Abstract
To improve the efficiency of animal production, livestock have been extensively selected or managed to reduce fat accumulation and increase lean growth, which reduces intramuscular or marbling fat content. To enhance marbling, a better understanding of the mechanisms regulating adipogenesis is needed. Vitamin A has recently been shown to have a profound impact on all stages of adipogenesis. Retinoic acid, an active metabolite of vitamin A, activates both retinoic acid receptors (RAR) and retinoid X receptors (RXR), inducing epigenetic changes in key regulatory genes governing adipogenesis. Additionally, Vitamin D and folates interact with the retinoic acid receptors to regulate adipogenesis. In this review, we discuss nutritional regulation of adipogenesis, focusing on retinoic acid and its impact on epigenetic modifications of key adipogenic genes.
Keywords: Adipogenesis, Vitamin A, Retinoic acid, Marbling, Meat, Progenitor cells
1. Introduction
There are four adipose depots: visceral, subcutaneous, intermuscular and intramuscular. The visceral and subcutaneous fat depots develop and mature prior to the other fat depots (Cianzio, Topel, Whitehurst, Beitz, & Self, 1985), accounting for the vast majority of body fat. Due to the low value of visceral and subcutaneous fat, meat animals have been selected for generations for their high lean/fat ratio, resulting in lean animals. However, the selection for high lean growth is negatively associated with intramuscular fat accumulation, or marbling, which is critical for the palatability of meat (Du et al., 2013a; Hausman, Basu, Du, Fernyhough-Culver, & Dodson, 2014; Kauffman, Carpenter, Bray, & Hoekstra, 1964). The use of implants and harvesting at increasingly younger ages are also contributing factors to the low marbling in beef cattle. As a result, marbling, together with tenderness, are consistently identified as the top issues related to beef quality (Garcia et al., 2008; McKenna et al., 2002), with only 2.1% of beef carcasses exhibiting the slightly abundant marbling necessary to grade as prime, the highest quality grade (Moore et al., 2012). While increasing the marbling present in beef would greatly enhance beef quality and consumer eating experiences, fat accumulation in the other fat depots is a waste. Thus, advanced strategies which can enhance marbling without increasing or even decreasing overall adiposity is needed.
Both adipocyte hyperplasia and hypertrophy contribute to adipose accumulation. Previous studies have focused on lipid metabolism, as lipid deposition accounts for adipose hypertrophy (Smith et al., 2009). On the other hand, adipogenesis, or the formation of new adipocytes, was less studied in livestock species. Adipogenesis can be separated into multiple stages, including adipogenic commitment, adipogenic differentiation and lipid accumulation. Vitamin A affects each stage of adipogenesis. Retinoic acid, an active metabolite of vitamin A, induces epigenetic changes in adipogenic genes, regulating their expression and adipocyte formation (Dani et al., 1997; Nebbioso et al., 2010; Wei, 2012). In addition, retinoic acid reduces lipid accumulation (Berry, DeSantis, Soltanian, Croniger, & Noy, 2012; Kawada, Kamei, & Sugimoto, 1996; Schwarz, Reginato, Shao, Krakow, & Lazar, 1997b). Other nutrients, such as vitamin D, interacts with retinoic acid receptor signaling to alter adipogenic differentiation and development (Hida, Kawada, Kayahashi, Ishihara, & Fushiki, 1998). Altogether, nutrients have profound impacts on gene expression and cell differentiation. Research in this field is becoming increasingly active, which forms an exciting new field of research, termed nutrigenomics.
2. Adipose tissue development
The formation of discernable adipocytes/adipose tissue begins before mid-gestation in beef cattle (Bonnet, Cassar-Malek, Chilliard, & Picard, 2010). In perirenal fat, adipocytes were detected as early as 80 days of gestation while adipocytes in the intermuscular fat are detectable at 180 days of gestation (Taga et al., 2011; Taga et al., 2012). Most adipocytes are formed during the fetal and early postnatal stages, and adipocyte hyperplasia largely ceases in perirenal fat after birth (Bonnet et al., 2010). In humans, the total number of adipocytes is set when reaching adolescence (Goessling et al., 2009). Though new adipocytes can be generated lifelong, such capacity attenuates as animals become older due to the reduction in the density of adipogenic progenitors (Du, Yin, & Zhu, 2010). Therefore, nutritional and physiological conditions during the fetal, postnatal and early postweaning stages have greater impact on adipogenesis compared to the fattening stage.
Adipogenesis is used to describe the de novo generation of adipocytes, which is roughly separated into two stages: commitment and differentiation (MacDougald & Mandrup, 2002). During the differentiation stage, peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer-binding proteins (C/EBPs) have critical regulatory roles (Avram, Avram, & James, 2007). C/EBPβ/δ is expressed in the very early stage of adipogenesis and triggers the expression of PPARγ (Fajas, Debril, & Auwerx, 2001), an essential transcription factor for adipogenic differentiation (Rosen & MacDougald, 2006; Spiegelman & Flier, 1996). The mechanisms underlying adipogenic commitment are much less studied. Recently, based on studies in mice, Zinc-finger protein 423 (Zfp423) was identified as a transcriptional factor responsible for the adipogenic commitment of progenitor cells (Gupta et al., 2010). The expression of Zfp423 commits progenitor cells to the adipogenic lineage and differentiate into pre-adipocytes, further inducing PPARγ expression, which results in the terminal differentiation of adipocytes (Gupta et al., 2010; Gupta et al., 2012). The importance of Zfp423 in bovine adipogenesis was further confirmed (Huang, Das, Yang, Zhu, & Du, 2012).
Sterol responsive element-binding protein-1c (SREBP-1c) is also an important regulator of adipogenesis, especially during terminal differentiation and lipid accumulation (Feve, 2005). It enhances adipose conversion by stimulating the generation of PPARγ ligands that in turn activate the transcriptional activity of PPARγ (Seo et al., 2004). Consistently, SREBP-1c induces the expression of adipocyte signature genes including fatty acid synthase and fatty acid binding protein (aP2). The expression of these genes leads to the rapid accumulation of lipids in adipocytes, allowing the adipocyte to expand in size. As a result of both the increased size of existing fat cells and the proliferation of preadipocyte cells, white adipose tissue deposition occurs rapidly after birth (Novakofski, 2004).
