Abstract
Human body sites represent ecological niches for microorganisms, each providing variations in microbial exposure, nutrient availability, microbial competition, and host immunological responses. In this study, we investigated the oral, anal, and cervical microbiomes from the same 20 sexually active adolescent females, using culture-independent, next-generation sequencing. DNA from each sample was amplified for the bacterial 16S rRNA gene and sequenced on an Illumina platform using paired-end reads. Across the three anatomical niches, we found significant differences in bacterial community composition and diversity. Overall anal samples were dominated with Prevotella and Bacteriodes, oral samples with Streptococcus and Prevotella, and cervical samples with Lactobacillus. The microbiomes of a few cervical samples clustered with anal samples in weighted principal coordinate analyses, due in part to a higher proportion of Prevotella in those samples. Additionally, cervical samples had the lowest alpha diversity. Our results demonstrate the occurrence of distinct microbial communities across body sites within the same individual.
Keywords: human microbiome, cervical microbiome, anal microbiome, oral microbiome, metacommunity theory, massively-parallel sequencing, next-generation sequencing
Introduction
The mucosal epithelia of the mouth, anus, and cervix comprise three ecological niches (or “patches”, in the language of metacommunity theory [1,2]) with the potential for highly variable exposure to microbial sources. In particular, these sites are frequently involved in sexual contact between individuals. In addition, potentially influential factors include frequency of exposure from the external environment, bacterial strains to which exposure occurs, and species selection due to nutrient availability, inter-microbial competition or immune system functions. All of these factors may vary across sites and between individuals. In addition, dispersal of organisms between these and other corporal niches most likely occurs and may vary between sites and individuals, especially during sexual contact. The extent to which any of these factors determine the local dynamics of these ecosystems or the global dynamics of the body’s meta-ecosystem remains unknown. Furthermore, all three are body sites for which disease states are common and can be serious. Oral, anal, and cervical cancer have all been found to have increased risk due to infections with human papillomaviruses (HPV), however associations with host bacteria are not yet clear [3,4]. Apart from some well-known pathogens (e.g. Helicobacter pylori), interactions between microbial communities and pathogenesis remain poorly understood. There may also be forensic value in understanding inter-personal and inter-site variability for anal, oral, and cervical microbiotas. Previous studies have characterized the cervical [3] and oral microbiomes [5-7], but few have characterized the anal microbiome [8] using modern culture-independent molecular techniques, to the best of our knowledge. Furthermore, apart from one report [9], there have been no analyses of microbes from all three niches from the same cohort of individuals.
Here, we present the oral, anal, and cervical microbiotas from a cohort of 20 sexually active adolescent women using culture-independent, next generation sequencing. The resultant bacterial communities were analyzed for variability across sites and individuals, and their compositions interpreted in relation to what is already know about human microbial niches.
Methods
Paired cervical, anal, and oral specimens from 20 women were used for this investigation and were selected from a cohort study of adolescent women conducted at the Mount Sinai Adolescent Health Center in New York [10]. Cervical cells were collected using an endocervical Cytobrush® placed in PreservCyt transport medium (ThinPrep®, Hologic, MA) medium. Anal cells were also collected in PreservCyt using a Dacron swab moistened in tap water. Oral cell samples were collected by oral rinse and gargle using a Scope® mouthwash (Proctor & Gamble, OH). Specimens were stored at -20ºC immediately following collection.
Written informed consent, approved by the Institutional Review Board at the Icahn School of Medicine at Mount Sinai, was collected from all subjects. Subjects were eligible to participate if they: 1) were between 12 to 19 years of age at time of consent, 2) had previously engaged in vaginal or anal intercourse, and 3) intended to get or had already received the quadrivalent HPV vaccine (Gardasil®). Potential participants who were pregnant at time of enrollment or who had either given birth or terminated a pregnancy within the prior 4 to 6 weeks were excluded.
