ABSTRACT
Bundibugyo virus (BDBV) is the etiological agent of a severe hemorrhagic fever in humans with a case-fatality rate ranging from 25 to 36%. Despite having been known to the scientific and medical communities for almost 1 decade, there is a dearth of studies on this pathogen due to the lack of a small animal model. Domestic ferrets are commonly used to study other RNA viruses, including members of the order Mononegavirales. To investigate whether ferrets were susceptible to filovirus infections, ferrets were challenged with a clinical isolate of BDBV. Animals became viremic within 4 days and succumbed to infection between 8 and 9 days, and a petechial rash was observed with moribund ferrets. Furthermore, several hallmarks of human filoviral disease were recapitulated in the ferret model, including substantial decreases in lymphocyte and platelet counts and dysregulation of key biochemical markers related to hepatic/renal function, as well as coagulation abnormalities. Virological, histopathological, and immunohistochemical analyses confirmed uncontrolled BDBV replication in the major organs. Ferrets were also infected with Ebola virus (EBOV) to confirm their susceptibility to another filovirus species and to potentially establish a virus transmission model. Similar to what was seen with BDBV, important hallmarks of human filoviral disease were observed in EBOV-infected ferrets. This study demonstrates the potential of this small animal model for studying BDBV and EBOV using wild-type isolates and will accelerate efforts to understand filovirus pathogenesis and transmission as well as the development of specific vaccines and antivirals.
IMPORTANCE The 2013-2016 outbreak of Ebola virus in West Africa has highlighted the threat posed by filoviruses to global public health. Bundibugyo virus (BDBV) is a member of the genus Ebolavirus and has caused outbreaks in the past but is relatively understudied, likely due to the lack of a suitable small animal model. Such a model for BDBV is crucial to evaluating vaccines and therapies and potentially understanding transmission. To address this, we demonstrated that ferrets are susceptible models to BDBV infection as well as to Ebola virus infection and that no virus adaptation is required. Moreover, these animals develop a disease that is similar to that seen in humans and nonhuman primates. We believe that this will improve the ability to study BDBV and provide a platform to test vaccines and therapeutics.
INTRODUCTION
Bundibugyo virus (BDBV) is a member of the Ebolavirus genus in the Filoviridae family, and infections by this virus result in severe hemorrhagic fever in humans. The virus was initially discovered in 2007 during an outbreak of hemorrhagic fever in the Bundibugyo district of Uganda that resulted in a case-fatality rate (CFR) of 25% (1, 2). A subsequent outbreak occurred in 2012 in the Democratic Republic of Congo, in which the CFR was 36% (3). Ebola virus (EBOV) is also a member of the Ebolavirus genus and is the best known of all filoviruses. EBOV has caused 12 outbreaks so far, with the first outbreak occurring during 1976 in Yambuku, Democratic Republic of the Congo, and the most recent outbreak taking place from 2014 to 2016 in Guinea, Sierra Leone, and Liberia. Outbreaks of EBOV disease can result in a CFR of up to 90%. Although BDBV is related to EBOV, phylogenetic analysis indicates that these two viruses are antigenically distinct, with only 60% sequence similarity for the glycoprotein at the amino acid level (2, 3). The natural reservoir for these viruses remains unknown; however, serological evidence suggests that fruit bats may serve as an animal host for EBOV (4, 5).
The development of small animal models for studying filoviruses has been a focus in the scientific community to advance our understanding of these pathogens (6). Since wild-type filovirus isolates are not lethal to immunocompetent rodents, host-adapted virus variants have been generated through sequential passaging in mice and guinea pigs to establish animal models for studying filovirus pathogenesis as well as evaluating vaccines and therapeutics. Using this method, small animal models have been developed for EBOV (7, 8), Ravn virus (9), and Marburg virus (10, 11), and recently a guinea pig model has been developed for Sudan virus (12). While these animal models have played a considerable role in the development of specific antivirals against filoviruses, the establishment of these animal models can be laborious and time-consuming, limiting the ability to study outbreak strains in a timely manner.
Domestic ferrets (Mustela putorius furo) are used mainly for modeling respiratory diseases, such as influenza virus. While these animals require special housing/husbandry requirements and may vary in individual immunological responses and the reagents to study T-cell immunity/cytokine responses are not yet widely available (13, 14), ferrets are nevertheless popular animals for studying the pathogenesis and transmission of many wild-type RNA viruses across the Mononegavirales order, including those in the Rhabdoviridae, Paramyxoviridae, Orthomyxoviridae, Togaviridae, Bunyaviridae, and Coronaviridae (15, 16) families. Since Filoviridae species also belong to Mononegavirales, it is reasonable to suspect that ferrets may also be susceptible to infection by members of the Ebolavirus genus without the need for host adaptation, facilitating the capacity for studying outbreak virus strains. To investigate this possibility, we challenged ferrets via multiple routes of infection with either a wild-type BDBV or an EBOV isolate from the recent outbreak in West Africa and monitored the clinical profile and pathology of the animals following infection.
MATERIALS AND METHODS
Ethics statement.
The animal work was performed in the biosafety level 4 (BSL4) facility at the Canadian Science Centre for Human and Animal Health (CSCHAH) in Winnipeg, Canada. All experiments were approved by the Animal Care Committee (ACC) of the CSCHAH, in accordance with guidelines from the Canadian Council on Animal Care (CCAC). Ferrets were acclimatized for 7 days prior to infection and were given food and water ad libitum. Animals were monitored twice daily. Environmental enrichment in the form of toys and plastic huts was also provided throughout the duration of the study.
Animals and viruses.
Six-month-old female ferrets (Mustela putorius furo), ranging in weight from 0.66 to 0.92 kg, were purchased from Marshall BioResources (North Rose, NY, USA). All animals had received vaccinations against distemper and rabies viruses and were given anticoccidials prior to the experiment. Animals were housed in caging units that provide negative pressure and HEPA-filtered air in the BSL4 facility at CSCHAH and were monitored twice daily for signs of disease during the course of the experiment.
Groups of six animals were infected intramuscularly (i.m.) with either 159 50% tissue culture infective doses (TCID50) of Bundibugyo virus/H.sapiens-wt/UGA/2007 (BDBV; GenBank accession number KR063673.1) (2), or 200 TCID50 of Ebola virus/H.sapiens-wt/GIN/2014/Makona-C07 (EBOV; GenBank accession number KT013257.3) via the i.m. or intranasal (i.n.) route. Both viruses were passaged once in VeroE6 cells to generate viral stocks. Following infection, clinical scoring was performed using a scoring system for ferrets previously approved by the ACC. Blood was drawn every 2 days to determine viral load, measure complete blood counts, and evaluate biochemical markers. Major organs, including the kidneys, spleen, and liver were collected at the time of euthanasia for virological, histopathological, and immunohistochemical analysis. The experiment was not blinded, and animals were randomized into the groups.
