated cells for 48 hours and the target gene expression [IGFBP-4, thrombospondin 1, S-adenosylmethionine decarboxylase (AMD1)] was quantitated by RT-PCR. OAZ1 (antizyme for ornithine decarboxylase) is not known to be a target for ER-α and was used as a control. D: Silencing of MGMT expression attenuates both ER-α and MGMT protein levels. MCF7 cells were transfected with a plasmid encoding MGMT shRNA (1μg) or with an MGMT siRNA (20 nmol/L). Forty eight hours later, the cell extracts were analyzed for the DNA repair activity of MGMT (upper panel) and ER-α and MGMT protein levels (lower panel). A mutual decrease in the levels of both proteins was clearly evident in two independent experiments. MGMT: O6-methylguanine DNA-methyltransferase; ER: estrogen receptor; BG: O6-benzylguanine; C; control; ICI: fulvestrant.
Diminished binding of ER to ERE in cells treated with BG or fulvestrant
To demonstrate that reduced ER-α protein levels observed in BG or fulvestrant treated breast cancer cells, EMSA was performed using a biotinylated consensus ERE sequence as described in Methods. Nuclear extracts were prepared 24 hours after inhibitor treatments served as the source of ER in DNA binding assays. Fig. 7B reveals a significant binding of ER with its target DNA (top band in lanes 2 and 3) in untreated control cells, which was inhibited in BG or fulvestrant cell treatments. The binding was specific because a non-biotinylated probe abolished the signal (not shown).
Downregulated transcription of ER-α target genes in BG or fulvestrant-treated breast cancer cells
A major finding of this study is that BG, a specific inhibitor of MGMT, can downregulate the ER –α protein in hormone-responsive breast cancer cells. All biological actions of estrogen are mediated by estrogen binding to its receptors; therefore, to confirm the anticipated attenuation of estrogenic signaling, we determined the expression levels of three genes known to be transactivated by the ER-α. These are thrombospondin (TSP; 36), IGF binding protein 4 (IGFBP4; 37) and AMD1 (S-adenosylmethionine decarboxylase; 38). The ornithine decarboxylase antizyme (OAZ1), a gene unresponsive to ER-α[38], was also assessed as a control. RT-PCR analysis was performed using the total RNA isolated from untreated, BG- and fulvestrant-treated MCF-7 cells. As shown in Fig. 7C, the transcripts for IGFBP4, TSP1 and AMD1 were suppressed significantly upon both fulvestrant and BG treatments, whereas the OAZ1 was unaffected. The results indeed substantiate a diminished ER-α mediated transcription by MGMT inhibition.
MGMT gene silencing in MCF-7 cells results in diminished ER-α protein levels
In view of the strong protein–protein association and co-degradation patterns observed, it was of interest to explore whether the MGMT and ER-α genes are expressed in a coordinate manner. For this, we silenced the MGMT gene in MCF-7 cells using specific siRNA and shRNA for 24 h and determined the levels of ER-α, MGMT proteins. The upper panel of Fig. 7D shows that both the RNA interference procedures selectively downregulated the DNA repair activity of MGMT by 70-85%. Consistent with the stoichiometric mechanism of MGMT, its protein levels were also reduced to similar extents by the siRNA and shRNA treatments. While these results were expected, it was intriguing to note a significant reduction of ER-α protein in cells treated with MGMT-specific siRNA or shRNAs (Fig. 7D lower panel). Probing for ER-α expression following RNAi intervention showed a 70% inhibition after shRNA transfection and a 40% inhibition following siRNA. Lack of gene expression changes in the MCF-7 cells transfected with the scrambled siRNA (not shown) pcDNA 3.1 vector (Fig. 7D) further validated the results obtained. The molecular mechanisms underlying this mutual dependent expression of ER-α and MGMT proteins remain unclear at present. However, such a coordinated and mutually–dependent maintenance of protein levels is known for a number of gene products. These include the production of the ribosomal proteins to maintain equivalent levels of different proteins that associate with the rRNA[53]. Other partners are the Gpn1 and Gpn3 GTPases[54] and Raptor and mTOR proteins[55], which all associate tightly and their steady-state protein levels share a reciprocal relationship. The significance of these associations has been discussed in the conclusion section.
