ABSTRACT
Interleukin 2 (IL-2) signaling through the IL-2 receptor alpha chain (CD25) facilitates HIV replication in vitro and facilitates homeostatic proliferation of CD25+ FoxP3+ CD4+ T cells. CD25+ FoxP3+ CD4+ T cells may therefore constitute a suitable subset for HIV infection and plasma virion production. CD25+ FoxP3+ CD4+ T cell frequencies, absolute numbers, and the expression of CCR5 and cell cycle marker Ki67 were studied in peripheral blood from HIV+ and HIV− study volunteers. Different memory CD4+ T cell subsets were then sorted for quantification of cell-associated HIV DNA and phylogenetic analyses of the highly variable EnvV1V3 region in comparison to plasma-derived virus sequences. In HIV+ subjects, 51% (median) of CD25+ FoxP3+ CD4+ T cells expressed the HIV coreceptor CCR5. Very high frequencies of Ki67+ cells were detected in CD25+ FoxP3+ memory CD4+ T cells (median, 27.6%) in comparison to CD25− FoxP3− memory CD4+ T cells (median, 4.1%; P < 0.0001). HIV DNA content was 15-fold higher in CD25+ FoxP3+ memory CD4+ T cells than in CD25− FoxP3− T cells (P = 0.003). EnvV1V3 sequences derived from CD25+ FoxP3+ memory CD4+ T cells did not preferentially cluster with plasma-derived sequences. Quasi-identical cell-plasma sequence pairs were rare, and their proportion decreased with the estimated HIV infection duration. These data suggest that specific cellular characteristics of CD25+ FoxP3+ memory CD4+ T cells might facilitate efficient HIV infection in vivo and passage of HIV DNA to cell progeny in the absence of active viral replication. The contribution of this cell population to plasma virion production remains unclear.
IMPORTANCE Despite recent advances in the understanding of AIDS virus pathogenesis, which cell subsets support HIV infection and replication in vivo is incompletely understood. In vitro, the IL-2 signaling pathway and IL-2-dependent cell cycle induction are essential for HIV infection of stimulated T cells. CD25+ FoxP3+ memory CD4 T cells, often referred to as regulatory CD4 T cells, depend on IL-2 signaling for homeostatic proliferation in vivo. Our results show that CD25+ FoxP3+ memory CD4+ T cells often express the HIV coreceptor CCR5, are significantly more proliferative, and contain more HIV DNA than CD25− FoxP3− memory CD4 T cell subsets. The specific cellular characteristics of CD25+ FoxP3+ memory CD4+ T cells probably facilitate efficient HIV infection in vivo and passage of HIV DNA to cell progeny in the absence of active viral replication. However, the contribution of this cell subset to plasma viremia remains unclear.
INTRODUCTION
AIDS is caused by human immunodeficiency virus (HIV) infection and is characterized by the failure of the immune system to control diverse opportunistic infections facilitated by the progressive loss of CD4 T cells. The rate of CD4 T cell depletion correlates with set point levels of HIV-1 viral load in plasma (1) and is critically dependent on ongoing viral replication. Antiretroviral therapy (ART) blocks viral replication, reverses CD4 T cell depletion (2), and reconstitutes immunity to most opportunistic pathogens. Replication of HIV within CD4 T cells significantly contributes to plasma viral load and thus to HIV disease progression (3). It is well established that intracellular HIV DNA loads in vivo are influenced by CD4 T cell differentiation (4–6), functional properties of CD4 T cells (7), and pathogen specificity (8–10) and that T cell activation and proliferation contribute to productive HIV infection of memory CD4 T cells (11–15). Together these results imply that, depending on their biological properties, different CD4 T cell subsets might differ in their susceptibilities to HIV infection and their contributions to virion production in vivo. Perhaps the best characterized CD4 T cell subset in this regard is follicular CD4 T helper cells (Tfh), which are essential for germinal center formation and which reside in the periphery of B cell follicles within secondary lymphoid organs (reviewed in reference 16). Recent data demonstrate that Tfh cells are a major reservoir for HIV replication in vivo (17, 18) and contribute to persistent simian immunodeficiency virus (SIV) virion production even in elite controller, aviremic macaques (19). In viremic macaques, virion production appears to be less restricted anatomically (19) and other cell subsets are likely to contribute.
One such cell subset may be memory CD4 T cells expressing the interleukin 2 (IL-2) receptor alpha chain (CD25). Interception of IL-2 signaling, which is required for antigen-specific proliferation and survival of CD4 T cells (reviewed in reference 20) almost completely abrogates productive HIV infection in cell cultures stimulated in vitro (13, 21–23). Moreover, expression of CD25 defines a CD4 T cell population that efficiently supports productive HIV infection in lymphoid tissue explants (10, 14). In vivo, CD25 expression is characteristic for CD4 T cells (24–26) coexpressing the transcription factor forkhead box P3 (FoxP3) often referred to as regulatory T cells (Tregs). CD25+ FoxP3+ CD4 T cells can suppress the activation, proliferation, and effector functions of a wide range of immune cells, including CD4 and CD8 T cells (reviewed in reference 27), activities shown essential for the maintenance of self-tolerance but which can also impede the clearance of chronic infections (28, 29). The vast majority (>80%) of circulating CD25+ FoxP3+ CD4 T cells express the memory marker CD45RO (30, 31), and high frequencies of these cells coexpress the cell cycle marker Ki67 in peripheral blood (10 to 20%) and even more so in secondary lymphoid tissue (40 to 80%) (30, 32), indicating high levels of in vivo proliferation. The doubling time of memory CD25+ FoxP3+ CD4 T cells in humans is only 8 days, which is 3-fold and 25-fold less than that of memory and naive CD4 T cells, respectively (33). These specific cell characteristics and the proposed mechanism of constant IL-2-dependent homeostatic replenishment of this cell subset (33, 34) support the hypothesis that CD25+ FoxP3+ CD4 T cells are particularly susceptible to HIV infection in vivo and may contribute to plasma virus production in viremic HIV progressors—potentially driven by IL-2 secreted by autoantigen-specific T cells (35).
To address this hypothesis, we analyzed peripheral blood samples of HIV-positive (HIV+) and -negative (HIV−) individuals for CD25+ FoxP3+ CD4 T cell numbers and frequencies, expression of HIV coreceptor CCR5, and the cell proliferation marker Ki67 in relation to HIV infection. We have also assessed the levels of cell-associated viral DNA and the phylogenetic relationship between cell- and plasma-derived HIV envelope sequences relative to those of other memory CD4 T cell subsets. Confirming previous reports (36), our data show that high proportions of circulating CD25+ FoxP3+ CD4 T cells express the HIV coreceptor CCR5. Furthermore, memory CD25+ FoxP3+ CD4 T cells from HIV+ subjects contained high frequencies of Ki67+ cells and higher levels of HIV DNA than memory CD4 T cells that were CD25− FoxP3−. However, a phylogenetic comparison of the highly variable HIV EnvV1V3 region between plasma- and cell-derived virus sequences did not allow definite conclusions about the cellular origin of plasma virions, because sequences from the two compartments behaved similarly and intermingled with no evidence of compartmentalization. Instead, we observed that the phylogenetic distance between plasma- and memory cell-derived viral sequences increases with the duration of HIV infection, with a simultaneous decrease in the proportion of detectable quasi-identical cell-plasma sequence pairs.
MATERIALS AND METHODS
Cohorts, study volunteers, and blood processing.
The WHIS cohort comprised 361 adult volunteers who were enrolled in a prospective cohort (WHIS) that studies the interaction between HIV-1 and helminth infection in the Mbeya region in southwestern Tanzania. The WHIS cohort study is described in detail elsewhere (37). HIV status was determined using the HIV 1/2 Stat-Pak assay (Chembio Diagnostics Systems), and positive results were confirmed using an enzyme-linked immunosorbent assay (ELISA; Bio-Rad). Discrepancies between HIV 1/2 Stat-Pak and ELISA results were resolved by Western blotting (MPD HIV blot 2.2; MP Biomedicals). Forty milliliters of venous blood was drawn from each participant by the use of anticoagulant tubes (citrate phosphate dextrose adenine [CPDA], EDTA; BD Vacutainer). Absolute CD4 T cell counts were determined in anticoagulated whole blood using the BD Multitest IMK kit (BD Biosciences) according to the manufacturer's instructions. Blood samples were processed in less than 6 h after the blood draw. Frequencies of CD25+ FoxP3+ CD4 T cells and surface CCR5 expression were determined in fresh, anticoagulated whole blood as described below. The absolute numbers of CD25+ FoxP3+ CD4 T cells in the peripheral blood was calculated from the total CD4 T cell counts and the percentage of CD25+ FoxP3+ CD4 T cells. Peripheral blood mononuclear cells (PBMCs) were isolated using the Ficoll centrifugation method and LeucoSep tubes (Greiner Bio One) according to standard protocols. For the HHECO and HISIS cohorts, PBMCs were isolated from 28 HIV-positive blood donors who were recruited from a previously described cohort (HHECO) at the Komfo Anokye Teaching Hospital in Kumasi, Ghana (38, 39), and PBMCs from the previously described HISIS cohort (40) were also isolated by centrifugation of heparinized venous blood on a Ficoll-Hypaque (Biocoll separating solution; Biochrom AG, Berlin, Germany) density gradient, prior to cryopreservation.
Ethics statement.
Ethical approvals for the WHIS and HISIS cohorts were obtained from the Mbeya Regional and National Ethics Committees of the Tanzanian National Institute for Medical Research (NIMR)/Ministry of Health in Dar es Salaam, Tanzania, and from the ethics committee of the University of Munich. The HHECO study was approved by the appropriate ethics committees of the Kwame Nkrumah University of Science and Technology (Ghana) and of the medical association in Hamburg (Germany) (38, 39). Signed informed consent was obtained from all participants.
Characterization of CD25+ FoxP3+ CD4 T cells in fresh whole blood.
