ABSTRACT
B virus (Macacine herpesvirus 1) can cause deadly zoonotic disease in humans. Molecular mechanisms of B virus cell entry are poorly understood for both macaques and humans. Here we investigated the abilities of clinical B virus isolates to use entry receptors of herpes simplex viruses (HSV). We showed that resistant B78H1 cells became susceptible to B virus clinical strains upon expression of either human nectin-2 or nectin-1. Antibody against glycoprotein D (gD) protected these nectin-bearing cells from B virus infection, and a gD-negative recombinant B virus failed to enter these cells, indicating that the nectin-mediated B virus entry depends on gD. We observed that the infectivity of B virus isolates with a single amino acid substitution (D122N) in the IgV-core of the gD ectodomain was impaired on nectin-1-bearing cells. Computational homology-based modeling of the B virus gD–nectin-1 complex revealed conformational differences between the structures of the gD-122N and gD-122D variants that affected the gD–nectin-1 protein-protein interface and binding affinity. Unlike HSV, B virus clinical strains were unable to use herpesvirus entry mediator (HVEM) as a receptor, regardless of conservation of the gD amino acid residues essential for HSV-1 entry via HVEM. Based on the model of the B virus gD-HVEM interface, we predict that residues R7, R11, and G15 are largely responsible for the inability of B virus to utilize HVEM for entry. The ability of B virus to enter cells of a human host by using a combination of receptors distinct from those for HSV-1 or HSV-2 suggests a possible mechanism of enhanced neuropathogenicity associated with zoonotic infections.
IMPORTANCE B virus causes brainstem destruction in infected humans in the absence of timely diagnosis and intervention. Nectins are cell adhesion molecules that are widely expressed in human tissues, including neurons and neuronal synapses. Here we report that human nectin-2 is a target receptor for B virus entry, in addition to the reported receptor human nectin-1. Similar to a B virus lab strain, B virus clinical strains can effectively use both nectin-1 and nectin-2 as cellular receptors for entry into human cells, but unlike HSV-1 and HSV-2, none of the clinical strains uses an HVEM-mediated entry pathway. Ultimately, these differences between B virus and HSV-1 and -2 may provide insight into the neuropathogenicity of B virus during zoonotic infections.
INTRODUCTION
B virus (Macacine herpesvirus 1) is an enveloped, double-stranded DNA virus belonging to the genus Simplexvirus of the subfamily Alphaherpesvirinae. B virus generally produces either mild disease or asymptomatic infection in monkeys of the Macaca genus, which are natural reservoir hosts. Similar to human herpes simplex viruses (HSV), B virus in natural host animals initially infects mucosal or skin epidermal and dermal cells and then enters nerve terminals of the sensory neurons subserving these sites. Subsequently, B virus travels in a retrograde direction to the dorsal root ganglion, where it can establish a latent lifelong infection with periodic reactivation (1, 2). B virus infections of the central nervous system (CNS) are extremely rare in the natural host and are usually associated with immunosuppression or intercurrent diseases (3, 4). In most human cases, B virus spreads to the CNS, causing an acute ascending paralysis and encephalomyelitis with an ∼80% mortality rate if not treated in a timely manner. Postmortem examinations reveal focal neuronal lesions occasionally seen in parietal neurons, but far more often in the brainstem and cervical spinal cord, which are primary sites of virus recovery (5–11). The molecular basis for the differences in neurovirulence between HSV and B virus in humans remains a mystery despite the fact that specific molecular differences between these two viruses have been identified (12–19).
B virus is genetically and immunologically closely related to HSV, and some aspects of cell entry and cell-to-cell transmission of B virus and HSV are conserved (14, 20–23). The specific interactions of glycoprotein D (gD) with cognate cellular receptors, viz., herpesvirus entry mediator (HVEM), nectin-1, and nectin-2, as well as one of the several isoforms of 3-O-sulfated heparan sulfate, play a key role in initiation of the cascade of events leading to HSV cell entry (24–32). Similarly, B virus can utilize nectin-1 as a cell entry receptor, and its interaction with B virus gD is required for virus entry into cells bearing nectin-1 as the sole entry receptor (13, 14). Unlike HSV, B virus cannot use either the human or monkey HVEM-mediated pathway for cell entry (14). Another substantial difference between HSV and B virus entry mechanisms is the ability of B virus to enter skin cells by using a gD-independent cell entry pathway(s) (13).
During in vivo infection, alphaherpesviruses likely utilize distinct sets of receptors to infect different cell types in different tissues. Nectin-1 is expressed on a variety of cells and tissues encountered during the progression of HSV pathogenesis; it serves as the primary receptor for infection of neuronal cells and is a predominant receptor on epithelial cells (33–37). The receptor HVEM is expressed on immune cells, including T and B lymphocytes and conventional dendritic cells, and thus functions as the primary receptor on these cell types (38–41). Human nectin-2, like nectin-1, is widely expressed in different cells and tissues, including the brain, synapse junctions, and endothelial, epithelial, fibroblast, and some hematopoietic cells (34, 42, 43). Nectin-2 can serve as a receptor for pseudorabies virus (PRV), bovine herpesvirus 1 (BHV-1), some HSV-2 strains, and HSV-1 gD mutant lab strains rid1, ANG, and HF (44–47).
Receptor usage has been described only for the B virus laboratory strain E2490, which has a long-term passage history in African green monkey kidney cells (Vero cells) (14). The entry receptors of this culture-adapted strain might be substantially different from those of B virus clinical strains. Also, human nectin-2 has never been studied as a potential receptor for B virus entry until now by our group. The present study was undertaken to determine the receptor tropism of low-passage-number clinical strains of B virus isolated from macaque monkeys and from humans with zoonotic infections. In addition to HVEM and human nectin-1, we examined human nectin-2 for the ability to mediate B virus entry. We also compared the efficiencies of receptor usage between B virus and HSV as well as between different B virus strains. Computer modeling of the B virus gD-HVEM interface was performed to study the structural basis for the failure of B virus to enter cells via HVEM. Further, we performed comparative modeling of the B virus gD–nectin-1 interaction to understand the molecular basis of experimentally observed differences in infectivity between B virus isolates carrying an asparagine versus an aspartic acid residue in the IgV-core gD domain on human nectin-1-bearing cells.
MATERIALS AND METHODS
Cells.
Vero cells (ATCC CCL-81; ATCC, Manassas, VA) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA) and 1% antibiotic-antimycotic solution (Invitrogen, Carlsbad, CA). Cells expressing HSV entry receptors were kindly provided by Gary H. Cohen and Roselyn J. Eisenberg (University of Pennsylvania, Philadelphia, PA). These cell lines were developed by transfecting murine melanoma B78H1 cells with pcDNA3.1 expression plasmids carrying the human HVEM (B78-HVEM), nectin-1 (B78-nectin-1), or nectin-2 (B78-nectin-2) gene or with empty vector (B78c) (48). The cells were maintained in DMEM supplemented with 5% FBS and 500 μg/ml Geneticin (G418 sulfate) (Invitrogen, Carlsbad, CA).
Virus strains and isolates.
HSV-1 strain KOS (ATCC VR-1493), HSV-2 strain G (ATCC VR-734), B virus laboratory strain E2490 (a kind gift from the late R. N. Hull, Eli Lilly, Indianapolis, IN), a B virus gD deletion mutant (BV-ΔgDZ) containing the β-galactosidase (β-Gal) gene under the control of a human cytomegalovirus promoter (13), and 19 B virus clinical isolates (obtained from the National B Virus Resource Laboratory, Atlanta, GA) were propagated in Vero cells in maintenance medium with DMEM containing 2% heat-inactivated FBS (2% DMEM). Virus stocks were titrated by plaque assay on Vero cells and were stored in aliquots at −80°C. During these investigations, B virus was categorized as a select agent by the Department of Homeland Security (DHS), and thus all experiments with low levels of virus were done in accordance with relevant Health and Human Services (HHS) (49, 50) and DHS regulations in the Viral Immunology Center biosafety level 3 (BSL-3) laboratory of Georgia State University prior to 2007 and in the BSL-4 laboratory after that date. Stocks and large quantities of B virus were prepared in the BSL-4 laboratory at all times, in accordance with the recommendations for risk group 4 agents as found in the 4th and 5th editions of Biosafety in Microbiological and Biomedical Laboratories (49, 50).
TCID50 assay.
Cell monolayers were prepared in 96-well culture plates. The B virus stock was serially 10-fold diluted in maintenance medium (2% DMEM). The culture medium was removed from the plates, and diluted virus at 25 μl/well was added to each well across the plate, from columns 1 to 9. One column (eight wells) per virus dilution was used for infection. One column of wells was left uninfected; 25 μl/well of maintenance medium was added to this column. The plates were then incubated for 1 h at 37°C with 5% CO2 in a humidified culture incubator. After 1 h of incubation, 100 μl of maintenance medium was added to each well. At 72 h postinfection (hpi), cytopathic effect (CPE) was examined by light microscopy and then by immunostaining using pooled sera obtained from B virus antibody-positive macaques. The 50% tissue culture infective dose (TCID50) of viral stocks, i.e., the dose which gave rise to CPE in 50% of inoculated cultures, was calculated using the Kärber algorithm as described by Hierholzer and Killington (51). Reported infective titers refer to the infective dose per milliliter of viral stock.