Fulvestrant induced MGMT repair-deficient state facilitates a greater DNA damage interstrand crosslinking in breast cancer cells
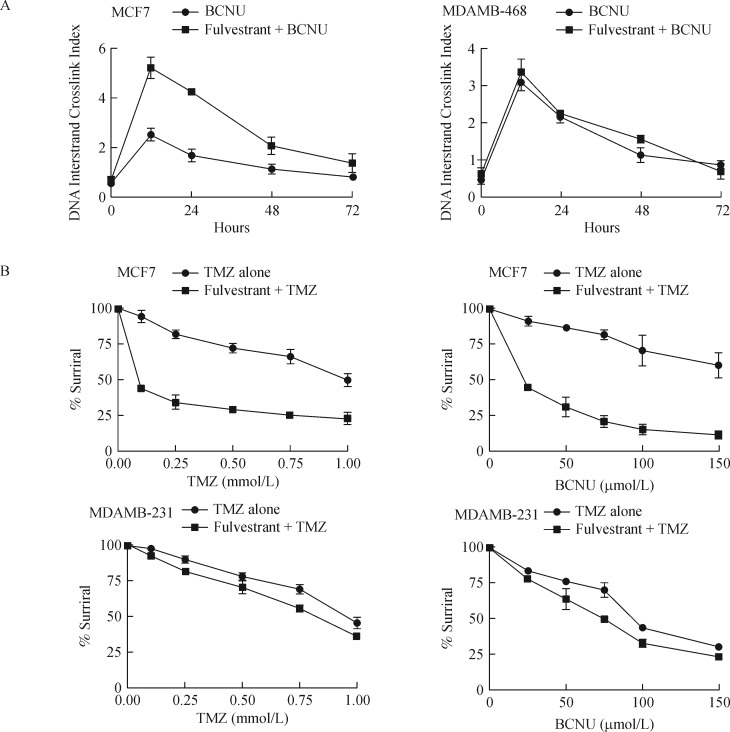
Fulvestrant-induced inhibition and the subsequent reduction of MGMT protein (Fig. 5) is expected to augment the levels of alkylation DNA damage and the interstrand cross-linking of DNA induced by bifunctional alkylating agents. This postulate was tested by the MCF-7 cells with fulvestrant for 48 hours followed by BCNU, a bifunctional alkylator known to induce G-C interstrand DNA crosslinking. An ethidium bromide fluorescence assay, well established in our laboratory, was used to determine the levels of cellular DNA crosslinking. MCF-7 cells treated with fulvestrant, and BCNU showed more than 2-fold increase in interstrand cross-links as compared with BCNU alone (Fig. 8A, left panel). However, fulvestrant was unable to increase the DNA crosslinks in the MGMT-proficient and ER-α-negative MDAMB-468 breast cancer cells (Fig. 8A, right panel). These data confirm that MGMT downregulation by fulvestrant was strictly ER-dependent.
Fig. 8. Fulvestrant increases the DNA damage induced by MGMT-targeted alkylating agents and triggers synergistic cytotoxicity in ER-α positive MCF7 cells, but not in ER- α negative MDAMB-468 cells.
