ABSTRACT
STING has emerged in recent years as a key player in orchestrating innate immune responses to cytosolic DNA and RNA derived from pathogens. However, the regulation of STING still remains poorly defined. In the present study, we investigated the mechanism of the regulation of STING expression in relation to the RIG-I pathway. Our data show that signaling through RIG-I induces STING expression at both the transcriptional and protein levels in various cell types. STING induction by the RIG-I agonist 5′triphosphorylated RNA (5′pppRNA) was recognized to be a delayed event resulting from an autocrine/paracrine mechanism. Indeed, cotreatment with tumor necrosis factor alpha and type I/II interferon was found to have a synergistic effect on the regulation of STING expression and could be potently decreased by impairing NF-κB and/or STAT1/2 signaling. STING induction significantly contributed to sustainment of the immune signaling cascade following 5′pppRNA treatment. Physiologically, this cross talk between the RNA- and DNA-sensing pathways allowed 5′pppRNA to efficiently block infection by herpes simplex virus 1 (HSV-1) both in vitro and in vivo in a STING-dependent fashion. These observations demonstrate that STING induction by RIG-I signaling through the NF-κB and STAT1/2 cascades is essential for RIG-I agonist-mediated HSV-1 restriction.
IMPORTANCE The innate immune system represents the first line of defense against invading pathogens. The dysregulation of this system can result in failure to combat pathogens, inflammation, and autoimmune diseases. Thus, precise regulation at each level of the innate immune system is crucial. Recently, a number of studies have established STING to be a central molecule in the innate immune response to cytosolic DNA and RNA derived from pathogens. Here, we describe the regulation of STING via RIG-I-mediated innate immune sensing. We found that STING is synergistically induced via proinflammatory and antiviral cytokine cascades. In addition, we show that in vivo protection against herpes simplex virus 1 (HSV-1) by a RIG-I agonist required STING. Our study provides new insights into the cross talk between DNA and RNA pathogen-sensing systems via the control of STING.
INTRODUCTION
Innate immunity is crucial for the host to defeat disease-causing pathogens. This is characterized by the rapid and efficient detection of pathogen-associated molecular patterns (PAMPs) via pattern recognition receptors (PRRs) (1, 2). Retinoic acid-induced gene I (RIG-I), a cytoplasmic PRR, is essential for recognizing viral RNA that contains either a 5′ triphosphate (ppp) or 5′ diphosphate (pp) signature (3–5). Upon RNA stimulation, RIG-I recruits the adaptor protein mitochondrial antiviral-signaling protein (MAVS; also known as IPS-1, Cardif, or VISA) to activate the TANK binding kinase 1 (TBK1)-I kappa B kinase ε (IKKε) complex and the I kappa B kinase α (IKKα)-I kappa B kinase β (IKKβ) complex, which are responsible for the activation of the transcription factor interferon (IFN) regulatory factor 3 (IRF3), the transcription factor IFN regulatory factor 7 (IRF7), and nuclear factor-kappa B (NF-κB) (6, 7). These transcription factors then translocate to the nucleus and coordinate the induction of type I IFN and proinflammatory cytokines (8, 9). Released alpha/beta IFN (IFN-α/β) bind to their cognate receptors and lead to the transcriptional activation of interferon-stimulated genes (ISG) by the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway (10, 11). The products of ISG are key effectors in limiting pathogen replication (12, 13).
A number of studies have established the role of stimulator of IFN genes (STING; also called MITA, TMEM173, MPYS, and ERIS) and have shown that it is a key element in the innate immune response elicited by pathogenic nucleic acids, including cytosolic DNA and cyclic dinucleotides (CDNs) (14–19). STING not only interacts with cytosolic single-stranded DNA/double-stranded DNA (dsDNA) sensors, including DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (DDX41) (20), IFN-γ-inducible protein 16 (IFI16) (21), and LSm14A (22), for type I IFN induction but also independently senses CDNs, such as cyclic dimeric GMP (c-di-GMP), cyclic dimeric AMP (c-di-AMP), and cyclic GMP-AMP (cGAMP) (19, 23–25). The recently identified general DNA sensor cGAMP synthase (cGAS), which is responsible for the recognition of DNA from viruses, bacteria, parasites, and retroviruses, employs STING as an adaptor for downstream IFN responses (19, 26–30).
The role of STING in the innate immune response to RNA viruses has become increasingly apparent. STING can function as a cofactor in the RIG-I-mediated IFN response to RNA viruses, such as Sendai virus (SeV), vesicular stomatitis virus (VSV), Newcastle disease virus (NDV), and Japanese encephalitis virus (JEV) (14, 16, 18, 31–34). Indeed, STING interacts with RIG-I and MAVS (14, 31), and its signaling is antagonized by several RNA viruses (34–37). Both DNA and RNA triggered immune response converge on the TBK1-IRF3 axis, where STING serves as a scaffold protein facilitating the phosphorylation of IRF3 (31, 38). Activated IRF3 and NF-κB cooperate in the production of type I IFN and proinflammatory cytokines.
Many host factors and viral proteins (34, 36, 39) have been implicated in modulating STING at the transcriptional level (40) or posttranslational level (32, 41–43), yet the regulatory mechanisms of STING have not been fully elucidated. In our previous work to gain system-wide insight into the RIG-I transcriptome, STING was identified to be a differentially expressed gene induced by the RIG-I agonist 5′ triphosphorylated RNA (5′pppRNA) (44). This observation suggested that RIG-I signaling was involved in STING expression. In the present study, we show that STING is a late RIG-I-inducible gene triggered by autocrine/paracrine signaling via tumor necrosis factor alpha (TNF-α) and IFN synergy. Furthermore, STING sustains the inflammatory/antiviral expression profile elicited by 5′pppRNA treatment and is required to protect mice from a lethal challenge with herpes simplex virus 1 (HSV-1). This report provides insight into the regulation of STING and demonstrates its manipulation by a RIG-I agonist to protect against a DNA viral pathogen in vivo.
MATERIALS AND METHODS
In vitro synthesis of 5′pppRNAs.
The sequence of 5′pppRNA was derived from the 5′ and 3′ untranslated regions (UTR) of the VSV genome as previously described (44). In vitro-transcribed RNA was prepared using an Ambion MEGAscript T7 high-yield transcription kit according to the manufacturer's instruction (Invitrogen, NY, USA). 5′pppRNA was purified using a Qiagen microRNA minikit (Qiagen). An RNA with the same sequence but lacking the 5′ppp moiety was purchased from Integrated DNA Technologies Inc. (IDT). This RNA generated results identical to those obtained with 5′pppRNA, which was dephosphorylated enzymatically with calf intestinal alkaline phosphatase (Invitrogen).
Cell culture and transfections.
