Abstract
Although radiation resistance is a common challenge in the clinical treatment of esophageal squamous cell carcinoma (ESCC), an effective treatment strategy has yet to be developed. Aberrant expression of microRNAs (miRNAs) is responsible for cancer sensitivity to radiation. In this study, we aimed to identify the miRNAs that are associated with radioresistance in ESCC. We used a miRNA microarray to perform a comparison of miRNA expression in both ESCC parental and acquired radioresistance cell lines. qRT-PCR was used to confirm the alterations. Cell radiosensitivity was determined with a survival fraction assay. Functional analyses of the identified miRNA in ESCC cells with regard to metastasis and apoptosis were performed by transwell assays and flow cytometry. The miRNA targets were identified with pathway analysis and confirmed with a luciferase assay. miR-98 was recognized as the most downregulated miRNA in established radioresistant cell line. AmiR-98 mimic enforced the expression of miRNA-98 and made ESCC cells sensitive to radiotherapy, while anti-miR-98 reversed this process. Optimal results were achieved by decreasing cellular proliferation, decreasing cell migration and inducing apoptosis. The luciferase target gene analysis results showed that the overexpression of miRNA-98 inhibited tumor growth and resistance tolerance by directly binding to the BCL-2 gene. Our study indicated that increasing miRNA-98 expression can be used as a potential radiosensitive therapeutic strategy for treating esophageal cancer cells.
Keywords: esophageal squamous cell carcinoma, microRNA, radiation, cell line, resistance
INTRODUCTION
Esophageal carcinoma (EC) remains one of the leading cancer-related causes of death, with a 5-year survival rate of <20%. Esophageal cancer is pathologically categorized into two types: esophageal adenocarcinomas and esophageal squamous cell carcinomas (ESCCs). Compared with adenocarcinomas, ESCCs are more prevalent among Asian populations and account for ~95% of all Chinese EC patients [1, 2]. Currently, radiotherapy is recognized as the standard clinical treatment protocol for ESCC throughout the world. Unfortunately, frequent recurrent cancer has been reported due to the presence of an inherently radioresistant cell line found in ESCC patients. Moreover, this tolerance has led to a more aggressive ESCC phenotype [3, 4]. Thus, studying radiation sensitivity, resistance processes, and their underlying molecular mechanisms may ultimately improve clinical outcomes and minimize the risk of recurrence in ESCC patients.
Many studies have discovered that a number of potential therapeutic target genes are involved in acquired radioresistance for various types of cancer [5, 6]. However, the progress of the reaction to cancer radiation treatment is usually regulated by multiple signaling pathways; thus, these results are not entirely affected by the alteration of a single gene [7]. Currently, microRNAs (miRNAs), a class of small non-coding RNA molecules, are becoming a popular research tool in the study of cancer radioresistance, as some miRNAs can simultaneously regulate multiple oncogenic pathways and influence the cell response to radiation [8, 9]. Generally, miRNAs can form miRNA–mRNA complexes, and they also have the ability to either completely or partially block translation, potentially leading to the inhibition of various target genes via mRNA degradation [10]. Because of their ability to bind to multiple targets, miRNAs can have both onco- and anti-onco effects and can regulate many cancer-associated behaviors, such as metastasis, invasion, angiogenesis, and the promotion of a chemoresistant phenotype [11, 12]. Several studies have demonstrated that the survival rates of cancer patients are associated with miRNA expression levels in tissue samples, and this may be involved in developing radioresistance to cancer [13, 14]. Despite research to date, there is little data on the miRNA expression profile of radioresistance in ESCC. Hence, it is necessary to investigate the underlying mechanisms of miRNAs as well as their potential functions.
In this study, we investigated global miRNA expression from established radioresistant esophageal cancer cell lines. Furthermore, we analyzed the miRNA–mRNA interaction in regard to radiation therapy. The goal of this work was to clarify the mechanisms of resistance in esophageal cancer cells as well as to identify specific miRNAs that are present in esophageal cells following radiation treatment.
MATERIAL AND METHODS
Cell lines and cell culture
Six human ESCC cell lines—TE1, ECA109, EC9706, KYSE30, KYSE150 and KYSE450—were purchased from American Type Culture Collection (Manassas, VA, USA). Based on their characteristics and a scholarly literature review, we selected these six cell lines for use in our current study to minimize the known differences that influence radiation response. All of these cell lines have been established in countries marked by a higher incidence of esophageal cancer and were histologically derived from squamous cell carcinomas. Additionally, none of the cell lines that were donated had been exposed to X-ray irradiation prior to surgery [15, 16]. All of these cells lines were maintained in their recommended growth medium and incubated in a humidified 5% CO2 atmosphere at 37°C. Each cell line was regularly observed and tested to avoid possible mycoplasma contamination.
