Abstract
The development of the neonatal gut microbiome is influenced by multiple factors, such as delivery mode, feeding, medication use, hospital environment, early life stress, and genetics. The dysbiosis of gut microbiota persists during infancy, especially in high-risk preterm infants who experience lengthy stays in the Neonatal intensive care unit (NICU). Infant microbiome evolutionary trajectory is essentially parallel with the host (infant) neurodevelopmental process and growth. The role of the gut microbiome, the brain-gut signaling system, and its interaction with the host genetics have been shown to be related to both short and long term infant health and bio-behavioral development. The investigation of potential dysbiosis patterns in early childhood is still lacking and few studies have addressed this host-microbiome co-developmental process. Further research spanning a variety of fields of study is needed to focus on the mechanisms of brain-gut-microbiota signaling system and the dynamic host-microbial interaction in the regulation of health, stress and development in human newborns.
Keywords: Gut Microbiome, Infant, Early life, Brain-Gut-Microbiota Axis, Host-Microbial Genomics
Introduction
The etiologies of infant health and neurodevelopmental outcomes are complex and multifactorial [1]. A growing body of evidence points to critical developmental periods during infancy and early childhood when environmental factors interact with genomic susceptibilities to increase the risk for chronic diseases in adulthood. In early life, the role of the microbiota in health and disease has taken center stage because of strong evidence that the gut microbiome can greatly influence many aspects of human bio-behaviors, such as brain function and stress responses through brain-gut communication [2]. Expanding research supports the function of the brain-gut-microbiota axis, in which intestinal microbiome plays a key role in early programming and regulation of the neuro-immune system [2]. Meanwhile, a metagenomic cross talk has recently been described suggesting that host genes are involved in determining the microbiome. The microbes that, together with the infant, constitute the “holobiont,” the combination of a host and its microbial communities that together provide the host with its capabilities, may be a key component contributing to health outcomes in early life [3]. In other words, infants may have genetic predispositions for certain gut microbiome patterns and functions, and in turn, the microbiota regulates host gene expression [4]. Thus, in the context of personalized treatment of vulnerable infants, the combination and interaction of the developing intestinal microbiome with host genomics may play a critical role in infant early development.
The gut microbiome refers to a collection of microorganisms and the habitat they colonize. It encompasses the collective genomes of its microbes, also known as the metagenome [5]. The culture-independent analysis of microbial community structure includes using either targeted regions such as the 16S rRNA gene to survey the composition of the microbiota and/or the shotgun sequencing of the metagenome to catalogue the genes that are present. The 16S rRNA gene-based sequencing can reveal gut microbiota patterns and diversity and the high-throughput metagenomic sequencing can precisely investigate the microbiota present, functional genes and microbial pathways [5]. The microbial content of the human gut is an environment dominated by bacteria, mainly strict anaerobes. The majority of human gut bacteria belongs to four phyla, Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, in which Firmicutes and Bacteriodetes are the two dominated phyla comprising the gut microbiome of healthy adults. The balance of gut microbial diversity is crucial for maintaining the health status of the host [2]. These gut bacteria are the key players in regulating gut metabolism, and are critical in understanding metabolism dysfunctions. Hence, dysbiosis, the imbalance or alteration of gut microbiota, has been associated with many acute and chronic health conditions, such as inflammatory bowel disease, obesity, and cardiovascular disease [2,6].
Factors Influence Infant Gut Microbial Community
The infant gut microbial community is a dynamic ecosystem and is influenced by multiple factors, such as delivery mode, feeding, medication use, hospital environment, other early life experiences, and host genetics [7,8]. In addition, infants’ demographic factors, i.e., gender and gestational age and postnatal age also contribute to the development of the gut microbiota [9]. These contributing factors need to be accounted for when the linkages of early life experience, host-microbial interplay, and health and developmental outcomes are investigated.
The full-term, breast-fed infant often serves as the health standard or the “norm” for newborns. Colonization begins with facultative anaerobic organisms, followed by the development of obligate anaerobes, including Bifidobacterium, Bacteroides, and Clostridium [10]. Contact with the vaginal flora of the mother during delivery fosters the development of diverse flora, and the subsequent diet of the infant influences further inoculation of microorganisms [11]. While full-term, breastfed infants demonstrate a predominance of obligate anaerobes, such as Bifidobacteria in the gastrointestinal (GI) tract [11], preterm infants demonstrate reduced levels of obligate anaerobes, as well as increased levels of facultative anaerobes such as Enterobacteriaceae and Enterococcaceae [12]. Researchers hypothesize that this delay of colonization by obligate anaerobes may impede the establishment of a mature immune system in preterm infants [12]. Thus, with an altered gut microbiome, preterm infants also exhibit a high abundance of Proteobacteria [13,14] and increased colonization by pathogens [15].