A: Kinetics of DNA interstrand crosslinks formed in MCF7 cells after treatment with 100 μmol/L BCNU. MDAMB-468 cells that do not express ER-α but possess MGMT, served as the control. Cells were treated or untreated with 1 μmol/L fulvestrant for 48 hours to deplete the MGMT protein, following which they were exposed to 100 μmol/L BCNU as described in Methods. At times specified, the cells were harvested, DNA isolated and the extent of interstrand crosslinking of DNA was determined by the ethidium bromide fluorescence assay. Values are mean±S.D. DNA crosslinking by BCNU in cells peaks around 6-12 hours; The 2-fold increased DNA crosslinking found at 12 hours in fulvestrant+ BCNU treated cells was significant at P < 0.05. B: Fulvestrant pre-exposure sensitized the MCF7 tumor cells (ER-α+ and MGMT+) but not the MDAMB-231 tumor cells (ER-α− and MGMT−) to the clinically used alkylation agents. Cells were treated with fulvestrant (1 μmol/L) for 48 hours following which they were treated with increasing concentrations of temozolomide (TMZ) or BCNU. Cells were then cultured for 3 days before performing the MTT assays. Since a 1 μmol/L concentration for Fulvestrant generated a less than 5% cell killing (not shown), this concentration was chosen to potentiate the cytotoxicity of alkylating drugs. The results assessed by Student's t-test were significant at P < 0.05.
Fulvestrant markedly sensitizes MGMT and ER-α proficient breast cancer cells to alkylating agents
To test whether the downregulation of MGMT caused by fulvestrant translates to increased efficacy of clinically used alkylating agents that generate O6-alkylguanines, the ER-α positive MCF-7 and –negative MDAMB 231 cells were pretreated with fulvestrant for 48 hours and then exposed to temozolomide or BCNU at varying concentrations. Cell survival assays using the MTT were performed. In this setting, TMZ (0–1000 mmol/L) + fulvestrant combination showed a 3-fold increased cytotoxicity compared with TMZ alone (Fig. 8B, upper left). For BCNU, the potentiation was about 4-fold (Fig. 8B, upper right). Fulvestant, however, did not potentiate the cytotoxicity of TMZ or BCNU in the ER-α non-expressing MDAMB 231 cells (Fig. 8B, lower panels) again suggesting a nexus between the receptor and the MGMT protein mediates the augmented alkylator cytotoxicity. The data points to a novel way of inhibiting the MGMT-mediated DNA repair during endocrine therapy and suggests that addition of alkylating agents therewith may enhance breast cancer treatment.
Discussion
Our studies uncovered several new and important elements of physical and functional relationship between the human MGMT and ER-α proteins. First, the presence and functionality of the EREs in MGMT promoter were demonstrated. Next, the existence of these proteins as specific complexes in breast cancer cells was shown. The data revealed a specific association of the MGMT and ER-α proteins in vivo, and this complex formation is likely to occur in the nuclei. Addition of the recombinant MGMT protein in EMSAs did not interfere with the binding of estrogen receptor with its recognition sequences (data not shown); therefore, the physical binding is unlikely to influence the ligand and DNA binding functions of the ER-α. Further, the steady-state levels of MGMT-ER-α protein pair also appeared to be mutually dependent as shown by the siRNA and shRNA studies (Fig. 7D); silencing of MGMT expression using RNA interference showed that decreased MGMT protein were accompanied by diminished ER-α protein levels. The data indicate a coordinated synthesis of the partner proteins.
More notably, the response of the MGMT/ER-α proteins to their respective inhibitors was interesting and significant. Both proteins underwent proteasomal degradation in breast cancer cells irrespective of whether the cells were exposed to an MGMT inhibitor such as the BG, or an ER inhibitor, fulvestrant. To explain this mutual co-destruction or synchronized proteolysis, we propose that the close association and proximity of the ER-α and MGMT plays a major role; functional inactivation of either of the proteins may result in the simultaneous targeting of both partners for ubiquitin conjugation, perhaps by a single ubiquitin-ligase, and subsequent proteasomal disposition (Fig. 9). Various lines of data, including the stabilization of the inactivated proteins through proteasome inhibition (Fig. 6) are consistent with this postulate. Indeed, there are several proteins that tightly associate with each other, whose steady-state levels are finely co-regulated as described for the MGMT and ER-α as described here. These include the GTPases Gpn1 and GP3 that function in nuclear targeting of RNA polymearase II[54], the Raptor and mTOR that regulate cell growth in response to the nutrient and energy status of the cell[55], the ARP2/3 and IFO-1 proteins that regulate the cell polarity[56]. There is also an example of a coordinated degradation that involves the MGMT itself; the BRCA2 protein has been reported to undergo co-degradation with MGMT in BG-treated human cells[57].
