ABSTRACT
The envelope glycoprotein (Env) is the major target for HIV-1 broadly neutralizing antibodies (bNAbs). One of the mechanisms that HIV has evolved to escape the host's immune response is to mask conserved epitopes on Env with dense glycosylation. Previous studies have shown that the removal of a particular conserved glycan at N197 increases the neutralization sensitivity of the virus to antibodies targeting the CD4 binding site (CD4bs), making it a site of significant interest from the perspective of vaccine design. At present, the structural consequences that result from the removal of the N197 glycan have not been characterized. Using native-like SOSIP trimers, we examine the effects on antigenicity and local structural dynamics resulting from the removal of this glycan. A large increase in the binding of CD4bs and V3-targeting antibodies is observed for the N197Q mutant in trimeric Env, while no changes are observed with monomeric gp120. While the overall structure and thermostability are not altered, a subtle increase in the flexibility of the variable loops at the trimeric interface of adjacent protomers is evident in the N197Q mutant by hydrogen-deuterium exchange mass spectrometry. Structural modeling of the glycan chains suggests that the spatial occupancy of the N197 glycan leads to steric clashes with CD4bs antibodies in the Env trimer but not monomeric gp120. Our results indicate that the removal of the N197 glycan enhances the exposure of relevant bNAb epitopes on Env with a minimal impact on the overall trimeric structure. These findings present a simple modification for enhancing trimeric Env immunogens in vaccines.
IMPORTANCE The HIV-1 Env glycoprotein presents a dense patchwork of host cell-derived N-linked glycans. This so-called glycan shield is considered to be a major protective mechanism against immune recognition. While the positions of many N-linked glycans are isolate specific, some are highly conserved and are believed to play key functional roles. In this study, we examine the conserved, CD4 binding site-proximal N197 glycan and demonstrate that its removal both facilitates neutralizing antibody access to the CD4 binding site and modestly impacts the structural dynamics at the trimer crown without drastically altering global Env trimer stability. This indicates that surgical glycosylation site modification may be an effective way of sculpting epitope presentation in Env-based vaccines.
INTRODUCTION
The trimeric HIV-1 envelope glycoprotein (Env), a trimer composed of gp120/gp41 heterodimers, is the primary antigenic feature on the virus and the sole target for neutralizing antibodies (1). Despite the extensive genetic diversity that exists among circulating HIV-1 variants, broadly neutralizing antibodies (bNAbs) capable of neutralizing a diverse panel of viral isolates have been identified in rare HIV-infected individuals. Elicitation of such bNAbs is thought to be a critical requirement for an effective HIV-1 vaccine (2, 3), but to date, HIV vaccine efforts have resulted in limited, narrow neutralization activity and have failed to elicit bNAbs (4–7).
Nearly 50% of the molecular mass of Env is contributed by host cell-derived N-linked glycans, and this dense glycan shield is considered to be a major protective mechanism against immune recognition (8, 9). Glycans play important roles in Env folding, viral assembly and infectivity, and modulating the immune response (10–12). Although glycans typically attenuate antigenicity by occluding polypeptide epitopes, several conserved glycans are actually targets for potent HIV-1 bNAbs (13–18). Specific glycans within variable loops, within conserved C2-C4 regions, and within gp41 were found to affect HIV-1 sensitivity to neutralizing antibodies (12, 15, 19). One of the most conserved epitopes on Env, the CD4 binding site (CD4bs), is a recessed pocket on gp120 surrounded by glycans (20). Removal of the glycans peripheral to the CD4 binding site results in increased sensitivity to neutralization (21–24).
Recent studies have intensified interest in glycan modification as a means of sculpting epitope accessibility in Env-based HIV-1 vaccine immunogens. For example, it has been demonstrated that the removal of the glycosylation site at N276 in the gp120 D-loop can increase the reactivity of germ line precursor forms of CD4bs bNAbs such as VRC01 (25–28). By and large, however, the consequence of glycosylation site modification on trimer structure and stability has not been directly probed. While glycans are often considered to be flexible decorations on a more structurally defined protein substrate, recent structures have revealed significant ordered density for glycan chains (29, 30) as well as interactions between glycan chains themselves and with the protein surface. It is conceivable that the removal of specific glycans can impact the presentation of others and may impact the stability of the trimer by removing favorable interactions.
The N197 glycosylation site is highly conserved (>92%) among HIV-1 Env sequences. It is located at the base of the V2 loop, proximal to the CD4bs (12). Removal of the N197 sequon was found to enhance the neutralization sensitivity of diverse HIV-1 isolates to CD4bs- and V3-specific antibodies and, more importantly, to induce higher levels of neutralizing antibodies with greater breadth in macaques (12, 21, 23, 24, 31). However, in the context of monomeric gp120, the removal of N197 had no significant effect on the binding of CD4 or neutralizing antibodies (24). Direct probing of the role of the N197 glycan in the context of trimeric Env has been hampered due to the difficulty in obtaining homogeneous preparations of native-like Env trimers for analysis (32, 33). Recently, a stabilized soluble Env trimer (“SOSIP”) was developed, which has structural and antigenic characteristics as well as a glycosylation pattern similar to those of native HIV-1 Env from virions (34–36). Although the stabilizing mutations are likely to alter some of the properties of the SOSIP Env trimer in solution, it presents the only available reagent for studying trimeric Env, and several studies have shown that antibody binding to SOSIPs correlates strongly with neutralization potency (34).
Here we used the SOSIP trimers to examine the structural and antigenic effects of N197 glycan removal in a native-like Env trimer. Biolayer interferometry (BLI) was used to measure binding kinetics for several bNAbs to track antigenic changes in gp120 monomers and the native-like trimer. Hydrogen-deuterium exchange mass spectrometry (HDX-MS) was applied to probe local structural dynamic changes resulting from the removal of N197. All-atom modeling of the glycosylated Env trimer was used to visualize glycan occlusion of various epitopes. Overall, this study shows that in the context of an Env trimer, the N197 glycan limits the accessibility of neutralizing antibodies targeting the CD4bs while also influencing the local dynamics of variable loops at the interface of adjacent protomers. Additionally, we find that global trimer stability was not significantly impacted by the removal of the glycan.
MATERIALS AND METHODS
Expression and purification of trimeric envelope glycoproteins.
The expression vector for trimeric BG505 SOSIP.664 Env and full-length furin expression vectors were kindly supplied by John P. Moore (34, 37–40). To remove the N-glycan at residue 197 of Env, the original asparagine (N) residue was replaced with glutamine (Q) by using a QuikChange II XL site-directed mutagenesis kit (Stratagene). The env gene for full-length PVO.4 (41) was synthesized by GeneArt Gene Synthesis Service (Invitrogen) and cloned into expression vector pPPI4 by using PstI and NotI sites (38). Mutations A501C, T605C, and I559P; six arginine residues replacing the REKR furin cleavage sequence; and a stop codon after residue 664 were introduced into the PVO.4 env gene to create the PVO.4 SOSIP vector. Mutation N197Q was then introduced to generate a PVO.4 SOSIP N197Q expression vector.
Env trimers were expressed in HEK293F cells by transient transfection using FreeStyle Max 293 (Life Technologies, Thermo Fisher Scientific). Six days after transfection, the supernatant was collected and passed through a 0.45-μm filter (Fisher Scientific). A cocktail of protease inhibitors (Roche Diagnostics) was added, and the supernatant was loaded onto a Galanthus nivalis lectin (GNA; Vector Laboratories) affinity column. The eluted protein from the GNA column was buffer exchanged into a solution containing 100 mM NaCl and 20 mM Tris-Cl (pH 8.0) and then loaded onto HiTrap DEAE Sepharose FF (GE Healthcare). The flowthrough from the DEAE column was collected, buffer exchanged into a solution containing 2 M (NH4)2SO4 and 0.1 M NaH2PO4 (pH 7.0) (buffer A), and loaded onto a proPac HIC-10 column (Thermo Scientific) equilibrated with buffer A. Trimeric Env was eluted from the hydrophobic interaction chromatography (HIC) column with a linear gradient from 0 to 100% buffer B (0.1 M NaH2PO4, pH 7.0). Subsequent purification of the Env trimer was done by gel filtration using a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with 1× phosphate-buffered saline (PBS) (pH 7.4) (1 mM EDTA, 0.02% NaN3) (89). Fractions (500 μl each) were collected and analyzed by using SDS-PAGE and blue native PAGE (BN-PAGE).
Dynamic light scattering (DLS).
The hydrodynamic radius (Rh) of Env trimer samples (1 mg/ml in PBS) was measured by using a Dynapro Nanostar instrument (Wyatt Technology, Santa Barbara, CA) from 20°C to 90°C in 1°C increments. A total of 5 acquisitions of 5 s were collected at each temperature. Dynamics software (Wyatt Technologies) was used to calculate the onset temperature at which Rh begins to change significantly and the midpoint of the transition curve (Tm).
Small-angle X-ray scattering.