Serum biochemistry, complete blood counts, and coagulation.
Serum biochemistry was evaluated with the VetScan VS2 analyzer (Abaxis, USA), in accordance with the manufacturer's instructions. To determine complete blood cell counts and measure the coagulation parameters, samples were analyzed using a VetScan HM5 hematology machine (Abaxis, USA) and a Start4 instrument (Diagnostica Stago), respectively, according to the manufacturers' instructions.
Quantification of viral loads.
Viral RNA from blood, oral and rectal swabs, and nasal washes was extracted using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions. Viral RNA was also extracted from approximately 30 mg of harvested organs using the RNeasy kit (Qiagen) according to the manufacturer's instructions. Reverse transcription-quantitative PCR (RT-qPCR) was used to determine viral loads and was performed using an ABI StepOnePlus instrument as adapted from a protocol described previously (12). Sequences of the BDBV primers and probes used in this study are as follows: BEBOVGP F, 5′-GRATTCTRCAATTGCCC-3′; BEBOVGP R, 5′-TGRAATAGGATTATTACCCAAACAA-3′; BEBOVGPP FAM, 5′-FAM-GTGARCGCTTCAGRAAAACATCTTTYT-BHQ1-3′. For EBOV, the primer and probe sequences are as follows: EBOVLF2, 5′-CAGCCAGCAATTTCTTCCAT-3′; EBOVLR2, 5′-TTTCGGTTGCTGTTTCTGTG-3′; EBOVLP2FAM, 5′-FAM-ATCATTGGCGTACTGGAGGAGCAG-BHQ1-3′.
Whole-genome sequencing of BDBV and EBOV.
For whole-genome sequencing, purified RNA was subject to cDNA synthesis using a Maxima H minus ds cDNA kit (Thermo Fisher Scientific, USA) as per manufacturer instructions. Following synthesis, the resultant double-stranded cDNA was purified using Agencourt Ampure XP magnetic beads (Beckman Coulter Canada, LP), resuspended in sterile water, and quantified fluorometrically via PicoGreen prior to library generation with a Nextera XT DNA Library Preparation kit using compatible primers from the Nextera XT Index kit (Illumina Inc., USA). Libraries were quantified following tagmentation and indexing with the Agilent 2200 Tapestation (Agilent Technologies, USA) according to the manufacturer's instructions. Libraries for each sample were normalized to 2 nM and pooled for sequencing with 1% phiX and 1% in-house control spikes. The final concentration of library pools for sequencing was 20 pM. The Illumina 300-cycle reagent kit with V2 chemistry was used on the MiSeq Illumina platform to generate paired-end sequence data and fed through an in-house bioinformatic pipeline. Briefly, the sequences for BDBV and EBOV were transformed to reference files using bowtie2 (ver 2.0.5) and then aligned and assembled using samtools (ver 1.1, htslib 1.1). Variant calling was done using freebayes (ver 0.9.8) with a minimum coverage of 20, a frequency threshold of 5%, and a minimum quality score of 30. For BDBV samples, the average coverage was 121-fold (median, 103-fold; interquartile range [IQR], 129-fold; range, 0- to 560-fold). For EBOV samples, the average coverage was 34-fold (median, 34-fold; IQR, 30-fold; range, 0- to 103-fold). Any sequences present in both the challenge virus stock and that isolated from ferrets after infection were considered not mutated, whereas any differences were considered mutations.
Data analysis.
Figures were generated and statistical analyses performed using GraphPad Prism 6.0 software. Differences between means were evaluated using the paired, two-tailed, Student t test and were deemed significant at P values of ≤0.05. Kaplan-Meier survival curves were analyzed by the log rank test.
Histopathology and immunohistochemistry.
Tissues were fixed in 10% neutral phosphate-buffered formalin, routinely processed, sectioned at 5 μm, and stained with hematoxylin and eosin (H&E) for histopathologic examination. For immunohistochemistry (IHC), paraffin-embedded tissue sections were quenched for 10 min in aqueous 3% hydrogen peroxide and then pretreated with proteinase K for 10 min. The primary antibody applied to the sections was a polyclonal anti-Ebola VP40 antibody produced in rabbits from IBT BioServices (USA), at a 1:750 dilution for 30 min. Sections were visualized using a horseradish peroxidase-labeled polymer, Envision + system (anti-rabbit) (Dako, USA), subjected to reaction with the chromogen diaminobenzidine (DAB), and counterstained with Gill's hematoxylin.
RESULTS
Survival and viral load of ferrets infected with BDBV.
Ferrets were challenged with 159 TCID50 BDBV via the intramuscular route. Infection with BDBV was found to be lethal to all animals, and the median time to death was 8 days (Fig. 1A). Viremia was first detectable at 4 days postinfection (dpi), and peak viremia of ∼107 to ∼108 genome equivalents per ml (GEQ/ml) or ∼105 to ∼107 TCID50/ml occurred at 8 dpi (Table 1). Sequencing of the virus isolated from ferrets during the experiment showed that the majority of the viral population had not mutated, suggesting that the ferrets were likely susceptible to the wild-type isolate. Interestingly, within this minority viral population a common mutation, Y28H, was detected in the GP gene in isolates from all infected animals (Table 2). Additionally, BDBV was detected at concentrations of up to ∼107 GEQ/ml or up to ∼103 TCID50/ml in oral and rectal swabs as well as nasal washes between 6 and 8 dpi (Table 1).
FIG 1.
Clinical parameters in BDBV-challenged ferrets. (A) Animals were monitored for survival. (B) Representative picture of petechial rash in an infected ferret. (C and D) Clinical scores (C) and temperatures (D) in BDBV-infected ferrets. (E) Weight loss over the course of infection in animals.
TABLE 1.
