ABSTRACT
Overcoming host resistance in gene-for-gene host-virus interactions is an important instance of host range expansion, which can be hindered by across-host fitness trade-offs. Trade-offs are generated by negative effects of host range mutations on the virus fitness in the original host, i.e., by antagonistic pleiotropy. It has been reported that different mutations in Pepper mild mottle virus (PMMoV) coat protein result in overcoming L-gene resistance in pepper. To analyze if resistance-breaking mutations in PMMoV result in antagonistic pleiotropy, all reported mutations determining the overcoming of L3 and L4 alleles were introduced in biologically active cDNA clones. Then, the parental and mutant virus genotypes were assayed in susceptible pepper genotypes with an L+, L1, or L2 allele, in single and in mixed infections. Resistance-breaking mutations had pleiotropic effects on the virus fitness that, according to the specific mutation, the host genotype, and the type of infection, single or mixed with other virus genotypes, were antagonistic or positive. Thus, resistance-breaking mutations can generate fitness trade-offs both across hosts and across types of infection, and the frequency of host range mutants will depend on the genetic structure of the host population and on the frequency of mixed infections by different virus genotypes. Also, resistance-breaking mutations variously affected virulence, which may further influence the evolution of host range expansion.
IMPORTANCE A major cause of virus emergence is host range expansion, which may be hindered by across-host fitness trade-offs caused by negative pleiotropy of host range mutations. An important instance of host range expansion is overcoming host resistance in gene-for-gene plant-virus interactions. We analyze here if mutations in the coat protein of Pepper mild mottle virus determining L-gene resistance-breaking in pepper have associated fitness penalties in susceptible host genotypes. Results show that pleiotropic effects of resistance-breaking mutations on virus fitness depend on the specific mutation, the susceptible host genotype, and the type of infection, single or mixed, with other virus genotypes. Accordingly, resistance-breaking mutations can have negative, positive, or no pleiotropic effects on virus fitness. These results underscore the complexity of host range expansion evolution and, specifically, the difficulty of predicting the overcoming of resistance factors in crops.
INTRODUCTION
Changes in virus host range affect virus ecology and epidemiology, condition virus emergence, and can compromise the success of strategies for the control of viral diseases (1–4). The acquisition of new hosts, that is, host range expansion, would provide a virus with more opportunities for transmission and survival. However, differential host-associated selection may result in across-host fitness trade-offs so that increasing the virus fitness in a new host will decrease its fitness in the original one, which will hinder host range expansion (4–6). The simplest mechanism generating across-hosts fitness trade-offs is antagonistic pleiotropy, in which mutations that increase fitness in one host are deleterious in another one (7). Evidence indicates that antagonistic pleiotropy is common in RNA viruses as a result of their small genomes, which are compacted with genetic information and encode few, multifunctional proteins (5, 8).
Host range evolution is particularly relevant for the sustainable use of genetic resistance to control viral diseases of crops. Disease control based on resistant cultivars is target specific and highly efficient but is not durable due to the appearance of new virus genotypes able to infect otherwise resistant host genotypes. The increase in frequency of resistance-breaking virus genotypes eventually renders resistance inefficient (9–11). Plant-virus interactions can often be explained by the gene-for-gene (GFG) model, under which direct or indirect recognition of viral proteins by those encoded by plant resistance genes (R genes) triggers plant defenses that limit virus multiplication to infection sites, thus triggering a hypersensitive response (HR) (12, 13). Mutations in the viral proteins that impair their recognition by R proteins result in resistance breaking so that the virus can infect a larger set of host genotypes, expanding its host range. Indeed, a key feature of GFG systems is that the capacity of pathogens to infect all host genotypes will evolve as a result of subsequent resistance-breaking events (14). The evolution of resistance breaking may be hindered by across-host fitness trade-offs if the resistance-breaking genotypes are less fit than the non-resistance-breaking ones in susceptible hosts, that is, if there are resistance-breaking costs. The relevance of resistance-breaking costs in plant-virus coevolution and as modulators of the durability of crop resistance has led to efforts for their detection and quantification. Analyses of virus multiplication in susceptible hosts have provided evidence for resistance-breaking costs in various plant-virus systems (15–21).
Our group has documented resistance-breaking costs in the tobamoviruses that infect pepper (Capsicum annuum L.) genotypes carrying different resistance alleles at the L locus. Tobamoviruses have single-stranded, messenger sense, monopartite RNA genomes encapsidated into rod-shaped particles and are transmitted in pepper crops through plant-to-plant contact or vertically through the seed (22). After plant harvest or death, infectious particles can survive for several months in the soil, a major source of inoculum for epidemics (23, 24). Four resistance alleles at the pepper L locus confer resistance to tobamoviruses (25, 26) according to the GFG model, and tobamovirus species and genotypes are classified into pathotypes according to their capacity to infect pepper genotypes carrying the various L alleles (27–29). Plants homozygous for allele L+ are susceptible to all described pathotypes (P0, P1, P1,2, P1,2,3, and P1,2,3,4), L1/− plants are resistant only to pathotype P0; L2/− plants are resistant to pathotypes P0 and P1; L3/− plants are resistant to pathotypes P0, P1, and P1,2, and so on. Pathotype P1,2,3,4 is able to infect pepper genotypes with each of the four known resistance alleles. L-gene resistance is elicited by the virus coat protein (CP), and the resulting HR is expressed by the appearance in inoculated leaves of necrotic local lesions at infection sites (30–33). The capacity to establish an infection or to elicit an HR depends on the alleles at the virus CP and the pepper L protein and is little affected by the host genetic background, except in the temporal rate of development of the hypersensitive reaction (25–33). Conclusive evidence for resistance-breaking costs in field isolates of pepper-infecting tobamoviruses was derived from long-term comparison of changes in pathotype frequency and in the fraction of the crop carrying the various resistance alleles in southeast (SE) Spain and by quantifying virus multiplication in the L+/L+ universally susceptible host, both in single infection and in competition (17). In the L+/L+ host, isolates of pathotype P1,2,3 were outcompeted by isolates of pathotype P1,2, and both were outcompeted by isolates of pathotype P0 (17). Only isolates of pathotypes P0, P1,2, and P1,2,3 were present infecting pepper in SE Spain; P0 isolates were Tobacco mild green mosaic virus (TMGMV) isolates, while P1,2 and P1,2,3 isolates were Pepper mild mottle virus (PMMoV) (17). Studies by Fraile et al. (17) were performed with field virus isolates that differed in genomic regions other than the CP gene, which precluded the analysis of the mechanisms generating the observed resistance-breaking costs.
The goal of the present work is the analysis of the mechanisms generating resistance-breaking fitness costs in pepper-infecting tobamoviruses. Since TMGMV and PMMoV differ in ≈30% of amino acid positions at the CP, the analysis was restricted to PMMoV. Most reported PMMoV isolates are of pathotype P1,2 (22), and different mutations at the CP have been reported as determinants of pathotypes P1,2,3 and P1,2,3,4 (27, 30, 34–36). We addressed the following specific questions: (i) if resistance-breaking mutations in PMMoV CP have associated fitness costs in susceptible hosts, (ii) if pleiotropic effects of resistance-breaking mutations vary according to the genotype of the susceptible host, and (iii) if pleiotropic effects of resistance-breaking mutations vary according to the type, single or mixed, of infection. In addition, we characterized the relationship between infectivity, that is, the capacity to infect a host (14), and virulence, that is, the negative effect of infection on the host fitness (2), which is an understudied topic (37). Results show pleiotropic effects of resistance-breaking mutations both on virus fitness and on virulence. However, the magnitude and sense of pleiotropy, antagonistic or positive, depended on the genotype of the susceptible host and on the type, single or mixed with other virus genotypes, of infection. Thus, across-host fitness trade-offs depend on the interactions of virus genotype versus host genotype and on the interactions of virus genotype versus type of infection, an observation which underscores the complexity of mechanisms underlying host range expansion in RNA viruses.
MATERIALS AND METHODS
Plants, inoculations, virus, and virulence quantification.