DNA Extraction, Amplification, and Sequencing
An aliquot of each sample was incubated with a proteinase K and sodium laureth-12 sulfate solution and DNA was then precipitated in a 0.825 M ammonium acetate/ethanol (AAE) solution, pelleted by centrifugation and resuspended in TE, as described previously [11,12]. Samples were PCR amplified using primers to an approximately 145 bp region spanning the V6 region of the bacterial 16S ribosomal RNA gene, using “universal” bacterial/archael primers (target primer sequences obtained from [13]). A unique 8 bp Hamming DNA barcode [14] was introduced to the PCR amplicons from each sample by the forward PCR primers [3]. Successful amplification of the predicted fragment size was confirmed and amplicon concentration estimated by relative band brightness against a control using gel electrophoresis [15].
Massively Parallel Sequencing
Prior to sending samples for NGS, barcoded PCR products from all clinical samples were pooled at approximately equimolar DNA concentrations and run on an agarose gel. The correct sized band was excised, the DNA was electroeluted, precipitated in ethanol and resuspended in TE buffer as previously described [15]. An aliquot of pooled, purified, barcoded DNA amplicons was sequenced on an Illumina HiSeq (Illumina Inc., San Diego, CA, USA) by the Epigenomics and Genomics Core Facility, Albert Einstein College of Medicine (Bronx, NY, USA) using 100 bp paired-end reads.
Bioinformatics
Illumina reads were demultiplexed using the unique sample barcode with novobarcode V1.00 (Novocraft Technologies Sdn Bhd). The 3’ end of the demultiplexed reads was trimmed with PrinSeq- lite V0.20.4 [16] to remove bases that had a PHRED quality score below 25. Processed reads were then merged with PANDASEQ 2.8.1 [17] if their overlap threshold exceeded 0.6. Reads were then quality filtered by using the usearch quality-filtering pipeline in QIIME 1.9 [18]: reads were sorted by length, de-replicated to ensure that only unique sequences were analyzed, sorted by abundance, and chimeras were removed by the implementation of usearch in QIIME. Taxonomy was assigned to the operational taxonomic unit (OTU) clusters with uclust using a custom database. This custom database was constructed using the 13.8 release of Greengenes [19], the Human Oral Microbiome Database (HOMD) [20] and also included species in the Vaginal Microbiome Consortium (VMC) [21]. Once created the database was pruned for redundancy in order to ensure that there were no sequences with sequence similarity of 97 percent or greater. The merging of these databases allowed for the identification of clinically relevant species like Gardnerella vaginalis. Using this database, we employed R 3.2.1 [22] with the Phyloseq package V1.12.2 in order to parse the QIIME generated biom table.
Statistical and Comparative Analyses
Statistical analyses were performed in R version 3.2.1 [22]. The phyloseq [23] R package was used to calculate the Shannon index rarefaction curves, with 1000 iterations of 100 steps between the smallest and largest sample read numbers, in order to determine the minimum sample read numbers for downstream analyses. Phyloseq was also used to compute the Simpson diversity index for all samples. Abundance based analyses were performed by rarefying the samples to an even depth (1,000 observations in this case) and aggregating the OTU counts for OTUs that were assigned the same genus level taxonomy. Error for these analyses was determined by calculating the median absolute deviation, a robust measure of variability in a sample, with R stat package’s mad function. The clustering for the heatmap was performed using the relative abundances of the samples in the Hierarchical cluster function, hclust, from the stat package using Euclidean distances and the ward.D2 clustering function. The plotting was performed with the heatmap.2 package. Plots for the boxplot abundance figures were constructed using ggplot2. Alpha diversity for the three body sites was analyzed by calculating the Simpson’s diversity index, which takes into account both the abundance and evenness of present OTUs in each sample, using the richness function from phyloseq. The cor.test function from the R stats package was used to perform a Spearman rank correlation on the Simpson’s alpha diversity indices between the various sites. Values were paired if the sample came from a different body site from the same individual. β-diversity was determined using Unifrac distance matrices and visualized using principle coordinate analysis (PCoA) plots with the phyloseq package. Pairwise significance between individual OTU abundances was calculated using the t.test function in R. The adonis function from vegan package V2.3 [24] was used in order to perform permutational multivariate analysis of variance calculations on the Unifrac distance matrices to determine whether there was significant site-specific variation based on phylogenic diversity. Weighted and unweighted PCoA plots were made with phyloseq and ggplot2 to show the extent to which abundance of unique taxa affected site clustering.