Infectivity assay with BV-ΔgDZ and X-Gal staining.
Cell monolayers were exposed to 10-fold serial dilutions of β-Gal-expressing BV-ΔgDZ diluted in Hanks' balanced salt solution (HBSS). After 1 h of incubation at 37°C with 5% CO2 in a humidified culture incubator, the virus inoculum was replaced by maintenance medium, and the cells were incubated for 48 to 72 h and then washed, fixed, and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as previously described (13).
Antisera.
Rabbit polyclonal antisera against B virus gD were obtained previously by DNA immunization (52). Pooled B virus antibody-positive and -negative macaque serum samples were prepared as described previously (53).
Neutralization assays.
Preimmune sera and antisera collected from immunized rabbits were heat inactivated at 56°C for 30 min. Diluted virus stock (100 PFU/100 μl or 100 TCID50/25 μl) was mixed with an equal volume of DMEM containing serial 2-fold dilutions of rabbit serum and incubated for 1 h at 37°C with 5% CO2 in a humidified culture incubator. The neutralization capacity of the rabbit antiserum on nectin-1-bearing and Vero cells was determined by a previously described plaque reduction assay (53). The cell monolayers were fixed with 100% methanol at 48 hpi and then stained with 0.2% crystal violet, and plaques were counted. The neutralization capacity of the serum dilution was calculated according to the following formula: % neutralization = (1 − PFU in a treated well/PFU in a control well) × 100%. The neutralization titer was expressed as the reciprocal of the serum dilution resulting in 50% plaque reduction. Because B virus does not produce well-defined plaques on nectin-2-expressing cells, a CPE inhibition assay was used to determine neutralization titers of the antisera for these cells, as follows. Each preincubated virus-serum mixture (50 μl) was combined with 100 μl of B78-nectin-2 cell suspension (2 × 105 cells/ml) and added to each well of a 96-well cell culture plate. Eight wells were prepared on a plate per dilution of serum. To simplify detection of infected cells, monolayers were fixed at 72 hpi and immunostained with the prepared pool of B virus-positive macaque serum samples. Neutralization titers were determined by the Reed and Muench method, with each neutralization titer expressed as the reciprocal of the highest antibody dilution at which half of the wells were negative for infection (54).
Immunostaining.
Immunostaining of infected cells with a B virus antibody pool was performed as previously described (52).
Sequence analysis of B virus gD.
PCR amplification of the gD gene of each B virus isolate from genomic DNA and gene sequencing were described earlier (53). The nucleotide sequences of the gD alleles were deposited in the GenBank database under accession numbers AF447074 to AF447094. Multiple-sequence alignments between B virus, HSV-1, and HSV-2 gD protein genes were performed and analyzed by using the DNASTAR MegAlign program (DNASTAR, Madison, WI).
Protein structure modeling.
The sequence of B virus gD (GenBank accession no. NP_851925) with the signal peptide deleted was used to prepare a model of a hypothetical complex between B virus gD and HVEM or nectin-1. The structures of the HSV-1 gD–HVEM complex (PDB code 1JMA) (55) and the HSV-1 gD–nectin-1 complex (PDB code 3U82) (56) were used as templates for computer modeling of the B virus gD-HVEM and B virus gD–nectin-1 complexes, respectively. The model structures were prepared using software approbated in the framework of Critical Assessment of Structure Prediction (CASP) conferences as described previously (57). The stability of the gD protein globule structure was calculated using the software package ECMMS (Energy Calculations for MacroMolecular Systems) for molecular mechanics modeling (58). Protein-protein interactions between gD and receptors in the models and in the template structures were analyzed using the INTG program, which calculates hydrogen bonds and hydrophobic interactions (59, 60). Models of the complexes were minimized using the multiple-run procedure as described previously (61). The multiple-run procedure is known to produce accurate side chain predictions (62, 63). The structural alignment of the gD models was prepared using the software package EVM (Expert Validation Modeling) as described previously (57). The illustrations of protein structures were prepared using DS Visualizer (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization.html; Accelrys Inc., CA). An atom-atom “clash” (a severe violation of geometry) was counted if the distance between any two nonbonded atoms was less than 2 Å.
RESULTS
Effectiveness of receptors for B virus, HSV-1, and HSV-2 entry.
The B78H1 murine cell line is resistant to infection by alphaherpesviruses because of the lack of suitable entry receptors (13, 64). We used recombinant B78H1 cell lines, each stably transfected with a transgene encoding one of the HSV entry receptors, for comparative analyses of receptor usage by B virus and HSV types 1 and 2 (48).
The lack of a created reporter wild-type B virus precluded us from using replication-independent cell entry assays. Instead, a calculated infectivity titer for each receptor-bearing cell line was used as a measure of effectiveness of the corresponding receptor for viral entry. Although they are less precise, infectivity assays were used to assess virus entry because reporter reagents were not available. The permissiveness of the B78-HVEM, B78-nectin-1, and B78-nectin-2 cell lines to B virus (lab strain E2490), HSV-1 (strain KOS), and HSV-2 (strain G) was tested by a standard plaque assay. To assess quantitative differences in receptor usage between B virus and HSV types 1 and 2, infectivity titers of each virus on the receptor-expressing cells were normalized to those on Vero cells (determined in parallel experiments). Cell monolayers were immunostained with pooled rhesus B virus antibody-positive serum to simplify counting of plaques and foci of infected cells.
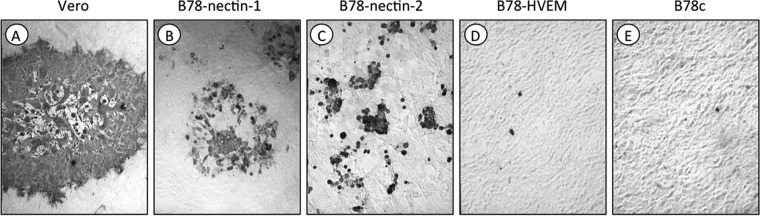
HVEM-expressing B78H1 cells were highly susceptible to HSV-1 and HSV-2 (Table 1); however, only a few immunostained cells were observed in the B78c and B78-HVEM cell monolayers infected with the highest dose of B virus (4.3 × 107 PFU/well) used in the experiments (Fig. 1D and E; Table 1). Thus, using a different cell line expressing human HVEM, B78H1 cells, instead of the Chinese hamster ovary cells used by Fan and colleagues (14), we confirmed their observation that HVEM is incapable of mediating entry of a B virus lab strain.
TABLE 1.
Comparison of entry receptor usages of B virus, HSV-1, and HSV-2
| Virus | Titer (PFU/ml) on cells |
Titer relative to Vero cell infectivity (%)a |
||||||
|---|---|---|---|---|---|---|---|---|
| Vero | B78-HVEM | B78-nectin-1 | B78-nectin-2 | B78c | B78-HVEM | B78-nectin-1 | B78-nectin-2 | |
| B virus E2490 | 7.5 × 107 | 0 | 3.0 × 106 | 1.3 × 106 | 0 | 0.0 | 4.0 | 1.7 |
| HSV-1 (KOS) | 1.8 × 108 | 1.5 × 107 | 8.2 × 106 | 0 | 0 | 8.3 | 4.5 | 0.0 |
| HSV-2 (G) | 1.3 × 107 | 9.5 × 106 | 3.0 × 106 | 1.0 × 103 | 1.0 × 103 | NA | NA | NA |
Calculated as follows: (titer on receptor-expressing cells/titer on Vero cells) × 100%. NA, HSV-2 can use an endogenous receptor to enter B78H1 cells, and therefore the effect of expressed receptors for HSV-2 entry is difficult to quantify precisely on this cell line.
FIG 1.
Susceptibility of murine B78H1 cells expressing HSV entry receptors to B virus infection. B virus E2490 was titrated on confluent monolayers of Vero (A), B78-nectin-1 (B), B78-nectin-2 (C), B78-HVEM (D), and B78c (E) cells. At 48 hpi, the cells were fixed and immunostained with rhesus B virus antibody-positive serum. Panels D and E represent images captured from culture wells infected with 4.3 × 107 PFU/well of B virus. Only a few immunostained cells were observed in the B78c (E) and B78-HVEM (D) cells. Panels C, B, and A represent selected wells that best reflect the CPE produced by B virus on the nectin-2-expressing, nectin-1-expressing, and Vero cells, respectively. Magnification, ×10.