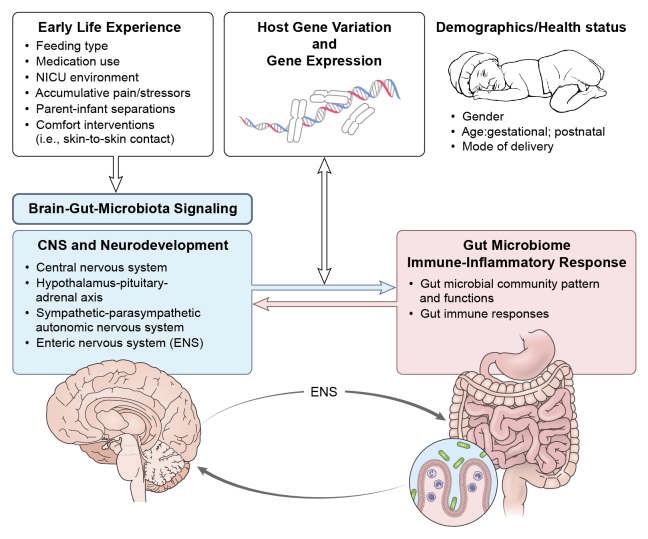
A summary of the literature that includes contributing factors to the infant gut microbiome are shown in Tables 1,2, 3, 4, and 5. A theoretical model, “Regulation of Infant Health and Development in Early Life by Host-Microbial Genomics” has been developed to propose that host-microbial interaction is involved in infant health and developmental mechanisms (Fig. 1).
Table 1. Literatures of Investigating Contributing Factors to Infant Gut Microbiome: Delivery Type.
| Author/Year | Country | Factors | Participants | Measures | Outcomes |
| Backhed, F., et. al., (2015). [44] | Sweden, Denmark, China, Saudi Arabia, Norway, Hong Kong | Mode of delivery; feeding; age | 98 mother-infant (full-term) pairs | Maternal stool sample at delivery; infant stool sample during the first days of life and at 4 and 12 months. | Bacteroides were absent in the C-section delivered newborns; Gut microbiota of infants delivered by C-section were less resemblance to their mothers, compared to vaginally delivered infants. |
| Biasucci G., et. al., (2010). [17] | Italy | Mode of delivery | 46 full-term infants: 23 - C-section delivered; 23 - vaginal delivered infants | Stool samples at first 3 days of life. | The gut flora of C-section and vaginally delivered infants were significantly different; C-section delivered infants had an absence of Bifidobacteria sp. |
Table 2. Literatures of Investigating Contributing Factors to Infant Gut Microbiome: Feeding Type.
| Author/Year | Country | Factors | Participants | Measures | Outcomes |
| Cong, X., et. al., (2016). [9] | United States | Feeding; gender | 29 stable preterm infants; 20 - mother’s own breastmilk fed infants; 9 - non-mother’s own breastmilk fed infants (human donor milk, formula) | Stool samples at first 30 days of life. | Infants fed mother’s own breastmilk had a higher diversity of gut microbiome and higher abundance in Clostridiales and Lactobacillales than infants. Female infants were more likely to have a higher abundance of Clostridiales, and lower abundance of Enterobacteriales than males during early life. |
| Backhed, F., et. al., (2015). [44] | Sweden | Mode of delivery; feeding; age | 98 mother-infant (full-term) pairs | Maternal stool sample at delivery; infant stool sample during the first days of life and at 4 and 12 months. | Bifidobacterium and Lactobacillus dominated the gut microbiota of breast-fed infants in the first year of life. |
| Moles, L., et. al. (2015). [27] | Spain | Feeding; (Probiotics isolated from human milk) | 5 preterm infant pairs (two pair of twins) | 14 stool samples and 10 blood samples; weekly of the first month of life. | Lactobacillus salivarius PS12934 were found in all infants’ stool samples while Bifidobacterium breve PS12929 is only present in part of the stool samples. |
| Biesbroek, G., et. al., (2014). [57] | Netherlands | Feeding | 202 full-term infants: 101 - exclusive breastfeeding; 101 - exclusive formula feeding | Nasopharyngeal swabs, at age 6 weeks and 6 months. | Breastfed infants had an increased Dolosigranulum and several Corynebacterium spp. in the respiratory tract. |
| Underwood, M. A., et. al., (2014). [42] | United States | Feeding; (Breastmilk; formula; prebiotics) | 27 preterm infants: 12 - formula group, 15 - breast milk group | Stool sample: formula-fed infants at baseline and weekly for 5 weeks; breastmilk–fed infants were collected at baseline and every 2 weeks for 6 weeks. | Low levels of Bifidobacteria and no measurable Lactobacilli were found in all the breast-milk-fed and formula-fed infants. None of the prebiotic interventions resulted in significant increases in Bifidobacteria. |
| Jost, T., et. al., (2012) [22] | Switzerland | Feeding | 7 mother - infant (full-term) pairs | Breast milk sample; maternal and infant stool samples; at 3 time points in the first month postpartum; total 63 samples. | Bifidobacterium was potentially vertically transferred via breast milk from mother to infant; obligate anaerobic genera were commonly present in maternal faeces and breast milk; facultative anaerobes were common in breast milk and neonatal faeces. |
| Bezirtzoglou, E., et. al., (2011). [20] | Greece; Netherlands | Feeding | 12 full-term infants: 6 breast-fed; 6 formula-fed | 12 infant stool samples. | Breastfed infants had increased Bifidobacterium than formula-fed infants; Breast-fed infants had more diverse microbiome community. |
| Solis, G., et. al., (2010). [25] | Spain | Feeding | 20 mother - infant (full-term) pairs | Breast-milk samples; infant stool samples; 3 times in the first 90 days after delivery. | Breast-milk contained viable Lactobacilli and Bifidobacteria that might contribute to the establishment and development of the microbiota in the newborn. |
Table 3. Literatures of Investigating Contributing Factors to Infant Gut Microbiome: Medication Use.
