ABSTRACT
Endogenous retroviruses (ERVs) are the remnants of ancient retroviral infections of germ cells. Previous work identified one of the youngest feline ERV groups, ERV-DC, and reported that two ERV-DC loci, ERV-DC10 and ERV-DC18 (ERV-DC10/DC18), can replicate in cultured cells. Here, we identified another replication-competent provirus, ERV-DC14, on chromosome C1q32. ERV-DC14 differs from ERV-DC10/DC18 in its phylogeny, receptor usage, and, most notably, transcriptional activities; although ERV-DC14 can replicate in cultured cells, it cannot establish a persistent infection owing to its low transcriptional activity. Furthermore, we examined ERV-DC transcription and its regulation in feline tissues. Quantitative reverse transcription-PCR (RT-PCR) detected extremely low ERV-DC10 expression levels in feline tissues, and bisulfite sequencing showed that 5′ long terminal repeats (LTRs) of ERV-DC10/DC18 are significantly hypermethylated in feline blood cells. Reporter assays found that the 5′-LTR promoter activities of ERV-DC10/DC18 are high, whereas that of ERV-DC14 is low. This difference in promoter activity is due to a single substitution from A to T in the LTR, and reverse mutation at this nucleotide in ERV-DC14 enhanced its replication and enabled it to persistently infect cultured cells. Therefore, ERV-DC LTRs can be divided into two types based on this nucleotide, the A type or T type, which have strong or attenuated promoter activity, respectively. Notably, ERV-DCs with T-type LTRs, such as ERV-DC14, have expanded in the cat genome significantly more than A-type ERV-DCs, despite their low promoter activities. Our results provide insights into how the host controls potentially infectious ERVs and, conversely, how ERVs adapt to and invade the host genome.
IMPORTANCE The domestic cat genome contains many endogenous retroviruses, including ERV-DCs. These ERV-DCs have been acquired through germ cell infections with exogenous retroviruses. Some of these ERV-DCs are still capable of producing infectious virions. Hosts must tightly control these ERVs because replication-competent viruses in the genome pose a risk to the host. Here, we investigated how ERV-DCs are adapted by their hosts. Replication-competent viruses with strong promoter activity, such as ERV-DC10 and ERV-DC18, were suppressed by promoter methylation in LTRs. On the other hand, replication-competent viruses with weak promoter activity, such as ERV-DC14, seemed to escape strict control via promoter methylation by the host. Interestingly, ERV-DCs with weak promoter activity, such as ERV-DC14, have expanded in the cat genome significantly more than ERV-DCs with strong promoter activity. Our results improve the understanding of the host-virus conflict and how ERVs adapt in their hosts over time.
INTRODUCTION
Endogenous retroviruses (ERVs) are resident DNA copies that abound in host chromosomal DNA and compromise ∼8 to 10% of human and mouse genomes (1, 2). They have been found in all vertebrates, including mammals, fish, birds, reptiles, and amphibians (3). ERVs were originally acquired through germ cell infections with exogenous retroviruses of ancestral host lineages and have thereafter been transmitted vertically from parent to offspring according to Mendelian inheritance patterns (4, 5). Consequently, mammalian genomes are littered with the signatures of ancient retroviral infections. Most ERVs have already been inactivated by accumulated mutations that disrupt viral genes or regulatory sequences.
Although most ERVs are inactivated, a few ERVs that still retain potential replication capacities have been observed in several animals (6), including mice (7), koalas (8, 9), pigs (10–14), and mule deer (15, 16). Some of these ERVs reside in the host genome as intact proviruses, while others appear to have been generated through reassortment or recombination among inactivated proviruses (17–19). The hosts have some silencing mechanisms against these potentially active ERVs; however, failure of their suppression can cause malignant diseases, such as leukemia or lymphoma (18–20) in mice, or can alter phenotypes in domestic cats (21) or in mice (22) by insertional mutagenesis.
Retroviruses, including ERVs, typically consist of gag, pol, and env genes with two flanking noncoding long terminal repeats (LTRs). The LTRs contain a transcriptional start site and various regulatory cis elements, which act as a promoter and an enhancer that determine viral transcriptional tropism (23). Hosts often target the ERV LTRs via epigenetic mechanisms, such as nucleotide CpG methylation or suppressive histone modification, to silence the ERVs (24, 25).
ERV in domestic cats (Felis silvestris catus) (26, 27), termed ERV-DC (28), is one of the youngest feline ERV groups. We previously identified and molecularly cloned 13 loci of ERV-DC (28). These ERVs belong to the gammaretrovirus genus, and they initially invaded the cat genome 2.8 million years ago (28). This invasion is ongoing, and today, cats generally have 7 to 16 copies of ERV-DC in their genomes (28). ERV-DCs are classified into three different genotypes: genotype I (ERV-DC1, -DC2, -DC3, -DC4, -DC8, -DC14, -DC17, and -DC19), genotype II (ERV-DC7 and -DC16), and genotype III (ERV-DC6, -DC10, and -DC18) (28). Notably, ERV-DC10 and ERV-DC18 reside in the feline genome and retain their capacities for infection and replication. ERV-DC10 and ERV-DC18 are located on chromosome C1q12-q21 and in the D4q14 region, respectively, and their sequences are completely identical except for one nucleotide in the U5 primer binding site (U5-PBS) of tRNAGly. ERV-DC18 is found in only a certain family of cats, and so this virus seems to have been generated by reinfection by ERV-DC10 (28). Thus, ERV-DCs are capable of giving rise to new provirus insertions in vivo. Furthermore, ERV-DCs can behave as donors and/or acceptors of viral recombination and generate novel exogenous recombinant viruses, termed feline leukemia virus (FeLV) subgroup D (FeLV-D) (28). Thus, ERV-DCs can still impact the lives of their hosts through both their potential replication activity and their contribution to the emergence of recombinant viruses.
In addition to those described above, the feline genome contain other types of ERV-DCs, ERV-DC7 and ERV-DC16. Both of these ERVs encode Refrex-1, a restriction factor against ERV-DC genotype I and FeLV-D, in their truncated env open reading frames (ORFs) (29). ERV “domestication” is a phenomenon in which ERVs gain a physiological function, such as placentation or viral resistance, and eventually evolve into host genes (30, 31). ERV domestication has occurred through multistep modifications of the viral genes, including mutations as well as allelic or nonallelic gene conversions (32, 33). Refrex-1 appears to be a typical example of ERV domestication, as are Fv1 (34), Fv4 (35), Rmcf (36), Rmcf2 (37), and endogenous Jaagsiekte sheep retrovirus (enJSRV) (33, 38), which all play a role in viral resistance.
In this study, we identified another replication-competent ERV, ERV-DC14. ERV-DC14 differs from ERV-DC10 and ERV-DC18 (ERV-DC10/DC18) in its phylogeny, receptor usage, and, especially, transcriptional activities. Thus, two distinct subsets of infectious ERVs reside in the feline genome. Furthermore, to explore how the host controls these infectious ERVs, we investigated ERV-DC transcription and its regulatory mechanism. Our results reveal that these two distinct types of infectious ERVs are controlled by two different mechanisms. These findings demonstrate the regulatory effects of the interplay between ERVs and hosts.
MATERIALS AND METHODS
Ethics approval.
Animal studies were conducted according to guidelines for the care and use of laboratory animals of the Ministry of Education, Culture, Sports, Science, and Technology, Japan (73). All experiments were approved by the Genetic Modification Safety Committee of Yamaguchi University.
Cells.
HepG2 (39), HeLa (40), MCF7 (41), Cos7 (42), HEK293T (43), AH927 (44), G355 (45, 46), CRFK (47), NIH 3T3 (48), Vero (49), BHK-21 (50), MDTF (51), 104C1 (52), KwDM (28), and MDBK (53) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. HepG2, HeLa, MCF7, and Cos7 cells were obtained from the Center for Gene Research, Yamaguchi University. HEK293T, AH927, G355, CRFK, and NIH 3T3 cells were obtained from Sandra Ruscetti (National Cancer Institute [NCI]). Vero and BHK-21 cells were obtained from the Cell Resource Center for Biomedical Research, Institute of Development, Aging, and Cancer, Tohoku University. MDTF cells were obtained from Yoshinao Kubo (Nagasaki University). 104C1 cells were obtained from the JCRB (Japanese Collection of Research Bioresources) cell bank. MDBK cells were obtained from Kenji Baba (Yamaguchi University). 293Lac cells (54) were established by transfecting HEK293T cells with the LacZ-coding retroviral vector pMxs-nLIP (28). The LacZ-coding retroviral vector is rescued in the presence of a replication-competent ERV in 293Lac cells. These cells were also maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
Viruses.
ERV-DC10 (28), ERV-DC18 (28), ERV-DC14 (28), and ERV-DC14TA were prepared by transfecting each plasmid into HEK293T cells or 293Lac cells by using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Culture supernatants were collected 3 days after transfection, filtered through 0.45-μm-pore-size filters, and stored as virus stocks at −80°C until use. ERV-DC14TA, which contains a substitution of T to A at positions 281 and 8598 in the LTRs of ERV-DC14, was constructed by using QuikChange II site-directed mutagenesis kits (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions, with the following complementary primers: DC8-mu6S (5′-CTC CAA GTT GCA TCA GCC GAG AGA AAC TCC-3′) and its complementary sequence (5′-GGA GTT TCT CTC GGC TGA TGC AAC TTG GAG-3′).
Infection assay.
Target cells were infected with the viral stocks in the presence of 8 μg/ml of Polybrene (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Two or three days after infection, the cells were stained with X-Gal (5-bromo-4-chloro-indolyl-β-d-galactopyranoside; Wako, Osaka, Japan), and viral titers were determined as infectious units (IU) per milliliter by counting blue-stained nuclei.
Virus purification.
Culture supernatants (5 ml) were collected, filtered through 0.45-μm-pore-size filters, and ultracentrifuged for 30 min at 29,000 × g at 4°C in an Optima Max-XP ultracentrifuge (Beckman Coulter KK, Ariake, Tokyo, Japan). The virions were resuspended in 30 μl of phosphate-buffered saline (PBS) and were used for Western blot analyses.
Immunoblotting.
Cells were lysed in buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 1 mM Na3VO4, and 1 μg/ml each of aprotinin and leupeptin), followed by incubation on ice for 20 min. Insoluble components were removed by centrifugation at 18,000 × g for 20 min at 4°C, and the protein concentrations of the resulting samples were determined by using a protein assay kit (Bio-Rad Laboratories, Carlsbad, CA, USA) according to the manufacturer's instructions. Simultaneously, culture supernatants were collected, and the viruses were purified as described above. Proteins were separated by electrophoresis on 7.5% or 10-to-20% gradient Tris-glycine minigels (Oriental Instruments Co., Ltd., Kanagawa, Japan) under reducing conditions (3.5 × 10−2 M 2-mercaptoethanol) and then transferred via electrophoresis onto nitrocellulose filters for Western blotting with goat anti-RD-114 (NCI, Frederick, MD, USA), goat anti-FeLV gp70 (NCI), and mouse anti-human β-actin (Santa Cruz Biotechnology). The blots were incubated with a horseradish peroxidase (HRP)-conjugated anti-goat IgG (Santa Cruz Biotechnology) or an anti-mouse IgG (GE Healthcare Japan, Tokyo, Japan) secondary antibody, followed by visualization using a 20× LumiGLO instrument (Cell Signaling Technology, Danvers, MA, USA).
Detection of ERV-DCs by reverse transcription-PCR (RT-PCR).
