ABSTRACT
Avian leukosis virus (ALV) induces tumors by integrating its proviral DNA into the chicken genome and altering the expression of nearby genes via strong promoter and enhancer elements. Viral integration sites that contribute to oncogenesis are selected in tumor cells. Deep-sequencing analysis of B-cell lymphoma DNA confirmed that the telomerase reverse transcriptase (TERT) gene promoter is a common ALV integration target. Twenty-six unique proviral integration sites were mapped between 46 and 3,552 nucleotides (nt) upstream of the TERT transcription start site, predominantly in the opposite transcriptional orientation to TERT. Transcriptome-sequencing (RNA-seq) analysis of normal bursa revealed a transcribed region upstream of TERT in the opposite orientation, suggesting the TERT promoter is bidirectional. This transcript appears to be an uncharacterized antisense RNA. We have previously shown that TERT expression is upregulated in tumors with integrations in the TERT promoter region. We now report that the viral promoter drives the expression of a chimeric transcript containing viral sequences spliced to exons 4 through 7 of this antisense RNA. Clonal expansion of cells with ALV integrations driving overexpression of the TERT antisense RNA suggest it may have a role in tumorigenesis.
IMPORTANCE The data suggest that ALV integrations in the TERT promoter region drive the overexpression of a novel antisense RNA and contribute to the development of lymphomas.
INTRODUCTION
Avian leukosis virus (ALV) is a simple retrovirus that does not carry a viral oncogene but induces tumors by insertional mutagenesis (1, 2). ALV typically induces B-cell lymphomas, but can also induce erythroblastomas, hemangiomas, and myeloid tumors (1–3). Proviral integration can upregulate the expression of nearby genes via strong enhancers and promoter elements in the viral long terminal repeat (LTR) sequences. An advantage of using ALV as an insertional-mutagenesis tool is its relatively random integration pattern, with only a slight preference for actively transcribed sites (4–6). A reduced integration bias allows us to map proviral integrations in many genomic locations and to observe the selection of integration sites with oncogenic potential. Previous studies have shown common integration sites in ALV-induced lymphomas in the MYC, MYB, BIC (miR 155 precursor), and telomerase reverse transcriptase (TERT) genes (6–10). High-throughput sequencing revealed multiple integration sites in a series of rapid-onset B-cell lymphomas (6). The TERT promoter region was identified as the most clonally expanded of these integrations, suggesting this is an early event in tumorigenesis (6). Twenty-six unique integration sites were identified in the region in multiple independent tumors (6).
Telomerase is a ribonucleoprotein complex that adds repeat sequences to chromosome ends. It contains a catalytic protein component, TERT, as well as a noncoding telomerase RNA template component (TERC). Elevated telomerase activity has been detected in more than 90% of all human cancers (11). In addition, many human tumors have a point mutation in the TERT promoter at nucleotide (nt) −124 or −146 upstream of the TERT translation start site (12). These mutations upregulate TERT expression (13–16). Elevated telomerase activity maintains telomere lengths and prevents apoptotic signaling, thus allowing continual proliferation and long-term viability of cancer cells (17). It has also been shown that TERT can promote oncogenesis independently of the reverse transcriptase function of telomerase (18).
Telomerase activity in most somatic cells is limited by the availability of the TERT protein, and expression of TERT is tightly regulated at the transcriptional level through epigenetic modifications in the promoter region (19, 20). In addition, extensive alternative splicing of the TERT transcript has been shown to generate inactive variants that decrease telomerase activity (21–23). Both human and chicken telomerase expression is downregulated in most normal somatic tissues (24, 25). Furthermore, human and chicken telomeres shorten with age, and telomerase activity is important for oncogenesis (26). In contrast, mice express telomerase in normal somatic cells and have longer telomeres than humans or chickens (27). Therefore, the chicken serves as a good model to study oncogenic events of TERT activation and signaling.
We previously reported that ALV integrations upstream of TERT cause 2- to 4-fold upregulation of TERT expression in rapid-onset B-cell lymphomas (10). However, these integrations were in the opposite transcriptional orientation to TERT, unlike most previously characterized common integration sites in ALV-induced tumors (7, 9, 28). In this work, we show that these integrations also drive the overexpression of a novel antisense transcript associated with the bidirectional TERT promoter, which we call TAPAS (TERT antisense promoter associated) RNA. The ALV integrations result in chimeric transcripts with ALV leader sequences spliced into exon 4 of the 7-exon TAPAS RNA.
MATERIALS AND METHODS
Tumor induction.
All of the B-cell lymphomas included in the study were rapid-onset lymphomas induced by either the wild type (WT) or variants of LR-9 virus infections in 10-day-old chicken embryos, as described previously (29). LR-9 is an ALV subgroup A recombinant virus consisting of gag, pol, and env genes derived from UR2-associated virus and LTRs derived from ring-necked pheasant virus (30). Tumors were collected from primary bursal (B) tumors or metastasized liver (L) tumors. A1B was induced by ΔLR-9, with a deletion in the gag gene, causing increased splicing to downstream genes (31). Tumors C2B, C2L, C6L, C7B, and C7L were induced by infection with LR-9 containing a silent mutation, G919A, which induces a higher incidence of rapid-onset lymphomas (29), probably due to increased readthrough and splicing to downstream genes (32). Tumor D2L was induced by WT LR-9 (30).
DNA and RNA isolation.