Results
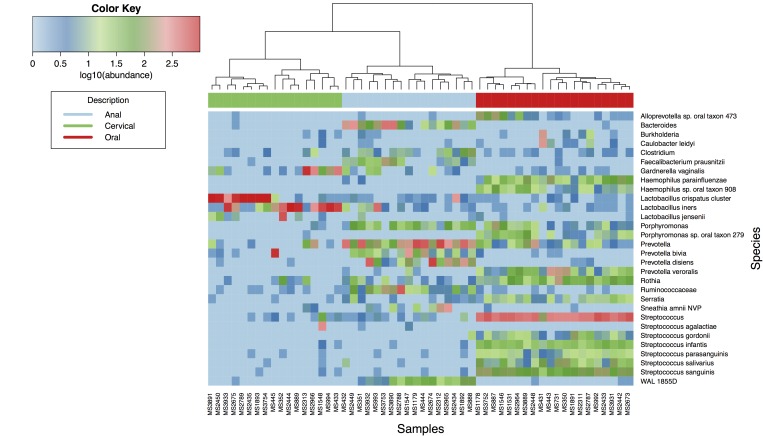
After quality filtering, a total of 286,103 reads were clustered into 2,422 OTUs with sequence similarity of 97 percent or less. This amounted to an average of 4,768 ± 3,207 reads per sample. Figure 1 shows the hierarchical clustering based on the 60 most prevalent OTUs across all samples. The information presented includes 20 oral, 17 anal, and 17 cervical samples that had sufficient read counts for analysis. A total of six samples yielded insufficient reads ( < 1,000) to adequately represent their respective microbiotas, based on Shannon rarefaction curves. Rarefaction analysis of counts assigned at the genus level for each anatomic site suggested that we had sampled sufficiently to capture the majority of bacterial genera expected in each site (data not shown). While a per-sample rarefaction analysis (data not shown) suggested that some specimens contained an underestimate of the true number of genera present, the narrow width and long tail of the community distributions and their similarity to more deeply sampled specimens suggested that the proportions shown in Figure 1 are sufficiently accurate to be compared.
Figure 1.
Heatmap and hierarchical clustering showing the microbiotas of subjects sampled from three anatomical niches (anal, cervical, and oral). Each column represents a sample with the niche shown by the colored bar across the top. Each row represents an operational taxonomic unit; where possible, reads were assigned down to the level of species. When a higher order taxon is named, the quantity of reads attributed to it is all those that could be mapped to that particular taxonomic level and not lower, i.e., reads that did map to lower taxa are not counted in the row corresponding to the parent taxon. The cladogram shown at the top represents hierarchical clustering.