Viral load and shedding of ferrets infected with BDBV, as determined by RT-qPCR and live virus titrationa
| Animal ID | Sample | Day 4 |
Day 6 |
Day 8 |
|||
|---|---|---|---|---|---|---|---|
| GEQ/ml | TCID50/ml | GEQ/ml | TCID50/ml | GEQ/ml | TCID50/ml | ||
| 70 | Blood | 1.49E4 | 1.47E1 | 1.50E7 | 1.47E5 | 8.17E7 | 6.81E6 |
| Nasal | Neg | — | Neg | — | 5.10E5 | Neg | |
| Oral | Neg | — | 6.43E4 | 3.16E2 | 1.77E6 | Neg | |
| Rectal | Neg | — | 2.06E4 | 5.62E2 | 8.46E5 | Neg | |
| 72 | Blood | 2.51E3 | 1.47E1 | 4.54E3 | 1.47E6 | 5.95E7 | 1.47E6 |
| Nasal | Neg | — | 1.51E5 | 3.16E2 | 0.00E0 | Neg | |
| Oral | Neg | — | Neg | — | 7.81E6 | Neg | |
| Rectal | Neg | — | 5.32E3 | 5.62E2 | 1.10E6 | Neg | |
| 101 | Blood | 3.85E3 | 1.47E1 | 1.33E7 | 1.47E5 | 9.62E7 | 3.16E6 |
| Nasal | Neg | — | Neg | — | 4.43E5 | Neg | |
| Oral | Neg | — | Neg | — | 1.01E6 | Neg | |
| Rectal | Neg | — | 5.39E2 | 3.16E2 | 1.64E6 | Neg | |
| 156 | Blood | 6.93E3 | 6.81E0 | 1.04E7 | 1.47E5 | 7.62E7 | 3.16E6 |
| Nasal | Neg | — | Neg | — | 9.61E4 | Neg | |
| Oral | Neg | — | 4.29E4 | Neg | 3.76E6 | Neg | |
| Rectal | Neg | — | Neg | — | 8.27E5 | Neg | |
| 157 | Blood | Neg | — | 6.87E6 | 1.47E5 | 1.80E7 | 1.47E7 |
| Nasal | Neg | — | 5.09E8 | 1.78E2 | 2.39E4 | Neg | |
| Oral | Neg | — | 1.06E5 | 1.78E3 | 1.92E6 | Neg | |
| Rectal | Neg | — | 1.27E4 | 1.78E2 | 6.41E6 | 1.78E2 | |
| 161 | Blood | 2.59E3 | 6.81E0 | 1.76E6 | 1.47E4 | 5.54E7 | 3.16E5 |
| Nasal | Neg | — | 1.62E4 | 3.16E2 | 4.87E5 | Neg | |
| Oral | Neg | — | 6.46E3 | 3.16E2 | 1.28E5 | Neg | |
| Rectal | Neg | — | 5.44E4 | 1.78E2 | 1.10E5 | Neg | |
Neg, negative for virus; —, not tested. Live virus titrations were not performed in samples that tested negative for BDBV by RT-qPCR.
TABLE 2.
Sequencing results of BDBV or EBOV reisolated from ferretsa
| Gene | Mutations found in virus reisolated from indicated animal |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BDBV |
EBOV |
||||||||||
| 70 | 72 | 101 | 156 | 157 | 161 | 140 | 157 | 163 | 145 | 138 | |
| GP gene | Y28H | Y28H | Y28H | Y28H | Y28H | Y28H | T435I, T544S | V277A, N278H, P279S, T435I, T544S | D78E, V79A, T435I, T544S | D78E, V79A, T435I, T544S | |
| VP24 gene | K163N | ||||||||||
| L gene | V128F, E133Q, R571H | Q1307H | M945K | K1938I, L1946S, F2116L | |||||||
No mutations were found for NP, VP35, VP40, or VP30 genes.
Clinical findings of ferrets infected with BDBV.
Examination of BDBV-infected ferrets revealed an observable petechial rash in 4 of 6 animals, evident by 8 dpi (Table 3; Fig. 1B). An increase in clinical scores, indicating signs of disease, was detectable only by 7 dpi, and loss in body weight began at 4 dpi (Fig. 1C and D). A majority of ferrets had developed fever by 7 dpi (Fig. 1E).
TABLE 3.
Clinical findings for ferrets infected with BDBV or EBOVa
| Animal ID | Virus | Route of infection | Clinical signs (dpi) | Fever onset (dpi) | Cutaneous finding(s) (dpi) | White blood cell finding (dpi) | Platelet finding(s) (dpi) | Time of death (dpi) |
|---|---|---|---|---|---|---|---|---|
| 140 | EBOV | i.m. | Depression (4), dark feces, diarrhea (4), blood in stool (5), increased respiratory rate (4) | 4 | Subcutaneous hemorrhage (5) | Leukocytosis (2) | 5 | |
| 159 | EBOV | i.m. | Depression (4), dark feces, diarrhea (4), increased respiratory rate (4) | 4 | Rash, subcutaneous hemorrhage (5) | Leukocytosis (4) | 5 | |
| 163 | EBOV | i.m. | Depression (4), dark feces, diarrhea (4), increased respiratory rate (4) | 4 | Rash, subcutaneous hemorrhage (6) | Leukocytosis (2, 4) | Thrombocytopenia (2) | 6 |
| 73 | EBOV | i.n. | Leukocytosis (2) | 4 | ||||
| 138 | EBOV | i.n. | Depression (6), increased respiratory rate (6) | 4 | Rash, subcutaneous hemorrhage (6) | Leukocytosis (4) | 6 | |
| 145 | EBOV | i.n. | Depression (6), decreased respiratory rate (6) | Rash (6) | 6 | |||
| 70 | BDBV | i.m. | Depression (7), dark feces, diarrhea (7), blood in stool (8), increased respiratory rate (7) | 6 | Rash (8) | Leukocytopenia (6) | 8 | |
| 101 | BDBV | i.m. | Depression (7), increased respiratory rate (7), dark feces, diarrhea (7), blood in stool (8) | Rash (8) | Leukocytopenia (4, 6) | Thrombocytopenia (2, 6) | 8 | |
| 156 | BDBV | i.m. | Depression (7), dark feces (7), liquid stool (8), increased respiratory rate (7), decreased respiratory rate (8) | 6 | Thrombocytopenia (2, 4, 6) | 8 | ||
| 161 | BDBV | i.m. | Dark feces, diarrhea (7), fecal blood in cage (9), increased respiratory rate (7) | 6 | Rash, subcutaneous hemorrhage (9) | Leukocytopenia (2, 4, 6) | Thrombocytopenia (2, 6) | 9 |
| 72 | BDBV | i.m. | Depression (7), dark feces, diarrhea (7), increased respiratory rate (7), decreased respiratory rate (8) | Rash, subcutaneous hemorrhage (8) | Leukocytopenia (6) | Thrombocytosis (2,4); thrombocytopenia (6) | 8 | |
| 157 | BDBV | i.m. | Dark feces, diarrhea (7), increased respiratory rate (7) | 6 | Leukocytosis (2, 4) | Thrombocytosis (2, 4, 6) | 8 |
Fever was defined as a temperature change of >2.5°C over baseline or 1.5°C over baseline and >39.7°C. Mild rash was defined as focal areas of petechiae covering <10% of the skin, moderate rash was defined as areas of petechiae covering 10 to 40% of the skin, and severe rash was defined as areas of petechiae and/or ecchymosis covering >40% of the skin. Leukocytopenia and thrombocytopenia were defined as a ≥30% decrease in the numbers of WBCs and platelets, respectively. Leukocytosis and thrombocytosis were defined as a 2-fold or greater increase in numbers of WBCs and platelets above baseline.