Development of acquired radioresistant cells
The acquired radioresistant cell line used in the current study was developed with continuous 2 Gy fractionated irradiation to a total of 20 Gy, as previously described [17]. EC9706 cells (1 × 106) were plated in 75-cm2 culture flasks, exposed to 6-MV X-rays (2100EX, Varian) and set to a field size of 25 × 25 cm2 with a source-to-surface distance of 100 cm. Cells were moved to the incubator after receiving radiation and were subsequently allowed to achieve ~70% confluence. The exposed flasks were placed between a water incubator and 15-mm-thick histoacryl to ensure uniform exposure to the radiation field. The parental cells were trypsinized, counted and passaged under the same conditions without irradiation. To identify possible genetic changes that occurred during the radioresistance developmental process, we used a short tandem repeat test that included eight core STR markers—D5S818, D13S317, D7S820, D16S539, vWA, TH01, TPOX and CSF1PO—to compare the genetic modification between the parental and acquired resistance cell line.
In vitro X-ray irradiation
Cells (1 × 106 cells/ml) were seeded into six-well plates and incubated at 37°C in a 5% CO2 incubator. X-ray irradiation was performed using 6-MV X-rays (2100EX, Varian) at a dose rate of 300 cGy/min.
Survival fraction assay
Cells were plated on six-well plates at a density of 1 × 106 until reaching 60% confluence. These logarithmic growth cells were exposed to X-rays with irradiation doses of 0, 2, 4, 6, 8 or 10 Gy. Following 14 days of incubation at 37°C, the colonies were stained with Giemsa. Colonies with more than 50 cells were counted. The surviving fraction (SF) was calculated based on the following equation: SF = the number of colonies formed/the total number of cells plated [13]. The colony formation assay was also conducted with transiently transfected cells. This experiment was repeated three times.
RNA extraction and microRNA array
Total RNA was extracted from resistant cells and their corresponding parental cells using an RNA extraction kit (Invitrogen, CA, USA) according to the manufacturer's instructions. The quantity and quality of extracted RNA samples were detected with a Thermo Nano-Drop1000 Spectrophotometer (Thermo, MA, USA) and they were then frozen at −80°C. For miRNA microarray analysis, we chose to use a miRCURYTM LNA Array (Exiqon, Denmark) because this array was designed based on the newly released miRBase version 19.0. The miRNA microarray was supplied and analyzed by KangChen Corporation (Shanghai, China).
Quantitative reverse transcription-polymerase chain reaction
Mature miRNA-98 expression in parental and resistant cells was detected by using TaqMan miRNA assays (Life Technologies, NY, USA) in an ABI 7500 real-time polymerase chain reaction (PCR) system. The RT and PCR primers for miRNA-98 were purchased from Life Science (Life Technologies, NY, USA). The cDNA transcription process was conducted using commercial kits (Takara, Tokyo, Japan). The conditions of the Taqman quantitative reverse transcription-polymerase chain reaction (qRT-PCR) were: 95°C for 10 min, 40 cycles for 15 s at 95°C and 60°C for 60 s. U6 was chosen as an internal control. The relative miRNA levels were calculated using the 2−ΔΔCt method.
Plasmid construction and cell transfection
A miRNA-98 mimic, anti-miR-98 and negative control (Life Technologies, NY, USA) were transfected into EC9706 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) to enhance or inhibit miRNA-98 expression in cells. qRT-PCR was utilized to validate the transfection efficiency of the miRNA-98.
Western blot analysis
Protein was prepared with a lysis buffer by adding protease and phosphatase inhibitors (Thermo Fisher Scientific, Boston, MA, USA) followed by SDS-PAGE separation. Protein was then transferred to nitrocellulose membranes under semi-dry system conditions (BioRad, Hercules, CA, USA). Next, the membranes were incubated with primary antibodies (caspase-3 [1:500 dilution], Bcl-2 [1:500] and β-actin [1:5000]) at 4°C overnight. Following two continuous washes, the membranes were then blocked with secondary antibodies. All of the antibodies were purchased from the Santa Cruz Biotechnology, Inc. Protein expression was visualized with a gel imaging system (Biorad).
Cell apoptosis assays
The cells (1 × 106 cells) were seeded in a flask and allowed to grow to 70% confluence. They then received irradiation treatment (8 Gy); at 8 h after irradiation they were collected (in the form of a single-cell suspension) for apoptosis detection. The apoptotic cell percentage was observed by flow cytometry (BD FACS Calibur, USA) using an annexin V-FITC apoptosis detection kit (Sigma). Only the ratio of early apoptotic cells was counted and considered for the test results. Hochest33324 was used to confirm apoptosis in all of the groups, following the manufacturer's guidance (Sigma).
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA) was used to conduct statistical comparisons between different treatment groups. Data are presented as the mean ± standard error (SEM) and were compared with one-way analysis of variance (ANOVA). P values of <0.05 were considered to be significant.