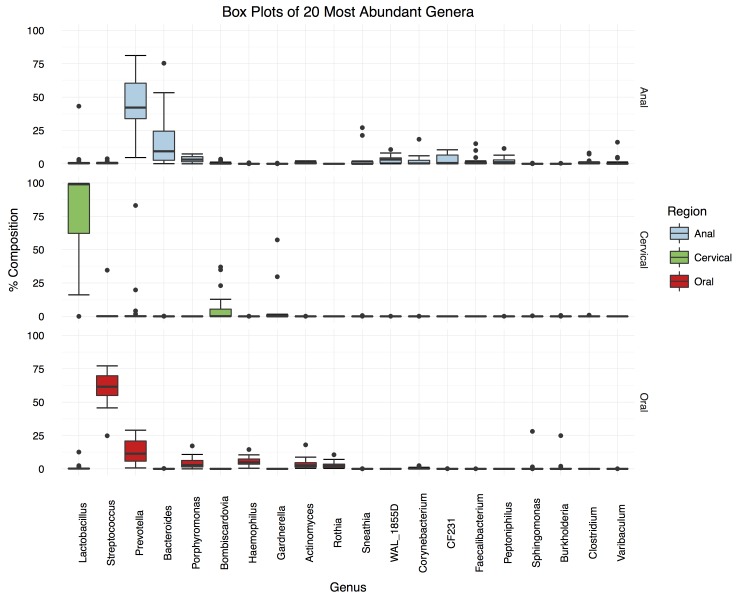
From the genus-level community distribution boxplots (Figure 2, top panel), it was observed that anal microbiotas were dominated by the genera Prevotella (median proportional abundance of 42.2 percent ± 24.2 percent) and Bacteroides (18.5 percent ± 11.6 percent), with other genera (WAL_1855D, Porphyromonas, Peptoniphilus, Clostridium, CF231, Faecalibacterium, Actinomyces) constituting proportions greater than a median of 1.0 percent. Prevotella and Bacteroides are closely related gram-negative, non-motile, biofilm-forming genera, considered normal constituents of the mouth, gastrointestinal and female reproductive tracts. Certain species (e.g. Bacteroides fragilis, Prevotella melaninogenica and Porphyromonas spp.) have been associated with perirectal abscesses [25], though it’s unclear whether this association is simply due to the co-occurrence of the pathogens with the normal anal flora or whether certain components of the normal flora can, under certain circumstances become opportunistic pathogens of the epidermis. Two of the three global gastrointestinal enterotypes previously identified [26], have as their main constituents Prevotella and Bacteroides, respectively though none apparently exhibit a mixture of the two, such as that seen here.
Figure 2.
Boxplots showing the distributions of proportions of the 20 most common genera observed at each anatomical niche across subjects. OTUs were condensed based on shared genus designation and plotted based on relative abundance within each niche. The genera were arranged in accordance to the sample sums across all niches to highlight the most prevalent representatives at each site. The median absolute deviation was used to estimate the error between samples and shown as box-and-whisker plots.
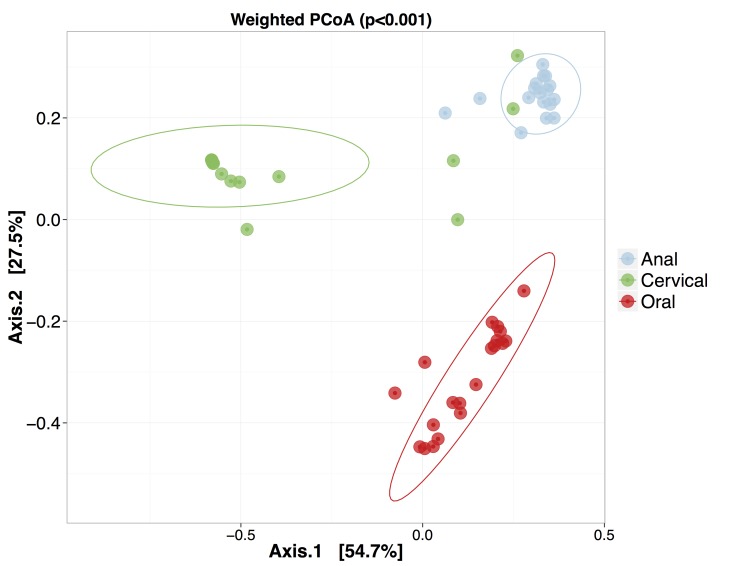
A weighted and an unweighted principal coordinate analysis on the matrix of pairwise Euclidean distances between all samples, coupled with a permutational multivariate calculation using physiological niche as the factor (Figure 3), indicated that statistically significant differences exist among the three sites (p < 0.001). It is also interesting to note that the cervical samples show a noticeably greater interpersonal variation than the other two sites. As also apparent from the clustering in Figure 1, some overlap occurs between anal and cervical microbiotas due to the presence of Prevotella spp. The pairwise comparison between anal and cervical clusters was not significantly different (p = 0.2), however, cervical with oral, or anal with oral were significantly different (p ≤ 0.001).
Figure 3.