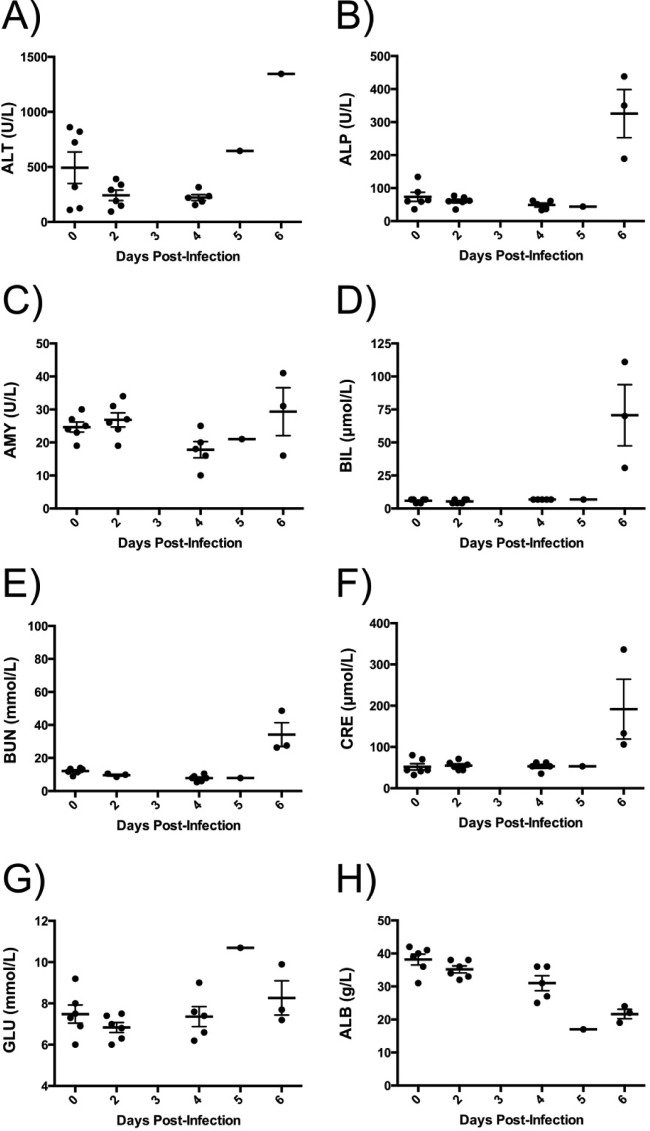
Abnormalities in several serum biochemistry markers were observed in all infected animals (Fig. 2), suggesting that multiple organs were impacted during the disease course. Specifically, increases in concentrations/activities of aminotransferase (ALT), alkaline phosphatase (ALP), amylase (AMY), and total bilirubin (BIL) were observed (Fig. 2A to D), indicating liver damage. Increased concentrations of blood urea nitrogen (BUN) and creatinine (CRE) in all animals during advanced BDBV disease indicate possible renal failure (Fig. 2E and F). Decreased glucose (GLU) concentrations suggest pancreatic disease (Fig. 2G), whereas reduced albumin (ALB) concentrations could be caused by hepatic or renal dysfunction.
FIG 2.
Biochemical parameters of ferrets infected with BDBV. Blood was drawn from animals at various time points during the course of infection with BDBV, and the concentrations of the following were measured: aminotransferase (ALT) (A); alkaline phosphatase (ALP) (B); amylase (AMY) (C); total bilirubin (BIL) (D); blood urea nitrogen (BUN) (E); creatine (CRE) (F); blood glucose (GLU) (G); serum albumin (ALB) (H). Data represent the means, and error bars represent the standard errors of the means.
Dysregulation of the coagulation pathway and depletion in the number of immune cells are both hallmarks of severe filoviral hemorrhagic disease in humans (17, 18). In all BDBV-infected animals, significant increases in coagulation parameters were observed. Over the course of BDBV disease, the activated partial thromboplastin time (APTT), thrombin time (TT), and prothrombin time (PT) were prolonged between 20 and 40 s (Fig. 3A). Increased fibrinogen levels were also observed postinfection, but this difference was not significant (Fig. 3A). Hematological analysis revealed a decrease in overall counts of white blood cells (WBC), lymphocytes (LYM), and platelets (PLT) in ferrets after infection (Fig. 3B to D). Overall, the disease process in ferrets showed many similarities to that which is observed in nonhuman primates (NHPs) (19, 20).
FIG 3.
Hematological parameters of ferrets infected with BDBV. Blood was drawn from animals at various time points during the course of infection with BDBV (n = 6). (A) Coagulation data collected preinfection and at the experimental endpoint (i.e., euthanasia), including fibrinogen levels, activated partial thromboplastin time (APTT), thrombin time (TT), and prothrombin activity (PT). (B to D) Lymphocytes (LYM) (B), white blood cells (WBC) (C), and platelets (D) were enumerated over the course of infection. Data represent the means, and error bars represent the standard errors of the means. Samples were compared using paired, two-tailed, Student's t test with significant differences indicated by an asterisk (*, P ≤ 0.05).
Histopathology and viral loads indicate that multiple organs are affected during disease.
The observed dysregulation of biochemical and hematological pathways suggested organ pathology as a result of viral infection. To investigate this, organs were harvested from ferrets after death and sectioned to examine the pathology. In the BDBV-infected group, it was observed that the livers of all animals showed severe pathology with diffuse vacuolar degeneration and loss of hepatocytes leading to disruption of normal architecture (Fig. 4A). Additionally, infiltration of inflammatory cells into the portal areas was noted, and viral inclusion bodies were detected (Fig. 4B). Immunohistochemical staining revealed viral antigen throughout the section, including within hepatocytes and sinusoidal lining cells, as well as free virus within sinusoids and blood vessels (Fig. 4C).
FIG 4.
Histopathology and immunohistochemistry findings in BDBV-infected ferrets. (A) Hepatocyte vacuolation and degeneration. (B) Cytoplasmic viral inclusion bodies in vacuolated hepatocytes (arrows). (C) Viral antigen in a section of the liver. (D) Lymphocytolysis (arrow) and hyperplasia of cells in red pulp areas (arrowhead) in the spleen. (E) Lymphoid depletion (arrow) associated with necrotic debris and pyknotic nuclei (arrowhead) in the germinal centers of the spleen. (F) Viral antigen in the spleen. Positive immunostaining of large macrophages is identified by arrows. (G) Dilation of tubules containing proteinaceous material (arrow) or granular casts (arrowhead). (H) Renal tubules contain eosinophilic granular casts and are lined by attenuated epithelial cells (arrow). Glomerular capillary walls (thickening indicated by arrowhead). (I) Viral antigen in the kidney. (J, K) Lung showing expanded alveolar walls due to cellular infiltration with occasional necrotic cells (arrows). (L) Viral antigen throughout the lung section, primarily in macrophages and within capillaries. Bar, 100 μm (A, C, D, F, G, I, J, and L), 20 μm (E, H, K), or 10 μm (B). (M) Tissues were collected, and mean viral titers from individual animals postmortem (n = 6) were determined by RT-qPCR. Each horizontal segment represents the mean for the group.