Small-angle X-ray scattering (SAXS) measurements were conducted on Beam Line 4-2 at the Stanford Synchrotron Radiation Laboratory with online size exclusion chromatography (SEC) as described previously (42, 43). Fifty microliters of the BG505 trimer (2 mg/ml) was injected over a precision S200 column (GE Healthcare) and eluted with PBS (pH 7.4) with a solution containing 1 mM EDTA and 0.02% sodium azide. The first 100 data points (before the void volume) were averaged as buffer scattering data and subtracted from the corresponding protein scattering data. SAXS patterns, the radius of gyration (Rg), the maximal particle dimension (Dmax), and the pairwise distance distribution histogram [P(r) plot] were analyzed by using the ATSAS software suite (44, 45).
Negative-stain electron microscopy (EM).
A 3-μl aliquot of the BG505 SOSIP.664 wild type (WT) or the N197Q mutant, diluted to 20 μg/ml in PBS, was applied for 60 s to glow-discharged C-Flat, 300-mesh, Cu grids (Electron Microscopy Sciences) and stained for an additional 60 s by using Nano-W (Nanoprobes). Data were collected by using an FEI Tecnai T12 transmission electron microscope operating at 120 keV. Images were collected by using a Gatan Ultrascan 4000 charge-coupled-device (CCD) camera at a magnification of ×52,000 and with a 1-μm defocus corresponding to a pixel size of 2.07 Å. Particle picking and classification were performed by using the EMAN2.1 image processing suite (46). In short, particles were selected by using interactive particle picking from 98 micrographs. A 2-fold binned, phase-flipped, contrast transfer function-corrected stack of ∼9,600 particles were created and subjected to reference-free two-dimensional (2D) classification to generate 50 2D classes. From these classes, a representative panel was chosen for illustrative purposes.
Biolayer interferometry.
An Octet RED 96 biolayer interferometry system (FortéBio, Menlo Park, CA) was used to measure the binding of each Env trimer to different antibodies. Antibodies 17b (47, 48); A32 (49); VRC01 (50); NIH45-46 (51); VRC03 (50); F105 (52); b12 (53); PG9 and PG16 (16); PGT121, PGT126, and PGT145 (54); 2G12 (55); and CD4-IgG2 (56) were obtained from the NIH AIDS Reagents Program. Anti-human IgG Fc capture (AHC) biosensors were hydrated for at least 10 min prior to antibody loading. Human anti-HIV antibody (10 μg/ml) in binding buffer (1× PBS [pH 7.4], 1% bovine serum albumin [BSA], 0.03% Tween 20, 0.02% NaN3) was then bound to the AHC tips for 240 s and washed in binding buffer for 60 s to reach a stable baseline. The AHC antibody tips were then dipped into Env trimer samples prepared with a 2-fold dilution series. After association for several minutes, the tips were then placed back into the baseline buffer to allow dissociation to occur. According to different antibodies, the times for loading of antibody, association, and dissociation were optimized. Data Analysis 7.0 (FortéBio) was used to analyze changes in the refractive index, and the association and dissociation curves were fit by using a 1:1 binding model. The final association rate (ka) and dissociation rate (kd) rate constants were used to calculate the ensemble dissociation constants (KD = kd/ka).
Hydrogen-deuterium exchange mass spectrometry.
Hydrogen-deuterium exchange was initiated by diluting 100 μg/ml Env trimer protein 10-fold into phosphate buffer (10 mM phosphate, 144 mM NaCl [pH 7.4]) with 85% D2O (Cambridge Isotope Labs). Hydrogen-deuterium exchange was carried out at room temperature for 3 s, 1 min, 30 min, and 20 h. At each time point, the solution was quenched by 1:1 dilution into prechilled 200 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 0.02% formic acid for a final pH of 2.5. The samples were frozen in liquid nitrogen and stored at −80°C. A zero-time-point sample and a fully deuterated sample were prepared as previously described (57). HDX measurements were performed by liquid chromatography-mass spectrometry (LC-MS) on a Synapt G2-Si instrument coupled with Waters HDX manager, keeping all loops, lines, and columns, at 1°C (Waters, Milford, MA). Samples were loaded over a 2.1- by 50-mm column filled with POROS-immobilized pepsin (58) at 150 μl/min; captured on a 2.1- by 5-mm, 1.7-μm ethyl-bridged hybrid (BEH) C18 trap column (Waters) for 3 min; and subsequently resolved over a 1- by 100-mm, 1.7-μm ethyl-bridged hybrid (BEH) C18 column (Waters). Optimized settings were used to circumvent source-induced loss of deuterium (59). Deuterium uptake was analyzed with HX-Express v2 (60), and percent exchange is reported relative to the fully deuterated and zero time points.
Modeling of fully glycosylated monomeric gp120 and SOSIP trimers.
Models of glycosylated BG505 SOSIP.664 trimers and gp120 monomers were generated by threading the BG505 sequence into the available crystal structure of the SOSIP.664 trimer (PDB accession number 4TVP) (61) by using MODELLER (62). In monomeric gp120, the N/C-terminal tails, the V1/V2 loop, and the V3 loop are highly dynamic (43, 57). For this reason, residues 32 to 41, 495 to 511, 114 to 205, and 299 to 326 were left unrestrained, with the exception of disulfide bond connectivities during modeling. Glycan chains (Man9 structures) were attached and treated as flexible residues during simulated annealing, as described previously (42). Forty models were generated for the trimer, and 120 models were generated for the gp120 monomers, using 5 rounds of refinement. Each round of refinement included simulated annealing from 1,300 K to 300 K in 8 steps with 1,000 iterations at each step. Each protomer was aligned to cocrystal structures of CD4-gp120 (PDB accession number 3JWD) (63), VRC01-gp120 (PDB accession number 3NGB) (64), VRC03-gp120 (PDB accession number 3SE8) (65), F105-gp120 (PDB accession number 3HI1) (66), b12-gp120 (PDB accession number 2NY7) (67), and PG9-V1/V2 (PDB accession number 3U4E) (68). Steric clashes (>1-Å overlap) between the various ligands and specific glycan chains in each model were assessed with PyMOL (69). The relative positions of PGT122 Fab and 2G12 Fab2 were obtained from data reported under PBD accession number 4TVP (61) and Electron Microscopy Data Bank accession number EMD-5982 (70), respectively.
RESULTS
Removal of the N197 glycan does not alter the global structure of SOSIP trimers.
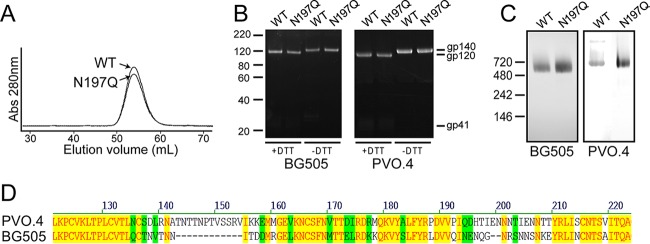
To examine the effects of N197 glycan removal within a native trimer, the N197Q mutation was introduced into the BG505 SOSIP construct, and both WT and N197Q trimers were transiently expressed in HEK293F cells. The two Env trimers were purified by a combination of lectin affinity chromatography, anion exchange chromatography, HIC, and a final SEC step to generate pure trimer preparations (89). The SEC elution profiles of the BG505 SOSIP.664 WT and the N197Q mutant showed the expected trimer peaks with identical elution volumes (Fig. 1A). The purity and homogeneity of the purified trimers were confirmed by SDS-PAGE and BN-PAGE (Fig. 1B and C).
FIG 1.
Purification of WT and N197Q SOSIP trimers. (A) Size exclusion profile of purified fully glycosylated WT and N197Q BG505 SOSIP expressed in HEK293F cells. Abs, absorbance. (B) SDS-PAGE analysis of purified BG505 (left) and PVO.4 (right) WT and N197Q mutant trimers under reducing and nonreducing conditions. DTT, dithiothreitol. (C) BN-PAGE analysis of WT and N197Q mutant trimers. (D) Sequence alignment of BG505 and PVO.4 at the V1/V2 region of Env. Protein alignment was performed by using Vector NTI AlignX, and the color scheme represents amino acid identity (yellow) and blocks of similarity (green) (88).
We next tested whether the removal of the N197 glycan affects thermal stability using dynamic light scattering (DLS) to monitor the hydrodynamic radius (Rh) with increasing temperatures. The Rh of the WT trimer was constant at 6.9 ± 0.04 nm up to 66°C (Fig. 2A). Between 66°C and 73°C, the Rh shifted to 9.1 nm, likely corresponding to a partial unfolding event. The Rh remained at 9.1 nm until ∼78°C, at which point it increased further, presumably due to further unfolding resulting in protein aggregation. The midpoint melting temperature (68.5°C) is consistent with the thermal denaturation midpoint of BG505 SOSIP (68.1°C) observed by differential scanning calorimetry (34). The N197Q mutant showed a very similar Rh and thermal profile, with a slightly lower midpoint unfolding temperature (67.6°C) (Fig. 2A). The nature of the aggregation appears different in the mutant, as seen in the change in the apparent Rh at temperatures above 78°C; however, the temperature of the transition was very similar. Therefore, the N197 glycan deletion appears to have only a very minor thermodynamic destabilizing effect on the trimer.