Genomic DNA was prepared by standard proteinase K digestion followed by phenol-chloroform extraction, as described previously (3). RNA was extracted from tissue homogenates using RNA Bee extraction agent (Tel-Test, Inc., Friendswood, TX).
Reverse transcription, PCR amplification, and Sanger sequencing.
Total RNA was reverse transcribed with Maxima H minus reverse transcriptase (ThermoFisher Scientific) and oligo(dT)18 primer and/or random hexamers, following the manufacturer's protocol. Tumor C2L was excluded from analysis due to poor RNA quality. All the PCR primers were commercially synthesized (Integrated DNA Technologies, Inc.). 3′ rapid amplification of cDNA ends (RACE) was performed with the lock-docking oligo(dT) primer (33). PCR amplification was performed with Phusion High-Fidelity DNA polymerase (New England BioLabs), following the manufacturer's instructions. The amplified fragments were gel purified using a gel extraction kit (Qiagen) and sequenced (Eurofins MWG Operon LLC).
Detection of splice variants and ALV-TAPAS RNA fusion transcripts.
Spliced variants were detected using primers in the antisense direction in TERT exon 1 and in exon 7 of TAPAS RNA (TGGCCTCGGCGTAGCAG and CAAATGGCTTGTCTGCATTTTCTTC). Fusion transcripts were amplified using a 5′ primer positioned immediately before the viral splice donor or complementary to sequences in the R region of the viral LTR (TCAAGCATGGAAGCCGTCATAAAG and GCCATTTGACCATTCACCACATTG) and a set of 3′ primers located throughout the TAPAS RNA gene sequence (CAAATGGCTTGTCTGCATTTTCTTC, CCAAAGCCACGGCTTCCATGTTAGTATC, and TAAGGTGGAGAATAAGACATAATAATATGAGATGAG).
High-throughput sequencing.
DNA libraries for deep sequencing were prepared and analyzed as previously described (3, 6). Transcriptome-sequencing (RNA-seq) libraries were prepared according to a previously published protocol (34). The reads were aligned to the chicken genome (galGal4) using TopHat (35). Splice junctions enabled the determination of the orientations of spliced transcripts. Additional RNA-seq data for analysis of tissue distribution and embryonic expression of TAPAS RNA and TERT in chickens were downloaded from the public Sequence Read Archive (SRA) database (SRA accession no. ERX697750 and DRX001564) (36). The abundances of transcripts (fragments per kilobase per million [FPKM]) were estimated and compared using Cufflinks (37).
Quantitative PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was performed using iQ SYBR green Supermix (Bio-Rad) according to the manufacturer's protocol on a Bio-Rad C1000 thermal cycler/CFX96 Real-Time System. The expression of TAPAS RNA in chicken tissue was measured using primers in exons 4 and 5 (CAGACTACTTTACCTCTTGACACAG and ATGGTGAGCCTTGTGTTGGC). TERT expression was measured using primers in exons 11 and 12 (AACATGAAATGCAAATTGACTGC and ACTGTCTGAAGGCTGTTGATCT). Expression was normalized to the expression of ribosomal protein L30 (RPL30) using exon junction primers (38). Quantitative PCR (qPCR) was performed in triplicate, with each sample present in technical duplicate during each run. The results were normalized to those for normal bursa using the comparative threshold cycle (CT) method.
Evolutionary analysis.
The sequence encompassing exons 4 to 6 and intervening introns of the chicken TAPAS gene was analyzed via BLASTN against the whole-genome shotgun database at the NCBI website (http://blast.ncbi.nlm.nih.gov/). All high-quality matches with an E value lower than 10−40 were retrieved and further analyzed. Sequences were aligned and plotted by the maximum-likelihood method with PhyML, utilizing the GTR substitution and aBayes branch support (39).
RESULTS
The TERT promoter is a common ALV integration site in B-cell lymphomas.
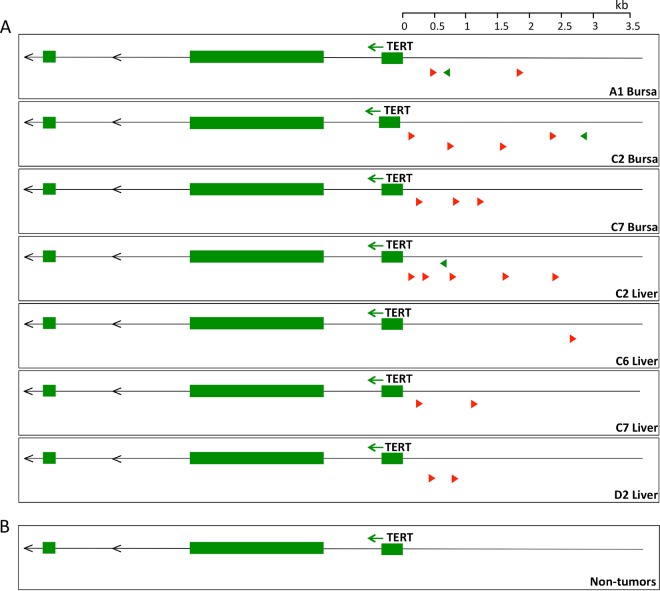
In order to identify genes contributing to the formation of ALV-A (subgroup A)-induced rapid-onset B-cell lymphomas, high-throughput sequencing of proviral-host DNA junctions was previously performed (6). Common integration sites in the host genome that contribute to tumorigenesis are present in multiple tumor cells and thus are overrepresented in the deep-sequencing data. The TERT promoter region was identified as the most clonally expanded common integration site, with integrations present in seven different lymphomas from five birds (Fig. 1A). We analyzed 19 of the most clonally expanded unique integrations from both primary bursal tumors (B) and tumors metastasized to the liver (L). Three of the clonally expanded integrations were present in multiple tissues from the same bird. The integration sites ranged from 46 nt to 3,552 nt upstream of the TERT transcription start site. The majority of the proviral integrations (16/19) were in the opposite transcriptional orientation to TERT. Four out of 7 lymphomas, termed C7B, C6L, C7L, and D2L, had integrations only in the opposite orientation. The remaining three tumors—A1B, C2B, and C2L—harbored integrations in the same orientation as TERT but also contained integrations in the opposite orientation that were more clonally expanded. In contrast, no integrations in the TERT promoter region were identified in any nontumor tissues of infected birds (Fig. 1B). The observation of proviral integrations in this region in multiple tumors suggests that ALV integration in the TERT promoter contributes to driving lymphomagenesis in these birds.