RESULTS
Determination of experimental ESCC cell lines by comparing the radiosensitivities of a variety of ESCC cancer cells
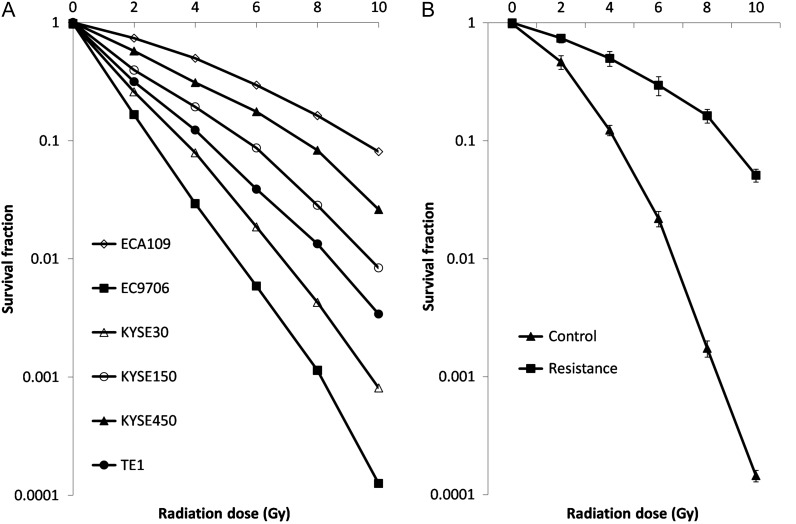
To select the appropriate cell lines for this experiment, survival fractions along with a series of radiation treatments were evaluated in all six ESCC cell lines (TE1, ECA109, EC9706, KYSE30, KYSE150 and KYSE450) to determine their radiosensitivity. The results showed that the radiosensitivity of the six ESCC cell lines in descending order were as follows: EC9706, KYSE30, KYSE450, TE1, ECA109 and KYSE150 (Fig. 1A). As this study focused on the radiotolerance of ESCC, EC9706 was selected as an appropriate candidate because of its high innate radiosensitivity. Acquired resistant cells were developed and named EC9706R. Short tandem repeat test confirmed there was no genetic modification between parent and resistant cell line. This cell line demonstrated significantly enhanced colony formation ability compared with its parent cell line (Fig. 1B). As expected, EC9706R cells also exhibited increased proliferation rates compared with EC9706 cells, indicating that they were more radioresistant than their counterparts.
Fig. 1.
Comparison of the radiosensitivities of the six ESCC cell lines. (A) Cells were exposed to X-rays at different irradiation doses (0, 2, 4, 6, 8 and 10 Gy) for 14 days, and the surviving fraction was evaluated. (B) The developed radioresistant cell line EC9706R demonstrated significantly decreased radiosensitivity compared with its parental cell line.
Comparison of differential miRNA expression profiles between ESCC parental cells and acquired resistance cells
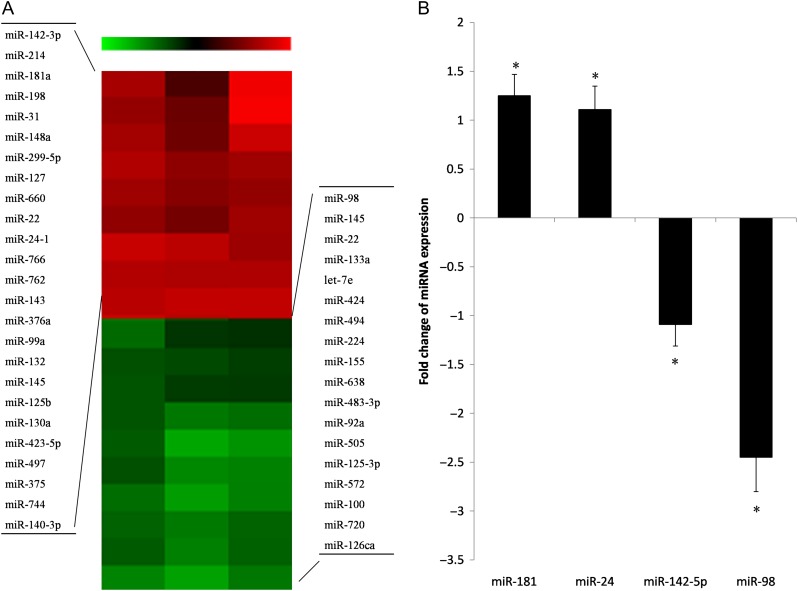
A miRNA expression array indicated that a total of 43 miRNAs were differentially expressed (changed in expression by more than 1.5-fold in both directions) in the resistant cells after radiotherapy compared with the parental cells; they included 29 upregulated and 14 downregulated miRNAs (Fig. 2A). Among them, 8 miRNAs from the upregulation group and 6 miRNAs from the downregulation group had changes in magnitude greater than 2-fold. Additionally, miR-98 showed the most downregulation in the resistant cells, with 4-fold change (Table 1). TaqMan quantitative real-time PCR analysis further confirmed the microarray data in two randomly selected highly upregulated and two randomly selected highly downregulated miRNAs with at least three repeated tests (Fig. 2B).