Species and cultivars of Capsicum plants differing in the alleles at the L resistance locus were used for characterizing the pathotypes of the different PMMoV genotypes and for assays of virus multiplication and virulence: C. annuum cv. Dulce Italiano (L+/L+), C. annuum cv. Yolo Wonder (L1/L1), C. frutescens cv. Tabasco (L2/L2), C. chinense PI 159236 (L3/L3), C. annuum cv. Ferrari (L3/L+), C. chacoense PI260429 (L4/L4), and C. annuum cv. Imperio (L4/L+). Note that we used the original Capsicum sp. genotype donors of the resistance alleles or widely grown cultivars; hence, the assayed hosts differed in their genetic makeup in addition to the differences at the L locus. However, for convenience, the different hosts will be identified by their genotypes at the L locus rather than by the full species or cultivar name. Plants were grown in 15-cm-diameter, 1.5-liter pots at 23 to 25°C and with a 16-h light photoperiod in a P2-level biological containment greenhouse. Plants were inoculated in the first two true leaves with 400 ng of freshly purified virus particles suspended in 0.1 M phosphate buffer, pH 7.2. Infection status was checked 10 days postinoculation (dpi) by reverse transcription-PCR (RT-PCR) amplification of the coat protein gene (primer sequences are available upon request).
PMMoV particles were purified as described in Bruening et al. (38), and virion RNA was extracted with phenol-chloroform after virus particles were disrupted in 0.1 M Tris-HCl (pH 9.0)–5% SDS.
Virus multiplication was estimated at 21 dpi as viral RNA accumulation in all systemically infected leaves. Leaf RNA was extracted using TRIzol reagent (Life Technologies), and viral RNA accumulation was determined by quantitative real-time RT-PCR, as described in Fraile et al. (39), using different sets of primers for each viral genotype (available upon request). Levels of viral RNA were deduced from comparison with standard curves produced using a set of serial dilutions of purified virion RNA. Virus fitness was estimated from virus RNA accumulation as described in Lalic et al. (40) using the Malthusian parameter m, which represents the population exponential growth rate at a time t after inoculation, calculated as m = (1/t) ln(Q), where Q is virus accumulation. Fitness, W, is then computed as W = em (41).
Virulence (V) was quantified as the effect of infection on the above-ground host plant biomass, calculated as V = 1 − (Pi/Pm), where Pi is the dry weight of each infected plant and Pm is the mean dry weight of mock-inoculated plants (42).
Virus isolates and mutants.
Inoculations were done with transcripts from full-length infectious cDNA clones derived from two PMMoV field isolates of the P1,2 pathotype. Thus, plasmid pTS, generating TS transcripts, was derived from isolate PMMoV-S and has been described previously (30). Plasmid pMG, generating MG transcripts, was obtained in this work, derived from isolate P84/8 (17) using primers complementary and identical to the 3′ and 5′ ends of the genomic RNA (available upon request); the 5′ primer included the sequence of the T7 transcription promoter, and a MluI restriction site was added to the 5′ of the 3′ primer. The full-length sequence of the insert of plasmid pMG was also determined (primer sequences are available upon request). Plasmids pTS and pMG and derived mutants were multiplied in Escherichia coli XL2-Blue MRF′ (Agilent Technologies).
Mutations were introduced in the coat protein gene of plasmids pTS and pMG by site-directed mutagenesis, generating the following set of genotypes of P1,2,3 pathotype: TS-(M138N) and MG-(M138N) in which the substitution M138N was introduced through the mutations T416A and G417T in pTS and pMG, respectively, using primers M138N-F and M138N-R; TS-(T43K+D50G) and MG-(T43K+D50G), in which the substitutions T43K and D50G were introduced through the mutations C131A and A152G in pTS and pMG, respectively, using primers T43KD50G-F and T43KD50G-R; TS-(L13F+G66V), in which the substitutions L13F and G66V were introduced through the mutations A42T and G200T in plasmid pTS using the primer pair L13F-F and L13F-R and the pair G66V-F and G66V-R, respectively.
The genotype MG-(A87G) of the P1,2,3,4 pathotype was also obtained, in which the substitution A87G was introduced through the mutation C260G using primers A87G-F and A87G-R. All primer sequences are available upon request. Nucleotide and amino acids mentioned above are numbered according to their positions in the CP gene or protein.
The pathotypes of all parental and mutant genotypes were checked by their inoculation in the cultivars and genotypes of Capsicum homozygous for the various L-gene alleles, described above (Table 1).
TABLE 1.
Description of PMMoV genotypes used in this work
| Genotype | CP gene profile |
Capacity to infect the indicated hostc |
Pathotyped | |||||
|---|---|---|---|---|---|---|---|---|
| Amino acid substitution(s)a | Nucleotide substitution(s)b | L1/L1 | L2/L2 | L3/L3 | L4/L+ | L4/L4 | ||
| TS | + | + | NLL | NLL | NLL | P1,2 | ||
| TS-(M138N) | M138N | T433A + G434T | + | + | + | NLL | NLL | P1,2,3 |
| TS-(T43K+D50G) | T43K + D50G | C131A + A152G | + | + | + | + | NLL | P1,2,3 |
| TS-(L13F+G66V) | L13F + G66V | A42T + G200T | + | + | + | NLL | NLL | P1,2,3 |
| MG | + | + | NLL | NLL | NLL | P1,2 | ||
| MG-(M138N) | M138N | T433A + G434T | + | + | + | NLL | NLL | P1,2,3 |
| MG-(T43K+D50G) | T43K + D50G | C131A + A152G | + | + | + | + | NLL | P1,2,3 |
| MG-(A87G) | A87G | G260C | + | + | + | + | + | P1,2,3,4 |
Change of amino acid in the CP from parental TS or MG genotype (pathotype P1,2).
Change of nucleotide in the CP gene from parental TS or MG genotype (pathotype P1,2).
NLL, necrotic local lesions; +, infection present.
Pathotype of the parental and mutant genotypes.
Statistical analyses.
The distribution of data of all analyzed variables was tested against the null hypothesis of normality using a Shapiro-Wilk test, and homogeneity of variances was analyzed by Levene's test. The distribution of data was normal for the fitness of 5 out of 7 virus genotypes in single infection, but distributions were not normal for fitness in mixed infection, for virus accumulation in single or mixed infection, or for virulence. Also, no data distribution was homoscedastic. Thus, differences in virus accumulation, relative fitness, and virulence were analyzed using generalized linear models (GzLM) as this method is robust with respect to data distribution and allows for unbalanced experimental designs. The significance of the differences among classes within a factor was tested using the least significant difference (LSD) test. The relationship between virus accumulation and virulence was analyzed by bivariate Spearman correlation. All statistical analyses were implemented in SPSS, version 17.0 (SPSS, Inc.), and are described in Sokal and Rohlf (43).
Accession number(s).
The complete genome sequence of the PMMoV isolate pMG was deposited in the GenBank under accession number KX063611.
RESULTS
Characterization of parental and mutant virus genotypes.
The sequence of the genomes of PMMoV isolates inserted in the full-length infectious clones pTS and pMG were determined (GenBank accession numbers NC_003630 and KX063611, respectively), with 6,357 nucleotides (nt) for TS and 6,361 nt for MG. TS and MG sequences differed at 19 positions: two were in the 5′ untranslated region (UTR), eight were in the 126-kDa protein open reading frame (ORF), three were in the read-through portion of the 183-kDa protein ORF, three were in the movement protein ORF, one was in the CP ORF, and two were in the 3′ UTR. All nucleotide substitutions within ORFs were silent.
The pathotypes of the parental genotypes TS and MG and of the derived CP mutants were determined by inoculation of plants of the indicator hosts C. annuum cv. Dulce Italiano (L+/L+), C. annuum cv. Yolo Wonder (L1/L1), C. frutescens cv. Tabasco (L2/L2), C. chinense PI 159236 (L3/L3), C. annuum cv. Ferrari (L3/L+), C. chacoense PI260429 (L4/L4), and C. annuum cv. Imperio, (L4/L+). Results are shown in Table 1; all genotypes had the expected pathotypes. However, it was found that TS-(T43K+D50G) and MG-(T43K+D50G) were able to systemically infect C. annuum cv. Imperio (L4/L+) plants but not C. chacoense PI260429 (L4/L4), thus partially overcoming allele L4 in addition to fully overcoming allele L3.