A549 cells and PC3 cells (American Type Culture Collection [ATCC]) were grown in F-12K medium (Wisent) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (Wisent). Human normal lung fibroblast MRC5 cells (kindly provided by Jerry Pelletier, McGill University, Canada), immortalized human hepatocytes (IHHs; kindly provided by Ranjit Ray, Saint Louis University, St. Louis, MO, USA) (45), Huh7.5 cells (ATCC), and Huh7 cells (ATCC) were grown in Dulbecco modified Eagle medium (DMEM; Wisent) supplemented with 10% FBS, 1% nonessential amino acids, and 1% antibiotics. U87 cells (ATCC) and mouse embryonic fibroblasts (MEFs) were grown in DMEM (Wisent) supplemented with 10% FBS and 1% antibiotics. Wild-type (WT) and RIG-I−/− MEFs were kind gifts from Shizou Akira (Osaka University, Osaka, Japan). The Lipofectamine RNAiMax reagent (Invitrogen) was used for transfections of 5′pppRNA in A549 cells according to the manufacturer's instructions. For knockdown with short interfering RNA (siRNA), A549 cells were transfected with human RIG-I (catalog number sc-61480; Santa Cruz Biotechnology), the IFN-α/β receptor (IFN-α/βR) α chain (catalog number sc-35637; Santa Cruz Biotechnology) and β chain (catalog number sc-40091; Santa Cruz Biotechnology), STAT1 (catalog number sc-44123; Santa Cruz Biotechnology), STING (catalog number sc-92042; Santa Cruz Biotechnology), control siRNA (catalog number sc-37007; Santa Cruz Biotechnology), RELA (catalog number SI02663094; Qiagen), IRF3 (catalog number SI00026418; Qiagen), TNFRSF1A (catalog number SI00301945; Qiagen), and control siRNA (catalog number sc-37007; Santa Cruz Biotechnology) using Lipofectamine RNAiMax (Life technologies) according to the manufacturer's guidelines.
Virus production, quantification, and infection.
Recombinant green fluorescent protein (GFP)-tagged VSV (VSV-GFP), which harbors the methionine 51 deletion in the matrix protein-coding sequence (VSVΔ51) (46), was kindly provided by J. Bell (Ottawa Health Research Institute, Canada). Virus stock concentrated from cell-free supernatants by centrifugation was grown in Vero cells and titrated by standard plaque assay (44). WT HSV-1 strain F expressing firefly luciferase (HSV-1–Luc) and WT HSV-1 strain F expressing GFP (HSV-1–GFP) were kindly provided by Chunfu Zheng (Soochow University, Suzhou, China). The WT HSV-1 strain was kindly provided by Karen Mossman (McMaster University, Canada). Virus was propagated in Vero cells, purified over a 36% sucrose cushion, and titrated by standard plaque assay. Cells were infected with virus at a multiplicity of infection (MOI) of 0.1 in a small volume of serum-free medium for 1 h at 37°C and then prior to analysis incubated with complete medium for the period of time indicated below. Infection with SeV (Charles River Laboratories) was performed at 40 hemagglutination units (HAU) per ml.
Quantitative real-time PCR.
DNase-treated total RNA from cells was prepared using an RNeasy kit (Qiagen). Total RNA was reverse transcribed using high-capacity cDNA reverse transcription (RT) kits from Applied Biosystems according to the manufacturer's instructions. Reverse transcription-quantitative PCR (RT-qPCR) was performed on a 7500 Fast system using SYBR green (Roche Diagnostics). All data presented are quantities with efficiency correction based on the level of expression of the target gene relative to that of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene as a reference and were analyzed using GraphPad Prism (version 5) software. The following primers were used in this study: TMEM173 forward, 5′-ATATCTGCGGCTGATCCTGC-3′; TMEM173 reverse, 5′-CTCAGGTTATCAGGCACCG-3′; DDX58 forward, 5′-GCAGAGGCCGGCATGAC-3′; DDX58 reverse, 5′-TGTAGGTAGGGTCCAGGG-3′; STAT2 forward, 5′-CTCGGAAGGTGGCTATTGT-3′; STAT2 reverse, 5′-AAAGGAGAGGCTGTGGGAAT-3′; GAPDH forward; 5′-AATCCCATCACCATCTTCCA-3′; GAPDH reverse; 5′-TGAGTCCTTCCACGATACCA-3′, IFNB1 forward, 5′-TTGTGCTTCTCCACTACAGC-3′; IFNB1 reverse, 5′-CTGTAAGTCTGTTAATGAAG-3′, IFIT1 forward, 5′-CAACCAAGCAAATGTGAG-3′; IFIT1 reverse, 5′-AGGGGAAGCAAAGAAAATGG-3′, IRF7 forward, 5′-CTTCGTGATGCTGCGAGATA-3′; IRF7 reverse, 5′-AAGCCCTTCTTGTCCCTGTC-3′; TNFAIP3 forward; 5′-ACCCCATTGTTCTCGGCTAT-3′; and TNFAIP3 reverse, 5′-CGGTCTCTGTTAACAAGTGGAA.
Immunoblot analysis.
Whole-cell lysates were resolved by 10% SDS-PAGE. After electrophoresis, proteins were transferred for 1 h at 100 V and 4°C to nitrocellulose membranes (pore size, 0.45 μm; Bio-Rad) in a buffer containing 25 mM Tris, 192 mM glycine, and 10% ethanol. Membranes were blocked for 1 h at room temperature in 5% (wt/vol) dried milk in phosphate-buffered saline (PBS) and 0.1% (vol/vol) Tween 20 (PBST) and were then probed with the following primary antibodies: anti-STING (Cell Signaling), anti-RIG-I (EMD-Millipore), anti-ISG56 (Thermo Fischer Scientific), anti-A20 (Santa Cruz Biotechnology), anti-STAT1 (Santa Cruz Biotechnology), antiactin (EMD Millipore), anti-IRF3 (IBL International), anti-RELA (Santa Cruz Biotechnology), anti-TNF receptor (anti-TNFR; Santa Cruz Biotechnology), and anti-GFP (Santa Cruz Biotechnology). Antibody signals were detected by chemiluminescence using secondary antibodies conjugated to horseradish peroxidase (Mandel Scientific) and an enhanced chemiluminescence detection kit (Thermo Scientific).
Flow cytometry analysis.
The percentage of cells infected with HSV-1 and VSVΔ51 was determined on the basis of the level of GFP expression. Cells were analyzed on a BD Fortessa flow cytometer (Becton, Dickinson). Compensation calculations and cell population analysis were done using FACSDiva software.
Stimulation with recombinant cytokines.
Recombinant IFN-α (Merck), IFN-β (PBI Assay Science), IFN-γ (PBI Assay Science), IFN-λ (R&D systems), interleukin-1β (IL-1β; R&D Systems), and TNF-α (Invitrogen) were used at a final concentration of 1,000 IU/ml (IFNs), 25 μg/ml (IL-1β), or 20 ng/ml (TNF-α).
ELISAs.
The levels of release of human IFN-α (PBL Biomedical Laboratories) and TNF-α (R&D Systems) into the culture supernatants of A549 cells were measured by enzyme-linked immunosorbent assays (ELISAs) according to the manufacturers' instructions.
In vivo administration of 5′pppRNA and HSV-1 infection model.
All animal experimentations were performed according to the guidelines of the Canadian Council on Animal Care and were approved by the McGill University Animal Care Committee (protocol 5816). C57BL/6 mice (age, 8 weeks) were obtained from Charles River Laboratories. MAVS−/− and WT (mixed 129/SvEv-C57BL/6 mouse background) were obtained from Z. Chen (Howard Hughes Medical Institute, USA). STING-deficient Goldenticket ([STINGgt/gt]) mice were purchased from The Jackson Laboratory. Ifnar1tm1Agt mice were kindly provided by Jörg Hermann Fritz. For intracellular delivery, 25 μg of 5′pppRNA was complexed with in vivo-jetPEI (PolyPlus, France) at an N/P ratio of 8 per the manufacturer's instruction and administered intravenously via tail vein injection. 5′pppRNA was administered on the day prior to infection (day −1) and on the day of infection (day 0). Mice were injected intravenously with the HSV-1 F strain. For determination of viral titers, organs were homogenized (20%, wt/vol) in DMEM, and titers were determined by standard plaque assay. For determination of protein expression levels, organs were homogenized (20%, wt/vol) in lysis buffer and subjected to immunoblot analysis.
RESULTS
RIG-I signaling induces STING expression.