Feline tissues were obtained from a specific-pathogen-free (SPF) cat (Kyoto-SPF1) described in our previous study (29). Peripheral blood samples were obtained from seven healthy domestic cats. Total RNA was extracted from cells or tissues with an RNAiso Plus kit (TaKaRa, Kyoto, Japan) according to the manufacturer's instructions, and cDNA was synthesized with a PrimeScript II first-strand cDNA synthesis kit (TaKaRa) according to the manufacturer's instructions for inclusion in a treatment with recombinant DNase I (TaKaRa). Viral RNA was isolated from cell culture supernatants by using the QIAamp viral RNA minikit (Qiagen, Tokyo, Japan) according to the manufacturer's instructions. PCR was performed by using a Kod Fx Neo kit (Toyobo Co., Osaka, Japan) with primer set Fe-13S (5′-CGG TCC AGC TAG CCA TCC CAG TC-3′) and Fe-18R (5′-ACA GAT CTG CCG CCG GGT TCG TAG TGG CC-3′) for env and primer set Fe-73S (5′-AAT TTG GAC CTC CGG TTA CCT TG-3′) and Fe-96R (5′-TTC CTT TCG GGG AAG GAC TA-3′) for pol, and for human β-actin, previously reported primers were used (28).
Qualitative PCR of ERV-DCs was conducted by using the cDNAs described above. The expression of ERV-DC was detected via PCR using primers Fe-184S (5′-CCT AGG RGC TTG GBT CCY AAC ATT TGG TG-3′) and Fe-168R (5′-GAA GRT AGG GTG GGG GTG TKT TAG TAA GCT A-3′), which were designed for the end of the 5′ LTR and the start of the 3′ LTR, respectively (29). PCR fragments were cloned into a pCR-Blunt vector (Invitrogen) and then sequenced. ERV-DC genotype-specific SYBR green-based quantitative PCR was established. cDNA from feline tissues was amplified with SYBR Premix Ex Taq II (Tli RNase H Plus; TaKaRa) in a CFX96 Touch real-time PCR detection system (Bio-Rad Laboratories) by using the following primer pairs: Fe228S (5′-GCT TGC ACT TCC ACC AGT TG-3′) and Fe205R (5′-ACC TGT TCC TGT CTT GCG TAG-3′) for ERV-DC genotype I, Fe232S (5′-GCC AGA TAC AAT CGA ATG AAA GG-3′) and Fe206R (5′-TGC CAA CTG GTT TTG TTA CTT ATG-3′) for ERV-DC genotype II, and Fe230S (5′-GCC TCC CTA CCC GAC TTC C-3′) and Fe207R (5′-AGG GGG TTT AGC CGT TAG G-3′) for ERV-DC genotype III. The gene for feline peptidyl prolyl isomerase A (PPIA) was used as a reference gene (55).
Transmission electron microscopy (TEM).
HEK293T cells transfected with ERV-DC14 or ERV-DC14TA were washed with PBS, fixed with 2% glutaraldehyde at 4°C for at least 2 h, washed with PBS again, and postfixed with 2% osmium tetroxide for 1.5 h. Cells were dehydrated through a graded ethanol series followed by propylene oxide and then embedded in Epon 812 (TAAB Laboratories Equipment Ltd., Aldermaston, Berks, United Kingdom) for 48 h at 60°C. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined with an electron microscope (JEM-1200EX; JEOL, Tokyo, Japan).
Fluorescence in situ hybridization analysis.
Feline peripheral blood mononuclear cell (PBMC) cultures stimulated with 3 μg/ml of concanavalin A (Sigma-Aldrich, St. Louis, MO, USA) were treated with 25 μg/ml E5-bromo-2′-deoxyuridine (Sigma-Aldrich) for 5.5 h, subsequently treated with 20 ng/ml of colcemid (Nacalai Tesque, Kyoto, Japan) for 0.5 h, and then harvested. Replication-banded chromosomes were obtained by exposure of the chromosome slides to UV light after staining with Hoechst 33258. pGM14L2 (2.0 kb of ERV-DC14 flanking sequences amplified by PCR with primers Fe-245S [5′-GAG CCC TGG TGC ACC AGT AAA GA-3′] and Fe-413R [5′-CCT AGT GCT CAT GGG GAA AA-3′]) was labeled with digoxigenin-11-dUTP by nick translation for use as probes. After hybridization, the slides were washed, and the probe signals were detected with Cy3-labeled antidigoxigenin. Fluorescence in situ hybridization (FISH) images were captured with the CW4000 FISH application program (Leica Microsystems, Wetzlar, Germany).
Bisulfite cytosine methylation analysis.
Genomic DNAs from feline blood and virus-infected HEK293T cells were isolated by using a QIAamp DNA blood minikit (Qiagen) according to the manufacturer's instructions. The ERV-DC LTR DNA methylation patterns were investigated by using a MethylEasy Xceed Rapid DNA bisulfite modification kit (TaKaRa) according to the manufacturer's instructions. The 5′ LTRs of ERV-DCs were amplified by using nested PCR with locus-specific or ERV-DC10/DC18-specific primer pairs (listed in Table 1) in ERV-DC10- or ERV-DC18-infected HEK293T cells in combination with bisulfite-treated genomic DNA (listed in Table 2). TaKaRa Taq Hot Start or TaKaRa EpiTaq HS DNA polymerases were used in these PCR assays according to the manufacturer's instructions. The amplicons were cloned into pGEM-T (Promega), and 12 clones for each sample were used for the determination of their sequences. The bisulfite data were also analyzed by using QUMA software (http://quma.cdb.riken.jp/). To make a statistical judgment on the methylation rate of each CpG position, we used a 90% confidence interval of the mean methylation rate value for each of the ERV-DC proviruses. When the methylation rate of a CpG position exceeded the upper limit of the 90% confidence interval, we considered the methylation rate of that CpG position to be significantly higher (P < 0.05). The calculation of the confidence interval was performed by using SAS 9.4 (SAS Institute, Inc., Cary, NC, USA).
TABLE 1.
PCR primers used for analysis of ERV-DC LTR DNA methylation patternsa
| 5′ LTR of ERV-DC | Forward primer | Forward primer sequence (5′–3′) | Reverse primer | Reverse primer sequence (5′–3′) |
|---|---|---|---|---|
| ERV-DC1 | Fe-155S | GTTAGGGAGAGGGTTTTATTGTTT | Fe-144R | ACAAAAATCTATCTTCAATCAATCC |
| Fe-156S | GGTTTTTGGAAATATGTTTTTAAA | Fe-145R | CAAAACATAAAACACAATACCTATATACAA | |
| ERV-DC3 | Fe-157S | TTTTAGAAATTAAAAAGAAGGTAGGAGAGT | Fe-146R | ACCTCAAAATTCACAATTCAAAAAC |
| Fe-158S | TGAGTATGTTTAATGTAATTTTTTAAATT | Fe-147R | CTTCAACACTCCTACTAAAATAACC | |
| ERV-DC4 | Fe-159S | GTTAATTTATTTTGGGTGTAGTGAT | Fe-144R | |
| Fe-160S | TGTGTATTTAGTAGTGGGTTTTTTAG | Fe-148R | AAAATTCACAATTCAAAAACAAAAC | |
| ERV-DC7 | Fe-161S | AGTAAATGGGTATTGAGTTGGGTAAT | Fe-149R | TATACAAACAAAAATACAAAAATCC |
| Fe-162S | TTAAGAAAGAATTTATTGAGAATATAA | Fe-150R | AATACAAAAACAAAACACAATACCT | |
| ERV-DC8 | Fe-163S | TTTTTAGGGTAATATTGGAGTTTAA | Fe-144R | |
| Fe-164S | TGGATAATAAATTTAGGATGTTAGAATTAT | Fe-151R | CAATTCAAAAACAAAACATAAAACAC | |
| ERV-DC10 | Fe-150S | TTTTAGTGGGGATTTTGTAAATTGGT | Fe-143R | AACATAAAACACAATACCTATATACAAACA |
| Fe-154S | TTTTGGTAGTTTTATAAGAGAAAAGAGAGA | Fe-142R | TCTATCCTCCTTCAACACTCCTACTAAAAT | |
| ERV-DC14 | Fe-165S | TTTAGATTTGTAGTTAAGGGATTGTG | Fe-147R | |
| Fe-166S | GTAGATTTGTTATTAAATTGGTTTTTAGG | Fe-152R | ATAACAAAAAAAACCTACCTTC | |
| ERV-DC16 | Fe-225S | AGAAATGGTTAAGGAAAGTTTTAGGTT | Fe-147R | |
| Fe-226S | AGGTTTAAATTTGAAAGATTTTTGTTTAGT | Fe-149R | ||
| ERV-DC17 | Fe-167S | ATATAGTTTGGGGGATGAGATTATTTATTA | Fe-153R | AAATTCTCCTACCTTCCCTCTCTT |
| Fe-168S | TTAAGAGTTTTAAAATTTGTTTTTG | Fe-144R | ||
| ERV-DC18 | Fe-152S | TAAGGAGAGTTTGAATAAGGTTAAGGTTG | Fe-142R | |
| Fe-153S | GAGTAGTGGGAGGTGTTTGGTTT | Fe-143R | ||
| ERV-DC19 | Fe-169S | TAGTGGGATTTTTAAATGTAATGTT | Fe-169R | CAAAAACACAAAAATCTATCTTCAATC |
| Fe-170S | GTTGATTAGAAGTTTGGTGGGTT | Fe-151R | ||
| 293T/ERV-DC10 | Fe-151S | TGAGAGATTTTTGGTTTAGTTTATTAAAAT | Fe-142R | |
| Fe-151S | Fe-143R | |||
| 293T/ERV-DC18 | Fe-151S | Fe-142R | ||
| Fe-151S | Fe-143R |
The 5′ LTRs of ERV-DCs were amplified by nested PCR using locus-specific or ERV-DC10/DC18-specific primer pairs.
TABLE 2.
Information on the 5′ LTRs of ERV-DC and DNA samples used for analysis of ERV-DC LTR DNA methylation patternsa
| ERV-DC | Length of 5′ LTR (bp) | No. of CpGs | Cat or cell line | Genotype | Viral source |
|---|---|---|---|---|---|
| ERV-DC1 | 550 | 22 | ON-C | +/−, GI | |
| ERV-DC3 | 550 | 22 | ON-C | +/−, GI | |
| ERV-DC4 | 550 | 22 | GF33 | +/−, GI | |
| ERV-DC7 | 545 | 25 | GF33 | +/+, GII | |
| ERV-DC8 | 550 | 21 | GF33 | +/−, GI | |
| ERV-DC10 | 551 | 26 | AH927 cells | +/+, GIII | |
| GF33 | +/−, GIII | ||||
| HK25 | +/−, GIII | ||||
| NS14 | +/−, GIII | ||||
| TK14 | +/+, GIII | ||||
| YG22 | +/+, GIII | ||||
| ON-C | +/+, GIII | ||||
| ON-H | +/+, GIII | ||||
| ON-T | +/−, GIII | ||||
| 293T/AH927 cells | Infection | AH927 | |||
| 293T/GF33 cells | Infection | GF33 | |||
| 293T/HK25 cells | Infection | HK25 | |||
| 293T/NS14 cells | Infection | NS14 | |||
| 293T/TK14 cells | Infection | TK14 | |||
| 293T/YG22 cells | Infection | YG22 | |||
| ERV-DC14 | 551 | 21 | SO38 | +/−, GI | |
| ERV-DC16 | 575 | 26 | GF33 | +/+, GII | |
| ERV-DC17 | 551 | 21 | ON-C | +/−, GI | |
| ERV-DC18 | 551 | 26 | ON-C | +/−, GIII | |
| ON-H | +/−, GIII | ||||
| ON-T | +/−, GIII | ||||
| 293T/ON-C cells | Infection | ON-C | |||
| 293T/ON-H cells | Infection | ON-H | |||
| 293T/ON-T cells | Infection | ON-T | |||
| ERV-DC19 | 555 | 20 | IS10 | +/−, GI |
DNA samples from cats or from feline AH927 cells were used in this study. The length of the 5′ LTR, number of CpG sites, and genotype of the ERV-DCs are shown. In the genotype column, +/− indicates heterozygous, +/+ indicates homozygous, and infection indicates that the corresponding viruses from cats can replicate in HEK293T cells. HEK293T cells infected with ERV-DC10 or ERV-DC18 were described in a previous study (28). GI, genotype I; GII, genotype II; GIII, genotype III.
Luciferase reporter assay.