Weighted Principal Coordinate Analysis using Unifrac Distances. The weighted unifrac distances were used to construct a weighted principle coordinate plot. The Adonis package was used to perform PERMANOVA statistical tests between each site medoids. The resulting p-value is shown at the top of the plot. Statistical ellipses represent 95 percent confidence of enclosing all samples.
The oral microbiotas were dominated by the genera Streptococcus (median proportional abundance of 61.6 percent ± 11.9 percent) and Prevotella (11.4 percent ± 10.7 percent), with Haemophilus, Porphyromonas, Actinomyces, and Rothia constituting medians of 4.9 percent, 2.9 percent, 2.6 percent and 2.3 percent, respectively (Figure 2, lower panel). In the mouth, Streptococcus intermedius, Prevotella intermedia, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans have long been associated with periodontal pathogenesis [27]. In addition, Group A Streptococci frequently cause throat infections, particularly in children, while, Group B Streptococci are a particular risk to neonates [28].
Cervical microbiotas (Figure 2, middle panel) were mostly in close agreement with previously observed cervical microbiotas, despite the cohort having different geographical and ethnic origins and coming from a different age group [3,29]. A total of 14 out of 17 samples were dominated by Lactobacillus (median proportional abundance of 99.0 percent ± 1.0 percent), which consisted of L. iners and L. crispatus at the species level, corresponding to community types III and I, respectively [3,29]. One sample (MS2313) was dominated by Gardnerella vaginalis (57.3 percent – community type IV), but apparently had sufficient Prevotella spp. to cluster with the anal samples. The remaining two samples (MS445 and MS2966) contained large proportions of Prevotella (83.1 percent and 19.8 percent respectively), Lactobacillus (21 percent, 29 percent, and 13 percent, respectively) and one (MS2966) also contained a large proportion of Gardnerella (29.7 percent). These latter three samples were unlike previously characterized cervical community types due to their large Prevotella component [3,29].
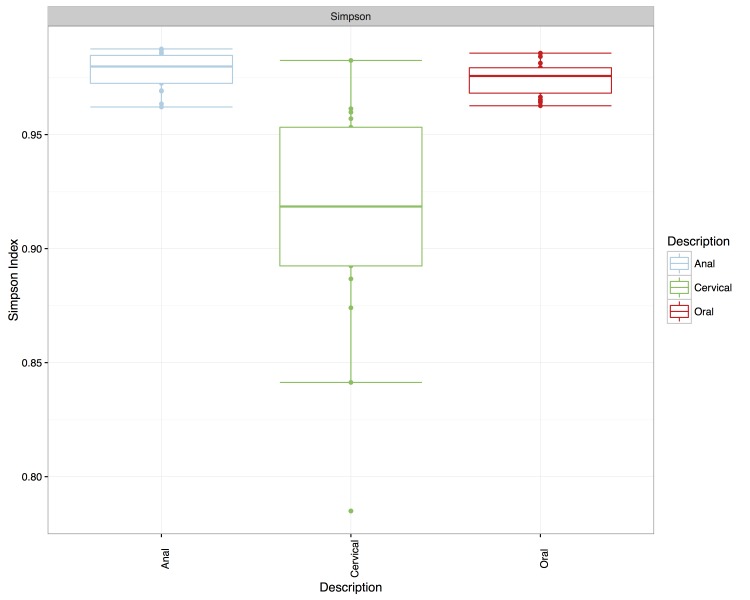
Anal samples exhibited the greatest alpha diversity (Simpson diversity index of 0.980 ± 0.00824), followed by the oral samples (Simpson diversity index of 0.976 ± 0.0113), and the cervical samples having the lowest diversity (Simpson diversity index of 0.917 ± 0.0540) (Figure 4). A Spearman rank correlation was performed on the Euclidean distances of samples from each site relative to the samples from a reference subject at the same site. Samples were paired according to subject and each pairwise comparison between sites was performed. This tests if the degree of similarity between the microbiotas of two subjects is independent across niches. In other words, if the microbiotas of two subjects are similar in one niche, are they also similar in another niche. The results indicated they were not; there was no correlation with an absolute value greater than 0.2 and none of the values was statistically significant (p > 0.90 in all cases) (data not shown).