Both nectin-1- and nectin-2-bearing cells were highly susceptible to B virus infection (Fig. 1B and C), but only nectin-1-expressing cells were susceptible to HSV-1 and HSV-2 (Table 1). The sensitivities of nectin-1-expressing cells to B virus and HSV-1 were comparable, indicating that nectin-1 is equally efficient at mediating entry of these two viruses (Table 1). Due to the partial susceptibility of B78H1 cells to HSV-2, we were unable to compare nectin-1 usage by B virus and HSV-2 accurately by using this approach. The CPE resulting from B virus infection was markedly different in nectin-1- versus nectin-2-bearing cells. Following B virus infection of B78-nectin-1 cell monolayers, syncytial plaques were approximately 2 to 3 times smaller than those produced in identically infected Vero cell monolayers (Fig. 1A and B). In the infected B78-nectin-2 cell monolayers, cells infected with B virus produced various morphological changes, ranging from single rounded cells or small grape-like clusters of rounded cells to small, nonsyncytial plaques that were smaller than and morphologically distinct from plaques on Vero and B78-nectin-1 cells (Fig. 1C). These results indicate that in addition to nectin-1, B virus can use nectin-2 as a cellular receptor for entry and cell-to-cell spread. The data also suggest that nectin-1 is a more efficient mediator of B virus lateral spread than nectin-2. Thus, both quantitative and qualitative differences exist in the use of entry receptors by B virus and HSV types 1 and 2.
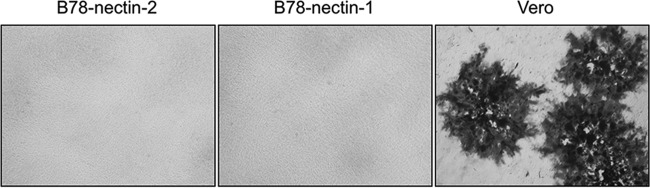
To determine if nectin-1- or nectin-2-mediated entry of B virus was dependent on gD, neutralization experiments using B virus strain E2490 and rabbit polyclonal serum against B virus gD and infectivity assays using the gD deletion mutant BV-ΔgDZ were performed on receptor-bearing cells. Neutralization experiments on Vero cells served as controls. The neutralization capacity of the gD antiserum was examined on B78-nectin-1 and Vero cells by using a plaque reduction assay and on B78-nectin-2 cells by using a CPE inhibition assay. The polyclonal gD antibody fully protected both nectin-1- and nectin-2-bearing cells from B virus infection (neutralization titers were 1:64 and 1:32, respectively), while preimmune serum offered no protection (Fig. 2A and B). In the control experiments, we observed a lack of B virus neutralization and efficient neutralization of HSV-1 infectivity on Vero cells by the gD antiserum, which is consistent with previously published results (52) (Fig. 2C). To assess the infectivity of the gD deletion mutant on nectin-2-bearing cells, BV-ΔgDZ was titrated on B78-nectin-2 cell monolayers, followed by X-Gal staining. We reported earlier that a gD-null B virus could not enter B78-nectin-1 cells but infected Vero cells as efficiently as the wild-type virus did (13). Therefore, for this study, we used Vero cells as a positive control and nectin-1-bearing cells as a negative control for BV-ΔgDZ infectivity. Similar to B78-nectin-1 cells, nectin-2-bearing cells were resistant to BV-ΔgDZ infection, whereas Vero cells were highly susceptible (Fig. 3). Together, these results provided evidence that gD was required by B virus for cell entry mediated by both nectin-1 and nectin-2.
FIG 2.
Blocking of virus infectivity by gD antibody. (A) B virus neutralization on B78-nectin-2 cells was performed using a CPE inhibition assay. The monolayers were fixed at 48 hpi and immunostained with rhesus B virus antibody-positive serum. Magnification, ×10. (B) B virus neutralization on B78-nectin-1 cells was assayed by a plaque reduction assay. The infected cells were fixed at 48 hpi and then stained with crystal violet to visualize viral plaques. The images represent culture wells of B78-nectin-1 cells infected with B virus-serum mixtures. (C) Neutralization curves for B virus and HSV-1 on Vero cells, representing negative and positive controls, respectively.
FIG 3.
Infection of cells with a gD-negative recombinant B virus (BV-ΔgDZ). Nectin-2-expressing, nectin-1-expressing (negative control), and Vero (positive control) cell monolayers were infected with BV-ΔgDZ. Cells were fixed at 48 hpi and then stained with X-Gal as described in Materials and Methods. Magnification, ×5.
Analysis of gD variability in clinical isolates.
Even a single amino acid substitution in HSV gD can influence the usage of cellular receptors (27, 65, 66). Thus, the inability of the B virus lab strain to use the HVEM receptor may have been due to the accumulation of lab strain-specific mutations in gD. To evaluate this possibility, we compared predicted amino acid sequences of gD extracellular domains between the prototype lab strain E2490 and 19 clinical strains isolated from macaques and/or human patients with confirmed B virus infections. The complete gD gene sequences of the strains were obtained as described previously (53) and then compared. The N-terminal domain of gD (amino acids [aa] 7 to 32), which is involved in the interaction of HSV-1 gD with HVEM, was conserved among all B virus isolates (55, 67). Naturally occurring substitutions (relative to the lab strain) at residues 122 and 143 were identified in the B virus gD IgV-core domain. In eight clinical strains, the D122N substitution generated a third potential N-glycosylation site, which was present in all reported gD-1 and gD-2 sequences (Fig. 4A). Natural gD polymorphism did not correlate with the species from which the virus was obtained, i.e., human or macaque. The frequencies of the three naturally occurring gD variants, designated gD-122D/143G, gD-122D/143S, and gD-122N/143S, and a list of B virus isolates are shown in Table 2. The fourth possible variant, gD-122N/143G, was not found among the clinical strains, while the frequencies of the other variants were comparable to each other.
FIG 4.
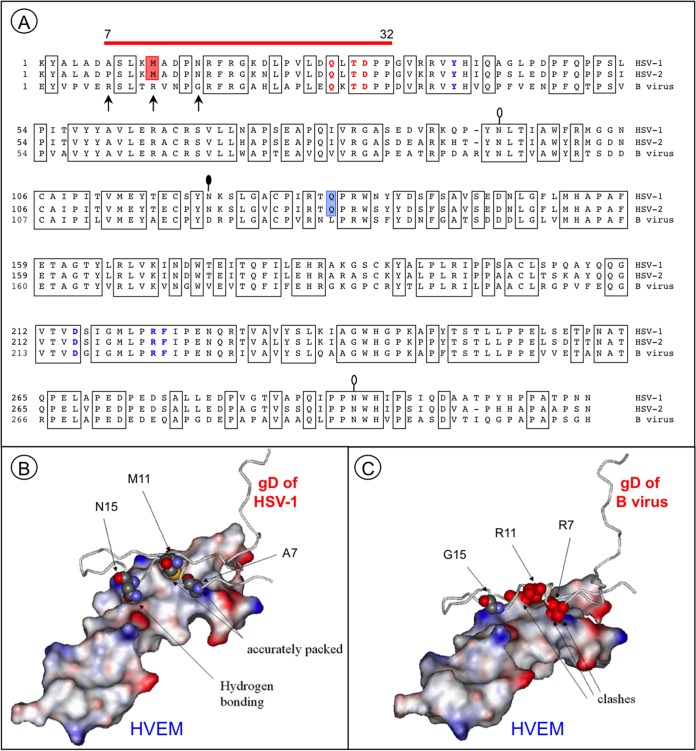
Essential differences in protein-protein interfaces involving HSV-1 and B virus gD proteins and HVEM. (A) Alignment of the gD ectodomain residues of HSV-1 (KOS), HSV-2 (HG52), and the B virus lab strain (E2490). Conserved amino acid residues are boxed. The red line above the gD sequences indicates HVEM contact residues. The residues essential for HVEM-mediated and nectin-1-mediated HSV-1 entry are shown in bold red and bold blue, respectively. Red shading indicates the M11 residue, which is involved in the interaction with the HVEM “hot spot” residue Y28. Blue shading indicates the only residue in the IgV-core, Q132, that directly interacts with the nectin-1 C″ strand. The arrows point to the residues that most probably distort the HVEM-B virus gD interface. Lollipops mark the predicted N-glycosylation sites. The black-filled lollipop marks an N-glycosylation site that is absent in the B virus lab strain. (B) Resolved structure of the HSV-1 gD and HVEM complex (PDB code 1JMA). Residues 1 to 50 of gD are shown. The residues A7, M11, and N15 (shown in space-filling format) are accurately packed at the molecular surface of the receptor. (C) Model of the HVEM and B virus gD complex. Severe steric clashes of R7 and R11 are predicted, with no interactions between G15 and the receptor.
TABLE 2.
Amino acid substitutions in the gD extracellular domain in B virus isolates from macaques and humans
| gD variant | Residue at position: |
B virus isolatesa | No. of isolates (% of total) | |
|---|---|---|---|---|
| 122 | 143 | |||
| gD-122D/143G | D | G | E2490, MR5, RP, A5, A6 | 5 (25) |
| gD-122D/143S | D | S | MR3, MR6, MR7, MJ1, MJ2, MJ3, A4 | 7 (35) |
| gD-122N/143S | Nb | S | MR1, MR2, MR4, MR8, A1, A2, A3, AR1 | 8 (40) |
| gD-122N/143G | N | G | Not found | 0 |
| Total | 20 | |||
Strains in bold indicate B virus strains isolated from infected humans.