Fitness of PMMoV genotypes in single infection of susceptible host genotypes.
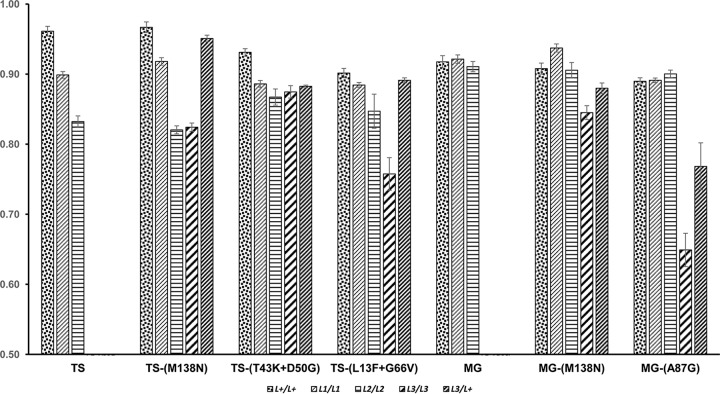
To analyze if the CP mutations that determine resistance breaking of alleles L3 and L4 are associated with fitness penalties in susceptible hosts, the parental genotypes TS and MG with the P1,2 pathotype, the mutant genotypes TS-(M138N), TS-(T43K+D50G), TS-(L13F+G66V), and MG-(M138N) with the P1,2,3 pathotype, and MG-(A87G) with the P1,2,3,4 pathotype were assayed in C. annuum cv. Dulce Italiano (L+/L+), C. annuum cv. Yolo Wonder (L1/L1), C. frutescens cv. Tabasco (L2/L2), C. chinense PI 159236 (L3/L3), and C. annuum cv. Ferrari (L3/L+). The assays followed a random block design with 35 virus genotype treatments versus host genotype treatments, plus the five mock-inoculated controls, with 10 replicated plants per treatment/control. Plants were harvested at 22 dpi, virus RNA was quantified in systemically infected leaves (data not shown), and fitness was estimated as described in Materials and Methods. Data are shown in Fig. 1.
FIG 1.
Fitness of seven PMMoV genotypes in single infection in five Capsicum host genotypes differing at the L locus. Data are means ± standard errors from 10 replicated plants.
To determine if pathotype and/or host genotype affected virus fitness, data were analyzed by a GzLM considering host genotype and virus pathotype fixed factors, and virus genotype nested to pathotype was considered a random factor in a full factorial model. Because genotypes of the P1,2 pathotype do not infect plants of L3/− genotypes, L3/− plants were excluded from the analysis. Virus fitness depended on pathotype, virus genotype nested to pathotype, and host genotype [χ2W(2) = 6.37, P = 0.041; χ2W(4) = 46.31, P = 1 × 10−4; χ2W(2) = 99.92, P = 1 × 10−4, respectively), and the interaction of pathotype versus host genotype and the interaction of virus genotype nested to pathotype versus host genotype were also significant (P = 1 × 10−4). To determine if the resistance-breaking mutations had an effect on virus fitness, a second GzLM analysis was done in which virus genotype and host genotype were considered fixed factors, which showed that virus genotype, host genotype, and their interaction had significant effects on virus fitness [χ2W(6) = 52.48, P = 1 × 10−4; χ2W(2) = 166.72, P = 1 × 10−4; χ2W(12) = 183.91, P = 1 × 10−4, respectively]. LSD analyses showed that, over hosts, the fitness of the three pathotypes ranked P1,2 > P1,2,3 = P1,2,3,4 (P < 0.023), and the fitness of the pooled set of virus genotypes significantly differed according to the host genotype (P = 1 × 10−4) in the order L+/L+ > L1/L1 > L2/L2. Over host genotypes, fitness levels of MG and MG-(M138N) were similar (P > 0.140) and higher than the fitness levels of TS, TS-(M138N), TS-(T43K+D50G), and MG-(A87G), which were also similar; TS-(L13F+G66V) was the least fit genotype (P < 0.026).
Then, the fitness levels of the P1,2,3 and P1,2,3,4 virus genotypes were analyzed over all host genotypes. A GzLM analysis was done in which host genotype and virus pathotype were considered fixed factors and virus genotype nested to pathotype was considered a random factor in a full factorial model. Again, virus fitness depended on pathotype, virus genotype nested to pathotype, and host genotype [χ2W(1) = 108.97, P = 1 × 10−4; χ2W(3) = 36.23, P = 1 × 10−4; χ2W(4) = 437.66, P = 1 × 10−4, respectively], and the interactions of pathotype versus host genotype and of virus genotype nested to pathotype versus host genotype were also significant (P = 1 × 10−4). LSD analyses showed that the pooled set of virus genotypes, with the exclusion of TS and MG, significantly differed in fitness according to the host genotypes in the order L+/L+ > L1/L1> L2/L2 = L3/L+ > L3/L3, and the fitness of P1,2,3 genotypes was higher than that of the P1,2,3,4 genotype MG-(A87G) (P < 0.025).
The fitness of each virus genotype in each host genotype was compared in new GzLM analyses considering host genotype a fixed factor. For all virus genotypes except MG, host genotype had a significant effect (P < 0.016) on virus fitness. LSD analyses ranked virus fitness over hosts differently according to virus genotype, as presented in Fig. 1, showing that the variation of fitness according to host genotype was specific for each virus genotype.
The significance of the interactions of virus genotype versus host genotype in all analyses above indicates host genotype-dependent pleiotropic effects of resistance-breaking mutations on the virus fitness. To quantify the magnitude of these pleiotropic effects, the difference between the fitness of each mutant (Wm) and that of the parental genotype (Wp) was computed in each host genotype, and its significance was analyzed by GzLM and LSD. Table 2 shows these results for host genotypes L+/L+, L1/L1, and L2/L2 as the parental genotypes TS and MG with the P1,2 pathotype did not infect plants with an L3/− genotype. In the genetic context of the TS genotype, the mutation M138N increased fitness in L1/L1 and had no effect on L+/L+ or L2/L2; the double mutation T43K+D50G decreased fitness in L+/L+, had no effect on L1/L1, and increased fitness in L2/L2; and the double mutation L13F+G66V decreased fitness in L+/L+ and L1/L1 and had no effect on L2/L2. In the genetic context of the MG genotype, the mutation M138N increased fitness in L1/L1 and had no effect on L+/L+ or L2/L2 as in the TS context, and the mutation A87G decreased fitness in L+/L+ and L1/L1 and had no effect on L2/L2. Thus, the magnitude of pleiotropy is significantly different from zero in 8/15 interactions of virus genotype versus host genotype, with five antagonistic and three positive interactions. Interestingly, the sense of pleiotropy was always antagonistic in plants of the L+/L+ genotype and antagonistic or positive in L1/L1 plants, and there was no pleiotropy or it was positive in L2/L2 plants.
TABLE 2.