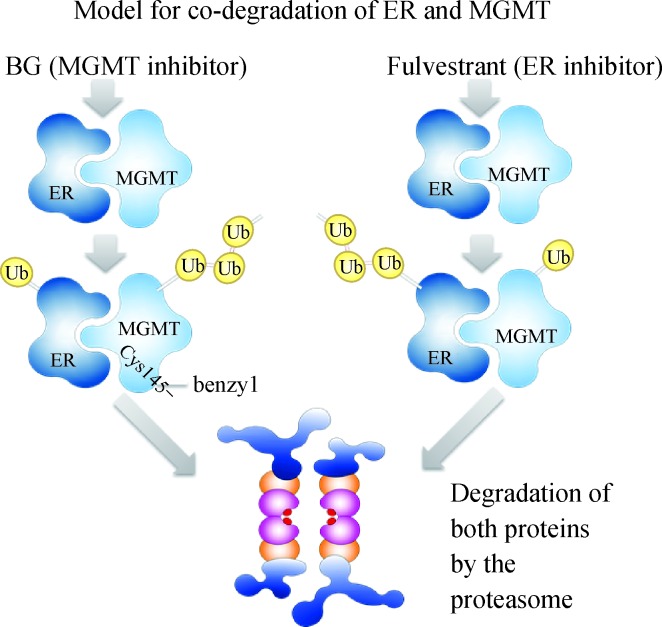
Fig. 9. Scheme showing a model for co-degradation of ER-α and MGMT proteins.
Based on the physical association of ER-α and MGMT, and other findings of this study, the inactivation of either protein in the complex is postulated to trigger the ubiquitinylation of both proteins followed by proteasomal digestion. Ub, ubiquitin.
A previous study implied that the alkylated (inactivated) human MGMT is a negative regulator of ER-mediated transcription following alkylation damage in DNA[26]. They postulated that since the ER-signaling promotes cell proliferation, to stop replication on an alkylated genome, the functionally inactivated MGMT would assume an altered conformation and bind with the ER-α to halt the estrogenic signaling[26]. In contrast, our experiments showed that the binding of ER-α with the functionally active MGMT in breast cancer cells and highlighted the versatile interplay between these regulators. The close in vivo association of ER-α and MGMT indicates specific physiologic functions for the complex, which, however, remains unclear. Despite this uncertainty, our findings of a co-degradation of these partners by fulvestrant have significant therapeutic implications. Fulvestrant (Faslodex®) is a SERD with a long half-life, administered once a month at 250 or 500 mg doses by intramuscular injections to postmenopausal women with ER- positive breast cancer[11]. SERMs such as the tamoxifen and toremifene used in endocrine therapy bind the ER and inhibit the activation function 2 (AF2), but fail to inhibit AF1 activity of the receptor, and thereby have partial estrogen agonist activity. Fulvestrant, as a pure anti-estrogen is able to inhibit both AF1 and AF2 and thus has no agonist activity at all. Structurally, fulvestrant alters and destabilizes the ER, completely blocking ER-mediated transcription and greatly accelerates the receptor degradation (Fig. 10). Although all ER-ligands including the tamoxifen and toremifene ultimately induce the receptor break-down through the uniquitin-proteolytic pathway[50–52], they seem to eliminate the receptor at slower rates than fulvestrant. We propose that the augmented turnover of ER in the ER-MGMT complex by fulvestrant also accounts for the enhanced degradation of MGMT we observed in breast cancer cells. The destruction of MGMT in the ER complexes does occur in tamoxifen treated cells, however, to a lower level, as has been implied by previous observations on the ability of BG to overcome tamoxifen resistance[28] and tamoxifen-induced ubiquitin degradation of MGMT in HT29 cells[42]. Therefore, our finding on the ability of fulvestrant to trigger MGMT deficiency can be rationally exploited for improved therapy by combining fulvestrant with MGMT-targeted alkylating agents to obtain greater anticancer efficacy. Work is underway to test and validate these significant observations in breast cancer xenografts.