Figure 4.
Boxplot of the Simpson diversity indices for each anatomical niche. Simpson diversity was estimated at each niche using the richness function in phyloseq. The median absolute deviation was used to construct the whiskers and highlight the richness spread at each site.
Conclusions
The bacterial communities inhabiting mucosal epithelia of the mouth, anus and cervix were sampled at the same visit in each of 20 sexually active adolescent women and characterized using 16S rRNA next- generation sequencing. A direct comparison across the three anatomical niches was possible for 16 of those participants and revealed significant variability both across sites and across patients. The group of oral microbiotas was significantly different form anal and cervical samples, whereas four of the cervical samples clustered with the anal samples, preventing the two groups (anal and cervical) from appearing statistically significantly different. The mediods of the cervical and anal samples on a principal component clustering were, however, at least as far apart as either was from that of the oral group. The most ecologically diverse niche was found to be the anal, followed by oral and then cervical, with cervical communities in most individuals exhibiting strong dominance by a single, but variable species, which is in agreement with previous findings in the majority of women [3,29,30].
The exception to cervical samples dominated by a single species is the group of four samples showing co-dominance by Gardnerella and Prevotella spp. Since anal samples were observed to have high proportions of Prevotella spp., the possibility of cross-sample contamination during collection is one possibility, however, given the stringency of the clinical protocols, another explanation is species dispersal from the anal to the cervical niche, for example, via sexual intercourse. As this community type appears rarely, if this was the case a dispersal event may have had occurred recently and subsequent environmental selection would revert the community to a more common type over time. To confirm a process of dispersal followed by selection, cross-sample contamination would first need to be ruled out, then time-resolved data spanning a time-period before and after the dispersal event would need to be analyzed. However, it is only among the Prevotella genus that these samples showed similarity to the anal samples – they didn’t for example contain Bacteroides. Furthermore, they appeared to have different distributions of Prevotella spp. than any of the anal samples (Figure 1). These factors indicate yet another possible explanation; that they may be of a distinct cervical microbiome community type that supports Prevotella spp. The co-occurrence of BVAB1 (bacterial vaginosis-associated bacteria 1) at relatively high levels in three of these four samples may suggest a state of bacterial dysbiosis [9] in these subjects, and therefore may also explain why they do not cluster with the more common cervical microbiome community types.
This study has strengths and limitations. The study of ecological niches frequently involved in sexual contact allowed us to compare across these sites in a number of young women. Nevertheless, the study was limited by the number of subjects studied and the reliance of using a single region of the 16S rRNA gene to characterize the microbiota.
In summary, the microbiotas inhabiting the mucosal epithelia of the mouth, anus, and cervix vary greatly across niches within the same individual and can vary, to a lesser extent, between individuals. Among the 16 subjects for which direct comparison was possible, there was no evidence to suggest that similarity between the microbiotas from two individuals for one niche ensures a similar degree of similarity between their microbiotas in another niche; a Spearman rank correlation of Euclidean distances showed no statistically significant correlations. Environmental contribution (e.g., eating, breathing) and dispersal (e.g., digestion, sexual intercourse) of microorganisms all occur for each of these sites, with some regularity. These results therefore could indicate strong environmental selection of species through factors specific to anatomical niches that are relatively similar across individuals.However, in order to understand the dynamics of bacterial ecosystems, and thereby discover how best to manipulate microbiota for treating and preventing disease, it is essential for studies to gather time-resolved, simultaneous cross-site samples. Single time-point snapshots of microbiota provide the ability to characterize them with high accuracy, but, as is apparent from this study, only hint at the details of their complex dynamics.