Magnitude and sense of fitness differences between mutant and parental genotypes
| Host genotype | Virus genotype | Wma | Wpb | Wm − Wp | Pc |
|---|---|---|---|---|---|
| L+/L+ | TS-(M138N) | 0.967 | 0.961 | 0.006 | 0.552 |
| L+/L+ | TS-(T43K+D50G) | 0.931 | 0.961 | −0.030 | 0.002* |
| L+/L+ | TS-(L13F+G66V) | 0.902 | 0.961 | −0.059 | 0.000* |
| L+/L+ | MG-(M138N) | 0.908 | 0.918 | −0.010 | 0.367 |
| L+/L+ | MG-(A87G) | 0.890 | 0.918 | −0.028 | 0.010* |
| L1/L1 | TS-(M138N) | 0.918 | 0.899 | 0.019 | 0.007* |
| L1/L1 | TS-(T43K+D50G) | 0.886 | 0.899 | −0.013 | 0.068 |
| L1/L1 | TS-(L13F+G66V) | 0.884 | 0.899 | −0.015 | 0.040* |
| L1/L1 | MG-(M138N) | 0.937 | 0.921 | 0.016 | 0.025* |
| L1/L1 | MG-(A87G) | 0.891 | 0.921 | −0.030 | <10−3* |
| L2/L2 | TS-(M138N) | 0.820 | 0.832 | −0.012 | 0.505 |
| L2/L2 | TS-(T43K+D50G) | 0.867 | 0.832 | 0.035 | 0.048* |
| L2/L2 | TS-(L13F+G66V) | 0.847 | 0.832 | 0.015 | 0.393 |
| L2/L2 | MG-(M138N) | 0.905 | 0.910 | −0.005 | 0.773 |
| L2/L2 | MG-(A87G) | 0.900 | 0.910 | −0.010 | 0.577 |
Wm is the average fitness of each mutant in each host at 22 dpi.
Wp is the average fitness of each parental genotype.
The asterisk indicates significance at a P value of ≤0.05 for fitness differences according to an LSD analysis.
Thus, results show that resistance-breaking mutations have a pleiotropic effect on virus fitness that depends on the host genotype, and this pleiotropy may be an important cause of the observed fitness in the interaction of virus genotype versus host genotype.
Virulence of PMMoV resistance-breaking genotypes in single infection of susceptible host genotypes.
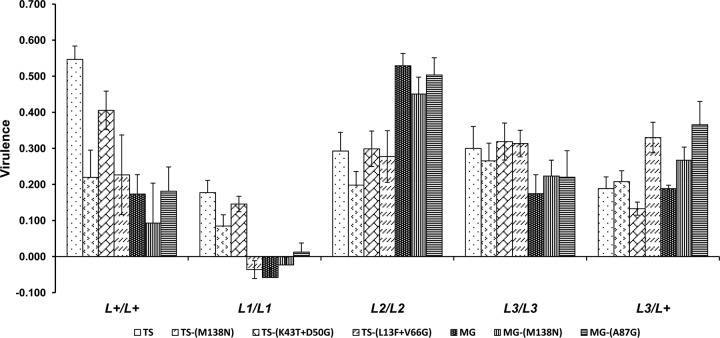
In the above-described experiment it was observed that the symptoms developed by infected plants differed according to virus genotype so that in general those induced by the P1,2,3,4 genotype were more severe than those induced by the P1,2,3 genotypes, which were more severe than those of the P1,2 genotype, suggesting differences in virulence. To further analyze the possible effect of resistance-breaking mutations on virulence, virulence was quantified as the effect of infection on plant biomass (data not shown), and the derived virulence values are shown in Fig. 2.
FIG 2.
Virulence of seven PMMoV genotypes in single infection in five Capsicum host genotypes differing at the L locus. Data are means ± standard errors from 10 replicated plants.
The possible effect of pathotype on virulence was analyzed by GzLM considering host genotype and virus pathotype fixed factors and virus genotype nested to pathotype a random factor in a full factorial model. Virulence depended on pathotype, virus genotype nested to pathotype, and host genotype [χ2W(2) = 8.38, P = 0.015; χ2W(4) = 22.47, P = 1 × 10−4; χ2W(4) = 143.156, P = 1 × 10−4, respectively], and the interactions of pathotype versus host genotype and of virus genotype nested to pathotype versus host genotype were also significant (P < 0.004). To determine if the resistance-breaking mutations had an effect on virulence, GzLM analyses were done considering host and virus genotypes fixed factors in a full factorial model. Both factors and their interaction had an effect on virulence [χ2W(4) = 150.23, P = 1 × 10−4; χ2W(6) = 32.36, P = 1 × 10−4; χ2W(24) = 83.65, P = 1 × 10−4]. LSD analyses showed that virulence ranked according to host genotype as L1/L1 < L+/L+ = L3/L+ = L3/L3 < L2/L2 (P ≤ 0.001 for differences). Over hosts the virulence of the three pathotypes ranked as P1,2 = P1,2,3,4 > P1,2,3, (P = 0.001). Virus genotypes ranked according to virulence as TS = TS-(T43K+D50G) > MG-(A87G) = TS-(L13F+G66V) = TS-(M138N) = MG-(M138N) = MG (P ≤ 0.012 for differences). The effect of the interaction of host genotype versus virus genotype on virulence is further shown in Fig. 2. When the virulence of the resistance-breaking genotypes was compared with that of their parental genotypes, it was found that TS-(M138N) and TS-(L13F+G66V) were less virulent than TS in L+/L+ and L1/L1 plants; all other comparisons were not significant (data not shown).
The fitness of the various virus genotypes was correlated with their virulence only in hosts L+/L+ and L2/L2 (ρ > 0.45, P = 1 × 10−4; Spearman test) (Fig. 3).
FIG 3.
Relationship between fitness and virulence in five susceptible hosts. Hosts are identified by their genotype at the L locus as L+/L+ (black diamonds), L1/L1 (empty squares), L2/L2 (gray triangles), L3/L+ (crosses), or L3/L3 (gray circles). Significant positive correlations between fitness and virulence are shown for L+/L+ (black) and L2/L2 (gray).
Fitness of PMMoV resistance-breaking genotypes in mixed infection of the universal susceptible host genotype L+/L+.
The fitness of the virus genotypes above was also analyzed in mixed infections, in which two different genotypes compete for the host resources. For this assay, the universally susceptible host genotype L+/L+ was chosen, and all the virus genotypes of the previous assay were included with the exception of TS-(L13F+G66V) as we were unable to develop a specific RT-quantitative PCR (RT-qPCR) for quantification of its accumulation in mixed infection. On the other hand, the double mutant MG-(T43K+D50G), previously unavailable, was included. In this experiment each virus genotype was assayed in single and in mixed infections so that each genotype of a P1,2,3 pathotype competed with its P1,2 parental genotype, and the genotype of the P1,2,3,4 pathotype competed with both P1,2 genotypes and with the P1,2,3 genotypes derived from the same parental genotype (Table 3). The assay followed a random block design with 23 virus infection treatments, plus the mock-inoculated controls, with 10 replicated plants per treatment/control. Plants were harvested at 22 dpi, virus RNA was quantified in systemically infected leaves, and fitness was estimated.
TABLE 3.
Fitness of PMMoV genotypes in single and mixed infections in L+/L+ host plants
| Genotype | Fitness in single infectiona | Fitness in mixed infection by pathotypea |
||||||
|---|---|---|---|---|---|---|---|---|
| P1,2 |
P1,2,3 |
P1,2,3,4 [MG-(A87G)] | ||||||
| TS | MG | TS-(M138N) | TS-(T43K+D50G) | MG-(M138N) | MG-(T43K+D50G) | |||
| TS | 0.96 ± 0.03 | 0.98 ± 0.98 | 0.92 ± 0.02 | 0.93 ± 0.01 | ||||
| MG | 0.99 ± 0.02 | 0.97 ± 0.01 | 0.97 ± 0.00 | 0.94 ± 0.02 | ||||
| TS-(M138N) | 0.96 ± 0.03 | 0.84 ± 0.01 | ||||||
| TS-(T43K+D50G) | 0.98 ± 0.01 | 0.84 ± 0.04 | ||||||
| MG-(M138N) | 0.99 ± 0.01 | 0.83 ± 0.01 | 0.79 ± 0.01 | |||||
| MG-(T43K+D50G) | 0.99 ± 0.01 | 0.90 ± 0.03 | 0.92 ± 0.01 | |||||
| MG-(A87G) | 0.98 ± 0.03 | 1.00 ± 0.02 | 0.97 ± 0.01 | 0.99 ± 0.01 | 1.02 ± 0.004 | |||
Data are average fitness (W) ± standard error for 10 replicated plants.