| Author/Year | Country | Factors | Participants | Measures | Outcomes |
| Arboleya, S., et. al., (2015). [45] | Spain, Italy | Perinatal antibiotics | 13 full-term, breast fed, vaginally delivered infants; 27 very low birth weight preterm infants. 12 infants received antibiotic therapy | Stool samples were collected between 24 and 48 hours of life and at 10, 30, and 90 days of life. | Intrapartum antimicrobial prophylaxis therapy had a large impact on the intestinal microbiome, but was not detected for some time post-delivery. Results suggest that this alteration is long lasting. |
| Dardas, M., et. al., (2014). [48] | United States | Postnatal antibiotics | 29 premature infants: 8 received 2 days of antibiotic treatment and 11 infants received at least 7 days of antibiotic treatment | Stool swabs were collected at 10 days of life and 13 days of life. | Infants who received at least 7 days of antibiotic use experienced a greater decrease in microbial diversity than those who received only 2 days of use. Both chronological age and antibiotic exposure impact gut microbiome. |
| Greenwood, C., et. al., (2014). [28] | United States | Empiric antibiotic use | 74 premature infants: 13 received 0 days of antibiotic treatment, 48 received 1-4 days of antibiotic treatment, and 13 received 5-7 days of antibiotic treatment | Stool samples were collected between 4 and 7 days of life, 10 and 16 days of life, and 20-23 days of life; 239 samples were collected. | Microbial diversity was decreased in infants who received any antibiotic therapy. Diversity was decreased by greater margins and for a longer period of time in infants who received 5-7 days of antibiotics when compared to those who received only 1-4 days of antibiotics. |
| Gupta, R., et. al., (2013). [50] | United States | Histamine-2 Receptor blockers (H2RB) | 76 premature infants: 25 received ranitidine for an average of 19 days; 51 infants never received H2RBs | One stool sample was collected from each neonate. | Decreased gastric acidity reduces the microbial diversity of the gut microbiome. The pattern shifted towards Proteobacteria in infants exposed to H2RBs. |
| Jenke, A., et. al., (2013). [49] | Germany, Australia | Antibiotic use and development of NEC | 68 extremely low birth weight infants: 12 subsequently developed NEC and 56 had no gastrointestinal disorders | Stool samples were collected on alternate days for the first 4 weeks of life (total of 248 samples). | Higher prevalence of antibiotic use in the first 7 days in infants who went on to develop NEC. Antibiotic therapy for > 48 hours in the first week of life was associated with increased C. diff proportions at 7 days of life. A large increase in total bacteria count and the proportion of E. coli was noted before the onset of NEC. |
Table 4. Literatures of Investigating Contributing Factors to Infant Gut Microbiome: Environmental Factors.
| Author/Year | Country | Factors | Participants | Measures | Outcomes |
| Brooks, B., et. al., (2014). [52] | United States | Environmental (NICU) surfaces | 2 very low birth weight preterm infants ( < 1500 g) | First month of life Stool samples; every 3 days; 33 swabs were collected each room surfaces. | Infant gut organisms were widely distributed throughout the room environment; Keyboards, mouse, and telephones had the lowest amount of colonizing organisms detected in the gut; Intubation and feeding tubing had the highest amount of colonizing organisms detected in the gut. |
| Torrazza, et. al., (2013) [54] | United States | Environmental (NICU) surfaces | 53 preterm: 18 NEC cases; 35 control cases; 3 NICUs | Stool samples at 2, 1, and 0 weeks, prior to the diagnosis of NEC. | Microbiota composition differed between the 3 NICUs; Abnormal patterns of Proteobacteria may be associated with a greater risk of developing NEC. |
| Conceicao, T., et. al., (2012). [53] | Portugal | Environmental surfaces, equipment, parental skin, health care provider skin | 16 infants (mean gestation age of 31 w) with Staphylococcus aureus hemo positive | Swabs from parents nares; NICU clinicians nares; mothers’ nipples; NICU environment. | Three major methicillin susceptible S. aureus clones were endemic in the NICU; Clinicians, plastic folders; and mothers' nipples might be potential vehicles of dissemination of S. aureus. |
| Ferraris, L., et. al., (2012) [55] | France | Environmental (NICU) surfaces | 76 preterm infants | Stool samples weekly during NICU stay; 3 NICUs. | 79% of infants were colonized by Clostridia; Clostridium sp. colonization, with a high diversity of species, increased throughout the hospitalization |
Table 5. Literatures of Investigating Contributing Factors to Infant Gut Microbiome: Comfort Interventions.
| Author/Year | Country | Factors | Participants | Measures | Outcomes |
| Hendricks-Munoz, K.D., et. al., (2015) [58] | United States | Skin to skin contact | 42 preterm infants ( < 32 weeks of gestation at birth) | Saliva swabs were collected at 1 month of life and/or at discharge. | Skin to skin care led to an increased pace of oral microbe repertoire maturity development and lower prevalence of organisms associated with intestinal dysfunction |
Figure 1.
Regulation of Infant Health and Development in Early Life by Host-Microbial Genomics. ENS = enteric nervous system.
Delivery Type
The infant microbiome evolves prenatally from exposure to amniotic fluid and during the mode of delivery [16]. Infants born vaginally have intestinal microbial content similar to the mother’s vaginal and intestinal flora including commensal bacteria, i.e., Bacteroides, Bifidobacterium, and Escherichia coli, whereas infants born via caesarean section have microbial flora similar to environmental microbes [17,18]. These initial microbes form the basis of nutrient utilization, gut barrier function, and immune development [19]. While the mode of delivery influences the initial microbiota, further interactions with the environment continue to impact the microbiome development.