3. Epigenetic regulation of adipose development
3.1. Epigenetic modifications
Stem cells and progenitor cells maintain their pluri- or multi-potency through reversible inhibition of lineage-specific genes while allowing genes for stem cell self-renewal to express. Conversely, lineage-specific genes are expressed while pluri-or multi-potency genes are inhibited during differentiation (Meissner et al., 2008; Mohn et al., 2008). Progenitor cell commitment to a specific lineage is often initiated by the expression of a key developmental gene, which induces the expression of a cascade of transcription factors and lineage specific genes (Reik, 2007). Key developmental genes possess CpG rich promoters, and their expression is primarily regulated by epigenetic modifications (Aloia, Di Stefano, & Di Croce, 2013), one of which is Zfp423 (Yang et al., 2013).
Epigenetic modifications refer to both histone modifications and DNA methylation. Polycomb repression complexes (PRCs) are mainly responsible for reversible inhibition of genes through catalyzing histone methylations. There are two well-characterized PRCs, namely PRC1 and PRC2. Enhancer of Zeste 2 (EZH2) is one of the core components of PRC2 (Margueron & Reinberg, 2011), which mediates histone 3 lysine 27 trimethylation (H3K27me3) (McCabe et al., 2012; Qi et al., 2012), a marker for gene silencing (Bernstein et al., 2006). A specific DNA binding element for PRC2 has not been previously identified, though PRC2 preferably binds to promoters with rich CpG sites, which subsequently attracts PRC1 binding (Mendenhall et al., 2010; Mohn et al., 2008). In the absence of stimulation to release PRCs, these promoters frequently become DNA methylated (Ko, Hsu, Shen, Chang, & Wang, 2008; Lorente et al., 2006; Mohn et al., 2008).
Trithorax group (trxG) catalyzes H3K4 trimethylation (H3K4me3), activating gene transcription. It appears that H3K4me3 is transient and only induced when gene expression is needed to counter the inhibitory effect of the Polycomb group (Eissenberg & Shilatifard, 2010; Schuettengruber, Martinez, Iovino, & Cavalli, 2011). Interestingly, H3K4me3 and H3K27me3 co-exist in key developmental genes which are highly enriched with CpG sites, forming a ‘bivalent state’ (Meissner et al., 2008; Mikkelsen et al., 2007), which positions genes for activation or inhibition. During differentiation, non-induced bivalent genes lost active H3K4me3 but kept repressive H3K27me3 mark (Schuettengruber & Cavalli, 2009), leading to generally permanent inhibition of gene expression by inducing DNA methylation (Mohn et al., 2008).
Recent studies also point to the importance of DNA demethylation in gene expression. Active DNA demethylation is mediated by ten-eleven translocation hydroxylases (TETs), including TET1, 2 and 3 (Ficz et al., 2011; Ito et al., 2010). In the reaction, TETs oxidize 5-methylcytosine (5mC) to form 5-hydromethylcytosine (5hmC) and further oxidation products. Oxidized cytosines are replaced by nucleotide or base excision repairs to achieve demethylation (Wu & Zhang, 2014), a process mediated by Growth arrest and DNA damage protein 45a (Gadd45a). It is a member of a stress response gene family which encode 18-kDa acidic histone fold proteins (Zhan et al., 1994). Gadd45a mediates nucleotide exchange DNA repair and thus demethylation (Barreto et al., 2007; Ma, Guo, Ming, & Song, 2009; Niehrs & Schafer, 2012). However, Gadd45a protein lacks a DNA binding domain and depends on tumor suppressor inhibitor of growth protein 1 (ING1) to recruit to promoters enriched with H3K4me3, which then triggers locus specific DNA demethylation (Schafer, Karaulanov, Stapf, Doderlein, & Niehrs, 2013). In short, epigenetic modifications include histone methylations, DNA methylation and demethylation, which coordinate to regulate lineage-specific gene expression.
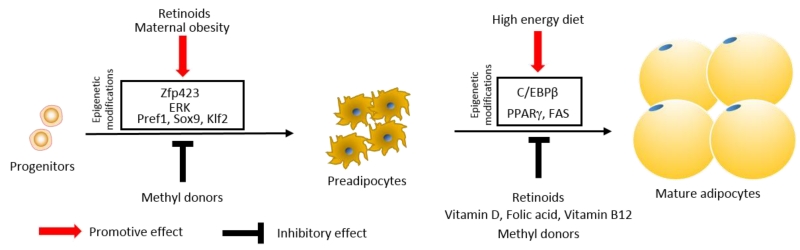
The dynamics of these epigenetic regulatory systems of key developmental genes are affected by both genetic and environmental factors. Gene polymorphisms in the promoters of key developmental genes affect the binding of complexes involved in epigenetic modifications, altering the lineage commitment of progenitor cells during development. Similarly, environmental factors and clues, including nutrients, alter cell signaling pathways or the recruitment of transcription factors which regulate epigenetic modifications to alter animal development, including adipogenesis (Fig. 1).
Fig. 1.
Genetic and environmental factors converge on the epigenome of progenitor cells to regulate adipogenesis during the early stage of animal development
3.2. Zfp423 epigenetic modifications and adipogenic commitment
There are accumulating evidence supporting the role of epigenetic modifications in key genes regulating adipogenesis. In our previous studies in sheep, we observed that adipogenic differentiation was enhanced in the fetuses of dams fed with a high energy diet (Yan et al., 2010; Zhu et al., 2008). The high energy diet is correlated with increased intramuscular fat content in offspring (Yan et al., 2011), as well as overall adiposity (Samuelsson et al., 2008; Tong et al., 2011). We further found that Zfp423 expression was enhanced in fetal tissue of over-fed mothers (Yang et al., 2013). We then analyzed epigenetic modifications in the Zfp423 promoter and found that maternal high energy diet reduced DNA methylation in the Zfp423 promoter by about 50% (Yang et al., 2013). Our data has been independently confirmed by another study in rats (Borengasser et al., 2013).
The Zfp423 promoter has exceptionally rich CpG sites, positioning PRC2 as a key mediator of Zfp423 expression and adipogenic commitment (Bernstein et al., 2006). Our data show that the H3K27me3 and EZH2 levels in the Zfp423 promoter were lower in obese compared to control fetal tissue, consistent with the lower DNA methylation and the high expression of Zfp423. Furthermore, the level of H3K4me3 in the Zfp423 promoter was slightly higher in OB fetal tissue, which indicates that trxG was also involved in the control of Zfp423 expression due to maternal high energy diet (Yang et al., 2013). Overall, maternal high energy diet regulates Zfp423 expression and adipogenesis during fetal adipose tissue development through inducing epigenetic modifications.
