ABSTRACT
The cellular endosomal sorting complex required for transport (ESCRT) was recently found to mediate important morphogenesis processes at the nuclear envelope (NE). We previously showed that the Epstein-Barr virus (EBV) BFRF1 protein recruits the ESCRT-associated protein Alix to modulate NE structure and promote EBV nuclear egress. Here, we uncover new cellular factors and mechanisms involved in this process. BFRF1-induced NE vesicles are similar to those observed following EBV reactivation. BFRF1 is ubiquitinated, and elimination of possible ubiquitination by either lysine mutations or fusion of a deubiquitinase hampers NE-derived vesicle formation and virus maturation. While it interacts with multiple Nedd4-like ubiquitin ligases, BFRF1 preferentially binds Itch ligase. We show that Itch associates with Alix and BFRF1 and is required for BFRF1-induced NE vesicle formation. Our data demonstrate that Itch, ubiquitin, and Alix control the BFRF1-mediated modulation of the NE and EBV maturation, uncovering novel regulatory mechanisms of nuclear egress of viral nucleocapsids.
IMPORTANCE The nuclear envelope (NE) of eukaryotic cells not only serves as a transverse scaffold for cellular processes, but also as a natural barrier for most DNA viruses that assemble their nucleocapsids in the nucleus. Previously, we showed that the cellular endosomal sorting complex required for transport (ESCRT) machinery is required for the nuclear egress of EBV. Here, we further report the molecular interplay among viral BFRF1, the ESCRT adaptor Alix, and the ubiquitin ligase Itch. We found that BFRF1-induced NE vesicles are similar to those observed following EBV reactivation. The lysine residues and the ubiquitination of BFRF1 regulate the formation of BFRF1-induced NE-derived vesicles and EBV maturation. During the process, a ubiquitin ligase, Itch, preferably associates with BFRF1 and is required for BFRF1-induced NE vesicle formation. Therefore, our data indicate that Itch, ubiquitin, and Alix control the BFRF1-mediated modulation of the NE, suggesting novel regulatory mechanisms for ESCRT-mediated NE modulation.
INTRODUCTION
The eukaryotic nuclear envelope (NE) is a specialized compartment composed of double lipid-bilayer membranes and an underlying proteinaceous lamina network and connected by membrane-integrating nuclear pore complexes (NPCs) that selectively regulate the nucleocytoplasmic transport of macromolecules. The NE not only provides an intact meshwork to protect the genome's integrity from cytoplasmic insults, but also serves as a natural barrier against most DNA viruses that replicate their genomes within the nucleus (1). DNA viruses thus evolve various strategies to modify the NE for efficient material transport and nuclear egress of viral nucleocapsids.
Epstein-Barr virus (EBV) is a gammaherpesvirus that infects most of the human population. After primary infection, EBV becomes latent in resting B cells and can be reactivated periodically for lytic replication and virus shedding. During lytic infection, several EBV gene products modulate the cellular environment to facilitate viral DNA replication and virion maturation. The Zta and Rta immediate-early genes not only induce a cascade of viral gene expression, but also cause cell cycle arrest at G1/S transition to accumulate the resources for viral DNA replication. In addition, EBV-encoded BGLF4 is a Ser/Thr kinase that can mimic cyclin-dependent kinase 1 to induce several prophase-like phenomena, such as chromosome condensation and partial disassembly of the nuclear lamina, for the nuclear egress of viral nucleocapsids (1). BGLF4 also modulates the transport preference of NPCs for the nuclear import of viral components (2).
EBV BFRF1 is a homolog of herpes simplex virus 1 (HSV-1) UL34 and plays a crucial role in regulating NE architecture and the primary egress of nucleocapsids (3). Expression of BFRF1 alone induces not only multiple nuclear membranes and cytoplasmic cisternal membrane structures, but also the redistribution of the inner nuclear membrane (INM) protein emerin. This phenomenon is unique to BFRF1 and not to other herpesviral homologs that need to cooperate with their UL31 homologs to induce vesicles derived from the NE (3). For BFRF1 function, the cellular endosomal sorting complex required for transport (ESCRT) machinery, a major membrane scission pathway involved in multivesicular body biogenesis and cytokinesis, is exploited through the recruitment of the ESCRT adaptor protein Alix by BFRF1 (3). Correspondingly, inhibition of ESCRT machinery by RNA interference or the expression of dominant-negative proteins induced the accumulation of viral DNA and capsid proteins in the nuclei of EBV-reactivated cells. This observation suggests BFRF1 serves as a newly identified viral ESCRT adaptor protein, likely performing functions similar to those carried out by the HIV Gag protein, including membrane curvature and the recruitment of ESCRTs (4, 5).
Interestingly, recent studies have found that NE-derived vesicles may be used for transporting large ribonucleoprotein granules from the nucleus into the cytoplasm (6, 7), suggesting that modification of the structure of the NE is important for cellular nucleocytoplasmic transport, as well as the virus maturation process. Of note, accumulating evidence indicates the contribution of ESCRT machinery in regulating events at the cell's NE. For example, ESCRT-III/Vps4 were found to participate in the degradation of defective nuclear pore complex assembly intermediates, providing surveillance of NPC assembly (8). Recently, ESCRT-III factors were shown to coordinate mitotic-spindle disassembly and NE sealing at late anaphase. Likewise, ESCRT-III, Vps4, and spastin cooperatively participate in NE reformation, using mechanisms similar to those involved in cytokinetic abscission (9, 10). Mechanisms underlying ESCRT-mediated NE modulation and the regulation of NE-derived vesicles in the transport of cellular or viral components remain to be elucidated.
Components of the ESCRT pathway promote the budding from the plasma membrane of various enveloped viruses, including retroviruses and several RNA viruses (4). The structural proteins of these viruses carry the so-called “late-budding (L) domains,” which contain three types of conserved motifs, (L)YPXL, PT/SAP, and PPXY. These sequences specifically recruit the ESCRT-associated Alix, TSG101, and Nedd4-like E3 ubiquitin (Ub) ligases, respectively (11–13). Previous studies indicated that ubiquitination mediated by membrane-associated Nedd4-like E3 ligases contributes to the release of enveloped viruses from the plasma membrane (14). These ubiquitin ligases belong to the HECT (homologous to E6-AP carboxyl terminus) domain-containing Nedd4-like E3 ligase family, which includes Nedd4-1, Nedd4-2/4L, WWP1, and Itch/AIP4, and they can directly catalyze substrate ubiquitination. Nedd4-like E3 ligases (E3s) typically associate with intracellular membranes through their calcium binding C2 domains and regulate ubiquitin-mediated trafficking of membrane proteins through monoubiquitination (14). Nedd4-like E3s also regulate lysosomal and proteasomal degradation and nuclear translocation of multiple proteins, such as membrane receptors and transcription factors, and thereby modulate important signaling pathways during tumorigenesis or latent virus infection (15, 16). The release of ESCRT interaction-defective HIV-1 mutants is potently enhanced by ectopic expression of Nedd4-2s (17, 18), suggesting Nedd4-like ubiquitin ligase-mediated ubiquitination potentially cooperates with ESCRT components to catalyze virion release from the plasma membrane.
The transient and sophisticated cellular regulation of NE envelope structure shares key features with the EBV BFRF1-induced modulation of the NE, making it an ideal model to identify cellular components and to elucidate mechanisms involved in ESCRT-dependent NE modulation. We previously showed that expression of EBV BFRF1 protein induces NE structural changes accompanied by various cytoplasmic vesicles of different sizes and properties (3). In the present study, we identify additional cellular factors and mechanisms involved in this process and uncover the important role of ubiquitination in BFRF1-mediated NE modulation and vesicle formation, using multiple approaches. We find that the ubiquitin ligase Itch, ubiquitin, and Alix control the BFRF1-mediated modulation of NE and EBV maturation, revealing novel mechanisms that regulate ESCRT-induced EBV capsid nucleocytoplasmic egress.
MATERIALS AND METHODS
Cell culture, virus induction, and transfection.
HeLa cells were derived from human cervical epithelial cells (ATCC CCL-2). NA is a recombinant Akata EBV-converted NPC-TW01 cell line (19). All the cells were cultured in Dulbecco's modified Eagle's medium (HyClone) supplemented with 10% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2. For EBV induction, NA cells were transfected with plasmid pRTS15 expressing the Rta transactivator (20), using Lipofectamine 2000 (Invitrogen) in Opti-MEM medium (GIBCO-BRL) according to the manufacturer's instructions. For small interfering RNA (siRNA) treatment, Itch siRNA (5′-CAAUAGGAGACUUGUCAAUdTdT-3′, where dT is deoxythymidine) or control siRNA was synthesized by Misson Predesigned siRNA (Sigma) and transfected twice (final concentration, 100 nM) into cultured cells. For protein expression analysis, HeLa cells were transfected with siRNA for 48 h and then cotransfected with siRNAs and hemagglutinin epitope-tagged BFRF1 (HA-BFRF1)-expressing plasmid for an additional 24 h.
Protein sequence analysis.
The putative functional domains of EBV BFRF1 were analyzed manually or using the MUSCLE program (http://www.ebi.ac.uk/Tools/msa/muscle/). The protein secondary structure was predicted by GOR IV secondary-structure prediction (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html) and DAS protein structure modeling (http://www.sbc.su.se/∼miklos/DAS/). The putative ubiquitination sites were analyzed manually or using the UbPred program (http://www.ubpred.org/).
Plasmids.