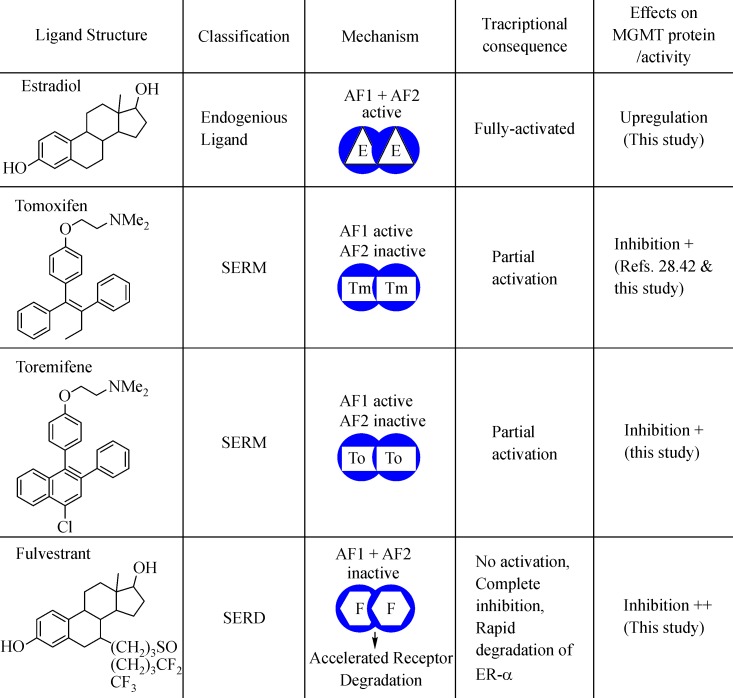
Fig. 10. Consequences of ligand binding with ER-α on transcription, receptor stability, and MGMT stability are summarized.
Binding of any ligand, be it may be the natural or synthetic estrogens, SERMs (tamoxifen, toremifene), or a SERD (fulvestrant) alters the structure and stability of the receptor, ultimately leading to its break-down through ubiquitination-dependent proteolysis. However, the rate and extent of this destabilization depend on the ligand. Fulvestrant deforms the receptor greatly and hastens its degradation; since MGMT forms a complex with ER-α, we propose that it is co-degraded with the receptor leading to the DNA repair deficiency and sensitization to O6-alkylguanine generating drugs. AF, activating function; E, estrogen, F, fulvestrant; Tm, tamoxifen, To, toremifene.
Acknowledgements
This work was supported by grants from the Cancer Prevention Research Institute of Texas (RP130266), the Carson-Leslie Foundation and the Association for Research of Childhood Cancer, all to K.S.S. We thank Drs. Sankar Mitra (Univ. of Texas Medical Branch) and Anil Jaiswal (Univ. of Maryland) for MGMT promoter constructs and Nrf2 expression vector respectively.