FIG 2.
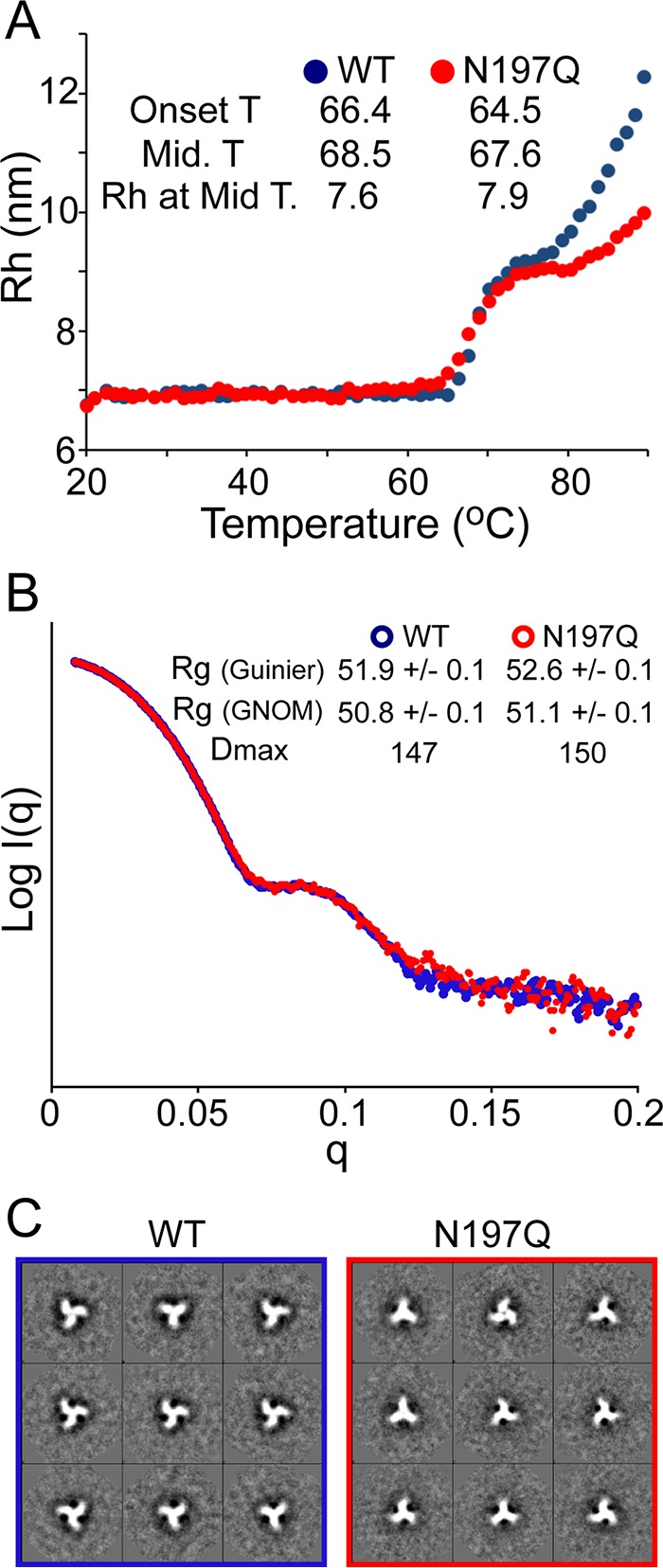
Biochemical and biophysical comparison of the purified BG505 SOSIP.664 WT and the BG505 SOSIP.664 N197Q mutant. (A) Thermal stability of the BG505 SOSIP.664 WT and the BG505 SOSIP.664 N197Q mutant analyzed by DLS. The BG505 SOSIP.664 N197Q mutant presents slightly lower onset and midpoint temperatures of melting, indicating slightly less stability than the wild-type trimer. (B) SAXS parameters for the BG505 SOSIP.664 WT and the BG505 SOSIP.664 N197Q mutant. Radii of gyration (Rg) from Guinier approximation and from the P(r) plot calculated by the program GNOM were in good agreement. (C) 2D class averages of EM reconstruction of the BG505 SOSIP.664 WT and the N197Q mutant. The two display 3-fold symmetry and are consistent with trimeric, native-like morphology.
SAXS and electron microscopy (EM) were used to examine any significant effect of the N197Q mutation on the overall morphology of BG505 SOSIP trimers. The SAXS profiles of both constructs were nearly superimposable, indicating that the structures are highly similar (Fig. 2B). However, the N197Q mutant showed a subtle but significant increase in the radius of gyration (Rg) (Fig. 2B, inset). Real-space analysis also showed a significantly higher Rg and suggested a slightly higher maximal particle dimension (Dmax) value. Class averages from negative-stain EM showed that the overall trimeric structure was not detectably altered by the removal of the N197 glycan (Fig. 2C), and both of the trimers appeared homogeneous and well ordered, as seen with high-quality trimers examined previously (34, 71).
Effects of the N197 glycan on the structural dynamics of Env trimers determined by HDX-MS.
To see if the removal of N197 affects structural flexibility, we compared the local structural dynamics of WT and N197Q SOSIP trimers by hydrogen-deuterium exchange with mass spectrometry (HDX-MS). A total of 50 peptic peptides were monitored, covering 83% of the BG505 SOSIP sequence (see Fig. S1 in the supplemental material). Due to heterogeneous glycosylation, we were unable to observe portions of V1/V2, V3, V4, and the gp41 loop. The overall exchange profiles for the WT and N197Q trimers were remarkably similar and consistent with data from previous HDX-MS studies of BG505 SOSIP trimers (Fig. 3A) (35). No differences in exchange within the V3 loop, the core of gp120, or gp41 were observed (Fig. 3B). A small but reproducible difference was observed at the base of the V1/V2 loop right at the trimeric interface of adjacent protomers. In the N197Q mutant, this region is slightly less protected, indicating that it is more flexible. Other subtle changes were observed at the bridging sheet, specifically at residues 112 to 128 and 416 to 426, which are also slightly more accessible in the N197Q mutant. Overall, the comparisons show that both trimers adopt stable trimeric conformations, but the N197Q mutation imparts a slight increase in structural flexibility around the quaternary interactions within V1/V2 and the bridging sheet segments.
FIG 3.
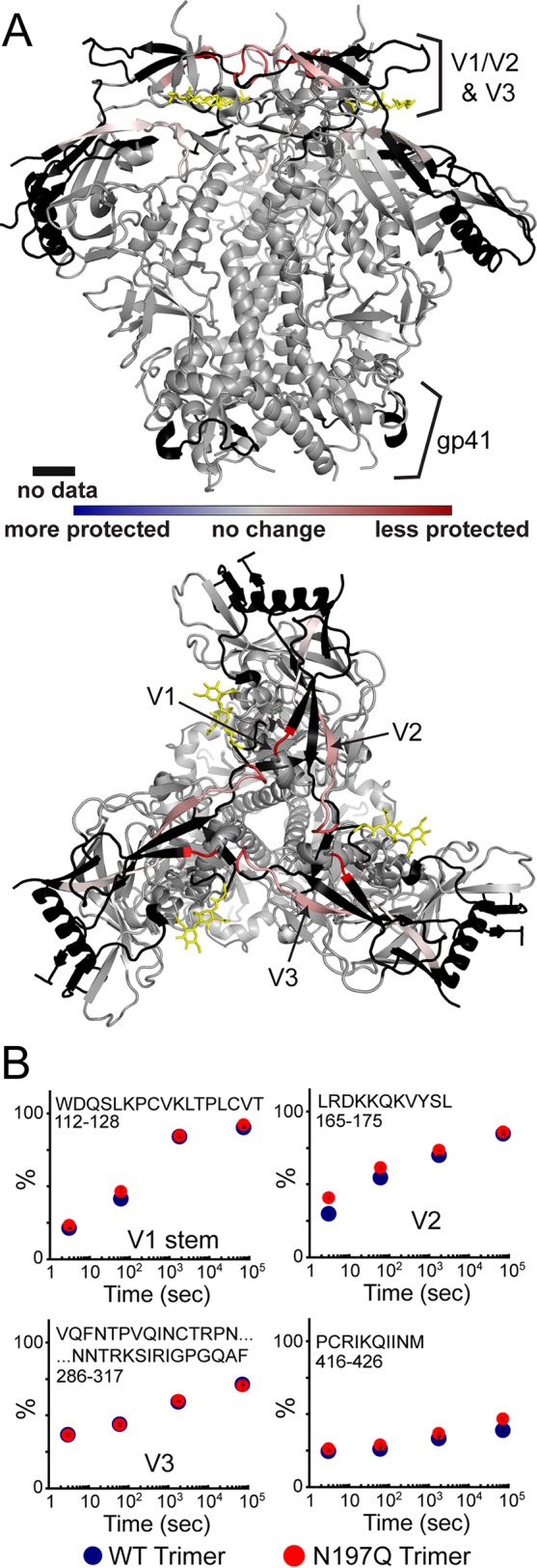
Local dynamic changes detected by HDX in the presence and absence of the N197 glycan. (A) Differences in deuterium exchange are plotted on the trimer crystal structure (PDB accession number 4TVP) (61). V1/V2 base and β20 bridging sheets are slightly less protected (red) in the BG505 SOSIP.664 N197Q trimer. The N197 glycan base is shown in yellow. (B) Individual deuterium uptake plots of selected peptides with error bars from replicate experiments. All uptake plots are shown in Fig. S1 in the supplemental material.