In a related study, we analyzed the role of Zfp423 in intramuscular adipogenesis and marbling in beef cattle. We sampled beef muscle for separation of stromal vascular cells. These cells were immortalized with pCI neo-hEST2 and individual clones were selected by G418. Three clones with high and low adipogenic potential respectively were selected for further analyses. The expression of Zfp423 was much higher in high adipogenic cells, which was correlated with lower DNA methylation in the Zfp423 promoter (Huang et al., 2012). In conclusion, the data suggest that Zfp423 is a critical regulator of adipogenesis in bovine stromal vascular cells and its expression is regulated epigenetically.
Besides Zfp423, epigenetic regulation of PPARγ and C/EBPα during adipogenesis was also observed (Ngo et al., 2014). The PPARγ2 promoter DNA demethylation was detected during 3T3-L1 adipogenesis (Kamstra et al., 2014). Histone H3K9 methylation suppresses PPARγ expression (Wang et al., 2013). Changes in histone acetylation in C/EBPα promoter was also observed during the adipogenic differentiation of bone marrow stromal cells (Zhao et al., 2013). Up to now, in livestock cells, epigenetic changes in these genes during adipogenic differentiation have not been examined and warrant further studies.
4. Nutrients and adipose development
4.1. Vitamin A, retinoic acid receptors, and adipogenesis
Nuclear receptors are intracellular receptors activated by lipid signaling molecules, including steroid hormones, thyroid hormones, retinoids, Vitamin D metabolites and several others. They are also ligand-activated transcription factors, which activate target gene expression through binding to their cognate DNA elements. In the following discussion, we summarize the effect of retinoids and Vitamin D metabolites on adipogenesis.
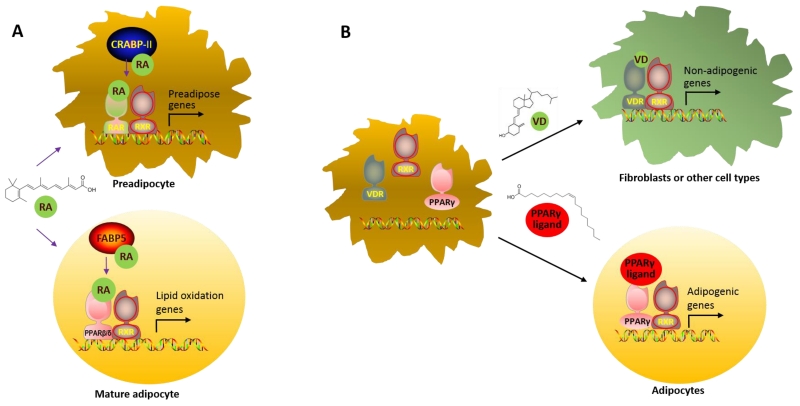
Dietary vitamin A is absorbed and converted into all-trans retinoic acid. Retinoic acid serves as a ligand for retinoic acid receptors (RARα, RARβ, and RARγ). They partner with retinoid X receptors (RXRα, RXRβ, and RXRγ) (Chawla, Repa, Evans, & Mangelsdorf, 2001) to bind to retinoic acid response elements (RAREs) on target gene loci (de The, Vivanco-Ruiz, Tiollais, Stunnenberg, & Dejean, 1990). Retinoic acid also activates orphan receptor PPARβ/δ, stimulating cell proliferation (Shaw, Elholm, & Noy, 2003) and lipid oxidation (Ravnskjaer et al., 2010). Thus, the partitioning of retinoic acid between RAR and PPARβ/δ determines the biological effects of retinoic acid. It appears that two cellular retinoic acid binding proteins regulate retinoic acid partitioning, with cellular retinoic acid binding protein II (CRABP-II) delivers retinoic acid to RAR, and fatty acid binding protein type 5 (FABP5) to PPARβ/δ. Adipogenic progenitor cells express a high ratio of CRABP-II/FABP5, resulting in the domination of RAR signaling (Noy, 2013). During adipogenic differentiation, however, CRABP-II and RAR down-regulate while FABP5 and PPARβ/δ up-regulate, which activate PPARβ/δ signaling in mature adipocytes (Berry, Soltanian, & Noy, 2010). Thus, retinoic acid affects progenitor cells and mature adipocytes differently due to the stage-specific expression of related transcription factors (Fig. 2a).
Fig. 2.
Choice of partnership determines fates. A. Delivery of retinoic acid to either retinoic acid receptor (RAR) or peroxisome proliferator-activated receptor β/δ (PPARβ/δ) determines its biological effects on adipose development; B. RAR and Vitamin D receptor (VDR) compete for retinoid X receptor (RXR) needed for partnering with PPARγ to initiate adipogenic differentiation.
As a metabolite of vitamin A, retinoic acid plays major roles in both preadipocyte commitment and terminal maturation of adipocytes. Decades ago, in an in vitro adipogenesis model using embryonic stem cells, retinoic acid was found to promote adipogenic commitment of embryonic stem cells (Dani et al., 1997). Consistently, retinoic acid treatment on stem cells derived from embryoid bodies leads to prolonged activation of the extracellular signal-regulated kinase-1 (ERK) pathway, required for adipogenic commitment (Bost et al., 2002). Retinoic acid is required for Zfp423 expression in adipocytes. In the mice lacking aldehyde dehydrogenase-1a1 (Aldh1a1), which catalyzes retinoic acid production from retinaldehyde, the expression of Zfp423, PPARg and Fabp4 were reduced by 70% (Reichert et al., 2011). Thus, retinoic acid promotes the adipogenic commitment. On the other hand, retinoic acid hampers terminal differentiation of adipocytes. In both 3T3-L1 and C3H10T1/2 cell lines, retinoic acid blocks terminal adipocyte maturation by enhancing preadipocyte gene expression (Berry et al., 2012) as well as inhibiting C/EBPβ mediated transcription (Schwarz, Reginato, Shao, Krakow, & Lazar, 1997a).
The promotion of retinoic acid on adipogenic commitment involves epigenetic modifications. Depending on the availability of RA, RAR/RXR heterodimers interact with nuclear co-repressor proteins including silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) and nuclear receptor corepressor (NCoR), or with coactivators such as (SRC)/p160 family and p300/CREB-binding protein (CBP) (Kashyap & Gudas, 2010). Nuclear co-repressor proteins elicit locus-specific changes in the chromatin structure which inhibits gene expression, while coactivators facilitate gene expression through recruiting ATP-dependent chromatin remodeling complex to loosen the structure, allowing RNA polymerase II to initiate gene expression. In the presence of retinoic acid, the PRC proteins rapidly dissociate from RAR target genes, forming permissive condition for gene expression (Kashyap & Gudas, 2010), which in turn, reduces DNA methylation in the corresponding promoters. This could explain the promotion effect of retinoic acid on preadipocyte gene expression (Berry et al., 2012).