HA-BFRF1 was generated by cloning XhoI-HA-BFRF1-NotI into pcDNA3.0 (Clontech) as described previously (3). All functional-domain deletion or amino acid substitution mutants of HA-BFRF1, F1(P75,78A), F1(ΔESR) (amino acid [aa] 180 to 313 deletion), F1(K49R), F1(K176R), F1(K253R), F1(K265,266R), F1(K120R), F1(3KR) (K120,265,266R), F1(4KR) (K120,253,265,266R), F1(5KR) (K120,176,253,265,266R), and F1(6KR) (K49,120,176,253,265,266R), were generated by a single primer-based site-directed-mutagenesis strategy (21) using pcDNA3.0-HA-BFRF1 as the template and the primers specified in Table 1. HA-DUb-BFRF1 and HA-DUb*-BFRF1 were generated by cloning in frame with BamHI-UL36 deubiquitinating enzyme (DUb) (N-terminal 15 to 260 residues) of herpes simplex virus type I or catalytic dead DUb*-EcoRI gene fragments (22) into HA-BFRF1, respectively. Plasmids expressing Flag-tagged HECT ubiquitin ligases, including Nedd4-1, Nedd4-2, WWP1, and Itch, were described by Jadwin et al. (23). Plasmids expressing glutathione S-transferase (GST)-fused Itch or Itch functional domains, including HECT, C2, PRD (proline-rich domain), and WW domains, were described previously (24).
TABLE 1.
Oligonucleotides, primers, and plasmid DNA templates used in this study
| HA-BFRF1 | Deletion or residue substitution | Sequence | Template |
|---|---|---|---|
| ΔESR | Δ(180–313) | 5′-ACG CCC AGA AGG CCT CGC GG ACA CCT TAT CTG GCA CGG GT-3′ | pcDNA3.0-HA-BFRFl |
| K49R | K49R | 5′-CCC AAC CCC TCT GCA CCG TGA GGC TGC GCC ACG GAC AGA TTT A-3′ | pcDNA3.0-HA-BFRFl |
| K120R | K120R | 5′-TCA ACG GAT GTT GAC CTA CCA AGG AAC TCC ATC ATT ATG CTG GGC-3′ | pcDNA3.0-HA-BFRFl |
| K176R | K176R | 5′-TTC AGG CCG CCA ACG CCC AGA GGG CCT CGC GGG TCA TGG ATA T-3′ | pcDNA3.0-HA-BFRFl |
| K253R | K253R | 5′-GTT ACG CTC CTT GGA GGG ACA GGG ACT CGT GGT CGG AAT CCG A-3′ | pcDNA3.0-HA-BFRFl |
| K265,266R | K265,266R | 5′-AAT CCG AGG CGG CGC CGT GGA GGA GGG AAC TCG TGA GGC ACC CCA T-3′ | pcDNA3.0-HA-BFRFl |
| 3KR | K120,265,266R | 5′-TCA ACG GAT GTT GAC CTA CCA AGG AAC TCC ATC ATT ATG CTG GGC-3′ | K265,266R |
| 4KR | K120,176,265,266R | 5′-TTC AGG CCG CCA ACG CCC AGA GGG CCT CGC GGG TCA TGG ATA T-3′ | 3KR |
| 5KR | K120,176,253,265,266R | 5′-GTT ACG CTC CTT GGA GGG ACA GGG ACT CGT GGT CGG AAT CCG A-3′ | 4KR |
| 6KR | K49,120,176,253,265,266R | 5′-CCC AAC CCC TCT GCA CCG TGA GGC TGC GCC ACG GAC AGA TTT A-3′ | 5KR |
| P75,78A | P75,78A | 5′-TAA GCT GAA GAA CTG CAA CTA CGC CTC CTC GGC CGT GTT TGT GAT ATC CAA CAA CG-3′ | pcDNA3.0-HA-BFRFl |
Live-cell imaging.
Live-cell imaging was performed on transfected HeLa cells grown on 6-well tissue culture dishes to reach 60 to 70% confluence. Cell images were visualized using a Zeiss Axiovert 200M inverted microscope and an environmental chamber (37°C with 5% CO2). The images were acquired with an LD Plan-Neofluar 40×/0.6 Korr Ph2 objective every 10 min from 6 to 24 h posttransfection. The images were processed with MetaMorph software (v7.7.8.0; Molecular Devices).
Indirect immunofluorescence.
For the detection of HA-BFRF1, emerin, Alix, or Itch, slide-cultured HeLa cells were transfected with plasmids expressing HA-BFRF1 or vector pcDNA3.0. The cells were fixed with 4% paraformaldehyde in PBS (145 mM NaCl, 1.56 mM Na2HPO4, 1 mM KH2PO4, pH 7.2) at 24 h posttransfection at room temperature (RT) for 20 min, washed with phosphate-buffered saline (PBS), and permeabilized with 0.1% Triton X-100 at RT for 5 min. The slides were then incubated with anti-HA (Covance or GeneTex), anti-emerin (Santa Cruz), anti-Alix (Sigma), or anti-Itch (BD BioSciences) at 37°C for 1.5 h. After washing with PBS for 5 min four times, the slides were incubated with rhodamine-, or fluorescein isothiocyanate (FITC)-conjugated anti-mouse or rabbit Ig antibodies (Cappel) at 37°C for 1 h. The DNA was stained with Hoechst 33258 at RT for 1 min. The staining patterns were observed under fluorescence or confocal microscopy (Ziess). All the experiments were repeated at least twice, and the predominant expressed pattern in each test was calculated in more than 50 cells. Protein colocalization (by Manders' coefficient [R]) was analyzed by ImageJ software (NIH), and the average diameters of vesicles or puncta were calculated in 5 randomly selected HA-positive structures per cell for a total of 35 cells in each set with ZEN software (Zeiss).
Protein preparation for mass spectrometric analysis.
To enrich HA-BFRF1 protein, HeLa cells (3 × 107) were transfected with plasmids expressing HA-BFRF1. At 24 h posttransfection, cells were harvested and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, complete protease inhibitor cocktail [Roche], and 1 mM Na3VO4). The cell lysates were then precleared and incubated with 6 μg anti-HA antibody (Covance) at 4°C for 2 h, and 300 μl of 20% protein A-Sepharose beads was added to pull down the immunocomplexes at 4°C for 1.5 h. The beads were washed extensively with RIPA buffer, resuspended in SDS sample buffer, and resolved in SDS-10% PAGE. For in-gel digestion, the target protein band in a Coomassie blue-stained gel was cut into 1-mm cubes. The gel slices were then washed with water and further destained with destaining buffer (40% acetonitrile [ACN], 50 mM NH4HCO3, pH 7.8). The gel pieces were dehydrated with 100% ACN, dried, and subjected to in-gel digestion with sequencing-grade modified trypsin (enzyme-to-substrate ratio, 1:50; Promega) in 50 mM NH4HCO3 overnight at 37°C. Peptides were extracted sequentially with 1% formic acid (FA), 60% ACN combined 0.1% FA and dried (25). To identify the protein modification by mass spectrometry (MS), tryptic-peptide preparation and data-dependent tandem MS (MS-MS) analysis were carried out as described previously (26). Protein fragments were then identified by automated searching (Mascot software) against the NCBI protein database.
Coimmunoprecipitation and pulldown assays.
To immunoprecipitate HA-BFRF1, ubiquitinated BFRF1, or Flag-tagged E3 ligases, HeLa cells (1 × 107) were transfected with plasmids expressing wild-type (WT) HA-BFRF1 or Lys residue substitution mutants alone or together with Flag-tagged E3 ligases (Nedd4-1, Nedd4-2, WWP1, or Itch) or a Myc-ubiquitin-expressing plasmid. The transfected cells were harvested 24 h posttransfection and lysed in RIPA buffer, and anti-HA (Covance) or anti-Flag (Sigma) antibodies were added for immunoprecipitation as described previously (3). In the ubiquitinated BFRF1 precipitations, the sulfhydryl alkylating agent N-ethylmaleimide (Sigma) was added to RIPA buffer to a final concentration of 20 mM to prevent unexpected deubiquitination.
For pulldown assays, equal amounts (∼2 μg) of GST and GST-fused Itch functional-domain fragments (full-length Itch, HECT, WW, PRD, and C2), which were expressed in transformed Escherichia coli BL21(DE3) by 1 mM IPTG (isopropylthiogalactopyranoside) induction at 25°C for 2.5 h, were incubated with glutathione-agarose beads (Sigma) at 4°C for 1.5 h. The beads were then pelleted, washed extensively with cold PBS with 1% Triton X-100, and finally incubated with cell lysates (300 to 500 μg) in binding buffer (20 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 1 mM EGTA, 10% glycerol, 1 mM dithiothreitol [DTT], complete protease inhibitor cocktail [Roche], and 1 mM phenylmethylsulfonyl fluoride [PMSF]) at 4°C for 16 h. The beads were then washed extensively with RIPA lysis buffer for immunoprecipitation analysis or with washing buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1% Nonidet P-40) for pulldown assays, resuspended in SDS sample buffer, and displayed in 10% SDS-PAGE for immunoblotting.
TEM analysis.