Removal of the N197 glycan increases binding of CD4 and CD4bs bNAbs.
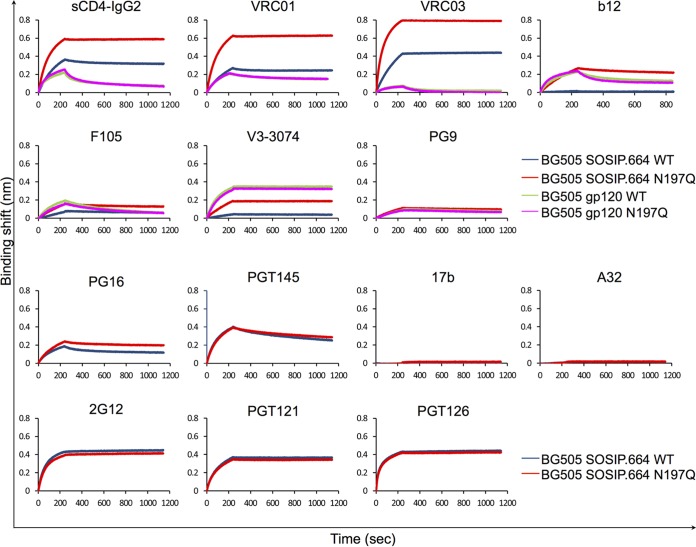
Having shown that the N197 glycan-deleted trimer retained a proper native-like trimeric conformation, we sought to investigate how the removal of this glycan results in the antigenic differences that were observed in previous studies (23, 24, 31). We compared the binding kinetics and affinities of CD4-IgG2 and various CD4bs antibodies for WT and N197Q mutant BG505 SOSIP trimers as well as the corresponding gp120 monomers by biolayer interferometry (BLI) (Fig. 4; see also Fig. S2 in the supplemental material). The N197Q trimer showed stronger binding to CD4-IgG2, with a higher (1.6-fold) association rate and a dramatically lower dissociation rate (860-fold lower). Similarly, the CD4bs bNAbs VRC01, NIH45-46, and VRC03 all showed higher association rates (1.8-, 1.5-, and 3.5-fold) and lower dissociation rates than with the WT (Table 1). In the context of gp120 monomers, the N197Q mutant did not affect the binding kinetics of CD4-IgG2 and had only minor effects on the binding of CD4bs bNAbs. The only notable effect was a 2-fold increase in the association rate for VRC03 (Table 1).
FIG 4.
Biolayer interferometry to assess antibody binding kinetics. Sensograms show the BG505 SOSIP.664 WT or the BG505 SOSIP.664 N197Q mutant and the WT BG505 gp120 monomer or the N197Q mutant BG505 gp120 monomer for binding to and unbinding from immobilized CD4 and antibodies targeting different epitopes on Env. The full concentration series of the sensograms are shown in Fig. S2 in the supplemental material. All analyses were performed at least in duplicate.
TABLE 1.
Octet biolayer interferometry binding parameters for various antibodies and Env constructsa
| Antibody and parameter | Mean value for construct ± SD |
|||||
|---|---|---|---|---|---|---|
| BG505 SOSIP.664 |
BG505 gp120 |
PVO.4 SOSIP.664 |
||||
| WT | N197Q | WT | N197Q | WT | N197Q | |
| CD4bs antibodies | ||||||
| CD4-IgG2 | ||||||
| ka (M−1 s−1) | (6.6 ± 3) × 104 | (1.08 ± 9) × 105 | (7.6 ± 2) × 104 | (7.0 ± 1) × 104 | (1.58 ± 9) × 103 | (5.9 ± 8) × 104 |
| kd (s−1) | (8.6 ± 6) × 10−5 | <1.0 × 10−7b | (5.2 ± 1) × 10−3 | (5.4 ± 1) × 10−3 | (8 ± 3) × 10−6 | (2.8 ± 2) × 10−5 |
| kD (nM) | 1.3 ± 2 | <1.0 × 10−3b | 68 ± 3 | 77 ± 3 | 5 ± 1 | (4.8 ± 1) × 10−1 |
| NIH45-46 | ||||||
| ka (M−1 s−1) | (3.05 ± 3) × 104 | (5.56 ± 8) × 104 | ||||
| kd (s−1) | (10.0 ± 5) × 10−5 | (1.37 ± 5) × 10−5 | ||||
| kD (nM) | 3.3 ± 2 | (2.5 ± 1) × 10−1 | ||||
| VRC01 | ||||||
| ka (M−1 s−1) | (6 ± 1) × 104 | (9 ± 1) × 104 | (9.6 ± 3) × 104 | (1.10 ± 3) × 105 | (1.97 ± 7) × 104 | (5.7 ± 3) × 104 |
| kd (s−1) | (1.9 ± 3) × 10−4 | <1.0 × 10−7 | (3.6 ± 3) × 10−4 | (3.16 ± 2) × 10−4 | (4 ± 1) × 10−5 | (4.2 ± 4) × 10−5 |
| KD (nM) | 3 ± 1 | <1.0 × 10−3 | 3.8 ± 2 | 2.86 ± 1 | 2.0 ± 6 | (7 ± 1) × 10−1 |
| VRC03 | ||||||
| ka (M−1 s−1) | (3.93 ± 4) × 104 | (1.4 ± 2) × 105 | (1.49 ± 8) × 104 | (3.1 ± 9) × 104 | ||
| kd (s−1) | <1.0 × 10−7 | <1.0 × 10−7 | (6.3 ± 2) × 10−3 | (7.2 ± 7) × 10−3 | ||
| KD (nM) | <1.0 × 10−3 | <1.0 × 10−3 | 423 ± 35 | 238 ± 53 | ||
| F105 | ||||||
| ka (M−1 s−1) | (2.8 ± 6) × 103 | (7 ± 1) × 103 | (3.26 ± 3) × 104 | (2.72 ± 4) × 104 | (2.7 ± 5) × 102 | (3.6 ± 1) × 103 |
| kd (s−1) | (2.3 ± 4) × 10−4 | (2.46 ± 7) × 10−4 | (1.04 ± 5) × 10−3 | (1.01 ± 2) × 10−3 | (4.14 ± 6) × 10−5 | (4.11 ± 2) × 10−4 |
| KD (nM) | 84 ± 4 | 36 ± 6 | 31.0 ± 6 | 37 ± 1 | 156 ± 6 | 114 ± 5 |
| b12 | ||||||
| ka (M−1 s−1) | NA | (5.14 ± 5) × 104 | (7.0 ± 1) × 104 | (1.18 ± 3) × 105 | (3.2 ± 8) × 104 | (5 ± 1) × 104 |
| kd (s−1) | NA | (1.88 ± 5) × 10−4 | (1.82 ± 4) × 10−3 | (3.5 ± 1) × 10−3 | (2.4 ± 6) × 10−3 | (1.0 ± 4) × 10−3 |
| KD (nM) | NA | 3.6 ± 1 | 25.8 ± 9) | 30 ± 2 | 73.3 ± 9 | 23 ± 2 |
| V3 specific antibody V3-3074 | ||||||
| ka (M−1 s−1) | (1.27 ± 2) × 104 | (3.28 ± 3) × 104 | (8.0 ± 5) × 104 | (6.7 ± 5) × 104 | (1.4 ± 2) × 105 | (1.33 ± 4) × 105 |
| kd (s−1) | (1.39 ± 8) × 10−4 | (4 ± 1) × 10−6 | <1.0 × 10−7 | <1.0 × 10−7 | (7 ± 1) × 10−5 | <1.0 × 10−7 |
| KD (nM) | 11.0 ± 8 | (1.2 ± 5) × 10−1 | <1.0 × 10−3 | <1.0 × 10−3 | (4.88 ± 4) × 10−1 | <1.0 × 10−3 |
| V1/V2-glycan antibodies | ||||||
| PG9 | ||||||
| ka (M−1 s−1) | (3.9 ± 2) × 104 | (2.88 ± 9) × 104 | (2.17 ± 1) × 104 | (2.91 ± 3) × 104 | (1.54 ± 4) × 105 | (1.7 ± 2) × 105 |
| kd (s−1) | (2.09 ± 6) × 10−4 | (1.27 ± 2) × 10−4 | (3.21 ± 7) × 10−4 | (3.3 ± 1) × 10−4 | (3.03 ± 2) × 10−4 | (5.0 ± 6) × 10−4 |
| KD (nM) | 5.4 ± 1 | 4.42 ± 7 | 14.8 ± 4 | 11.4 ± 5 | 2.30 ± 1 | 2.97 ± 4 |
| PG16 | ||||||
| ka (M−1 s−1) | (9.8 ± 1) × 104 | (7.42 ± 6) × 104 | ||||
| kd (s−1) | (1.25 ± 5) × 10−3 | (4.8 ± 2) × 10−4 | ||||
| KD (nM) | 12.7 ± 7 | 6.4 ± 4 | ||||
| PGT145 | ||||||
| ka (M−1 s−1) | (1.6 ± 2) × 105 | (1.4 ± 4) × 105 | (2.8 ± 2) × 105 | (2.6 ± 4) × 105 | ||
| kd (s−1) | (4.9 ± 1) × 10−4 | (3.22 ± 4) × 10−4 | (4.0 ± 3) × 10−4 | (3.8 ± 7) × 10−4 | ||
| KD (nM) | 3.2 ± 5 | 2.3 ± 6 | 1.4 ± 2 | 1.45 ± 3 | ||
| V3-glycan antibodies | ||||||
| PGT121 | ||||||
| ka (M−1 s−1) | (1.214 ± 4) × 105 | (1.571 ± 8) × 105 | ||||
| kd (s−1) | <1.0 × 10−7 | <1.0 × 10−7 | ||||
| KD (nM) | <1.0 × 10−3 | <1.0 × 10−3 | ||||
| PG9T126 | ||||||
| ka (M−1 s−1) | (1.10 ± 9) × 106 | (1.1 ± 2) × 106 | ||||
| kd (s−1) | <1.0 × 10−7 | <1.0 × 10−7 | ||||
| KD (nM) | <1.0 × 10−3 | <1.0 × 10−3 | ||||
| Mannose-dependent neutralizing antibody 2G12 | ||||||
| ka (M−1 s−1) | (1.040 ± 2) × 106 | (1.006 ± 6) × 106 | ||||
| kd (s−1) | <1.0 × 10−7 | <1.0 × 10−7 | ||||
| KD (nM) | <1.0 × 10−3 | <1.0 × 10−3 | ||||
Sensograms are shown in Fig. S2 and S3 in the supplemental material. ka, rate of association; kd, rate of dissociation; KD, equilibrium dissociation constant. Values are means ± standard deviations. NA, not applicable (due to weak binding, kinetics could not be fit).