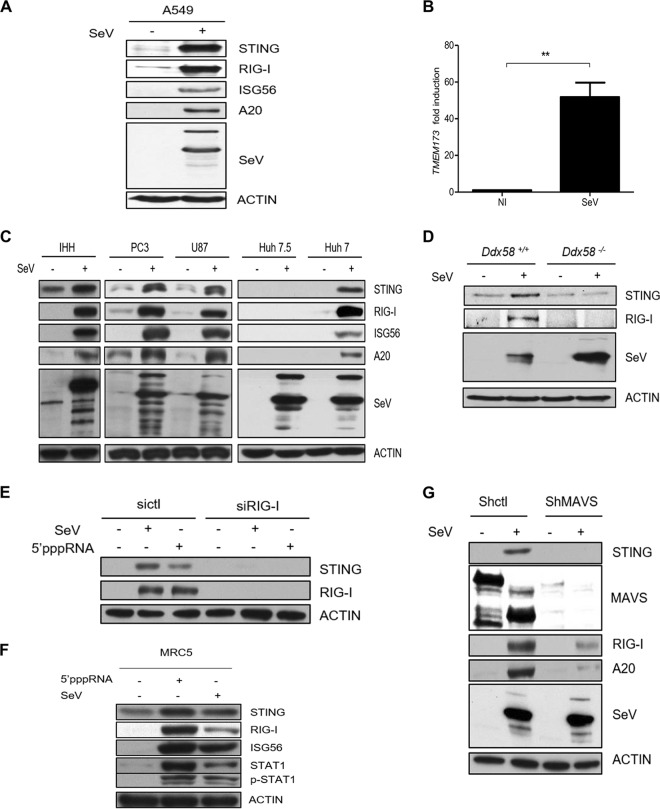
Our previous study indicated that STING is upregulated upon RIG-I agonist stimulation, as demonstrated by microarray analysis (44). To validate this finding, we examined whether SeV—a potent trigger of RIG-I signaling—could induce the expression of STING. We measured an increase in STING expression at both the protein and mRNA levels in A549 cells (Fig. 1A and B), as well as in IHHs and PC3, U87, and Huh7 cells (Fig. 1C). Importantly, Huh7.5 cells, which are derived from Huh7 cells but which harbor a mutation in RIG-I leading to RIG-I signaling deficiency, failed to induce STING in response to SeV infection (Fig. 1C).
FIG 1.
Expression of STING is upregulated upon RIG-I signaling. A549 cells (A, B), IHHs, PC3 cells, U87 cells, Huh7.5 cells, or Huh7 cells (C), Ddx58+/+ or Ddx58−/− MEFs (D), and shctl- and shMAVS-transfected A549 cells (F) were uninfected or infected with SeV (40 HAU/ml) for 24 h. (A and C to G) Whole-cell extracts were prepared, resolved by SDS-PAGE, and analyzed by immunoblotting for SeV, RIG-I, ISG56, A20, MAVS, STING, and actin. Results are from a representative experiment; all immunoblots were of the same samples. (B) The level of STING mRNA expression was analyzed by qPCR. NI, noninfected cells. (E) A549 cells were transfected with control siRNA (sictl) or siRNA directed against RIG-I (siRIG-I). Cells were then treated with SeV or 5′pppRNA (10 ng/ml) for 24 h. (F) MRC5 cells were either transfected with 5′pppRNA or infected with SeV for 24 h. Whole-cell extracts were subjected to immunoblotting to examine the levels of expression of STING, RIG-I, ISG56, phosphorylated STAT1 (p-STAT1), STAT1, and actin protein. Statistical analysis was performed by Student's test (**, P < 0.01).
To address whether STING upregulation is exclusively activated by RIG-I, mouse embryonic fibroblasts (MEFs) from wild-type (Ddx58+/+) and knockout (Ddx58−/−) mice were challenged with SeV. Despite the high level of endogenous expression of STING in MEFs, Ddx58+/+ MEFs harvested at 24 h postchallenge displayed considerable increases in STING levels compared with those for the Ddx58−/− MEFs (Fig. 1D). Similarly, we found that 5′pppRNA, which specifically activates RIG-I for full antiviral responses (44), could induce STING only in cells treated with control siRNA (sictl) and not in cells treated with siRNA directed against RIG-I (siRIG-I) (Fig. 1E). Similarly, high levels of endogenous expression of STING in human normal lung fibroblast (MRC5) cells was further induced upon RIG-I activation.
MAVS is an important adaptor protein for RIG-I signaling; it directly associates with RIG-I to coordinate downstream activation of TBK1-IKKε for type I IFN production. To investigate whether MAVS is involved in virus-mediated STING expression, A549 cells in which MAVS was stably knocked down with shRNA against MAVS (shMAVS) were generated. These MAVS-knockdown cells failed to induce STING during SeV infection, whereas cells transfected with control shRNA (shctl) were able to induce STING (Fig. 1F). Taken together, these results demonstrate that RIG-I is a key determinant of RNA virus-mediated STING induction.
RIG-I agonist 5′pppRNA-induced STING expression results from autocrine/paracrine signaling.
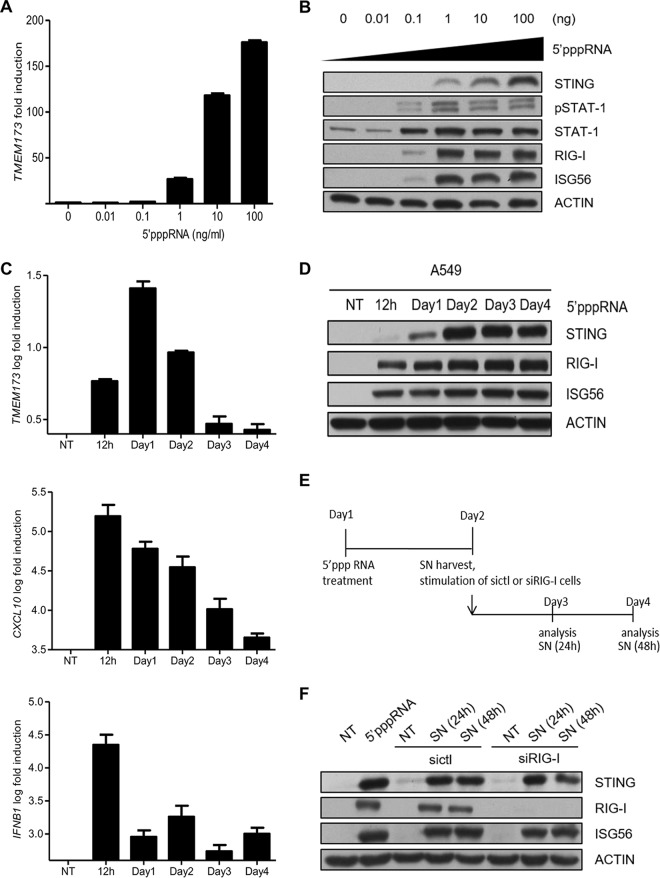
In order to explore the mechanisms and kinetics behind STING expression, we first treated A549 cells with increasing doses of a RIG-I agonist. We found that 5′pppRNA increased the levels of both STING mRNA and protein expression in a dose-dependent manner (Fig. 2A and B). Next, the STING expression profile was characterized at various time points. Quantitative PCR (qPCR) analyses revealed a significant increase in STING levels at 24 h posttreatment (Fig. 2C, top), while classic early antiviral genes, such as the genes for chemokine C-X-C motif ligand 10 (CXCL10) (Fig. 2C, middle) and interferon beta 1 (INFB1) (Fig. 2C, bottom), were significantly upregulated earlier at 12 h posttreatment. STING protein expression was robustly induced at 48 h posttreatment and was sustained for the 4-day examination period (Fig. 2D). Thus, STING induction by 5′pppRNA should be recognized as a late event.