Analysis of the spleen and kidney demonstrated that multiple organs were affected during the course of disease. Lesions in the spleens were characterized by hyperplasia of the red pulp, with an increase in macrophages and multifocal areas of necrosis as well as lymphoid depletion associated with necrotic debris and pyknotic nuclei (Fig. 4D and E). Moreover, vasculitis was also observed. Abundant amounts of viral antigen were detected within macrophages, as well as throughout the red pulp areas and multifocally within germinal centers (Fig. 4F). Within the kidney, there was tubular degeneration with vacuolation and attenuation of tubular epithelial cells and proteinaceous or granular casts within the tubular lumens (Fig. 4G and H). Glomerular capillary walls appeared to be slightly thickened possibly due to endothelial or epithelial swelling. A moderate amount of viral antigen was detected within glomerular and interstitial capillaries as well as occasionally in tubular epithelial cells.
In 4 of 5 animals, the lungs were diffusely affected by a mild inflammatory process characterized by the expansion of alveolar walls by inflammatory cells and edema (Fig. 4J). Occasionally, pyknotic nuclei were observed, indicating necrosis of cells within alveolar walls (Fig. 4K). Immunohistochemistry revealed abundant amounts of viral antigen throughout approximately 75% of the lung section (Fig. 4L). Moreover, viral antigen was detected within macrophages and endothelial cells, as well as within capillaries and larger vessels. For all BDBV-infected animals, viral titers in each of the affected organs were determined, and the highest titers (up to 1010 GEQ/gram of tissue or 106 TCID50/gram of tissue) as well as live virus were found in the liver, followed by the kidney, spleen, lung, and then heart (Fig. 4M and N).
Results for ferrets infected with EBOV.
To demonstrate that infection was not unique to BDBV, ferrets were also challenged with 200 TCID50 EBOV via either the i.m. or the i.n. route. The course of disease in all EBOV-infected ferrets, regardless of infection route, was similar to that observed with BDBV. The animals reached peak viremia of up to ∼107 GEQ/ml or ∼109 TCID50/ml by 5 or 6 dpi (Table 4) and also showed evidence of viral shedding from the oral, nasal, and rectal cavities, but shedding of live virus was detected only sporadically (Table 4). The median time to death was 5 or 6 dpi, respectively, for the EBOV i.m. and i.n. groups (Fig. 5A), which was slightly shorter than in ferrets infected with BDBV. A petechial rash weight loss and fever were observed, corresponding with an increase in clinical scores (Table 3 and Fig. 5B to E). Notably, weight loss was less pronounced in animals infected with EBOV than in those infected with BDBV.
TABLE 4.
Viral load and shedding of ferrets infected with EBOV, as determined by RT-qPCR and live virus titrationa
| Animal ID | Sample | Day 4 |
Day 5 |
Day 6 |
|||
|---|---|---|---|---|---|---|---|
| GEQ/ml | TCID50/ml | GEQ/ml | TCID50/ml | GEQ/ml | TCID50/ml | ||
| 73 | Blood | 1.63E6 | 1.78E3 | ||||
| Nasal | Neg | — | |||||
| Oral | Neg | — | |||||
| Rectal | Neg | — | |||||
| 138 | Blood | 3.28E6 | 1.78E5 | — | — | 2.05E7 | 3.16E9 |
| Nasal | 1.67E6 | Neg | — | — | 3.86E6 | Neg | |
| Oral | 1.21E6 | Neg | — | — | 4.51E6 | Neg | |
| Rectal | Neg | — | — | — | 1.25E6 | 6.81E2 | |
| 140 | Blood | 7.09E6 | 3.16E6 | 1.22E7 | 5.62E8 | ||
| Nasal | Neg | — | 9.46E5 | Neg | |||
| Oral | Neg | — | 2.24E6 | Neg | |||
| Rectal | 1.04E6 | Neg | 2.43E6 | Neg | |||
| 145 | Blood | 3.43E6 | 3.16E4 | — | — | 2.17E7 | 3.16E9 |
| Nasal | 1.44E6 | Neg | — | — | 1.52E6 | Neg | |
| Oral | Neg | — | — | — | 4.08E6 | Neg | |
| Rectal | 3.16E6 | Neg | — | — | 3.16E6 | Neg | |
| 159 | Blood | 6.42E6 | 1.78E5 | 1.87E7 | 5.62E7 | ||
| Nasal | Neg | — | Neg | — | |||
| Oral | 7.94E5 | 6.81E2 | Neg | — | |||
| Rectal | Neg | — | Neg | — | |||
| 163 | Blood | 4.83E6 | 3.16E4 | 1.26E7 | 1.78E6 | 1.85E7 | 3.16E7 |
| Nasal | Neg | — | 8.02E5 | 1.47E0 | 1.98E6 | Neg | |
| Oral | Neg | — | 1.01E6 | Neg | 4.56E6 | Neg | |
| Rectal | Neg | — | 1.72E6 | Neg | 2.50E6 | Neg | |
Neg, negative for virus; —, not tested. Live virus titrations were not performed in samples that tested negative for EBOV by RT-qPCR.
FIG 5.
Clinical parameters in EBOV-challenged ferrets. (A) Animals were monitored for survival. (B) Representative picture of petechial rash in an infected ferret. (C and D) Clinical scores (C) and temperatures (D) in EBOV-infected ferrets. (E) Weight loss over the course of infection in animals.
Biochemical analyses were similar to those observed for human EBOV disease. Specifically, increases in concentrations/activities of ALT, ALP, and BIL were observed (Fig. 6A to D), indicating liver damage. Investigation of renal markers indicated increased levels of BUN and CRE in all animals (Fig. 6E and F). The elevated levels of these biomarkers indicate that kidney dysfunction occurred as a result of infection. Furthermore, GLU and ALB abnormalities were also observed during advanced EBOV disease in these animals (Fig. 6G and H), and decreased levels of ALB are predicted to be the cause of edema during the course of EBOV disease, as previously observed in infected nonhuman primates (21).
FIG 6.
Biochemical parameters of ferrets infected with EBOV. Blood was drawn from animals at various time points during the course of infection with EBOV, and the concentrations of the following were measured: aminotransferase (ALT) (A); alkaline phosphatase (ALP) (B); amylase (AMY) (C); total bilirubin (BIL) (D); blood urea nitrogen (BUN) (E); creatine (CRE) (F); blood glucose (GLU) (G); serum albumin (ALB) (H). Data represent the means, and error bars represent the standard errors of the means.