Fresh anticoagulated whole blood samples from the WHIS cohort were incubated for 30 min using the following fluorochrome-labeled monoclonal antibodies (MAbs) for cell surface staining; CD3-Pacific Blue (BD), CD4-peridinin chlorophyll (PerCP)-Cy5.5 (eBioscience), CD25-phycoerythrin (PE)-Cy7 (eBioscience), and CCR5-allophycocyanin (APC)-Cy7 (BD). Red blood cells in samples were then lysed by incubating and washing the samples twice for 10 min with 1× cell lysis solution (BD). Intracellular FoxP3 was detected with FoxP3-Alexa Fluor 647 (eBioscience) according to the manufacturer's instructions. Cells were finally fixed with 2% paraformaldehyde prior to acquisition. Acquisition was performed on a FACSCanto II system (BD). Compensation was performed with antibody capture beads (BD) stained separately with the individual antibodies used in the test samples. Flow cytometry data were analyzed using FlowJo (version 9.5.3; Tree Star, Inc.).
Characterization of CD25+ FoxP3+ memory CD4 T cells.
Cell surface markers of immune regulation and cell proliferation/cell turnover were stained on cryopreserved PBMCs of individuals from the HHECO cohort using anti-CD3-PerCP, anti-CD4-Pacific Blue, anti-CD45RA-Alexa Fluor 700, and anti-CD25-PE-Cy7 (BD Biosciences, Germany). The stained cells were later fixed and permeabilized (FoxP3 staining buffer set; eBiosciences) for intracellular staining using anti-FoxP3-PE (Biolegend, Germany) and anti-Ki67-Alexa Fluor 647 (BD Biosciences, Germany). Flow cytometric data were acquired with an LSRII flow cytometer (BD Biosciences, Germany). Compensation was performed with antibody capture beads (BD CompBeads set anti-mouse Igκ; BD Biosciences, Germany), stained separately with the individual fluorochrome-conjugated monoclonal antibodies used in all samples. Flow cytometry measurements were analyzed using FlowJo version 9.6.2 (Tree Star, San Carlos, USA).
Cell sorting.
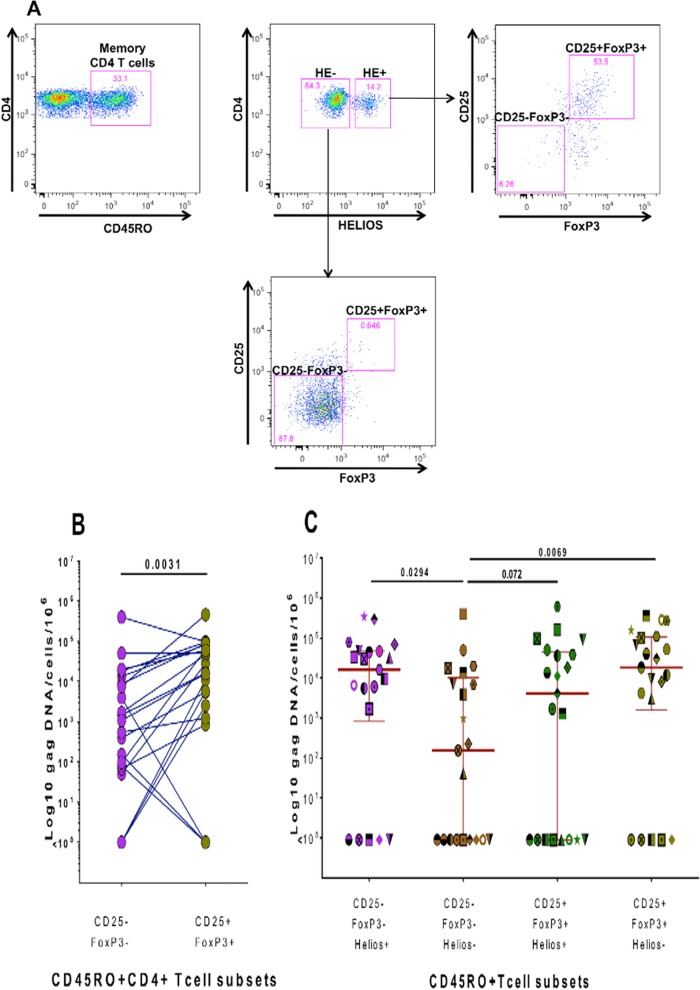
Cryopreserved PBMCs from HIV+ WHIS (n = 15) and HISIS (n = 6) participants were thawed and washed twice in prewarmed (37°C) complete medium (RPMI medium plus 10% heat-inactivated fetal bovine serum [GIBCO]) that was supplemented with Benzonase (5 U/ml; Novagen). Surface staining was performed with CD3-Pacific Blue, CD4-PerCP-Cy5.5, CD25-Pe-Cy7, and CD45RO-PE (BD) for 30 min in the dark at room temperature; intracellular staining was performed with FoxP3-Alexa Fluor 647 (eBioscience) and Helios-fluorescein isothiocyanate (FITC) (BioLegend) according to the CD25+ FoxP3+ CD4 T cell staining protocol mentioned above. Cell sorting was performed on a FACSAria cell sorter (BD) after gating on CD3+ CD4+ CD45RO+ cells into “regulatory T cell populations” (CD25+ FoxP3+ Helios+ and CD25+ FoxP3+ Helios−) and memory populations (CD25− FoxP3− Helios+ and CD25− FoxP3− Helios−) (see Fig. 4A). Between 293 and 750,000 fixed CD4 T cells from each of the four different populations were collected, depending on the number of PBMCs available from each individual. Cells were collected on fluorescence-activated cell sorter (FACS) buffer consisting of phosphate-buffered saline (PBS) mixed with 0.5% bovine serum albumin (BSA; Sigma), 2 mM EDTA, and 0.2% sodium azide at pH 7.45. The median counts of fixed cells collected for each population were as follows: CD25+ FoxP3+ Helios+ cells, 9,017 (median) and 3,931 to 14,412 (interquartile range [IQR]); CD25+ FoxP3+ Helios− cells, 4,381 (median) and 1,579 to 9,799 (IQR); CD25− FoxP3− Helios+ cells, 2,646 (median) and 1,336 to 5,644 (IQR); and CD25− FoxP3− Helios− cells, 185,000 (median) and 79,000 to 315,000 (IQR). Sorted cells were then centrifuged at 13,000 rpm for 3 min, and the supernatant was removed. The cell pellet was stored at −80°C until further analysis.
FIG 4.
Quantification of cell-associated HIV gag DNA in sorted memory CD4 T cell subsets. (A) Gating/sorting strategy used to sort different memory CD4 T cell populations delineated by Helios, CD25, and FoxP3 expression. (B) Numbers of gag DNA copies/106cells detected in CD25− FoxP3− and CD25+ FoxP3+ memory CD4 T cells from 21 different subjects. (C) Numbers of gag DNA copies/106cells detected in these memory CD4 T cell subsets further delineated by Helios expression. gag DNA within different CD4 T cell populations of the same subject was quantified during the same real-time (RT)-PCR run. Cryopreserved PBMCs from the WHIS and HISIS cohorts were used for cell sorting. Statistical analysis was performed using the Wilcoxon rank matched-pairs test.
Quantification of cell-associated HIV gag DNA.
Quantification of cell-associated HIV gag DNA was performed as previously described (8) with minor modifications. Sorted CD4 T cell subsets were lysed in 30 μl of 0.1 mg/ml proteinase K (Roche) containing 10 mM Tris-Cl (pH 8) (Sigma) for 1 h at 56°C, followed by a proteinase K inactivation step for 10 min at 95°C. Cell lysates were then used to quantify cell-associated HIV DNA by quantitative PCR (qPCR) as previously described, with some modifications (10). Briefly, the gag primers and probe used were as follows: 783gag, forward primer, 5′-GAG AGA GAT GGG TGC GAG AGC GTC-3′ (Tm > 60); 895gag, reverse primer, 5′-CTK TCC AGC TCC CTG CTT GCC CA-3′ (Tm > 60); 6-carboxyfluorescein (FAM)-labeled probe 844gagPr, 5′-ATT HGB TTA AGG CCA GGG GGA ARG AAA MAA T-3′. They had been designed to optimally cover subtypes A, C, and D, which prevalent in the Mbeya Region (10). To quantify the cell number in each reaction mix, the human prion gene copy number was also assessed by qPCR. Prion primers and probe sequences were as follows: prion forward primer, 5′TGC TGG GAA GTG CCA TGA G-3′; prion reverse primer, 5′CGG TGC ATG TTT TCA CGA TAG-3′; probe, 5′FAM-CAT CAT ACA TTT CGG CAG TGA CTA TGA GGA CC-6-carboxytetramethylrhodamine (TAMRA) (41). Five microliters of lysate was used in a total reaction volume of 25 μl containing 0.8 μM gag primers or 0.4 μM prion primers, 0.4 μM probe, a 0.2 mM concentration of each deoxynucleoside triphosphate, 3.5 mM MgCl2, and 0.65 U Platinum Taq in the supplied buffer. Standard curves were generated using the HIV-1 gag gene (provided by Brenna Hill, Vaccine Research Center, NIH, Bethesda, MD) and prion gene-containing plasmids. Real-time PCR was performed in a Bio-Rad cycler CFX96 (Bio-Rad): 5 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 60°C. To ensure comparability of the results, cell-associated gag DNA from the four different memory CD4 T cell subsets, which were sorted from one patient, were always quantified simultaneously. Cell-associated gag DNA in memory CD25+ FoxP3+ CD4 T cells and CD25− FoxP3− memory CD4 T cells independent of Helios expression was calculated as follows: ∑gag DNA load (Helios+ cells + Helios− cells)/∑sorted cells in 5 μl lysate (Helios+ cells + Helios− cells).
Amplification and phylogenetic comparison of HIV envelope sequences from plasma and sorted cell populations.