Feeding Type
Mother’s breast milk promotes the development of gut microbiome by introducing probiotics and prebiotics, and providing protection against pathogens. Different bacteria genera that are dominant in breast milk include Bifidobacterium, Lactobacillus, Staphylococcus, Bacteroides, Enterococcus, Streptococcus, and Clostridium [20-25]. These genera have been found present both in the mother’s breast milk and the infant’s stool, indicating vertical transmission of bacteria from breastfeeding [20,23,25-27]. The composition of the microbiome in breast milk changes over time, and is influenced by maternal genetics, maternal weight, and delivery mode [23,26]. While there is still little known about the influence of feeding type on the development of gut microbiota in preterm infants, our recent findings suggest that preterm infants fed mother’s own breast milk had a significantly more diverse gut microbiome than those fed donor milk and/or formula [9]. Infants fed mother’s own breast milk had a higher abundance of Clostridiales and Lactobacillales, whereas infants fed donor milk and/or formula had a gut microbial community extensively dominated by Enterobacteriales [9]. Increased diversity of the gut microbiome with high abundance of Bifidobacterium, Clostridiales and Lactobacillales may protect preterm infants against devastating diseases, such as necrotizing enterocolitis (NEC) that affects mostly the intestine of premature infants [28].
Mothers who give birth to preterm infants produce breast milk with higher protein, fat, carbohydrate, and immunological composition, but less diverse and mature HMOs [29-32] compared to term milk. Thus, the “premature” human milk may impact the premature gut through different mechanisms [33,34]. Human milk oligosaccharides (HMOs), the third most abundant component in human milk, function as a prebiotic, which promotes the growth and activity of beneficial microorganisms in the gut, such as Bifidobacterium spp. and Lactobacillus. These gut bacteria play important roles in inhibiting the growth of pathogens [31,35-37]. Full-term infants exclusively breastfed tend to have more than twice the amount of Bifidobacterium [20], but possess relatively less diverse gut microbiome compared to formula-fed infants [38-40]. Formula introduction leads to decreased abundance of Bifidobacterium, and increased number of Bacteroides [20]. However, the interaction between the gut and HMOs are also altered among preterm infants in comparison to full-term infants [41,42]. The variation of HMOs and the unresponsiveness of the gut to HMOs may lead to the alteration of the microbiome community among preterm infants, which composites an increased number of facultative anaerobes and Enterobacteriaceae, as well as a reduced level of strict anaerobes, including Bifidobacterium, Bacteroides, and Atopobium [36,43]. Cessation of breastfeeding and introduction of solid food at 12 months of age leads to the shift of microbiome community similar to the composition of an adult’s gut [44]. Future studies will explore the depth to which variations in feeding type affect the developing microbiome.
Medication Use
The administration of antibiotics is nearly a universal protocol for the care of high-risk infants, such as preterm neonates in the NICU. Mothers of these infants also commonly receive intrapartum antibiotics, however, the lasting effects of such use on infant health and development are still largely unknown. Antibiotics can induce dysbiosis, including loss of keystone taxa, altered metabolic capacity, low diversity, and blooms of pathogens that typically are less abundant in a healthful state [45-47]. Different microbial community patterns between preterm and full-term infants may be potentially moderated by the higher rate of antibiotic use in preterm neonates.
Recent research shows that antibiotic use leads to reduced levels of total bacteria [45] as well as decreased microbial diversity of the gut [28]. In infants exposed to antibiotics, either pre- or postnatally, the diversity of the microbiota was drastically decreased, consisting of only a few phyla as opposed to the microbial community that makes up the microbiome of full-term, healthy infants [28]. It is likely that the length of exposure to the antibiotics has a role in determining the severity of the changes of the gut microbial community. Gut microbial diversity has been found to be significantly decreased at ten days of life in infants who received an extensive of antibiotics, in comparison to those who received a brief course of antibiotics [48]. Investigation on the use of Ampicillin and Gentamicin has also shown that there were milder changes in the microbiomes of infants with brief antibiotic use than when compared to infants who underwent intensive antibiotic use [28]. Additionally, infants subjected only to a brief course of antibiotics were able to partially recover the diversity lost by three weeks after the administration of antibiotics [28].
The composition of the gut microbial species also changes along with the administration of antibiotics. Preterm infants who were subjected to an intensive courses of antibiotics pre- or postnatally, had microbiomes composed of only Enterobacter, Escherichia, Enterococcus, and Staphylococcus, whereas, infants who were not exposed to antibiotics developed a wider variety of bacteria [28]. In other studies with similar findings, this relative dominance of Enterbacteriaceae lasted up to three months in some neonates, and has the potential to lead to increased pathogenic Enterobacteria [45]. Additionally, Clostridium difficile species were detected at seven days of life in infants who experienced more than 48 hours of antibiotics during their first week [49]. However, it should be noted that these species decreased as antibiotic use decreased [49].
Histamine-2 Receptor blockers (H2RBs) are also commonly used in neonates to decrease the acidity in the stomach, which may play a role in altering the gut microbiota [50]. One study demonstrated a decreased microbial diversity in fecal samples from neonates treated with H2RBs, when compared to neonates who were not [50]. More specifically, the mean relative abundance of Proteobacteria, a phyla containing many pathogenic strains of bacteria, was increased in infants who were administrated H2RBs [50]. However, these results are confounded by the use of antibiotics in the majority of the sample population [50]. Antibiotics may also play a role in decreasing microbial diversity, and therefore, it is difficult to determine which medication induced the microbial alteration.