Due to slow dissociation, dissociation rates or binding constants could not be fit reliably.
b12 and F105 also target the CD4bs but lack breadth and bind poorly to the BG505 SOSIP trimer (34). Accordingly, we detected weak binding of the BG505 SOSIP WT trimer to F105 and virtually no binding to b12 (Fig. 4). The removal of N197 significantly increased the rate of association with F105 and yielded observable binding to b12, with a KD of 3.6 nM (Table 1). With gp120 monomers, the removal of N197 resulted in a minor increase in association (1.6-fold) but also a 2-fold higher rate of dissociation from b12, resulting in similar overall KD values between the WT and the N197Q mutant. No difference in the association with or affinity for F105 was observed in the context of gp120 monomers.
To assess whether the removal of N197 has similar antigenic phenotypes in other Env isolates, we generated and purified WT and N197Q SOSIP trimers from isolate PVO.4, a clade B tier 3 virus (72). PVO.4 SOSIP produced native-like trimers that could be purified to homogeneity (Fig. 1B and C) but with lower yields (Verkerke et al., unpublished), which limited the number of antibodies examined in this study. Similarly to BG505 SOSIP trimers, the removal of the N197 glycan in PVO.4 trimers resulted in stronger binding to CD4-IgG2 by an order of magnitude. The N197Q mutant also showed a 2.9-fold increase in the association rate for CD4bs antibody VRC01 (Table 1; see also Fig. S3 in the supplemental material). Therefore, the removal of the N197 glycan within a trimer appears to generally enhance the binding of ligands targeting the CD4 binding site, predominantly by increasing the association rates.
Antigenic changes beyond the CD4 binding site after removal of the N197 glycan.
The crown of the V3 loop is buried under the N197 glycan from an adjacent protomer (37), and V3-specific antibodies are generally weakly neutralizing, although some can still bind SOSIP.664 trimers (34). By BLI, we observed significant binding of antibody V3-3074, which targets the tip of the V3 loop (73), to BG505 SOSIP WT trimers, with a KD of 11 nM. The removal of N197 increased the overall level of binding, with an association rate more than twice that of the WT trimer and a much lower dissociation rate, resulting in a KD of 0.1 nM (Table 1). A similar increase in affinity was observed with PVO.4 SOSIP trimers but without the increased association rate. V3-3074 showed stronger and faster binding to gp120 monomers, but there were no obvious differences in binding or association rates between WT and N197Q gp120 monomers.
Antibodies PG9, PG16, and PGT145 target quaternary epitopes in V1/V2 involving N-linked glycans (68, 74, 75). Both WT and N197Q BG505 trimers displayed strong binding to PG9/PG16/PGT145 with no obvious differences in association rates (Fig. 4 and Table 1). The only notable change was a 2-fold-lower dissociation rate for PG16 in the N197Q mutant. The PVO.4 N197Q trimer displayed the same binding to PG9 as that of the WT. The removal of N197 had no effect on BG505 gp120 monomers. Overall, the removal of the N197 glycan has little effect on the binding of V1/V2-directed antibodies.
CD4-induced (CD4i) antibodies, such as 17b and A32, are largely nonneutralizing, and both antibodies are unable to bind to BG505 SOSIP trimers by an enzyme-linked immunosorbent assay (ELISA) in the absence of CD4 (34, 48). By BLI, the binding of the BG505 WT SOSIP trimer to 17b or A32 was too weak to measure, even at a higher trimer concentration of 500 μM (Fig. 4). No increase in binding was observed with the N197Q mutant, suggesting that no CD4-induced conformational changes result from the removal of the N197 glycan. Finally, we compared the binding of glycan-dependent bNAbs 2G12, which targets a conserved glycan patch on gp120 (70), and PGT121 and PGT126, which recognize the base of V3 (54). All of these antibodies bound to both the WT and N97Q trimers with identical affinities and kinetics (Table 1).
Steric occlusion of the CD4 binding site by the N197 glycan.
As the removal of N197 has little effect the overall structure of the envelope trimer, we investigated whether the effect of the N197 glycan on antigenicity is due to steric occlusion of neutralizing epitopes. To define the spatial position of the N197 glycan relative to the epitopes of neutralizing antibodies, an ensemble of fully glycosylated BG505 trimer models was generated (see Materials and Methods). It is readily apparent that the N197 glycan position partially occludes the CD4bs in the Env trimer (Fig. 5A and C).
FIG 5.
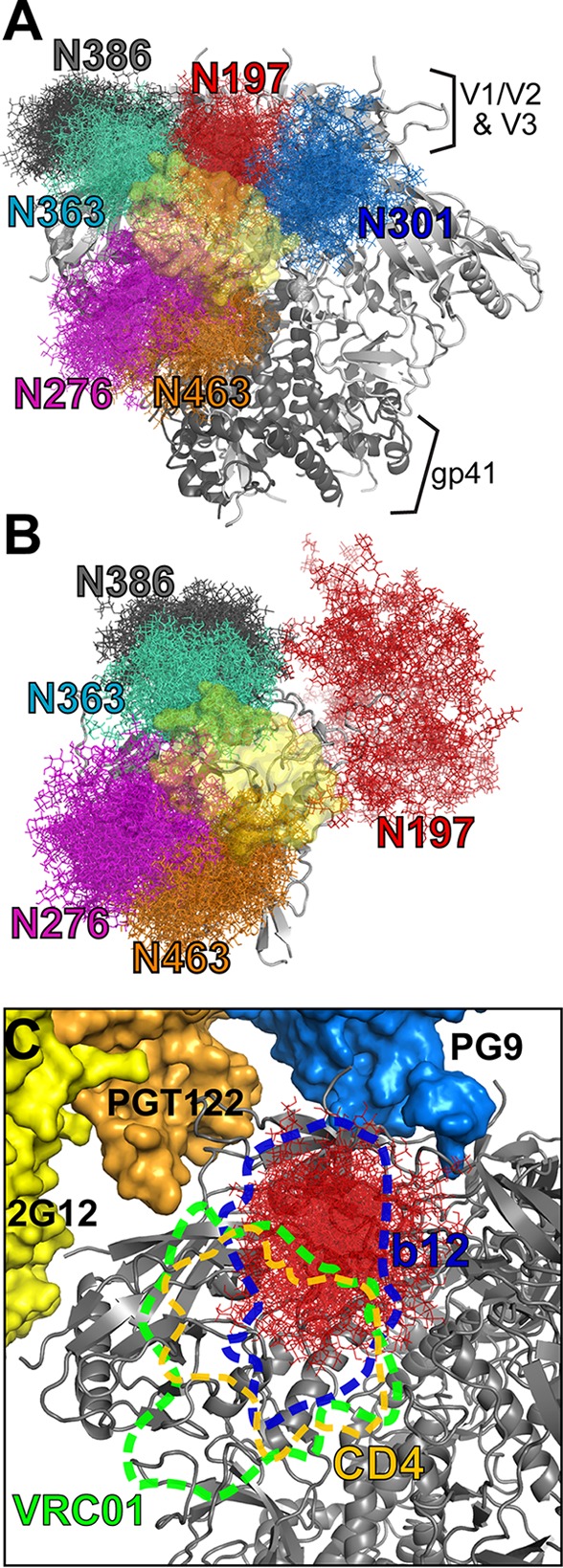
Position of the N197 glycan in the Env trimer. (A and B) The spatial occupancy from the ensemble of glycosylated Env models shows the positions of the N197, N276, N301, N363, N386, and N463 glycans, which were found to limit access to the CD4bs (yellow surface) on trimeric Env (A) and monomeric gp120 (B). (C) Position of the N197 glycan relevant to various epitopes on the trimer. The binding surface is indicated in dashed lines for CD4, b12, and VRC01. The positions of other antibodies relevant to this study are shown, including PG9 (blue), PGT122 (which binds a similar epitope as PGT121) (orange), and 2G12 (yellow).