A highly variable envelope region spanning the V1 to V3 region (EnvV1V3; Hxb 6559 to 7320) was amplified using a nested-PCR strategy from 10 μl of lysed sorted cells (described above) or from plasma virus cDNA. HIV RNA was extracted with sample preparation system RNA on the m24sp automatic extraction instrument (Abbott Molecular, USA) in accordance with the manufacturer's instructions. The HIV cDNA was synthesized from 3 μl of extracted RNA using the reverse primer ACD_Env7521R (5′ATGGGAGGGGCATAYATTGC) and the Superscript III reverse transcriptase (Life Technologies, Darmstadt, Germany) according to the manufacturer's instructions. Newly designed PCR primer pairs optimized for the detection of subtypes A, C, and D were used to amplify the EnvV1V3 region. The first-round PCR was performed with 10 μl of template in a 50-μl reaction mixture (0.5 μl [5 U] Platinum Taq [Life Technologies, Darmstadt, Germany], 2.0 mM primers [ACD_Env6420F, 5′CATAATGTCTGGGCYACACATGC; ACD_Env7521R, 5′ATGGGAGGGGCATAYATTGC], 3.5 mM MgCl2, 4 μl of deoxynucleoside triphosphates [dNTPs]) at 95°C for 10 min, followed by 45 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s, and finally 7 min at 72°C. The second-round PCR was performed with 2 μl of first-round PCR product in a 50-μl reaction mixture (0.25 μl [2.5 U] AmpliTaq Gold [Life Technologies, Darmstadt, Germany], 2.0 mM ACD_Env6559F [5′GGGAYSAAAGCCTAAARCCATGTG] and ACD_Env7320R [GTTGTAATTTCTRRR TCCCCTCC], 2.0 mM MgCl2, 4 μl of dNTPs) at 95°C for 10 min, followed by 45 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 90 s, and finally 7 min at 72°C. The second-round PCR products were extracted from agarose gel and then cloned using the TOPO-TA Cloning kit for sequencing (Life Technologies, Darmstadt, Germany), including the precut vector pCR4.1 and One Shot chemically competent E. coli according to the manufacturer's instructions. EnvV1V3 sequences from 11 to 23 clones per population per subject were then sequenced unidirectionally using Mnrev primers at Eurofins Genomics (Ebersberg, Germany). In total, 384 EnvV1V3 sequences from 6 subjects were analyzed.
To assess the error rate of the applied nested-PCR strategy, the positive-control template (Du422, clone 1 [SVPC5]) (42) was endpoint diluted using a 10-fold dilution series and amplified as described above. The EnvV1V3 product from the last detectable dilution step was then cloned as described above. Sequences from 21 clones were analyzed and compared to the original Du422 template sequence.
Phylogenetic analyses.
Nucleotide sequences were aligned with respect to the predicted amino acid sequence of the reference alignment extracted from the Los Alamos HIV database (http://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html) as previously described (43) Evolutionary analyses were conducted using MEGA6 software (44). The evolutionary history is inferred by using the maximum-likelihood method based on the general time-reversible substitution model (GTR+G) (45) and is rooted in previous outbreaks. Upon each analysis, the tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together is presented next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value.
Next-generation sequencing (NGS).
Library preparation from EnvV1V3 PCR second-round products was done using a TruSeq DNA PCR-free sample preparation kit (Illumina, Inc., San Diego, CA, USA) with 550 bp as the insert size in accordance with the manufacturer's instruction. The libraries were controlled with an Agilent Bioanalyzer high-sensitivity chip (Agilent Technologies) and sequenced using a MiSeq desktop sequencer (Illumina, Inc.) and MiSeq reagent kits v3 (Illumina, Inc.). The sequencing was done to 250 cycles in both directions. The produced reads were processed through a quality control pipeline that removed all reads containing unresolved positions or had a mean quality below 20. Furthermore, poly(A) tails and low-quality read ends were trimmed away. All reads that had lengths below 30 nucleotides (nt) after trimming were also excluded from further analysis. An initial mapping was created for each sample by placing the reads onto the HIV HXB2 reference sequence (GenBank accession no. K03455.1 [46]) using segemehl (version 0.1.6) (47). The difference parameter was set to 2 in order to increase the sensitivity, given that the origin of the sequences was a highly variable viral genome. Using an adapted samtools (version 0.1.19) (48) pipeline, we created a consensus sequence for each sample from the initial mapping to use as an individual reference for a second round of alignments. This was necessary, as the official HIV reference sequence is very divergent from our set of reads, and thus the initial mapping was only able to place an unsatisfyingly low number of reads onto this sequence. The second individual mapping was able to use a higher number of reads and to create sufficient alignments, which were used as input for the quasispecies reconstruction tool QuasiRecomb (49). This tool uses an expectation maximization algorithm not only to reconstruct the single sequences present in the viral population but also to assign their relative proportions.
Statistical analysis.
Data analyses were performed using Prism version 4.0 software (GraphPad, Inc.). Comparisons of two groups were performed using the Mann-Whitney test. Comparisons of paired groups were performed using the Wilcoxon matched-pairs test. For correlation analyses, the Spearman r test, the Pearson two-tailed statistical test, or linear regression was used. Differences were considered significant at P values of <0.05. Tests used for statistical analysis are described in the figure legends.
Accession number(s).
Newly determined sequences were submitted to GenBank under accession numbers KX689364 to KX689748.
RESULTS
Study subjects.
Table 1 provides an overview of the subjects included in this study. A total of 258 HIV-negative and 103 HIV-positive adults (mean age, 34.3 years) from the WHIS cohort (37) were included in this study, of which 217 (60%) were female. The vast majority of HIV+ subjects from the WHIS cohort were treatment naive (97%), with a median CD4+ T cell count of 396.3 cells/μl and median log10 plasma viral load of 4.7 copies/ml. Twenty-eight subjects from the previously described HHECO cohort were included for the in-depth characterization of memory CD25+ FoxP3+ CD4 T cells (38, 39) (Table 1). PBMCs from 6 viremic HIV+ subjects from the HISIS cohort (40) were used for the characterization of HIV infection within different memory T cell subsets.
TABLE 1.
Characteristics of study subjects from different cohorts
| Parameter | Value for indicated cohort |
||
|---|---|---|---|
| WHIS | HHECO | HISIS | |
| No. of subjects | 361 | 28 | 6 |
| No. of HIV-positive subjects | 103 | 28 | 6 |
| No. of females | 217 | 25 | 6 |
| Age (yr), mean (SD) | 34.3 (11.05) | 38.8 (7.5) | 28 (3.2) |
| Median no. of CD4 cells/μl (IQR)a | 396 (265–603) | 629 (444–900) | 496 (231–707) |
| Median log no. of pVL copies/ml (IQR)a | 4.67 (3.74–5.23) | 1.59 (1.59–3.82) | 4.9 (4.4–5.5) |
| No. of subjects (%) on ARTa | 3 (0.8) | 20 (71.4) | 0 (0) |
Data given for HIV-positive subjects only.
Correlation between CD4 T cells and CD25+ FoxP3+ CD4 T cell counts in HIV-infected subjects.
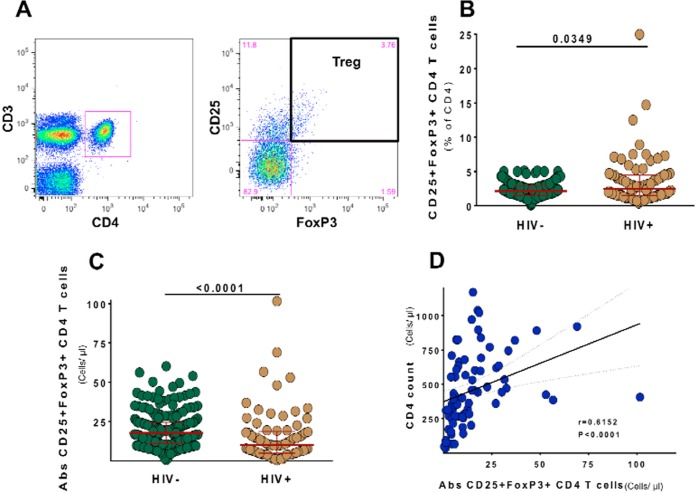
We first determined and compared the frequencies and absolute numbers of CD25+ FoxP3+ CD4+ T cells in fresh anticoagulated peripheral blood of HIV+ (treatment naive, n = 100) and HIV− (n = 258) subjects from the WHIS cohort. A representative dot plot and gating of CD25+ FoxP3+ CD4+ T cells are shown in Fig. 1A. In HIV+ individuals, compared to HIV− individuals, CD25+ FoxP3+ CD4+ T cell frequencies were moderately increased (Fig. 1B) (for HIV+, median, 2.5%; IQR, 1.5% to 4.5%; versus for HIV−, median, 2.1%; IQR, 1.5% to 2.9%; P = 0.03), but absolute numbers of CD25+ FoxP3+ CD4+ T cells were significantly decreased, with median counts of 10.16 cells/μl (IQR, 4.88 to18.57 cells/μl) in HIV+ subjects and 17.75 cells/μl (IQR, 11.06 to 24.56 cells/μl) in HIV− subjects (P < 0.0001) (Fig. 1C). Within HIV+ subjects, there was a positive correlation between CD25+ FoxP3+ CD4+ T cell counts and CD4 T cell counts (P < 0.0001, r = 0.6152) (Fig. 1D). Confirming previous reports (50–54), our data show that the depletion of CD25+ FoxP3+ CD4+ T cells is closely linked to the loss of CD4 T cells.
FIG 1.
Frequencies and absolute numbers of CD25+ FoxP3+ CD4 T cells in the peripheral blood in relation to HIV infection. (A) Representative dot plots and gating strategy for the detection of regulatory T cells through CD25 and FoxP3 expression on CD3+ CD4+ T cells from fresh anticoagulated whole blood of WHIS subjects. (B and C) CD25+ Foxp3+ CD4 T cell frequencies (B) and absolute numbers (C) are compared between HIV− and HIV+ subjects. (D) Correlation analysis of absolute CD4 counts and CD25+ Foxp3+ CD4 T cell counts. Statistical analysis was performed using the Mann-Whitney test for comparison of groups and the Spearman r statistical test for correlation analyses.