FIG 2.
5′pppRNA induces STING expression through secreted proteins. (A, B) A549 cells were transfected with increasing amount of 5′pppRNA, and STING mRNA and protein levels were analyzed by qPCR and Western blotting, respectively. (C, D) A549 cells were transfected with 5′pppRNA (10 ng/ml), and whole-cell extracts were prepared at different times after transfection (time zero to day 4) and then subjected to SDS-PAGE and probed for STING, RIG-I, ISG56, and actin (C) or qPCR analysis (D). (E) Schematic outline of the timeline used for experiments whose results are shown in panel F. (F) A549 cells were transfected with 5′pppRNA for 24 h. Fresh A549 cells were transfected with control siRNA or RIG-I siRNA and then incubated with supernatants from 5′pppRNA-transfected cells for 24 h or 48 h. Cells were collected, and whole-cell extracts were prepared and subjected to immunoblot analyses. Statistical analysis was performed by Student's test. NT, not treated; SN, supernatants.
This delayed induction suggests the possibility of necessary de novo protein synthesis and/or the secretion of regulatory factors. To test this possibility, supernatants of 5′pppRNA-transfected A549 cells were collected at 24 h and placed on freshly plated cells. These new cells were treated with siRNA against RIG-I (or sictl) prior to incubation of the supernatants to control for RIG-I activation by the residual 5′pppRNA in the supernatants (Fig. 2E). As shown in Fig. 2F, stimulation of fresh cells with supernatants for 24 h and 48 h still led to an increase in STING levels to levels comparable to those obtained after the initial 5′pppRNA transfection. It is important to note that supernatant-mediated STING induction was independent of RIG-I. Altogether, these results indicate that STING induction by 5′pppRNA is mediated through an autocrine/paracrine mechanism.
TNF-α and type I IFNs synergize to induce STING expression.
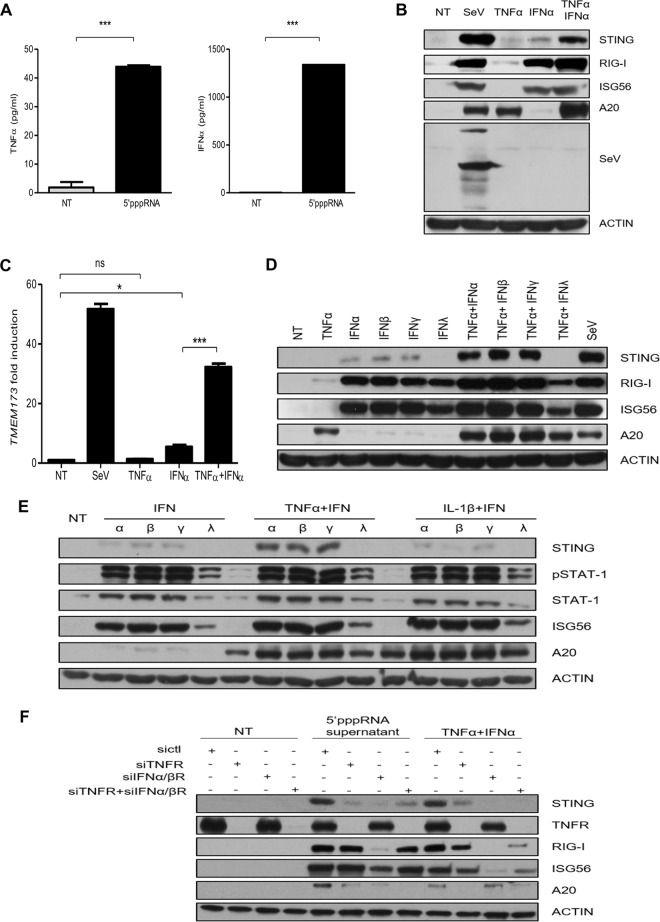
We next set out to identify the secreted factor(s) which contributed to 5′pppRNA-mediated STING induction. It was reported that 5′pppRNA stimulation of the RIG-I pathway activates both the IFN and NF-κB pathways (44), which can be activated by secreted type I IFNs and TNF-α, respectively. TNF-α is known to synergize with IFN signaling to induce the expression of delayed type I IFN response genes via an autocrine loop (47, 48). As an ELISA confirmed the presence of both IFN-α and TNF-α in the supernatants of 5′pppRNA-transfected cells (Fig. 3A), we sought to determine if STING induction could be driven by these two factors. A549 cells were incubated with SeV, IFN-α, TNF-α, or both IFN-α and TNF-α for 24 h, and STING levels were assessed. As shown in Fig. 3B and C, IFN-α triggered a weak induction of STING, whereas TNF-α alone had no effect. However, the cotreatment with IFN-α and TNF-α led to a significant increase in STING levels similar to those achieved with SeV infection.
FIG 3.
STING is induced through costimulation of TNF-α and IFN-α. (A) A549 cells were transfected with 5′pppRNA (10 ng/ml), and IFN-α and TNF-α levels in the supernatants were measured by ELISA. (B, C) A549 cells were incubated with or without SeV (40 HAU/ml), TNF-α (20 ng/ml), IFN-α (1,000 IU/ml), or both TNF-α (20 ng/ml) and IFN-α (1,000 IU/ml) for 24 h. (B) Whole-cell extracts were analyzed by SDS-PAGE and immunoblotted for SeV, RIG-I, ISG56, A20, STING, and actin. (C) The level of STING mRNA expression was analyzed by qPCR. (D, E) A549 cells were stimulated with different combinations of cytokines for 24 h. The levels of phosphorylated STAT1, STAT1, RIG-I, ISG56, A20, STING, and actin protein expression were analyzed by immunoblot analyses. α, β, γ, and λ, IFN-α, IFN-β, IFN-γ, and IFN-λ, respectively. (F) A549 cells were transfected with control siRNA, siRNA targeting IFN-α/βR (siIFNα/βR), siRNA targeting TNFR (siTNFR), or siRNA targeting IFN-α/βR and siRNA targeting TNFR. Cells were then incubated with either the supernatants from 5′pppRNA-transfected cells or TNF-α plus IFN-α for 24 h. Cells were collected, and whole-cell extracts were prepared and subjected to immunoblot analyses. Statistical analysis was performed by Student's test (*, P < 0.05; ***, P < 0.001; ns, not statistically significant).
Various types of IFN, including IFN-β, IFN-γ, and IFN-λ, were also tested, either alone or in combination with TNF-α, for their ability to induce STING expression. STING expression was weakly induced by IFN-α, IFN-β, and IFN-γ alone but was significantly enhanced by the addition of TNF-α (Fig. 3D). IFN-λ alone or IFN-λ combined with TNF-α was ineffective in inducing STING. IL-1β, another potent activator of the NF-κB signaling pathway, failed to induce STING expression and did not exhibit any synergic effect with IFN-α, IFN-β, IFN-γ, or IFN-λ (Fig. 3E). Several combinations within three types of IFNs were also tested, but none of them resulted in effective STING induction (data not shown). As demonstrated in Fig. 3F, knocking down of the TNF-α receptor (TNFR) and/or type I IFN receptor (IFN-α/βR) significantly reduced the levels of STING expression induced by the supernatants from 5′pppRNA-treated cells. However, the level of STING expression induced by the supernatants was not completely diminished by siRNA targeting TNFR (siTNFR) and siRNA targeting IFN-α/βR (siIFN-α/βR), indicating that other factors could also drive STING induction. Collectively, these results demonstrate that the 5′pppRNA induction of STING results from an autocrine or paracrine mechanism, depending on the synergistic activities between TNF-α and type I IFN.