Predicted N-glycosylation site.
Receptor usage by B virus clinical isolates.
To establish receptor preferences, six B virus clinical isolates (macaque and/or human origin) representing various natural gD variants, as well as the prototype lab strain, were evaluated for entry into the specific receptor-expressing cells. In these experiments, the infectivity of each B virus strain was evaluated using a TCID50 assay, which is more sensitive (although less precise) than plaque assay (68, 69). To compare strain infectivities by using different cell lines, each B virus strain was simultaneously titrated on parallel cultures of B78-nectin-1, B78-nectin-2, B78-HVEM, and B78c cells. The infected monolayers were fixed at 72 hpi and stained with pooled rhesus B virus antibody-positive serum samples, and viral titers (TCID50 per milliliter) on each cell line were calculated.
A few sporadic singly stained cells were observed in both infected B78c (control) and infected B78-HVEM cells. These singly infected cells were most likely a result of entry via murine nectin-1, as suggested previously for HSV-1 (70). Thus, similar to the lab strain, none of the clinical isolates were able to infect B78-HVEM or B78c cell monolayers. The TCID50 titers of the clinical isolates on B78-nectin-2 cells were ∼4 times lower, on average, than those on B78-nectin-1 cell monolayers (Table 3). These findings demonstrated that B virus primary isolates, regardless of the gD variant, can use both nectin-1 and nectin-2, but not HVEM, as cellular receptors for entry into human cells. Nectin-1 was more effective than nectin-2 in mediating this process. The inability of any of the tested strains to utilize HVEM for entry could not be attributed to the identified amino acid variability in B virus gD. Further, since none of the isolates utilized HVEM for entry, the inability of B virus to use the HVEM receptor was independent of the variability in B virus gD as originally hypothesized.
TABLE 3.
Use of HSV entry receptors by B virus clinical isolates as determined by TCID50 assay
| gD variant | Isolate | Infectivity titer (TCID50/ml)a on cells expressing: |
Nectin-1/nectin-2 titer ratio | ||
|---|---|---|---|---|---|
| HVEM | Nectin-1 | Nectin-2 | |||
| gD-122D/143G | E2490 | 0 | 7.1 × 105 | 4.0 × 105 | 1.8 |
| A5 | 0 | 4.0 × 107 | 7.1 × 106 | 5.6 | |
| gD-122D/143S | A4 | 0 | 7.1 × 105 | 2.2 × 105 | 3.2 |
| MR7 | 0 | NT | NT | ||
| gD-122N/143S | AR1 | 0 | 4.0 × 106 | 1.3 × 106 | 3.1 |
| A1 | 0 | 4.0 × 106 | 7.1 × 105 | 5.6 | |
| MR4 | 0 | NT | NT | ||
NT, not tested.
Next, we compared the levels of effectiveness of nectin-1 usage by the selected B virus strains. To enable an infectivity comparison, the virus titers (PFU per milliliter) of each clinical strain were measured by plaque assays on parallel cultures of B78-nectin-1 and Vero cells (Table 4). The relative infectivity titers of all three gD-122N/143S-carrying strains on B78-nectin-1 cells were 5 to 10 times lower than the titers of gD-122D/143G- and gD-122D/143S-carrying strains, suggesting that residue 122 plays a critical role in the interaction of B virus gD with nectin-1.
TABLE 4.
Use of nectin-1 for entry by B virus clinical isolates as determined by plaque assay
| gD variant | Isolate | Infectivity titer (PFU/ml) on cells |
Vero/B78-nectin-1 titer ratio | |
|---|---|---|---|---|
| Vero | B78-nectin-1 | |||
| gD-122D/143G | E2490 | 1.4 × 107 | 6.4 × 105 | 22 |
| A5 | 3.2 × 107 | 6.5 × 105 | 50 | |
| gD-122D/143S | A4 | 1.0 × 107 | 2.2 × 105 | 45 |
| MR7 | 9.0 × 107 | 3.0 × 106 | 30 | |
| gD-122N/143S | AR1 | 7.0 × 107 | 3.5 × 105 | 200 |
| A1 | 3.0 × 107 | 1.4 × 105 | 214 | |
| MR4 | 3.4 × 107 | 9.0 × 104 | 378 | |
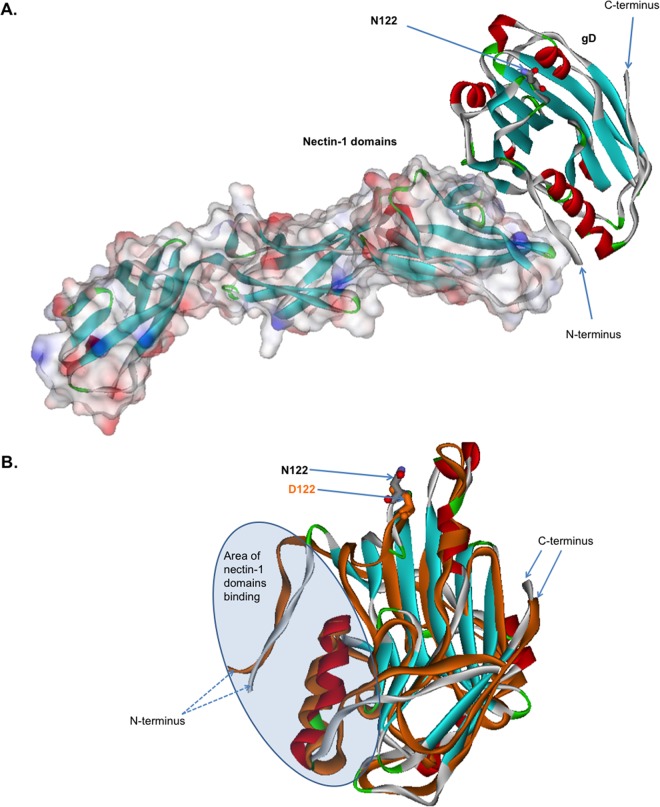
Structure modeling of the complex between B virus gD and nectin-1.
For comparative modeling, we used the X-ray crystallographic structure of the HSV-1 gD ectodomain (gD285) bound to human nectin-1 (PDB code 3U82) (56). B virus gD isolates contain either asparagine or aspartic acid at position 122, whereas HSV-1 gD isolates contain an asparagine residue at the corresponding position. To investigate the molecular details of the role of B virus gD residue 122 in the gD–nectin-1 complex, we prepared models of the structure of this complex by using software that was extensively tested during CASP conferences, as described in detail by Torshin (57) (Fig. 5A). The analysis showed that the N122 residue is located more than 15 Å from the nectin-1–gD protein-protein interface and thus is not involved in the direct interaction between gD and nectin-1. Next, we created a computer model of nectin-1-liganded gD-122D and performed a structural alignment of this gD variant and gD-122N by using the software package EVM as described previously (57) (Fig. 5B). Energy calculations were performed using the software package ECMMS, and the results showed that the N122D substitution increased the stability of the gD globular structure by 0.23 kcal/mol (58). The N122D replacement also resulted in a conformational change of the entire globule of the gD protein (root mean square deviance of 3.41 Å compared to the gD-122N structure). As a result, the energy of gD binding to nectin-1 was reduced by 0.16 kcal/mol. We suggest that the N122D substitution-induced conformational changes in the globule of the gD protein can lead to destabilization of the gD–nectin-1 interface and thus affect B virus entry.
FIG 5.
Computational modeling and comparison of the structures of B virus gD-122N and gD-122D variants liganded with nectin-1. (A) Computational modeling of B virus N122-carrying gD and nectin-1. The solved crystal structure of HSV-1 gD bound to human nectin-1 (PDB code 3U82) was used as a template. B virus gD and nectin-1 are represented as a stereo ribbon and van der Waals surfaces, respectively. The N122 residue is located more than 15 Å from the protein-protein nectin-1–gD interface and thus is not involved in the direct interaction between gD and nectin-1. (B) Overlay of the B virus gD-122N and gD-122D structures. The arrows mark N122 (gray) and D122 (orange). Note the considerable conformational changes of the globule in the area of the nectin-1 binding domain. These changes lead to a reduction in the energy of binding of the gD-122D variant to nectin-1, by ∼0.16 kcal/mol, compared to that of the gD-122N variant.
Full atomic model of B virus gD-HVEM interfaces.
The gD contact residues for HVEM were identified in the N terminus of the HSV-1 gD molecule by X-ray crystallography (55, 71). The alignment of the gD amino acid sequences of HSV-1, HSV-2, and B virus (Fig. 4A) showed that these contact residues are also present in B virus gD, but 9 of 18 contact residues differ. Nevertheless, the three residues in this region of HSV-1 gD (Q27, T29, and D30) essential for both HVEM binding and cell entry are conserved in all B virus isolates (66, 72). These data suggest that the inability of B virus to use HVEM is likely attributable to other residue substitutions that influence accessibility of the critical regions required for gD interactions with human HVEM.