High frequencies of CD25+ FoxP3+ CD4 T cells express HIV coreceptor CCR5 and the cell cycle marker Ki67.
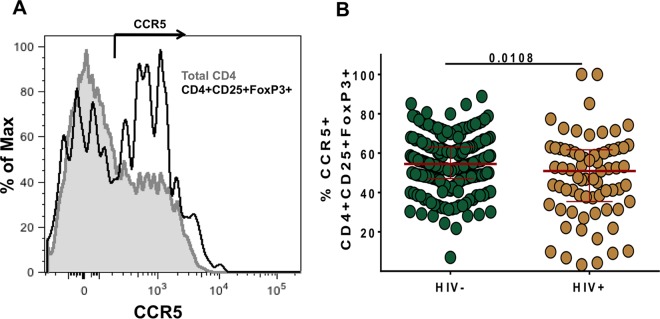
In order to determine whether CD25+ FoxP3+ CD4+ T cells could potentially support entry of HIV, we assessed the expression of the HIV coreceptor CCR5. Fresh anticoagulated whole blood was used for improved CCR5 staining. A representative plot is shown in Fig. 2A. A considerable proportion of CD25+ FoxP3+ CD4+ T cells expressed CCR5 (median, 53.7%), which was higher than that previously observed in total memory CD4 T cells (median, 40%; data not shown). HIV infection was associated with a moderate decrease in the frequency of CCR5-positive CD25+ FoxP3+ CD4+ T cells (Fig. 2B) (median, 50.9%, compared to 54.5%; P = 0.01).
FIG 2.
Ex vivo HIV coreceptor (CCR5) expression on CD25+ Foxp3+ CD4 T cells. (A) A histogram overlay of CCR5 expression on total CD4 T cells (gray) and CD25+ Foxp3+ CD4 T cells (black). (B) The frequencies of CCR5-expressing CD25+ Foxp3+ CD4 T cells are compared between HIV-negative and -positive subjects. For maximum staining sensitivity, fresh anticoagulated whole blood of individuals from the WHIS cohort was used to determine CCR5 expression on CD4 T cells. Statistical analysis was performed using the Mann-Whitney test.
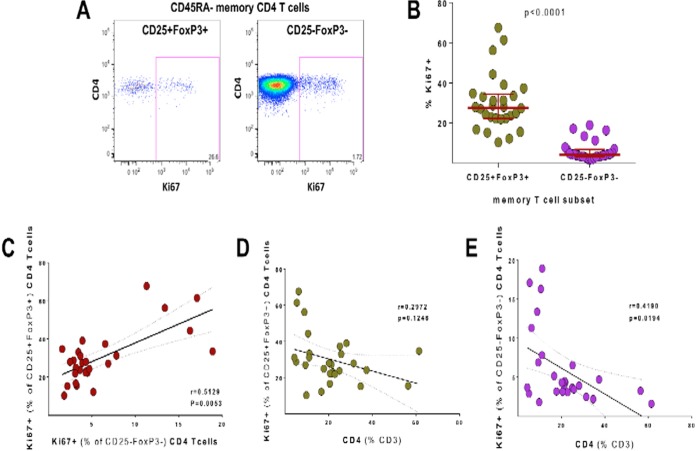
We next studied the cell cycle status of memory CD25+ FoxP3+ and CD25− FoxP3− CD4+ T cells in HIV-infected subjects and analyzed cellular Ki67 expression using cryopreserved PBMC samples (n = 28 from HHECO cohort [Table 1]). The representative dot plots for Ki67 staining in memory (CD45RA−) CD25+ FoxP3+ and CD25− FoxP3− CD4+ T cells are shown in Fig. 3A. HIV-infected study participants had very high frequencies of Ki67+ memory CD25+ FoxP3+ CD4 T cells (median, 27.6%) (Fig. 3B) despite the majority of subjects from the HHECO cohort being on ART. Importantly, the frequencies of Ki67+ cells detected were 6.7-fold higher in CD25+ FoxP3+ memory CD4+ T cells than in CD25− FoxP3− memory CD4+ T cells (median, 4.1%; P < 0.0001), consistent with high in vivo proliferation of memory CD25+ FoxP3+ CD4 T cells. Correlation analysis demonstrated a close association between the proportions of Ki67+ CD25+ FoxP3+ and Ki67+ CD25− FoxP3− memory CD4 T cells (P = 0.005, r = 0.51) (Fig. 3C), which is linked to the level of CD4 T cell depletion in HIV+ subjects (P = 0.1, r = −0.3 [Fig. 3D], and P = 0.02, r = −0.4 [Fig. 3E]). Memory CD25+ FoxP3+ CD4 T cells may therefore potentially support CCR5-mediated viral entry and subsequent steps of the viral life cycle due to their high in vivo proliferation. The correlation between the frequency of Ki67+ memory T cells and CD25+ FoxP3+ CD4 memory T cells and the fact that the losses of these cell subsets are closely linked support the proposed mechanism of constant replenishment of memory CD25+ FoxP3+ CD4 T cells from the memory CD4 T cell pool (30) also during HIV infection.
FIG 3.
Ki67 expression in CD25+ FoxP3+ and CD25− FoxP3− memory CD4 T cells in HIV+ subjects. (A) Representative dot plots for Ki67 staining. (B) Comparison of the frequencies of Ki67+ cells in CD25+ FoxP3+ and CD25− FoxP3− memory CD4 T cells in HIV+ subjects. (C) Correlation analysis of the frequency of Ki67+ cells between CD25+ FoxP3+ (y axis) and CD25− FoxP3− (x axis) memory CD4 T cells. (D and E) Correlation analysis of the frequency of Ki67+ cells among CD25+ FoxP3+ (D) and CD25− FoxP3− (E) CD4 T cells versus CD4 T cell frequencies (% of CD3). The analysis was done using cryopreserved PBMC samples from HIV+ HHECO study participants. Memory status of CD4 T cells was determined by CD45RA staining. Statistical analysis was performed using the Mann-Whitney test for comparison of groups and the Spearman r statistical test for correlation analyses.
Memory Helios+ and Helios− CD25+ FoxP3+ CD4 T cells are frequent targets for HIV infection in vivo.
To determine in vivo HIV infection rates of memory CD25+ FoxP3+ CD4 T cells, we sorted four different subsets of CD45RO+ memory CD4 T cells on the basis of their Helios, CD25, and FoxP3 expression (Fig. 4A) for 22 subjects (the WHIS cohort plus 6 subjects from the HISIS cohort [Table 1]) and quantified HIV gag DNA within the sorted subsets. Helios, an Ikaros transcriptional factor family member that is critical for the regulatory function of CD25+ FoxP3+ CD4 T cells (55–58), is a negative regulator of IL-2 signaling in CD25+ FoxP3+ CD4 T cells (59). A large fraction of CD25+ FoxP3+ CD4 T cells expressed the memory marker CD45RO in HIV+ subjects (median, 87.3%; IQR, 71.85% to 93.55%), and most of these expressed Helios (median, 76.30%; IQR, 69.18% to 84.43%; data not shown), consistent with a regulatory cell function of this subset. In contrast, only a minor fraction of CD25− FoxP3− memory CD4 T cells expressed Helios (median, 1.65%; IQR, 1.15% to 2.75%). HIV gag DNA was detected in >80% of memory CD25+ FoxP3+ and CD25− FoxP3− CD4 T cells, with a 15-fold-higher median gag DNA load in CD25+ FoxP3+ memory CD4 T cells than in CD25− FoxP3− memory CD4 T cells [∑(Helios+ cells + Helios− cells), 16,072 versus 1,074 copies/106 cells, respectively; P = 0.003] (Fig. 4B). From 16 subjects, we also determined the plasma viral load (pVL) and found a correlation between log cell-associated DNA gag in memory CD25− FoxP3− memory CD4 T cells and log pVL (P = 0.025, r = 0.56; data not shown). No such association was detected for memory CD25+ FoxP3+ memory CD4 T cells (P = 0.1, r = 0.39; data not shown).
Figure 4C shows the levels of HIV gag DNA within these memory CD4 T cell subsets further delineated by Helios expression. In comparison to the largest sorted memory CD4 T cell population in the blood (FoxP3− CD25− Helios−), which contained a median of 154.4 HIV copies/106 cells (IQR, 0 to 10,241 copies/106 cells), the levels of HIV gag DNA were substantially increased in the other subsets: the FoxP3+ CD25+ Helios− CD4 T cells (119-fold increase; median, 18,407 copies/106 cells; IQR, 1,556 to 106,067 copies/106 cells; P = 0.007), the FoxP3− CD25− Helios+ CD4 T cells (104-fold increase; median, 16,096 copies/106 cells; IQR, 837.9 to 47,903 copies/106 cells, P = 0.029), and the FoxP3+ CD25+ Helios+ CD4 T cells (26-fold increase; median, 4,106 copies/106 cells; IQR, 0 to 44,612 copies/106 cells; P = 0.072). Together these data demonstrate that CD25+ FoxP3+ memory CD4 T cells and in particular the small Helios− population contain high HIV DNA levels in vivo. Likewise, the small CD25− FoxP3− Helios+ memory CD4 T cell population contained a substantially increased number of HIV DNA copies. In comparison, the main CD25− FoxP3− Helios− memory CD4 T cell subset (>90% of memory CD4 T cells in peripheral blood), of which high cell numbers were sorted for all 22 subjects, contained few and often a surprisingly undetectable number of gag DNA copies. Together these data suggest that CD25+ FoxP3+ and also CD25− FoxP3− Helios+ memory CD4 T cells are frequent targets for HIV infection. However, the lack of correlation between plasma viral load and gag DNA loads in CD25+ FoxP3+ memory CD4 T cells is inconsistent with the hypothesis of significant plasma virus production by this cell subset.