For cells with replicating EBV, a total of 3 × 105 NA cells were transfected with plasmid expressing Rta or vector plasmid and harvested at 72 h posttransfection. For cells expressing HA-BFRF1, the same number of HA-BFRF1-transfected HeLa cells were harvested at 24 h posttransfection and processed for transmission electron microscopy (TEM) analysis, as previously described (3). Samples were imaged using a Hitachi H-7100 transmission electron microscope, and images were acquired using an Advanced Microscopy Techniques (AMT; Woburn, MA) camera system. The images were taken at a final magnification of ×10,000. More than 30 cells were observed, and representative nuclear and cytoplasmic regions of transfected HeLa cells are shown.
Establishment of DOX-inducible cells containing a wild-type or BFRF1-knockout (KO) EBV bacmid.
The wild-type p2089 EBV bacmids were kindly provided by Henri-Jacques Delecluse (DKFZ unit F100, Germany) (27). p2089BFRF1-KO was described previously (28). 293TREx_Flag_EZTA (293TetEZ) cells were seeded in a 6-well culture dish (5 × 105 cells/well) and transfected with 10 μg of p2089 or p2089BFRF1-KO using Lipofectamine 2000. At 72 h posttransfection, the transfected cells were split into two 10-cm culture dishes and selected with hygromycin B (100 μg/ml; Invitrogen) for 1 month. Four to 6 green fluorescent protein (GFP)-positive cell colonies were picked up to obtain pool clones. The selected 293TetEZ/p2089 and 293TetEZ/p2089BFRF1-KO pool clones were treated with doxycycline (DOX) (50 ng/ml for 24 h) to confirm successful lytic induction by immunoblotting.
EBV DNA extraction and quantitative real-time PCR (qPCR) analysis.
EBV-harboring 293TetEZ/p2089 and 293TetEZ/p2089BFRF1-KO cells were transfected with plasmids expressing HA-BFRF1 WT, F1(6KR), or the vector pcDNA3.0 for 6 h, followed by 50 ng/ml doxycycline treatment for an additional 48 or 72 h. The culture supernatants were collected and subjected to centrifugation at 10,000 × g for 30 min at 4°C to remove cell debris. The intracellular viral DNA and secreted virion DNA were harvested and analyzed as described previously (3).
RESULTS
Expression of EBV BFRF1 induces NE modulation and cytoplasmic vesicles similar to those observed following EBV reactivation.
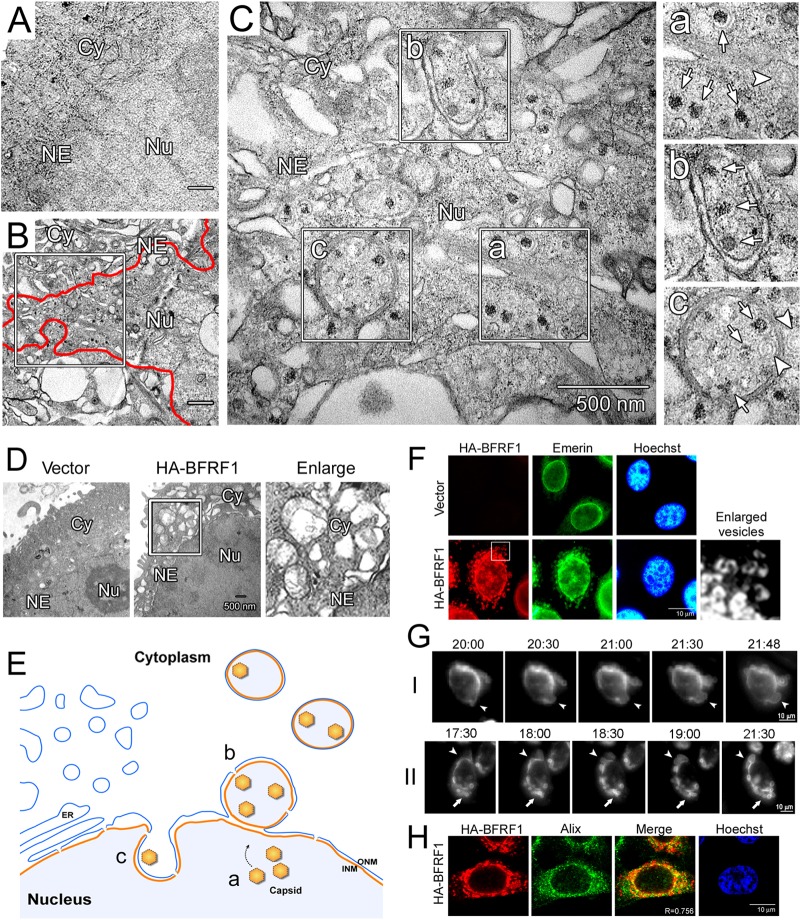
To further understand the molecular mechanism of BFRF1-mediated NE modulation and vesicle formation, we compared these morphological changes to subcellular ultrastructures observed in epithelial cells upon EBV reactivation. An EBV-positive epithelial cell line, NA, was transfected with a plasmid expressing the viral transactivator Rta for 72 h and processed for electron microscopy (EM) analysis. Compared to vector-transfected cells (Fig. 1A), EBV replication induced obvious ultrastructures in the cytoplasm and dramatic changes in the NE (Fig. 1B). Within the nuclei of EBV-infected replicating cells, two types of viral capsids were seen in the nucleus or in the perinuclear region: viral-DNA-containing C-type nucleocapsids and empty A-type capsids (Fig. 1C, a to c, indicated by arrows and arrowheads, respectively). Some capsids were parceled by double-layered membranes in the juxtanuclear region close to the nuclear membrane (Fig. 1C, b) or in vesicle-like ultrastructures (Fig. 1C, c). In comparison, the NE-derived vesicles in transiently BFRF1-transfected HeLa cells were similar in size and location but appeared more irregular at times, with some multiple-layered structures (compare Fig. 1C and D), suggesting other viral components also contribute to vesicle formation in cells following virus replication. These data indicate that EBV reactivation induces NE changes and cytoplasmic ultrastructures that are strikingly reminiscent of those observed following the ectopic expression of BFRF1 (Fig. 1D and E), which likely facilitate the nuclear egress of EBV nucleocapsids, as we and others previously reported (3, 28).
FIG 1.
EBV reactivation or BFRF1 expression induces reorganization of the NE and NE-derived cytoplasmic vesicles. (A) Negatively stained EM images of vector plasmid-transfected NA cells. Cy, cytoplasm; Nu, nucleus; NE, nuclear envelope. (B) EM images of Rta-transfected NA cells at 72 h posttransfection, with EBV capsid production. The red line indicates the edge of the nuclear membrane, and the boxed region is enlarged in panel C. (C) EBV replication induced not only the production of viral C-type (a, arrows) or A-type (a, arrowhead) capsids in the nucleus, but also the formation of viral-capsid-containing perinuclear vesicles (b) and invagination (c) from the nuclear envelope. The images on the right are enlargements of the boxed regions on the left. Arrows, C-type capsids; arrowheads, A-type capsids. (D) EM images of HA-BFRF1- or vector plasmid-transfected HeLa cells at 24 h posttransfection. The boxed region is enlarged on the right. (E) Subcellular ultrastructure in NA cells with replicating EBV. a, b, and c refer to the enlarged regions in panel C. ONM, outer nuclear membrane. Reactivation of EBV induced reorganization of the nuclear membrane and the formation of capsid-containing vesicles. (F) Confocal images of emerin in HA-BFRF1-expressing HeLa cells. HA-BFRF1 and emerin were visualized by indirect immunofluorescence at 24 h posttransfection. The boxed region is enlarged on the right. (G) Live-cell imaging of BFRF1-induced vesicle formation. HeLa cells transfected with CFP-BFRF1 were subjected to time-lapse fluorescent imaging analysis from 20 to 22 h posttransfection at 10-min intervals (indicated as hour:minute. The expression of CFP-BFRF1 induced a large vesicle protruding from the NE (I and II, arrowheads), which subsequently moved to the cytoplasm (II, arrows; see Movie S1 in the supplemental material). (H) Confocal images of Alix in HA-BFRF1-expressing HeLa cells. HA-BFRF1 and Alix were visualized by indirect immunofluorescence at 24 h posttransfection.
To further observe the NE structure changes, along with BFRF1 expression, HeLa cells were next transfected with HA-tagged BFRF1 and monitored for the distribution of BFRF1 and the INM-anchored protein emerin in fluorescence microscopy. In contrast to the typical nuclear-rim-staining pattern of emerin in the vector-transfected cells, expression of HA-BFRF1 caused cytoplasmic distribution of dispersive emerin-containing vesicles (Fig. 1F), indicating that the vesicles observed with EM are derived from the NE. To further examine the dynamic distribution pattern of BFRF1 in these vesicles at a single-cell level, cyan fluorescent protein (CFP)-tagged BFRF1 was expressed in HeLa cells and recorded under time lapse fluorescence microscopy (Fig. 1G; see Movie S1 in the supplemental material). We found that ectopically expressed CFP-BFRF1 became visible as early as 18 h posttransfection and induced the reorganization of the NE to form large cytoplasmic vesicles. The initial NE-associated protrusions were gradually enlarged (Fig. 1G, I and II, arrowheads), and some membrane fusion or scission was simultaneously observed to form cytoplasmic vesicles (nearly 22 h posttransfection) (Fig. 1G, II, arrows).