FIG 1.
The TERT promoter region is a common site of ALV proviral integration in lymphomas. (A) Schematic of the most clonally expanded ALV integration sites near TERT in 7 tumors, shown with the first 3 exons of the TERT gene. The tumor names correspond to the bird and tissue from which the tumor was collected. All of the integrations clustered within 3.5 kb upstream of the TERT transcriptional start site. The integrations are predominantly in the opposite orientation (red) with respect to TERT transcription. (B) Schematic of integrations near TERT in 6 nontumor tissues from infected chickens.
Novel antisense (TAPAS) RNA is transcribed from the TERT bidirectional promoter.
In order to assess the effects of proviral integrations on host gene expression, deep sequencing of the transcriptomes of selected ALV-induced lymphomas and normal-bursa controls was performed. This analysis revealed a 9-kb transcribed region upstream of and in the opposite transcriptional orientation to TERT in the normal-bursa controls (Fig. 2A). This suggests that the TERT promoter is bidirectional. With the use of TopHat bioinformatics tools, a number of putative introns were identified and confirmed by sequence analysis of exon junctions. This analysis suggested a 3.6-kb spliced transcript containing 7 exons. RT-PCR studies confirmed 2.2 kb of this transcript containing exons 1 through 7 (Fig. 2A). The RT-PCR experiments were not able to amplify the last 1,050 nt of exon 7. Two putative poly(A) sites were identified by 3′ RACE at positions 1051 and 1114 of exon 7.
FIG 2.
Schematic of the TAPAS gene. (A) Representative RNA-seq (Bedgraph) from normal bursa tissue showing normalized transcription coverage. Coverage on the plus (green) and minus (blue) strands is shown. The primary transcript observed by RNA-seq in normal chicken bursa is approximately 9 kb. The principal form of the spliced transcript is 3.6 kb and contains 7 exons. The confirmed region of the shared exon 1 is hatched in blue. A portion of exon 7 that could not be verified by RT-PCR is indicated in light blue. Transcripts confirmed by RT-PCR are 2 to 3 kb. The two main 3′ ends identified are indicated by vertical orange lines and are located at nucleotides 1051 and 1114 of exon 7. (B) Multiple alternatively spliced variants of TAPAS RNA were also identified in normal bursa by RT-PCR.
Strand-specific RNA-seq data indicated that the first exon of TERT and the associated bidirectional transcript overlap (Fig. 2A). RT-PCR verified that at least the first 161 nt of TERT exon 1 are shared with TAPAS RNA. It is possible that more of exon 1 is shared between the two genes; however, this could not be confirmed by RT-PCR, likely due to the high GC content of TERT exon 1. Additionally, a number of alternatively spliced transcripts were detected, including some skipping exon 2 and others skipping both exons 2 and 3 (Fig. 2B).
There is a small open reading frame (ORF) (258 nt) that spans exons 4 and 5 and two longer ORFs, of 408 and 375 nt, present in exon 7. However, one of these ORFs is located within the unverified region at the 3′ end of the transcript beyond the main transcription termination sites discussed above. Further, we observed that exon 7 is poorly conserved between most avian species (data not shown). Moreover, no protein domain homology was observed in any region of the transcript (40), implicating the transcript as a putative long noncoding RNA (lncRNA).
The recent release of the Gallus gallus 5 whole-genome assembly predicts an antisense transcript upstream of TERT (gene ID LOC107052651). The predicted transcript variant (XR_001465267.1) corresponds to exons 2 through 7 of TAPAS RNA (Fig. 2A). This variant contains 643 nt more of exon 7 and does not share any sequence with TERT exon 1, unlike the transcript reported here. Another transcript variant with retention of an intron between our exons 2 and 3 is also predicted (XR_001465266.1). Further, the NCBI eukaryotic gene prediction tool, Gnomon, annotates the predicted transcript as an lncRNA (accession no. NC_006089).
TAPAS RNA expression is elevated in tumors with integrations in the TERT promoter.