Abbreviations
- DNA
deoxyribonucleic acid
- rRNA
ribosomal RNA
- HPV
human papillomaviruses
- TE buffer
Tris-EDTA buffer
- NCBI
National Center for Biotechnology Information
- MAD
median absolute deviation
- OTU
operational taxonomic unit
- BVAB1
bacterial vaginosis-associated bacteria 1
Author Contributions
Author Contributions: Benjamin C. Smith (design, experiments, analyses, manuscript preparation), Christine P. Zolnik (analyses, manuscript preparation), Mykhaylo Usyk (bioinformatics analyses, manuscript preparation), Zigui Chen (design, experiments, analyses, manuscript preparation), Katherine Kaiser (analyses, manuscript preparation), Anne Nucci-Sack (design, clinical component), Ken Peake (design, clinical component), Angela Diaz (design, clinical component), Shankar Viswanathan (analyses), Howard D. Strickler (design, analyses), Nicolas F. Schlecht (design, analyses, manuscript preparation), Robert D. Burk (design, experiments, analyses, manuscript preparation).
References
- Costello EK, Stagaman K, Dethlefsen L. et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336(6086):1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N. et al. The metacommunity concept: a framework for multi-scale community ecology. Ecology Letters. 2004;7(7):601–613. [Google Scholar]
- Smith BC, McAndrew T, Chen Z. et al. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PloS One. 2012;7(7):e40425. doi: 10.1371/journal.pone.0040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Liu L, Zhang F. et al. Human papillomavirus type 16 exists in bacteria isolated from cervical cancer biopsies. J Int Med Res. 2009;37(4):1065–1074. doi: 10.1177/147323000903700411. [DOI] [PubMed] [Google Scholar]
- Ahn J, Yang L, Paster BJ. et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PloS One. 2011;6(7):e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK. et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS pathogens. 2010;6(1):e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Whiteson K, Huse S. et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79(3):266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Fadrosh D, Ma B. et al. Anal microbiota profiles in HIV-positive and HIV-negative MSM. Aids. 2014;28(5):753–760. doi: 10.1097/QAD.0000000000000154. [DOI] [PubMed] [Google Scholar]
- Marrazzo JM, Fiedler TL, Srinivasan S. et al. Extravaginal reservoirs of vaginal bacteria as risk factors for incident bacterial vaginosis. J Infect Dis. 2012;205(10):1580–1588. doi: 10.1093/infdis/jis242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht NF, Burk RD, Nucci-Sack A. et al. Cervical, anal and oral HPV in an adolescent inner-city health clinic providing free vaccinations. PloS One. 2012;7(5):e37419. doi: 10.1371/journal.pone.0037419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle PE, Schiffman M, Gravitt PE. et al. Comparisons of HPV DNA detection by MY09/11 PCR methods. J Med Virol. 2002;68(3):417–423. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- Herrero R, Schiffman MH, Bratti C. et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa Rica: the Guanacaste Project. Rev Panam Salud Publica. 1997;1(5):362–375. doi: 10.1590/s1020-49891997000500005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Qian P-Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PloS One. 2009;4(10):e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK. et al. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5(3):235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM. et al. PANDAseq: paired-end assembler for illumina sequences. BMC bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J. et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettweis JM, Serrano MG, Sheth NU. et al. Species-level classification of the vaginal microbiome. BMC Genomics. 2012;13(8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Vegan: Community Ecology Package. R package version 2.0-10. 2013. 2015. Available from: https://cran.r-project.org/web/packages/vegan/index.html .
- Brook I, Frazier EH. The aerobic and anaerobic bacteriology of perirectal abscesses. J Clin Microbiol. 1997;35(11):2974–2976. doi: 10.1128/jcm.35.11.2974-2976.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E. et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche WJ. The antimicrobial treatment of periodontal disease: changing the treatment paradigm. Crit Rev Oral Biol Med. 1999;10(3):245–275. doi: 10.1177/10454411990100030101. [DOI] [PubMed] [Google Scholar]
- Brzychczy-Włoch MM, Pabian W, Majewska E. et al. Dynamics of colonization with group B streptococci in relation to normal flora in women during subsequent trimesters of pregnancy. New Microbiol. 2014;37(3):307–319. [PubMed] [Google Scholar]
- Ravel J, Gajer P, Abdo Z. et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer P, Brotman RM, Bai G. et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]