To study possible theoretical structural bases for the inability of B virus gD to utilize HVEM, we performed comparative homology modeling of the B virus gD-HVEM interface by using the structure of the HSV-1 gD–HVEM complex (Fig. 4B and C). In the template structure (HSV-1 gD–HVEM; PDB code 1JMA), the protein-protein interface is tightly packed, with no steric violations and with many hydrogen bonds stabilizing protein-protein interactions (Fig. 4B and Table 5). In the model (B virus gD-HVEM), however, the protein packing is characterized by a large number of atom-atom clashes, primarily due to the introduction of bulky arginine side chains at gD positions 7 and 11 (Fig. 4C and Table 5), which result in considerable steric hindrance due to the gross shape mismatch. The presence of these severe atom-atom clashes in the protein-protein interface suggests that the hypothetical complex cannot possibly be formed. In addition, arginine side chains cannot be accommodated at the molecular surface of the receptor without significant distortions of the binding geometry. Further, a number of hydrogen bonds stabilizing the complex are absent in the structure of the hypothetical complex. For example, the two hydrogen bonds formed by asparagine at position 15 in HSV-1 gD are lost in B virus gD due to the N15G substitution. The presence of atom-atom clashes, disruption of the tight packing, and loss of almost half of the hydrogen bonds stabilizing the protein-protein complex suggest that B virus gD and the human receptor HVEM likely do not form a stable protein-protein complex.
TABLE 5.
Residues of the protein-protein interfaces in gD-receptor complexes
| Amino acid position | HSV-1 gD (solved structure)a |
B virus gD (model)b,e |
Remarks | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | No. of H bondsc | No. of hydrophobic contacts | No. of clashesd | Residue | No. of H bondsc | No. of hydrophobic contacts | No. of clashesd | ||
| 7 | A | 1 | 2 | 0 | R | NA | NA | 5 | Too many clashes in the model suggest that the residues will disrupt the complex |
| 11 | M | 0 | 10 | 0 | R | NA | NA | 5 | Too many clashes in the model suggest that the residues will disrupt the complex |
| 12 | A | 1 | 0 | 0 | V | 1 | 0 | 0 | |
| 14 | P | 1 | 11 | 0 | P | 0 | 9 | 0 | A hydrogen bond is lost |
| 15 | N | 2 | 1 | 0 | G | 0 | 0 | 0 | Hydrogen bonding interactions are lost |
| 24 | V | 0 | 3 | 0 | P | 0 | 0 | 0 | Protein-protein packing is disrupted |
| 26 | D | 2 | 1 | 0 | E | 1 | 3 | 1 | Hydrogen bonding interactions are lost |
| 27 | Q | 2 | 1 | 0 | Q | 1 | 0 | 1 | A hydrogen bond is lost |
| 28 | L | 0 | 4 | 0 | K | 0 | 4 | 1 | |
| 29 | W | 2 | 2 | 0 | Y | 3 | 1 | 0 | |
| 31 | P | 0 | 2 | 0 | P | 0 | 2 | 0 | |
Downloaded from the Protein Data Bank (PDB entry 1JMA).
Based on the complex of HSV-1 gD and HVEM.
Residues of the interfaces were identified using lists of intersubunit hydrogen bonds and van der Waals contacts as described earlier (59, 60).
An atom-atom “clash” was registered if the distance between two nonbonded atoms was less than 2 Å.
NA, not applicable (too many clashes in this region).
DISCUSSION
In the present study, we set forth to identify which of the known human HSV receptors are utilized by low-passage-number B virus clinical isolates obtained from infected macaques and humans in order to better understand the interactions between B virus and human cells during viral entry. The experimental data presented in this paper revealed that B virus utilizes nectin-1 and, less effectively, nectin-2 to gain entry into human cells. We also showed that, similar to the situation with HSV, nectin-mediated pathways of B virus cell entry are gD dependent, based on our experiments showing that polyclonal gD antibodies blocked virus entry and that a gD-negative recombinant B virus was unable to enter human nectin-1- or nectin-2-expressing murine cells.
For the first time, to our knowledge, we show the inability of clinical isolates from macaques and humans to infect HVEM-bearing cells. In marked contrast, all HSV primary isolates (n = 60) evaluated used both HVEM and nectin-1 for entry, with similar efficiencies, whereas only a few strains used nectin-2, as demonstrated by Krummenacher and colleagues (47). There are, however, several HSV-1 laboratory strains (e.g., rid1 and rid2) with a unique receptor tropism not previously reported for clinical isolates. Examination of these strains by multiple independent groups revealed gD mutations (L25P, Q27R, or Q27P) that are responsible for the ability of these mutants to utilize nectin-2 instead of HVEM for cell entry (45, 72–74). Although B virus resembles these HSV-1 mutants with respect to receptor usage, conservation of L25 and Q27 in all 20 sequenced strains of B virus indicates that the similarity in receptor tropism for B virus gD and these HSV mutants was not due to mutations at these positions. Residues T29 and D30, which were revealed by alanine mutagenesis of the gD-1 contact region to be essential for use of HVEM and nectin-2 for entry (66, 72), were also conserved in B virus gD. Additionally, B virus gD lacks the amino acid substitutions (M11P, P14R, G43P, L28G, and L28P) that correlated with loss of HSV-1 entry via HVEM following random mutagenesis of HSV-1 gD (75). Thus, the structural modifications in B virus gD that changed the B virus receptor usage appear to be different from those observed in HSV-1 gD.
We predicted that the following three key residues in the N terminus of B virus gD are responsible for the inability of B virus to enter cells via HVEM: R7, R11, and G15. These predictions were based on the proposed full atomic model of the B virus gD-HVEM interface, prepared using the crystallographic structure of HSV-1 gD–HVEM (55) as a template (Fig. 4C). For proteins with conserved sequence and structural similarities, such homology modeling of three-dimensional structures can be generated with a precision sufficient to make biologically relevant conclusions, as previously demonstrated independently by multiple investigators (76, 77). According to our model, two bulky arginine residues in B virus gD, corresponding to A7 and M11 in HSV-1 gD, notably alter the geometry of the gD contact surface. The fact that the protruding side chains of arginine residues interfere with the putative fit of gD onto the HVEM binding interface, coupled with the loss of stabilizing hydrogen bonds (Table 5), suggested that a stable interaction between B virus gD and HVEM is unlikely. Our hypothesis does not contradict the results of HSV-1 gD alanine scanning mutagenesis studies. In these studies, the substitutions M11A and N15A did not abrogate HSV-1 entry but did reduce gD binding to HVEM to undetectable levels (65, 66, 78), while the M11P substitution resulted in a complete loss of the ability of the HSV-1 mutant to enter cells via HVEM (75).
Our computational modeling data are in agreement with the in silico model of the B virus gD-HVEM complex described previously by Li and colleagues (79). Their explanation for the inability of gD to adapt HVEM as a receptor was based on the low shape complementarity value and small interface area for the B virus gD-HVEM complex compared to those for the HSV-1 gD–HVEM complex. We extended their observations, and by using a different computer modeling approach, we were able to identify specific amino acids (R7, R11, and G15) producing steric clashes between HVEM and B virus gD that prevent complex formation (Fig. 4).
In addition to influencing cell tropism, the inability of B virus to interact with HVEM may also abrogate B virus interactions with some immune cells of the human host. HVEM performs multiple immunomodulatory functions and is expressed on both T and B lymphocytes (39, 80–83). HSV-1 gD interaction with HVEM on immune cells results in production of proinflammatory chemokines, and this has been suggested to play an essential role in the recruitment of adaptive immune defenses (84–87). Whether the inability to interact with HVEM contributes to the remarkable differences in B virus pathogenesis in humans versus macaques requires further study. Remarkably, B virus infections in macaques, similar to HSV infections in humans, are predominantly subclinical. Therefore, it is reasonable to suggest that B virus either utilizes an unknown receptor that results in a similar HVEM-mediated signaling pathway or has evolved unique mechanisms to modulate the natural host defenses. Pollara and colleagues suggest that delays in adaptive immune responses permit the virus to enter nerve endings to evade immune defenses (88).
The ability to effectively use nectin-2 is another feature that distinguishes B virus from HSV-1 clinical strains that either were unable to utilize nectin-2 for entry or used this receptor inefficiently (47). The reasons for the enhanced ability of B virus gD to use nectin-2 are unknown, in part due to a limited understanding of the actual HSV gD and nectin-2 structural interactions. Nectin-2 is widely expressed in various cells and tissues, including the brain and synaptic junctions (34, 42, 43, 47). Interestingly, several HSV-1 clinical isolates from the CNS of patients with encephalitis can efficiently use nectin-2 for entry (47). In view of the fact that untreated B virus human infections often result in encephalitis, the ability of B virus to efficiently use nectin-2 as an entry receptor may play a significant role in the neuroinvasiveness of B virus in humans.