Fig. 2.
Differential miRNA expression profiles between EC9706 parental and resistant cells. (A) The different expression of miRNAs was determined by a miRNA expression array between EC9706 parental and resistant cells (EC9706R). The red color represents an increased expression level of miRNAs in EC9706R compared with the parental line. The green color represents a decreased expression level of miRNAs compared with the parental line. (B) Four randomly selected differentially expressed miRNAs from the array data were confirmed with qRT-PCR. U6 served as an internal control. Asterisk indicates P < 0.05 compared with parental cells. The results are presented in triplicate.
Table 1.
Differential expression of miRNA in resistance cell line as compared with non-resistant cells
| No. | miRNA name | Mean fold change | P-value |
|---|---|---|---|
| 1 | miR-142-3p | 3.89 | 0.0151 |
| 2 | miR-214 | 3.76 | 0.0069 |
| 3 | miR-181a | 3.68 | 0.0051 |
| 4 | miR-198 | 3.41 | 0.0068 |
| 5 | miR-31 | 2.89 | 0.0394 |
| 6 | miR-148a | 2.75 | 0.0178 |
| 7 | miR-299-5p | 2.67 | 0.0076 |
| 8 | miR-127 | 2.12 | 0.0020 |
| 9 | miR-98 | −5.62 | 0.0096 |
| 10 | miR-145 | −3.51 | 0.0411 |
| 11 | miR-22 | −3.29 | 0.0260 |
| 12 | miR-133a | −3.17 | 0.0145 |
| 13 | Let-7e | −2.95 | 0.0279 |
| 14 | miR-424 | −2.48 | 0.0411 |
Positive and negative fold change scores mean significant downregulation and upregulation, respectively, in resistance cells.
Enhanced miRNA-98 expression increased the radiosensitivity of ESCC cells and suppressed cell migration
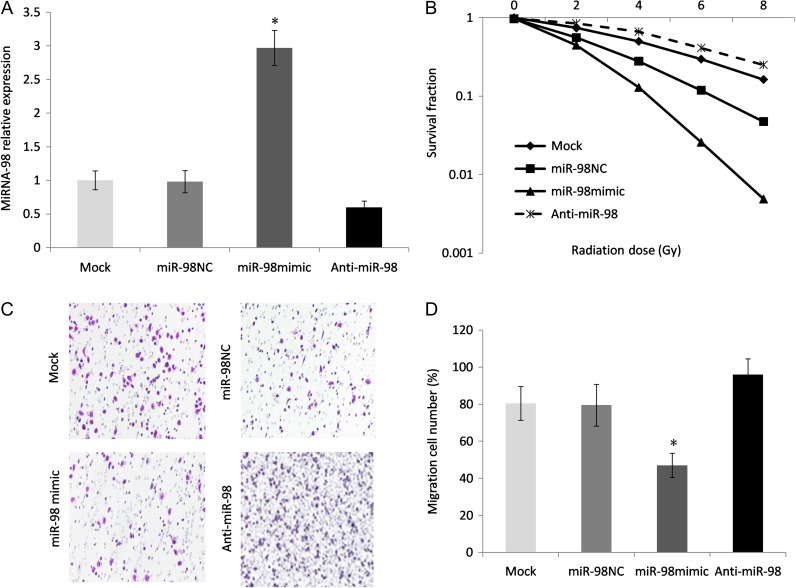
According to microarray results, miR-98 was recognized as the most downregulated miRNA. Next, we transfected the miRNA-98 mimic and anti-miR-98 vector into radioresistant EC9706 cells. The transfection efficiency was demonstrated by qRT-PCR and showed that miRNA-98 was successfully upregulated or downregulated in the cell lines (P < 0.05; Fig. 3A). Radiation colonogenic survival assays were performed after the expression of miRNA-98 had taken place. The miRNA-98 precursor–transfected cells displayed a significantly decreased survival fraction compared with the negative control cells (Fig. 3B), reflecting increased radiosensitivity, while miRNA-98 inhibition reversed this radiosensitivity. In addition, cell transwell assays revealed significantly reduced migration by overexpressing miRNA-98 in EC9706R cell lines (Fig. 3C and D). This provides evidence for the negative feedback functional role of miRNA-98 in the aggression of ESCC cells.
Fig. 3.