The reporter plasmids pLTR DC1-Luc, pLTR DC4-Luc, pLTR DC7-Luc, pLTR DC8-Luc, pLTR DC10-Luc, pLTR DC14-Luc, pLTR DC16-Luc, pLTR DC17-Luc, and pLTR DC19-Luc were constructed as described below. The 5′ LTR of each ERV-DC provirus (28, 29) was amplified by using PCR with the specific primers listed in Table 3. The forward primers bind upstream of the 5′ LTR and contain a KpnI site, and the reverse primer (Fe-157R) binds downstream of the 5′ LTR and has a SacI linker. Each resulting KpnI and SacI fragment of the LTR was cloned upstream of the luciferase gene in the pGL4.10[luc2] vector (Promega, Madison, WI, USA).
TABLE 3.
PCR primers used in the construction of the reporter plasmidsa
| Reporter plasmid | Primer | Sequence (5′–3′) |
|---|---|---|
| pLTR DC1-Luc | Fe-174S | AAAGGTACCAAACATGGCAGCATTTGTGG |
| Fe-157R | TTTGAGCTCACCGACGGGGCCGGGGTTCTC | |
| pLTR DC4-Luc | Fe-175S | AAAGGTACCCAATTTACCTTGGGTGCAGTGA |
| Fe-157R | ||
| pLTR DC7-Luc | Fe-199S | AAAGGTACCTGCAATTTGGGAGACACAGACT |
| Fe-157R | ||
| pLTR DC8-Luc | Fe-176S | AAAGGTACCGCCTTCACACTGGGAGAGATTT |
| Fe-157R | ||
| pLTR DC10-Luc | Fe-190S | AAAGGTACCATCCTGTGAGTTGGTGAGAGACC |
| Fe-157R | ||
| pLTR DC14-Luc | Fe-178S | AAAGGTACCTGCAGTTAAGGGACTGTGGACT |
| Fe-157R | ||
| pLTR DC16-Luc | Fe-223S | AAAGGTACCGCCACGGTCATGAAAATAAAAA |
| Fe-157R | ||
| pLTR DC17-Luc | Fe-179S | AAAGGTACCCAGCTTGGGGGATGAGATTATT |
| Fe-157R | ||
| pLTR DC19-Luc | Fe-180S | AAAGGTACCGAAACCTCCCACAAAAGTCCAC |
| Fe-157R | ||
| pLTR DC8-mu6 | DC8-mu6S | CTCCAAGTTGCATCAGCCGAGAGAAACTCC |
| DC8-mu6R | GGAGTTTCTCTCGGCTGATGCAACTTGGAG | |
| pLTR DC8-mu7 | DC8-mu7S | CTCTCTGCCGAAGTCGGAGTGCCGTGTGGT |
| DC8-mu7R | ACCACACGGCACTCCGACTTCGGCAGAGAG | |
| pLTR DC8-mu8 | DC8-mu8S | CGTGTGGTTCTTTGCACCAACTCTCATTCCATAG |
| DC8-mu8R | CTATGGAATGAGAGTTGGTGCAAAGAACCACACG | |
| pLTR DC10A281T | DC10-mu1S | GGCTCCAAGTTGCATCTGCCAAAAGAAACTTCA |
| DC10-mu1R | TGAAGTTTCTTTTGGCAGATGCAACTTGGAGCC | |
| pLTR DC19A285T | DC19-mu1S | CTCCAAGTTGCATCTGCCGAGAGAAACTCC |
| DC19-mu1R | GGAGTTTCTCTCGGCAGATGCAACTTGGAG |
Plasmids pLTR DC1-Luc, pLTR DC4-Luc, pLTR DC7-Luc, pLTR DC8-Luc, pLTR DC10-Luc, pLTR DC14-Luc, pLTR DC16-Luc, pLTR DC17-Luc, and pLTR DC19-Luc were constructed by PCR amplification with specific primer pairs. Reporter plasmids pLTR DC8-mu6, pLTR DC8-mu7, pLTR DC8-mu8, pLTR DC10-A281T, and pLTR DC19-A285T containing a point mutation in the LTR were generated by site-directed mutagenesis using complementary primers.
Chimeric pLTR DC8/DC19-luc was generated by the ligation of the KpnI and SmaI fragments of pLTR DC8-luc with the SmaI and SacI fragments of pLTR DC19-luc in the pGL4.10[luc2] vector. Chimeric pLTR DC19/DC8-luc was similarly generated by the ligation of the KpnI and SmaI fragments of pLTR DC19-luc with the SmaI and SacI fragments of pLTR DC8-luc in the pGL4.10[luc2] vector.
Reporter plasmids with a point mutation in the LTR, pLTR DC8-mu6, pLTR DC8-mu7, pLTR DC8-mu8, pLTR DC10-A281T, and pLTR DC19-A285T, were generated by site-directed mutagenesis using pLTR DC8-Luc, pLTR DC10-Luc, and pLTR DC19-Luc as the templates along with the complementary primers listed in Table 3.
Each LTR reporter plasmid (1 μg) together with Renilla luciferase-expressing plasmid phRL-CMV (50 ng) were cotransfected into cells cultured in 12-well plates by using Lipofectamine 2000 (Invitrogen). Luciferase and Renilla luciferase activity levels were measured from the cell lysates ∼48 h after transfection by using the Dual Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Statistical analyses of the obtained results were performed by using Student's t tests.
In vitro methylation of reporter plasmids.
Reporter plasmids (3 μg) were treated with either CpG methyltransferase (M.SssI) or heat-inactivated M.SssI (20 min at 65°C) supplemented with 160 μM S-adenosylmethionine (SAM) for 4 h at 37°C, and the plasmids were then extracted with phenol-chloroform, followed by ethanol precipitation. To determine the methylation levels, the DNAs were treated with the CpG methylation-sensitive restriction enzyme BstUI, separated by gel electrophoresis, and then visualized by ethidium bromide staining.
Phylogenetic analysis of ERV-DC LTRs.
We constructed a phylogenetic tree of the LTRs harbored by ERV-DC proviruses based on the following sequences: the ERV-DC1 5′ LTR (GenBank accession number AB674439.1), the ERV-DC2 5′ LTR (accession number AB674452.1), the ERV-DC3 5′ LTR (accession number AB674440.1), the ERV-DC4 5′ LTR (accession number AB674441.1), the ERV-DC6 3′ LTR (accession number AB674450.1), the ERV-DC7 5′ LTR (accession number AB807599.1), the ERV-DC8 5′ LTR (accession number AB674443.1), the ERV-DC10 5′ LTR (accession number AB674444.1), the ERV-DC14 5′ LTR (accession number AB674445.1), the ERV-DC16 5′ LTR (accession number AB807600.1), the ERV-DC17 5′ LTR (accession number AB674446.1), the ERV-DC18 5′ LTR (accession number AB674447.1), and the ERV-DC19 5′ LTR (accession number AB674448.1). Multiple-sequence alignments of LTRs were generated by using MUSCLE (56). Nucleotide substitution models based on a Kimura 2-parameter model (57) were selected based on their Bayesian information criterion scores (58). A phylogenetic tree was constructed by using the maximum likelihood method, and its robustness was evaluated by bootstrapping (1,000 times).
We next obtained ERV-DC LTRs from a reference genome of F. catus (ICGSC Felis_catus 6.2/felcat5) by using the BLAT program implemented in the UCSC (University of California—Santa Cruz) genome browser (http://genome.ucsc.edu/), and we conducted phylogenetic analyses with them. The 5′ LTRs of ERV-DC14, -DC7, and -DC10 were used as the search queries. LTRs containing deletions or missing data were eliminated from this analysis. Additionally, RD-114 LTRs were also eliminated based on the 5′ portions of their LTR sequences, which have a large insertion/deletion between ERV-DC and RD-114. In this search, 92 ERV-DC LTRs were identified, and most of them were solo LTRs. Multiple LTR alignments were generated by using MUSCLE (56). Nucleotide substitution models based on the Kimura 2-parameter model (57) with an inferred proportion of invariable sites (+I = 0.552039) were selected based on their Bayesian information criterion scores (58). A phylogenetic tree was constructed by using the maximum likelihood method, and its robustness was evaluated by bootstrapping (1,000 times). All programs used in this study were packaged in MEGA6 (59). The statistical analysis of ERV-DC invasion of the cat genome was performed by using a chi-squared test.
RESULTS
ERV-DC14 is an infectious virus.
We previously identified 13 ERV-DCs in domestic cats. Of these proviruses, ERV-DC10 and ERV-DC18 have now been established as replication-competent viruses (28), but although ERV-DC14 also has intact open reading frames, our previous work did not include an in-depth study of the viral infectivity of this provirus. Therefore, we began this study by determining whether or not ERV-DC14 is replication competent and infectious.
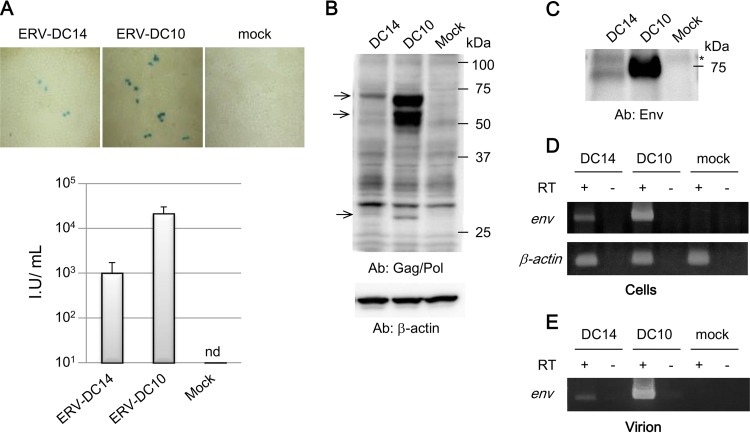
We initially established indicator cells, termed 293Lac cells (54), by stably transfecting a LacZ-expressing retroviral vector, pMxs-nlLacZ, into HEK293T cells to monitor viral infection and replication. The supernatant from 293Lac cells transfected with a full-length ERV-DC14 plasmid was filtered and then used to treat fresh HEK293T cells for 2 days, after which LacZ staining was performed. As shown in Fig. 1A, treatment with the supernatant from ERV-DC14-transfected cells produced a LacZ titer of ∼103 IU/ml. Treatment with the supernatant from ERV-DC10-transfected cells produced a much higher viral titer (∼2 × 104 IU/ml) than did treatment with the supernatant from ERV-DC14-transfected cells. As expected, HEK293T cells treated with the supernatant from mock-transfected cells were not LacZ positive (Fig. 1A).
FIG 1.
Assessment of the replication competence of ERV-DC14. (A) 293Lac cells were transfected with plasmids containing ERV-DC10 or ERV-DC14 or were mock transfected, and the supernatants were then collected and used to infect HEK293T cells. Cells were fixed at 48 h postinfection and visualized by X-Gal staining (top). The viral titers are expressed as infectious units (IU) per milliliter (bottom) with standard deviations. nd, not detected. (B) Western blot analysis, using an anti-RD-114 (an anti-Gag/Pol) antibody (Ab), of cell lysates from HEK293T cells at 48 h posttransfection that had been transfected with plasmids containing ERV-DC10 or ERV-DC14 or that had been mock transfected. The membranes from the original Western blot assay were stripped and reanalyzed by using an anti-β-actin antibody. Arrows indicate ERV-DC-specific bands. (C) Western blot analysis of purified virions using anti-Env antibody. Virions were purified at 48 h posttransfection from the culture supernatants of HEK293T cells that had been transfected with plasmids containing ERV-DC10 or ERV-DC14 or that had been mock transfected. The asterisk indicates a nonspecific band. (D) Detection of ERV-DC env mRNA in HEK293T cells 48 h after infection with ERV-DC10, ERV-DC14, or mock virus by RT-PCR. (E) Detection of viral RNA in the supernatant of HEK293T cells 48 h after infection with ERV-DC10, ERV-DC14, or mock virus by RT-PCR. DNA was visualized with ethidium bromide staining, and cDNA was synthesized with (+) or without (−) reverse transcriptase (RT).