We previously reported that expression of EBV BFRF1 recruits Alix and ESCRT machinery to modulate the NE structure and produce NE-derived cytoplasmic vesicles (3). Here, we noticed that the ESCRT-bridging Alix partially colocalized with HA-BFRF1 in the region near the nuclear rim at 24 h posttransfection (Fig. 1H) (with a Manders coefficient [R] of 0.756). Considering the data obtained from these varied and complementary imaging techniques, we concluded that expression of BFRF1 alone provides a simple system that recapitulates EBV-induced and Alix/ESCRT-dependent NE morphological vesicular modulation and is therefore suitable for the identification of new cellular factors and mechanisms involved in these processes that are pertinent to nucleocytoplasmic EBV capsid egress.
EBV BFRF1 recruits cellular Alix transiently for NE modulation.
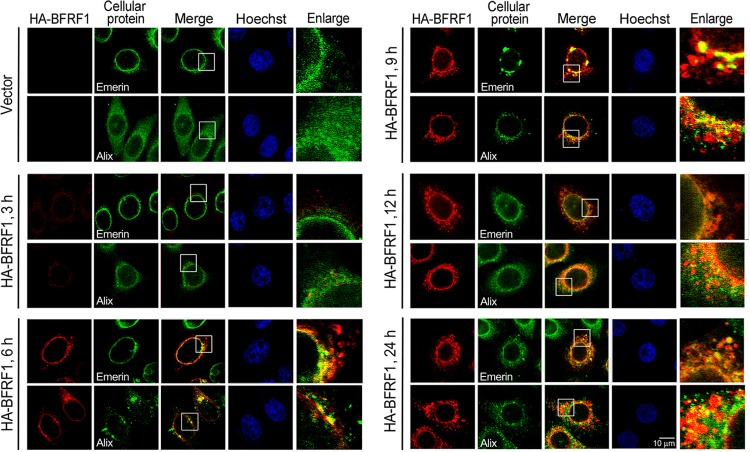
As ESCRT-associated Alix partially colocalized with HA-BFRF1 at the nuclear rim and in perinuclear vesicles (Fig. 1H) and was crucial for BFRF1-induced vesicle formation (3), we next monitored the sequential events of BFRF1-induced alteration of the NE and vesicle formation. By monitoring the distribution of INM-anchored emerin and Alix at different time points after transfection by confocal microscopy, cells with the predominant phenotypes (>70% of the cell population) were displayed (Fig. 2). We found that HA-BFRF1 protein expression was detected as early as 3 h posttransfection. Small cytoplasmic HA-BFRF1-containing puncta without detectable emerin or with nuclear-rim emerin partially colocalized with BFRF1 were visible at 6 h posttransfection, suggesting BFRF1 is capable of inducing small cytoplasmic puncta directly from cytoplasmic membrane materials. HA-BFRF1 then induced an obvious aggregation of emerin at the nuclear rim at 9 h posttransfection, accompanied by the formation of large HA-BFRF1-containing vesicles adjacent to the nucleus, implying the BFRF1-mediated modulation of INM at this stage. Perinuclear colocalized HA-BFRF1 and emerin became obvious at the early time points from 6 to 9 h posttransfection (Fig. 2, Merge and Enlarge); emerin was consistently detected in the cytoplasmic BFRF1-containing vesicles after 9 h posttransfection.
FIG 2.
Cellular Alix is recruited by BFRF1 to modulate the nuclear envelope. Shown are confocal images of HA-BFRF1- and vector-transfected HeLa cells. HA-BFRF1 (red) and emerin or Alix (green) were visualized by indirect immunofluorescence at 3, 6, 9, 12, 18, and 24 h posttransfection. The boxed regions are enlarged on the right.
Simultaneously, cellular Alix was detected along with the progression of BFRF1-induced vesicle formation. Showing a diffuse cytoplasmic distribution in vector plasmid-transfected cells, Alix was redistributed close to the nuclear rim at 3 h posttransfection and appeared to have relatively strong HA-BFRF1 colocalization in the perinuclear region at 6 h posttransfection in HA-BFRF1-expressing cells. The sequential nuclear recruitment of Alix at 3 h posttransfection and the formation of BFRF1-containing cytoplasmic puncta at 6 h posttransfection suggested a transient progression of NE modulation by BFRF1, with Alix playing an important role in the formation of BFRF1-containing cytoplasmic structures. Interestingly, the colocalization signal of Alix-BFRF1 was gradually attenuated following the formation of large cytoplasmic vesicles after 9 h posttransfection (Fig. 2, Merge and Enlarge). When large vesicles containing BFRF1 and emerin were formed in the cytoplasm at a later stage (24 h posttransfection), a large portion of Alix became diffused into the cytoplasm, suggesting that sequential intermolecular interactions among BFRF1-interacting proteins may temporally and spatially regulate BFRF1-induced vesicle formation.
Ubiquitination regulates BFRF1 induction of nucleocytoplasmic vesicles.
It is known that ubiquitination plays important roles in regulating cellular and viral ESCRT-dependent processes (29). Similarly, Ub conjugation to Gag assembly sites was found to be critical for ESCRT-dependent HIV-1 budding from the plasma membrane (22, 29). To explore whether ubiquitination contributes to BFRF1-induced nuclear egress of cytoplasmic vesicles, BFRF1-expressing cells were treated with the proteasome inhibitor MG132, which inhibits the function of the proteasome core particle and depletes the intracellular pool of free ubiquitin (30, 31). Using Myc-tagged Ub coexpression with HA-BFRF1 and anti-HA antibodies in immunoprecipitation assays, the ubiquitinated BFRF1 signal did not increase in MG132-treated cells, suggesting ubiquitination does not regulate BFRF1 for degradation (Fig. 3A, lanes 8 and 12). In line with this, the ubiquitinated BFRF1 signal was reduced in cells expressing the myc-Ub Lys-48 (K48) mutant Ub48R, which is defective for poly-Ub chain-mediated proteasomal degradation (Fig. 3A, lanes 9 and 13). In addition, immunofluorescence staining showed that HA-BFRF1 expression induced cytoplasmic-vesicle formation (in 89% of HA-BFRF1-expressing cells under dimethyl sulfoxide [DMSO] treatment) and redistributed a portion of emerin from the nuclear rim to the cytoplasm of transfected cells (Fig. 3B, Emerin). Coexpressed Myc-Ub enhanced the accumulation of cytoplasmic BFRF1-containing vesicles (Fig. 3B, HA-BFRF1, bottom). Following MG132 treatment, the formation of BFRF1-containing vesicles was significantly reduced (only 6% of HA-BFRF1-expressing cells), accompanied by the accumulation of BFRF1 in the perinuclear region (Fig. 3B, Hoechst), suggesting free ubiquitins are required for BFRF1-induced vesicle formation. For cells treated with MG132, overexpression of Myc-Ub restored the cytoplasmic appearance of BFRF1-containing vesicles in 85% of HA-BFRF1-expressing cells (Fig. 3B, Merge, bottom). However, these vesicles were smaller, with limited colocalization of emerin, indicating that most vesicles may be derived from cytoplasmic membranous materials rather than the NE. Taken together, even though MG132's inhibitory effect was not fully restored by trans-complementation of free Ub, these data indicate that ubiquitin plays important roles in the formation of NE-derived BFRF1-containing vesicles.
FIG 3.
Treatment of MG132 diminishes the NE modulation and vesicle formation abilities of EBV BFRF1. (A) The steady-state expression level of EBV BFRF1 was not enhanced by MG132 treatment. HeLa cells were transfected individually or cotransfected with HA-BFRFl-, Myc-Ub-, or Myc-Ub48R-expressing plasmids. At 6 h posttransfection, the cells were treated with DMSO solvent or 10 μM MG132 for 18 h. The cell lysates were harvested and immunoprecipitated (IP) using antibody against HA or GST as a negative control. The immunocomplexes were detected by antibodies against Myc and HA. (B) Confocal images of HeLa cells transfected with HA-BFRF1, together with vector control or Myc-Ub, and treated with DMSO solvent (left) or 10 μM MG132 (right) for 18 h. HA-BFRF1 and emerin were visualized by indirect immunofluorescence. The boxed regions are enlarged below. (C) Overexpressed HA-BFRF1 protein in HeLa cells was partially purified by immunoprecipitation with HA antibody, separated by SDS-PAGE, and subjected to mass spectrometric analysis. IgH and IgL indicate the immunoglobulin heavy and light chains. M, molecular mass marker. (D) The recovered BFRF1 fragments were identified by Mascot software against the NCBI virus protein database. The coverage rate of the BFRF1 fragment was 63% (red letters). Shown is ubiquitination of Lys-120 (K120) in the recovered BFRF1 103 to 120 fragment.
Multiple lysine residues are important for regulating BFRF1-induced vesicle formation.
Considering the importance of ubiquitin described above, we next sought to identify possible ubiquitination sites in BFRF1 using immunoprecipitated HA-BFRF1 proteins from transfected HeLa cells and mass spectrometry (Fig. 3C). With 63% recovery of BFRF1 fragments, Lys-120 was identified as a dominant ubiquitin receptor residue in BFRF1 (Fig. 3D).