Additionally, disruption of the coagulation pathways was also observed and was similar to that seen after infection with BDBV (Fig. 7). Notably, prolonged activated partial thromboplastin time (APTT) and thrombin time (TT), an increase in fibrinogen levels, and a decrease in prothrombin activity percentage (PT) were observed. Collectively, this suggests that ferrets may model disseminated intravascular coagulation (Fig. 7A). Hematological analysis revealed abnormalities in the counts of WBC, LYM, and PLT in ferrets after EBOV infection (Fig. 7B to D). Interestingly, ferrets infected with EBOV had decreased levels of serum albumin after challenge, which is predicted to be the cause of edema during the course of disease, as previously observed in infected nonhuman primates (21).
FIG 7.
Hematological parameters of ferrets infected with EBOV. Blood was drawn from animals at various time points during the course of infection with EBOV (n = 3 per route of infection). (A) Coagulation data collected preinfection and at the experimental endpoint, including fibrinogen levels, activated partial thromboplastin time (APTT), thrombin time (TT), and prothrombin activity (PT). (B to D) Lymphocytes (LYM) (B), white blood cells (WBC) (C), and platelets (PLT) (D) were enumerated over the course of infection. Data represent the means, and error bars represent the standard errors of the means. Samples were compared using paired, two-tailed, Student's t test with significant differences indicated by an asterisk (*, P ≤ 0.05).
Histopathological changes observed in the organs of all EBOV-infected animals were consistent with severe damage stemming from uncontrolled virus replication (Fig. 8 and 9). However, in the lungs of the EBOV i.n. group, there was a diffuse and severe necrotizing pneumonia, bronchiolitis, and perivasculitis (Fig. 9A and B). There was more viral antigen detected than in the EBOV i.m. groups, with abundant antigen detected around blood vessels (Fig. 9C). Pathology in the kidneys, liver, spleen, and lungs were similar to what was seen in BDBV-infected animals (Fig. 9D to L). Additionally, viral titers in each of the affected organs were up to 100-fold higher in EBOV-infected animals than in BDBV-infected animals (Fig. 9M and N).
FIG 8.
Histopathology and immunohistochemistry findings in EBOV i.m.-infected ferrets. (A) Widespread hepatocyte vacuolation and degeneration with portal inflammation (arrow). (B) Cytoplasmic viral inclusion bodies (arrows), vacuolated hepatocytes, and degenerating/necrotic cells (arrowheads). (C) Viral antigen in a section of the liver. (D) Lymphocytolysis (arrow) and hyperplasia of cells in red pulp areas (arrowhead) in the spleen. (E) Lymphoid depletion associated with necrotic debris and pyknotic nuclei (arrow) in the germinal centers of the spleen. (F) Viral antigen in the spleen. Note positive immunostaining within germinal center (arrow). (G) Dilation of renal tubules containing proteinaceous material or granular casts (arrows). (H) Renal tubules contain eosinophilic proteinaceous material and are lined by vacuolated epithelial cells. (I) Moderate amounts of viral antigen observed in the kidney. (J, K) Expanded alveolar walls in lungs due to cellular infiltration and necrotic cells (arrows). (L) Viral antigen observed multifocally throughout the lung section, primarily in macrophages and within capillaries. Bar, 100 μm (A, C, D, F, G, I, J, L), 20 μm (E, H, K), or 10 μm (B). Pathology was examined for three animals, and representative pictures are shown.
FIG 9.
Histopathology and immunohistochemistry findings in EBOV i.n.-infected ferrets. (A) Hepatocyte vacuolation and degeneration and portal inflammation (arrow). (B) Cytoplasmic viral inclusion bodies within vacuolated hepatocytes. (C) Viral antigen in a liver section. (D) Lymphocytolysis (arrow) and hyperplasia of cells in red pulp areas (arrowhead) in the spleen. (E) Lymphocytolysis within the germinal centers of the spleen (arrow). (F) Viral antigen in the spleen. Note clusters of immunopositive macrophages (arrows). (G) Dilation of tubules containing proteinaceous material or granular casts (arrows). (H) Renal tubules contain granular casts and are lined by attenuated epithelial cells (arrow). (I) Viral antigen in the kidney. (J, K) Lung showing diffuse interstitial pneumonia with intense infiltration of inflammatory cells within alveolar walls and extensive necrosis. Note numerous necrotic cells within alveolar walls (arrows) and perivasculitis (arrowhead). (L) Viral antigen in the lung section and in perivascular areas (arrow). Bar, 100 μm (A, C, D, F, G, I, J, L), 20 μm (E, H, K), or 10 μm (B). Pathology was examined for three animals, and representative pictures are shown. (M) Tissues were collected, and mean viral titers from individual animals postmortem (n = 3 per route of infection) were determined by RT-qPCR. Each horizontal segment represents the mean for each group.
DISCUSSION
At present, several immunocompetent small animal models for studying filoviruses are available, including mice (7), guinea pigs (8), and hamsters (22), and infection with these host-adapted viruses resembles human disease to various levels (23). Until now, the use of wild-type virus isolates in small animal models has been restricted to immunodeficient mice, such as STAT-1 and interferon α/β receptor knockout (KO) mice (24, 25). While these mouse models offer a means of investigating the role of different host pathways on disease, they are not ideal for vaccine and therapeutic research. For example, infection of STAT-1 KO mice with EBOV was uniformly lethal, but immunization with a vaccine shown to result in full protection in immunocompetent mice failed to induce full protection in STAT-1 KO animals (25). In contrast, infection of interferon α/β receptor KO mice with BDBV resulted in transient viremia, but the animals did not succumb to infection (24). As such, nonhuman primates are currently the only recognized immunocompetent animal model for studying BDBV (20, 26), which complicates efforts to gather sufficient preclinical data to advance experimental vaccines or therapeutics through the pipeline and limits the ability to study viral pathogenesis. In addition, despite the availability of rodent models for EBOV, only preliminary investigations into EBOV transmission have been studied in guinea pigs (27). Since the ferrets in our study are susceptible to infection with wild-type virus, subsequent studies should focus on the efficiency of virus transmission from an infected host to naive animals (either by direct or by indirect contact) to confirm the translatability of previous findings in guinea pigs with regard to transmission.