Upregulation of miRNA-98 led to increased radiation sensitivity in radioresistant ESCC cells. (A) EC9706R cells were transfected with a miRNA-98 precursor vector, anti-miR-98 or a control vector using Lipofectamine. miRNA-98 expression was verified by qRT-PCR; U6 was chosen as an internal control. Asterisk indicates statistical significance (P < 0.05). (B) Radiation-induced surviving fraction changes were investigated in EC9706R and parental cells that were transfected with anti-miR-98, controls or a miRNA-98 mimic. (C) Cell transwell assays were conducted to explore the influence of miRNA-98 on the migration ability of ES9706R resistant cells. (D) The number of cells that migrate through the transwell membrane.
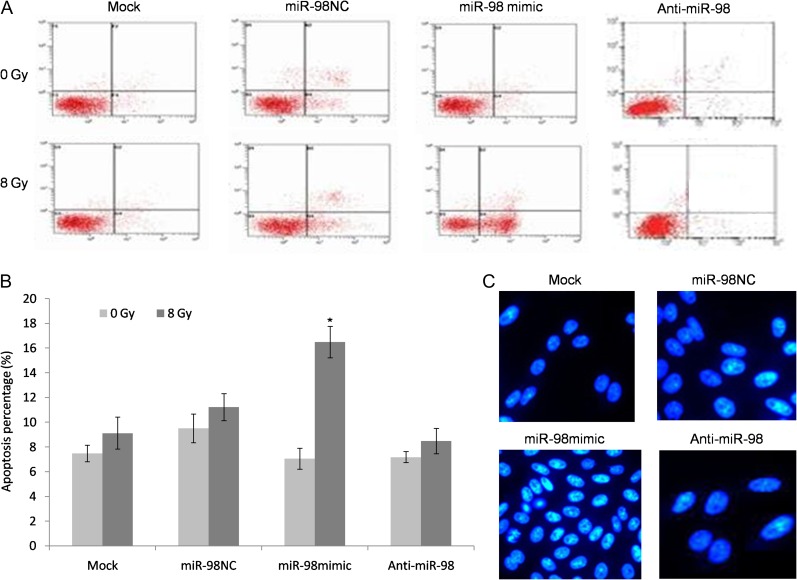
miR-98 promoted X-ray–induced apoptosis in EC9706R cells
The major therapeutic effect induced by radiation is apoptosis. miR-98–induced apoptosis was further investigated in the miR-98 mimic, anti-miR-98 (miR-98 inhibitor) and negative controls. The sublethal dose was calculated based on cell proliferation, as described previously [18], and 8 Gy of radiation was calculated to be the sublethal dose. 8 Gy of radiation was chosen as the standard dose because it not only effectively kills cells, but also leaves a fair number of survivors. Following 48 h of incubation, the cells were exposed to 0 and 8 Gy of radiation and were subjected to Annexin V/PI double staining and FACS analysis. The results showed that miR-98 overexpression significantly increased X-ray–induced apoptosis in cells in contrast to non-transfected cells and negative control mimics. However, cells transfected with anti-miR-98 showed no response to X-ray treatment. Moreover, Hoechst 33342 staining confirmed apoptosis induced by miR-98 expression (Fig. 4).
Fig. 4.
miR-98 promoted X-ray–induced apoptosis of ESCC resistant cells. (A) A flow cytometry assay was used to determine the apoptotic percentage of EC9706R cells following transfection with miR-98 mimics, anti-miR-98 or negative control. (B) Hoechst 3332–stained apoptotic cells. (C) Percentage of apoptotic cells in anti-miR-98, negative controls and miR-98 mimic–treated EC9706R cells before and after 8 Gy irradiation. Data are presented as the means ± SEM. (Asterisk indicates P < 0.05.)