The predominance of proviral integrations in the opposite orientation to TERT, as well as the identification of a bidirectional transcript, suggested that the integrations might also drive increased expression of TAPAS RNA. To test this hypothesis, we performed qRT-PCR to determine TAPAS RNA expression levels in tumors containing integrations in the TERT promoter region (Fig. 3A). Normal liver has 148- and 5-fold less expression than normal bursa for TAPAS and TERT RNAs, respectively. Compared to normal liver, tumors with integrations in the TERT promoter had significantly increased expression of the TAPAS RNA. Expression of the bidirectional TAPAS RNA was upregulated approximately 250- to 3,858-fold relative to normal liver tissue. In contrast, TERT was upregulated 4- to 42-fold relative to normal liver tissue (Fig. 3B). This suggests that the observed integrations in the TERT promoter drive expression of a bidirectional lncRNA. These findings were also confirmed by RNA-seq analysis of liver tumors C6, C7, and D2 (data not shown).
FIG 3.
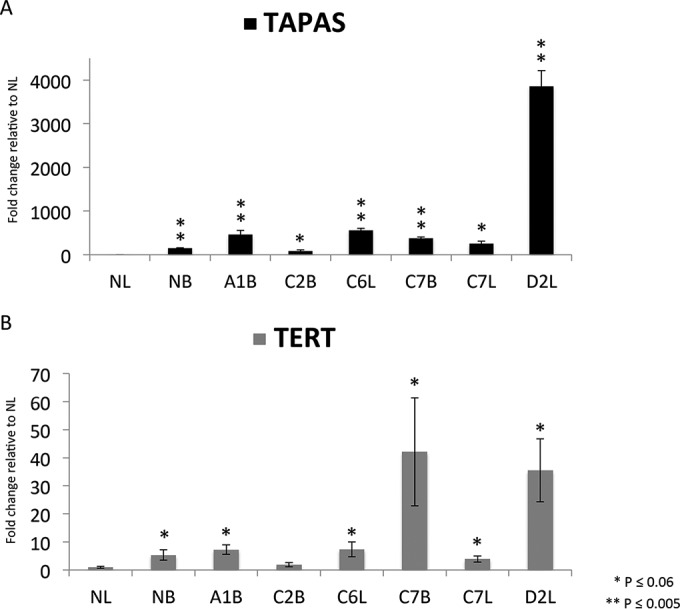
Expression of TAPAS RNA and TERT in ALV-induced B-cell lymphomas. qRT-PCR was performed to determine TAPAS RNA (A) and TERT (B) expression in seven ALV-induced tumors, as well as normal bursa (NB) and normal liver (NL) controls. Expression of both transcripts was significantly higher in 5 of the 6 tumors than in NL. The P values are representative of Bonferroni corrections for multiple comparisons. The error bars indicate standard deviations.
Viral transcripts splice into exon 4 of TAPAS RNA.
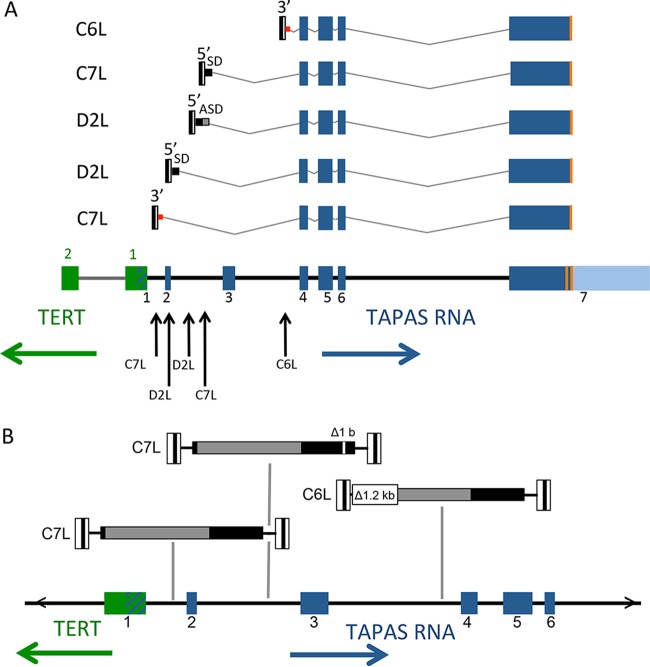
Retroviruses can induce overexpression of host genes by multiple mechanisms (1, 41). For instance, insertion of viral enhancer elements in the vicinity of host gene promoters can induce overexpression (1). Alternatively, the viral promoter can drive expression of the host gene directly, if both are in the same orientation, by readthrough of the viral poly(A) site (28). If the promoter in the viral 5′ LTR is driving expression, the viral RNA transcript can splice via the gag splice donor into the cellular mRNA (28). Alternatively, if the promoter in the 3′ LTR is used, readthrough into the adjacent host genomic region occurs (7). To determine the mechanism by which proviral integrations affect TAPAS RNA expression, we analyzed metastasized tumors with integrations in the same transcriptional orientation as the TAPAS RNA. We performed RT-PCR using LTR-specific primers and primers within the TAPAS RNA exons to obtain and sequence viral TAPAS RNA fused transcripts (Fig. 4A).
FIG 4.