4. 2. Synergism of Vitamin D metabolites with retinoic acid receptor signaling to regulate adipogenesis
Beyond the traditional role in calcium homoeostasis and bone metabolism, Vitamin D is now recognized as a regulator of adipogenesis. Obesity, a metabolic disorder associated with excessive accumulation of adipocytes, is frequently associated with Vitamin D deficiency (Yao et al., 2015). Polymorphisms in genes involved in Vitamin D metabolism and signaling increase the susceptibility to obesity and diabetes (Jiang et al., 2007; Ye et al., 2001). Vitamin D consistently demonstrates inhibitory effects on adipogenesis in both in vivo and in vitro studies. The active metabolite of Vitamin D, 1,25-dihydroxyvitamin D, inhibits 3T3-L1 preadipcyte differentiation in a dose dependent manner (Ishida, Taniguchi, & Baba, 1988), which reduces the expression of adipogenic genes (Hida et al., 1998; Kong & Li, 2006). It has been further demonstrated that 1,25-dihydroxyvitamin D regulates adipogenesis by activating the vitamin D receptor (VDR). In the presence of 1,25-dihydroxyvitamin D, VDR blocks adipogenesis by down-regulating C/EBPβ expression (Blumberg et al., 2006).
The exact reasons for the inhibitory effect of VDR on adipogenesis remain to be clarified. Because RXR is a heterodimeric partner for both VDR and PPARγ, it is possible that VDR competes with PPARγ for RXR, which reduces adipogenesis. Supportively, in 3T3-L1 cells, VDR expression reduced PPARγ expression while simultaneous RXR ectopic expression abolishes this inhibition, confirming the competitive relationship between VDR and PPARγ for RXR (Kong & Li, 2006). This also raises the possibility that activated VDR depletes RXR needed for the formation of RXR/RAR complex, thus inhibiting adipogenic commitment (Fig. 2b). These notions were confirmed by a recent study in the inhibitory effect of vitamin D on adipogenesis (Ji, Doumit, & Hill, 2015).
4.3. Methyl donors and B vitamins
Epigenetic modifications include both histone and DNA methylations. For methylation to occur, the presence of methyl donors is indispensable. DNA methyltransferases catalyze the transfer of a methyl group to DNA using S-adenosyl methionine (SAM) as the methyl donor. SAM is converted to S-adenosylhomocysteine (SAH) after the methyl group is transferred to DNA or histone. In this process, dietary nutrients including folic acid, vitamin B12, choline and betaine are involved. Dietary methyl supplementation of folic acid, vitamin B12, choline and betaine consistently increases DNA and histone methylations of genes by contributing to SAM synthesis (Waterland & Jirtle, 2003).
There is accumulating evidence supporting the inhibitory roles of methyl donor supplementation in suppressing adipogenesis both in vivo and in vitro. Folate increases overall CpG methylation in C/EBPa promoter, and inhibits its expression and adipogenesis in chicken preadipocytes (Yu et al., 2014). In 3T3-L1 cells, treatment of folic acid and vitamin B12 attenuates adipogenic differentiation by upregulating Wingless and Int (Wnt)10b (Bellner et al., 2015). Eight months of methyl donor (choline, betamine, vitamin B12 and folic acid) supplementation reverts DNA hypomethylation in the fatty acid synthase promoter induced by a high fat diet, suppressing weight gain (Cordero, Gomez-Uriz, Campion, Milagro, & Martinez, 2013). On the contrary, maternal dietary folate and vitamin B12 restrictions increases visceral adiposity in rat offspring (Kumar et al., 2013). These data show that methyl donors affect adipogenesis through altering epigenetic changes in genes associated with adipogenesis.
5. Retinoic acid and lipid accumulation in mature adipocytes
While retinoic acid promotes adipogenic commitment, in mature adipocytes, vitamin A reduces lipid accumulation. Because vitamin A metabolite, retinoic acid, activates PPARα and PPARβ/δ in mature adipocytes, which induces fatty acid oxidation and lipid catabolism (Berry & Noy, 2009; Berry et al., 2010; Noy, 2013), it is not surprising that vitamin A reduces both lipid accumulation and adipocyte hypertrophy (Amengual, Petrov, Bonet, Ribot, & Palou, 2012; Amengual, Ribot, Bonet, & Palou, 2008; Cook, Yeldandi, Rao, Hashimoto, & Reddy, 2000). In addition, retinoic acid may also affect PPARγ transcription activity via enhancing the partnership of RAR with RXR, depriving RAR/PPARγ required for late stage adipogenesis (Dawson & Xia, 2012; Ziouzenkova & Plutzky, 2008). Retinoic acid blocks late-stage adipogenesis by inhibiting C/EBPβ-mediated transcription (Schwarz et al., 1997b) and PPARγ transcriptional activity (Berry et al., 2012; Kawada et al., 1996), and thus terminal differentiation of adipocytes. Therefore, reducing vitamin A intake has been used to enhance intramuscular lipid accumulation and marbling in finishing beef cattle (Arnett et al., 2009; Gorocica-Buenfil, Fluharty, Bohn, Schwartz, & Loerch, 2007; Pickworth, Loerch, & Fluharty, 2012a, 2012b; Smith et al., 2009; Ward, McKinnon, Hendrick, & Buchanan, 2012). However, there are also reports showing that vitamin A deficiency has no effect on lipid metabolism and marbling (Bryant et al., 2010; Gorocica-Buenfil, Fluharty, Bohn, et al., 2007; Gorocica-Buenfil, Fluharty, & Loerch, 2008; Gorocica-Buenfil, Fluharty, Reynolds, & Loerch, 2007; Pickworth et al., 2012a, 2012b). Such discrepancy could be due to the difference in the degree of deficiency, time and duration of deficiency, as well as cattle nutrition and genetics.
6. Complications and Conclusions
Available studies suggest that nutrients such as vitamin A promote early adipogenic commitment, which may promote later formation of adipocytes during the fattening stage, likely enhancing marbling. Vitamin D metabolites by depriving RXR needed for adipogenesis, reduce adipocyte formation during early adipose development. Methyl donors also affects adipogenesis by regulating epigenetic modifications during early adipose development. On the other hand, vitamin A supplementation to feedlot beef cattle promotes lipid oxidation and thus reduces adipocyte hypertrophy and marbling.