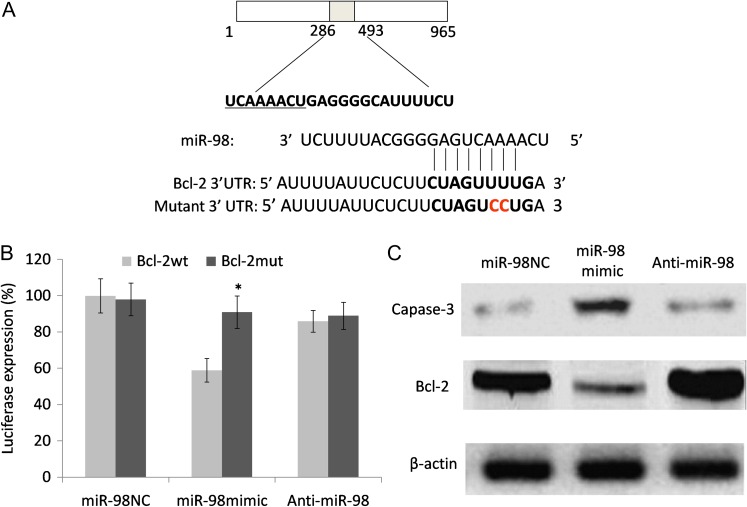
Identification of the direct target of miR-98 and its influence on its target gene
To explore the influence of miR-98 on the development of radioresistance in ESCC, we searched for potential targets of miR-98 using three online prediction tools (miRanda, PicTar and TargetScan) and identified a putative miR-98–binding site at position 56901–575 in the 3′-UTR of the BCL-2 gene. Then, a luciferase reporter was constructed in which BCL-2/3′-UTR nucleotides complementary to miR-98 were inserted into the pLUC vector. Additionally, a mutant reporter was generated in which two nucleotides in the miR-98 seed region's complementary sites were mutated (Fig. 5A). We then cotransfected the appropriate plasmids with either the negative control miR-NC, anti-miR-98 or miR-98 mimics into EC9706R cells to measure luciferase activity. The results showed that miR-98 significantly inhibited luciferase activity in miR-98–transfected cells compared with negative controls and anti-miR-98–transfected cells (Fig. 5B), but there were no observed differences in the activity of the mutation reporter vector. This suggests that miR-98 directly interacts with the 3′-UTR of the BCL-2 gene. Furthermore, the expression of pro-apoptotic protein caspase-3 and anti-apoptotic protein Bcl-2 was determined via western blot. At 48 h after transfection, the cells were subjected to 8 Gy radiation and collected 48 h after irradiation. As shown in Fig. 5C, miR-98 increased the expression of caspase-3 and decreased the expression of Bcl-2 in radiation-resistant cells. In contrast, miR-98 inhibition had no such effect on the caspase-3 and Bcl-2 protein levels. Overall, these results demonstrated that miR-98 enhanced X-ray irradiation–induced apoptosis in esophageal cells by regulating the expression of apoptosis-related proteins.
Fig. 5.
miR-98 binds to the Bcl-2 gene and influences the Bcl-2 expression level. (A) A luciferase reporter system vector was constructed that included a wild-type 3′-UTR fragment of Bcl-2 or a mutant miR-98–binding sequence. (B) Luciferase activity was detected in cells that were cotransfected with both a luciferase reporter system and either miR-98, anti-miR-98 or miR-NC. (C) Following 8 Gy X-ray irradiation, apoptotic-related proteins caspase-3 and Bcl-2 from different treatments of ESCC cells were evaluated via western blot analysis. β-actin was used as an internal control. Each experiment was performed at least in triplicate. Asterisk indicates statistical significance (P < 0.05) compared with the negative control.
DISCUSSION
In the present study, we successfully established radioresistance cells in the selected ESCC cell line and found differential expression profiles for miRNAs. miRNA-98 was identified as being the most downregulated miRNA and was associated with ESCC radiosensitivity. In vitro functional studies further demonstrated that the overexpression of miRNA-98 increased the radiosensitivity of ESCC cells, promoted cell apoptosis, and inhibited cell migration ability. The overexpression of miRNA-98 conferred radiosensitivity and increased inhibition in radioresistant ESCC cells by targeting the Bcl-2 pathway. For years, radioresistance has been a difficult clinical challenge for researchers treating ESCC [19, 20] because the process of clinically acquired radioresistance is complex and involves multiple molecular mechanisms [21]. Recently, miRNAs that have been functionally analyzed in tumors were found to be associated with various malignant tumor cell behaviors, including radioresistance [22–24]. These results are encouraging and require further research to investigate the role of miRNA expression in ESCC radioresistance [11, 19].
Among the significant upregulated and downregulated miRNAs in this study, some have been previously reported to be associated with ESCC progress and clinical prognosis. miR-142-3p [25], miR-181a [26], miR-214-3p [27] and miR-198 [28] were reported to be associated with poor prognosis and an advanced tumor-node-metastasis (TNM) stage in ESCC patients. While miR-22 expression was found to inhibit ESCC and was responsible for inhibiting cell migration and invasion, miR-145a and miR-133a were reported to be downregulated in ESCC patients. Moreover, miR-133a was found to be potentially associated with chemotherapy resistance [29–31]. In addition, several population-based research studies reported a correlation between miRNAs and clinical outcomes in esophageal cancer. Liu et al. [32] reported that altered expression of miR-143 and miR-145 was a risk factor for esophageal cancer patients. Another study found significant upregulation of miRNA-25 in 60 ESCC tissues samples, compared with the matched adjacent non-cancerous normal tissues, and this upregulation was also revealed to significantly contribute to the worsened status of lymph node metastasis and advanced TNM status [33]. The study also determined the aggressive migration and invasion ability of ESCC cells. Kurashiqe et al. [34] observed significantly elevated serum levels of miRNA-21 in ESCC patients, in contrast to the corresponding healthy controls. Moreover, a significant reduction in serum miRNA-21 levels in post-operative samples compared with pre-operative samples was observed in the same study. Other miRNAs, such as miRNA-223 and let-7, were also found to have an effect on the progress of ESCC [35, 36].