Viral RNAs splice into exon 4 of TAPAS RNA. (A) Splicing of viral transcripts was determined by RT-PCR of tumor RNA. All proviruses shown were in the opposite transcriptional orientation to TERT. The arrows indicate the genomic locations of proviral integration. Despite the presence of upstream exons, all the viral transcripts analyzed spliced into exon 4 of the TAPAS RNA either from a canonical gag splice donor (SD) or via alternate splice donor (ASD) sites. Readthrough transcription from the 3′ LTR is depicted by the red square. (B) Sequence analysis showing mutations in proviruses C6L and one of the C7L integrations. Three integrated proviruses, two in tumor C7L and one in tumor C6L, were sequenced. The viral LTRs are depicted as white boxes at the termini of the viral genome with the U3, R, and U5 direct repeats. gag-pol are depicted in gray and env in black. The 1.2-kb deletion in C6L removes the canonical viral splice donor and induces transcription from the 3′ LTR. The exon positions of TERT and TAPAS RNA are shown for reference.
Proviruses in tumor D2L use the 5′ LTR to drive expression of TAPAS RNA. One splice variant used the canonical 5′ viral splice donor site in gag (nucleotide 398). Transcripts were also detected in which an alternative splice donor site in the viral gag gene (nucleotide 857) spliced into exon 4 of TAPAS RNA. In tumor C7L, a provirus integrated in intron 2 also spliced into exon 4 of TAPAS RNA from the canonical 5′ viral splice donor site.
Alternatively, transcripts in which the viral 3′ LTR served as the promoter were observed in tumor C6L. These transcripts contained 63 nucleotides of host DNA adjacent to the 3′ LTR. It appears that a cryptic splice donor site present in this intronic region may be used to splice into the downstream exon 4. Sequencing of the provirus C6L revealed a large deletion that included the viral splice donor site in gag (Fig. 4B); this would prevent its splicing into the TAPAS RNA if it initiated in the 5′ LTR. However, the deletion would probably also prevent transcription initiation at the 5′ LTR, as previously observed with ALV integrations in MYC (42). A similar 3′ LTR-initiated transcript was observed from the C7L proviral integration in intron 1 of TAPAS RNA. This variant spliced from a cryptic splice donor site 28 nucleotides downstream of the provirus into exon 4 of the TAPAS RNA.
The majority of proviral integrations are located between exon 1 and exon 4 of the TAPAS gene (Fig. 4A). However, regardless of the integration site location, all of the viral transcripts analyzed invariantly spliced into exon 4 of the TAPAS RNA. For most of the viral transcripts, this means that nearby exons are skipped and splicing is preferentially to exon 4. While the 5′ spliced viral leader sequence contains a bit of the ALV gag gene, the analyzed chimeric transcripts have a termination codon before the AUG of the ORF in exon 4 of the TAPAS RNA. Thus, no hybrid protein is predicted. Consistent splicing into exon 4 suggests the possible functional importance of this region of the TAPAS RNA.
TAPAS RNA is expressed in normal chicken tissues and during development.
To further characterize the expression of the TAPAS RNA, we analyzed publicly available RNA-seq data sets from the SRA database (36). TAPAS RNA and TERT expression was measured in various chicken tissues of an 18-day embryo. In addition, transcriptomes of total embryos were analyzed at different time points up to 12 days of development (SRA accession no. ERX697750 and DRX001564) (36). Quantification of transcript expression was performed by calculating the fragment count normalized to the transcript length and the total number of reads (FPKM). This analysis showed that the TAPAS RNA is expressed in some normal chicken embryo tissues. TAPAS RNA was expressed at particularly high levels in bursa, testes, and kidney and was undetectable in muscle and heart tissue (Fig. 5A). TAPAS RNA expression was higher than that of TERT in bursa and testes but more comparable in other tissues and in total early embryos.
FIG 5.
TERT and TAPAS RNA are expressed at comparable levels in adult tissues and during chick development. (A) RNA-seq data on expression of TAPAS RNA in normal 18-day chicken embryo tissues. TAPAS RNA is expressed in various tissues at levels similar to those of TERT. In bursa, TAPAS RNA expression is approximately 3-fold higher than that of TERT (Pearson correlation coefficient = 0.66; P = 0.07). (B) The expression of TERT and TAPAS RNA also seem to be correlated throughout development (Pearson correlation coefficient = 0.92; P = 0.001). The error bars indicate standard deviations.
Furthermore, both TAPAS RNA and TERT expression were elevated early in chick development and progressively decreased with time (Fig. 5B). Thus, the tissue-specific and developmental expression of TAPAS RNA correlates with TERT expression. This suggests that the TAPAS RNA may have a role in regulating TERT expression or that the expression of the two transcripts is coregulated.
TAPAS RNA is conserved.