Phylogenetic sequence analyses of the highly variable EnvV1V3 region in plasma virus and sorted memory CD4 T cell populations.
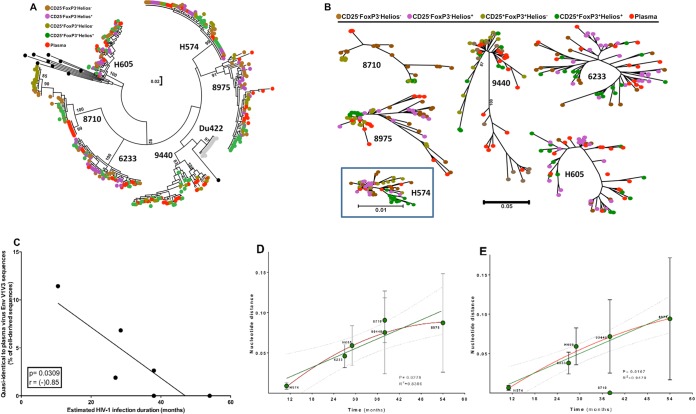
To assess whether memory CD25+ FoxP3+ CD4 T cells could potentially contribute to plasma virion production, we compared the highly variable envelope V1V3 regions from cell DNA sequences (CD25+ FoxP3+ and CD25− FoxP3− memory CD4 T cell subsets) and plasma virus sequences in seven viremic subjects. The estimated HIV infection durations varied, from 9 months (subject H574), 27 to 30 months (subject H605), 1.3 to 3.3 years (subject 6233K12), above 3.2 years (subjects 3806A11, 8710U11, and 9440A11), and above 4.5 years (subject 8975T11). PCR-related sequence background variation was controlled for by using an endpoint diluted molecular clone of the subtype isolate Du422 clone 1. Ten of the 21 Du422 sequences did not contain any nucleotide substitutions compared to the template sequence, seven sequences had one substitution, and three sequences had two substitutions. Hence, the PCR protocol introduced only two or fewer nucleotide substitutions and no insertions or deletions in 95% of the amplicons. We therefore considered up to four substitutions between cell- and plasma-derived sequence variants as quasi-identical. EnvV1V3 amplicons containing clones from 6 of the 7 subjects were subjected to Sanger sequencing, and clonal sequences were analyzed using the maximum-likelihood method (Fig. 5A and B). In 4 of these 6 subjects (H574, H605, 6233K12, 9440A11) we found quasi-identical cell- and plasma-derived EnvV1V3 sequence pairs (Table 2). For subject H574 (HIV infected for 9 months), viral sequences were closely related to each other and sequences from all four sorted cell populations were closely related to those of plasma virus (Fig. 5B; Table 2). Of the cell-derived sequences, 11.4% (8 of 70) were quasi-identical to plasma-derived sequence variants. For subject H605 (infected 27 to 30 months), the closest sequence was derived from the “dominant” memory CD4 T cell subset (CD25− FoxP3− Helios−; 3 substitutions) and 6.8% (3 of 44) of cell-derived sequences were quasi-identical to plasma-derived sequence variants. For subject 6233K12 (infected 16 to 38 months), only 1 of 53 EnvV1V3 sequences was quasi-identical to a plasma-derived sequence variant and was derived from the CD25− FoxP3− Helios+ memory T cell subset. The three subjects, i.e., 8710U11, 8975T11, and 9440A11, were infected for at least 3.2 years, and the closest cell-derived sequences to a plasma-derived sequence variant had 32, 54, and 4 substitutions, respectively. Hence, we detected a single “quasi-identical pair” between cell- and plasma-derived EnvV1V3 sequences (derived from CD25+ Helios+ memory CD4 T cells) in only one of these three subjects. In summary, sequences derived from CD25+ FoxP3+ memory CD4 T cells (or those derived from the other sorted memory CD4 T cell subsets) were not preferentially clustering with plasma-derived sequence variants. Quasi-identical cell- and plasma-derived EnvV1V3 sequence pairs were generally infrequent, and their proportion decreased with HIV infection duration (Fig. 5C; P = 0.03, r = −0.85) as the nucleotide distances between cell- and plasma-derived sequences (Fig. 5D; P = 0.02, r2 = 0.84) and between individual plasma-derived sequences (Fig. 5E; P = 0.02, r2 = 0.95) increased. To ascertain the relatedness of the plasma sequences and the sequences isolated from the cell fractions, we estimated the nucleotide variation within each fraction. The estimation was performed using the neighbor-joining model with the Kimura 2-parameter method. The sequence diversity analyses showed that the sequence diversity in plasma was not different from the estimated diversity between the plasma- and cell-derived sequences (data not shown).
FIG 5.
Phylogenetic relationship of HIV envelope sequences derived from plasma and sorted memory CD4 T cell populations. Plasma- and cell-derived sequences of the highly variable EnvV1V3 region (Hxb 6559 to 7320) were amplified, cloned, sequenced (n = 384, Sanger method), and analyzed for 6 viremic subjects from the WHIS and HISIS cohorts with differing HIV infection durations (H574, H605, 6233K12 [6233], 9440A11 [9440], 8710U11 [8710], 8975T11 [8975]). (A and B) The phylogenetic relationship was inferred by the maximum-likelihood method based on the general time-reversible substitution model (GTR+G). (C) Correlation between frequency of cell-derived sequences that were quasi-identical to plasma-derived sequences and the estimated infection duration. (D and E) Linear regression analysis (green line) between the distance of the EnvV1V3 sequences derived from plasma to the sequences extracted from the corresponding cellular fractions and the estimated duration of infection (D) and plasma sequence diversity plotted against the estimated duration of infection (E). The red line indicates a nonlinear analysis performed using a second-order polynomial equation taking into account the best-fit values. The evolutionary distances were computed using the Kimura 2-parameter method (77) and are given in units of the number of base substitutions per site, including both transitions and transversions. The rate variation among sites was modeled with a gamma distribution. The analysis was conducted in MEGA6 (44). No sequence diversity was observed in the subject 8710 plasma fraction, probably because the number of viruses sampled in each PCR was very low (Table 2). We therefore excluded the results for subject 8710 from the linear regression analysis. P and r values were calculated with the Pearson two-tailed statistical test.
TABLE 2.
Key data of the EnvV1V3 phylogenetic studies and HIV infection duration for 6 viremic subjects
| Subject ID | HIV infection duration (mo) | % of cell-derived sequences quasi-identical to plasma-derived sequences (n) | Mean no. of nt substitutions between plasma and cell-derived sequences | Cellular origin of closest sequence | No. of nt substitutions | Cellular origin of most distant sequence | No. of nt substitutions |
|---|---|---|---|---|---|---|---|
| H574 | 9–12 | 11.4 (8 of 70) | 6 | CD25+ FoxP3+ Helios− | 1 | CD25+ FoxP3+ Helios+ | 16 |
| CD25+ FoxP3+ Helios+ | 1 | ||||||
| CD25− FoxP3− Helios+ | 1 | ||||||
| CD25− FoxP3− Helios− | 1 | ||||||
| H605 | 27–30 | 6.8 (3 of 44) | 39 | CD25− Foxp3− Helios− | 3 | CD25+ FoxP3+ Helios+ | 32 |
| 6233K12 | 16–38 | 1.9 (1 of 53) | 30 | CD25− FoxP3− Helios+ | 2 | CD25− FoxP3− Helios+ | 30 |
| 9440A11a | >38 | 2.6 (1 of 38) | 46 | CD25+ Helios+ | 4 | CD25+ Helios+ | 76 |
| 8710U11 | >38 | 0 (0 of 39) | 57 | CD25− FoxP3− Helios− | 32 | CD25− FoxP3− Helios− | 67 |
| 8975T11 | >54 | 0 (0 of 55) | 53 | CD25− FoxP3− Helios+ | 54 | CD25− FoxP3− Helios− | 86 |
Cells (PBMCs) from this subject were sorted into four populations only on the basis of CD25 and Helios expression on memory (CD45RO) CD4 T cells.
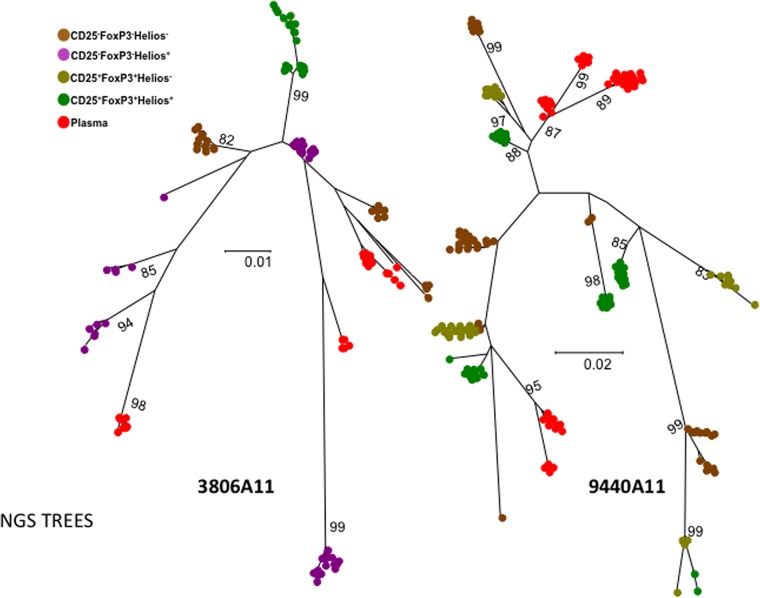
We also analyzed plasma- and cell-derived EnvV1V3 amplicons from two HIV+ subjects (3806A11 and 9440A11) infected for more than 3.2 years by using next-generation sequencing to detect “rare” quasi-identical sequence pairs that we might have missed in the previous analyses. Between 780 and 10,000 EnvV1V3 sequences were first reconstructed using QuasiRecomb (49). The 50 most frequent sequences/population were aligned, and sequences were compared (Fig. 6). The closest cell-associated and plasma sequences were 6 and 14 nucleotide substitutions apart for subjects 3806A11 and 9440A11, respectively, inconsistent with a major contribution of the sorted peripheral memory CD4+ T cell subsets to plasma virus production. BLAST searches of all plasma sequence variants against the 150 highest-frequency cell-derived variants (per sorted cell subset) identified the closest pairs as 4 (subject 3806A11, CD25− FoxP3− Helios+) and 10 (subject 9440A11, CD25+ Helios+) nucleotides apart.