Environmental Factors
High-risk infants including preterm infants are typically cared for in the NICU for weeks or months after delivery to provide time for the infant’s neuro-immune and cardiopulmonary development and to prevent and manage complications. A recent systematic review of the NICU environment demonstrated a myriad of sources that influence the development of the preterm infant’s microbiome [51], including the parental and healthcare provider’s skin and surfaces of equipment and workspace. The studies reviewed support that the infant microbiome is colonized differently on the basis of the composition of the NICU environment [52-55]. Factors such as intubation, feeding tube placement, incubator temperatures and humidity were found to influence the type and amount of colonizing organisms. While this body of research is not conclusive as to which exposure sources are detrimental to neurodevelopment of the infant, ongoing research on how the infant’s developing microbiome in the NICU impacts neuro-immune development and health outcomes may provide further insight. The ability to reduce colonization of environmental bacteria on parental/provider skin, work surfaces and equipment with disinfectant solutions and protection of the infant’s epidermal barrier are important measures to prevent exposure to pathological organisms.
Comfort Interventions
Few studies have been conducted to investigate the effects of different comfort measures on infant gut health and microbial development. It is widely understood that direct breastfeeding provides numerous benefits to the mother-infant relationship, including improvements in milk production, increased parent satisfaction, and decreased pain perception [56]. In older infants, it has been found that exclusive direct breastfeeding may protect the infant's respiratory system through microbial colonization [57]. However, there is very limited research related to the effects of direct breastfeeding and other comfort interventions, such as skin-to-skin contact (Kangaroo Mother Care), touch and direct communication, on the development of the infant microbiome in the NICU. One recent study determined that skin-to-skin contact between the infant and mother is associated with an increased pace of oral microbe composition maturity amongst preterm infants [58]. The skin-to-skin contact reduced stress among the treated infants and thus facilitated the acceleration of mature oral microbial repertoire and a lower relative prevalence of organisms associated with intestinal dysfunction, in comparison to the untreated infants [58]. The aforementioned study had several limitations, including small sample size, varied amount of skin-to-skin care, and additional stress reduction methods that were not taken into account, however it suggests an overarching link between skin-to-skin contact and microbial colonization.
Ongoing research on the topic of comfort interventions and its effects will help to establish a better understanding of the relationship between these measures and the microbiome. These findings may assist in preventing future intestinal issues and diseases for the infant as well as development of a healthful adult microbiome.
The Brain-Gut-Microbiota Axis in Regulation of Early Life
The brain-gut-microbiota axis has been recognized recently as an important mechanism in the regulation of health and stress response. The components of the axis includes the central nervous system (CNS), endocrine-immune system, hypothalamus-pituitary-adrenal (HPA) axis, sympathetic-parasympathetic autonomic nervous system, enteric nervous system, and intestinal microbiota [6]. This bidirectional communication network enables top-down signaling from the brain to influence the motor, sensory and secretory modalities of the GI tract, and conversely, bottom-up signaling from the gut to affect brain function, especially the hypothalamus and amygdala that are devoted to emotion and stress. Physical and emotional stress including maternal separation results in alterations of the functions of the GI tract [59,60]. Increasing evidence suggests that the GI microbiota plays a key role in this axis. A recent study using functional MRI first shows that ingestion of probiotic bacteria alters brain function in humans, demonstrating an apparent link between the gut microbiota and human health [61,62].
Stressful Early Life Experienced by High-risk Infants in the NICU
Because high-risk infants including preterm neonates are developmentally immature, physiological limitations force these infants to depend on sophisticated medical technology and rigorous nursing interventions for survival [63]. On average, neonates are subjected to 10 to 16 invasive, painful/stressful procedures per day in the NICU, and very preterm infants (gestational age < 32 weeks) may require hundreds of painful procedures during their NICU stay [2]. For instance, the most common interventions include heel prick blood sampling, endotracheal suction, and intravenous cannula insertion. In addition to these routine invasive procedures, maternal-infant separation is another stressor that infants must endure in the NICU. Numerous animal and human studies show that deprivation of maternal care results in altered neuronal, hormonal, genetic, and bio-behavioral outcomes during the sensitive growth and developmental period [64,65]. However, the effects of early life stress, including prolonged and accumulative painful/stressful procedures, parent-infant separation, infection, antibiotic use and feeding intolerance, on modulating the gut microbiome and neurobehavioral outcomes remain largely unexplored.
The vulnerability of preterm infants during early life stressful or painful experiences may yield long term, often permanent effects on subsequent neurobiological and behavioral development [64]. For instance, a recent study demonstrates that greater numbers of painful procedures directly predict lower head circumference and brain function in very preterm infants [66]. The study also indicates that repeated pain during a vulnerable period may activate a downstream cascade of stress signaling that affects later growth [66].
Regulation of Early Life Stress by the Brain-Gut-Microbiota Signaling System
Even though studies using animal models have provided solid support for the relationship between physical and psychological stress and behavior and gut microbiota through the brain-gut signaling system [67], there have been a limited number of human studies. However, the body of research in animal models suggests that the gut microbiota contributes to developmental programming and there is a “window of vulnerability” within which the gut microbiota can affect physiological function, with potentially life-long consequences [68]. Since the rapidly developing nervous system of immature preterm neonates differs from term infants, they are particularly vulnerable to the effects of stress/pain. Stress activates the HPA axis and sympathetic nervous system, thereby increasing gut permeability which allows bacteria and bacterial antigens to cross the epithelial barrier, activate the mucosal immune response, and alter the microbiome composition [67]. Research also suggests that the oxidative stress on the intestine modulates the process of microbiome establishment in preterm infants [10], however, the exact causality of preterm infant gut distortions has not yet been proven. Continual adaptation to repeated pain appears to induce functional changes in stress/pain processing systems [69]. Furthermore, repeated pain may contribute to long-term changes in generalized stress systems, including altered stress hormone levels, long after NICU discharge [70]. However, the mechanisms underlying long-term consequences of neonatal noxious insults on brain function and pain sensitivity are still unknown.