The results from this study demonstrate that ferrets are indeed susceptible to challenge with multiple filovirus species. Interestingly, infection of ferrets with BDBV or EBOV results in disease that resembles that observed in humans, including petechial rash and coagulation disorders, although it should be noted that the ferret model likely represents only one form of clinical disease and that signs of BDBV or EBOV infections in humans may manifest in more ways than were observed by this study, as indicated by recent reports describing the ability of the virus to infect the central nervous system (28, 29). The results of our study are in agreement with those from another report that was recently published (30), in which the authors infected ferrets i.n. using 1,000 PFU of BDBV. The disease course for BDBV infection of ferrets via i.n. is similar to an i.m. challenge in this study, with comparable times to death (7 versus 8 dpi), uncontrolled viral replication/shedding, rash, abnormalities in LYM and WBC counts, and evidence of multiple-organ failure (30). The combined results from these two studies show that ferrets are susceptible to multiple routes of BDBV infection, regardless of whether they consisted of an accidental needle-stick injury (i.m.) or exposure via a potential physiological route of infection (i.n.).
The authors of the same study (30) also infected ferrets i.n. using 1,000 PFU of EBOV, Kikwit variant, which was isolated from a patient during the 1995 EBOV outbreak in the Democratic Republic of the Congo, Central Africa. The disease course for EBOV infection is similar to that observed in our study using isolates from West Africa, with similar times to death (6 dpi), uncontrolled viral replication/shedding, rash, abnormalities in LYM and WBC counts, and evidence of multiple-organ failure (30). Interestingly, in our study the ferrets infected with EBOV showed a substantial increase in PLT counts after infection. This may be indicative of the reactive thrombocytosis that is attributed to hemorrhaging observed during advanced EBOV disease. The combination of the results from these studies shows that ferrets are susceptible to multiple routes of EBOV infection, as well as different variants (Kikwit versus C07).
Interestingly, Cross and colleagues did not look for mutations in the viral sequences following infection (30). In contrast, we noted mutations in a minority population (<10%) of the viruses isolated from the animals, the majority of which were in the GP gene for both viruses. It was observed that BDBV had fewer variants than did EBOV, although the Y28H mutation was found in a small number of viruses in all animals. However, no one mutation was common to all EBOV isolates. Future studies investigating these changes using an EBOV reverse genetics system (31) could help determine what role, if any, these mutations may play.
In the future, ferrets may be of comparable value to nonhuman primates in predicting the efficacy of candidate vaccines and therapeutics in humans. Recently, pan-Ebolavirus monoclonal antibodies with demonstrated in vitro activity have been identified (32–34), and these experimental candidates could be screened in ferrets in vivo. Other advantages with using ferrets as a potential animal model include better availability, smaller body sizes, less stringent housing requirements, and reduced costs. Future follow-up studies to determine the 50% lethal dose (LD50) will be necessary, as will be the testing of previously validated vaccines and antibodies in ferrets to determine the translatability of candidate medical countermeasures between different animal species and humans.
ACKNOWLEDGMENTS
We thank Kevin Tierney, Anders Leung, Estella Moffat, and Brad Collignon for their technical assistance.
This work was supported by the Public Health Agency of Canada. We declare no conflict of interests.
Author contributions: X.Q. conceived and designed the experiment. R.K., S.H., A.K., M.-A.D.L.V., C.U., K.A., G.W., and X.Q. performed experiments; R.K., M.-A.D.L.V., J.A., C.E.-H., G.W, G.P.K., and X.Q. analyzed the data; R.K., G.W., C.E.-H., G.P.K., and X.Q. wrote the paper.
Funding Statement
This work was funded by the Public Health Agency of Canada.
Gary Wong is a recipient of a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research (CIHR) and of a fellowship from the Chinese Academy of Sciences (CAS) through the President's International Fellowship Initiative.
REFERENCES
- 1.Roddy P, Howard N, Van Kerkhove MD, Lutwama J, Wamala J, Yoti Z, Colebunders R, Palma PP, Sterk E, Jeffs B, Van Herp M, Borchert M. 2012. Clinical manifestations and case management of Ebola haemorrhagic fever caused by a newly identified virus strain, Bundibugyo, Uganda, 2007-2008. PLoS One 7:e52986. doi: 10.1371/journal.pone.0052986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, Quan PL, Lipkin WI, Downing R, Tappero JW, Okware S, Lutwama J, Bakamutumaho B, Kayiwa J, Comer JA, Rollin PE, Ksiazek TG, Nichol ST. 2008. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog 4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albariño CG, Shoemaker T, Khristova ML, Wamala JF, Muyembe JJ, Balinandi S, Tumusiime A, Campbell S, Cannon D, Gibbons A, Bergeron E, Bird B, Dodd K, Spiropoulou C, Erickson BR, Guerrero L, Knust B, Nichol ST, Rollin PE, Stroher U. 2013. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in Uganda and the Democratic Republic of the Congo in 2012. Virology 442:97–100. doi: 10.1016/j.virol.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Delicat A, Paweska JT, Gonzalez JP, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa H, Miyamoto H, Nakayama E, Yoshida R, Nakamura I, Sawa H, Ishii A, Thomas Y, Nakagawa E, Matsuno K, Kajihara M, Maruyama J, Nao N, Muramatsu M, Kuroda M, Simulundu E, Changula K, Hang'ombe B, Namangala B, Nambota A, Katampi J, Igarashi M, Ito K, Feldmann H, Sugimoto C, Moonga L, Mweene A, Takada A. 2015. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J Infect Dis 212(Suppl 2):S101–S108. doi: 10.1093/infdis/jiv063. [DOI] [PubMed] [Google Scholar]
- 6.van der Groen G, Jacob W, Pattyn SR. 1979. Ebola virus virulence for newborn mice. J Med Virol 4:239–240. doi: 10.1002/jmv.1890040309. [DOI] [PubMed] [Google Scholar]
- 7.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. 1998. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 178:651–661. doi: 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- 8.Connolly BM, Steele KE, Davis KJ, Geisbert TW, Kell WM, Jaax NK, Jahrling PB. 1999. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis 179(Suppl 1):S203–S217. doi: 10.1086/514305. [DOI] [PubMed] [Google Scholar]