Western blot analyses showed that viral proteins corresponding to Gag and Env were detected with 73S-045 (anti-Gag/Pol) and 81S-210 (anti-Env) antisera, respectively, in the cell lysates (Fig. 1B) and in the supernatants (Fig. 1C) from cells transfected with ERV-DC14 or ERV-DC10. Furthermore, when the supernatants from HEK293T cells that had been transfected with ERV-DC14 or ERV-DC10 were used to treat uninfected HEK293T cells for 2 days, ERV-DC14 and -DC10 viral mRNA and virions were detected by RT-PCR with ERV-DC-specific env primers in the cells themselves (Fig. 1D) as well as in the supernatants of the cells (Fig. 1E). These results demonstrate that ERV-DC14 is a replication-competent virus and is capable of being transmitted among cells. The levels of viral transcription, production, replication, and transmission of ERV-DC14 were all lower than those of ERV-DC10.
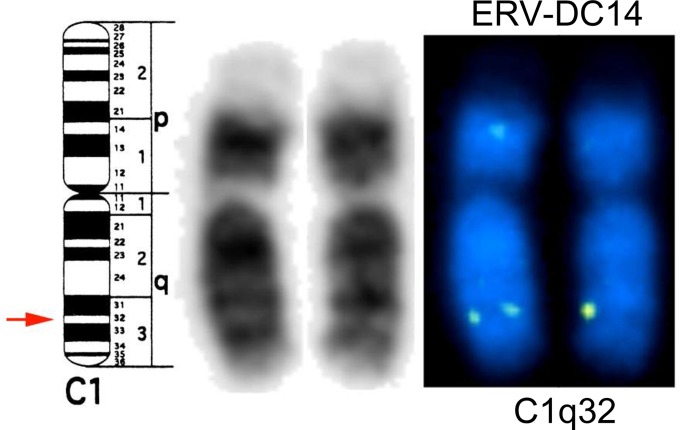
To define the ERV-DC14 locus, we conducted fluorescence in situ hybridization (FISH) analysis using the 2.0-kb ERV-DC14 flanking sequences that had been amplified by PCR as a FISH probe. ERV-DC14 was mapped to feline chromosome C1q32 (Fig. 2). These results indicate that ERV-DC14 and -DC10 (located on chromosome C1q12-21) (28) are located on the same chromosome but are completely distinct from each other.
FIG 2.
FISH analysis of ERV-DC14. (Left and middle) Cytogenetic map (left) and G banding (middle) of feline chromosome C1. (Right) Results of FISH performed by using 2.0-kb ERV-DC14 flanking sequences as a probe.
Expression of ERV-DCs in feline tissues.
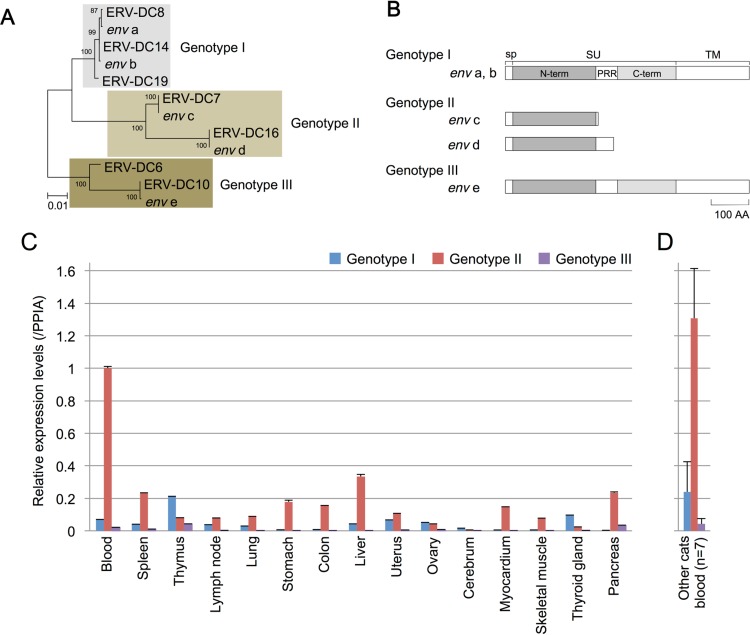
Although we have previously shown that Refrex-1, which is encoded by both the ERV-DC7 and ERV-DC16 loci, is expressed in domestic cats (29), it remained unclear whether or not the other ERV-DCs were also expressed in feline tissues. Therefore, we attempted to determine the overall expression of ERV-DCs in an SPF cat. cDNA was obtained from the tissues of an SPF cat, and ERV-DC transcripts were amplified by using RT-PCR with the ERV-DC-specific primers Fe-184S and Fe-168R, which correspond to the 5′ and 3′ LTRs, respectively, and these amplicons were then cloned and sequenced. Sequence analysis indicated that the 2.5-kb fragments were env subgenomic RNA. Several different transcripts of env genes were detected from feline tissues, and they were genotyped by constructing a phylogenetic tree (Fig. 3A). A schematic of Env encoded in these transcripts is shown in Fig. 3B. All three ERV genotypes (genotypes I, II, and III) were expressed in feline tissues (Fig. 3C). Three of the env genes (env a, env b, and env e) encode full-length Env, while two of them (env c and env d) encode truncated Env proteins corresponding to Refrex-1, which is derived from the ERV-DC7 and ERV-DC16 env genes (29). The env a and env b sequences are highly similar to the env sequences for ERV-DC8 and -DC14, respectively, both of which are classified as genotype I ERV-DCs. The env e sequence is highly similar to that of the ERV-DC10 (a genotype III ERV-DC) env gene. The env genes differ by 0 to 3 bp in comparison with the reference sequences. Notably, the SPF cat used in this study did not have ERV-DC19.
FIG 3.
Expression of ERV-DCs in feline tissues. (A) A phylogenetic tree of the env gene nucleotide sequences was constructed by using the maximum likelihood method, and its robustness was evaluated by bootstrapping (1,000 times) as previously described (29). ERV-DC env genes, env a to e, were detected in feline tissues. (B) Schematic of the structure of Env encoded in the identified transcripts of ERV-DCs. sp, signal peptide; SU, surface unit; N-term, N-terminal region of the surface unit; PRR, proline-rich region; C-term, C-terminal region of the surface unit; TM, transmembrane. (C) Quantifications of ERV-DC transcript levels determined by qRT-PCR with primer pairs that were specific for ERV-DCs belonging to genotype I, genotype II, or genotype III. The total gene expression level of each ERV-DC genotype was adjusted according to the expression level of PPIA (55). (D) Expression of ERV-DC genotype I, genotype II, or genotype III in blood samples from seven healthy domestic cats. The total gene expression level of each ERV-DC genotype was adjusted according to the expression level of PPIA.
Next, we performed quantitative RT-PCR (qRT-PCR) assays with primer pairs that are specific for ERV-DCs belonging to genotype I, II, or III. The total gene expression level of each ERV-DC genotype was adjusted based on the PPIA expression level (Fig. 3C). The expression level of ERV-DC genotype II, which includes Refrex-1, was high in PBMCs, and this genotype was broadly expressed in most tissues (Fig. 3C). In contrast, ERV-DC in genotypes I and III showed limited expression in all of the tested feline tissues (Fig. 3C). These results show that the expression levels of ERV-DCs differ among their genotypes and loci, indicating that their promoter activities also vary. Notably, the expression level of genotype III was very low, even though this group contains ERV-DC10, which can replicate in cultured cells at high levels.
Additionally, we examined ERV-DC expression levels in PBMCs from seven healthy domestic cats. We observed the same trends in these cells that we saw in feline tissues: the expression level of genotype II is high, whereas that of genotype III is low (Fig. 3C and D). Although ERV-DC insertional polymorphism exists in domestic cats, a preference for promoter activity may affect gene expression in feline tissues. In summary, ERV-DCs are expressed in normal tissues, and most ERV-DC transcripts belong to genotype II.
CpG methylation status of the ERV-DC LTRs.
The ERV-DC expression levels in feline tissues differed among the three genotypes. High expression levels were observed for genotype II ERV-DC, which includes the Refrex-1-coding loci ERV-DC7 and ERV-DC16, whereas low expression levels were observed for genotype III ERV-DCs, despite the inclusion of a highly replication-competent provirus, ERV-DC10/DC18, in this group. These findings indicate that the transcriptional regulation of viral genes or genomes differs among ERV-DC loci. Therefore, we next investigated how ERV-DC expressions are regulated.
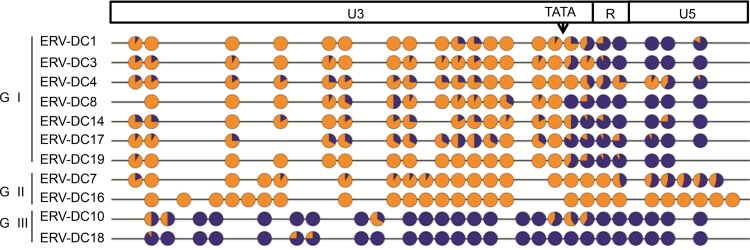
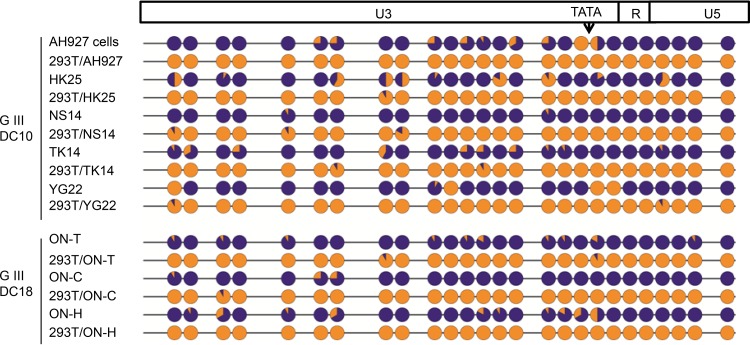
We first examined the methylation status of CpG nucleotides in the 5′ LTR of each ERV-DC provirus, which functions as the viral promoter and enhancer. For the analysis of CpG methylation, we used the sodium bisulfite sequencing technique and analyzed the genomic DNA from the blood of eight different cats as well as from one feline fibroblast cell line, AH927. We selected blood samples for use in this experiment because, compared to their expression levels in other tissue types, all three ERV-DC types are relatively well expressed in blood cells (Fig. 3C). The samples, 5′-LTR lengths, numbers of CpG sites within the 5′ LTR, and basic information for each of the ERV-DC loci are summarized in Table 2.
As shown in Fig. 4 and 5, the ERV-DC10 5′ LTR in feline genomic DNA from the tested samples was densely methylated. The methylation status was not dependent on whether the ERV-DC10 locus was a heterozygote or a homozygote. The ERV-DC18 5′ LTR also exhibited strong methylation in all three ERV-DC18-positive samples. The ERV-DC10 and ERV-DC18 LTR sequences are identical, and they each contain 26 positions of CpG dinucleotides in their 5′ LTRs. The methylation rates of ERV-DC10 and ERV-DC18 ranged from 84.3 to 96.2% in nine samples (data for three samples are not shown) and from 91.7 to 97.8% in three samples, respectively. Thus, the ERV-DC10 and -DC18 promoters are strongly methylated in the feline genome. These highly methylated states were not observed in replicating ERV-DC10 and ERV-DC18 in HEK293T cells (Fig. 5). In contrast to ERV-DC10/DC18, the methylation levels of ERV-DC7 and -DC16, both of which are the loci encoding Refrex-1, were extremely low; the methylation rates of CpG nucleotides in the 5′ LTRs of ERV-DC7 and -DC16 were 16.7% and 0%, respectively (Fig. 4).
FIG 4.
CpG methylation statuses of the ERV-DC provirus 5′ LTRs from blood samples. The CpG sites in the 5′ LTRs of ERV-DCs were aligned. The CpG methylation of each of the 5′ LTRs from the 11 ERV-DC loci in feline blood genomic DNA was analyzed by using the sodium bisulfite sequencing technique. One representative result from ERV-DC10 (GF33) and ERV-DC18 (ON-C) is shown. Orange indicates the unmethylated frequency, and royal blue indicates the methylated frequency. The arrow indicates the position of the TATA box.