For a more quantitative assessment of how various glycans peripheral to the CD4bs might occlude various antibodies, we analyzed the occurrence of steric clashes between glycans and the position of CD4 or various bNAbs in the 40 fully glycosylated models. Glycans at N197, N276, N301, N363, N386, and N463 were identified to potentially clash with the position of 2-domain soluble CD4 (sCD4) when it is bound (Fig. 5A). Of these glycans, the N197 glycan was most likely to clash with CD4, as seen in 70% of the models (Tables 2 and 3). The nonneutralizing CD4bs antibodies F105 and b12 clashed with the N197 glycan in 100% of the models as well as with the N301 glycan from the neighboring protomer. With antibodies VRC01 and VRC03, the major clashes were seen with N276 and N463, occurring in >97% of the models, but clashes with N197 still occurred in 73% of the models. In contrast, the N197 glycan clashed with the position of PG9 in only 30% of the models (Fig. 5C).
TABLE 2.
Steric clashes between various ligands and glycans on the Env trimer as percentage of models in the ensemblea
| Glycan | % of models with steric clash on Env trimer |
|||||
|---|---|---|---|---|---|---|
| sCD4 | VCR01 | VRC03 | F105 | b12 | PG9 | |
| N197 | 70 | 73 | 78 | 100 | 100 | 30 |
| N276 | 43 | 98 | 100 | 22 | 17 | |
| N301b | 11 | 43 | 42 | 100 | 100 | |
| N363 | 56 | 64 | 57 | 32 | 41 | |
| N386 | 12 | 11 | 7 | 23 | 46 | |
| N463 | 24 | 98 | 97 | 7 | 7 | |
See also Fig. 5.
Clashes from the N301 glycan were predominantly from the neighboring protomer.
TABLE 3.
Steric clashes between various ligands and glycans on gp120 monomers as a percentage of models in the ensemblea
| Glycan | % of models with steric clash on gp120 monomer |
||||
|---|---|---|---|---|---|
| sCD4 | VCR01 | VRC03 | F105 | b12 | |
| N197 | 1 | 2 | 2 | 13 | 11 |
| N276 | 53 | 99 | 98 | 20 | 19 |
| N301 | 0 | 0 | 0 | 9 | 3 |
| N363 | 68 | 74 | 59 | 58 | 60 |
| N386 | 3 | 3 | 1 | 12 | 49 |
| N463 | 33 | 98 | 98 | 19 | 13 |
See also Fig. 5.
To compare how glycan clashes differ in the context of a gp120 monomer, we generated an ensemble of BG505 gp120 models. The N/C-terminal regions, bridging sheet, and V1/V2 and V3 loops were largely unrestrained, as they have been shown to be highly disordered in solution (43, 57). For glycans at N276, N363, N386, and N463, the clashes are similar to what was observed with the trimers (Fig. 5B). N301, which in the trimers resulted in clashes with CD4 from an adjacent protomer, showed no clashes with CD4 in monomeric gp120. Interestingly, unlike in the trimer, N197 glycan clashes with sCD4 and VRC01 in gp120 are very rare (1 to 2% of models). These observations indicate that the N197 glycan occludes the CD4 binding site only within the context of a native Env trimer.
DISCUSSION
Effects of N197 glycan removal are observed only in the context of an Env trimer.
The N197 glycan was found to modulate tier 2 cross-reactivity (76), and removal of N197 has been shown to increase neutralization sensitivity to CD4bs and V3-specific antibodies (12, 22–24). Promising results have shown that N197-deleted Env elicited a stronger neutralizing antibody response in vivo (23), indicating improved immunogenicity. However, when tested with monomeric gp120, the N197 deletion had negligible effects on CD4 binding (24). In light of our binding data, it is clear that the N197 glycan has significant effects on the interactions with CD4 and CD4bs antibodies only in the context of a native-like Env trimer (Fig. 6). The removal of N197 led to significantly increased binding of CD4 and CD4bs antibodies. At the same time, the binding of bNAbs targeting other epitopes (2G12, PGT121, and PGT126) was unaffected, indicating that the N197Q trimers were otherwise antigenically similar to the WT. Computational modeling of the glycosylated Env trimer suggests that the spatial occupancy of the N197 glycan impedes access to the CD4bs. This steric occlusion explains why the removal of this particular glycan results in a large increase in the association rates with CD4bs-targeting ligands. Beyond the enhanced association rate, we also observed a slower dissociation of CD4 and CD4bs antibodies in the N197Q trimer. The entropic penalty of positioning glycans away from the CD4bs and restricting their spatial sampling may result in a lower net free energy of binding, which in turn would result in a higher rate of dissociation of ligands from the N197 glycan-deleted trimer. Therefore, the presence of the N197 glycan may not only limit epitope access (resulting in slow antibody association) but also destabilize certain Env-antibody complexes (resulting in faster antibody dissociation).
FIG 6.
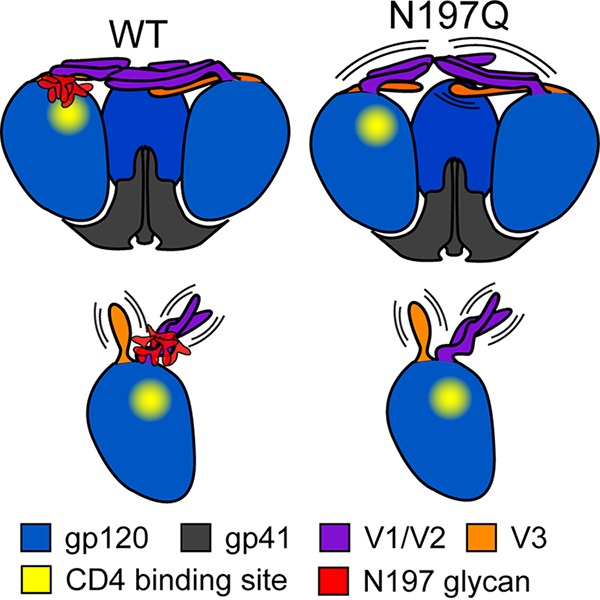
Effects of N197 glycan removal in the Env trimer or gp120 monomer. In the trimer (top), the N197 glycan partially occludes the CD4 binding site. Removal of the N197 glycan increases the exposure of the CD4 binding site while also making the V1/V2 loop more flexible and increasing the accessibility of V3. In gp120 monomers (bottom), without tertiary and quaternary structural constrains, the V1/V2/V3 loops are relatively disordered. Since the N197 glycan does not occlude the CD4 binding site, its removal has no significant influence on the binding of CD4bs or V3-specific antibodies.
With gp120 monomers, there were no significant differences in binding to CD4bs antibodies or V3-specific antibodies between the WT and the N197Q mutant. Without the quaternary constraints of a trimer, gp120 monomers are relatively dynamic molecules with a high degree of flexibility at V1/V2 and V3 (43, 57, 77). In our models of glycosylated monomeric gp120, the N197 glycan was predominantly oriented away from the CD4 binding site, and clashes with most CD4bs antibodies were seldom observed. However, with F105 and b12, there were still some minor clashes with the N197 glycan, which can explain the slightly higher on-rate of binding of b12 to the gp120 N197Q mutant (Table 1). As the V3 loop is readily exposed in gp120 monomers, it is not surprising that the removal of the N197 glycan does not affect the binding of V3-specific antibodies. The stark differences in the phenotypes between trimeric Env and monomeric gp120 highlight the importance of evaluating Env modifications in the context of a native-like trimer to assess full effects on antigenicity.