STING induction is mediated by the STAT and NF-κB pathways.
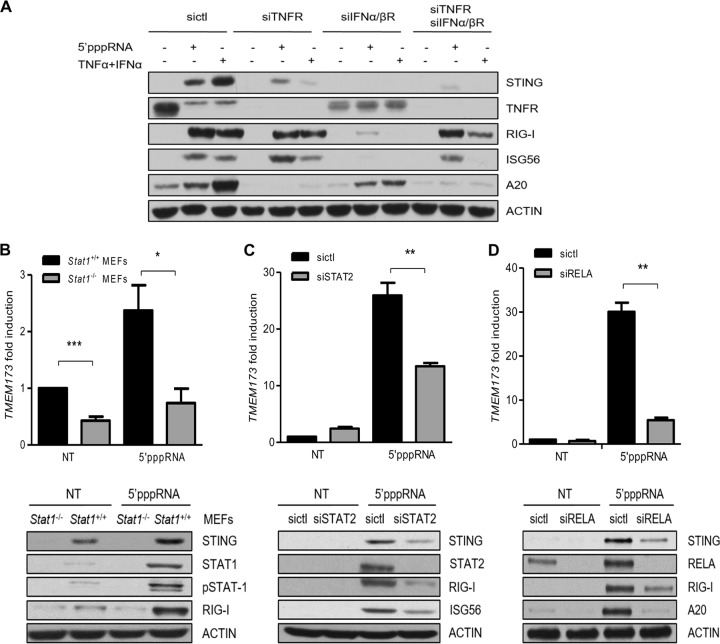
To validate the importance of TNF-α and IFN during STING expression, A549 cells were transfected with siRNA directed against TNFR and/or IFN-α/βR (Fig. 4A). Knocking down of the receptors alone or in combination reduced the level of STING induction by 5′pppRNA or TNF-α and IFN-α cotreatment compared to that achieved with control cells. We next explored further downstream mechanisms that control STING expression. Since STAT1/2 and RELA (p65) are crucial elements in the activation of IFN and NF-κB signaling, respectively, we silenced these transcription factors and evaluated their potential roles in STING induction. We found that the depletion of STAT1, STAT2, and RELA significantly diminished the 5′pppRNA induction of STING (Fig. 4B to D). Thus, STAT1/2 and RELA together mediate the IFN- and TNF-α-dependent induction of STING in response to 5′pppRNA stimulation.
FIG 4.
STING induction is regulated by STAT1/2 and RELA. (A) A549 cells were transfected with control siRNA, siRNA targeting IFN-α/βR, and/or siRNA targeting TNFR. After 48 h, cells were transfected with 5′pppRNA or treated with TNF-α and IFN-α for 24 h. Whole-cell extracts were analyzed by SDS-PAGE and subjected to immunoblotting. (B to D) Stat1+/+ MEFs and Stat1−/− MEFs (B) and A549 cells transfected with control siRNA or siRNA targeting STAT2 (siSTAT2) or RELA (siRELA) (C, D) were treated with 5′pppRNA at 48 h after transfection. STING, STAT1, pY701STAT1 (pSTAT1), STAT2, RELA, RIG-I, A20, and actin protein levels were analyzed by Western blotting. Statistical analysis was performed by Student's test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
STING contributes to sustained expression of interferon/inflammatory response genes.
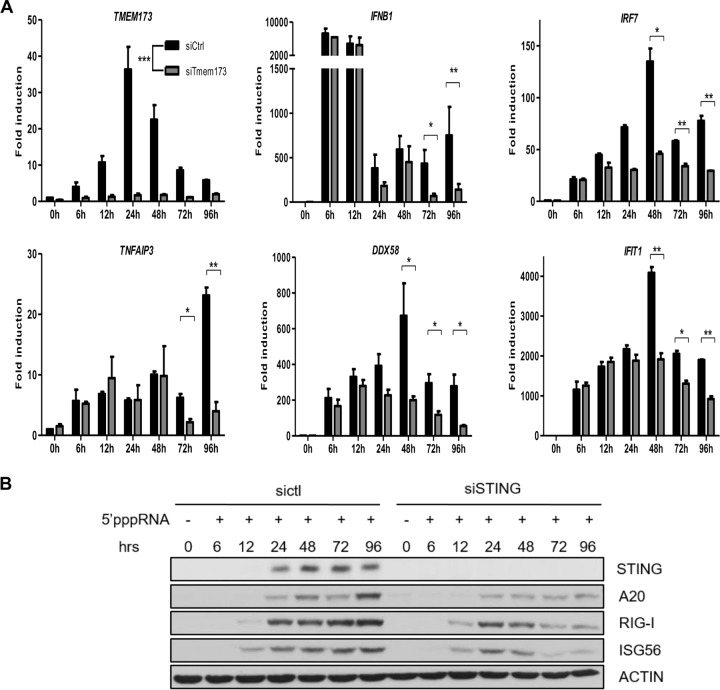
Given that STING plays a key role in the establishment of the antiviral state during PAMP detection, we explored whether it contributed to the immune response following its induction by 5′pppRNA. A549 cells in which STING was silenced were stimulated with 5′pppRNA at various time points, and the mRNAs of several immune response genes were monitored by qPCR. STING mRNA was inhibited, as expected, when TMEM173-specific siRNA was used. Interestingly, the levels of mRNA for IFNB1, IRF7, TNFAIP3, DDX58, and IFIT1 were not initially affected by the absence of STING at early time points (6 h, 12 h, and 24 h) but were significantly reduced at 48 h, 72 h, and 96 h posttreatment (Fig. 5A). The levels of the RIG-I, ISG56, and A20 proteins at late time points were also reduced when STING was silenced (Fig. 5B). Altogether, these results suggest that the induction of STING contributes to a sustained interferon and inflammatory response.
FIG 5.
STING sustains interferon-inducible gene expression. (A, B) A549 cells were transfected with sictl or siRNA targeting STING prior to 5′pppRNA transfection. Cell samples were collected at the indicated time points for qPCR (A) or Western blotting (B). Statistical analysis was performed by Student's test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
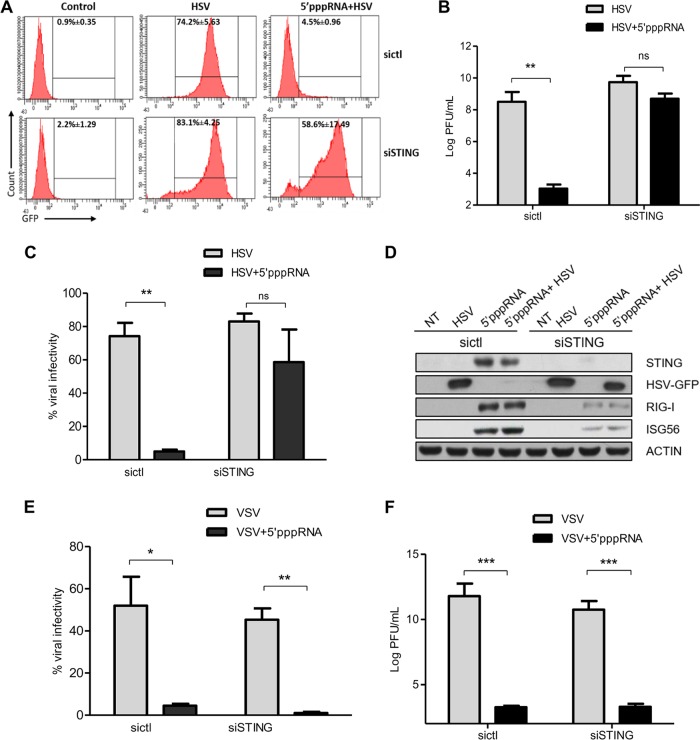
STING is essential for 5′pppRNA-mediated protection against HSV-1 infection in vitro.