FIG 5.
CpG methylation statuses of ERV-DC10 and ERV-DC18 5′ LTRs. The CpG methylation statuses of the ERV-DC10 or ERV-DC18 5′ LTRs in cat blood or in HEK293T cells that were persistently infected with ERV-DC10 or ERV-DC18 from either feline AH927 cells or blood from healthy cats (28) were analyzed by using the sodium bisulfite sequencing technique. Each result was derived from 12 sequences produced by sodium bisulfite sequencing. Orange indicates the unmethylated frequency, and royal blue indicates the methylated frequency. The arrow indicates the position of the TATA box.
In PBMCs, the CpG nucleotides in the 5′ LTRs of ERV-DC14, a member of genotype I, were partially methylated (37.7%) (Fig. 4). The CpG nucleotide methylation rate in genotype I ERV-DCs ranged between 23.1 and 47.6%. Interestingly, this group exhibited lower levels of CpG methylation in the U3 regions but exhibited higher levels of CpG methylation in the R and U5 regions (Fig. 4); in other words, the region downstream of TATA was significantly more methylated (P < 0.05) than the region upstream of TATA. Overall, our results show that the methylation status of 5′ LTRs corresponds to the expression levels of ERV-DCs in the blood (Fig. 3C and D). Moreover, our data show that methylation in the ERV-DC 5′ LTRs can be separated into three levels: high, medium, and low. Additionally, these methylation levels correlate with their genotypes: genotypes I, II, and III exhibit medium, low, and high methylation levels, respectively.
Basal promoter activity of ERV-DC LTRs.
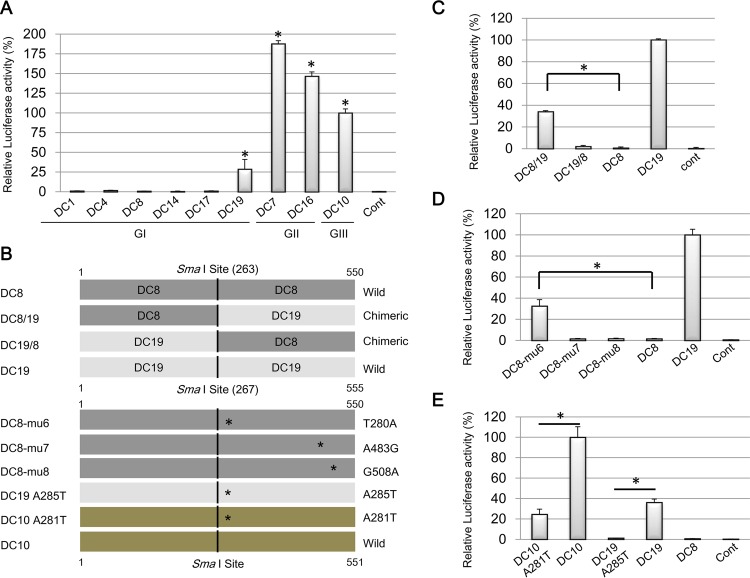
Next, we examined the intrinsic promoter activity of the ERV-DC 5′ LTRs by using luciferase reporter assays. We constructed nine LTR-Luc reporter plasmids by inserting a 5′ LTR from each provirus into a separate promoterless firefly luciferase reporter plasmid: pLTR DC1(DC3)-Luc, pLTR DC4-Luc, pLTR DC7-Luc, pLTR DC8-Luc, pLTR DC10(DC18)-Luc, pLTR DC14-Luc, pLTR DC16-Luc, pLTR DC17-Luc, and pLTR DC19-Luc. The LTR sequences for ERV-DC3 and -DC18 were the same as those for ERV-DC1 and -DC10, respectively, and thus, to avoid redundancy, we made only one reporter plasmid for each of these identical pairs. Each of the LTR-Luc plasmids were transfected into AH927 and HEK293T cells, and the luciferase activities were measured and adjusted according to the Renilla luciferase activity. As shown in Fig. 6A, the LTRs from ERV-DC7, -DC10, -DC19, and -DC16 exhibited promoter activity in HEK293T cells (P < 0.05). Similar results were obtained with AH927 cells (data not shown). ERV-DC7 and -DC16 LTRs had the highest promoter activities (Fig. 6A). The promoter activities of ERV-DC7, ERV-DC16, and ERV-DC19 were ∼1.9-fold (5.1-fold in AH927 cells), 1.5-fold (4.6-fold in AH927 cells), and 0.3-fold (0.7-fold in AH927 cells) higher than that of ERV-DC10. No promoter activity was detected in HEK293T cells (Fig. 6A) transfected with ERV-DC1(DC3), -DC4, -DC8, -DC14, or -DC17 in comparison with those in cells transfected with an empty vector. The promoter activity of ERV-DC14 was detected at very low levels in HepG2 cells (data not shown). Among genotype I ERV-DCs, only the ERV-DC19 LTR exhibited detectable promoter activity (Fig. 6A).
FIG 6.
Luciferase reporter assay for basal promoter activity of ERV-DC LTRs and identification of LTR cis elements. (A) The basal promoter activity levels of wild-type LTRs were measured by using a luciferase reporter assay. Genotype I (GI) includes DC1 (DC3), DC4, DC8, DC14, DC17, and DC19; genotype II (GII) includes DC7 and DC16; and genotype III (GIII) includes DC10 (DC18). (B) Schematic of chimeric and mutant ERV-DC LTRs that were used to investigate the presence of a cis element in ERV-DC LTRs. The name of each construct is indicated on the left, and the wild-type status or the chimeric or point mutation of the LTRs is indicated on the right. Asterisks indicate a point mutation of the nucleotide. The SmaI restriction enzyme site is located in the LTRs of ERV-DC8 and ERV-DC19 at positions 263 and 267, respectively. (C) The promoter activity levels of chimeric LTRs between the DC19 LTR and DC8 LTR (DC8/19 and DC19/8 LTRs) were measured by a luciferase reporter assay. (D) The promoter activity levels of DC8-mu6 (T280A substitution), DC8-mu7 (A483G substitution), and DC8-mu8 (G508A substitution), which are all DC8 LTRs containing a point mutation, were measured by a luciferase reporter assay. (E) The promoter activity levels of DC10 A281T (A281T substitution) and DC19 A285T (A285T substitution), which are the DC10 and DC19 LTRs with a point mutation, respectively, were measured by a luciferase reporter assay. LTR-Luc reporter plasmids were cotransfected with Renilla luciferase-expressing phRL-CMV into HEK293T cells. The cells were harvested at 48 h posttransfection, and luciferase activity was measured. The control (Cont) was a pGL4.10[luc2] empty vector. The luciferase activity of each LTR was adjusted according to the Renilla luciferase activity. The results from three replicates were collected and statistically analyzed by Student's t test and one-way analysis of variance (*, P < 0.05).
Identification of a cis element in ERV-DC LTRs.
Of the genotype I ERV-DCs, only ERV-DC19 showed any obvious promoter activity in HEK293T cells; the others did not exhibit detectable promoter activity in the tested cells. To investigate the reason for this difference, two chimeric LTRs (the DC8/19 LTR and the DC19/8 LTR) were initially constructed by using the SmaI restriction enzyme site located in the LTRs of ERV-DC8 and -DC19 at positions 263 and 267, respectively, (Fig. 6B), and the promoter activity of these chimeras was assessed by using luciferase reporter assays. As shown in Fig. 6C, the DC8/19 LTR exhibited promoter activity, while the DC19/8 LTR did not. This indicates that the region downstream of the SmaI site is responsible for the difference in promoter activity between ERV-DC8 and ERV-DC19. A sequence comparison of this region between ERV-DC8 and ERV-DC19 revealed the presence of three nucleotide differences: thymine (T) at nucleotide position 280, adenine (A) at position 483, and guanine (G) at position 508 in the ERV-DC8 5′ LTR all differ from the nucleotides at these positions in the ERV-DC19 5′ LTR. Next, we constructed three ERV-DC8 5′-LTR mutants: the DC8-mu6 LTR (T-to-A replacement at position 280 in the ERV-DC8 5′ LTR), the DC8-mu7 LTR (A-to-G replacement at position 483 in the ERV-DC8 5′ LTR), and DC8-mu8 LTR (G-to-A replacement at position 508 in the ERV-DC8 5′ LTR) (Fig. 6B). When these LTR plasmids were transfected into HEK293T cells, only the DC8-mu6 LTR significantly exhibited any promoter activity (P < 0.05). No promoter activity was observed in HEK293T cells transfected with DC8-mu7 or DC8-mu8 LTRs (Fig. 6D). These results indicate that the thymine at nucleotide position 280 of the ERV-DC8 5′ LTR is responsible for the reduced or absent promoter activity. Furthermore, we investigated whether or not the promoter activities of the 5′ LTRs from ERV-DC10 and ERV-DC19 were reduced by the converse mutations at nucleotides 281 and 285, respectively, both of which correspond to nucleotide position 280 in the ERV-DC8 5′ LTR. We constructed two mutants, the DC10 A281T LTR (A-to-T substitution at nucleotide position 281 of the DC10 5′ LTR) and the DC19 A285T LTR (A-to-T substitution at nucleotide position 285 of the DC19 5′ LTR). These two mutants exhibited significantly reduced promoter activity levels (P < 0.05) in HEK293T cells (Fig. 6E). Thus, we determined that the nucleotide substitution responsible for the difference in promoter activities among the 5′ LTRs of ERV-DCs is an A-to-T substitution corresponding to nucleotide 280 in the ERV-DC8 5′ LTR. This A-to-T substitution may disrupt an important cis element at this region that influences the magnitude of the ERV-DC basal promoter activity.
ERV-DC LTR-driven transcription is suppressed by CpG methylation.
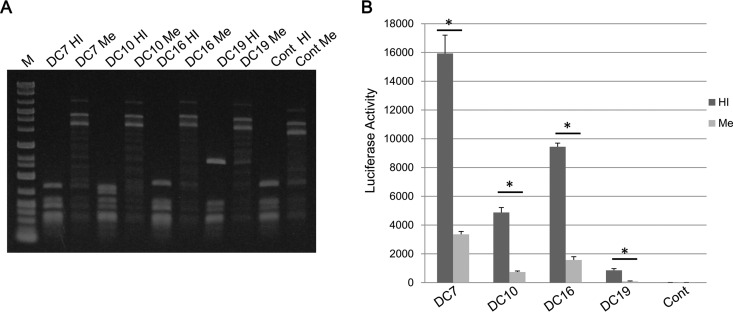
Generally, LTR promoter activity is reduced by a suppressive epigenetic status, such as CpG methylation, so we tested whether or not the promoter activities of ERV-DC 5′ LTRs were affected by their methylation states in vitro. We induced CpG methylations of the LTR reporter plasmids of ERV-DC7, -DC10, -DC16, and -DC19 with either M.SssI or heat-inactivated M.SssI and confirmed their methylation levels by digesting them with BstUI, which is a CpG methylation-sensitive restriction enzyme. As shown in Fig. 7A, all of the plasmid DNA was successfully methylated.
FIG 7.
ERV-DC LTR-driven transcription in the presence of CpG methylation. (A) The LTR reporter plasmids (DC7, DC10, DC16, DC19, and empty vector) were methylated in vitro by either M.SssI (Me) or heat-inactivated M.SssI (HI). The DNA was treated with BstUI, separated by gel electrophoresis, and visualized by ethidium bromide staining. M indicates the DNA marker, and Cont indicates the control pGL4.10[luc2] empty vector. (B) The promoter activity levels of methylated or unmethylated (treated with heat-inactivated M.SssI) ERV-DC LTRs were measured by a luciferase reporter assay. The LTR-Luc reporter plasmids were cotransfected with Renilla luciferase-expressing phRL-CMV into HEK293T cells, the cells were harvested at 48 h posttransfection, and luciferase activity was measured. The control was the pGL4.10[luc2] empty vector. The luciferase activity of each LTR was adjusted according to the Renilla luciferase activity level. These results include data from three independent replicates and were statistically analyzed by Student's t test (*, P < 0.05).