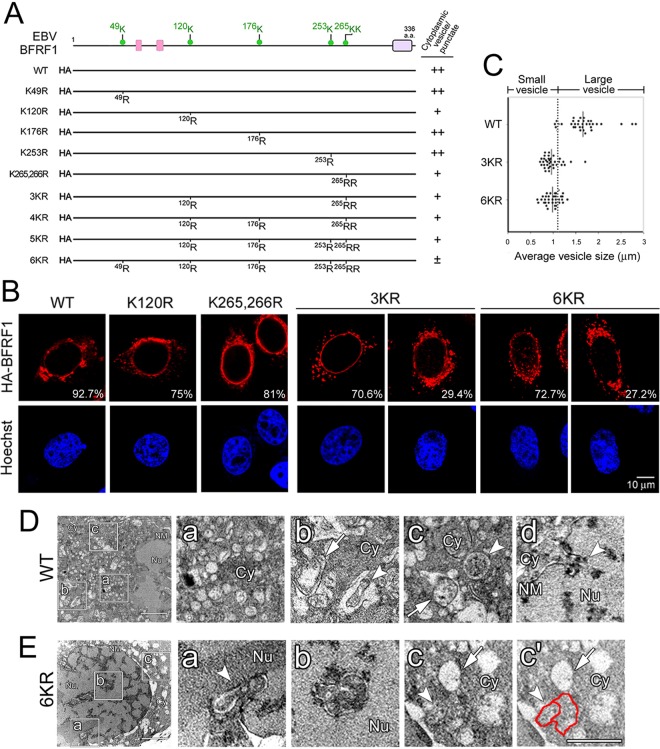
Because ubiquitin can be added to other lysines in proteins when the main or predominant site(s) becomes unavailable, we opted to mutate all potentially ubiquitinated lysine residues in BFRF1 to test the ability of ubiquitination-defective mutants to induce NE-derived cytoplasmic vesicles. The UbPred program (32) helped identify residues at positions Lys-49, -120, -176, -253, -265, and -266 in BFRF1 as potential ubiquitination sites (Fig. 4A). Single and multiple Lys-to-Arg substitutions were generated, expressed, and tested for vesicle formation in HeLa cells (Fig. 4A and B). Expression of F1(K49R), F1(K176R), and F1(K253R) induced cytoplasmic vesicles, similar to WT BFRF1 (data not shown). In contrast, cells with F1(K120R) and F1(K265,266R) showed reduced numbers of vesicles, accompanied by the appearance of cytoplasmic puncta, suggesting that ubiquitination of lysine at positions 120, 265, and 266 in BFRF1 is involved in the regulation of vesicle formation (Fig. 4B, WT). Compared with the WT, which induced vesicles with a diameter of 1.64 ± 0.42 μm, F1(3KR) (K120,265,266R) induced smaller cytoplasmic vesicles (0.95 ± 0.21 μm). With additional lysine mutations, F1(6KR) (K49,120,176,253,265,266R) induced a vesicle size (0.95 ± 0.17 μm) similar to that induced by F1(3KR) (Fig. 4B and C). This result indicated that disruption of key ubiquitin-conjugating lysine residues at Lys-120, -265, and -266 interfered with BFRF1's ability to modulate/deform the NE, while the possibility of protein structure change caused by Lys-to-Arg substitutions could not be excluded.
FIG 4.
Lys-to-Arg substitution of EBV BFRF1 influences its NE-modulating and vesicle-forming abilities. (A) Schematic representation of HA-tagged BFRF1 mutants with multiple Lys-to-Arg substitutions. Putative late domains (pink boxes) and a transmembrane domain (purple box) were predicted manually or by the MUSCLE and DAS programs. Six putative ubiquitination sites (green dots), Lys-49, -120, -176, -253, -265, and -266, on BFRF1 were identified by the UbPred program. The abilities of BFRF1 mutants to induce cytoplasmic vesicles, or puncta, and intranuclear puncta are summarized on the right. Pluses and minuses indicate the relative qualifications of specific phenotypes in each experiment. (B) Confocal images of HA-BFRF1 WT-, F1(120R)-, F1(K265,266R)-, F1(3KR)-, and F1(6KR)- or control vector-transfected HeLa cells immunostained for HA and emerin. The vesicle morphology and emerin distribution in HA-BFRF1 WT- and mutant-expressing cells were observed. The percentages of cells with specific BFRF1 vesicle expression patterns were obtained from three independent experiments with more than 100 cells containing HA-BFRF1. (C) The average diameters of cytoplasmic vesicles in BFRF1 WT-, 3KR-, and 6KR-transfected cells was calculated in randomly selected sections with more than 20 cells in each set. Thirty-five randomly selected vesicles/puncta are shown with the average sizes indicated. The cutoff value for the average vesicle size was defined as 1.1 μm. (D and E) NE and vesicle ultrastructure in HA-BFRF1 WT- and F1(6KR)-expressing HeLa cells. (D) Expression of HA-BFRF1 WT altered the nuclear membrane (NM) structure and induced multiple irregular vesicles in the cytoplasm. The accumulation of cytoplasmic vesicles (a) or invagination of the NM (d, arrowhead) was observed in the HA-BFRF1 WT-transfected cells. Empty vesicles with single- or double-layer membranes (b and c, arrows) or vesicles containing fuzzy inclusions (b and c, arrowheads) were also found at higher magnification in cells transfected with HA-BFRF1. (E) Expression of F1(6KR) induced protrusion of the NM into the nucleus and multiple vesicles in the cytoplasm. The modulated NM (a, arrowhead), intranuclear vesicle with double membranes (b), and cytoplasmic vesicle without (c and c′, arrows) or with (c and c′, arrowheads and red line) fuzzy inclusions are shown at higher magnification. The boxed regions in the left image are enlarged on the right.
To further evaluate the cellular ultrastructures between WT BFRF1 and F1(6KR), HeLa cells expressing either protein were examined by transmission electron microscopy (Fig. 4D and E). The results showed that expression of WT BFRF1 induced multiple cytoplasmic vesicles (Fig. 4D, a to c) and an irregular nuclear membrane (Fig. 4D, d). Parts of cytoplasmic vesicles contained double-layered membranes (4% of randomly selected vesicles) or fuzzy inclusions (13%) (Fig. 4D, b to d). In contrast, expression of F1(6KR) induced some inward invaginations of a double-layered nuclear membrane structure into the nucleus (Fig. 4E, a and b), which resembled the intranuclear puncta observed in immunostained images (Fig. 4B). The F1(6KR) mutant still induced irregular cytoplasmic vesicles, but without double-layered membranes. Empty vesicles with fuzzy margins (71% in randomly selected vesicles) or fused irregular vesicles with inclusions (27%) were also observed in the cytoplasm (Fig. 4E, c and c′), suggesting that disruption of ubiquitination sites in BFRF1 interfered with the formation of typical nucleocytoplasmic vesicles. Overall, these data indicate that the ability of BFRF1 to induce NE-derived vesicles is regulated by lysine residues at positions 120, 265, and 266 and/or possible ubiquitination of these sites.
Ubiquitin is a key regulator of BFRF1-mediated vesicle formation at the NE.
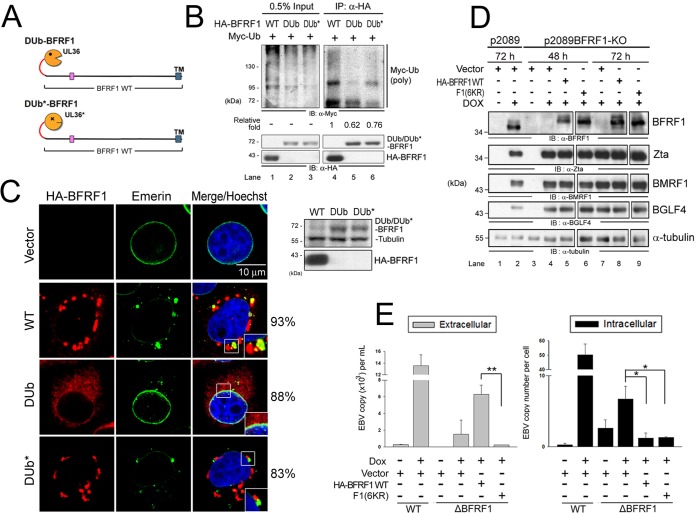
To directly assess the role of ubiquitination in vesicle formation of BFRF1 at the NE, we fused the catalytic domain of HSV-1 UL36 DUb or its inactive counterpart (DUb*) (22, 33) with BFRF1 to generate a ubiquitination-resistant DUb-BFRF1 protein (Fig. 5A). In comparison to WT BFRF1, polyubiquitin signal was obviously attenuated in cells expressing HA-tagged DUb-BFRF1 (62% of WT), but not in its DUb*-BFRF1 inactive counterpart, as indicated by the amounts of Myc-Ub conjugated to BFRF1 in immunoprecipitation analysis (Fig. 5B).
FIG 5.
Ubiquitination is required for BFRF1 function in vesicle induction and virus replication. (A) Schematic representation of fusion of the DUb of HSV-1 UL36 and its inactive form (DUb*) onto BFRF1. (B) Lysates of HeLa cells with expression of HA-BFRF1 WT, HA-DUb-BFRF1, or HA-DUb*-BFRF1 and Myc-Ub were harvested and immunoprecipitated with antibody against HA. The immunocomplexes were then detected by antibodies against Myc and HA by immunoblotting. (C) Confocal images of HA-BFRF1- and DUb- or DUb*-HA-BFRF1-transfected HeLa cells immunostained for HA (red) and emerin (green). The boxed regions are enlarged in the insets. The protein expression of BFRF1 WT, DUb-BFRF1, and DUb*-BFRF1 was also confirmed by immunoblotting using HA antibody. Tubulin served as a protein-loading control. The percentages of cells showing the representative pattern are indicated. (D) EBV-positive 293TetEZ/p2089 and 293TetEZ/p2089BFRF1-KO cells transfected with HA-BFRF1 WT or F1(6KR) or vector were mock treated or treated with DOX to induce Rta expression and harvested at 72 h postinduction. Viral protein expression was detected by immunoblotting with antibodies against BFRF1 (E10), Zta (4F10), BMRF1 (88A9), and BGLF4 (2616). α-Tubulin served as a protein-loading control. (E) Viral DNAs in the culture supernatant (extracellular) and transfected cells (intracellular) were harvested simultaneously and subjected to qPCR analysis targeting the EBV DNA BamHI W fragment. Statistically significant differences between HA-BFRF1 WT and F1(6KR) expression levels in supernatant and transfected cells are indicated by the asterisks. *, P < 0.05; **, P < 0.01.