Table 3 shows the fitness values of each virus genotype in single and in mixed infection. To determine if the infection type, single or mixed, had an effect on viral fitness according to the virus pathotype, data were analyzed by a GzLM in which infection type and pathotype were considered fixed factors and virus genotype was considered a random factor nested to pathotype in a full factorial model. Infection type, pathotype, and virus genotype nested to pathotype had significant effects on fitness [χ2W(1) = 21.42, P = 1 × 10−4; χ2W(2) = 36.72, P = 1 × 10−4, χ2W(4) = 11.92, P = 0.018, respectively], as well as the interaction of pathotype versus infection type [χ2W(2) = 36.60, P = 1 × 10−4]. LSD analyses showed that fitness of pathotypes P1,2 and P1,2,3 (P < 0.007) but not P1,2,3,4 (P = 0.098) was higher in single than in mixed infection and that in mixed infection the fitness of the three pathotypes ranked P1,2,3,4 > P1,2 > P1,2,3 (P ≤ 0.005).
To determine if the resistance-breaking mutations had an effect on the virus fitness in mixed infection, a GzLM analysis was performed in which infection type and virus genotype were considered fixed factors in a full factorial model. Infection type, virus genotype, and their interaction had significant effects on fitness [χ2W(1) = 59.49, P = 1 × 10−4; χ2W(6) = 47.51, P = 1 × 10−4; χ2W(6) = 44.00, P = 1 × 10−4, respectively]. Similar analyses were done considering only the set of genotypes that competed with TS or those that competed with MG. In both cases the interaction of infection type verus genotype was significant (P ≤ 0.050), and results showed that the fitness of genotypes of then P1,2 (TS and MG) or P1,2,3,4 [MG-(A87G)] pathotype did not depend on the type of infection, while that of genotypes of the P1,2,3 pathotype [TS-(M138N), TS-(T43K+D50G), MG-(M138N), and MG-(T43K+D59G)] was lower in mixed than in single infection (P < 0.003). Also, the fitness of genotypes of the P1,2,3 pathotype in mixed infection was lower than that of their P1,2 parental genotypes (P < 0.036). Last, fitness levels of the P1,2,3 pathotype genotypes MG-(M138N) and MG-(T43K+D50G) were significantly lower in mixed infection with the P1,2,3,4 genotype MG-(A87G) than in single infection and were lower than the fitness of MG-(A87G) (P < 0.002), while fitness levels did not differ in single infection. Thus, virus fitness depends on the interaction of genotype versus environment (G versus E), considering the type of infection, single or mixed, as the environment.
To quantify the magnitude of these pleiotropic effects, the difference between the fitness of each mutant and that of the parental genotype was computed, and its significance was analyzed by GzLM and LSD. Fitness differences were significant only for the magnitude of the pleiotropic effects of the mutations determining the P1,2,3 pathotype (M138N and T43K+D50G), which showed antagonistic pleiotropy in mixed versus single infection (Table 4). The magnitude of the antagonistic pleiotropy was higher for the M138N mutation than for the T43K+D50G double mutation, and, interestingly, the magnitude was independent of the genetic context, TS or MG (Table 4).
TABLE 4.
Magnitude and sense of fitness differences between mutant and parental genotypes in competition in L+/L+ host plants
| Parental genotype | Mutant genotype | Wma | Wpb | Wm − Wp | Pc |
|---|---|---|---|---|---|
| TS | TS-(M138N) | 0.845 | 0.986 | −0.141 | 0.001* |
| TS | TS-(T43K+D50G) | 0.848 | 0.926 | −0.079 | 0.036* |
| MG | MG-(M138N) | 0.835 | 0.970 | −0.136 | <10−3* |
| MG | MG-(T43K+D50G) | 0.900 | 0.980 | −0.080 | 0.013* |
| MG | MG-(A87G) | 0.966 | 0.945 | 0.021 | 0.506 |
Wm is the average fitness of the mutant.
Wp is the average fitness of the parental genotype.
The asterisk indicates a significant difference according to an LSD analysis.
Thus, regardless of the relative accumulation in single infection, in mixed infection the genotypes of P1,2,3 pathotype were outcompeted by those of the P1,2 or P1,2,3,4 pathotype, showing that mutations determining the P1,2,3 pathotype have a negative effect on virus fitness in the susceptible host. This is not the case for the mutation determining the P1,2,3,4 pathotype.
DISCUSSION
It is often considered that RNA viruses have a high potential to adapt to new hosts because their high census numbers and the high error rates of RNA-dependent RNA polymerases will translate into a strong potential to generate host range mutants (1, 5, 44). However, RNA viruses have small, information-compact genomes encoding few multifunctional proteins, which will favor pleiotropic effects of mutations (5, 45). A consequence can be the generation of across-host fitness trade-offs so that mutants that perform well in one host will perform poorly in another one, limiting host range expansion and, more generally, virus emergence (45, 46). It is thought that the major cause of across-host fitness trade-offs in viruses is antagonistic pleiotropy, that is, the negative fitness effect in the original host of mutations that increase fitness in the new one (7), expressed as the interaction of the virus genotype versus the host environment (G versus E) (5, 40). The role of antagonistic pleiotropy in limiting host range expansion has been well documented in viruses infecting bacteria, animals, and plants, often from host adaptation experiments (e.g., 40, 47–54).
A particular case of host range expansion is the overcoming of host resistance in GFG host-pathogen interactions, which compromises the control of crop viral diseases. In GFG systems there is a hierarchy of infectivity alleles in the pathogen and of resistance alleles in the host so that some pathogen infectivity alleles are intrinsically better than others, conferring the capacity to infect and multiply in a larger set of host genotypes, and, conversely, some host resistance alleles confer the capacity to resist a larger set of pathogen genotypes (14, 55). In GFG systems the pathogen is predicted to evolve by successive steps of resistance breaking until it infects and multiplies in all host genotypes (14). Host-pathogen coevolution under the GFG model has been extensively modeled; models assume that resistance-breaking mutations have fitness costs in the susceptible host (56, 57), thus generating across-host fitness trade-offs. Evidence for resistance-breaking fitness costs derives mostly from analyses of the multiplication of resistance-breaking and wild-type virus genotypes in a susceptible host genotype. Previous reports refer to different plant-virus systems (16, 18, 58–60), including the pepper-PMMoV system analyzed here (17).
The interaction of PMMoV genotypes with pepper genotypes carrying different resistance alleles at the L locus is according to the GFG model (30–33). Most PMMoV field isolates are of a P1,2 pathotype; i.e., they infect plants of the universally susceptible L+/L+ genotype and those carrying resistance alleles L1 and L2, but P1,2,3 isolates, which also infect L3 plants, also occur in pepper crops. Analyses of field PMMoV isolates of P1,2 and P1,2,3 pathotypes have shown fitness costs associated with L3 resistance breaking (17). To identify the mechanisms that generate such fitness costs, different mutations in the virus CP reported as determinants of resistance breaking of the L3 or L3 and L4 alleles (P1,2,3 or P1,2,3,4 pathotype, respectively) (27, 30, 34, 36) were introduced into two PMMoV P1,2 isolates by site-directed mutagenesis of infectious cDNA clones. Mutation M138N is present in all characterized P1,2,3 isolates from Spain and the Mediterranean basin (17, 30), while mutations T43K+D50G and L13F+V66G have been reported only in the P1,2,3 isolates PMMoV-Ij and PMMoV-Is, respectively, from Japan (34, 36). Mutation A87G has been reported as determining the P1,2,3,4 pathotype in a PMMoV isolate from Israel, together with mutation L47Q (27). The parental genotype MG has a Q at position 47 of the CP, rendering the introduction of mutation L47Q unnecessary. All mutants had the expected pathotype, confirming that the introduced CP mutations are responsible for the viral pathotype, regardless of the genotype of the P1,2 isolate in which they were introduced.