FIG 6.
Phylogenetic analyses of HIV envelope sequences derived from plasma and sorted memory CD4 T cell populations using next-generation sequencing. Shown are the phylogenetic analyses of EnvV1V3 sequences from the 50 most frequently detected sequences derived from either plasma or the different sorted memory CD4 T cell subsets for two viremic subjects of the WHIS cohort. The phylogenetic relationship was inferred by the maximum-likelihood method based on the general time-reversible substitution model (GTR+G). EnvV1V3 amplicons were directly subjected to next-generation sequencing. Quasispecies reconstruction was performed using the software QuasiRecomb. The methods used are described in detail in Materials and Methods.
DISCUSSION
HIV plasma viremia predicts the rate of HIV disease progression (1, 60) and depends on active HIV replication in CD4+ cells. Memory CD4 T cells are most probably the primary substrate for virus replication (11, 61–63). HIV infection rates differ substantially between different CD4 T cell subsets (4–6, 64). Recent data show that follicular T helper (Tfh) cells are a prime target for virus replication and contribute to virion production even in elite controller rhesus macaques (19) and most probably to plasma viremia (17). To what extent other CD4+ cell subsets contribute to plasma virus production in viremic progressors is unclear. In various in vitro infection models, HIV replication is associated with IL-2 signaling and CD25 expression on stimulated CD4 T cells (10, 13, 14, 21–23). Because IL-2 is important for the homeostatic proliferation of the CD25+ FoxP3+ CD4 T cells (35, 65), and because of the high in vivo proliferation rates of this subset (32), we hypothesized that CD25+ FoxP3+ CD4 T cells constitute a prime target for HIV infection and may contribute to plasma virion production in vivo.
Consistent with a previous report, we showed that a large fraction of CD25+ FoxP3+ CD4 T cells express the HIV coreceptor CCR5 (35), potentially supporting viral entry. Although frequencies of CD25+ FoxP3+ CD4 T cells were slightly elevated in viremic, HIV+ subjects, absolute cell numbers of this subset were significantly depleted, which confirms previously published data (50, 52, 66). A greater proportion of CD25+ FoxP3+ memory CD4 T cells from HIV+ subjects expressed Ki67+, with almost one-third of these cells “cycling” at any given time. This pattern, i.e., depleted cell counts despite increased fractions of Ki67+ “cycling” cells, demonstrates that homeostasis of CD25+ FoxP3+ CD4 T cells is heavily perturbed by HIV infection. Furthermore, expression of CCR5 and high proportions of cycling cells within CD25+ FoxP3+ CD4 T cells should support both cell entry and reverse transcription of HIV, which is supported by the increased HIV DNA loads observed in memory CD25+ FoxP3+ CD4 T cells observed in this study (12, 36). Other reports show discrepant results regarding in vivo levels of HIV DNA in “regulatory” CD4 T cells that are typically defined by a CD25high phenotype, instead of the definition using coexpression of CD25 and FoxP3 that we used (67–69). Tran et al. observed a higher infection rate in CD25high than CD25− CD4 T cells (70) but did not exclude naive CD4 T cells, which are not susceptible to CCR5-tropic strains which predominate throughout most of the infection course. Of note, high in vivo proliferation of memory CD25+ FoxP3+ CD4 T cells might also potentially pass on proviral HIV DNA to the cell progeny in the absence of productive HIV infection during ART. Previous studies reported that CD25high T cells (which were >99% FoxP3+) release virus upon in vitro restimulation and have ∼3-fold-higher HIV infection rates than other CD4 T cells upon in vitro activation (36, 70). Together these data suggest that CD25+ FoxP3+ CD4 T cells are a prime cellular target for HIV infection that might serve as an important HIV reservoir during ART.
We next wanted to address whether memory CD25+ FoxP3+ CD4 T cells could potentially contribute to plasma virion production. Because cell fixation complicates analyses of HIV transcription in sorted cell populations defined by intranuclear transcription factors (such as FoxP3), we decided to study the phylogenetic relationship between plasma- and cell-derived sequences within the highly variable EnvV1V3 region; if CD25+ FoxP3+ memory CD4 T cells significantly contribute to plasma virion production, EnvV1V3 DNA sequences derived from this cell population should often be quasi-identical or preferentially cluster with plasma-derived sequences. A previous study had reported rapid replacement of cell- and plasma-derived HIV sequences by an incoming superinfecting HIV strain (71), implying a highly dynamic exchange between these two compartments. In our study, the detection of quasi-identical sequence pairs derived from cells and plasma was rare and their fraction further decreased with infection duration, which is consistent with the broadening of the viral reservoir with time. There was no clear pattern of phylogenetic clustering of the plasma virus with any of the cell subset-derived sequences we had sorted. In fact, cell-derived sequences did not “behave differently” from plasma-derived sequences, and sequences from both compartments intermingled. Our phylogenetic data therefore do not allow definite conclusions about the cellular origin of plasma virions. The high variability between individual plasma-derived sequences during chronic infection emphasizes that a huge number of infected cells must contribute to plasma virion production at any given time during chronic infection. It might therefore be difficult to determine the exact cellular origins of plasma virus through phylogenetic sequence analyses. Nonetheless, in our analyses of individual sequences, we did find several quasi-identical sequence pairs between plasma and CD25+ FoxP3+ CD4 T cells, indicating that they may contribute to the plasma viremia. One limitation of our study was that we used comparatively small amounts of PBMCs and plasma (compared to the total body amount) for phylogenetic analyses, and we therefore probably included insufficient numbers for detection of clusters of cell- and plasma-derived sequences. Virus sequences from very large amounts of specimen will need to be analyzed and will optimally include material from secondary lymphoid tissues for more conclusive answers. Secondary lymphoid tissues are thought to constitute the primary site for virion production (reviewed in reference 61). After ART interruption, the onset of viral RNA transcription in lymph nodes coincides with a rise in plasma viral load (72). CD25+ FoxP3+ CD4 T cells in secondary lymphoid organs contain high frequencies of Ki67+ “cycling” cells with a significant capacity for IL-2 production and often express a CD69+ “recently activated” phenotype (32), thereby differing from those in peripheral blood. A recent study detected colocalization of SIV p27 and FoxP3 expression in intestinal tissues using confocal microscopy (73). We therefore consider it likely that CD25+ FoxP3+ CD4 T cells in lymphoid tissues are targeted by HIV, but additional studies will be needed to define the role of CD25+ FoxP3+ CD4 T cells for plasma virion production in vivo.
We also sorted memory CD4 T cell populations depending on their Helios expression. Helios, an Ikaros transcriptional factor family member, is critical for the regulatory function of CD25+ FoxP3+ CD4 T cells (55–57) and for the prevention of autoimmunity (58). Helios modulates cell cycle progression and sustained cell survival through the regulation of genes involved in IL-2 signaling (58, 59). Helios expression is also linked to expression of a range of suppressive T cell markers and can be induced in CD4 T cells upon in vitro activation (74, 75). In vitro, dividing CD25+ FoxP3+ CD4 T cells coexpress Helios, while nondividing regulatory T cells lose expression of FoxP3 and Helios, suggesting that Helios is a marker of recently divided cells. In the same set of in vitro experiments, CD25− Helios+ CD4 T cells were composed of highly activated “effector” memory cells (74). We detected higher median gag DNA loads in both Helios-positive (26-fold increased) and -negative (119-fold increased) CD25+ FoxP3+ memory CD4 T cells as well as in CD25− FoxP3− Helios+ memory CD4 T cells (104-fold increased) than in FoxP3− CD25− Helios− memory CD4 T cells. It is remarkable that we often did not detect HIV DNA in this “dominant” memory CD4 T cell subset. A history of more frequent or recent cell divisions within CD25− FoxP3− Helios+ memory CD4 T cells might have contributed to high HIV susceptibility in this memory cell subset, whereas removal of such cells in the sorted CD25− FoxP3− Helios− memory CD4 T cells may potentially explain the low HIV infection rates observed in this memory cell subset. “Nonactivated” circulating memory CD4 T cells are probably less susceptible and accumulate less HIV DNA over time than other memory CD4 T cell subsets with a history of in vivo proliferation. Helios-deficient regulatory CD4 T cells exhibit an activated phenotype, i.e., an increased capacity to secrete gamma interferon (IFN-γ) and to develop into nonanergic cells under inflammatory conditions (58, 76). The increased responsiveness to cellular activation in comparison to their Helios+ counterparts' signaling may potentially explain the higher HIV DNA levels in CD25+ FoxP3+ Helios− memory CD4 T cells than in their Helios+ counterparts. These data show that Helios and CD25/FoxP3 expression patterns are linked to different cellular HIV infection rates, consistent with a role of the IL-2 signaling pathway for HIV infection in vivo.
In conclusion, we find that homeostasis of CD25+ FoxP3+ CD4 T cells is heavily perturbed during HIV infection. High expression of HIV coreceptor CCR5 and in vivo proliferation potentially facilitate efficient HIV infection of memory CD25+ FoxP3+ CD4 T cells. Furthermore, high proliferative activity of this cell subset is likely to passage HIV DNA to cell progeny in the absence of active viral replication. This subset could therefore serve as an important viral reservoir during ART. Neither circulating memory CD25+ FoxP3+ CD4 T cells nor any of the other memory CD4 T cell subset-derived EnvV1V3 sequences preferentially clustered with plasma-derived sequences. Instead, sequences from the two compartments intermingled, and the genetic distance between and within the two compartments increased with infection duration, precluding a definite conclusion about the cellular origin of the plasma virus in this study.