Using immunofluorescence imaging, we also found that, compared to the WT BFRF1, DUb-BFRF1 lost nuclear-rim localization and the ability to redistribute nuclear emerin and induce cytoplasmic vesicles at the NE (as indicated by the representative cell pattern that was observed in 88% of cells) (Fig. 5C). In contrast, the DUb* fusion of BFRF1 still induced cytoplasmic vesicles with partial perinuclear emerin accumulation (in 83% of Dub*-BFRF1-expressing cells), indicating that the deubiquitinase activity regulates BFRF1-mediated vesicle formation from the NE. Altogether, the data described here suggest that ubiquitination of BFRF1 and/or a BFRF1-associated complex is important for its NE modulation and induction of cytoplasmic vesicle formation there.
Replacement of multiple lysines in BFRF1 attenuates EBV maturation.
We previously found that BFRF-1 recruits Alix/ESCRTs to generate NE-derived vesicles, a process known to contribute to the translocation of EBV nucleocapsids from the nucleus to the cytoplasm for subsequent maturation (3). Here, we explored a role for BFRF1 ubiquitination in regulating EBV maturation. Recombinant WT EBV p2089 or the BFRF1-lacking bacmid p2089BFRF1-KO (28) was used to establish EBV Zta-expressing clonal 293T-TetEZ cells. The cells were subsequently transfected to express the WT BFRF1, F1(6KR), or the control vector and treated with doxycycline for Zta expression to reactivate EBV replication. The intracellular EBV protein expression, including that of the DNA polymerase processivity factor BMRF1 and the protein kinase BGLF4, was detected by immunoblotting (Fig. 5D). In p2089-containing cells, the intracellular EBV genomes increased about 50-fold and the extracellular released virions about 13.5-fold after induction, as indicated by qPCR (Fig. 5E). In comparison, p2089-BFRF1-KO cells contained less intracellular viral DNA and released very small numbers of EBV virions into the medium, demonstrating that BFRF1 also contributes to viral DNA replication and is essential for virion release. In cells trans-complemented with WT BFRF1, virion release was rescued, while conversely, no virion release was detected with cells trans-complemented with the ubiquitination-defective F1(6KR) mutant (Fig. 5E). Taken together, these results support a critical role of BFRF1 ubiquitination or the lysine residues at positions 120, 265, and 266 in EBV DNA replication and virion release.
Nedd4-like ubiquitin ligases are recruited by BFRF1.
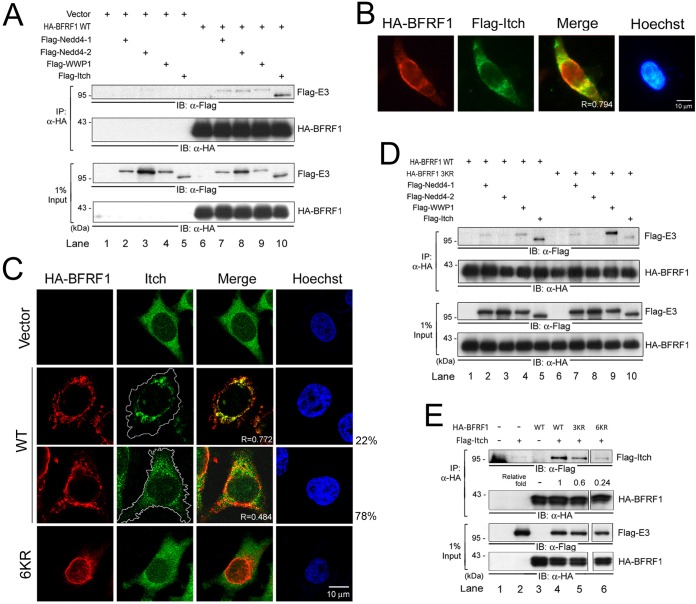
The data described above indicated that the ubiquitination of BFRF1 plays important roles in its ability to modulate the NE. BFRF1-mediated modulation of the NE is also dependent on members of the ESCRT pathway, whose function is known to be regulated by Nedd4-like ubiquitin ligases (34). We therefore probed for BFRF1 association with members of the Nedd4-like ubiquitin ligase family. The abilities of the Flag-tagged Nedd4-like ligases Nedd4-1, Nedd4-2, WWP1, and Itch to interact with HA-BFRF1 were evaluated in coimmunoprecipitation experiments using an anti-HA antibody. While all the Nedd4-like E3 ligases tested displayed certain levels of binding to HA-BFRF1, Itch consistently exhibited the highest binding intensity (Fig. 6A, lanes 6 to 10). In line with this, partial colocalization (R = 0.794) of Itch with HA-BFRF1 at the nuclear rim and in the perinuclear region was observed in Flag-Itch-transfected cells (Fig. 6B). When endogenous Itch distribution was analyzed by confocal imaging, approximately 22% of HA-BFRF1-expressing cells showed obvious Itch colocalization (R = 0.772) (Fig. 6C). The limited colocalization signal (R = 0.484) observed in most (78%) of the population of transfected cells implies that the association between BFRF1 and Itch may be transient, similar to BFRF1-Alix interaction.
FIG 6.
Wild-type BFRF1 associates predominantly with Nedd4-like Itch ligase. (A) Lysates of HeLa cells transfected with HA-BFRF1 and Flag-tagged Nedd4-1, Nedd4-2, WWP1, or Itch were harvested and immunoprecipitated with HA antibody. The immunocomplexes were then detected by antibodies against Flag and HA by immunoblotting (IB). (B) Immunofluorescence images of HA-BFRF1/Flag-Itch-coexpressing HeLa cells stained for HA and Flag. The results were obtained from three independent experiments with more than 100 cells containing HA-BFRF1, and the predominant protein expression images (>85% of the cells) are shown. (C) Confocal images of HeLa cells expressing WT HA-BFRF1 or F1(6KR) and immunostained for HA and endogenous Itch. The white lines indicate the cell margins. The percentages of cells with specific Itch expression in more than 100 cells with HA-BFRF1 expression are shown. (D) Lysates of HeLa cells transfected with HA-BFRF1 WT or 3KR, and Flag-tagged Nedd4-1, Nedd4-2, WWP1, or Itch were harvested and immunoprecipitated with HA antibody. The immunocomplexes were then detected by antibodies against Flag and HA by immunoblotting. (E) Lysates of HeLa cells transfected with HA-BFRF1 WT or 3KR and Flag-Itch were harvested and immunoprecipitated with HA antibody. The immunocomplexes were then detected by antibodies against Flag and HA by immunoblotting.
Considering that the key lysine residues on BFRF1 are important for its vesicle induction, we next explored the association abilities among BFRF1 lysine mutants and Nedd4-like ligases (Fig. 6D). While WT BFRF1 predominantly associated with Itch, F1(3KR) preferred to associate with WWP1 ligase (Fig. 6D, lanes 2 to 5 and 7 to 9). This suggests that lysine residues or lysine-mediated ubiquitination on BFRF1 determines its ubiquitin E3 ligase preference. Indeed, the coimmunoprecipitation analysis indicated that the abilities of F1(3KR) and F1(6KR) to interact with Itch were weaker than that of WT HA-BFRF1 (Fig. 6E, lanes 4 to 6). The immunostaining of F1(6KR) and Itch also showed weak colocalization (R = 0.449) (Fig. 6C, bottom). Taken together, these results indicate that lysine residues (or related ubiquitination) on BFRF1 are crucial for Nedd4-like ubiquitin ligase association.
The EBV-specific region of BFRF1 is crucial for Itch-Alix complex association.