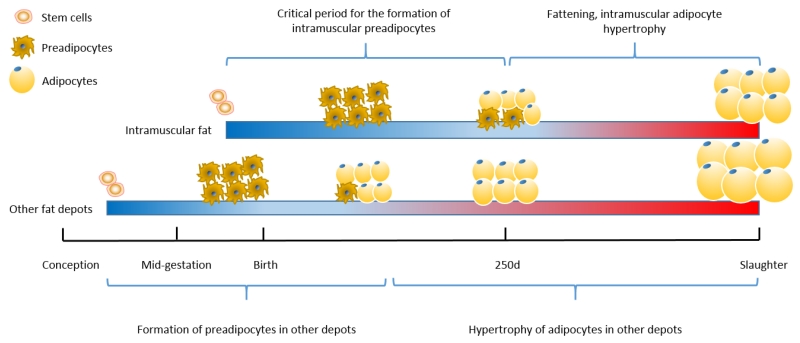
The remaining question is how to specifically promote intramuscular adipogenesis without increase in adipogenesis in the visceral and subcutaneous fat. This problem can be partially addressed by targeting a period critical for intramuscular adipocyte formation. The four adipose depots of cattle do not develop at the same time; the formation of visceral fat occurs earliest in beef cattle, the formation of subcutaneous adipocytes occurs slightly later and prolongs to the early weaning stage in cattle, while the formation of intramuscular adipocytes is estimated to occur mainly during the late fetal/neonatal stage to about 250 days of age in beef cattle (Du et al., 2013b); the dwindling presence of progenitor cells as animals age is the major reason for the declining ability to generate new adipocytes (Du et al., 2010). Due to the sequential formation of adipocytes in different tissues, nutrient supplementation such as vitamin A during the neonatal to pre-weaning stages may specifically enhance intramuscular adipocyte formation, leading to more intramuscular preadipocytes that will provide sites for lipid accumulation (hypertrophy) during the ‘fattening’ stage, enhancing marbling without overall increase in carcass fatness (Fig. 3).
Fig. 3.
Approximate timelines for adipose tissue development of beef cattle. Nutrient supplementation during a period critical for intramuscular adipocyte formation enhances preadipocyte formation and adipocyte hyperplasia, which provide sites for later lipid accumulation to enhance marbling, without significantly increase fat mass in other fat depots (Du, Wang, Fu, Yang, & Zhu, 2015).
Highlights.
Nutrigenomics is a new direction in adipogenic research.
Retinoic acid enhances adipogenic commitment through altering epigenetic modifications in the promoters of key adipogenic genes.
Retinoic acid alters the partnership of retinoid X receptors with other nuclear receptors to regulate adipogenesis.
Methyl donors inhibit adipogenesis through promoting histone and DNA methylation.
Acknowledgements
This project was supported by National Research Initiative Competitive Grant no. 2015-67015-23219 and 2016-68006-24634 from the USDA National Institute of Food and Agriculture.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140(12):2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- Amengual J, Petrov P, Bonet ML, Ribot J, Palou A. Induction of carnitine palmitoyl transferases 1 and fatty acid oxidation by retinoic acid in HepG2 cells. International Journal of Biochemistry and Cell Biology. 2012;44(11):2019–2027. doi: 10.1016/j.biocel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Amengual J, Ribot J, Bonet ML, Palou A. Retinoic acid treatment increases lipid oxidation capacity in skeletal muscle of mice. Obesity (Silver Spring) 2008;16(3):585–591. doi: 10.1038/oby.2007.104. [DOI] [PubMed] [Google Scholar]
- Arnett AM, Dikeman ME, Daniel MJ, Olson KC, Jaeger J, Perrett J. Effects of vitamin A supplementation and weaning age on serum and liver retinol concentrations, carcass traits, and lipid composition in market beef cattle. Meat Science. 2009;81(4):596–606. doi: 10.1016/j.meatsci.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Avram MM, Avram AS, James WD. Subcutaneous fat in normal and diseased states 3. Adipogenesis: from stem cell to fat cell. Journal of the American Academy of Dermatology. 2007;56(3):472–492. doi: 10.1016/j.jaad.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445(7128):671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bellner L, Nichols A, Pandey V, Vanella L, Gilliam C, Gupte R, Abraham N. Effect of Vitamin B12 and Nutrients on Adipogenesis-adipogenic Markers in 3T3 Cells. The FASEB Journal. 2015;29(1 Supplement) 996.992. [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Berry DC, DeSantis D, Soltanian H, Croniger CM, Noy N. Retinoic acid upregulates preadipocyte genes to block adipogenesis and suppress diet-induced obesity. Diabetes. 2012;61(5):1112–1121. doi: 10.2337/db11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor β/δ and retinoic acid receptor. Molecular and cellular biology. 2009;29(12):3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DC, Soltanian H, Noy N. Repression of cellular retinoic acid-binding protein II during adipocyte differentiation. Journal of Biological Chemistry. 2010;285(20):15324–15332. doi: 10.1074/jbc.M110.110635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg JM, Tzameli I, Astapova I, Lam FS, Flier JS, Hollenberg AN. Complex role of the vitamin D receptor and its ligand in adipogenesis in 3T3-L1 cells. Journal of Biological Chemistry. 2006;281(16):11205–11213. doi: 10.1074/jbc.M510343200. [DOI] [PubMed] [Google Scholar]
- Bonnet M, Cassar-Malek I, Chilliard Y, Picard B. Ontogenesis of muscle and adipose tissues and their interactions in ruminants and other species. Animal. 2010;4(7):1093–1109. doi: 10.1017/S1751731110000601. [DOI] [PubMed] [Google Scholar]
- Borengasser SJ, Zhong Y, Kang P, Lindsey F, Ronis MJ, Badger TM, Gomez-Acevedo H, Shankar K. Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology. 2013;154(11):4113–4125. doi: 10.1210/en.2012-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost F, Caron L, Marchetti I, Dani C, Le Marchand-Brustel Y, Binétruy B. Retinoic acid activation of the ERK pathway is required for embryonic stem cell commitment into the adipocyte lineage. Biochemical Journal. 2002;361:621–627. doi: 10.1042/0264-6021:3610621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant TC, Wagner JJ, Tatum JD, Galyean ML, Anthony RV, Engle TE. Effect of dietary supplemental vitamin A concentration on performance, carcass merit, serum metabolites, and lipogenic enzyme activity in yearling beef steers. Journal of Animal Science. 2010;88(4):1463–1478. doi: 10.2527/jas.2009-2313. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Cianzio DS, Topel DG, Whitehurst GB, Beitz DC, Self HL. Adipose tissue growth and cellularity: changes in bovine adipocyte size and number. Journal of Animal Science. 1985;60(4):970–976. doi: 10.2527/jas1985.604970x. [DOI] [PubMed] [Google Scholar]