Next, the methylated or unmethylated LTR plasmids were transfected into HEK293T cells, and their promoter activities were assessed. As shown in Fig. 7B, the promoter activity levels of the methylated plasmids were significantly lower than those of controls in all tested LTRs (P < 0.05) (Fig. 7B). Thus, the high methylation status of CpG nucleotides in the ERV-DC LTRs results in a reduction of promoter activity in vitro.
Viral properties of ERV-DC14 and the ERV-DC14TA mutant.
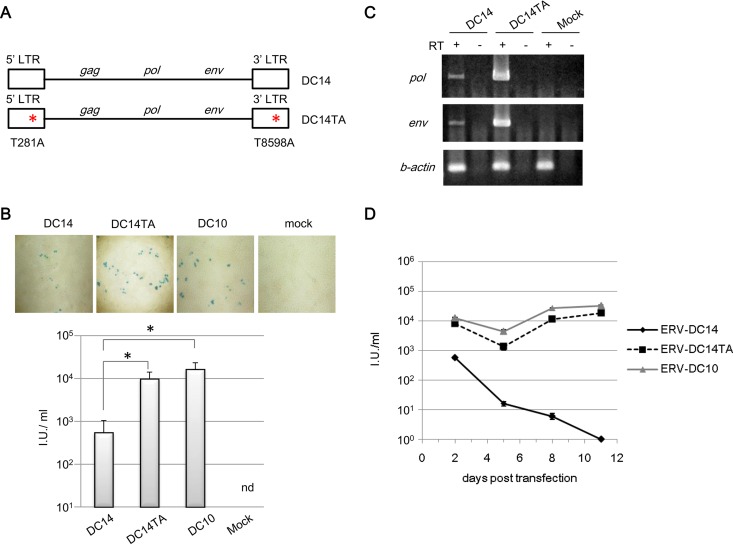
As shown in Fig. 1, the levels of viral transcription, production, replication, and transmission of ERV-DC14 are lower than those of ERV-DC10. Notably, although ERV-DC14 can replicate in HEK293T cells, it cannot establish a persistent infection in these cells. We speculated that the low replication level of ERV-DC14 is a result of its low promoter activity level, so we constructed a mutant of ERV-DC14, termed ERV-DC14TA, that has point mutations of T to A at nucleotide 281 in its 5′ LTR and at nucleotide 8598 in its 3′ LTR (Fig. 8A). This plasmid was transfected into 293Lac cells or HEK293T cells. The viral titer in the supernatant or virus-infected cells was then tested by LacZ staining, and the expression of viral mRNA was assessed by RT-PCR. As shown in Fig. 8B, ERV-DC14TA induced more LacZ staining foci than did ERV-DC14, and the infection titer of ERV-DC14TA was ∼10-fold higher than that of ERV-DC14 at 2 days postinfection. Additionally, when HEK293T cells were infected with ERV-DC14 or ERV-DC14TA for 2 days, we were able to detect their pol and env genes by RT-PCR (Fig. 8C). Thus, ERV-DC14TA, like ERV-DC14, is replication competent, and it can produce more infectious virions than ERV-DC14.
FIG 8.
Viral properties of ERV-DC14 and of the ERV-DC14TA mutant. (A) ERV-DC14TA, which contains substitutions of T to A at positions 281 and 8598 in the 5′ and 3′ LTRs, respectively, of ERV-DC14, was constructed. (B) 293Lac cells were transfected with ERV-DC14, ERV-DC14TA, or ERV-DC10 or were mock transfected, and the resulting supernatants were collected and used to infect HEK293T cells. (Top) Cells were fixed at 48 h postinfection and visualized by X-Gal staining. (Bottom) Viral titers are expressed in IU per milliliter. nd, not detected. The results were collected from at least six independent experiments with ERV-DC14 (n = 6), ERV-DC14TA (n = 7), and ERV-DC10 (n = 7). Pictures from one of them are shown. The results were statistically analyzed by one-way analysis of variance (*, P < 0.05). (C) The expression levels of pol, env, and β-actin mRNAs in HEK293T cells 2 days after mock infection or infection with ERV-DC14 or ERV-DC14TA were assessed by RT-PCR. (D) Growth kinetics of ERV-DCs. 293Lac cells were transfected with equal amounts of ERV-DC14, ERV-DC14TA, or ERV-DC10. To monitor viral growth, culture supernatants were collected periodically after transfection as indicated, and viruses filtered through a 0.45-μm filter were used to inoculate HEK293T cells. The resulting viral titers were determined by X-Gal staining.
Next, we monitored viral infections of ERV-DC14, -DC14TA, and -DC10 in HEK293T cells over the course of several days. 293Lac cells were transfected with each plasmid, and the resulting supernatants were used to infect HEK293T cells. As shown in Fig. 8C, the viral titers of ERV-DC14TA and ERV-DC10 were ∼10-fold higher than that of ERV-DC14 at 2 days postinfection. Subsequently, the ERV-DC14TA and ERV-DC10 infections became persistent, with viral titers of ∼104 IU/ml, whereas ERV-DC14 gradually disappeared from 293Lac cells after 10 days and was unable to maintain viral replication in HEK293T cells. Additionally, we detected infectious ERV-DC14TA in the supernatants from persistently infected cells by measuring viral infectivity (data not shown), indicating that, similarly to ERV-DC10, ERV-DC14TA was persistently produced in HEK293T cells. Thus, although ERV-DC14 cannot establish a persistent infection in HEK293T cells, this defect can be recovered by T-to-A substitutions at positions 281 and 8598 in the 5′ and 3′ LTRs, respectively.
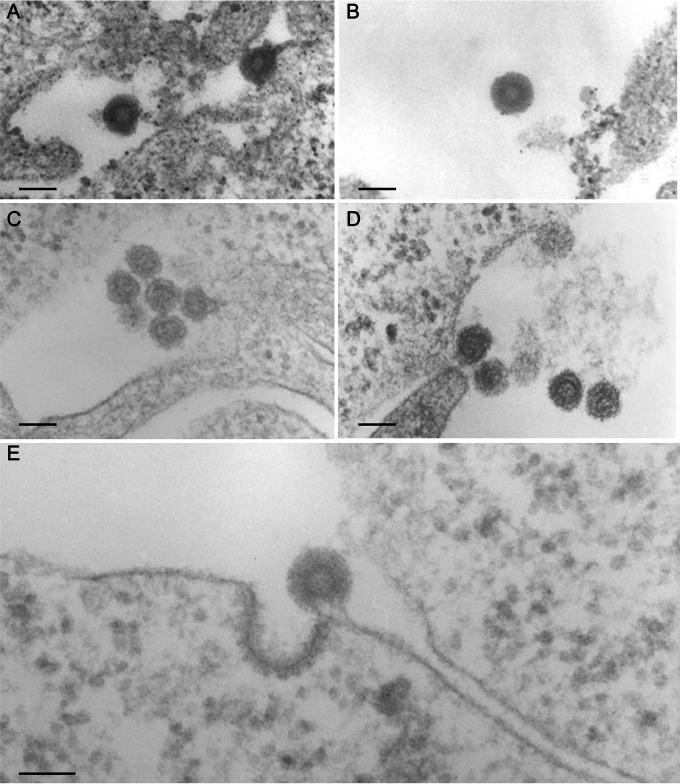
We examined ERV-DC14 and ERV-DC14TA by transmission electron microscopy (TEM) in HEK293T cells. Both ERV-DC14 (Fig. 9A and B) and ERV-DC14TA (Fig. 9C to E) particles were detected by TEM and morphologically were retrovirus type C. Figure 9E shows a late stage of budding. Overall, we confirmed that ERV-DC14 is a replication-competent virus and found that the ancestral mutation (thymine) of the ERV-DC14 LTR cis element reduced its viral replication. Furthermore, these results suggest that this ancestral mutation in the LTR affects the interplay between the host and the viruses.
FIG 9.
ERV-DC14 and ERV-DC14TA mutant virions. Shown are representative images from TEM of retroviral particles in HEK293T cells that were transfected with molecular clones of ERV-DC14 (A and B) or ERV-DC14TA (C to E). Bars, 100 nm.
ERV-DC14 and ERV-DC10 tropism.
Our previous study demonstrated that ERV-DC14 did not interfere with ERV-DC10, indicating that these viruses use different viral receptors (29, 32). In this study, we compared the host ranges of ERV-DC14 and ERV-DC10 in detail by using the ERV-DC14TA mutant so that it would be easier to monitor viral infection. As shown in Table 4, ERV-DC14TA broadly infected all of the tested cell lines, except for the mouse and hamster cell lines, while ERV-DC10 infected only a limited subset of the tested cell lines. In these experiments, we washed Refrex-1 out of the feline cell cultures because ERV-DC14 infection is affected by the presence of Refrex-1 (29). Most of the tested human cell lines were similar to one another in their sensitivities to ERV-DC14TA and ERV-DC10, but MCF7 cells had a somewhat lower viral titer, suggesting that they are less sensitive to these viruses and may contain viral resistance factors. These results indicate that the receptor usages of ERV-DC14 and ERV-DC10 are different from one another. Moreover, ERV-DC14 broadly infects many species, while ERV-DC10 induced only a limited infection in the tested cells.
TABLE 4.
ERV-DC10 and ERV-DC14 tropismsa
| Species | Cell line | Mean viral titer (IU/ml) ± SD |
|
|---|---|---|---|
| ERV-DC14TA | ERV-DC10 | ||
| Human | HepG2 | (2.1 ± 0.1) × 104 | (1.1 ± 0.0) × 104 |
| HeLa | (1.0 ± 0.1) × 104 | (7.4 ± 0.0) × 103 | |
| HEK293T | (1.1 ± 0.2) × 104 | (4.4 ± 0.5) × 104 | |
| MCF7 | (9.0 ± 1.4) × 102 | 6.7 ± 2.9 | |
| Cat | AH927 | (3.8 ± 0.9) × 103 | 0 |
| CRFK | (4.4 ± 1.7) × 103 | (5.6 ± 2.5) × 101 | |
| Monkey | Vero | (6.0 ± 2.5) × 102 | (1.4 ± 1.0) × 101 |
| Cos7 | (3.3 ± 0.7) × 103 | 2.0 ± 1.4 | |
| Mouse | NIH 3T3 | 0 | 0 |
| MDTF | 0 | 0 | |
| Hamster | BHK21 | 0 | 0 |
| Guinea pig | 104C1 | (1.7 ± 0.7) × 103 | 0 |
| Dog | KwDM | (1.8 ± 0.3) × 104 | (1.4 ± 0.1) × 104 |
| Cow | MDBK | (5.3 ± 1.9) × 103 | 0 |
Human, monkey, feline, canine, bovine, murine, hamster, and guinea pig cells were infected with the viruses generated by the transfection of viral molecular clones of ERV-DC14TA or ERV-DC10 into 293Lac cells. At 2 days posttransfection, LacZ-positive cells were quantified microscopically. These results are representative of data from at least three independent experiments.
ERV-DC invasion in the cat genome.
As described above, we identified an A-to-T substitution in the LTR, which is commonly observed in a certain clade of ERV-DC genotype I, and this substitution is responsible for inducing reductions in viral promoter activities and for the low replication levels of ERV-DC14. Therefore, ERV-DC LTRs can be divided into two groups, A-type LTRs and T-type LTRs (Fig. 10A). A-type LTRs (ERV-DC10/DC18, -DC7, -DC16, and -DC19) have strong or ubiquitous promoter activities, whereas T-type LTRs (ERV-DC1, -DC4, -DC8, -DC14, and -DC17) have weak or attenuated promoter activities (Fig. 6A).