The fitness of the mutants in single infection was compared to that of the parental genotypes in five hosts with different alleles at the L locus (Fig. 1). The virus fitness depended on the host genotype, the virus genotype, and the virus pathotype, but increasing infectivity is not correlated with increasing fitness penalties. This analysis also showed that virus fitness depended on the interaction between virus and host genotypes, i.e., on a genotype per environment (G versus E) interaction, which is evidence of pleiotropic effects of the resistance-breaking mutations (5, 40). When the sense and magnitude of the pleiotropy, whether negative (i.e., antagonistic) or positive, were analyzed for the various resistance-breaking mutants, they were found to depend on both the specific resistance-breaking mutation and on the host genotype. Most (3/5) resistance-breaking mutants showed antagonistic pleiotropy in the universally susceptible host L+/L+. Interestingly, only 1/5 mutants showed antagonistic pleiotropy in the L1/L1 host, and none showed antagonistic pleiotropy in L2/L2; conversely, 2/5 mutants showed positive pleiotropy in L1/L1, and the only detected pleiotropy in L2/L2 was positive. Hence, the negative effects of resistance-breaking mutations on virus fitness are reduced, or inverted, in the susceptible hosts carrying L alleles of a higher hierarchy (Table 3). It should be underscored that the genotype at the L locus of the assayed hosts is the major determinant of the infectivity of each virus genotype (25–33), but since the assayed hosts were different pepper cultivars or species that will differ in many other genes, host-dependent differences in fitness among virus genotypes cannot be attributed only to their interaction with the L alleles. Still, it could be speculated that the broader the L allele resistance, the lower the antagonistic pleiotropy effects of resistance-breaking mutations.
Antagonistic pleiotropy has been shown to explain resistance-breaking-associated fitness costs in several plant viruses (16, 17, 58, 59), as shown here for some interactions. Our results also provide evidence of resistance breaking without cost, as occasionally reported (48, 61–64). What is novel in our results and highly significant is the evidence of the host-dependent positive pleiotropy of resistance-breaking mutations, which may be costly or favorable in different susceptible hosts.
Mixed infection of hosts by different virus species or genotypes is frequent in nature (65–68) and may be relevant for virus evolution, including the evolution of resistance breaking. Comparison of virus fitness levels in mixed versus single infection in the susceptible L+/L+ host also showed interactions of G versus E, where E represents the type of infection, single or mixed. Thus, resistance-breaking mutations have pleiotropic effects on fitness that depend on the type of infection. Mutations determining a P1,2,3 pathotype, including M138N, involve fitness penalties in mixed infection in the L+/L+ host, as shown by significant magnitudes of antagonistic pleiotropy (Table 4). Fitness costs due to pleiotropic effects of resistance-breaking mutations were best unraveled under competition with the parental genotype than in single infection, in agreement with reports from field isolates of PMMoV and with other systems (16, 17). Interestingly, the P1,2,3,4-determining A87G mutation had no fitness costs in mixed infection, which may increase the risks of breaking L4 allele resistance, presently bred into most hybrid cultivars of bell pepper. It is relevant to stress that the interaction of G versus E according to the type of infection would result in trade-offs across infection types. Consequently, the frequency of mixed infections would be a further determinant of the evolution of host range expansion in virus populations, a topic that to our knowledge has not been analyzed previously.
Resistance-breaking mutations could also affect other components of the virus life history, such as virulence. We analyzed the virulence of the parental and the mutant genotypes in the five susceptible hosts in single infection. Plant biomass correlates with both fecundity and survival (37, 69, 70); thus, the effect of infection on biomass is a proxy to virulence. As was the case for fitness, virulence depended on the interaction of G versus E, revealing a new pleiotropic effect of resistance-breaking mutations. Pleiotropic effects on virulence were mutation specific and were unrelated to those on fitness, and, overall, virulence and fitness did not correlate, as is often the case for plant viruses (71, 72). It is noteworthy that although pathotype has a significant effect on virulence, increased infectivity does not necessarily translate into virulence changes. This is an interesting result as the relationship between infectivity and virulence has rarely been analyzed either experimentally or theoretically (but see reference 73). Our data are consistent with the only other analysis we know off, in which it was shown that rice yellow mottle virus genotypes differing in infectivity on the Rymv-2 resistance allele did not differ in virulence (74); that is, virulence and infectivity were not linked. Virulence may also be an important determinant of resistance-breaking evolution in the pepper-PMMoV system because growers eliminate infected plants when detected (22), and detection will be easier in plants infected by more virulent virus genotypes.
The present results thus significantly contribute to understanding the complexity of the mechanisms that determine the evolution of resistance breaking and host range expansion in RNA viruses. One novel result of this work is that pleiotropic effects on virus multiplication of host range mutations depend on the specific mutation and on the specific susceptible host. Host range mutations are not always pleiotropic on fitness, and when they are, pleiotropy can be positive or negative, favoring or hindering, respectively, host range expansion. Thus, the fate of host range mutations in the virus population will depend on not only the frequency of resistant versus susceptible hosts genotypes, as often considered (75), but also the frequency of the different susceptible host genotypes. A second novel result is that the pleiotropic effects on the virus fitness of host range mutations differ in single and in mixed infections so that trade-offs across infection types (single or mixed) are expected to occur, similarly to across-host effects. Hence, the evolution of host range expansion will also be determined by the frequency and type of mixed infections. Last, pleiotropic effects of host range mutations on other virus life history traits, such as survival (39) or virulence (this work), may also affect host range expansion. All of these factors should be considered in modeling the evolution of host range expansion and, specifically, the evolution of resistance breaking in crops.
ACKNOWLEDGMENTS
We thank Antolín López Quirós and Miguel Angel Mora for excellent technical assistance.
REFERENCES
- 1.Elena SF, Fraile A, García-Arenal F. 2014. Evolution and emergence of plant viruses. Adv Virus Res 88:161–191. doi: 10.1016/B978-0-12-800098-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 2.Frank SA. 1996. Models of parasite virulence. Q Rev Biol 71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- 3.Lajeunesse MJ, Forbes MR. 2002. Host range and local parasite adaptation. Proc Biol Sci 269:703–710. doi: 10.1098/rspb.2001.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse MEJ, Taylor LH, Haydon DT. 2001. Population biology of multihost pathogens. Science 292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 5.Bedhomme S, Hillung J, Elena SF. 2015. Emerging viruses: why they are not jacks of all trades? Curr Opin Virol 10:1–6. doi: 10.1016/j.coviro.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Kirchner JW, Roy BA. 2002. Evolutionary implications of host-pathogen specificity: fitness consequences of pathogen virulence traits. Evol Ecol Res 4:27–48. [Google Scholar]