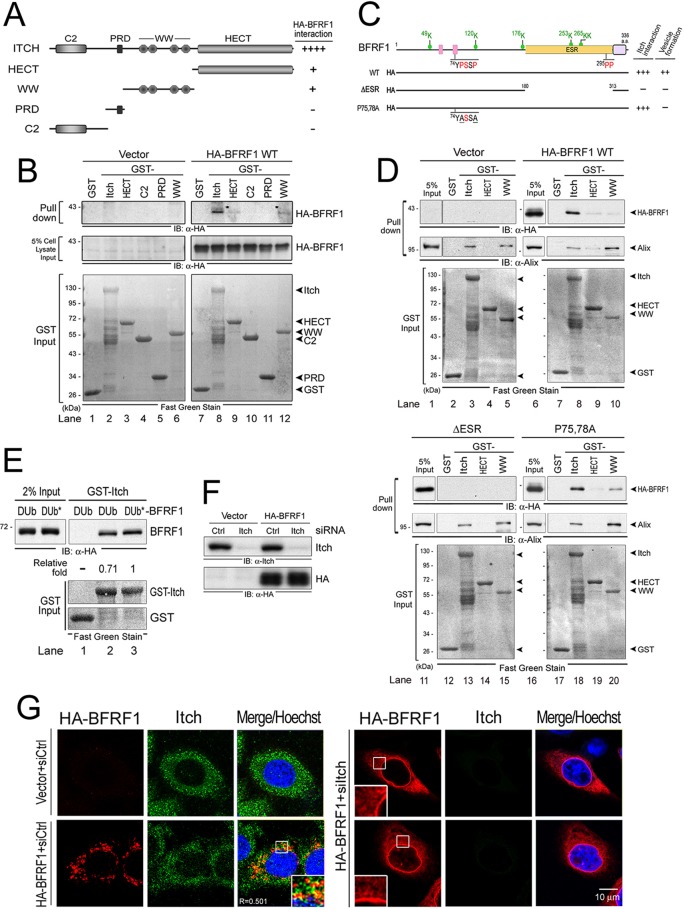
We next proceeded to map the interacting domains between Itch and BFRF1 using protein pulldown analysis. Itch consists of an N-terminal C2 domain, a PRD, four WW domains, and a C-terminal catalytic HECT domain, and therefore, it belongs to the HECT E3 ubiquitin ligase family (24, 35). Using similar amounts of bacterially expressed GST fusion proteins, GST-Itch, -WW, and -HECT pulled down HA-BFRF1 (Fig. 7A and B, lanes 8, 9, and 12), indicating that both Itch WW and HECT domains mediate interaction with BFRF1.
FIG 7.
The ESR of BFRF1 mediates Itch complex association. (A) Schematic representation of GST-fused Itch fragments. The relative association intensities between the GST-fused Itch fragments and HA-BFRF1 are summarized on the right. Pluses and minuses indicate the relative qualifications of specific phenotypes in each experiment. (B) Characterization of the BFRF1-interacting domain on Itch in vitro. Equal amounts of bacterially expressed GST, GST-fused full-length Itch, HECT, WW, PRD, and the C2 domain fragment were captured by glutathione beads and incubated with the HA-BFRF1-expressing cell lysates. Inputs of HA-BFRF1 and GST-fused proteins were detected by HA antibody and Fast Green staining, respectively. (C) Schematic representation of HA-tagged BFRF1 mutants for mapping the Itch-interacting region. Putative late domains (pink boxes), the transmembrane domain (purple box), and putative ubiquitination sites (green dot) are predicted. The Itch-interacting abilities of BFRF1 mutants in GST pulldown analysis are indicated on the right. Red letters in the sequences indicate the putative Itch-targeting residues; underlined letters indicate mutated residues. (D) Bacterially expressed GST-Itch, GST-HECT, GST-WW, and GST fragment were captured by glutathione beads and incubated individually with the lysates from HA-BFRF1 WT-, F1(ΔESR)-, F1(P75,78A)-, or vector plasmid-transfected HeLa cells. The pulled down protein complexes were then detected by antibodies against HA and Alix. The GST protein inputs were detected by Fast Green staining. (E) Bacterially expressed GST-Itch and the GST fragment were captured by glutathione beads and incubated individually with the lysates from DUb- or Dub*-BFRF1 plasmid-transfected HeLa cells. The pulled down protein complexes were then detected by antibodies against HA. The GST protein inputs were detected by Fast Green staining. (F) Lysates from HeLa cells transfected with Itch or control (Ctrl) siRNA, together with a plasmid expressing HA-BFRF1 or a control vector, were harvested at 72 h posttransfection and subjected to immunoblot analysis using antibodies against Itch and HA. (G) HeLa cells were transfected with Itch (siItch) or control (siCtrl) siRNA for 48 h and cotransfected with siRNAs and a plasmid expressing HA-BFRF1 for an additional 24 h. HA (red) and Itch (green) were visualized by indirect immunofluorescence and observed by confocal microscopy. The boxed regions are enlarged in the insets.
While no ubiquitin E3 ligase association has been reported among the herpesviral homologs of EBV BFRF1, we first explored the possible contribution of the EBV-specific region (ESR) of BFRF1 (Fig. 7C). The ESR was identified as an EBV-unique region in our previous study based on sequence alignment of all human herpesviral BFRF1 homologs (3). Using purified GST-Itch, wild-type BFRF1 showed strong binding with full-length Itch and detectable but much weaker binding to the HECT or WW fragment alone (Fig. 7D, lanes 8 to 10), confirming that both domains contribute to Itch-BFRF1 interaction. Deletion of the ESR, but not an irrelevant mutant, F1(P75,78A), reduced Itch-BFRF1 association significantly (Fig. 7D, compare lanes 13 to 15 to lanes 18 to 20), identifying the ESR domain in BFRF1 as the main contributor to Itch-BFRF1 interaction. Since BFRF1 binds Alix simultaneously, we checked for Alix association with Itch and found that Alix associated with the Itch WW region (Fig. 7D, lanes 3 and 5). Itch and the WW fragment captured similar amounts of Alix, indicating that the WW domain mediates Itch-Alix interaction(s). In the GST-Itch pulldown analysis, a reduced Itch association was observed using DUb-BFRF1 protein compared to that with DUb*-BFRF1 (Fig. 7E, lanes 2 and 3), suggesting that the removal of ubiquitination on BFRF1 or BFRF1-interacting protein would attenuate Itch binding. Taken together, these results indicate that the ESR, as well as the ubiquitination status of BFRF1, is crucial for ubiquitin-Itch ligase association.
Itch is required for BFRF1-mediated vesicle formation.
Finally, we addressed whether Itch plays a direct role in BFRF1-induced vesicle formation by siRNA depleting cellular Itch (Fig. 7F). In contrast to the predominantly cytoplasmic distribution of Itch in control cells, Itch translocated to the perinuclear vesicles in cells expressing BFRF1 (Fig. 7G, inset). Knockdown of Itch induced a diffuse distribution of BFRF1 at the nuclear rim or a cytoplasmic reticular pattern, similar to that observed in cells expressing DUb-BFRF1 (Fig. 5C and 7D) and Alix knockdown cells (Lee et al. [3], Fig. 4F). No cytoplasmic vesicles were observed in Itch-depleted cells, confirming that Itch (and/or Itch-mediated ubiquitin conjugation) is required for BFRF1-mediated NE modulation and vesicle formation.
DISCUSSION
Herpesvirus remodeling of the NE for virus maturation is a complex process that involves protein trafficking between the cytoplasm and nucleus and the modification of NE-associated factors. In this study, we have provided evidence that ubiquitination and the ubiquitin ligase Itch regulate ESCRT-mediated and EBV BFRF1-induced modulation of the NE. We found that EBV BFRF1 induces the reorganization of the NE and cytoplasmic-vesicle formation in a manner similar to that observed in EBV-reactivated cells (Fig. 1). During these processes, Alix is recruited at an early stage at 6 h posttransfection, and the ubiquitin is involved in BFRF1-mediated NE modulation and the formation of cytoplasmic punctate vesicles (Fig. 2). Disruption of key Lys residues at positions 120, 265, and 266 in BFRF1 or its fusion with a DUb enzyme interfered with its ability to deform the NE (Fig. 4 and 5), revealing that lysine residues or related ubiquitin conjugation regulates BFRF1-mediated NE modulation and induction of nucleocytoplasmic vesicles. We obtained evidence that the ubiquitin ligase Itch is preferentially recruited by the BFRF1-Alix complex, while multiple Nedd4-like ubiquitin ligases associated weakly with WT BFRF1 (Fig. 6 and 7). Even though WWP1 interacts with F1(3KR), it appeared not to be able to compensate for the Itch function in BFRF1-induced vesicle formation (Fig. 6D). Accordingly, knockdown of cellular Itch abolished BFRF1-induced vesicle formation (Fig. 7G), indicating that Itch is preferentially selected among other cellular Nedd4-like ligases to specifically play a critical role in BFRF1-induced changes of the NE. In our EM analysis of EBV-replicating NA cells, the double-membrane invaginations (Fig. 1) were similar to large vesicular structures containing multiple NPCs observed in the electron tomography or EM images of cells replicating murine gammaherpesvirus 68 (MHV-68), HSV-1, or pseudorabies virus (PrV) (36–38), suggesting other herpesviruses may also use similar mechanisms during viral replication. An advanced three-dimensional imaging approach may be helpful to further examine these intricate NE-derived ultrastructures in EBV-replicating or BFRF1-expressing cells.
Regarding the contribution of ESCRT components, Alix-dependent cytoplasmic-vesicle formation is supported by our previous findings that siAlix caused the accumulation of BFRF1 on the membrane of an endoplasmic reticulum (ER)-associated apparatus in the cytoplasm (3). Because most of the ESCRT components are distributed in the cytoplasm, the recruitment of cytoplasmic ESCRT components is believed to facilitate the formation of small cytoplasmic vesicles at an early stage of BFRF1 expression (Fig. 2, 6 h posttransfection). In contrast to vesicles derived from cytoplasmic-membrane materials, the reassembly of BFRF1 with ESCRT components on the INM is believed to be essential for nucleocytoplasmic transport of nuclear cargos, as well as viral nucleocapsids. However, further studies are warranted to elucidate how ESCRT proteins were transported onto the INM or whether the same ESCRT proteins participate in BFRF1-mediated transport on both sides of the NE. In terms of ubiquitin-mediated regulation of BFRF1, we found that overexpression of Myc-Ub restored cytoplasmic-vesicle formation, but not NE modulation by BFRF1, in the presence of MG132, suggesting that MG132 may have had pleiotropic effects or affected more than one factor involved in BFRF1 modulation of the NE (Fig. 3B). A more targeted method using DUb-BFRF1 was used to confine the deubiquitination of EBV BFRF1 and its binding partners without affecting the cell. DUb*-BFRF1 induced vesicles (albeit with smaller sizes), while fusion of an active DUb enzyme completely abolished the NE targeting and the vesicle formation of BFRF1 (Fig. 5C), emphasizing the critical role of ubiquitination in the function(s) of EBV BFRF1 at the NE. Of note, DUb-BFRF1 showed a phenotype distinct from that of ubiquitination-defective F1(6KR) (Fig. 4B and 5C), suggesting ubiquitination might control the BFRF1-mediated NE modulation at multiple steps and/or factors, in line with Myc-Ub's ability to only partially restore BFRF1-induced function(s) in MG132-treated cells (Fig. 3B). While the DUb fusion performs both cis- and trans-deubiquitination of BFRF1, the F1(6KR) mutant represents only the cis-ubiquitination scenario. Combining the two approaches will allow further molecular dissection of the role of ubiquitin in BFRF1 modulation of the NE.