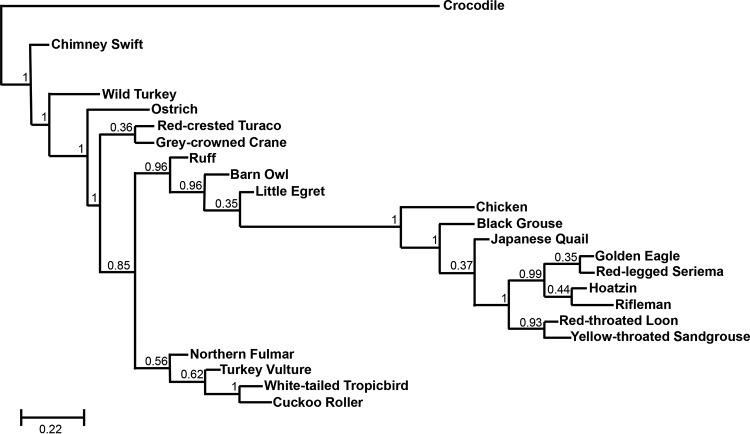
To determine if the novel TAPAS RNA is conserved, we performed phylogenetic analysis in multiple avian species. Exons 4, 5, and 6 were used for the analysis because this region was found in all alternatively spliced transcripts, as well as in all viral TAPAS RNA fused transcripts, suggesting it may have functional importance. Regions homologous to the TAPAS gene exons 4 to 6 and intervening introns were identified in various avian genomes by BLASTN, and the sequences were aligned by ClustalX. It was observed that the region is highly conserved at the sequence level in many, but not all, avian lineages (Fig. 6). Additionally, based on transcriptome analysis, there exists a predicted lncRNA in the TERT promoter region in the most recent genome assemblies for chicken (LOC107052651), turkey (LOC104910189), and Japanese quail (LOC107309454) (data not shown). The chicken sequence shared 95% identity with the corresponding genomic region of the closely related black grouse (Lyrurus tetrix) and 72 to 76% identity with the genomes of various neoavian lineages. The turkey and chicken sequences share limited similarities in exons 4 to 6 but have significant sequence homology in exon 7. The only perching bird species in which this region was conserved was the most basal species of New Zealand wren (Acanthisitta chloris), suggesting the possibility that these sequences underwent rapid evolutionary changes in Passeriformes. The ORF in exons 4 and 5 was not conserved in most birds (data not shown). Further, we did not find any regions homologous to the chicken TAPAS RNA in mammalian genomes at the sequence level. Exon 6 was the most conserved in avian species, with 72 to 96% identity. The splice donor sites of exon 5, as well as the donor and acceptor sites of exon 6, are perfectly conserved in all species analyzed (data not shown).
FIG 6.
The TAPAS gene is conserved in avian species. Shown is phylogenetic analysis of exons 4 to 6 and the intervening introns of the TAPAS gene in several different avian species. Crocodile is depicted as an outgroup. The numbers at nodes of the tree are posterior probability values from a maximum-likelihood approach as described in Materials and Methods.
DISCUSSION
In order to better understand tumor pathogenesis, we analyzed the distribution of ALV integration sites in chicken B-cell lymphomas using a high-throughput sequencing strategy. We identified numerous clonally expanded proviral integrations in the TERT promoter region associated with elevated TERT expression, suggesting these integrations may promote tumorigenesis (6, 10). Previously, ALV subgroup J proviral integrations have been observed near TERT in chicken myeloid tumors (43). Other studies have reported integrations of hepatitis B virus and human papillomavirus DNAs in the TERT promoter region in human liver and cervical tumors, resulting in elevated TERT expression (44). While sense and antisense retroviral activation has been previously reported (1, 41), to our knowledge, this is the first report of retroviral activation of a bidirectional promoter-associated lncRNA. In this work, we show that most of the proviral integrations in our tumors are in the antisense direction relative to TERT and drive the overexpression of a novel lncRNA transcribed from a bidirectional promoter. The prevalence of integrations in the same orientation as the TAPAS RNA in multiple tumors suggests that overexpression of the transcript may have contributed to oncogenic transformation. We further characterize this novel transcript and show that, in addition to being conserved among some avian lineages, expression of the transcript is correlated with TERT expression in various tissues and throughout development in chickens.
Deep sequencing of transcriptomes from various species, including humans, revealed the presence of a transcribed region upstream of TERT (data not shown). We found that in chickens it corresponds to an alternatively spliced, polyadenylated, antisense lncRNA, leading us to propose that the TERT promoter is bidirectional. Bidirectional promoters have been found to be pervasive in mammalian genomes, with more than 10% of known protein-coding genes being transcribed from such promoters (45–47). In humans, many lncRNAs are transcribed from bidirectional promoters. Approximately 65% of identified human lncRNAs originate within 2 kb of transcription start sites, and almost all of them are transcribed in the antisense direction relative to the associated protein-coding genes (48). Furthermore, bidirectional promoters are significantly more likely to regulate DNA repair genes and genes implicated in somatic cancers (49).
The chicken TERT promoter has many features characteristic of bidirectional promoters, such as the absence of a TATA box and high GC content. The most important feature of a bidirectional promoter seems to be the presence of binding sites for an Ets family transcription factor, GABP (46, 50). Interestingly, recurrent promoter mutations present in the human TERT promoter have been shown to introduce a de novo Ets binding site, which has been shown to drive TERT expression (15, 16, 51). It has recently been shown that it is the specific Ets factor, GABP, that selectively binds the mutant TERT promoter (51). Thus, it is possible that, because GABP selectively binds the promoter mutation, it may induce bidirectional transcription of an antisense transcript, similar to what has been observed in chickens. There is evidence for antisense transcription in the TERT promoter region in humans from deep-sequencing data (data not shown). However, there is no conservation between this putative transcript and the chicken transcript. Further, RNA abundance in the region is much lower relative to TERT than what is observed in chickens.