It has been demonstrated that 5′pppRNA stimulation protects cells against a broad spectrum of DNA and RNA viruses (5, 44). We therefore investigated whether this protection was mediated through de novo STING expression. A549 cells were transfected with TMEM173-specific siRNA (siSTING) or control siRNA (sictl), pretreated with 5′pppRNA for 48 h, and then challenged with HSV-1–GFP for a 2-day period. As assessed by flow cytometry (Fig. 6A and B), HSV-1 infectivity was decreased significantly in 5′pppRNA-pretreated sictl-transfected cells but remained high in cells lacking STING. Similar results were found when we investigated the release of infectious HSV-1 particles via a standard plaque essay. The HSV-1 viral titer was significantly reduced by 5′pppRNA pretreatment in sictl-transfected cells, but siSTING-transfected cells pretreated or not pretreated with the RIG-I agonist 5′pppRNA produced comparable levels of virus (Fig. 6C). Consistent with our earlier data (Fig. 5), HSV-1-infected, siSTING-transfected cells also displayed immune responses weaker than those of infected, sictl-transfected ones (Fig. 6D). Altogether, these data unveiled the essential contribution of STING to the establishment of the 5′pppRNA-induced antiviral responses during HSV-1 infection.
FIG 6.
STING contributes to the 5′pppRNA-induced antiviral responses during HSV-1 infection in vitro. (A to F) A549 cells were transfected with sictl or siRNA targeting STING. Cells were then pretransfected with 5′pppRNA for 48 h, followed by infection with HSV-1–GFP (MOI, 0.1) or VSV-GFP (0.1MOI) for 2 days. (A, C, E) The percentage of HSV-1- or VSV-infected cells was determined by flow cytometry analysis of GFP expression. (B, F) HSV-1 or VSV viral titers in cell culture supernatants were determined by standard plaque assay. (D) Whole-cell extracts were subjected to immunoblotting to examine the levels of expression of STING, HSV-1–GFP, VSV-GFP, RIG-I, ISG56, and actin protein. Data are means ± SDs from a representative experiment performed in triplicate. Statistical analysis was performed by Student's test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not statistically significant).
We also investigated whether STING induction by 5′pppRNA influences RNA virus replication. To pursue this, a set of experiments similar to those described above was carried out using VSV-GFP instead of HSV-1–GFP. VSV replication was inhibited by 5′pppRNA pretreatment, whether or not STING was expressed (Fig. 6E). Correspondingly, the VSV viral titer was also reduced by 5′pppRNA regardless of the presence of STING (Fig. 6F). This suggests that despite STING inhibition, 5′pppRNA pretreatment in the context of VSV infection is sufficient to prevent viral replication.
5′pppRNA protection against HSV-1 infection in mice requires STING.
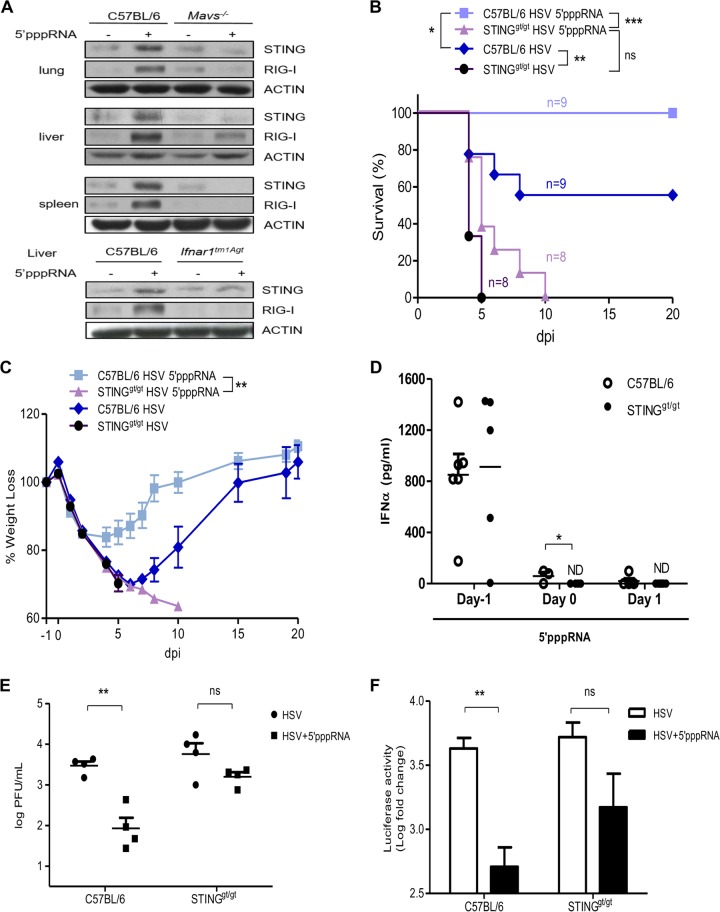
To determine the potential of 5′pppRNA-mediated STING induction in vivo, C57BL/6 mice were inoculated intravenously with 5′pppRNA (25 μg) in complex with the in vivo-jetPEI transfection reagent. As expected, 5′pppRNA treatment led to the upregulation of STING production in the lungs, liver, and spleen, while MAVS-deficient mice failed to induce STING in these organs (Fig. 7A, top). IFNAR-deficient mice (Ifnar1tm1Agt) also displayed lower STING levels following 5′pppRNA treatment (Fig. 7A, bottom).
FIG 7.
STING is essential for 5′pppRNA-mediated protection from HSV-1 infection in vivo. (A) Six-week-old C57BL/6J mice, Mavs−/− mice, or Ifnar−/− mice were injected intravenously with 25 μg of 5′pppRNA in complex with in vivo-jetPEI. Immunoblot analyses were performed to measure the levels of STING, RIG-I, and actin protein expression in the indicated organs at 24 h posttreatment. (B to D) C57BL/6 mice and STINGgt/gt mice were treated with 5′pppRNA on the day prior to lethal HSV-1–Luc infection (2 × 107 PFU, day −1) and the day of infection (day 0). Percent survival (B) and percent weight loss (C) were monitored. (D) Serum IFN-α was quantified by ELISA. ND, not detectable. (E, F) C57BL/6 mice and STINGgt/gt mice were treated with 5′pppRNA on the day prior to nonlethal HSV-1–Luc infection (1 × 106 PFU, day −1) and the day of infection (day 0). Lung viral titers (E) and viral luciferase activity (F) in lung were measured on day 3 postinfection. Error bars represent SEMs of the results for four different animals. Statistical analysis was performed by Student's test, log-rank tests, or two-way analysis of variance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not statistically significant).
We next evaluated if STING induction by 5′pppRNA was necessary for HSV-1 resistance in vivo, as it was in vitro. C57BL/6 mice and STING-deficient mice (STINGgt/gt mice) were inoculated with 5′pppRNA 24 h before infection (day −1) and on the day of infection (day 0) with a lethal inoculum of luciferase-expressing HSV-1 (HSV-1–Luc).