Nectin-2 is also a ligand of the activating natural killer (NK) cell receptor DNAX accessory molecule 1 (DNAM-1) (43, 89–92). Work in PRV and HSV-2 showed that following cellular transfection with gD or viral infection, DNAM-1 was downregulated to prevent NK cell-mediated cytolysis of infected cells (93). Because B virus can interact with nectin-2, B virus may have a similar mechanism to evade NK cell-mediated cytolysis. The immune aspect in the context of interactions between B virus gD and cognate cellular receptors remains to be investigated.
Our study also shows, for the first time, that B virus clinical isolates use nectin-1 for entry as effectively as HSV-1 and -2 do, which is not surprising given that nectin-1 is currently considered a principal entry receptor for most alphaherpesviruses (94, 95). The essential nectin-1 binding residues are conserved in the B virus lab strain and each of the 19 human and macaque clinical strains we studied. Remarkably, all isolates that showed reduced infectivity on nectin-1-expressing cells (Table 4) contained D122N and G143S substitutions in gD relative to the gD sequence of the lab strain. Because the plating efficiencies of the gD-122D/143G- and gD-122D/143S-carrying isolates were comparable on nectin-1-expressing cells, we concluded that the G143S substitution does not affect B virus entry, and therefore the D122N substitution is likely to be solely responsible for the reduced infectivity of the gD-123N/143S-carrying strains.
Notably, the D122N substitution created a third N-glycosylation site in B virus gD, which was absent in the lab strain and 11 of the 19 clinical isolates examined (Table 2). The addition of an N-linked carbohydrate to the N122 residue may lead to a putative conformational change in B virus gD, potentially affecting the interaction of nectin-1 β-strand C″ with the gD Ig fold that was described by Di Giovine and colleagues (96). For HSV-1, however, the glycosylation status of the residue at position 121 (corresponding to position 122 in B virus gD) did not affect virus infectivity (97, 98) or gD binding to either HVEM (73) or nectin-1 (46).
A structural model of the B virus gD–nectin-1 complex was proposed earlier by Li and colleagues (79). Herein, using a comparative homology modeling approach, we illustrated putative conformational and structural differences at the atomic level between B virus gD-122N and gD-122D upon nectin-1 binding. The N122D replacement may result in a destabilization of the gD–nectin-1 protein-protein interface due to conformational changes in the globule of the gD protein. Destabilization of the nectin-1–gD-122D protein-protein interface may change the flexibility of gD, thus affecting specific structural rearrangements needed for the downstream interaction of the gD C terminus with glycoproteins gB and gH/gL that is essential for cell entry (31, 67, 99, 100). Making mutant viruses that have changes in the predicted amino acids will indicate whether the predictions are accurate.
In summary, our study shows, for the first time, that low-passage-number B virus isolates effectively utilized human nectin-1 and nectin-2 for entry and cell-to-cell spread but that none were able to use HVEM, regardless of the host (macaque or human) from which each was isolated. Based on our homology model of the B virus gD-HVEM interface, we predict that the combined effect of three residues—R7, R11, and G15—in the N terminus of B virus gD is largely responsible for the observation that B virus does not interact with HVEM. Additionally, based on energy calculations and conformational changes in gD associated with the N122D substitution, we concluded that the natural occurrence of the mutation of amino acid 122 alters the interaction of gD with nectin-1, affecting the infectivity of B virus on nectin-1-expressing cells. The results presented here suggest that B virus depends on a distinct combination of human cellular receptors for entry into cells. These data may advance our understanding of the molecular mechanisms of pathogenicity of zoonotic B virus infections.
ACKNOWLEDGMENTS
We thank Gary H. Cohen and Roselyn J. Eisenberg for providing B78H1 mouse cells expressing HSV entry receptors.
This work was supported by Public Health Service grants R01 RR03163 and P40 RR05162 from the NIH's National Center for Research Resources and by the generous and continued support of the Georgia Research Alliance.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Boulter EA. 1975. The isolation of monkey B virus (Herpesvirus simiae) from the trigeminal ganglia of a healthy seropositive rhesus monkey. J Biol Stand 3:279–280. doi: 10.1016/0092-1157(75)90031-1. [DOI] [PubMed] [Google Scholar]
- 2.Vizoso AD. 1975. Recovery of herpes simiae (B virus) from both primary and latent infections in rhesus monkeys. Br J Exp Pathol 56:485–488. [PMC free article] [PubMed] [Google Scholar]
- 3.Kirschstein RL, Van Hoosier GLJ. 1961. Virus B infection of the central nervous system of monkeys used for the poliomyelitis vaccine safety test. Am J Pathol 38:119–124. [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson CS, O'Sullivan MG, Jayo MJ, Anderson DK, Harber ES, Jerome WG, Bullock BC, Heberling RL. 1997. Fatal disseminated cercopithecine herpesvirus 1 (herpes B) infection in cynomolgus monkeys (Macaca fascicularis). Vet Pathol 34:405–414. doi: 10.1177/030098589703400504. [DOI] [PubMed] [Google Scholar]
- 5.Sabin AB, Wright WM. 1934. Acute ascending myelitis following a monkey bite, with isolation of a virus capable of reproducing the disease. J Exp Med 59:115–136. doi: 10.1084/jem.59.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gay FP, Holden M. 1933. Isolation of herpes virus from several cases of epidemic encephalitis. Proc Soc Exp Biol Med 30:1051–1053. doi: 10.3181/00379727-30-6788. [DOI] [Google Scholar]
- 7.Davidson WL, Hummeler K. 1960. B virus infection in man. Ann N Y Acad Sci 85:970–979. [DOI] [PubMed] [Google Scholar]
- 8.Hull RN. 1973. The simian herpesviruses, p 389–425. In Kaplan AS. (ed), The herpesviruses. Academic Press, New York, NY. [Google Scholar]
- 9.Palmer AE. 1987. B virus, Herpesvirus simiae: historical perspective. J Med Primatol 16:99–130. [PubMed] [Google Scholar]
- 10.Whitley R, Hilliard J. 2007. Cercopithecine herpesvirus (B virus), p 2888–2903. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 11.Holmes GP, Hilliard JK, Klontz KC, Rupert AH, Schindler CM, Parrish E, Griffin DG, Ward GS, Bernstein ND, Bean TW, Ball MR Sr, Brady JA, Wilder MH, Kaplan JE. 1990. B virus (Herpesvirus simiae) infection in humans: epidemiologic investigation of a cluster. Ann Intern Med 112:833–839. doi: 10.7326/0003-4819-112-11-833. [DOI] [PubMed] [Google Scholar]
- 12.Vasireddi M, Hilliard J. 2012. Herpes B virus, Macacine herpesvirus 1, breaks simplex virus tradition via major histocompatibility complex class I expression in cells from human and macaque hosts. J Virol 86:12503–12511. doi: 10.1128/JVI.01350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perelygina L, Patrusheva I, Vasireddi M, Brock N, Hilliard J. 2015. B virus (Macacine herpesvirus 1) glycoprotein D is functional but dispensable for virus entry into macaque and human skin cells. J Virol 89:5515–5524. doi: 10.1128/JVI.03568-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan Q, Amen M, Harden M, Severini A, Griffiths A, Longnecker R. 2012. Herpes B virus utilizes human nectin-1 but not HVEM or PILRalpha for cell-cell fusion and virus entry. J Virol 86:4468–4476. doi: 10.1128/JVI.00041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black D, Ritchey J, Payton M, Eberle R. 2014. Role of the virion host shutoff protein in neurovirulence of monkey B virus (Macacine herpesvirus 1). Virol Sin 29:274–283. doi: 10.1007/s12250-014-3495-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall LV, Zwartouw HT, Kelly DC. 1989. Discrimination between twenty isolates of herpesvirus simiae (B virus) by restriction enzyme analysis of the viral genome. Virus Res 12:283–296. doi: 10.1016/0168-1702(89)90044-0. [DOI] [PubMed] [Google Scholar]
- 17.Ohsawa K, Black D, Ohsawa M, Eberle R. 2014. Genome sequence of a pathogenic isolate of monkey B virus (species Macacine herpesvirus 1). Arch Virol 159:2819–2821. doi: 10.1007/s00705-014-2130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lees DN, Baskerville A, Cropper LM, Brown DW. 1991. Herpesvirus simiae (B virus) antibody response and virus shedding in experimental primary infection of cynomolgus monkeys. Lab Anim Sci 41:360–364. [PubMed] [Google Scholar]
- 19.Amen MA, Griffiths A. 2011. Identification and expression analysis of herpes B virus-encoded small RNAs. J Virol 85:7296–7311. doi: 10.1128/JVI.00505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilliard JK, Eberle R, Lipper SL, Munoz RM, Weiss SA. 1987. Herpesvirus simiae (B virus): replication of the virus and identification of viral polypeptides in infected cells. Arch Virol 93:185–198. doi: 10.1007/BF01310973. [DOI] [PubMed] [Google Scholar]