Many lncRNAs are known to play a role in transcriptional regulation. For example, XIST acts in cis to recruit the polycomb repressive complex 2 (PRC2) to chromosome X, causing gene silencing (52). HOTAIR, on the other hand, acts in trans to repress the expression of genes in the HoxD gene cluster (53). It has been proposed that antisense lncRNAs transcribed from bidirectional promoters may be involved in the regulation of the associated sense transcripts (47, 54). Such an arrangement may allow tighter transcriptional regulation. Based on the correlation between TERT and lncRNA expression in normal tissues and tumors, we hypothesize that the lncRNA may regulate TERT expression. However, it is also possible that TERT expression and lncRNA expression are only correlated due to mutual regulation by the same bidirectional promoter. This raises the possibility that the lncRNA may have an independent function apart from TERT regulation.
Given the important role of lncRNAs in transcriptional regulation, it comes as no surprise that many lncRNAs have been implicated in cancer and disease. With the advent of deep sequencing, many novel lncRNAs have been found to be associated with cancer pathogenesis. For example, HOTAIR, MALAT1, PCAT-1, PCGEM1, and TUC338 were reported to be oncogenes, while GAS5, MEG3, and PTENP1 were reported to be tumor suppressors (55). Since lncRNA functions range from cell growth to cancer development, they represent important biological players that merit further research.
In this study, we identified a novel antisense lncRNA transcribed from the TERT promoter. We show that this lncRNA is upregulated in tumors and is implicated in tumorigenesis. Further characterization of the structural and functional motifs of this TAPAS RNA is required to better understand its mechanistic and functional roles in cancer signaling. This will help elucidate possible gene-regulatory mechanisms of lncRNAs and may provide further insight into the role lncRNAs play in cancer pathogenesis.
ACKNOWLEDGMENTS
We thank Paul E. Neiman and the late Sandra J. Bowers, Fred Hutchinson Cancer Research Center, Seattle, WA, for generation of the tumors. We also thank Elizabeth Lake Potter for help with bioinformatics analysis of the avian lineages.
REFERENCES
- 1.Beemon K, Rosenberg N. 2012. Mechanisms of oncogenesis by avian and murine retroviruses, p 677–704. In Robertson ES. (ed), Cancer associated viruses. Springer, New York, NY. [Google Scholar]
- 2.Justice J, Beemon KL. 2013. Avian retroviral replication. Curr Opin Virol 3:664–669. doi: 10.1016/j.coviro.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Justice J, Malhotra S, Ruano M, Li Y, Zavala G, Lee N, Morgan R, Beemon K. 2015. The MET gene is a common integration target in avian leukosis virus subgroup J-induced chicken hemangiomas. J Virol 89:4712–4719. doi: 10.1128/JVI.03225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J Virol 78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. 2005. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J Virol 79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Justice JF, Morgan RW, Beemon KL. 2015. Common viral integration sites identified in avian leukosis virus-induced B-cell lymphomas. mBio 6:e01863–15. doi: 10.1128/mBio.01863-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayward WS, Neel BG, Astrin SM. 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 8.Baba TW, Humphries EH. 1986. Selective integration of avian leukosis virus in different hematopoietic tissues. Virology 155:557–566. doi: 10.1016/0042-6822(86)90216-3. [DOI] [PubMed] [Google Scholar]
- 9.Clurman BE, Hayward WS. 1989. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol 9:2657–2664. doi: 10.1128/MCB.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Xian RR, Li Y, Polony TS, Beemon KL. 2007. Telomerase reverse transcriptase expression elevated by avian leukosis virus integration in B cell lymphomas. Proc Natl Acad Sci U S A 104:18952–18957. doi: 10.1073/pnas.0709173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shay JW, Wright WE. 2011. Role of telomeres and telomerase in cancer. Semin Cancer Biol 21:349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. 2014. TERT promoter mutations in cancer development. Curr Opin Genet Dev 24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. 2015. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He T-C, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ, Rasheed BA, Riggins GJ, Rosenquist TA, Schiffman M, Shih I-M, Theodorescu D, Torbenson MS, Velculescu VE, Wang T-L, Wentzensen N, Wood LD, Zhang M, McLendon RE, Bigner DD, Kinzler KW, Vogelstein B, Papadopoulos N, Yan H. 2013. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. 2013. Highly recurrent TERT promoter mutations in human melanoma. Science 339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. 2013. TERT promoter mutations in familial and sporadic melanoma. Science 339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 17.Blasco MA. 2005. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet 6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 18.Koh CM, Khattar E, Leow SC, Liu CY, Muller J, Ang WX, Li Y, Franzoso G, Li S, Guccione E, Tergaonkar V. 2015. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J Clin Invest 125:2109–2122. doi: 10.1172/JCI79134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J, Zhao Y, Wang S. 2010. Chromatin and epigenetic regulation of the telomerase reverse transcriptase gene. Protein Cell 1:22–32. doi: 10.1007/s13238-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delany ME, Daniels LM. 2004. The chicken telomerase reverse transcriptase (chTERT): molecular and cytogenetic characterization with a comparative analysis. Gene 339:61–69. doi: 10.1016/j.gene.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Withers JB, Ashvetiya T, Beemon KL. 2012. Exclusion of exon 2 is a common mRNA splice variant of primate telomerase reverse transcriptases. PLoS One 7:e48016. doi: 10.1371/journal.pone.0048016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hrdlicková R, Nehyba J, Bose HR. 2012. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol Cell Biol 32:4283–4296. doi: 10.1128/MCB.00550-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saebøe-Larssen S, Fossberg E, Gaudernack G. 2006. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol Biol 7:26. doi: 10.1186/1471-2199-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins K, Mitchell JR. 2002. Telomerase in the human organism. Oncogene 21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 25.Taylor HA, Delany ME. 2000. Ontogeny of telomerase in chicken: impact of downregulation on pre- and postnatal telomere length in vivo. Dev Growth Differ 42:613–621. doi: 10.1046/j.1440-169x.2000.00540.x. [DOI] [PubMed] [Google Scholar]