- Cook WS, Yeldandi AV, Rao MS, Hashimoto T, Reddy JK. Less extrahepatic induction of fatty acid beta-oxidation enzymes by PPAR alpha. Biochemical and Biophysical Research Communications. 2000;278(1):250–257. doi: 10.1006/bbrc.2000.3739. [DOI] [PubMed] [Google Scholar]
- Cordero P, Gomez-Uriz AM, Campion J, Milagro FI, Martinez JA. Dietary supplementation with methyl donors reduces fatty liver and modifies the fatty acid synthase DNA methylation profile in rats fed an obesogenic diet. Genes & Nutrition. 2013;8(1):105–113. doi: 10.1007/s12263-012-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C, Smith AG, Dessolin S, Leroy P, Staccini L, Villageois P, Darimont C, Ailhaud G. Differentiation of embryonic stem cells into adipocytes in vitro. Journal of Cell Science. 1997;110:1279–1285. doi: 10.1242/jcs.110.11.1279. [DOI] [PubMed] [Google Scholar]
- Dawson MI, Xia Z. The retinoid X receptors and their ligands. Biochimica et Biophysica Acta. 2012;1821(1):21–56. doi: 10.1016/j.bbalip.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343(6254):177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- Du M, Huang Y, Das AK, Yang Q, Duarte MS, Dodson MV, Zhu M-J. MEAT SCIENCE AND MUSCLE BIOLOGY SYMPOSIUM: Manipulating mesenchymal progenitor cell differentiation to optimize performance and carcass value of beef cattle. Journal of Animal Science. 2013a;91(3):1419–1427. doi: 10.2527/jas.2012-5670. [DOI] [PubMed] [Google Scholar]
- Du M, Huang Y, Das AK, Yang Q, Duarte MS, Dodson MV, Zhu MJ. Meat Science and Muscle Biology Symposium: manipulating mesenchymal progenitor cell differentiation to optimize performance and carcass value of beef cattle. Journal of Animal Science. 2013b;91(3):1419–1427. doi: 10.2527/jas.2012-5670. [DOI] [PubMed] [Google Scholar]
- Du M, Wang B, Fu X, Yang Q, Zhu MJ. Fetal programming in meat production. Meat Science. 2015;109:40–47. doi: 10.1016/j.meatsci.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Du M, Yin J, Zhu MJ. Cellular signaling pathways regulating the initial stage of adipogenesis and marbling of skeletal muscle. Meat Science. 2010;86(1):103–109. doi: 10.1016/j.meatsci.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Developmental Biology. 2010;339(2):240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Debril MB, Auwerx J. Peroxisome proliferator-activated receptor-gamma: from adipogenesis to carcinogenesis. Journal of Molecular Endocrinology. 2001;27(1):1–9. doi: 10.1677/jme.0.0270001. [DOI] [PubMed] [Google Scholar]
- Feve B. Adipogenesis: cellular and molecular aspects. Best Practice & Research Clinical Endocrinology & Metabolism. 2005;19(4):483–499. doi: 10.1016/j.beem.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Garcia LG, Nicholson KL, Hoffman TW, Lawrence TE, Hale DS, Griffin DB, Savell JW, Vanoverbeke DL, Morgan JB, Belk KE, Field TG, Scanga JA, Tatum JD, Smith GC. National Beef Quality Audit-2005: survey of targeted cattle and carcass characteristics related to quality, quantity, and value of fed steers and heifers. Journal of Animal Science. 2008;86(12):3533–3543. doi: 10.2527/jas.2007-0782. [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, Zon LI. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136(6):1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorocica-Buenfil MA, Fluharty FL, Bohn T, Schwartz SJ, Loerch SC. Effect of low vitamin A diets with high-moisture or dry corn on marbling and adipose tissue fatty acid composition of beef steers. Journal of Animal Science. 2007;85(12):3355–3366. doi: 10.2527/jas.2007-0172. [DOI] [PubMed] [Google Scholar]
- Gorocica-Buenfil MA, Fluharty FL, Loerch SC. Effect of vitamin A restriction on carcass characteristics and immune status of beef steers. Journal of Animal Science. 2008;86(7):1609–1616. doi: 10.2527/jas.2007-0241. [DOI] [PubMed] [Google Scholar]
- Gorocica-Buenfil MA, Fluharty FL, Reynolds CK, Loerch SC. Effect of dietary vitamin A restriction on marbling and conjugated linoleic acid content in Holstein steers. Journal of Animal Science. 2007;85(9):2243–2255. doi: 10.2527/jas.2006-781. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464(7288):619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, Spiegelman BM. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metabolism. 2012;15(2):230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausman GJ, Basu U, Du M, Fernyhough-Culver M, Dodson MV. Intermuscular and intramuscular adipose tissues: Bad vs. good adipose tissues. Adipocyte. 2014;3(4):242–255. doi: 10.4161/adip.28546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida Y, Kawada T, Kayahashi S, Ishihara T, Fushiki T. Counteraction of retinoic acid and 1,25-dihydroxyvitamin D3 on up-regulation of adipocyte differentiation with PPARgamma ligand, an antidiabetic thiazolidinedione, in 3T3-L1 cells. Life Science. 1998;62(14):PL205–211. doi: 10.1016/s0024-3205(98)00059-9. [DOI] [PubMed] [Google Scholar]
- Huang Y, Das AK, Yang QY, Zhu MJ, Du M. Zfp423 promotes adipogenic differentiation of bovine stromal vascular cells. PLoS One. 2012;7(10):e47496. doi: 10.1371/journal.pone.0047496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Taniguchi H, Baba S. Possible involvement of 1 alpha,25-dihydroxyvitamin D3 in proliferation and differentiation of 3T3-L1 cells. Biochemical and Biophysical Research Communications. 1988;151(3):1122–1127. doi: 10.1016/s0006-291x(88)80482-0. [DOI] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Doumit ME, Hill RA. Regulation of Adipogenesis and Key Adipogenic Gene Expression by 1, 25-Dihydroxyvitamin D in 3T3-L1 Cells. PLoS One. 2015;10(6):e0126142. doi: 10.1371/journal.pone.0126142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xiong DH, Guo YF, Shen H, Xiao P, Yang F, Chen Y, Zhang F, Recker RR, Deng HW. Association analysis of vitamin D-binding protein gene polymorphisms with variations of obesity-related traits in Caucasian nuclear families. International Journal of Obesity. 2007;31(8):1319–1324. doi: 10.1038/sj.ijo.0803583. [DOI] [PubMed] [Google Scholar]