The only structural effect on the trimer observed from the N197 deletion was a subtle increase in the dynamics of the base of the V1/V2 loop and the bridging sheet near the trimeric interface by HDX-MS (Fig. 6). The increased flexibility at V1/V2 could explain the slightly larger radius of gyration of N197Q trimers as observed by SAXS (Fig. 2B). This can also explain the slight decrease in the association rate for antibodies PG9 and PG16 targeting V1/V2 in the N197Q mutant (Table 1), as the epitope would be less conformationally restrained. Since the V3 loop is buried under V1/V2 in the trimer, increased flexibility at the crown of the trimer can also result in increased V3 exposure, providing a rationale for why some V3-specific antibodies are able to bind faster to the N197 glycan-deleted trimer. The subtle effects on the conformational dynamics of the trimer by N197 glycan removal would have largely been invisible by many other structural techniques (i.e., X-ray crystallography), illustrating the utility of HDX-MS for probing all of the structural effects that arise from Env modifications. We note that the full scale of structural effects imparted by the N197Q mutant may be underrepresented in this study due to the stabilizing mutations in the SOSIP trimer, and there may be other consequences in the context of full-length virus-associated Env that we cannot detect here.
Two recent reports touched upon the role of the N197 glycan in trimeric Env. A detailed glycan analysis of BG505 SOSIP trimers reported that N197 removal reduced bNAb recognition of V1/V2 apex-targeting antibodies, including PGT145 and PG9, as reflected by changes in the mutant's neutralization sensitivity (78). It was suggested that changes at N197 influence the nearby glycans at the trimer apex. The recent crystal structure of the glycosylated Env SOSIP revealed ordered density across the surface of the trimer crystal structure (29). For N197 in a clade G SOSIP trimer (X1193.c1; G459C mutant), the only ordered density that was observed was the core Man1GlcNac2, suggesting that it is relatively free of interactions with adjacent Env structural elements. However, it is possible that even without stable structural constrains, the absence of N197 increases the exposure of nearby glycans to glycosidases and glycosyltransferases, resulting in different glycosylation profiles. It should be noted that in the crystal structure, the trimer was complexed with 2 Fabs and the Fv portion of VRC01, and such interactions could perturb the native configuration of glycan chains, including N197.
From the observable glycopeptides used for HDX-MS comparison, we observed no apparent differences in the glycosylation patterns between the WT and the N197Q mutant. However, we could not monitor the glycosylation patterns within V1/V2 or the C-terminal base of V3, which is where the major antigenic effects of glycan alterations are expected to occur. Thus, while our study addresses how a targeted glycan deletion affects the steric occlusion of epitopes and the conformational dynamics of the trimer, the influence on the glycosylation patterns of nearby glycans remains to be directly addressed.
Isolate-specific effects of the N197 glycan.
Like with BG505, the N197Q PVO.4 trimer displayed stronger binding to CD4bs antibodies than the WT, with a higher association rate for CD4, VRC01, and F105. However, the association with b12 was not significantly faster, and the stronger binding affinity was due mostly to a lower dissociation rate. This is a distinct effect from what was observed for the BG505 trimer, which displayed detectable binding to b12 only after the removal of the N197 glycan. PVO.4 has a much longer V1/V2 loop than BG505, with 12 extra amino acids and additional glycosylation sites at the base of V1 (Fig. 1D). b12 makes contacts with a portion of V1/V2, and binding leads to a partial opening of the V1/V2 loop (79, 80). The longer V1/V2 loop might provide more steric hindrance for b12 accessing the Env trimer, even without the N197 glycan, explaining why the association of the N197Q trimer with b12 was not much faster than that of the wild type. The extra amino acids in the V1/V2 loop may also introduce additional contacts with b12 that are affected by the presence of the N197 glycan, explaining the changes in the dissociation rates. As with b12, the rates of association of the PVO.4 N197Q mutant and the WT with V3-3074 were similar, and both exhibited higher association rates than the BG505 trimers. It appears that the V3-3074 epitope is intrinsically more exposed in the PVO.4 SOSIP, and as a result, the removal of N197 has a negligible effect on the accessibility of V3.
Based upon the available data, we infer that the functional effects of the N197 glycan vary between different isolates in large part due to the large differences in V1/V2 sequences. This may provide a rationale for the isolate-dependent effects of N197 removal that were observed in previous studies. For example, the removal of the N197 sequon in some isolates has been reported to produce an enhancement in CD4-independent entry (81). In other cases, the magnitude of changes in antigenicity resulting from N197 modification differed among isolates (24). This was attributed to differences in V1/V2, but no obvious specific characteristic (i.e., length or number of glycans) correlated with the level of neutralization changes with N197 removal. Several studies have shown that isolates vary in the level of “openness” of Env (80, 82). Tier 1 or laboratory-adapted isolates are very susceptible to neutralization and are described as having more “open” conformations, while difficult-to-neutralize isolates have more “closed” Env conformations. We hypothesize that the level of openness will likely govern how much of an effect N197 removal will have, and much of this is determined by specific characteristics of V1/V2 and V3 that have yet to be understood in detail.
In general, the BLI measurements of antibody binding correlated well with the observed changes in neutralization; namely, CD4 binding site and V3 binding and neutralization sensitivities increase when the N197 glycan is removed. For example, isolate PVO.4 lacking N197 was ∼23-fold more susceptible to neutralization by VRC01 and 3 orders of magnitude more susceptible to V3-targeting 447-52D than the WT (24). From our binding assay, the N197Q SOSIP trimer from isolate PVO.4 showed 3-fold-stronger binding to VRC01 and 100-fold-stronger binding to 447-52D than the WT. Pritchard et al. reported a 10-fold increase in neutralization sensitivity to PGV04 (an antibody with a binding footprint very similar to that of VRC01) resulting from N197 glycan deletion in BG505 (36). In accord, we observe that the N197Q mutant trimer has an ∼1,000-fold increase in affinity for VRC01 compared to the WT. The relatively few differences between data from neutralization assays and binding may be due to the SOSIP-based stabilization of the soluble trimer constructs that we have available for analysis as well as the presence of the membrane-proximal external region (MPER), the transmembrane anchor, and the cytoplasmic tail on full-length Env on pseudotyped viruses used in neutralization assays.
Significance of the N197 glycan in vaccine design.
The structural and antigenic features of Env are a major focus of HIV-1 vaccine design. Conserved glycosylation sites are of particular interest since many bNAbs recognize conserved carbohydrates in combination with other protein elements (8, 15, 16, 54, 68, 74, 83–85). On the other hand, glycans also mask conserved polypeptide epitopes, and deletion of glycans to better expose neutralizing epitopes is another strategy in vaccine design (86). The BG505 SOSIP trimer is a promising immunogen candidate with stable native-like trimeric characters, but so far, it has been capable of eliciting only autologous tier 2 neutralizing antibodies, with tier 1 cross-reactivity biased to the V3 epitope (87). Our results give direct evidence for a specific modification for enhancing the binding of CD4bs antibodies while maintaining other relevant bNAb epitopes (e.g., PG9 and PGT122), without disrupting the overall quaternary structure of the trimer. Trimeric Env constructs modified by the deletion of one single glycan, like N197, are likely to provide improvements to the current trimer Env-based vaccine approaches, particularly for the elicitation of CD4bs antibodies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tsutomu Matsui and the support staff at the Stanford Synchrotron Radiation Lightsource (SSRL) for assistance with SAXS data collection. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: CD4-IgG2 was obtained from Progenics Pharmaceuticals, Inc.; HIV-1 gp120 monoclonal antibodies 17b and A32 were obtained from James E. Robinson; VRC01 and VRC03 were obtained from John Mascola; NIH45-46 was obtained from Pamela Bjorkman; F105 was obtained from Marshall Posner and Lisa Cavacini; b12 was obtained from Dennis Burton and Carlos Barbas; and PG9, PG16, PGT121, PGT126, and PGT145 were obtained from the International AIDS Vaccine Initiative (IAVI).
Portions of the work were carried out at the SSRL and were supported by the Structural Molecular Biology Program, Department of Energy, Office of Biological and Environmental Research, under NIH grants P41-GM103393 and P41-RR001209.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01116-16.