- 26.Delany ME, Krupkin AB, Miller MM. 2000. Organization of telomere sequences in birds: evidence for arrays of extreme length and for in vivo shortening. Cytogenet Cell Genet 90:139–145. doi: 10.1159/000015649. [DOI] [PubMed] [Google Scholar]
- 27.Hackett JA, Greider CW. 2002. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene 21:619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 28.Kanter MR, Smith RE, Hayward WS. 1988. Rapid induction of B-cell lymphomas: insertional activation of c-myb by avian leukosis virus. J Virol 62:1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polony TS, Bowers SJ, Neiman PE, Beemon KL. 2003. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J Virol 77:9378–9387. doi: 10.1128/JVI.77.17.9378-9387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon MC, Neckameyer WS, Hayward WS, Smith RE. 1987. Genetic determinants of neoplastic diseases induced by a subgroup F avian leukosis virus. J Virol 61:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith MR, Smith RE, Dunkel I, Hou V, Beemon KL, Hayward WS. 1997. Genetic determinant of rapid-onset B-cell lymphoma by avian leukosis virus. J Virol 71:6534–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Sullivan CT, Polony TS, Paca RE, Beemon KL. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses SRC mRNA splicing and promotes polyadenylation. Virology 302:405–412. doi: 10.1006/viro.2002.1616. [DOI] [PubMed] [Google Scholar]
- 33.Borson ND, Salo WL, Drewes LR. 1992. A lock-docking oligo(dT) primer for 5′ and 3′ RACE PCR. PCR Methods Appl 2:144–148. doi: 10.1101/gr.2.2.144. [DOI] [PubMed] [Google Scholar]
- 34.Nagalakshmi U, Waern K, Snyder M. 2010. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol Chapter 4:Unit 4.11.1-13. doi: 10.1002/0471142727.mb0411s89. [DOI] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leinonen R, Sugawara H, Shumway M. 2011. The sequence read archive. Nucleic Acids Res 39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Lei X, Rodriguez-Palacios A, Tang C, Yue H. 2013. Selection of reference genes for quantitative real-time PCR analysis in chicken embryo fibroblasts infected with avian leukosis virus subgroup J. BMC Res Notes 6:402. doi: 10.1186/1756-0500-6-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokol M, Wabl M, Ruiz IR, Pedersen FS. 2014. Novel principles of gamma-retroviral insertional transcription activation in murine leukemia virus-induced end-stage tumors. Retrovirology 11:36. doi: 10.1186/1742-4690-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodenow MM, Hayward WS. 1987. 5′ long terminal repeats of myc-associated proviruses appear structurally intact but are functionally impaired in tumors induced by avian leukosis viruses. J Virol 61:2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Liu X, Yang Z, Xu C, Liu D, Qin J, Dai M, Hao J, Feng M, Huang X, Tan L, Cao W, Liao M. 2014. The MYC, TERT, and ZIC1 genes are common targets of viral integration and transcriptional deregulation in avian leukosis virus subgroup J-induced myeloid leukosis. J Virol 88:3182–3191. doi: 10.1128/JVI.02995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferber MJ, Montoya DP, Yu C, Aderca I, McGee A, Thorland EC, Nagorney DM, Gostout BS, Burgart LJ, Boix L, Bruix J, McMahon BJ, Cheung TH, Chung TKH, Wong YF, Smith DI, Roberts LR. 2003. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene 22:3813–3820. doi: 10.1038/sj.onc.1206528. [DOI] [PubMed] [Google Scholar]
- 45.Trinklein NT, Force Aldred S, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. 2004. An abundance of bidirectional promoters in the human genome. Genome Res 14:62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orekhova AS, Rubtsov PM. 2013. Bidirectional promoters in the transcription of mammalian genomes. Biochemistry 78:335–341. doi: 10.1134/S0006297913040020. [DOI] [PubMed] [Google Scholar]
- 47.Wakano C, Byun JS, Di LJ, Gardner K. 2012. The dual lives of bidirectional promoters. Biochim Biophys Acta 1819:688–693. doi: 10.1016/j.bbagrm.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. 2013. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A 110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang MQ, Koehly LM, Elnitski LL. 2007. Comprehensive annotation of bidirectional promoters identifies co-regulation among breast and ovarian cancer genes. PLoS Comput Biol 3:733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collins PJ, Kobayashi Y, Nguyen L, Trinklein ND, Myers RM. 2007. The ets-related transcription factor GABP directs bidirectional transcription. PLoS Genet 3:e208. doi: 10.1371/journal.pgen.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell RJA, Rube HT, Kreig A, Mancini A, Fouse SF, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, Wiencke JK, Wrensch MR, Chang SM, Walsh KM, Myong S, Song JS, Costello JF. 2015. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348:1036–1039. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei W, Pelechano V, Järvelin AI, Steinmetz LM. 2011. Functional consequences of bidirectional promoters. Trends Genet 27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huarte M. 2015. The emerging role of lncRNAs in cancer. Nat Med 21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]