References
- [1].Heldring N, Pike A, Andersson S, et al. Estrogen receptors: how do they signal and what are their targets[J]. Physiol Rev, 2007, 87(3): 905–931. [DOI] [PubMed] [Google Scholar]
- [2].Klinge CM, Jernigan SC, Mattingly KA, et al. Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors alpha and beta by coactivators and corepressors[J]. J Mol Endocrinol, 2004, 33(2): 387–410. [DOI] [PubMed] [Google Scholar]
- [3].Dechering K, Boersma C, Mosselman S. Estrogen receptors alpha and beta: two receptors of a kind[J]? Curr Med Chem, 2000, 7(5): 561–576. [DOI] [PubMed] [Google Scholar]
- [4].Huang B, Omoto Y, Iwase H, et al. Differential expression of estrogen receptor α, β1, and β2 in lobular and ductal breast cancer[J]. Proc Natl Acad Sci USA, 2014, 111(5): 1933–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Masood S. Use of monoclonal antibody for assessment of estrogen and progesterone receptors in malignant effusions[J]. Diagn Cytopathol, 1992, 8(2): 161–166. [DOI] [PubMed] [Google Scholar]
- [6].Sommer S, Fuqua SA. Estrogen receptor and breast cancer[J]. Semin Cancer Biol, 2001, 11(5): 339–352. [DOI] [PubMed] [Google Scholar]
- [7].Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance[J]. Nat Rev Cancer, 2002, 2(2): 101–112. [DOI] [PubMed] [Google Scholar]
- [8].Burstein HJ, Temin S, Anderson H, et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: american society of clinical oncology clinical practice guideline focused update[J]. J Clin Oncol, 2014, 32(21): 2255–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nazarali SA, Narod SA. Tamoxifen for women at high risk of breast cancer[J]. Breast Cancer (Dove Med Press), 2014, 6: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McDonnell DP, Wardell SE, Norris JD. Oral selective estrogen receptor downregulators (SERDs), a breakthrough endocrine therapy for breast cancer[J]. J Med Chem, 2015, 58(12): 4883–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Osborne CK, Wakeling A, Nicholson RI. Fulvestrant: an oestrogen receptor antagonist with a novel mechanism of action[J]. Br J Cancer, 2004, 90(Suppl 1): S2–S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Miller K. Estrogen and DNA damage: the silent source of breast cancer[J]? J Natl Cancer Inst, 2003, 95(2): 100–102. [DOI] [PubMed] [Google Scholar]
- [13].Caldon CE. Estrogen signaling and the DNA damage response in hormone dependent breast cancers[J]. Front Oncol, 2014, 4: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Walerych D, Napoli M, Collavin L, et al. The rebel angel: mutant p53 as the driving oncogene in breast cancer[J]. Carcinogenesis, 2012, 33(11): 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Allison KH. Molecular pathology of breast cancer: what a pathologist needs to know[J]. Am J Clin Pathol, 2012, 138(6): 770–780. [DOI] [PubMed] [Google Scholar]
- [16].Cayre A, Penault-Llorca F, De Latour M, et al. O(6)-methylguanine-DNA methyl transferase gene expression and prognosis in breast carcinoma[J]. Int J Oncol, 2002, 21(5): 1125–1131. [PubMed] [Google Scholar]
- [17].Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases[J]. Mutat Res, 2000, 462(2-3): 83–100. [DOI] [PubMed] [Google Scholar]
- [18].Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage[J]. Chem Rev, 2006, 106(2): 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gerson SL. Clinical relevance of MGMT in the treatment of cancer[J]. J Clin Oncol, 2002, 20(9): 2388–2399. [DOI] [PubMed] [Google Scholar]
- [20].Kaina B, Margison GP, Christmann M. Targeting O6-methylguanine-DNA methyltransferase with specific inhibitors as a strategy in cancer therapy[J]. Cell Mol Life Sci, 2010, 67(21): 3663–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Srivenugopal KS, Yuan XH, Friedman HS, et al. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea[J]. Biochemistry, 1996, 35(4): 1328–1334. [DOI] [PubMed] [Google Scholar]