- 7.Whitlock MC. 1996. The Red Queen beats the jack-of-all-trades: the limitations on the evolution of phenotypic plasticity and the niche breadth. Am Nat 148:S65–S77. doi: 10.1086/285902. [DOI] [Google Scholar]
- 8.Elena SF, Agudelo-Romero P, Lalic J. 2009. The evolution of viruses in multi-host fitness landscapes. Open Virol J 3:1–6. doi: 10.2174/1874357900903010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraile A, García-Arenal F. 2010. The coevolution of plants and viruses: Resistance and pathogenicity. Adv Virus Res 76:1–32. doi: 10.1016/S0065-3527(10)76001-2. [DOI] [PubMed] [Google Scholar]
- 10.Kang B-C, Yeam I, Jahn MM. 2005. Genetics of plant virus resistance. Annu Rev Phytopathol 43:581–621. doi: 10.1146/annurev.phyto.43.011205.141140. [DOI] [PubMed] [Google Scholar]
- 11.Maule AJ, Caranta C, Boulton MI. 2007. Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8:223–231. doi: 10.1111/j.1364-3703.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 12.de Ronde D, Butterbach P, Kormelink R. 2014. Dominant resistance against plant viruses. Front Plant Sci 5:307. doi: 10.3389/fpls.2014.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moffett P. 2009. Mechanisms of recognition in R gene mediated resistance. Adv Virus Res 75:1–33. doi: 10.1016/S0065-3527(09)07501-0. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal A, Lively CM. 2002. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res 4:91–107. [Google Scholar]
- 15.Ayme V, Souche S, Caranta C, Jacquemond M, Chadoeuf J, Palloix A, Moury B. 2006. Different mutations in the genome-linked protein VPg of Potato virus Y confer virulence on the pvr23 resistance in pepper. Mol Plant Microbe Interact 19:557–563. doi: 10.1094/MPMI-19-0557. [DOI] [PubMed] [Google Scholar]
- 16.Ishibashi K, Mawatari N, Miyashita S, Kishino H, Meshi T, Ishikawa M. 2012. Coevolution and hierarchical interactions of Tomato mosaic virus and the resistance gene Tm-1. PLoS Pathog 8:e1002975. doi: 10.1371/journal.ppat.1002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraile A, Pagán I, Anastasio G, Sáez E, García-Arenal F. 2011. Rapid genetic diversification and high fitness penalties associated with pathogenicity evolution in a plant virus. Mol Biol Evol 28:1425–1437. doi: 10.1093/molbev/msq327. [DOI] [PubMed] [Google Scholar]
- 18.Janzac B, Montarry J, Palloix A, Navaud O, Moury B. 2010. A point mutation in the polymerase of Potato virus Y confers virulence toward the pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol Plant Microbe Interact 23:823–830. doi: 10.1094/MPMI-23-6-0823. [DOI] [PubMed] [Google Scholar]
- 19.Montarry J, Cartier E, Jacquemond M, Palloix A, Moury B. 2012. Virus adaptation to quantitative plant resistance: Erosion or breakdown? J Evol Biol 25:2242–2252. doi: 10.1111/j.1420-9101.2012.02600.x. [DOI] [PubMed] [Google Scholar]
- 20.Poulicard N, Pinel-Galzi A, Hebrard E, Fargette D. 2010. Why Rice yellow mottle virus, a rapidly evolving RNA plant virus, is not efficient at breaking rymv1-2 resistance. Mol Plant Pathol 11:145–154. doi: 10.1111/j.1364-3703.2009.00582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulicard N, Pinel-Galzi A, Traore O, Vignols F, Ghesquiere A, Konate G, Hebrard E, Fargette D. 2012. Historical contingencies modulate the adaptability of Rice yellow mottle virus. PLoS Pathog 8:e1002482. doi: 10.1371/journal.ppat.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moury B, Verdin E. 2012. Viruses of pepper crops in the Mediterranean basin: a remarkable stasis, p 127–162. In Loebenstein G, Lecoq H (ed), Viruses and virus diseases of vegetables in the Mediterranean basin. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 23.Broadbent L. 1976. Epidemiology and control of tomato mosaic virus. Annu Rev Phytopathol 14:75–96. doi: 10.1146/annurev.py.14.090176.000451. [DOI] [Google Scholar]
- 24.Sakamoto M, Tomita R, Hamada H, Iwadate Y, Munemura I, Kobayashi K. 2008. A primer-introduced restriction analysis-PCR-based method to analyse Pepper mild mottle virus populations in plants and field soil with respect to virus mutations that break L3 gene-mediated resistance of Capsicum plants. Plant Pathol 57:825–833. doi: 10.1111/j.1365-3059.2008.01835.x. [DOI] [Google Scholar]
- 25.Boukema IW. 1980. Allelism of genes controlling resistance to TMV in Capsicum L. Euphytica 29:433–439. doi: 10.1007/BF00025143. [DOI] [Google Scholar]
- 26.Boukema IW. 1984. Resistance to TMV in Capsicum chacoense Hunz is governed by allele of the L-locus. Capsicum Newsl 3:47–48. [Google Scholar]
- 27.Antignus Y, Lachman O, Pearlsman M, Maslenin L, Rosner A. 2008. A new pathotype of Pepper mild mottle virus (PMMoV) overcomes the L4 resistance genotype of pepper cultivars. Plant Dis 92:1033–1037. doi: 10.1094/PDIS-92-7-1033. [DOI] [PubMed] [Google Scholar]
- 28.Genda Y, Kanda A, Hamada H, Sato K, Ohnishi J, Tsuda S. 2007. Two amino acid substitutions in the coat protein of Pepper mild mottle virus are responsible for overcoming the L4 gene-mediated resistance in Capsicum spp. Phytopathology 97:787–793. doi: 10.1094/PHYTO-97-7-0787. [DOI] [PubMed] [Google Scholar]
- 29.Rast ATB. 1988. Pepper tobamoviruses and pathotypes used in resistance breeding. Capsicum Newsl 7:20–24. [Google Scholar]
- 30.Berzal-Herranz A, De La Cruz A, Tenllado F, Diaz-Ruiz JR, López L, Sanz AI, Vaquero C, Serra MT, García-Luque I. 1995. The capsicum L3 gene-mediated resistance against the tobamoviruses is elicited by the coat protein. Virology 209:498–505. doi: 10.1006/viro.1995.1282. [DOI] [PubMed] [Google Scholar]
- 31.de la Cruz A, López L, Tenllado F, Díaz-Ruiz JR, Sanz AI, Vaquero C, Serra MT, García-Luque I. 1997. The coat protein is required for the elicitation of the capsicum L2 gene-mediated resistance against the tobamoviruses. Mol Plant Microbe Interact 10:107–113. doi: 10.1094/MPMI.1997.10.1.107. [DOI] [PubMed] [Google Scholar]
- 32.Gilardi P, García-Luque I, Serra MT. 2004. The coat protein of tobamovirus acts as elicitor of both L2 and L4 gene-mediated resistance in Capsicum. J Gen Virol 85:2077–2085. doi: 10.1099/vir.0.80017-0. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto K, Sawada H, Matsumoto K, Hamada H, Yoshimoto E, Ito T, Takeuchi S, Tsuda S, Suzuki K, Kobayashi K, Kiba A, Okuno T, Hikichi Y. 2008. The coat protein gene of tobamovirus P0 pathotype is a determinant for activation of temperature-insensitive L1a gene-mediated resistance in Capsicum plants. Arch Virol 153:645–650. doi: 10.1007/s00705-008-0032-y. [DOI] [PubMed] [Google Scholar]
- 34.Hamada H, Tomita R, Iwadate Y, Kobayashi K, Munemura I, Takeuchi S, Hikichi Y, Suzuki K. 2007. Cooperative effect of two amino acid mutations in the coat protein of Pepper mild mottle virus overcomes L3-mediated resistance in Capsicum plants. Virus Genes 34:205–214. doi: 10.1007/s11262-006-0049-9. [DOI] [PubMed] [Google Scholar]
- 35.Hamada H, Matsumura H, Tomita R, Terauchi R, Suzuki K, Kobayashi K. 2008. SuperSAGE revealed different classes of early resistance response genes in Capsicum chinense plants harboring L3-resistance gene infected with Pepper mild mottle virus. J Gen Plant Pathol 74:313–321. doi: 10.1007/s10327-008-0106-4. [DOI] [Google Scholar]
- 36.Tsuda S, Kirita M, Watanabe Y. 1998. Characterization of a Pepper mild mottle tobamovirus strain capable of overcoming the L3 gene-mediated resistance, distinct from the resistance-breaking Italian isolate. Mol Plant Microbe Interact 11:327–331. doi: 10.1094/MPMI.1998.11.4.327. [DOI] [PubMed] [Google Scholar]
- 37.Sacristán S, García-Arenal F. 2008. The evolution of virulence and pathogenicity in plant pathogen populations. Mol Plant Pathol 9:369–384. doi: 10.1111/j.1364-3703.2007.00460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruening G, Beachy RN, Scalla R, Zaitlin M. 1976. In vitro and in vivo translation of the ribonucleic acids of a cowpea strain of tobacco mosaic virus. Virology 71:498–517. doi: 10.1016/0042-6822(76)90377-9. [DOI] [PubMed] [Google Scholar]