- 9.Cross RW, Fenton KA, Geisbert JB, Ebihara H, Mire CE, Geisbert TW. 2015. Comparison of the pathogenesis of the Angola and Ravn strains of Marburg Virus in the outbred guinea pig model. J Infect Dis 212(Suppl 2):S258–S270. doi: 10.1093/infdis/jiv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu X, Wong G, Audet J, Cutts T, Niu Y, Booth S, Kobinger GP. 2014. Establishment and characterization of a lethal mouse model for the Angola strain of Marburg virus. J Virol 88:12703–12714. doi: 10.1128/JVI.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warfield KL, Bradfute SB, Wells J, Lofts L, Cooper MT, Alves DA, Reed DK, VanTongeren SA, Mech CA, Bavari S. 2009. Development and characterization of a mouse model for Marburg hemorrhagic fever. J Virol 83:6404–6415. doi: 10.1128/JVI.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong G, He S, Wei H, Kroeker A, Audet J, Leung A, Cutts T, Graham J, Kobasa D, Embury-Hyatt C, Kobinger GP, Qiu X. 2016. Development and characterization of a guinea pig-adapted Sudan virus. J Virol 90:392–399. doi: 10.1128/JVI.02331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meunier I, Embury-Hyatt C, Stebner S, Gray M, Bastien N, Li Y, Plummer F, Kobinger GP, von Messling V. 2012. Virulence differences of closely related pandemic 2009 H1N1 isolates correlate with increased inflammatory responses in ferrets. Virology 422:125–131. doi: 10.1016/j.virol.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Rutigliano JA, Doherty PC, Franks J, Morris MY, Reynolds C, Thomas PG. 2008. Screening monoclonal antibodies for cross-reactivity in the ferret model of influenza infection. J Immunol Methods 336:71–77. doi: 10.1016/j.jim.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enkirch T, von Messling V. 2015. Ferret models of viral pathogenesis. Virology 479-480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis T, Magill TP. 1935. Rift Valley fever: a report of three cases of laboratory infection and the experimental transmission of the disease to ferrets. J Exp Med 62:433–448. doi: 10.1084/jem.62.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortepeter MG, Bausch DG, Bray M. 2011. Basic clinical and laboratory features of filoviral hemorrhagic fever. J Infect Dis 204(Suppl 3):S810–S816. doi: 10.1093/infdis/jir299. [DOI] [PubMed] [Google Scholar]
- 18.Twenhafel NA, Mattix ME, Johnson JC, Robinson CG, Pratt WD, Cashman KA, Wahl-Jensen V, Terry C, Olinger GG, Hensley LE, Honko AN. 2013. Pathology of experimental aerosol Zaire ebolavirus infection in rhesus macaques. Vet Pathol 50:514–529. doi: 10.1177/0300985812469636. [DOI] [PubMed] [Google Scholar]
- 19.Hensley LE, Mulangu S, Asiedu C, Johnson J, Honko AN, Stanley D, Fabozzi G, Nichol ST, Ksiazek TG, Rollin PE, Wahl-Jensen V, Bailey M, Jahrling PB, Roederer M, Koup RA, Sullivan NJ. 2010. Demonstration of cross-protective vaccine immunity against an emerging pathogenic Ebolavirus species. PLoS Pathog 6:e1000904. doi: 10.1371/journal.ppat.1000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mire CE, Geisbert JB, Marzi A, Agans KN, Feldmann H, Geisbert TW. 2013. Vesicular stomatitis virus-based vaccines protect nonhuman primates against Bundibugyo ebolavirus. PLoS Negl Trop Dis 7:e2600. doi: 10.1371/journal.pntd.0002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebihara H, Zivcec M, Gardner D, Falzarano D, LaCasse R, Rosenke R, Long D, Haddock E, Fischer E, Kawaoka Y, Feldmann H. 2013. A Syrian golden hamster model recapitulating Ebola hemorrhagic fever. J Infect Dis 207:306–318. doi: 10.1093/infdis/jis626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama E, Saijo M. 2013. Animal models for Ebola and Marburg virus infections. Front Microbiol 4:267. doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brannan JM, Froude JW, Prugar LI, Bakken RR, Zak SE, Daye SP, Wilhelmsen CE, Dye JM. 2015. Interferon alpha/beta receptor-deficient mice as a model for Ebola virus disease. J Infect Dis 212(Suppl 2):S282–S294. doi: 10.1093/infdis/jiv215. [DOI] [PubMed] [Google Scholar]
- 25.Raymond J, Bradfute S, Bray M. 2011. Filovirus infection of STAT-1 knockout mice. J Infect Dis 204(Suppl 3):S986–S990. doi: 10.1093/infdis/jir335. [DOI] [PubMed] [Google Scholar]
- 26.Falzarano D, Feldmann F, Grolla A, Leung A, Ebihara H, Strong JE, Marzi A, Takada A, Jones S, Gren J, Geisbert J, Jones SM, Geisbert TW, Feldmann H. 2011. Single immunization with a monovalent vesicular stomatitis virus-based vaccine protects nonhuman primates against heterologous challenge with Bundibugyo ebolavirus. J Infect Dis 204(Suppl 3):S1082–S1089. doi: 10.1093/infdis/jir350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong G, Qiu X, Richardson JS, Cutts T, Collignon B, Gren J, Aviles J, Embury-Hyatt C, Kobinger GP. 2015. Ebola virus transmission in guinea pigs. J Virol 89:1314–1323. doi: 10.1128/JVI.02836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CIDRAP, University of Minnesota. 15 October 2015, posting date. Neuro complications cited in UK nurse's Ebola case. http://www.cidrap.umn.edu/news-perspective/2015/10/neuro-complications-cited-uk-nurses-ebola-case. [Google Scholar]
- 29.Wong G, Qiu X, Bi Y, Formenty F, Sprecher A, Gao GF, Kobinger GP. 2016. More challenges from Ebola: the infection of the central nervous system. J Infect Dis doi: 10.1093/infdis/jiw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cross RW, Mire CE, Borisevich V, Geisbert JB, Fenton KA, Geisbert TW. 2016. The domestic ferret (Mustela putorius furo) as a lethal infection model for 3 species of Ebolavirus. J Infect Dis 214:565–569. doi: 10.1093/infdis/jiw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albarino CG, Wiggleton Guerrero L, Lo MK, Nichol ST, Towner JS. 2015. Development of a reverse genetics system to generate a recombinant Ebola virus Makona expressing a green fluorescent protein. Virology 484:259–264. doi: 10.1016/j.virol.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, Branchizio A, King H, Brown L, Bryan C, Davidson E, Doranz BJ, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Saphire EO, Ward AB, Bukreyev A, Crowe JE Jr. 2016. Cross-reactive and potent neutralizing antibody responses in human survivors of natural Ebolavirus infection. Cell 164:392–405. doi: 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuyama W, Marzi A, Nanbo A, Haddock E, Maruyama J, Miyamoto H, Igarashi M, Yoshida R, Noyori O, Feldmann H, Takada A. 2016. Discovery of an antibody for pan-ebolavirus therapy. Sci Rep 6:20514. doi: 10.1038/srep20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtsberg FW, Shulenin S, Vu H, Howell KA, Patel SJ, Gunn B, Karim M, Lai JR, Frei JC, Nyakatura EK, Zeitlin L, Douglas R, Fusco ML, Froude JW, Saphire EO, Herbert AS, Wirchnianski AS, Lear-Rooney CM, Alter G, Dye JM, Glass PJ, Warfield KL, Aman MJ. 2016. Pan-ebolavirus and pan-filovirus mouse monoclonal antibodies: protection against Ebola and Sudan viruses. J Virol 90:266–278. doi: 10.1128/JVI.02171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]