In agreement with the findings described in a previous publication (18) and as demonstrated by the results of the Kaplan-Meier curve analysis shown in Fig. 7B, C57BL/6 mice were significantly more likely to survive than STINGgt/gt mice when subjected to HSV-1 lethal infection (rates of survival, 66.7% versus 0%; P = 0.0044, log-rank tests). C57BL/6 mice treated with 5′pppRNA exhibited rates of survival superior to those for the nontreated group (P = 0.0270). However, while death was delayed to day 10 in 5′pppRNA-treated STINGgt/gt mice, all nontreated mice succumbed to infection by day 5; thus, 5′pppRNA treatment ultimately did not improve survival (P was not statistically significant and P = 0.0566). The log-rank test revealed a significant survival advantage in C57BL/6 mice treated with 5′pppRNA compared with that in STINGgt/gt mice treated with 5′pppRNA (P = 0.0002).
When both C57BL/6 and STINGgt/gt mice were subjected to lethal HSV-1 infections, the mice lost weight rapidly before day 5 (Fig. 7C). The C57BL/6 mice then started to regain weight overall and completely recovered in 20 days. 5′pppRNA treatment significantly reversed the weight loss of C57BL/6 mice, whereas it did not in STINGgt/gt mice (two-way analysis of variance, P < 0.001).
As reported previously, 5′pppRNA triggered the robust release of IFN-α in the serum of C57BL/6 mice on day −1 (Fig. 7D) (44). The absence of STING did not affect the amount of 5′pppRNA-induced IFN-α released at an early time (6 h on day −1). However, a significant decrease in the serum IFN-α level was detected in STINGgt/gt mice compared to that in C57BL/6 mice on day 0. The serum IFN-α level in both groups was almost undetectable after day 1. HSV-1 replication in the lungs was monitored by plaque assay at day 3 after nonlethal infection. HSV-1 titers were 1.5 log units lower in 5′pppRNA-treated C57BL/6 mice than in the controls, while STING-deficient mice displayed no significant reduction (Fig. 7E). Similar results were obtained when we investigated HSV-1 luciferase activity (Fig. 7F). Overall, these results correlated well with the in vitro data obtained and presented in Fig. 6 and highlight the necessity of STING induction during 5′pppRNA treatment to protect against HSV-1 replication. Taken together, 5′pppRNA-mediated STING induction appears to be regulated by the synergistic activities of type I IFN and TNF-α signaling (Fig. 8).
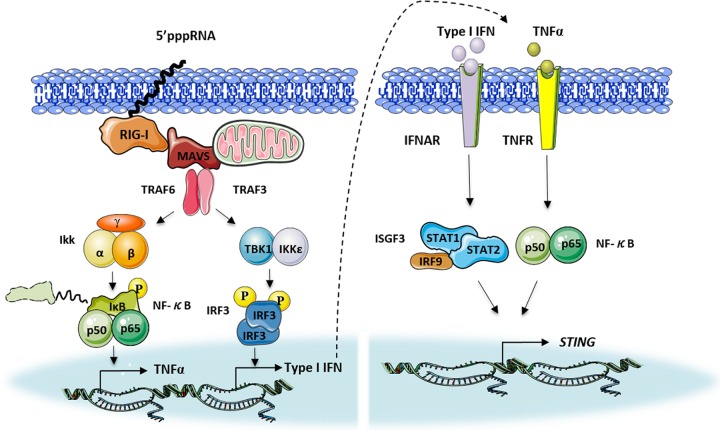
FIG 8.
Model of the regulation of STING via RIG-I signaling. The RIG-I agonist 5′pppRNA triggers the secretion of type I IFN and TNF-α. Type I IFN and TNF-α synergistically mediate STING induction through transcription factors STAT1/STAT2 and NF-κB.
DISCUSSION
In recent years, STING has been established to be a critical player in mediating cytosolic nucleic acid detection (14, 23, 25, 31). STING also plays an important role in the pathogenesis of inflammatory and autoimmune diseases and is linked to both protective and detrimental processes in vivo (49–53). However, knowledge of the regulatory mechanism of STING is still limited. In the present study, we demonstrated that (i) STING was induced following RIG-I activation in various cell models and multiple animal tissues, (ii) 5′pppRNA-mediated STING induction depended on the synergistic activities of TNF-α/IFN-α and STAT1/STAT2/RELA, (iii) STING contributed to the sustained expression of interferon/inflammatory response genes, and (iv) STING was essential for RIG-I agonist-mediated protection against HSV-1 infection in vitro and in vivo.
To date, the regulation of STING mediated by host factors or viral proteins has mainly been investigated at the posttranslational level. TRIM56 and TRIM32 can positively mediate the K63-linked ubiquitination of STING, which leads to its dimerization and the subsequent recruitment of TBK1 (43, 54). The phosphorylation of STING at Ser358 has also been reported following dsDNA stimulation (38) and SeV infection (31). Conversely, STING degradation can be mediated following K48-linked ubiquitination by RNF5 (32) or Ser366 phosphorylation by ATG1 (41). STING is also often targeted by various RNA viruses for immune invasion. For example, the hepatitis C virus (HCV) nonstructural protein NS4B colocalizes with STING and impairs its interaction with MAVS (55); dengue virus (DENV) cleaves STING by its NS2B3 proteinase, thereby inhibiting IFN production (35); and human coronavirus (HCoV) NL63 disrupts STING dimerization and ubiquitination (56). Previous studies have characterized STING as an ISG, as it is inducible by interferons (40). By employing multiple RIG-I-deficient or pathway-impaired cell lines, we showed for the first time that RIG-I signaling through MAVS leads to an increase in STING mRNA and protein levels in many cells types and tissues. These observations provide new evidence for potential cross talk between RNA and DNA signaling pathway regulation.
Various cytokines were tested for their ability to induce STING expression in order to mimic SeV and RIG-I agonist stimulation. Both type I IFNs (IFN-α and IFN-β) and the type II IFN (IFN-γ) were able to induce STING expression slightly when used alone, but this was not the case for the type III IFN (IFN-λ). The inductivity obtained by treatment with a single IFN was not amplified with the simple addition of an IFN mixture, but inductivity was significantly boosted when a single IFN was combined with TNF-α. IL-1β, which is another NF-κB trigger, does not engage with IFNs for the synergistic induction of STING. These findings highlight the distinct impact of TNF-α in the synergistic regulation of STING, suggesting that multiple regulatory mechanisms are probably involved in this synergy. It is worth noting that the supernatants from 5′pppRNA-treated cells contain a variety of soluble factors aside from TNF-α and IFNs. Whether they act individually or synergistically, those factors could also drive STING induction.
This phenomenon of cytokine synergy was first observed in 1986 when treatment with TNF-α and IFN combined was demonstrated to block both RNA and DNA viral replication (57). The responses to cytokines in combination are more potent than the sum of the individual responses but remain poorly characterized due to the technical difficulties with the deconstruction of the responses of cytokine mixtures and the extreme complexity of the molecular events mediated by synergistic states. However, it is known that the combination of type I IFN and TNF-α induces a powerful antiviral state distinct from the one induced by either cytokine alone (47). Furthermore, TNF-α can activate a feed-forward loop which sustains the expression of inflammatory genes and induces late interferon response genes (48).
A previous analysis of the promoters of hundreds of genes synergistically induced by TNF-α and IFN did not return either the enrichment of any transcription factors or defined transcription factor binding sites (47). However, it has recently been shown that the STING 5′ UTR contains STAT1 binding sites that are required for type I IFN induction of STING (58). In the study described in this report, we demonstrated that both STAT1 and STAT2 are critical mediators of STING induction. Importantly, the NF-κB subunit RELA was also required for the effective expression of STING. By searching the UCSC genome browser, we also discovered NF-κB consensus binding motifs on the human STING promoter. Still, further work is needed to confirm these binding sites and to identify the binding subunit(s).