ACKNOWLEDGMENTS
We thank Brenna Hill from the Vaccine Research Center, NIH, Bethesda, MD, for providing the HIV gag DNA standard and Andreas Wieser from the Max von Pettenkofer Institute, Medical Center of the University of Munich (LMU), for advice to include a PCR error control. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from D. Montefiori, F. Gao, C. Williamson, and S. Abdool Karim: Du422, clone 1 (SVPC5).
Funding Statement
The WHIS study was funded by the German Research Foundation (DFG) (SA 1878/1-1) with additional support by the European Commission's Seventh Framework Programme (FP7/2007-2013 and FP7/2007-2011 under EC-GA nu 241642) and by the German Center for Infection Research (DZIF), while the HHECO study was supported by a German Federal Ministry of Education and Research grant to T. Feldt (EDCTP project 01KA1102). Deep sequencing was supported by the Transvac European Network and funded by the European Commission's Seventh Framework Programme (FP7). The funders had no role in study design, data collection and analysis, preparation of the manuscript, or the decision to submit the work for publication.
REFERENCES
- 1.Mellors JW, Rinaldo CR, Gupta P, White RM, Todd JA, Kingsley LA. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 2.Li TS, Tubiana R, Katlama C, Calvez V, Ait Mohand H, Autran B. 1998. Long-lasting recovery in CD4 T-cell function and viral-load reduction after highly active antiretroviral therapy in advanced HIV-1 disease. Lancet 351:1682–1686. doi: 10.1016/S0140-6736(97)10291-4. [DOI] [PubMed] [Google Scholar]
- 3.Verhofstede C, Reniers S, Van Wanzeele F, Plum J. 1994. Evaluation of proviral copy number and plasma RNA level as early indicators of progression in HIV-1 infection: correlation with virological and immunological markers of disease. AIDS 8:1421–1427. doi: 10.1097/00002030-199410000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Ganesan A, Chattopadhyay PK, Brodie TM, Qin J, Mascola JR, Michael NL, Follmann DA, Roederer M. 2010. Immunological and virological events in early HIV infection predict subsequent rate of progression. J Infect Dis 201:272–284. doi: 10.1086/649430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casazza JP, Brenchley JM, Hill BJ, Ayana R, Ambrozak D, Roederer M, Douek DC, Betts MR, Koup RA. 2009. Autocrine production of beta-chemokines protects CMV-specific CD4 T cells from HIV infection. PLoS Pathog 5:e1000646. doi: 10.1371/journal.ppat.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 9.Geldmacher C, Koup RA. 2012. Pathogen-specific T cell depletion and reactivation of opportunistic pathogens in HIV infection. Trends Immunol 33:207–214. doi: 10.1016/j.it.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geldmacher C, Ngwenyama N, Schuetz A, Petrovas C, Reither K, Heeregrave EJ, Casazza JP, Ambrozak DR, Louder M, Ampofo W, Pollakis G, Hill B, Sanga E, Saathoff E, Maboko L, Roederer M, Paxton WA, Hoelscher M, Koup RA. 2010. Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J Exp Med 207:2869–2881. doi: 10.1084/jem.20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, Reinhart TA, Rogan M, Cavert W, Miller CJ, Veazey RS, Notermans D, Little S, Danner SA, Richman DD, Havlir D, Wong J, Jordan HL, Schacker TW, Racz P, Tenner-Racz K, Letvin NL, Wolinsky S, Haase AT. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 12.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213–222. doi: 10.1016/0092-8674(90)90802-L. [DOI] [PubMed] [Google Scholar]
- 13.Chou CS, Ramilo O, Vitetta ES. 1997. Highly purified CD25− resting T cells cannot be infected de novo with HIV-1. Proc Natl Acad Sci U S A 94:1361–1365. doi: 10.1073/pnas.94.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biancotto A, Iglehart SJ, Vanpouille C, Condack CE, Lisco A, Ruecker E, Hirsch I, Margolis LB, Grivel JC. 2008. HIV-1 induced activation of CD4+ T cells creates new targets for HIV-1 infection in human lymphoid tissue ex vivo. Blood 111:699–704. doi: 10.1182/blood-2007-05-088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maenetje P, Riou C, Casazza JP, Ambrozak D, Hill B, Gray G, Koup RA, de Bruyn G, Gray CM. 2010. A steady state of CD4+ T cell memory maturation and activation is established during primary subtype C HIV-1 infection. J Immunol 184:4926–4935. doi: 10.4049/jimmunol.0903771. [DOI] [PubMed] [Google Scholar]
- 16.Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pallikkuth S, Sharkey M, Babic DZ, Gupta S, Stone GW, Fischl MA, Stevenson M, Pahwa S. 2015. Peripheral T follicular helper cells are the major HIV reservoir within central memory CD4 T cells in peripheral blood from chronic HIV infected individuals on cART. J Virol 90:2718–2728. doi: 10.1128/JVI.02883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, Morcock D, Swanson T, Legasse AW, Axthelm MK, Hesselgesser J, Geleziunas R, Hirsch VM, Edlefsen PT, Piatak M, Estes JD, Lifson JD, Picker LJ. 2015. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyman O, Sprent J. 2012. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol 12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 21.Finberg RW, Wahl SM, Allen JB, Soman G, Strom TB, Murphy JR, Nichols JC. 1991. Selective elimination of HIV-1-infected cells with an interleukin-2 receptor-specific cytotoxin. Science 252:1703–1705. doi: 10.1126/science.1904628. [DOI] [PubMed] [Google Scholar]
- 22.Ramilo O, Bell KD, Uhr JW, Vitetta ES. 1993. Role of CD25+ and CD25− T cells in acute HIV infection in vitro. J Immunol 150:5202–5208. [PubMed] [Google Scholar]
- 23.Goletti D, Weissman D, Jackson RW, Graham NM, Vlahov D, Klein RS, Munsiff SS, Ortona L, Cauda R, Fauci AS. 1996. Effect of Mycobacterium tuberculosis on HIV replication. Role of immune activation. J Immunol 157:1271–1278. [PubMed] [Google Scholar]
- 24.Baecher-Allan C, Brown JA, Freeman GJ, Hafler DA. 2003. CD4+ CD25+ regulatory cells from human peripheral blood express very high levels of CD25 ex vivo. Novartis Found Symp 252:67–91, 106–114. doi: 10.1002/0470871628.ch6. [DOI] [PubMed] [Google Scholar]
- 25.Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, Landay A, Solomon M, Selby W, Alexander SI, Nanan R, Kelleher A, Fazekas de St Groth B. 2006. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med 203:1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hori S, Takahashi T, Sakaguchi S. 2003. Control of autoimmunity by naturally arising regulatory CD4+ T cells. Adv Immunol 81:331–371. doi: 10.1016/S0065-2776(03)81008-8. [DOI] [PubMed] [Google Scholar]
- 27.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. 2008. Regulatory T cells and immune tolerance. Cell 133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Aandahl EM, Michaëlsson J, Moretto WJ, Hecht FM, Nixon DF. 2004. Human CD4+ CD25+ regulatory T cells control T-cell responses to human immunodeficiency virus and cytomegalovirus antigens. J Virol 78:2454–2459. doi: 10.1128/JVI.78.5.2454-2459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinter AL, Horak R, Sion M, Riggin L, McNally J, Lin Y, Jackson R, O'shea A, Roby G, Kovacs C, Connors M, Migueles SA, Fauci AS. 2007. CD25+ regulatory T cells isolated from HIV-infected individuals suppress the cytolytic and nonlytic antiviral activity of HIV-specific CD8+ T cells in vitro. AIDS Res Hum Retroviruses 23:438–450. doi: 10.1089/aid.2006.0162. [DOI] [PubMed] [Google Scholar]
- 30.Booth NJ, McQuaid AJ, Sobande T, Kissane S, Agius E, Jackson SE, Salmon M, Falciani F, Yong K, Rustin MH, Akbar AN, Vukmanovic-Stejic M. 2010. Different proliferative potential and migratory characteristics of human CD4+ regulatory T cells that express either CD45RA or CD45RO. J Immunol 184:4317–4326. doi: 10.4049/jimmunol.0903781. [DOI] [PubMed] [Google Scholar]
- 31.Antons AK, Wang R, Oswald-Richter K, Tseng M, Arendt CW, Kalams SA, Unutmaz D. 2008. Naive precursors of human regulatory T cells require FoxP3 for suppression and are susceptible to HIV infection. J Immunol 180:764–773. doi: 10.4049/jimmunol.180.2.764. [DOI] [PubMed] [Google Scholar]
- 32.Peters JH, Koenen HJPM, Fasse E, Tijssen HJ, Ijzermans JNM, Groenen PJTA, Schaap NPM, Kwekkeboom J, Joosten I. 2013. Human secondary lymphoid organs typically contain polyclonally-activated proliferating regulatory T cells. Blood 122:2213–2223. doi: 10.1182/blood-2013-03-489443. [DOI] [PubMed] [Google Scholar]