Exposure to maternal separation in newborn animals also shows significantly increased adrenocorticotropic hormone levels and altered gut microbiota balance [71], whereas, maternal skin-to-skin contact with human preterm infants has been found to facilitate the pace of oral microbial maturity [47,58]. The microbial content of the gut, potentially influenced by factors such as maternal contact, is critical to the development of an appropriate stress response in early life as well as later development. There is a narrow window in early life where colonization must occur to ensure normal development of the HPA axis [6]. The establishment of a healthy microbiota has a profound effect on the future well-being of the individual [72]. Microbial patterns and functions in immune naïve premature infants may lay the groundwork for life-long health/disease status.
In the brain-gut-microbiota signaling system, S100 family of calcium binding proteins, including calprotectin (S100A8/A9), calgranulin C (S100A12) and S100B can serve as non-invasive immune and stress markers. Fecal calprotectin and calgranulin C have been found in phagocytes and in intestinal epithelial cells related to inflammatory GI disease [73,74], and likewise correlate with the fecal microbial community in preterm infants that demonstrates the intestinal innate immune response to the bacterial colonization [49]. The S100B protein is specifically expressed by enteric glial cells and has been considered as a pro-inflammatory factor in the gut [75]. Interestingly, S100B in blood and cerebrospinal fluid have been found to be a novel neonatal brain injury and stress biomarkers [76,77], whereas, the link of fecal S100B with brain function is unknown.
Host-Microbial Genomic Interaction
Host-Microbial Genomic Interaction has been found recently as an important mechanism associated with many aspects of human health and disease. The interplay of host-microbial genomes has been referred to as the holobiont, which entails the concept of the host in conjunction with its commensal microbes. Their collective genomes forge a “hologenome,” the dynamic interplay among the gut microbiota, host genes and the transcriptional response. As such, host-microbial genomics hold equivalent significance in the origin of new holobiont individuals [78,79].
Research reveals a metagenomic cross-talk that host genes are involved in shaping the microbiome, and in turn, the microbiota regulates host gene expression [4]. One study of adult twin-pairs shows that the abundances of many microbial taxa are influenced by host genetics [80]. Another study of children less than 10 years old reports that the highest levels of microbial similarity are shown in genetically identical twins compared to fraternal twins and unrelated controls, suggesting a strong genetic influence over the composition of the fecal microbes [81]. Mining the Human Microbiome Project data, Blekhman and colleagues analyzed the composition of the human microbiome and host genetic variation and found significant associations between host genetic variation and microbiome composition at different body sites [82]. Results also show that the host-microbial associations are driven by host genetic variation in immunity-related pathways, which are related to microbiome-related disorders including inflammatory bowel disease and obesity. The research provides a starting point to investigate the role of host genetic variation in shaping the microbial composition [82].
Increasing evidence also implicates host-microbe interactions in the etiology of anxiety, stress related behaviors, and neurodevelopmental disorders. Stilling and colleagues applied genome-wide transcriptional profiling to determine gene expression in the amygdala of germ-free (GF) mice, devoid of any microbiota throughout maturation [83]. They found that absence of microbes in GF mice was significantly associated with upregulation of synaptic plasticity-related genes and genes that promote transcription, which are associated with neural activities and established during early life. Their research suggested that downregulation of immune system-related genes may reflect some effects of microbiota on brain physiology and behavioral responses through the immune system [83]. The modulation of the gut microbiota by host genes and the regulation of the microbiome on host gene expression throughout molecular epigenetic processes are still largely unknown. Large scale and detailed studies in humans are needed to identify candidate genes that may decode the mechanism of host-microbial interactions in shaping brain function and neurodevelopment and can guide future microbiota-targeted diagnosis and interventions [8].
Conclusion
In summary, the dysbiosis of gut microbiota persists during infancy, especially in preterm infants, and then may reach a stable configuration at age 2 to 3 years, toward the characteristic microbiota of adults [4,84]. The development of the neonatal gut microbiome is influenced by multiple factors and further research is needed to investigate the potential contributing factors, such as early life stress. Not only are preterm infants exposed to an abundance of invasive interventions, but they are also especially vulnerable to stressful events during this highly technical care period in the NICU. The role of the gut microbiome, the brain-gut signaling system, and its interaction with the host genes have been shown to be related to both short and long term health and bio-behavioral development, as such infant microbiome evolutionary trajectory is essentially parallel with the host (infant) neurodevelopmental process and growth. The investigation of potential dysbiosis patterns in early childhood is still lacking and few studies have addressed this host-microbiome co-developmental process, therefore, the theoretical model that proposed herein is innovative and highly demanded [84,85]. Further studies are needed to focus on the multi-omic mechanisms of brain-gut-microbiota signaling system and the dynamic host-microbial interaction in the regulation of health, stress and development in human infants.
Acknowledgments
This publication was supported by the National Institute of Nursing Research of the National Institutes of Health (NIH-NINR) under award number 1K23NR014674-01-02 and Affinity Research Collaboratives award through University of Connecticut Institute for Systems Genomics to Dr. Cong. Additional Support provided to Dr. Henderson (NIH-NINR) award number 1ZIANR000018-01-05). Illustration by Alan Hoofring from the National Institutes of Health Medical Arts.