Several studies have demonstrated that a lack of STING results in the evolution of deficient type I IFN responses during RNA virus infection (16, 31, 59). Our data revealed that RIG-I agonist 5′pppRNA-mediated STING induction contributed to a significant persistence of innate immune gene expression at late time points. It is possible that the treatment with 5′pppRNA for days may have caused the leakage of mitochondrial DNA or genomic DNA which activated STING and prolonged the immune response. We also investigated whether RIG-I-mediated STING induction contributed to viral resistance. Under normal circumstances, 5′pppRNA treatment protects cells against numerous RNA and DNA viruses (44, 60). Here we demonstrated that 5′pppRNA-treated A549 cells were also resistant to HSV-1 infection when STING was expressed. It is important to note that the immune response following HSV-1 exposure was also severely dampened in STING-deficient cells. Taken together, these results indicate that the protection against DNA viruses like HSV-1 conferred by 5′pppRNA is STING dependent, which enhances innate immune detection and signaling. In contrast, we showed that VSV infection was inhibited by 5′pppRNA, regardless of the presence of STING. Overall, these data reinforce the notion that STING induction plays an active role in 5′pppRNA-mediated protection against HSV-1.
We further established the induction of STING in vivo, as 5′pppRNA-treated mice displayed increased STING levels in many organs via RIG-I signaling. In agreement with the findings of previous studies, STING is essential for protection against lethal challenge with HSV-1 (18, 61). All STINGgt/gt mice succumbed rapidly following HSV-1 infection. Intravenous administration of 5′pppRNA stimulated potent immune protection in C57BL/6 animals, decreased the virus load in the lungs, and completely prevented the mortality associated with HSV-1 infection. In contrast, all STINGgt/gt animals inoculated with 5′pppRNA succumbed to HSV-1 infection, though their survival was prolonged compared to that of the group only infected with HSV-1.
Of note, although 5′pppRNA did not exhibit a statistically significant HSV-1 inhibition in STING-deficient mouse models, there was a trend toward reduced viral infection. Considering the fact that RIG-I-mediated inhibition of HSV-1 is a complex situation coordinated by numerous elements, these antiviral factors, whether they were affected or not by the absence STING, could still exert a moderate effort in HSV-1 inhibition. In addition, HSV-1 has evolved multiple strategies to evade the host immune response. The deficiency of STING may have provided an environment in favor of HSV-1 replication. Overall, our data demonstrated that 5′pppRNA's anti-HSV-1 activity was largely dependent on the presence of STING.
In conclusion, our study proposes a model in which STING is regulated by the transcriptional synergy of STAT1/2 and NF-κB. We also unveil the key function of STING in the 5′pppRNA-mediated restriction of HSV-1. Thus, our work reveals a novel mechanism of RNA antiviral signaling that leads to the inhibition of DNA virus infection.
ACKNOWLEDGMENTS
We thank Tatiana Shorstova, Ryuhjin Ahn, and Cedric Darini (McGill University) and Michael Witcher (McGill University, Lady Davis Institute-Jewish General Hospital) for advice and technical assistance and Karen Mossman (McMaster University, Canada), Ranjit Ray (Saint Louis University, St. Louis, MO), and Jerry Pelletier (McGill University, Canada) for kindly providing HSV-1 and cell lines.
We declare that we have no conflict of interest.
This research work was supported by a Canadian Institutes of Health Research grant (MOP130401) to Rongtuan Lin. David Olagnier is supported by a Peter Quinlan McGill postdoctoral fellowship. Alexander Sze is supported by a Fond de Recherche Santé Quebec (FRSQ) fellowship award.
REFERENCES
- 1.Kumar H, Kawai T, Akira S. 2011. Pathogen recognition by the innate immune system. Int Rev Immunol 30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 4.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 5.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, Iskarpatyoti JA, Barchet W, Ludwig J, Dermody TS, Hartmann G, Reis e Sousa C. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 7.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 9.Kawai T, Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci 1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 10.Liu SY, Sanchez DJ, Cheng G. 2011. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol 23:57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang BX, Fish EN. 2012. The yin and yang of viruses and interferons. Trends Immunol 33:190–197. doi: 10.1016/j.it.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. 2011. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol 187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Chen R, Zhou Q, Xu Z, Li C, Wang S, Mao A, Zhang X, He W, Shu HB. 2012. LSm14A is a processing body-associated sensor of viral nucleic acids that initiates cellular antiviral response in the early phase of viral infection. Proc Natl Acad Sci U S A 109:11770–11775. doi: 10.1073/pnas.1203405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. 2012. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol 19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 25.Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. 2013. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. 2013. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam E, Stein S, Falck-Pedersen E. 2014. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J Virol 88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Nazmi A, Mukhopadhyay R, Dutta K, Basu A. 2012. STING mediates neuronal innate immune response following Japanese encephalitis virus infection. Sci Rep 2:347. doi: 10.1038/srep00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitta S, Sakamoto N, Nakagawa M, Kakinuma S, Mishima K, Kusano-Kitazume A, Kiyohashi K, Murakawa M, Nishimura-Sakurai Y, Azuma S, Tasaka-Fujita M, Asahina Y, Yoneyama M, Fujita T, Watanabe M. 2013. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57:46–58. doi: 10.1002/hep.26017. [DOI] [PubMed] [Google Scholar]
- 35.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. 2014. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell 5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, Lin YL. 2012. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog 8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A, Cheng G. 2015. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol 194:1545–1554. doi: 10.4049/jimmunol.1402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konno H, Konno K, Barber GN. 2013. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287:28646–28655. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. 2013. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog 9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray RB, Meyer K, Ray R. 2000. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology 271:197–204. doi: 10.1006/viro.2000.0295. [DOI] [PubMed] [Google Scholar]
- 46.Stojdl DF, Lichty BD, ten Oever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. doi: 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 47.Bartee E, Mohamed MR, Lopez MC, Baker HV, McFadden G. 2009. The addition of tumor necrosis factor plus beta interferon induces a novel synergistic antiviral state against poxviruses in primary human fibroblasts. J Virol 83:498–511. doi: 10.1128/JVI.01376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. 2008. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol 9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 49.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu YX, Weichselbaum RR. 2014. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klarquist J, Hennies CM, Lehn MA, Reboulet RA, Feau S, Janssen EM. 2014. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J Immunol 193:6124–6134. doi: 10.4049/jimmunol.1401869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre ML, Gajewski TF. 2014. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee CC, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, et al. 2014. Activated STING in a vascular and pulmonary syndrome. N Engl J Med 371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. 2012. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Q, Cao X, Lu J, Huang B, Liu YJ, Kato N, Shu HB, Zhong J. 2013. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol 59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 56.Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, Chen Z. 2012. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One 7:e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong GH, Goeddel DV. 1986. Tumour necrosis factors alpha and beta inhibit virus replication and synergize with interferons. Nature 323:819–822. doi: 10.1038/323819a0. [DOI] [PubMed] [Google Scholar]
- 58.Ma F, Li B, Yu Y, Iyer SS, Sun M, Cheng G. 2015. Positive feedback regulation of type I interferon by the interferon-stimulated gene STING. EMBO Rep 16:202–212. doi: 10.15252/embr.201439366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 60.Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, He Z, Lin R, Snijder EJ, van Hemert MJ, Hiscott J. 2014. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol 88:4180–4194. doi: 10.1128/JVI.03114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker ZM, Murphy AA, Leib DA. 2015. Role of the DNA sensor STING in protection from lethal infection following corneal and intracerebral challenge with herpes simplex virus 1. J Virol 89:11080–11091. doi: 10.1128/JVI.00954-15. [DOI] [PMC free article] [PubMed] [Google Scholar]