The nuclear-membrane-associated components and structural integrity of the NE are known to play important roles in various cellular processes, such as cytoskeleton reorganization and cell cycle progression (39). Recent studies have found that activated Torsin AAA-ATPase complexes play important roles in both nuclear and endoplasmic reticulum membrane-remodeling processes, and Torsin complexes appear to help the formation of nascent vesicles budding from the INM into the perinuclear space (6, 7). It was also reported that the nuclear vesiculation pathway might be responsible for the transport of large cellular or viral cargoes from the nucleus into the cytoplasm (40). Most recently, ESCRT-III and the AAA-ATPase Vps4 were found to destabilize and clear defective NPC assembly intermediates on the NE of Saccharomyces cerevisiae, suggesting novel roles of ESCRT machinery in modifying NE under physiological conditions (8). In mammalian cells, ESCRT-III components are required for the sealing of NE during late mitosis (9, 10); however, it remains unclear how ESCRTs are recruited onto the NE. These observations reveal novel roles of the ESCRT machinery in modifying the NE and also indicate that differential usage of ESCRT components is employed in diverse cellular functions (41). EBV appears to hijack parts of the ESCRT machinery to facilitate the transport of cytoplasmic components toward the nucleus and then remodels the INM to deliver the EBV nucleocapsids back to the cytoplasm, possibly by usurping cellular mechanisms and factors naturally used to load cellular (or viral) cargo back toward the cytoplasm.
In summary, the current study extends our previous findings that EBV BFRF1 alone is capable of recruiting Alix and ESCRTs to remodel the structure of the NE by recruiting the ubiquitin ligases and using ubiquitination to regulate BFRF1-mediated NE modulation. Because ESCRT functions at the NE have just begun to emerge, it will be interesting to use BFRF1 and EBV nuclear modulation of the NE to further elucidate the interplay of ESCRT and ubiquitination there. Furthermore, as NE integrity has been used as an important diagnostic parameter for malignancies, the modulation of the NE by BFRF1 may have implications for the pathogenesis of EBV-associated diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mei-Tzu Su and Yu-Chen Tsang for discussions of experiments and for technical support; Jiing-Guang Chuang at the Graduate Institute of Medical Genomics and Proteomics, National Taiwan University College of Medicine, for proteomic analysis; and Ji-Ying Huang and Yun-Yu Sun at the Image Core Laboratory, National Taiwan University Hospital, and Hua-Man Hsu and Cheng-Yen Huang at the First Core Facility, National Taiwan University College of Medicine, for technical assistance with confocal imaging.
This study was supported by grants NHRI-EX104-10201BI from the National Health Research Institutes and MOST102-2320-B-227-002 and MOST103-2320-B-227-001-MY3 from the Ministry of Science and Technology, Taiwan, and by a National Taiwan University ïntramural grant.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01235-16.
REFERENCES
- 1.Lee CP, Chen MR. 2010. Escape of herpesviruses from the nucleus. Rev Med Virol 20:214–230. doi: 10.1002/rmv.643. [DOI] [PubMed] [Google Scholar]
- 2.Chang CW, Lee CP, Su MT, Tsai CH, Chen MR. 2015. BGLF4 kinase modulates the structure and transport preference of the nuclear pore complex to facilitate nuclear import of Epstein-Barr virus lytic proteins. J Virol 89:1703–1718. doi: 10.1128/JVI.02880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CP, Liu PT, Kung HN, Su MT, Chua HH, Chang YH, Chang CW, Tsai CH, Liu FT, Chen MR. 2012. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLoS Pathog 8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercenne G, Alam SL, Arii J, Lalonde MS, Sundquist WI. 29 January 2015. Angiomotin functions in HIV-1 assembly and budding. eLife 4. doi: 10.7554/eLife.03778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, Moore MJ, Budnik V. 2012. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. 2013. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep 3:988–995. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster BM, Colombi P, Jager J, Lusk CP. 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522:231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 10.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. 2015. ESCRT-III controls nuclear envelope reformation. Nature 522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc Natl Acad Sci U S A 98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699. doi: 10.1016/S0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 13.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A 98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernassola F, Karin M, Ciechanover A, Melino G. 2008. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K. 2004. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23:6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- 16.Winberg G, Matskova L, Chen F, Plant P, Rotin D, Gish G, Ingham R, Ernberg I, Pawson T. 2000. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol Cell Biol 20:8526–8535. doi: 10.1128/MCB.20.22.8526-8535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol 82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Usami Y, Popov S, Popova E, Gottlinger HG. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol 82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y, Tung CH, Huang YT, Lu J, Chen JY, Tsai CH. 1999. Requirement for cell-to-cell contact in Epstein-Barr virus infection of nasopharyngeal carcinoma cells and keratinocytes. J Virol 73:8857–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarisky RT, Gao Z, Lieberman PM, Fixman ED, Hayward GS, Hayward SD. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol 70:8340–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova O, Kamberov E, Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970–972. [DOI] [PubMed] [Google Scholar]
- 22.Sette P, Nagashima K, Piper RC, Bouamr F. 2013. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10:79. doi: 10.1186/1742-4690-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. 2010. Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch. J Virol 84:704–715. doi: 10.1128/JVI.01319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A. 2006. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem 281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- 25.Chuang JG, Su SN, Chiang BL, Lee HJ, Chow LP. 2010. Proteome mining for novel IgE-binding proteins from the German cockroach (Blattella germanica) and allergen profiling of patients. Proteomics 10:3854–3867. doi: 10.1002/pmic.201000348. [DOI] [PubMed] [Google Scholar]
- 26.Wen CL, Chen KY, Chen CT, Chuang JG, Yang PC, Chow LP. 2012. Development of an AlphaLISA assay to quantify serum core-fucosylated E-cadherin as a metastatic lung adenocarcinoma biomarker. J Proteomics 75:3963–3976. doi: 10.1016/j.jprot.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farina A, Feederle R, Raffa S, Gonnella R, Santarelli R, Frati L, Angeloni A, Torrisi MR, Faggioni A, Delecluse HJ. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J Virol 79:3703–3712. doi: 10.1128/JVI.79.6.3703-3712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shields SB, Piper RC. 2011. How ubiquitin functions with ESCRTs. Traffic 12:1306–1317. doi: 10.1111/j.1600-0854.2011.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Bettignies G, Coux O. 2010. Proteasome inhibitors: dozens of molecules and still counting. Biochimie 92:1530–1545. doi: 10.1016/j.biochi.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Garcia MD, Meertens L, Bonazzi M, Cossart P, Arenzana-Seisdedos F, Amara A. 2011. Appraising the roles of CBLL1 and the ubiquitin/proteasome system for flavivirus entry and replication. J Virol 85:2980–2989. doi: 10.1128/JVI.02483-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. 2010. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stringer DK, Piper RC. 2011. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol 192:229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng B, Lever AM. 2013. Wrapping up the bad news: HIV assembly and release. Retrovirology 10:5. doi: 10.1186/1742-4690-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz SE, Rosa JL, Scheffner M. 1998. Characterization of human hect domain family members and their interaction with UbcH5 and UbcH7. J Biol Chem 273:12148–12154. doi: 10.1074/jbc.273.20.12148. [DOI] [PubMed] [Google Scholar]
- 36.Hofemeister H, O'Hare P. 2008. Nuclear pore composition and gating in herpes simplex virus-infected cells. J Virol 82:8392–8399. doi: 10.1128/JVI.00951-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm KS, Klupp BG, Granzow H, Muller FM, Fuchs W, Mettenleiter TC. 2012. Analysis of viral and cellular factors influencing herpesvirus-induced nuclear envelope breakdown. J Virol 86:6512–6521. doi: 10.1128/JVI.00068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maric M, Haugo AC, Dauer W, Johnson D, Roller RJ. 2014. Nuclear envelope breakdown induced by herpes simplex virus type 1 involves the activity of viral fusion proteins. Virology 460-461:128–137. doi: 10.1016/j.virol.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chow KH, Factor RE, Ullman KS. 2012. The nuclear envelope environment and its cancer connections. Nat Rev Cancer 12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCullough J, Sundquist WI. 2014. Putting a finger in the ring. Nat Struct Mol Biol 21:1025–1027. doi: 10.1038/nsmb.2928. [DOI] [PubMed] [Google Scholar]
- 41.Hurley JH. 2015. ESCRTs are everywhere. EMBO J 34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.