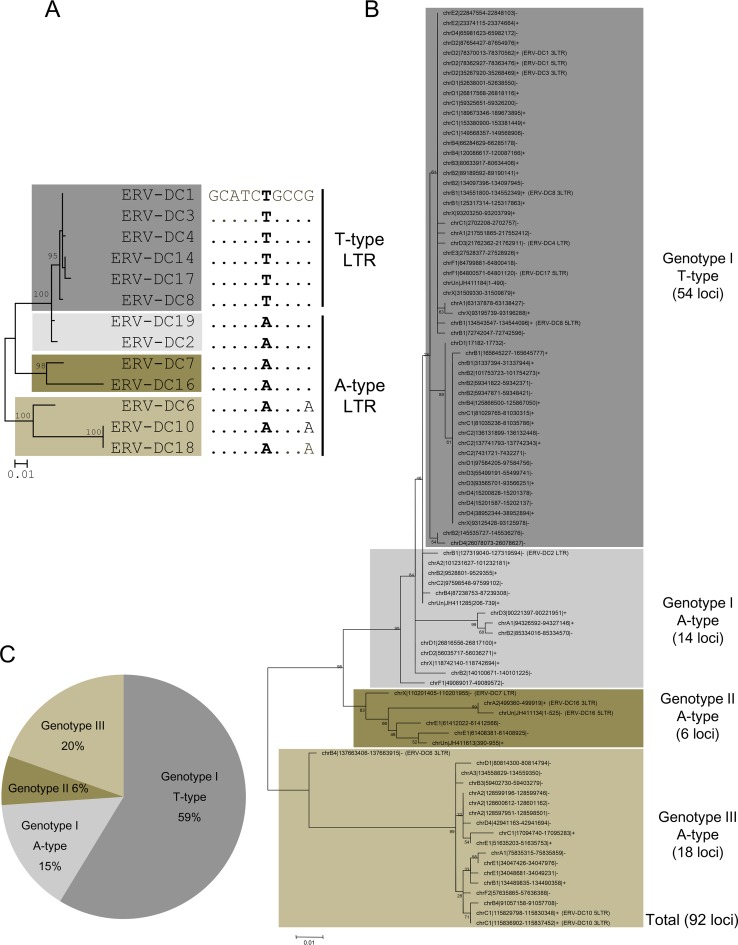
FIG 10.
ERV-DC invasion of the cat genome. (A) The single nucleotide polymorphism of either A or T in the cis element within ERV-DC LTRs was termed either an A-type LTR, which confers higher-level promoter activity, or a T-type LTR, which confers lower-level promoter activity. (B) ERV-DC LTRs were extracted from the UCSC feline genome database. A phylogenetic tree with 92 of these ERV-DC LTRs was constructed from an in silico analysis using the maximum likelihood method, and its robustness was evaluated by bootstrapping (1,000 times). The results were separated into genotypes I (68 LTRs), II (6 LTRs), and III (18 LTRs). (C) The ratio of each group, genotype I with T-type LTRs, genotype I with A-type LTRs, genotype II, and genotype III, is represented in a pie chart. A chi-squared test was conducted to assess the statistical significance of these results.
To gain insight into which LTR type is more adaptive to surviving within the host genome, we investigated which LTR type is more dominant in the ERV-DC population using an in silico analysis. First, ERV-DC LTRs were extracted from the feline reference genome (ICGSC Felis_catus 6.2/felcat5) by using the BLAT program implemented in the UCSC genome browser. Second, the LTR type (A or T type) of each locus was checked in a multiple-sequence alignment. Third, a phylogenetic tree of the identified LTRs was constructed by using the maximum likelihood method with bootstrapping (1,000 times). As a result, we identified a total of 92 ERV-DC LTR loci: 54 of genotype I/type T, 14 of genotype I/type A, 6 of genotype II, and 18 of genotype III (Fig. 10B and C). All of the genotype II and III ERV-DCs had A-type LTRs (Fig. 10B). Finally, we compared the copy numbers of LTRs among groups. This comparison revealed that the proportion of T-type LTRs is significantly higher than those of the others, both within the genotype I population (54/68; P < 0.001) and within the total ERV-DC population (54/92; P = 0.027) (Fig. 10C). These results indicate that genotype I ERV-DCs bearing an A-to-T substitution (T-type LTRs) have invaded the feline genome significantly more than other types have, and they support the idea that ERV-DCs with a substitution that causes promoter attenuation are more adaptive to surviving within host genomes.
DISCUSSION
ERV-DC10 and ERV-DC18 are the first feline ERVs that were identified as replication-competent proviruses residing within the host genome (32, 60, 61). In this study, we newly identified ERV-DC14 as another replication-competent ERV and characterized this virus in detail. Although ERV-DC14 is replication competent, its level of replication is very low, so this virus does not establish persist infection in cultured cells. This low level of ERV-DC14 replication results from its low promoter activity level, which is caused by one A-to-T substitution in each of its LTRs. The lack of persistent ERV-DC14 infection is the reason why we did not detect the replication-competent ability of ERV-DC14 in our previous study (28). In this study, we used a previously reported strategy to identify ERVs (62, 63), establishing 293Lac indicator cells, which allowed us to recognize the replication competency of ERV-DC14. Additionally, we observed ERV-DC14 virions using TEM. The essential attributes of ERV-DC14 match those of a type C retrovirus. This type of virus is morphologically similar to gammaretroviruses, such as ERV-DC10 and ERV-DC18, and has characteristic core, envelope, and virus budding (28). Thus, ERV-DC14 is an authentic replication-competent provirus residing within the feline genome.
ERV-DCs are divided into three genotypes: genotype I includes ERV-DC14, genotype III includes ERV-DC10/DC18, and genotype II includes ERV-DC7 and ERV-DC16, both of which encode Refrex-1, a known viral restriction factor. In addition to phylogeny, replication activity, and promoter activity, the ERV-DC14 receptor usage is distinct from that of ERV-DC10/DC18, as determined by the results of viral interference assays (29). In this study, we determined the infectious tropism of ERV-DC14 and ERV-DC10; in the absence of Refrex-1, ERV-DC14 broadly infects several species, including cats, while ERV-DC10 exhibits more limited infection. In other words, the infectivity of ERV-DC14 is nonecotropic, whereas that of ERV-DC10/DC18 appears to be xenotropic.
As described above, two distinct types of infectious ERVs reside within the feline genome, ERV-DC10/DC18 and ERV-DC14. They differ in their phylogenies, receptor usages, and transcriptional activities. Additionally, the feline genome also contains domesticated (noninfectious) ERV-DCs, such as ERV-DC7 and ERV-DC16. We were interested in which mechanisms the host uses to control these ERVs with distinctly different properties, so we investigated their transcriptional regulations.
First, we examined ERV-DC transcription in feline tissues. Although RD-114 feline ERV is expressed in both normal and malignant tissues in domestic cats (64, 65), the RT-PCR assays that we used to detect ERV-DCs cannot detect this ERV. Qualitative and quantitative analyses demonstrated that env genes corresponding to ERV-DC genotypes I, II, and III were expressed in feline tissues. All three genotypes were relatively well expressed in hematopoietic tissues such as blood, spleen, and thymus tissues. Genotype II, which includes Refrex-1, was dominantly expressed across broad tissue types, in agreement with data from our previous study (29). Genotype I was also expressed in feline tissues. This is consistent with the observation that FeLV-D is generated by the transduction of the ERV-DC genotype I env gene (29) through a template switch during reverse transcription (66). Genotype III had a lower level of expression than the others, even though this group includes ERV-DC10, which can replicate to high levels in cultured cells. These findings agree with our previous observation that ERV-DC production was not detected in the tested domestic cat PBMCs (28). However, this study did not determine whether or not ERV-DC14 is expressed in vivo. Owing to the very low frequency of ERV-DC14 provirus in cats, it is difficult to obtain RNA samples with ERV-DC14.
Second, we examined the CpG methylation status of the 5′ LTRs for each ERV-DC locus by performing a sodium bisulfite sequence analysis on blood from healthy cats. The CpG methylation of an LTR generally suppresses its viral promoter activity, and we confirmed that this trend also applies to ERV-DC LTRs (Fig. 7). The 5′ LTRs of ERV-DC10/DC18, which belong to genotype III, were hypermethylated (84 to 98%), whereas ERV-DC7 and ERV-DC16, which belong to genotype II, were hypomethylated (0 and 17%, respectively). These results agree with the low and high expression levels of genotype III and genotype II, respectively, in blood tissues. In cells persistently infected with titers of at least 104 IU/ml of ERV-DC10 or ERV-DC18, the viruses exhibited hypomethylation in their LTRs. The methylation state of ERV-DC7 may be affected by genome imprinting, because this provirus is located on feline chromosome X, and we used a female sample for this analysis. The LTRs of genotype I ERV-DCs, which are expressed at medium levels in some tissues, had correspondingly medium levels of methylation (23 to 47%). Interestingly, the methylation in the LTRs of genotype I specifically occurs downstream of the TATA box, indicating the importance of the methylation level in the transcriptional initiation region. In summary, the methylation levels of LTRs are different among ERV-DC loci, and these patterns are correlated with their genotypes. One of the major functions of DNA methylation is thought to be the silencing of ERVs (67–69), and ERV demethylation and expression cause an immune response (69, 70). A similar phenomenon may occur in the association with ERV-DC gene regulation. In this experiment, 5′ LTRs were investigated for their methylation status because 5′ LTRs, but not 3′ LTRs, are thought to be promoters for ERV-DCs. However, it would be interesting to analyze further elements, including 3′ LTRs, to determine the region-specific methylation within ERVs.
The key finding of this study is that the basal promoter activity of ERV-DC LTRs can be classified into two groups, A-type LTRs and T-type LTRs, according to the nucleotide present at position 281 in the ERV-DC14 LTR. A-type LTRs exhibit high levels of promoter activity, while T-type LTRs exhibit low or attenuated levels of promoter activity (no promoter activity detected in the reporter assay) (Fig. 6). Although promoter activities of T-type LTRs are attenuated, they appear to retain a minimum level of activity for productive viral replication based on the observed ERV-DC14 replication capacity and the expansion of T-type LTRs in the host genome. This is supported by our detection of T-type LTR genotype I ERV-DC expression in feline tissues (Fig. 3A). The T-type LTRs are commonly observed in a certain clade of ERV-DC genotype I. Given that genotype II is the oldest subgroup of ERV-DCs, T-type LTRs were likely generated from A-type LTRs through a preintegration substitution that occurred during the genomic invasion of genotype I.
Interestingly, RD-114, a virus with a high level of sequence similarity to ERV-DCs in its LTRs, has an A-type LTR. Furthermore, the region that includes the nucleotide corresponding to the 281st nucleotide of ERV-DC14 duplicated and formed direct repeats in RD-114 LTRs (71, 72). Previous studies also showed that this repeated region functions as a viral enhancer in the RD-114 virus (71, 72). Together with those results, our findings suggest that that A-type LTRs of ERV-DCs and RD-114 have a transcriptional cis element that enhances viral replication in this region and that T-type LTRs were generated from A-type LTRs through a disruption of their cis elements caused by an A-to-T substitution.
Our in silico analysis shows that the proportion of T-type LTRs is significantly higher in both the genotype I population and the total ERV-DC population (Fig. 10C). In other words, ERV-DCs with T-type LTRs have invaded the feline genome significantly more than other populations have, despite their low level of promoter activity. We hypothesize that this observation reflects a viral strategy for succeeding in endogenization: ERV-DCs with T-type LTRs appear to have attenuated their promoter activities in order to escape from strong suppressions or negative selections by the host. In fact, the 5′ LTR of ERV-DC14 refrains from severe methylation in blood cells, although this virus retains a minimal replication capacity. Thus, we consider that disruption of the cis element in the LTR and reduction of its promoter activity led to the preferential invasion of ERV-DCs with T-type LTRs in the host genome.
This study revealed the presence of two distinct types of infectious ERVs residing within the cat genome as well as how the hosts control these potentially infectious proviruses. Infectious ERV-DCs are regulated by two mechanisms: methylation of the promoter and mutation of the promoter. ERV-DC10 and ERV-DC18 expression levels are regulated by promoter methylation, while the ERV-DC14 expression level is low due to the attenuation of its promoter caused by a mutation in its LTR. Furthermore, the domesticated ERVs ERV-DC7 and ERV-DC16 are highly expressed in tissues, and they are regulated by a different mechanism than either of those used to control infectious ERVs; these domesticated ERVs retain the strongest promoter activity and the lowest methylation levels among ERV-DCs. In summary, various types of ERV-DCs are present in the host feline genome, and they are controlled via several different mechanisms. These different ways of controlling ERVs seem to have been formed through host-virus conflict.