The results from several other groups have confirmed the involvement of ~45 miRNAs in the radioresistance of human cancers [10]. In esophageal carcinomas, only three miRNAs, including miRNA-21, miRNA-22 and miRNA-31, were found to contribute to radioresistance by targeting different downstream genes. Su et al. [11] recently found a set of miRNAs that were aberrantly expressed in acquired radioresistant ESCC cells and identified miRNA-301a as a promoter in the sensibilization of radiotherapy through the Wnt/β-catenin signaling pathway. These previous reports indicate the role of miRNAs in the radioresistance of ESCC. miR-98 is a member of the let-7 family, which has been well documented as regulating many cellular biological responses, including cell migration and apoptosis [37, 38]. miR-98 is also involved in the RAS oncogene pathway, and its expression has been found to be decreased in various types of tumors [37, 38]. Previous studies have reported the association between the downregulation of let-7 and an unfavorable clinical outcome in lung cancer patients [39, 40]. These studies have also shown that IR-induced inhibition of let-7a and let-7b expression is closely related to both the p53 and ATM signaling pathways [39, 40].
Through miRNA target database research, KEGG pathway analysis and a literature review, multiple target genes were identified along with miRNA-98, including the TGF-β signaling pathway and the NF-kappa B signaling pathway, two well-known cancer-related regulation pathways. The observed changes in the cell migration ability in radioresistant cell lines may involve these two pathways, which influence the cell epithelial–mesenchymal transition process following long-term radiation treatment. It is commonly accepted that ionizing radiation leads to DNA damage. In the meantime, a DNA damage signal could trigger a cell cycle checkpoint and lead to either successful DNA repair or cell death [5–7]. Thus, we only focused on apoptosis genes. Based on the differential expression profiles of the miRNAs that were associated with the radioresistance of ESCC, we confirmed the important roles of miRNA in radioresistance and revealed that inhibition of miRNA-98 may promote radioresistance of ESCC in vitro, possibly by directly targeting Bcl-2 protein expression.
One weakness of our study was that the initial screening of the miRNAs used in the current study did not evaluate the full list of miRNAs that have been used in cancer research. This means that some existing unprofiled miRNAs may provide more value in clinical application than those examined here. Our current study showed that the reduced expression of miRNA-98 in ESCC radioresistance cells and the ectopic overexpression of miR-98 was able to sensitize ESCC cells to radiation treatment. We also characterized the functions of miRNA-98 in tumor progression and showed that it negatively regulated cell migration, invasion and apoptosis by targeting the anti-apoptosis gene Bcl-2. Thus, we identified a novel microRNA that acts as a key regulator of radiosensitivity in ESCC.
ACKNOWLEDGEMENTS
We thank all of the researchers who participated in this study.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, He Y, Zheng R, et al. Esophageal cancer incidence and mortality in China, 2009. J Thorac Dis 2013;5:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JJ, Tannock IF.. Repopulation of cancer cells during therapy: an important cause of treatment failure. Nat Rev Cancer 2005;5:516–25. [DOI] [PubMed] [Google Scholar]
- 4.Zhou S, Ye W, Ren J, et al. MicroRNA-381 increases radiosensitivity in esophageal squamous cell carcinoma. Am J Cancer Res 2014;5:267–77. [PMC free article] [PubMed] [Google Scholar]
- 5.Hein AL, Ouellette MM, Yan Y.. Radiation-induced signaling pathways that promote cancer cell survival (review). Int J Oncol 2014;45:1813–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rycaj K, Tang DG.. Cancer stem cells and radioresistance. Int J Radiat Biol 2014;90:615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vordermark D. Ten years of progress in radiation oncology. BMC Cancer 2011;11:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su H, Jin X, Zhang X, et al. Identification of microRNAs involved in the radioresistance of esophageal cancer cells. Cell Biol Int 2014;38:318–25. [DOI] [PubMed] [Google Scholar]
- 9.Zhao L, Bode AM, Cao Y, et al. Regulatory mechanisms and clinical perspectives of miRNA in tumor radiosensitivity. Carcinogenesis 2012;33:2220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balça-Silva J, Sousa Neves S, Gonçalves AC, et al. Effect of miR-34b overexpression on the radiosensitivity of non–small cell lung cancer cell lines. Anticancer Res 2012;32:1603–9. [PubMed] [Google Scholar]
- 11.Asuthkar S, Velpula KK, Chetty C, et al. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget 2012;3:1439–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Wang H, Ng WL, et al. Radiosensitizing effects of ectopic miR-101 on non–small cell lung cancer cells depend on the endogenous miR-101 level. Int J Radiat Oncol Biol Phys 2011;81:1524–9. [DOI] [PubMed] [Google Scholar]