- [22].Srivenugopal KS, Rawat A, Niture SK, et al. Posttranslational regulation of O6-methylguanine-DNA methyltransferase (MGMT) and new opportunities for treatment of brain cancers.See comment in PubMed Commons below[J]. Mini Rev Med Chem, 2016, 16(6): 455–464. [DOI] [PubMed] [Google Scholar]
- [23].Chang BD, Beattie CW, Hussain RA, et al. Estrous cycle modulation of O6-alkylguanine-DNA alkyltransferase expression in rat mammary epithelial cells[J]. Cancer Lett, 1993, 75(1): 11–18. [DOI] [PubMed] [Google Scholar]
- [24].Dutta-Choudhury TA, Bak BH, Guzman RC. Cellular levels of O6-methylguanine-DNA methyltransferase in mammary epithelial cells and liver from virgin, pregnant and pituitary grafted mice[J]. Carcinogenesis, 1991, 12(10): 1795–1800. [DOI] [PubMed] [Google Scholar]
- [25].Henderson BE, Feigelson HS. Hormonal carcinogenesis[J]. Carcinogenesis, 2000, 21(3): 427–433. [DOI] [PubMed] [Google Scholar]
- [26].Teo AK, Oh HK, Ali RB, et al. The modified human DNA repair enzyme O(6)-methylguanine-DNA methyltransferase is a negative regulator of estrogen receptor-mediated transcription upon alkylation DNA damage[J]. Mol Cell Biol, 2001, 21(20): 7105–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Musarrat J, Wilson JA, Abou-Issa H, et al. O(6)-alkylguanine DNA alkyltransferase activity levels in normal, benign and malignant human female breast[J]. Biochem Biophys Res Commun, 1995, 208(2): 688–696. [DOI] [PubMed] [Google Scholar]
- [28].Bobustuc GC, Smith JS, Maddipatla S, et al. MGMT inhibition restores ERα functional sensitivity to antiestrogen therapy[J]. Mol Med, 2012, 18(8): 913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Citron M, Schoenhaus M, Rothenberg H, et al. O6-methylguanine-DNA methyltransferase in normal and malignant tissue of the breast[J]. Cancer Invest, 1994, 12(6): 605–610. [DOI] [PubMed] [Google Scholar]
- [30].Isono S, Fujishima M, Azumi T, et al. O(6)-methylguanine-DNA methyltransferase as a prognostic and predictive marker for basal-like breast cancer treated with cyclophosphamide-based chemotherapy[J]. Oncol Lett, 2014, 7(6): 1778–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hansen RJ, Ludeman SM, Paikoff SJ, et al. Role of MGMT in protecting against cyclophosphamide-induced toxicity in cells and animals[J]. DNA Repair (Amst), 2007, 6(8): 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Niture SK, Doneanu CE, Velu CS, et al. Proteomic analysis of human O6-methylguanine-DNA methyltransferase by affinity chromatography and tandem mass spectrometry[J]. Biochem Biophys Res Commun, 2005, 337(4): 1176–1184. [DOI] [PubMed] [Google Scholar]
- [33].Paranjpe A, Zhang R, Ali-Osman F, et al. Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage[J]. Carcinogenesis, 2014, 35(3): 692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Klinge CM. Estrogen receptor interaction with estrogen response elements[J]. Nucleic Acids Res, 2001, 29(14): 2905–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Velu CS, Niture SK, Doneanu CE, et al. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress[J]. Biochemistry, 2007, 46(26): 7765–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harada H, Nakagawa K, Saito M, et al. Introduction of wild-type p53 enhances thrombospondin-1 expression in human glioma cells[J]. Cancer Lett, 2003, 191(1): 109–119. [DOI] [PubMed] [Google Scholar]
- [37].Sato H, Yazawa T, Suzuki T, et al. Growth regulation via insulin-like growth factor binding protein-4 and-2 in association with mutant K-ras in lung epithelia[J]. Am J Pathol, 2006, 169(5): 1550–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gross JA, Fiori LM, Labonté B, et al. Effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide[J]. J Psychiatr Res, 2013, 47(4): 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jiang C, Guo J, Wang Z, et al. Decursin and decursinol angelate inhibit estrogen-stimulated and estrogen-independent growth and survival of breast cancer cells[J]. Breast Cancer Res, 2007, 9(6): R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Niture SK, Rao US, Srivenugopal KS. Chemopreventative strategies targeting the MGMT repair protein: augmented expression in human lymphocytes and tumor cells by ethanolic and aqueous extracts of several Indian medicinal plants[J]. Int J Oncol, 2006, 29(5): 1269–1278. [PubMed] [Google Scholar]