- Kamstra JH, Hruba E, Blumberg B, Janesick A, Mandrup S, Hamers T, Legler J. Transcriptional and epigenetic mechanisms underlying enhanced in vitro adipocyte differentiation by the brominated flame retardant BDE-47. Environmental Science & Technology. 2014;48(7):4110–4119. doi: 10.1021/es405524b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Gudas LJ. Epigenetic regulatory mechanisms distinguish retinoic acid-mediated transcriptional responses in stem cells and fibroblasts. Journal of Biological Chemistry. 2010;285(19):14534–14548. doi: 10.1074/jbc.M110.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman RG, Carpenter ZL, Bray RW, Hoekstra WG. Biochemical Properties of Pork and Their Relationship to Quality II. Intramuscular Fat. Journal of Food Science. 1964;29(1):70–74. [Google Scholar]
- Kawada T, Kamei Y, Sugimoto E. The possibility of active form of vitamins A and D as suppressors on adipocyte development via ligand-dependent transcriptional regulators. International Journal of Obesity and Related Metabolic Disorders. 1996;20(Suppl 3):S52–57. [PubMed] [Google Scholar]
- Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. Journal of Biological Chemistry. 2008;283(45):30919–30932. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J, Li YC. Molecular mechanism of 1,25-dihydroxyvitamin D3 inhibition of adipogenesis in 3T3-L1 cells. American Journal of Physiology - Endocrinology and Metabolism. 2006;290(5):E916–924. doi: 10.1152/ajpendo.00410.2005. [DOI] [PubMed] [Google Scholar]
- Kumar KA, Lalitha A, Pavithra D, Padmavathi IJ, Ganeshan M, Rao KR, Venu L, Balakrishna N, Shanker NH, Reddy SU. Maternal dietary folate and/or vitamin B 12 restrictions alter body composition (adiposity) and lipid metabolism in Wistar rat offspring. The Journal of nutritional biochemistry. 2013;24(1):25–31. doi: 10.1016/j.jnutbio.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Lorente M, Perez C, Sanchez C, Donohoe M, Shi Y, Vidal M. Homeotic transformations of the axial skeleton of YY1 mutant mice and genetic interaction with the Polycomb group gene Ring1/Ring1A. Mechanisms of Development. 2006;123(4):312–320. doi: 10.1016/j.mod.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8(10):1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougald OA, Mandrup S. Adipogenesis: forces that tip the scales. Trends in Endocrinology & Metabolism. 2002;13(1):5–11. doi: 10.1016/s1043-2760(01)00517-3. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- McKenna DR, Roebert DL, Bates PK, Schmidt TB, Hale DS, Griffin DB, Savell JW, Brooks JC, Morgan JB, Montgomery TH, Belk KE, Smith GC. National Beef Quality Audit-2000: survey of targeted cattle and carcass characteristics related to quality, quantity, and value of fed steers and heifers. Journal of Animal Science. 2002;80(5):1212–1222. doi: 10.2527/2002.8051212x. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genetics. 2010;6(12):e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Molecular Cell. 2008;30(6):755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Moore MC, Gray GD, Hale DS, Kerth CR, Griffin DB, Savell JW, Raines CR, Belk KE, Woerner DR, Tatum JD, Igo JL, VanOverbeke DL, Mafi GG, Lawrence TE, Delmore RJ, Jr., Christensen LM, Shackelford SD, King DA, Wheeler TL, Meadows LR, O’Connor ME. National Beef Quality Audit-2011: In-plant survey of targeted carcass characteristics related to quality, quantity, value, and marketing of fed steers and heifers. Journal of Animal Science. 2012;90(13):5143–5151. doi: 10.2527/jas.2012-5550. [DOI] [PubMed] [Google Scholar]
- Nebbioso A, Dell’Aversana C, Bugge A, Sarno R, Valente S, Rotili D, Manzo F, Teti D, Mandrup S, Ciana P, Maggi A, Mai A, Gronemeyer H, Altucci L. HDACs class II-selective inhibition alters nuclear receptor-dependent differentiation. Journal of Molecular Endocrinology. 2010;45(4):219–228. doi: 10.1677/JME-10-0043. [DOI] [PubMed] [Google Scholar]
- Ngo S, Li X, O’Neill R, Bhoothpur C, Gluckman P, Sheppard A. Elevated S-adenosylhomocysteine alters adipocyte functionality with corresponding changes in gene expression and associated epigenetic marks. Diabetes. 2014;63(7):2273–2283. doi: 10.2337/db13-1640. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends in Cell Biology. 2012;22(4):220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Novakofski J. Adipogenesis: usefulness of in vitro and in vivo experimental models. Journal of Animal Science. 2004;82(3):905–915. doi: 10.2527/2004.823905x. [DOI] [PubMed] [Google Scholar]
- Noy N. The one-two punch: Retinoic acid suppresses obesity both by promoting energy expenditure and by inhibiting adipogenesis. Adipocyte. 2013;2(3):184–187. doi: 10.4161/adip.23489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickworth CL, Loerch SC, Fluharty FL. Effects of timing and duration of dietary vitamin A reduction on carcass quality of finishing beef cattle. Journal of Animal Science. 2012a;90(8):2677–2691. doi: 10.2527/jas.2011-4756. [DOI] [PubMed] [Google Scholar]
- Pickworth CL, Loerch SC, Fluharty FL. Restriction of vitamin A and D in beef cattle finishing diets on feedlot performance and adipose accretion. Journal of Animal Science. 2012b;90(6):1866–1878. doi: 10.2527/jas.2010-3590. [DOI] [PubMed] [Google Scholar]
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang JH, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskjaer K, Frigerio F, Boergesen M, Nielsen T, Maechler P, Mandrup S. PPARdelta is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. Journal of Lipid Research. 2010;51(6):1370–1379. doi: 10.1194/jlr.M001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert B, Yasmeen R, Jeyakumar SM, Yang F, Thomou T, Alder H, Duester G, Maiseyeu A, Mihai G, Harrison EH, Rajagopalan S, Kirkland JL, Ziouzenkova O. Concerted action of aldehyde dehydrogenases influences depot-specific fat formation. Molecular Endocrinology. 2011;25(5):799–809. doi: 10.1210/me.2010-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nature Review Molecular Cell Biology. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Samuelsson AM, Matthews PA, Argenton M, Christie MR, McConnell JM, Jansen EH, Piersma AH, Ozanne SE, Twinn DF, Remacle C, Rowlerson A, Poston L, Taylor PD. Diet-induced obesity in female mice leads to offspring hyperphagia, adiposity, hypertension, and insulin resistance: a novel murine model of developmental programming. Hypertension. 2008;51(2):383–392. doi: 10.1161/HYPERTENSIONAHA.107.101477. [DOI] [PubMed] [Google Scholar]