In conclusion, our results provide insights into how the host controls potentially infectious ERVs and, conversely, how ERVs adapt to and invade the host genome. Thus, cats provide a unique system for studying these mechanisms in field animals. Studying these ancestral infectious viruses might uncover previously unrecognized features or properties of retroviruses.
ACKNOWLEDGMENTS
We are grateful to Kunio Nagashima (Leidos Biomedical Research, Inc.) and Alan Rein (National Cancer Institute-Frederick) for their helpful suggestions.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. 1998. Retroviral diversity and distribution in vertebrates. J Virol 72:5955–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin JM. 2004. Evolution of retroviruses: fossils in our DNA. Proc Am Philos Soc 148:264–280. [PubMed] [Google Scholar]
- 5.Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 6.Weiss RA. 2006. The discovery of endogenous retroviruses. Retrovirology 3:67. doi: 10.1186/1742-4690-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak CA. 2015. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to gibbon ape leukemia virus. J Virol 74:4264–4272. doi: 10.1128/JVI.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong JA, Porterfield JS, De Madrid AT. 1971. C-type virus particles in pig kidney cell lines. J Gen Virol 10:195–198. doi: 10.1099/0022-1317-10-2-195. [DOI] [PubMed] [Google Scholar]
- 11.Todaro GJ, Benveniste RE, Lieber MM, Sherr CJ. 1974. Characterization of a type C virus released from the porcine cell line PK(15). Virology 58:65–74. doi: 10.1016/0042-6822(74)90141-X. [DOI] [PubMed] [Google Scholar]
- 12.Moennig V, Frank H, Hunsmann G, Ohms P, Schwarz H, Schafer W. 1974. C-type particles produced by a permanent cell line from a leukemic pig. II. Physical, chemical, and serological characterization of the particles. Virology 57:179–188. [DOI] [PubMed] [Google Scholar]
- 13.Wilson CA, Wong S, Muller J, Davidson CE, Rose TM, Burd P. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol 72:3082–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Tissier P, Stoye JP, Takeuchi Y, Patience C, Weiss RA. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- 15.Aaronson SA, Tronick SR, Stephenson JR. 1976. Endogenous type C RNA virus of Odocoileus hemionus, a mammalian species of New World origin. Cell 9:489–494. doi: 10.1016/0092-8674(76)90094-5. [DOI] [PubMed] [Google Scholar]
- 16.Fabryova H, Hron T, Kabickova H, Poss M, Elleder D. 2015. Induction and characterization of a replication competent cervid endogenous gammaretrovirus (CrERV) from mule deer cells. Virology 485:96–103. doi: 10.1016/j.virol.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ Jr, Coffin JM, Pathak VK. 2011. Recombinant origin of the retrovirus XMRV. Science 333:97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. 2012. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu P, Lubben W, Slomka H, Gebler J, Konert M, Cai C, Neubrandt L, Prazeres da Costa O, Paul S, Dehnert S, Dohne K, Thanisch M, Storsberg S, Wiegand L, Kaufmann A, Nain M, Quintanilla-Martinez L, Bettio S, Schnierle B, Kolesnikova L, Becker S, Schnare M, Bauer S. 2012. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Triviai I, Ziegler M, Bergholz U, Oler AJ, Stubig T, Prassolov V, Fehse B, Kozak CA, Kroger N, Stocking C. 2014. Endogenous retrovirus induces leukemia in a xenograft mouse model for primary myelofibrosis. Proc Natl Acad Sci U S A 111:8595–8600. doi: 10.1073/pnas.1401215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David VA, Menotti-Raymond M, Wallace AC, Roelke M, Kehler J, Leighty R, Eizirik E, Hannah SS, Nelson G, Schaffer AA, Connelly CJ, O'Brien SJ, Ryugo DK. 2014. Endogenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3 (Bethesda) 4:1881–1891. doi: 10.1534/g3.114.013425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins NA, Copeland NG, Taylor BA, Lee BK. 1981. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature 293:370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- 23.Rabson AB, Graves BJ. 1997. Synthesis and processing of viral RNA. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 24.Robbez-Masson L, Rowe HM. 2015. Retrotransposons shape species-specific embryonic stem cell gene expression. Retrovirology 12:45. doi: 10.1186/s12977-015-0173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slotkin RK, Martienssen R. 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 26.Beyer W, Mohring R, Drescher B, Notzel U, Rosenthal S. 1987. Molecular cloning of an endogenous cat retroviral element (ECE 1)—a recombinant between RD-114 and FeLV-related sequences. Brief report. Arch Virol 96:297–301. doi: 10.1007/BF01320971. [DOI] [PubMed] [Google Scholar]
- 27.van der Kuyl AC, Dekker JT, Goudsmit J. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEV(Gag-Pol)/baboon endogenous virus BaEV(Env) recombinant. J Virol 73:7994–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. doi: 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. doi: 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci 368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malfavon-Borja R, Feschotte C. 2015. Fighting fire with fire: endogenous retrovirus envelopes as restriction factors. J Virol 89:4047–4050. doi: 10.1128/JVI.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito J, Baba T, Kawasaki J, Nishigaki K. 2016. Ancestral mutations acquired in Refrex-1, a restriction factor against feline retroviruses, during its cooption and domestication. J Virol 90:1470–1485. doi: 10.1128/jvi.01904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnaud F, Caporale M, Varela M, Biek R, Chessa B, Alberti A, Golder M, Mura M, Zhang YP, Yu L, Pereira F, Demartini JC, Leymaster K, Spencer TE, Palmarini M. 2007. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog 3:e170. doi: 10.1371/journal.ppat.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda H, Sugimura H. 1989. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol 63:5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung YT, Lyu MS, Buckler-White A, Kozak CA. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J Virol 76:8218–8224. doi: 10.1128/JVI.76.16.8218-8224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu T, Yan Y, Kozak CA. 2005. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J Virol 79:9677–9684. doi: 10.1128/JVI.79.15.9677-9684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A 101:11117–11122. doi: 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB. 1979. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 40.Gey GO, Coffman MT, Kubicek WD. 1952. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res 12:264–265. [Google Scholar]
- 41.Soule HD, Vazguez J, Long A, Albert S, Brennan M. 1973. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst 51:1409–1416. [DOI] [PubMed] [Google Scholar]
- 42.Gluzman Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell 23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 43.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7:379–387. doi: 10.1128/MCB.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasheed S, Gardner MB. 1980. Characterization of cat cell cultures for expression of retrovirus, FOCMA and endogenous sarc genes, p 393–400. In Hardy WD Jr, Essex M, McClelland AJ (ed), Proceedings of the Third International Feline Leukemia Virus Meeting Elsevier, North-Holland Publishing Co, New York, NY. [Google Scholar]
- 45.Haapala DK, Robey WG, Oroszlan SD, Tsai WP. 1985. Isolation from cats of an endogenous type C virus with a novel envelope glycoprotein. J Virol 53:827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bassin RH, Ruscetti S, Ali I, Haapala DK, Rein A. 1982. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology 123:139–151. doi: 10.1016/0042-6822(82)90301-4. [DOI] [PubMed] [Google Scholar]
- 47.Crandell RA, Fabricant CG, Nelson-Rees WA. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176–185. doi: 10.1007/BF02618435. [DOI] [PubMed] [Google Scholar]
- 48.Jainchill JL, Aaronson SA, Todaro GJ. 1969. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol 4:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasumura Y, Kawakita Y. 1963. Studies on SV40 virus in tissue culture cells. Nippon Rinsho 21:1201–1215. [Google Scholar]
- 50.Macpherson I, Stoker M. 1962. Polyoma transformation of hamster cell clones—an investigation of genetic factors affecting cell competence. Virology 16:147–151. doi: 10.1016/0042-6822(62)90290-8. [DOI] [PubMed] [Google Scholar]
- 51.Lander MR, Chattopadhyay SK. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol 52:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans CH, DiPaolo JA. 1975. Neoplastic transformation of guinea pig fetal cells in culture induced by chemical carcinogens. Cancer Res 35:1035–1044. [PubMed] [Google Scholar]
- 53.Madin SH, Darby NB Jr. 1958. Established kidney cell lines of normal adult bovine and ovine origin. Proc Soc Exp Biol Med 98:574–576. doi: 10.3181/00379727-98-24111. [DOI] [PubMed] [Google Scholar]
- 54.Miyake A, Watanabe S, Hiratsuka T, Ito J, Ngo MH, Makundi I, Kawasaki J, Endo Y, Tsujimoto H, Nishigaki K. 2016. Novel feline leukemia virus interference group based on env gene. J Virol 90:4832–4837. doi: 10.1128/JVI.03229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Veron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci U S A 109:E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz G. 1978. Estimating the dimension of a model. Ann Stat 6:461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- 59.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roca AL, Pecon-Slattery J, O'Brien SJ. 2004. Genomically intact endogenous feline leukemia viruses of recent origin. J Virol 78:4370–4375. doi: 10.1128/JVI.78.8.4370-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimode S, Nakagawa S, Miyazawa T. 2015. Multiple invasions of an infectious retrovirus in cat genomes. Sci Rep 5:8164. doi: 10.1038/srep08164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delviks-Frankenberry K, Paprotka T, Cingoz O, Wildt S, Hu WS, Coffin JM, Pathak VK. 2013. Generation of multiple replication-competent retroviruses through recombination between PreXMRV-1 and PreXMRV-2. J Virol 87:11525–11537. doi: 10.1128/JVI.01787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kearney MF, Lee K, Bagni RK, Wiegand A, Spindler J, Maldarelli F, Pinto PA, Linehan WM, Vocke CD, Delviks-Frankenberry KA, Devere White RW, Del Prete GQ, Mellors JW, Lifson JD, Kewalramani VN, Pathak VK, Coffin JM, Le Grice SF. 2011. Nucleic acid, antibody, and virus culture methods to detect xenotropic MLV-related virus in human blood samples. Adv Virol 2011:272193. doi: 10.1155/2011/272193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niman HL, Stephenson JR, Gardner MB, Roy-Burman P. 1977. RD-114 and feline leukemia virus genome expression in natural lymphomas of domestic cats. Nature 266:357–360. doi: 10.1038/266357a0. [DOI] [PubMed] [Google Scholar]
- 65.Niman HL, Gardner MB, Stephenson JR, Roy-Burman P. 1977. Endogenous RD-114 virus genome expression in malignant tissues of domestic cats. J Virol 23:578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu WS, Temin HM. 1990. Retroviral recombination and reverse transcription. Science 250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 67.Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 68.Bestor TH, Tycko B. 1996. Creation of genomic methylation patterns. Nat Genet 12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 69.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Buhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Mergoub T, Chan TA, Baylin SB, Strick R. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurst TP, Magiorkinis G. 2015. Activation of the innate immune response by endogenous retroviruses. J Gen Virol 96:1207–1218. doi: 10.1099/jgv.0.000017. [DOI] [PubMed] [Google Scholar]
- 71.Spodick DA, Ghosh AK, Parimoo S, Roy-Burman P. 1988. The long terminal repeat of feline endogenous RD-114 retroviral DNAs: analysis of transcription regulatory activity and nucleotide sequence. Virus Res 9:263–283. doi: 10.1016/0168-1702(88)90035-4. [DOI] [PubMed] [Google Scholar]
- 72.Ghosh AK, Roy-Burman P. 1989. Characterization of enhancer elements and their mutations in the long terminal repeat of feline endogenous RD-114 proviruses. J Virol 63:4234–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Science Council of Japan. 2006. Guidelines for proper conduct of animal experiments Science Council of Japan, Tokyo, Japan: http://www.scj.go.jp/ja/info/kohyo/pdf/kohyo-20-k16-2e.pdf. [Google Scholar]