REFERENCES
- 1.Crooks ET, Jiang P, Franti M, Wong S, Zwick MB, Hoxie JA, Robinson JE, Moore PL, Binley JM. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364–378. doi: 10.1016/j.virol.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu Rev Immunol 28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 3.Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu Rev Med 61:135–152. doi: 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- 4.Haynes BF, Moody MA, Alam M, Bonsignori M, Verkoczy L, Ferrari G, Gao F, Tomaras GD, Liao HX, Kelsoe G. 2014. Progress in HIV-1 vaccine development. J Allergy Clin Immunol 134:3–10. doi: 10.1016/j.jaci.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JH, Rerks-Ngarm S, Excler JL, Michael NL. 2010. HIV vaccines: lessons learned and the way forward. Curr Opin HIV AIDS 5:428–434. doi: 10.1097/COH.0b013e32833d17ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley T, Fera D, Bhiman J, Eslamizar L, Lu X, Anasti K, Zhang R, Sutherland LL, Scearce RM, Bowman CM, Stolarchuk C, Lloyd KE, Parks R, Eaton A, Foulger A, Nie X, Karim SS, Barnett S, Kelsoe G, Kepler TB, Alam SM, Montefiori DC, Moody MA, Liao HX, Morris L, Santra S, Harrison SC, Haynes BF. 2016. Structural constraints of vaccine-induced tier-2 autologous HIV neutralizing antibodies targeting the receptor-binding site. Cell Rep 14:43–54. doi: 10.1016/j.celrep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scanlan CN, Offer J, Zitzmann N, Dwek RA. 2007. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature 446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 9.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 10.Haggerty S, Dempsey MP, Bukrinsky MI, Guo L, Stevenson M. 1991. Posttranslational modifications within the HIV-1 envelope glycoprotein which restrict virus assembly and CD4-dependent infection. AIDS Res Hum Retroviruses 7:501–510. doi: 10.1089/aid.1991.7.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Rey-Cuille MA, Hu SL. 2001. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res Hum Retroviruses 17:1473–1479. doi: 10.1089/08892220152644179. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Nie J, Prochnow C, Truong C, Jia Z, Wang S, Chen XS, Wang Y. 2013. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology 10:14. doi: 10.1186/1742-4690-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julien JP, Lee PS, Wilson IA. 2012. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev 250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crispin M, Doores KJ. 2015. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr Opin Virol 11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCaffrey RA, Saunders C, Hensel M, Stamatatos L. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J Virol 78:3279–3295. doi: 10.1128/JVI.78.7.3279-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 21.Huang X, Jin W, Hu K, Luo S, Du T, Griffin GE, Shattock RJ, Hu Q. 2012. Highly conserved HIV-1 gp120 glycans proximal to CD4-binding region affect viral infectivity and neutralizing antibody induction. Virology 423:97–106. doi: 10.1016/j.virol.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol 75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Cleveland B, Klots I, Travis B, Richardson BA, Anderson D, Montefiori D, Polacino P, Hu SL. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J Virol 82:638–651. doi: 10.1128/JVI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Townsley S, Li Y, Kozyrev Y, Cleveland B, Hu SL. 2016. The conserved role of an N-linked glycan on the surface antigen of HIV-1 modulating virus sensitivity to broadly neutralizing antibodies against the receptor and coreceptor binding sites. J Virol 90:829–841. doi: 10.1128/JVI.02321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE Jr, Johnson PR, Schief WR. 2014. Proof of principle for epitope-focused vaccine design. Nature 507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, Menis S, Jones M, Kubitz M, Spencer S, Adachi Y, Burton DR, Schief WR, Nemazee D. 2015. HIV-1 vaccines. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, Pietzsch J, Gray MD, Cupo A, van Gils MJ, Yao KH, Liu C, Gazumyan A, Seaman MS, Bjorkman PJ, Sanders RW, Moore JP, Stamatatos L, Schief WR, Nussenzweig MC. 2015. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell 161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi CW, Davison JR, Georgiev IS, Joyce MG, Kwon YD, Pancera M, Taft J, Yang Y, Zhang B, Shivatare SS, Shivatare VS, Lee CC, Wu CY, Bewley CA, Burton DR, Koff WC, Connors M, Crispin M, Baxa U, Korber BT, Wong CH, Mascola JR, Kwong PD. 2016. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, de Val N, Lyumkis D, Ward AB. 2015. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure 23:1943–1951. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utachee P, Nakamura S, Isarangkura-Na-Ayuthaya P, Tokunaga K, Sawanpanyalert P, Ikuta K, Auwanit W, Kameoka M. 2010. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. J Virol 84:4311–4320. doi: 10.1128/JVI.02619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M, Lee KK. 2013. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J Virol 87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien JP, Wilson IA, Klasse PJ, Ward AB, Moore JP. 2013. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A 110:18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guttman M, Garcia NK, Cupo A, Matsui T, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2014. CD4-induced activation in a soluble HIV-1 Env trimer. Structure 22:974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard LK, Harvey DJ, Bonomelli C, Crispin M, Doores KJ. 2015. Cell- and protein-directed glycosylation of native cleaved HIV-1 envelope. J Virol 89:8932–8944. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khayat R, Lee JH, Julien JP, Cupo A, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Structural characterization of cleaved, soluble HIV-1 envelope glycoprotein trimers. J Virol 87:9865–9872. doi: 10.1128/JVI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klasse PJ, Depetris RS, Pejchal R, Julien JP, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. 2013. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol 87:9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cloyd MW, Moore BE. 1990. Spectrum of biological properties of human immunodeficiency virus (HIV-1) isolates. Virology 174:103–116. doi: 10.1016/0042-6822(90)90059-Z. [DOI] [PubMed] [Google Scholar]
- 42.Guttman M, Weinkam P, Sali A, Lee KK. 2013. All-atom ensemble modeling to analyze small-angle X-ray scattering of glycosylated proteins. Structure 21:321–331. doi: 10.1016/j.str.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davenport TM, Guttman M, Guo W, Cleveland B, Kahn M, Hu SL, Lee KK. 2013. Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120. J Virol 87:10855–10873. doi: 10.1128/JVI.01535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun DI. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25:495–503. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- 45.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36:1277–1282. doi: 10.1107/S0021889803012779. [DOI] [Google Scholar]
- 46.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore JP, Thali M, Jameson BA, Vignaux F, Lewis GK, Poon SW, Charles M, Fung MS, Sun B, Durda PJ, Åkerblom L, Wahren B, Ho DD, Sattentau QJ, Sodroski J. 1993. Immunochemical analysis of the gp120 surface glycoprotein of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J Virol 67:4785–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diskin R, Scheid JF, Marcovecchio PM, West AP Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquir Immune Defic Syndr 6:7–14. [PubMed] [Google Scholar]
- 53.Burton DR, Barbas CF III, Persson MA, Koenig S, Chanock RM, Lerner RA. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci U S A 88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 56.Garlick RL, Kirschner RJ, Eckenrode FM, Tarpley WG, Tomich CS. 1990. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res Hum Retroviruses 6:465–479. doi: 10.1089/aid.1990.6.465. [DOI] [PubMed] [Google Scholar]
- 57.Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. 2012. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol 86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Pan H, Smith DL. 2002. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics 1:132–138. doi: 10.1074/mcp.M100009-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Guttman M, Wales TE, Whittington D, Engen JR, Brown JM, Lee KK. 2016. Tuning a high transmission ion guide to prevent gas-phase proton exchange during H/D exchange MS analysis. J Am Soc Mass Spectrom 27:662–668. doi: 10.1007/s13361-015-1330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guttman M, Weis DD, Engen JR, Lee KK. 2013. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom 24:1906–1912. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb B, Sali A. 2014. Protein structure modeling with MODELLER. Methods Mol Biol 1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 63.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 70.Murin CD, Julien JP, Sok D, Stanfield RL, Khayat R, Cupo A, Moore JP, Burton DR, Wilson IA, Ward AB. 2014. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol 88:10177–10188. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cadogan M, Austen B, Heeney JL, Dalgleish AG. 2008. HLA homology within the C5 domain promotes peptide binding by HIV type 1 gp120. AIDS Res Hum Retroviruses 24:845–855. doi: 10.1089/aid.2007.0194. [DOI] [PubMed] [Google Scholar]
- 74.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, Cupo A, Crispin M, Lee KK, Desaire H, Ward AB, Klasse PJ, Moore JP. 2015. Influences on the design and purification of soluble, recombinant native-like HIV-1 envelope glycoprotein trimers. J Virol 89:12189–12210. doi: 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O'Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. 2015. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog 11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang Y, Guttman M, Davenport TM, Hu SL, Lee KK. 2016. Probing the impact of local structural dynamics of conformational epitopes on antibody recognition. Biochemistry 55:2197–2213. doi: 10.1021/acs.biochem.5b01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, Struwe WB, Cupo A, Kumar A, Zitzmann N, Seabright GE, Kramer HB, Spencer DI, Royle L, Lee JH, Klasse PJ, Burton DR, Wilson IA, Ward AB, Sanders RW, Moore JP, Doores KJ, Crispin M. 2016. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep 14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. 2015. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun 6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol 75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB III, Kwong PD, Blanchard SC, Mothes W. 2014. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, Schief WR, Stamatatos L. 2013. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med 210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu G, Moriyama EN. 2004. Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinformatics 5:378–388. doi: 10.1093/bib/5.4.378. [DOI] [PubMed] [Google Scholar]
- 89.Verkerke HP, Williams JA, Guttman M, Simonich CA, Liang Y, Filipavicius M, Hu S-L, Overbaugh J, Lee KK. 2016. Epitope-independent purification of native-like envelope trimers from diverse HIV-1 isolates. J Virol 90:9471–9482. doi: 10.1128/JVI.01351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.