- Schafer A, Karaulanov E, Stapf U, Doderlein G, Niehrs C. Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes & Development. 2013;27(3):261–273. doi: 10.1101/gad.186916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136(21):3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nature Review Molecular Cell Biology. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Schwarz EJ, Reginato MJ, Shao D, Krakow SL, Lazar MA. Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription. Molecular and Cellular Biology. 1997a;17(3):1552–1561. doi: 10.1128/mcb.17.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz EJ, Reginato MJ, Shao D, Krakow SL, Lazar MA. Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription. Molecular and Cellular Biology. 1997b;17(3):1552–1561. doi: 10.1128/mcb.17.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park MG, Steffensen KR, Stulnig TM, Gustafsson JA, Park SD, Kim JB. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor gamma expression. Molecular and Cellular Biology. 2004;24(8):3430–3444. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw N, Elholm M, Noy N. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. Journal of Biological Chemistry. 2003;278(43):41589–41592. doi: 10.1074/jbc.C300368200. [DOI] [PubMed] [Google Scholar]
- Smith SB, Kawachi H, Choi CB, Choi CW, Wu G, Sawyer JE. Cellular regulation of bovine intramuscular adipose tissue development and composition. Journal of Animal Science. 2009;87(14 Suppl):E72–82. doi: 10.2527/jas.2008-1340. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87(3):377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- Taga H, Bonnet M, Picard B, Zingaretti MC, Cassar-Malek I, Cinti S, Chilliard Y. Adipocyte metabolism and cellularity are related to differences in adipose tissue maturity between Holstein and Charolais or Blond d’Aquitaine fetuses. Journal of Animal Science. 2011;89(3):711–721. doi: 10.2527/jas.2010-3234. [DOI] [PubMed] [Google Scholar]
- Taga H, Chilliard Y, Meunier B, Chambon C, Picard B, Zingaretti MC, Cinti S, Bonnet M. Cellular and molecular large-scale features of fetal adipose tissue: is bovine perirenal adipose tissue brown? Journal of Cell Physiology. 2012;227(4):1688–1700. doi: 10.1002/jcp.22893. [DOI] [PubMed] [Google Scholar]
- Tong JF, Yan X, Zhao JX, Zhu MJ, Nathanielsz PW, Du M. Metformin mitigates the impaired development of skeletal muscle in the offspring of obese mice. Nutrition and Diabetes. 2011;1(e7) doi: 10.1038/nutd.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, Ge K. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. EMBO Journal. 2013;32(1):45–59. doi: 10.1038/emboj.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AK, McKinnon JJ, Hendrick S, Buchanan FC. The impact of vitamin A restriction and ADH1C genotype on marbling in feedlot steers. Journal of Animal Science. 2012;90(8):2476–2483. doi: 10.2527/jas.2011-4404. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular Cell Biology. 2003;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LN. Chromatin remodeling and epigenetic regulation of the CrabpI gene in adipocyte differentiation. Biochimica et Biophysica Acta. 2012;1821(1):206–212. doi: 10.1016/j.bbalip.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156(1-2):45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Huang Y, Zhao JX, Long NM, Uthlaut AB, Zhu MJ, Ford SP, Nathanielsz PW, Du M. Maternal obesity-impaired insulin signaling in sheep and induced lipid accumulation and fibrosis in skeletal muscle of offspring. Biology of Reproduction. 2011;85(1):172–178. doi: 10.1095/biolreprod.110.089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhu MJ, Xu W, Tong JF, Ford SP, Nathanielsz PW, Du M. Up-regulation of Toll-like receptor 4/nuclear factor-kappaB signaling is associated with enhanced adipogenesis and insulin resistance in fetal skeletal muscle of obese sheep at late gestation. Endocrinology. 2010;151(1):380–387. doi: 10.1210/en.2009-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QY, Liang JF, Rogers CJ, Zhao JX, Zhu MJ, Du M. Maternal obesity induces epigenetic modifications to facilitate zfp423 expression and enhance adipogenic differentiation in fetal mice. Diabetes. 2013;62(11):3727–3735. doi: 10.2337/db13-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Zhu L, He L, Duan Y, Liang W, Nie Z, Jin Y, Wu X, Fang Y. A meta-analysis of the relationship between vitamin D deficiency and obesity. International Journal of Clinical and Experimental Medicine. 2015;8(9):14977–14984. [PMC free article] [PubMed] [Google Scholar]
- Ye WZ, Reis AF, Dubois-Laforgue D, Bellanne-Chantelot C, Timsit J, Velho G. Vitamin D receptor gene polymorphisms are associated with obesity in type 2 diabetic subjects with early age of onset. European Journal of Endocrinology. 2001;145(2):181–186. doi: 10.1530/eje.0.1450181. [DOI] [PubMed] [Google Scholar]
- Yu X, Liu R, Zhao G, Zheng M, Chen J, Wen J. Folate supplementation modifies CCAAT/enhancer-binding protein α methylation to mediate differentiation of preadipocytes in chickens. Poultry Science. 2014;93(10):2596–2603. doi: 10.3382/ps.2014-04027. [DOI] [PubMed] [Google Scholar]
- Zhan Q, Lord KA, Alamo I, Jr., Hollander MC, Carrier F, Ron D, Kohn KW, Hoffman B, Liebermann DA, Fornace AJ., Jr. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Molecular Cell Biology. 1994;14(4):2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QH, Wang SG, Liu SX, Li JP, Zhang YX, Sun ZY, Fan QM, Tian JW. PPARgamma forms a bridge between DNA methylation and histone acetylation at the C/EBPalpha gene promoter to regulate the balance between osteogenesis and adipogenesis of bone marrow stromal cells. FEBS Journal. 2013;280(22):5801–5814. doi: 10.1111/febs.12500. [DOI] [PubMed] [Google Scholar]
- Zhu MJ, Han B, Tong J, Ma C, Kimzey JM, Underwood KR, Xiao Y, Hess BW, Ford SP, Nathanielsz PW, Du M. AMP-activated protein kinase signalling pathways are down regulated and skeletal muscle development impaired in fetuses of obese, over-nourished sheep. Journal of Physiology. 2008;586(10):2651–2664. doi: 10.1113/jphysiol.2007.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziouzenkova O, Plutzky J. Retinoid metabolism and nuclear receptor responses: New insights into coordinated regulation of the PPAR-RXR complex. FEBS Letters. 2008;582(1):32–38. doi: 10.1016/j.febslet.2007.11.081. [DOI] [PubMed] [Google Scholar]