- 39.Fraile A, Hily J-M, Pagán I, Pacios LF, García-Arenal F. 2014. Host resistance selects for traits unrelated to resistance-breaking that affect fitness in a plant virus. Mol Biol Evol 31:928–939. doi: 10.1093/molbev/msu045. [DOI] [PubMed] [Google Scholar]
- 40.Lalic J, Cuevas JM, Elena SF. 2011. Effect of host species on the distribution of mutational fitness effects for an RNA virus. PLoS Genet 7:e1002378. doi: 10.1371/journal.pgen.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crow JF, Kimura M. 1970. An introduction to population genetics theory. Harper and Row, New York, NY. [Google Scholar]
- 42.Pagan I, Alonso-Blanco C, Garcia-Arenal F. 2007. The relationship of within-host multiplication and virulence in a plant-virus system. PLoS One 2:e786. doi: 10.1371/journal.pone.0000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokal RR, Rohlf FJ. 1995. Biometry, 3rd ed. W. H. Freeman and Company, New York, NY. [Google Scholar]
- 44.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elena SF, Sanjuán R. 2007. Virus evolution: insights from an experimental approach. Annu Rev Ecol Evol Syst 38:27–52. doi: 10.1146/annurev.ecolsys.38.091206.095637. [DOI] [Google Scholar]
- 46.Gandon S, Hochberg ME, Holt RD, Day T. 2013. What limits the evolutionary emergence of pathogens? Philos Trans R Soc Lond B Biol Sci 368:20120086. doi: 10.1098/rstb.2012.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agudelo-Romero P, de la Iglesia F, Elena SF. 2008. The pleiotropic cost of host-specialization in Tobacco etch potyvirus. Infect Genet Evol 8:806–814. doi: 10.1016/j.meegid.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Bedhomme S, Lafforgue G, Elena SF. 2012. Multihost experimental evolution of a plant RNA virus reveals local adaptation and host-specific mutations. Mol Biol Evol 29:1481–1492. doi: 10.1093/molbev/msr314. [DOI] [PubMed] [Google Scholar]
- 49.Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. 2008. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci U S A 105:6970–6975. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deardorff ER, Fitzpatrick KA, Jerzak GV, Shi PY, Kramer LD, Ebel GD. 2011. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS Pathog 7:e1002335. doi: 10.1371/journal.ppat.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duffy S, Turner PE, Burch CL. 2006. Pleiotropic costs of niche expansion in the RNA bacteriophage ϕ6. Genetics 172:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hillung J, Cuevas JM, Valverde S, Elena SF. 2014. Experimental evolution of an emerging plant virus in host genotypes that differ in their susceptibility to infection. Evolution 68:2467–2480. doi: 10.1111/evo.12458. [DOI] [PubMed] [Google Scholar]
- 53.Vasilakis N, Deardorff ER, Kenney JL, Rossi SL, Hanley KA, Weaver SC. 2009. Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog 5:e1000467. doi: 10.1371/journal.ppat.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallis CM, Stone AL, Sherman DJ, Damsteegt VD, Gildow FE, Schneider WL. 2007. Adaptation of plum pox virus to a herbaceous host (Pisum sativum) following serial passages. J Gen Virol 88:2839–2845. doi: 10.1099/vir.0.82814-0. [DOI] [PubMed] [Google Scholar]
- 55.Dybdahl MF, Jenkins CE, Nuismer SL. 2014. Identifying the molecular basis of host-parasite coevolution: merging models and mechanisms. Am Nat 184:1–13. doi: 10.1086/676591. [DOI] [PubMed] [Google Scholar]
- 56.Brown JKM. 2015. Durable resistance of crops to disease: a Darwinian perspective. Annu Rev Phytopathol 53:513–539. doi: 10.1146/annurev-phyto-102313-045914. [DOI] [PubMed] [Google Scholar]
- 57.Brown JKM, Tellier A. 2011. Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu Rev Phytopathol 49:345–367. doi: 10.1146/annurev-phyto-072910-095301. [DOI] [PubMed] [Google Scholar]
- 58.Goulden MG, Kohm BA, Cruz SS, Kavanagh TA, Baulcombe DC. 1993. A feature of the coat protein of Potato virus X affects both induced virus-resistance in potato and viral fitness. Virology 197:293–302. doi: 10.1006/viro.1993.1590. [DOI] [PubMed] [Google Scholar]
- 59.Jenner CE, Tomimura K, Ohshima K, Hughes SL, Walsh JA. 2002. Mutations in Turnip mosaic virus P3 and cylindrical inclusion proteins are separately required to overcome two Brassica napus resistance genes. Virology 300:50–59. doi: 10.1006/viro.2002.1519. [DOI] [PubMed] [Google Scholar]
- 60.Jenner CE, Wang X, Ponz F, Walsh JA. 2002. A fitness cost for Turnip mosaic virus to overcome host resistance. Virus Res 86:1–6. doi: 10.1016/S0168-1702(02)00031-X. [DOI] [PubMed] [Google Scholar]
- 61.Coffey LL, Vignuzzi M. 2011. Host alternation of chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. J Virol 85:1025–1035. doi: 10.1128/JVI.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cooper LA, Scott TW. 2001. Differential evolution of eastern equine encephalitis virus populations in response to host cell type. Genetics 157:1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. 2005. Effect of alternating passage on adaptation of Sindbis virus to vertebrate and invertebrate cells. J Virol 79:14253–14260. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novella IS, Hershey CL, Escarmis C, Domingo E, Holland JJ. 1999. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol 287:459–465. doi: 10.1006/jmbi.1999.2635. [DOI] [PubMed] [Google Scholar]
- 65.Gómez P, Sempere RN, Amari K, Gomez-Aix C, Aranda MA. 2010. Epidemics of Tomato torrado virus, Pepino mosaic virus and Tomato chlorosis virus in tomato crops: do mixed infections contribute to torrado disease epidemiology? Ann Appl Biol 156:401–410. doi: 10.1111/j.1744-7348.2010.00397.x. [DOI] [Google Scholar]
- 66.Gómez P, Sempere RN, Elena SF, Aranda MA. 2009. Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J Virol 83:12378–12387. doi: 10.1128/JVI.01486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim MS, Kim MJ, Hong JS, Choi JK, Ryu KH. 2010. Patterns in disease progress and the influence of single and multiple viral infections on pepper (Capsicum annuum L.) growth. Eur J Plant Pathol 127:53–61. doi: 10.1007/s10658-009-9570-8. [DOI] [Google Scholar]
- 68.Malpica JM, Sacristan S, Fraile A, García-Arenal F. 2006. Association and host selectivity in multi-host pathogens. PLoS One 1:e41. doi: 10.1371/journal.pone.0000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Escriu F, Fraile A, Garcia-Arenal F. 2003. The evolution of virulence in a plant virus. Evolution 57:755–765. doi: 10.1111/j.0014-3820.2003.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 70.Doumayrou J, Leblaye S, Froissart R, Michalakis Y. 2013. Reduction of leaf area and symptom severity as proxies of disease-induced plant mortality: The example of the Cauliflower mosaic virus infecting two Brassicaceae hosts. Virus Res 176:91–100. doi: 10.1016/j.virusres.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Alizon S, Hurford A, Mideo N, Van Baalen M. 2009. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol 22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 72.García-Arenal F, Fraile A. 2013. Trade-offs in host range evolution of plant viruses. Plant Pathol 62:2–9. doi: 10.1111/ppa.12104. [DOI] [Google Scholar]
- 73.Van den Bosch F, Akudibilah G, Seal S, Jeger M. 2006. Host resistance and the evolutionary response of plant viruses. J Appl Ecol 43:506–516. doi: 10.1111/j.1365-2664.2006.01159.x. [DOI] [Google Scholar]
- 74.Sorho F, Pinel A, Traore O, Bersoult A, Ghesquiere A, Hebrard E, Konate G, Sere Y, Fargette D. 2005. Durability of natural and transgenic resistances in rice to Rice yellow mottle virus. Eur J Plant Pathol 112:349–359. doi: 10.1007/s10658-005-6607-5. [DOI] [Google Scholar]
- 75.Lo Iacono G, van den Bosch F, Gilligan CA. 2013. Durable resistance to crop pathogens: an epidemiological framework to predict risk under uncertainty. PLoS Comput Biol 9:e1002870. doi: 10.1371/journal.pcbi.1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]