- 21.Eberle R, Black D, Hilliard JK. 1989. Relatedness of glycoproteins expressed on the surface of simian herpes-virus virions and infected cells to specific HSV glycoproteins. Arch Virol 109:233–252. doi: 10.1007/BF01311084. [DOI] [PubMed] [Google Scholar]
- 22.Hilliard JK, Black D, Eberle R. 1989. Simian alphaherpesviruses and their relation to the human herpes simplex viruses. Arch Virol 109:83–102. doi: 10.1007/BF01310520. [DOI] [PubMed] [Google Scholar]
- 23.Perelygina L, Zhu L, Zurkuhlen H, Mills R, Borodovsky M, Hilliard JK. 2003. Complete sequence and comparative analysis of the genome of herpes B virus (cercopithecine herpesvirus 1) from a rhesus monkey. J Virol 77:6167–6177. doi: 10.1128/JVI.77.11.6167-6177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol 10:305–319. [DOI] [PubMed] [Google Scholar]
- 25.Geraghty RJ, Jogger CR, Spear PG. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147–158. doi: 10.1006/viro.1999.0157. [DOI] [PubMed] [Google Scholar]
- 26.Spear PG, Longnecker R. 2003. Herpesvirus entry: an update. J Virol 77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spear PG, Manoj S, Yoon M, Jogger CR, Zago A, Myscofski D. 2006. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 344:17–24. doi: 10.1016/j.virol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 28.Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A 104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol 84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. 2005. Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochem J 385:451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. 1999. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with Afadin, a PDZ domain-containing protein. J Cell Biol 145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizoguchi A, Nakanishi H, Kimura K, Matsubara K, Ozaki-Kuroda K, Katata T, Honda T, Kiyohara Y, Heo K, Higashi M, Tsutsumi T, Sonoda S, Ide C, Takai Y. 2002. Nectin: an adhesion molecule involved in formation of synapses. J Cell Biol 156:555–565. doi: 10.1083/jcb.200103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takai Y, Nakanishi H. 2003. Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci 116:17–27. doi: 10.1242/jcs.00167. [DOI] [PubMed] [Google Scholar]
- 36.Shukla D, Scanlan PM, Tiwari V, Sheth V, Clement C, Guzman-Hartman G, Dermody TS, Valyi-Nagy T. 2006. Expression of nectin-1 in normal and herpes simplex virus type 1-infected murine brain. Appl Immunohistochem Mol Morphol 14:341–347. doi: 10.1097/00129039-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 37.Guzman G, Oh S, Shukla D, Engelhard HH, Valyi-Nagy T. 2006. Expression of entry receptor nectin-1 of herpes simplex virus 1 and/or herpes simplex virus 2 in normal and neoplastic human nervous system tissues. Acta Virol 50:59–66. [PubMed] [Google Scholar]
- 38.Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436. doi: 10.1016/S0092-8674(00)81363-X. [DOI] [PubMed] [Google Scholar]
- 39.Kwon BS, Tan KB, Ni J, Oh KO, Lee ZH, Kim KK, Kim YJ, Wang S, Gentz R, Yu GL, Harrop J, Lyn SD, Silverman C, Porter TG, Truneh A, Young PR. 1997. A newly identified member of the tumor necrosis factor receptor superfamily with a wide tissue distribution and involvement in lymphocyte activation. J Biol Chem 272:14272–14276. doi: 10.1074/jbc.272.22.14272. [DOI] [PubMed] [Google Scholar]
- 40.Duhen T, Pasero C, Mallet F, Barbarat B, Olive D, Costello RT. 2004. LIGHT costimulates CD40 triggering and induces immunoglobulin secretion; a novel key partner in T cell-dependent B cell terminal differentiation. Eur J Immunol 34:3534–3541. doi: 10.1002/eji.200425598. [DOI] [PubMed] [Google Scholar]
- 41.De Trez C, Schneider K, Potter K, Droin N, Fulton J, Norris PS, Ha SW, Fu YX, Murphy T, Murphy KM, Pfeffer K, Benedict CA, Ware CF. 2008. The inhibitory HVEM-BTLA pathway counter regulates lymphotoxin receptor signaling to achieve homeostasis of dendritic cells. J Immunol 180:238–248. doi: 10.4049/jimmunol.180.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. 1998. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood 92:4602–4611. [PubMed] [Google Scholar]
- 43.Pende D, Castriconi R, Romagnani P, Spaggiari GM, Marcenaro S, Dondero A, Lazzeri E, Lasagni L, Martini S, Rivera P, Capobianco A, Moretta L, Moretta A, Bottino C. 2006. Expression of the DNAM-1 ligands, Nectin-2 (CD112) and poliovirus receptor (CD155), on dendritic cells: relevance for natural killer-dendritic cell interaction. Blood 107:2030–2036. doi: 10.1182/blood-2005-07-2696. [DOI] [PubMed] [Google Scholar]
- 44.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 45.Warner MS, Geraghty RJ, Martinez WM, Montgomery RI, Whitbeck JC, Xu R, Eisenberg RJ, Cohen GH, Spear PG. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 46.Krummenacher C, Nicola AV, Whitbeck JC, Lou H, Hou W, Lambris JD, Geraghty RJ, Spear PG, Cohen GH, Eisenberg RJ. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol 72:7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krummenacher C, Baribaud F, Ponce De Leon M, Baribaud I, Whitbeck JC, Xu R, Cohen GH, Eisenberg RJ. 2004. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322:286–299. doi: 10.1016/j.virol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Miller CG, Krummenacher C, Eisenberg RJ, Cohen GH, Fraser NW. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol Ther 3:160–168. doi: 10.1006/mthe.2000.0240. [DOI] [PubMed] [Google Scholar]
- 49.Chosewood LC, Wilson DE (ed). 2009. Biosafety in microbiological and biomedical laboratories, 5th ed US Department of Health and Human Services, Public Health Service, Centers for Disease Control and Prevention, National Institutes of Health, Washington, DC. [Google Scholar]
- 50.Richmond JY, McKinney RW (ed). 1999. Biosafety in microbiological and biomedical laboratories, 4th ed US Government Printing Office, Washington, DC. [Google Scholar]
- 51.Hierholzer JC, Killington RA. 1996. Virus isolation and quantitation, p 25–46. In Mahy BWJ, Kangro HO (ed), Virology methods manual. Academic Press, London, United Kingdom. [Google Scholar]
- 52.Perelygina L, Patrusheva I, Zurkuhlen H, Hilliard JK. 2002. Characterization of B virus glycoprotein antibodies induced by DNA immunization. Arch Virol 147:2057–2073. doi: 10.1007/s00705-002-0889-0. [DOI] [PubMed] [Google Scholar]
- 53.Perelygina L, Zurkuhlen H, Patrusheva I, Hilliard JK. 2002. Identification of a herpes B virus-specific glycoprotein D immunodominant epitope recognized by natural and foreign hosts. J Infect Dis 186:453–461. doi: 10.1086/341834. [DOI] [PubMed] [Google Scholar]
- 54.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]
- 55.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell 8:169–179. doi: 10.1016/S1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 56.Zhang N, Yan J, Lu G, Guo Z, Fan Z, Wang J, Shi Y, Qi J, Gao GF. 2011. Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat Commun 2:577. doi: 10.1038/ncomms1571. [DOI] [PubMed] [Google Scholar]
- 57.Torshin IY. 2006. Bioinformatics in the post-genomic era: the role of biophysics. Nova Biomedical Books, New York, NY. [Google Scholar]
- 58.Torshin IY. 2004. Computed energetics of nucleotides in spatial ribozyme structures: an accurate identification of functional regions from structure. Sci World J 4:228–247. doi: 10.1100/tsw.2004.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torshin I. 1999. Activating oligomerization as intermediate level of signal transduction: analysis of protein-protein contacts and active sites in several glycolytic enzymes. Front Biosci 4:D557–D570. [DOI] [PubMed] [Google Scholar]
- 60.Torshin IY. 2002. Functional maps of the junctions between interglobular contacts and active sites in glycolytic enzymes—a comparative analysis of the biochemical and structural data. Med Sci Monit 8:BR123–BR135. [PubMed] [Google Scholar]
- 61.Torshin I. 2000. Direct and reversed amino acid sequence pattern analysis: structural reasons for activity of reversed sequence sites and results of kinase site mutagenesis. Biochem J 345:733–740. [PMC free article] [PubMed] [Google Scholar]
- 62.Caves LS, Evanseck JD, Karplus M. 1998. Locally accessible conformations of proteins: multiple molecular dynamics simulations of crambin. Protein Sci 7:649–666. doi: 10.1002/pro.5560070314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Torshin IY, Weber IT, Harrison RW. 2000. Randomized and multiple model approaches to homology modeling and ab initio modeling, abstr 058. Abstr 4th CASP Meet, Asilomar, CA: http://predictioncenter.org/casp4/abstracts/casp4-abstracts-cm.html#19. [Google Scholar]
- 64.Sawitzky D, Hampl H, Habermehl KO. 1990. Entry of pseudorabies virus into CHO cells is blocked at the level of penetration. Arch Virol 115:309–316. doi: 10.1007/BF01310539. [DOI] [PubMed] [Google Scholar]
- 65.Stump JD, Sticht H. 2014. Mutations in herpes simplex virus gD protein affect receptor binding by different molecular mechanisms. J Mol Model 20:2192. doi: 10.1007/s00894-014-2192-x. [DOI] [PubMed] [Google Scholar]