- [41].Ali-Osman F, Rairkar A, Young P. Formation and repair of 1,3-bis-(2-chloroethyl)-1-nitrosourea and cisplatin induced total genomic DNA interstrand crosslinks in human glioma cells[J]. Cancer Biochem Biophys, 1995, 14(4): 231–241. [PubMed] [Google Scholar]
- [42].Kuo CC, Liu JF, Shiah HS, et al. Tamoxifen accelerates proteasomal degradation of O6-methylguanine DNA methyltransferase in human cancer cells[J]. Int J Cancer, 2007, 121(10): 2293–2300. [DOI] [PubMed] [Google Scholar]
- [43].Harris LC, Potter PM, Tano K, et al. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene[J]. Nucleic Acids Res, 1991, 19(22): 6163–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Biswas T, Ramana CV, Srinivasan G, et al. Activation of human O6-methylguanine-DNA methyltransferase gene by glucocorticoid hormone[J]. Oncogene, 1999, 18(2): 525–532. [DOI] [PubMed] [Google Scholar]
- [45].Niture SK, Velu CS, Smith QR, et al. Increased expression of the MGMT repair protein mediated by cysteine prodrugs and chemopreventative natural products in human lymphocytes and tumor cell lines[J]. Carcinogenesis, 2007, 28(2): 378–389. [DOI] [PubMed] [Google Scholar]
- [46].Al-Sawaf O, Clarner T, Fragoulis A, et al. Nrf2 in health and disease: current and future clinical implications[J]. Clin Sci (Lond), 2015, 129(12): 989–999. [DOI] [PubMed] [Google Scholar]
- [47].Kansanen E, Kuosmanen SM, Leinonen H, et al. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer[J]. Redox Biol, 2013, 1(1): 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yao Y, Brodie AM, Davidson NE, et al. Inhibition of estrogen signaling activates the NRF2 pathway in breast cancer[J]. Breast Cancer Res Treat, 2010, 124(2): 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Carlson RW. The history and mechanism of action of fulvestrant[J]. Clin Breast Cancer, 2005, 6(Suppl 1): S5–S8. [DOI] [PubMed] [Google Scholar]
- [50].Yeh WL, Shioda K, Coser KR, et al. Fulvestrant-induced cell death and proteasomal degradation of estrogen receptor α protein in MCF-7 cells require the CSK c-Src tyrosine kinase[J]. PLoS ONE, 2013, 8(4): e60889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wijayaratne AL, McDonnell DP. The human estrogen receptor-α is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators[J]. J Biol Chem, 2001, 276(38): 35684–35692. [DOI] [PubMed] [Google Scholar]
- [52].Alarid ET, Bakopoulos N, Solodin N. Proteasome-mediated proteolysis of estrogen receptor: a novel component in autologous down-regulation[J]. Mol Endocrinol, 1999, 13(9): 1522–1534. [DOI] [PubMed] [Google Scholar]
- [53].de la Cruz J, Karbstein K, Woolford JL., Jr Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo[J]. Annu Rev Biochem, 2015, 84: 93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Méndez-Hernández LE, Pérez-Mejía AE, Lara-Chacón B, et al. Gpn1 and Gpn3 associate tightly and their protein levels are mutually dependent in mammalian cells[J]. FEBS Lett, 2014, 588(21): 3823–3829. [DOI] [PubMed] [Google Scholar]
- [55].Kim DH, Sabatini DM. Raptor and mTOR: Subunits of a nutrient-sensitive complex[J]. In: TOR target of rapamycin. 2004, Thomas G, et al. Springer-Verlag, Berlin Heidelberg, 260–269. [DOI] [PubMed] [Google Scholar]
- [56].Carberry K, Wiesenfahrt T, Geisler F, et al. The novel intestinal filament organizer IFO-1 contributes to epithelial integrity in concert with ERM-1 and DLG-1[J]. Development, 2012, 139(10): 1851–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Philip S, Swaminathan S, Kuznetsov SG, et al. Degradation of BRCA2 in alkyltransferase-mediated DNA repair and its clinical implications[J]. Cancer Res, 2008, 68(23): 9973–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]