Glossary
- GI
Gastrointestinal
- HMOs
Human milk oligosaccharides
- NICU
Neonatal intensive care unit
- H2RBs
Histamine-2 Receptor blockers
- CNS
Central nervous system
- HPA
Hypothalamus-pituitary-adrenal
- NEC
Necrotizing enterocolitis
- GF
Germ-free
References
- Behrman R.E., Butler A.S., editors. Committee on Understanding Premature Birth and Assuring Healthy Outcomes, I.o.M.o.t.N.A. Preterm Birth: Causes, Consequences, and Prevention. Washington, D.C.: The National Academies Press; 2007. [PubMed] [Google Scholar]
- Cong X. et al. Early Life Experience and Gut Microbiome: The Brain-Gut-Microbiota Signaling System. Adv Neonatal Care. 2015;15(5):314–323. doi: 10.1097/ANC.0000000000000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. A holobiont birth narrative: the epigenetic transmission of the human microbiome. Front Genet. 2014;5:282. doi: 10.3389/fgene.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Elinav E. Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med. 2015;7(1):120. doi: 10.1186/s13073-015-0249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489(7415):250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG, Cryan JF. Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology. 2012;37(9):1369–1378. doi: 10.1016/j.psyneuen.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Reid G. et al. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat Rev Microbiol. 2011;9(1):27–38. doi: 10.1038/nrmicro2473. [DOI] [PubMed] [Google Scholar]
- Rodriguez JM. et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X. et al. Gut Microbiome Developmental Patterns in Early Life of Preterm Infants: Impacts of Feeding and Gender. PLoS One. 2016;11(4):e0152751. doi: 10.1371/journal.pone.0152751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya S. et al. Assessment of intestinal microbiota modulation ability of Bifidobacterium strains in in vitro fecal batch cultures from preterm neonates. Anaerobe. 2013;19:9–16. doi: 10.1016/j.anaerobe.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Harmsen HJ. et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30(1):61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- Berrington JE. et al. The neonatal bowel microbiome in health and infection. Curr Opin Infect Dis. 2014;27(3):236–243. doi: 10.1097/QCO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- Aujoulat F. et al. Temporal dynamics of the very premature infant gut dominant microbiota. BMC Microbiol. 2014;14:325. doi: 10.1186/s12866-014-0325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3(8):944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshvildadze M. et al. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2010;156(1):20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince AL. et al. The microbiome and development: a mother's perspective. Semin Reprod Med. 2014;32(1):14–22. doi: 10.1055/s-0033-1361818. [DOI] [PubMed] [Google Scholar]
- Biasucci G. et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Marques TM. et al. Programming infant gut microbiota: influence of dietary and environmental factors. Curr Opin Biotechnol. 2010;21(2):149–156. doi: 10.1016/j.copbio.2010.03.020. [DOI] [PubMed] [Google Scholar]
- Collado MC. et al. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. 2012;3(4):352–365. doi: 10.4161/gmic.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezirtzoglou E, Tsiotsias E, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe. 2011;17(6):478–482. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Collado MC. et al. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48(5):523–528. doi: 10.1111/j.1472-765X.2009.02567.x. [DOI] [PubMed] [Google Scholar]
- Jost T. et al. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7(8):e44595. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodayar-Pardo P. et al. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014;34(8):599–605. doi: 10.1038/jp.2014.47. [DOI] [PubMed] [Google Scholar]
- Martin R. et al. Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol. 2007;103(6):2638–2344. doi: 10.1111/j.1365-2672.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- Solis G. et al. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16(3):307–310. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R. et al. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96(3):544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- Moles L. et al. Administration of Bifidobacterium breve PS12929 and Lactobacillus salivarius PS12934, two strains isolated from human milk, to very low and extremely low birth weight preterm infants: a pilot study. J Immunol Res. 2015;2015:538171. doi: 10.1155/2015/538171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood C. et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of Enterobacter. J Pediatr. 2014;165(1):23–29. doi: 10.1016/j.jpeds.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Gerss J. Longitudinal analysis of macronutrients and minerals in human milk produced by mothers of preterm infants. Clin Nutr. 2011;30(2):215–220. doi: 10.1016/j.clnu.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Castellote C. et al. Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J Nutr. 2011;141(6):1181–1187. doi: 10.3945/jn.110.133652. [DOI] [PubMed] [Google Scholar]
- De Leoz ML. et al. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res. 2012;11(9):4662–4672. doi: 10.1021/pr3004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molto-Puigmarti C. et al. Differences in fat content and fatty acid proportions among colostrum, transitional, and mature milk from women delivering very preterm, preterm, and term infants. Clin Nutr. 2011;30(1):116–123. doi: 10.1016/j.clnu.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Gabrielli O. et al. Preterm milk oligosaccharides during the first month of lactation. Pediatrics. 2011;128(6):e1520–e1531. doi: 10.1542/peds.2011-1206. [DOI] [PubMed] [Google Scholar]