- 66.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Cohen GH, Eisenberg RJ. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J Virol 77:8127–8140. doi: 10.1128/JVI.77.14.8127-8140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campadelli-Fiume G, Menotti L, Avitabile E, Gianni T. 2012. Viral and cellular contributions to herpes simplex virus entry into the cell. Curr Opin Virol 2:28–36. doi: 10.1016/j.coviro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Smither SJ, Lear-Rooney C, Biggins J, Pettitt J, Lever MS, Olinger GG Jr. 2013. Comparison of the plaque assay and 50% tissue culture infectious dose assay as methods for measuring filovirus infectivity. J Virol Methods 193:565–571. doi: 10.1016/j.jviromet.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 69.Grigorov B, Rabilloud J, Lawrence P, Gerlier D. 2011. Rapid titration of measles and other viruses: optimization with determination of replication cycle length. PLoS One 6:e24135. doi: 10.1371/journal.pone.0024135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menotti L, Lopez M, Avitabile E, Stefan A, Cocchi F, Adelaide J, Lecocq E, Dubreuil P, Campadelli-Fiume G. 2000. The murine homolog of human Nectin1delta serves as a species nonspecific mediator for entry of human and animal alpha herpesviruses in a pathway independent of a detectable binding to gD. Proc Natl Acad Sci U S A 97:4867–4872. doi: 10.1073/pnas.97.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J 24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoon M, Zago A, Shukla D, Spear PG. 2003. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J Virol 77:9221–9231. doi: 10.1128/JVI.77.17.9221-9231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitbeck JC, Peng C, Lou H, Xu R, Willis SH, Ponce de Leon M, Peng T, Nicola AV, Montgomery RI, Warner MS, Soulika AM, Spruce LA, Moore WT, Lambris JD, Spear PG, Cohen GH, Eisenberg RJ. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol 71:6083–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Campadelli-Fiume G. 2000. Nectin2alpha (PRR2alpha or HveB) and nectin2delta are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J Virol 74:1267–1274. doi: 10.1128/JVI.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon M, Spear PG. 2004. Random mutagenesis of the gene encoding a viral ligand for multiple cell entry receptors to obtain viral mutants altered for receptor usage. Proc Natl Acad Sci U S A 101:17252–17257. doi: 10.1073/pnas.0407892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. 2000. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 77.Chou KC. 2004. Structural bioinformatics and its impact to biomedical science. Curr Med Chem 11:2105–2134. doi: 10.2174/0929867043364667. [DOI] [PubMed] [Google Scholar]
- 78.Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Eisenberg RJ, Cohen GH. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J Virol 76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L, Qiu Z, Li Y, Liang F, Ye H, Cai Y, Guo W, Li Y, Yue J. 2014. Herpes B virus gD interaction with its human receptor—an in silico analysis approach. Theor Biol Med Model 11:27. doi: 10.1186/1742-4682-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morel Y, Schiano de Colella JM, Harrop J, Deen KC, Holmes SD, Wattam TA, Khandekar SS, Truneh A, Sweet RW, Gastaut JA, Olive D, Costello RT. 2000. Reciprocal expression of the TNF family receptor herpes virus entry mediator and its ligand LIGHT on activated T cells: LIGHT down-regulates its own receptor. J Immunol 165:4397–4404. doi: 10.4049/jimmunol.165.8.4397. [DOI] [PubMed] [Google Scholar]
- 81.Tan KB, Harrop J, Reddy M, Young P, Terrett J, Emery J, Moore G, Truneh A. 1997. Characterization of a novel TNF-like ligand and recently described TNF ligand and TNF receptor superfamily genes and their constitutive and inducible expression in hematopoietic and non-hematopoietic cells. Gene 204:35–46. doi: 10.1016/S0378-1119(97)00509-X. [DOI] [PubMed] [Google Scholar]
- 82.Sedy J, Bekiaris V, Ware CF. 2015. Tumor necrosis factor superfamily in innate immunity and inflammation. Cold Spring Harb Perspect Biol 7:a016279. doi: 10.1101/cshperspect.a016279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ware CF, Sedy JR. 2011. TNF superfamily networks: bidirectional and interference pathways of the herpesvirus entry mediator (TNFSF14). Curr Opin Immunol 23:627–631. doi: 10.1016/j.coi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lasaro MO, Tatsis N, Hensley SE, Whitbeck JC, Lin SW, Rux JJ, Wherry EJ, Cohen GH, Eisenberg RJ, Ertl HC. 2008. Targeting of antigen to the herpesvirus entry mediator augments primary adaptive immune responses. Nat Med 14:205–212. doi: 10.1038/nm1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoon M, Kopp SJ, Taylor JM, Storti CS, Spear PG, Muller WJ. 2011. Functional interaction between herpes simplex virus type 2 gD and HVEM transiently dampens local chemokine production after murine mucosal infection. PLoS One 6:e16122. doi: 10.1371/journal.pone.0016122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stiles KM, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C. 2010. Herpes simplex virus glycoprotein D interferes with binding of herpesvirus entry mediator to its ligands through downregulation and direct competition. J Virol 84:11646–11660. doi: 10.1128/JVI.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Soroosh P, Doherty TA, So T, Mehta AK, Khorram N, Norris PS, Scheu S, Pfeffer K, Ware C, Croft M. 2011. Herpesvirus entry mediator (TNFRSF14) regulates the persistence of T helper memory cell populations. J Exp Med 208:797–809. doi: 10.1084/jem.20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pollara G, Speidel K, Samady L, Rajpopat M, McGrath Y, Ledermann J, Coffin RS, Katz DR, Chain B. 2003. Herpes simplex virus infection of dendritic cells: balance among activation, inhibition, and immunity. J Infect Dis 187:165–178. doi: 10.1086/367675. [DOI] [PubMed] [Google Scholar]
- 89.Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, Cantoni C, Grassi J, Marcenaro S, Reymond N, Vitale M, Moretta L, Lopez M, Moretta A. 2003. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med 198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S, Lanier LL, Shibuya A. 2004. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). Int Immunol 16:533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- 91.Pende D, Bottino C, Castriconi R, Cantoni C, Marcenaro S, Rivera P, Spaggiari GM, Dondero A, Carnemolla B, Reymond N, Mingari MC, Lopez M, Moretta L, Moretta A. 2005. PVR (CD155) and Nectin-2 (CD112) as ligands of the human DNAM-1 (CD226) activating receptor: involvement in tumor cell lysis. Mol Immunol 42:463–469. doi: 10.1016/j.molimm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 92.Liu J, Qian X, Chen Z, Xu X, Gao F, Zhang S, Zhang R, Qi J, Gao GF, Yan J. 2012. Crystal structure of cell adhesion molecule nectin-2/CD112 and its binding to immune receptor DNAM-1/CD226. J Immunol 188:5511–5520. doi: 10.4049/jimmunol.1200324. [DOI] [PubMed] [Google Scholar]
- 93.Grauwet K, Cantoni C, Parodi M, De Maria A, Devriendt B, Pende D, Moretta L, Vitale M, Favoreel HW. 2014. Modulation of CD112 by the alphaherpesvirus gD protein suppresses DNAM-1-dependent NK cell-mediated lysis of infected cells. Proc Natl Acad Sci U S A 111:16118–16123. doi: 10.1073/pnas.1409485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci 65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krummenacher C, Carfi A, Eisenberg RJ, Cohen GH. 2013. Entry of herpesviruses into cells: the enigma variations. Adv Exp Med Biol 790:178–195. doi: 10.1007/978-1-4614-7651-1_10. [DOI] [PubMed] [Google Scholar]
- 96.Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. 2011. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog 7:e1002277. doi: 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sodora DL, Eisenberg RJ, Cohen GH. 1991. Characterization of a recombinant herpes simplex virus which expresses a glycoprotein D lacking asparagine-linked oligosaccharides. J Virol 65:4432–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tal-Singer R, Eisenberg RJ, Valyi-Nagy T, Fraser NW, Cohen GH. 1994. N-linked oligosaccharides on herpes simplex virus glycoprotein gD are not essential for establishment of viral latency or reactivation in the mouse eye model. Virology 202:1050–1053. doi: 10.1006/viro.1994.1437. [DOI] [PubMed] [Google Scholar]
- 99.Gianni T, Salvioli S, Chesnokova LS, Hutt-Fletcher LM, Campadelli-Fiume G. 2013. alphavbeta6- and alphavbeta8-integrins serve as interchangeable receptors for HSV gH/gL to promote endocytosis and activation of membrane fusion. PLoS Pathog 9:e1003806. doi: 10.1371/journal.ppat.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cooper RS, Heldwein EE. 2015. Herpesvirus gB: a finely tuned fusion machine. Viruses 7:6552–6569. doi: 10.3390/v7122957. [DOI] [PMC free article] [PubMed] [Google Scholar]