- 33.Vukmanovic-Stejic M, Zhang Y, Cook JE, Fletcher JM, McQuaid A, Masters JE, Rustin MHA, Taams LS, Beverley PCL, Macallan DC, Akbar AN. 2006. Human CD4+ CD25hi Foxp3+ regulatory T cells are derived by rapid turnover of memory populations in vivo. J Clin Invest 116:2423–2433. doi: 10.1172/JCI28941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vukmanovic-Stejic M, Agius E, Booth N, Dunne PJ, Lacy KE, Reed JR, Sobande TO, Kissane S, Salmon M, Rustin MH, Akbar AN. 2008. The kinetics of CD4+ Foxp3+ T cell accumulation during a human cutaneous antigen-specific memory response in vivo. J Clin Invest 118:3639–3650. doi: 10.1172/JCI35834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakaguchi S, Ono M, Setoguchi R, Yagi H, Hori S, Fehervari Z, Shimizu J, Takahashi T, Nomura T. 2006. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev 212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 36.Oswald-Richter K, Grill SM, Shariat N, Leelawong M, Sundrud MS, Haas DW, Unutmaz D. 2004. HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol 2:E198. doi: 10.1371/journal.pbio.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chachage M, Podola L, Clowes P, Nsojo A, Bauer A, Mgaya O, Kowour D, Froeschl G, Maboko L, Hoelscher M, Saathoff E, Geldmacher C. 2014. Helminth-associated systemic immune activation and HIV co-receptor expression: response to albendazole/praziquantel treatment. PLoS Negl Trop Dis 8:e2755. doi: 10.1371/journal.pntd.0002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarfo FS, Eberhardt KA, Dompreh A, Kuffour EO, Soltau M, Schachscheider M, Drexler JF, Eis-Hübinger AM, Häussinger D, Oteng-Seifah EE, Bedu-Addo G, Phillips RO, Norman B, Burchard G, Feldt T. 2015. Helicobacter pylori infection is associated with higher CD4 T cell counts and lower HIV-1 viral loads in ART-naïve HIV-positive patients in Ghana. PLoS One 10:e0143388. doi: 10.1371/journal.pone.0143388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberhardt KA, Sarfo FS, Dompreh A, Kuffour EO, Geldmacher C, Soltau M, Schachscheider M, Drexler JF, Eis-Hubinger AM, Haussinger D, Bedu-Addo G, Phillips RO, Norman B, Burchard GD, Feldt T. 2015. Helicobacter pylori coinfection is associated with decreased markers of immune activation in ART-naive HIV-positive and in HIV-negative individuals in Ghana. Clin Infect Dis 61:1615–1623. doi: 10.1093/cid/civ577. [DOI] [PubMed] [Google Scholar]
- 40.Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, Njovu L, Geis S, Hoffmann O, Maboko L, Williamson C, Birx D, Meyerhans A, Cox J, Hoelscher M. 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol 81:2440–2448. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann D, Wolfarth B, Hörterer HG, Halle M, Reichhuber C, Nadas K, Tora C, Erfle V, Protzer U, Schätzl HM. 2010. Elevated Epstein-Barr virus loads and lower antibody titers in competitive athletes. J Med Virol 82:446–451. doi: 10.1002/jmv.21704. [DOI] [PubMed] [Google Scholar]
- 42.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BTM, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J Virol 80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollakis G, Baan E, van Werkhoven MB, Berkhout B, Bakker M, Jurriaans S, Paxton WA. 2015. Association between gp120 envelope V1V2 and V4V5 variable loop profiles in a defined HIV-1 transmission cluster. AIDS 29:1161–1171. doi: 10.1097/QAD.0000000000000692. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 46.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2009. GenBank. Nucleic Acids Res 37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermüller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Töpfer A, Zagordi O, Prabhakaran S, Roth V, Halperin E, Beerenwinkel N. 2013. Probabilistic inference of viral quasispecies subject to recombination. J Comput Biol 20:113–123. doi: 10.1089/cmb.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angin M, Kwon DS, Streeck H, Wen F, King M, Rezai A, Law K, Hongo TC, Pyo A, Piechocka-Trocha A, Toth I, Pereyra F, Ghebremichael M, Rodig SJ, Milner DA, Richter JM, Altfeld M, Kaufmann DE, Walker BD, Addo MM. 2012. Preserved function of regulatory T cells in chronic HIV-1 infection despite decreased numbers in blood and tissue. J Infect Dis 205:1495–1500. doi: 10.1093/infdis/jis236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulze Zur Wiesch J, Thomssen A, Hartjen P, Tóth I, Lehmann C, Meyer-Olson D, Colberg K, Frerk S, Babikir D, Schmiedel S, Degen O, Mauss S, Rockstroh J, Staszewski S, Khaykin P, Strasak A, Lohse AW, Fätkenheuer G, Hauber J, van Lunzen J. 2011. Comprehensive analysis of frequency and phenotype of T regulatory cells in HIV infection: CD39 expression of FoxP3+ T regulatory cells correlates with progressive disease. J Virol 85:1287–1297. doi: 10.1128/JVI.01758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Presicce P, Orsborn K, King E, Pratt J, Fichtenbaum CJ, Chougnet CA. 2011. Frequency of circulating regulatory T cells increases during chronic HIV infection and is largely controlled by highly active antiretroviral therapy. PLoS One 6:e28118. doi: 10.1371/journal.pone.0028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montes M, Lewis DE, Sanchez C, Lopez de Castilla D, Graviss EA, Seas C, Gotuzzo E, White AC. 2006. Foxp3+ regulatory T cells in antiretroviral-naive HIV patients. AIDS 20:1669–1671. doi: 10.1097/01.aids.0000238415.98194.38. [DOI] [PubMed] [Google Scholar]
- 54.Simonetta F, Lecuroux C, Girault I, Goujard C, Sinet M, Lambotte O, Venet A, Bourgeois C. 2012. Early and long-lasting alteration of effector CD45RA(-)Foxp3(high) regulatory T-cell homeostasis during HIV infection. J Infect Dis 205:1510–1519. doi: 10.1093/infdis/jis235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, Sakaguchi S. 2006. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol 18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 56.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. 2010. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Getnet D, Grosso JF, Goldberg MV, Harris TJ, Yen HR, Bruno TC, Durham NM, Hipkiss EL, Pyle KJ, Wada S, Pan F, Pardoll DM, Drake CG. 2010. A role for the transcription factor Helios in human CD4(+)CD25(+) regulatory T cells. Mol Immunol 47:1595–1600. doi: 10.1016/j.molimm.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, Kaygusuz Y, Meissner T, Holderried TAW, Chan S, Kastner P, Haining WN, Cantor H. 2015. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science 350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baine I, Basu S, Ames R, Sellers RS, Macian F. 2013. Helios induces epigenetic silencing of Il2 gene expression in regulatory T cells. J Immunol 190:1008–1016. doi: 10.4049/jimmunol.1200792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ioannidis JP, Cappelleri JC, Lau J, Sacks HS, Skolnik PR. 1996. Predictive value of viral load measurements in asymptomatic untreated HIV-1 infection: a mathematical model. AIDS 10:255–262. doi: 10.1097/00002030-199603000-00003. [DOI] [PubMed] [Google Scholar]
- 61.Haase AT. 1999. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol 17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 62.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 63.Schacker T, Little S, Connick E, Gebhard K, Zhang ZQ, Krieger J, Pryor J, Havlir D, Wong JK, Schooley RT, Richman D, Corey L, Haase AT. 2001. Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J Infect Dis 183:555–562. doi: 10.1086/318524. [DOI] [PubMed] [Google Scholar]
- 64.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. 2005. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med 201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nilsson J, Boasso A, Velilla PA, Zhang R, Vaccari M, Franchini G, Shearer GM, Andersson J, Chougnet C. 2006. HIV-1-driven regulatory T-cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood 108:3808–3817. doi: 10.1182/blood-2006-05-021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA. 2009. Human regulatory T cells are targets for human immunodeficiency Virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J Virol 83:12925–12933. doi: 10.1128/JVI.01352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunham RM, Cervasi B, Brenchley JM, Albrecht H, Weintrob A, Sumpter B, Gordon S, Klatt NR, Frank I, Donald L, Douek DC, Paiardini M, Silvestri G, Engram J, Sodora DL, Sodora L. 2008. CD127 and CD25 expression defines CD4+ T cell subsets that are differentially depleted during HIV infection. J Immunol 180:5582–5592. doi: 10.4049/jimmunol.180.8.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chase AJ, Yang HC, Zhang H, Blankson JN, Siliciano RF. 2008. Preservation of FoxP3+ regulatory T cells in the peripheral blood of human immunodeficiency virus type 1-infected elite suppressors correlates with low CD4+ T-cell activation. J Virol 82:8307–8315. doi: 10.1128/JVI.00520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tran TA, de Goër de Herve MG, Hendel-Chavez H, Dembele B, Le Névot E, Abbed K, Pallier C, Goujard C, Gasnault J, Delfraissy JF, Balazuc AM, Taoufik Y. 2008. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One 3:e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCutchan FE, Hoelscher M, Tovanabutra S, Piyasirisilp S, Sanders-Buell E, Ramos G, Jagodzinski L, Polonis V, Maboko L, Mmbando D, Hoffmann O, Riedner G, von Sonnenburg F, Robb M, Birx DL. 2005. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J Virol 79:11693–11704. doi: 10.1128/JVI.79.18.11693-11704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, Schmidt TE, Thorkelson A, Reilly C, Perkey K, Reimann TG, Utay NS, Nganou Makamdop K, Stevenson M, Douek DC, Haase AT, Schacker TW. 2015. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci 112:E1126–E1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Xu H, Shen C, Alvarez X, Liu D, Pahar B, Ratterree MS, Doyle-Meyers LA, Lackner AA, Veazey RS. 2015. Profound loss of intestinal Tregs in acutely SIV-infected neonatal macaques. J Leukoc Biol 97:391–400. doi: 10.1189/jlb.4A0514-266RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. 2011. Helios expression is a marker of T cell activation and proliferation. PLoS One 6:e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verhagen J, Wraith DC. 2010. Comment on “Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells.” J Immunol 185:7129. doi: 10.4049/jimmunol.1090105. [DOI] [PubMed] [Google Scholar]
- 76.Sebastian M, Lopez-Ocasio M, Metidji A, Rieder SA, Shevach EM, Thornton AM. 2016. Helios controls a limited subset of regulatory T cell functions. J Immunol 196:144–155. doi: 10.4049/jimmunol.1501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]