- 13.Wang XC, Wang W, Zhang ZB, et al. Overexpression of miRNA-21 promotes radiation resistance of non–small cell lung cancer. Radiat Oncol 2013;8:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Liu GL, Liu SH, et al. MicroRNA-148b enhances the radiosensitivity of non-Hodgkins lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res 2012;53:516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishihira T, Hashimoto Y, Katayama M, et al. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol 1993;119:441–9. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z. The identification, regulation and function study of the differentially expressed genes in human esophageal squamous cell carcinoma In: Kinner HK. (ed.) Esophageal Cancer Research Developments. New York: Nova Science Publisher Inc., 2007: 1–97. [Google Scholar]
- 17.Xie L, Song X, Yu J, et al. Fractionated irradiation induced radio-resistant esophageal cancer EC109 cells seem to be more sensitive to chemotherapeutic drugs. J Exp Clin Cancer Res 2009;28:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Liu F, Sun Z, et al. The enhancement of radiosensitivity in human esophageal carcinoma cells by thalidomide and its potential mechanism. Cancer Biother Radiopharm 2011;26:219–27. [DOI] [PubMed] [Google Scholar]
- 19.Wang XC, Zhang ZB, Wang YY, et al. Increased miRNA-22 expression sensitizes esophageal squamous cell carcinoma to irradiation. J Radiat Res 2013;54:401–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li HZ, Gao XS, Xiong W, et al. Identification of differentially expressed genes related to radioresistance of human esophageal cancer cells. Chin J Cancer 2010;29:882–8. [DOI] [PubMed] [Google Scholar]
- 21.Huber SM, Butz L, Stegen B, et al. Ionizing radiation, ion transports, and radioresistance of cancer cells. Front Physiol 2013;4:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John-Aryankalayil M, Palayoor ST, Makinde AY, et al. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res 2012;178:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babar IA, Czochor J, Steinmetz A, et al. Inhibition of hypoxia-induced miR-155 radiosensitizes hypoxic lung cancer cells. Cancer Biol Ther 2011;12:908–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi H, Liang B, Jia J, et al. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett 2013;587:436–43. [DOI] [PubMed] [Google Scholar]
- 25.Lin RJ, Xiao DW, Liao LD, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol 2012;105:175–82. [DOI] [PubMed] [Google Scholar]
- 26.Xiang Z, Dong X, Sun Q, et al. Clinical significance of up-regulated miR-181a in prognosis and progression of esophageal cancer. Acta Biochim Biophys Sin (Shanghai) 2014;46:1007–10. [DOI] [PubMed] [Google Scholar]
- 27.Phatak P, Byrnes KA, Mansour D, et al. Overexpression of miR-214-3p in esophageal squamous cancer cells enhances sensitivity to cisplatin by targeting survivin directly and indirectly through CUG-BP1. Oncogene 2016;35:2087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi B, Yao WJ, Zhao BS.. Involvement of microRNA-198 overexpression in the poor prognosis of esophageal cancer. Asian Pac J Cancer Prev 2013;14:5073–6. [DOI] [PubMed] [Google Scholar]
- 29.Chen G, Peng J, Zhu W, et al. Combined downregulation of microRNA-133a and microRNA-133b predicts chemosensitivity of patients with esophageal squamous cell carcinoma undergoing paclitaxel-based chemotherapy. Med Oncol 2014;31:263. [DOI] [PubMed] [Google Scholar]
- 30.Wu BL, Xu LY, Du ZP.. MiRNA profile in esophageal squamous cell carcinoma: downregulation of miR-143 and miR-145. World J Gastroenterol 2011;17:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C, Ning SQ, Li ZY, et al. miR-22 is down-regulated in esophageal squamous cell carcinoma and inhibits cell migration and invasion. Cancer Cell Int 2014;14:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Liao J, Yang M, et al. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS One 2012;7:e33987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X, Chen Z, Zhao X, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun 2012;421:640–5. [DOI] [PubMed] [Google Scholar]
- 34.Kurashige J, Kamohara H, Watanabe M, et al. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol 2012;106:188–92. [DOI] [PubMed] [Google Scholar]
- 35.Kurashige J, Watanabe M, Iwatsuki M, et al. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer 2012;106:182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimura K, Miyata H, Tanaka K, et al. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res 2012;18:5144–53. [DOI] [PubMed] [Google Scholar]
- 37.Huang SD, Yuan Y, Zhuang CW, et al. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer 2012;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siragam V, Rutnam ZJ, Yang W, et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting active in receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget 2012;3:1370–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Yuan C, Lv K, et al. Lin28 mediates radiation resistance of breast cancer cells via regulation of caspase, H2AX and Let-7 signaling. PLoS One 2013;8:e67373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickey JS, Zemp FJ, Martin OA, et al. The role of miRNA in the direct and indirect effects of ionizing radiation. Radiat Environ Biophys 2011;50:491–9. [DOI] [PubMed] [Google Scholar]