- Underwood MA. et al. Human milk oligosaccharides in premature infants: absorption, excretion, and influence on the intestinal microbiota. Pediatr Res. 2015;78(6):670–677. doi: 10.1038/pr.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonia S. et al. Human Milk Oligosaccharides Inhibit Candida albicans Invasion of Human Premature Intestinal Epithelial Cells. J Nutr. 2015;145(9):1992–1998. doi: 10.3945/jn.115.214940. [DOI] [PubMed] [Google Scholar]
- Pacheco AR. et al. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci. 2015;3:419–445. doi: 10.1146/annurev-animal-022114-111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poroyko V. et al. Diet creates metabolic niches in the "immature gut" that shape microbial communities. Nutr Hosp. 2011;26(6):1283–1295. doi: 10.1590/S0212-16112011000600015. [DOI] [PubMed] [Google Scholar]
- Barile D, Rastall RA. Human milk and related oligosaccharides as prebiotics. Curr Opin Biotechnol. 2013;24(2):214–219. doi: 10.1016/j.copbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Bode L. Human milk oligosaccharides: prebiotics and beyond. Nutr Rev. 2009;67(2):S183–S191. doi: 10.1111/j.1753-4887.2009.00239.x. [DOI] [PubMed] [Google Scholar]
- Mueller NT. et al. The infant microbiome development: mom matters. Trends Mol Med. 2015;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groer MW. et al. Development of the preterm infant gut microbiome: a research priority. . Microbiome. 2014;2:38. doi: 10.1186/2049-2618-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MA. et al. Prebiotic oligosaccharides in premature infants. J Pediatr Gastroenterol Nutr. 2014;58(3):352–360. doi: 10.1097/MPG.0000000000000211. [DOI] [PubMed] [Google Scholar]
- Arboleya S. et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol. 2012;79(3):763–772. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- Backhed F. et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Arboleya S. et al. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr. 2015;166(3):538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- Vangay P. et al. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe. 2015;7(5):553–564. doi: 10.1016/j.chom.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groer MW. et al. The very low birth weight infant microbiome and childhood health. Birth Defects Res C Embryo Today. 2015;105(4):252–264. doi: 10.1002/bdrc.21115. [DOI] [PubMed] [Google Scholar]
- Dardas M. et al. The impact of postnatal antibiotics on the preterm intestinal microbiome. Pediatr Res. 2014;76(2):150–158. doi: 10.1038/pr.2014.69. [DOI] [PubMed] [Google Scholar]
- Jenke AC. et al. S100A12 and hBD2 correlate with the composition of the fecal microflora in ELBW infants and expansion of E. coli is associated with NEC. Biomed Res Int. 2013;2013:150372. doi: 10.1155/2013/150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RW. et al. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J Pediatr Gastroenterol Nutr. 2013;56(4):397–400. doi: 10.1097/MPG.0b013e318282a8c2. [DOI] [PubMed] [Google Scholar]
- Hartz LE, Bradshaw W, Brandon DH. Potential NICU Environmental Influences on the Neonate's Microbiome: A Systematic Review. Adv Neonatal Care. 2015;15(5):324–335. doi: 10.1097/ANC.0000000000000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2014;2(1):1. doi: 10.1186/2049-2618-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao T. et al. Staphylococcus aureus reservoirs and transmission routes in a Portuguese Neonatal Intensive Care Unit: a 30-month surveillance study. Microb Drug Resist. 2012;18(2):116–124. doi: 10.1089/mdr.2011.0182. [DOI] [PubMed] [Google Scholar]
- Torrazza RM. et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLoS One. 2013;8(12):e83304. doi: 10.1371/journal.pone.0083304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris L. et al. Clostridia in premature neonates' gut: incidence, antibiotic susceptibility, and perinatal determinants influencing colonization. PLoS One. 2012;7(1):e30594. doi: 10.1371/journal.pone.0030594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baley J. Skin-to-Skin Care for Term and Preterm Infants in the Neonatal ICU. Pediatrics. 2015;136(3):596–599. doi: 10.1542/peds.2015-2335. [DOI] [PubMed] [Google Scholar]
- Biesbroek G. et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190(3):298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- Hendricks-Munoz KD. et al. Skin-to-Skin Care and the Development of the Preterm Infant Oral Microbiome. Am J Perinatol. 2015;32(13):1205–1216. doi: 10.1055/s-0035-1552941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek PC, Brzozowski T, Konturek SJ. Stress and the gut: pathophysiology, clinical consequences, diagnostic approach and treatment options. J Physiol Pharmacol. 2011;62(6):591–599. [PubMed] [Google Scholar]
- O'Mahony SM. et al. Maternal separation as a model of brain-gut axis dysfunction. Psychopharmacology (Berl) 2011;214(1):71–88. doi: 10.1007/s00213-010-2010-9. [DOI] [PubMed] [Google Scholar]
- Tillisch K. et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144(7):1394–1401. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SM, Bercik P. Gut microbiota: Intestinal bacteria influence brain activity in healthy humans. Nat Rev Gastroenterol Hepatol. 2013;10(6):326–327. doi: 10.1038/nrgastro.2013.76. [DOI] [PubMed] [Google Scholar]
- Lin HC. et al. Relationship between energy expenditure and stress behaviors of preterm infants in the neonatal intensive care unit. J Spec Pediatr Nurs. 2014;19(4):331–338. doi: 10.1111/jspn.12087. [DOI] [PubMed] [Google Scholar]
- Anand KJ, Scalzo FM. et al. Can adverse neonatal experiences alter brain development and subsequent behavior? Biol Neonate. 2000;77(2):69–82. doi: 10.1159/000014197. [DOI] [PubMed] [Google Scholar]
- Higham JP. et al. Mu-opioid receptor (OPRM1) variation, oxytocin levels and maternal attachment in free-ranging rhesus macaques Macaca mulatta. Behav Neurosci. 2011;125(2):131–136. doi: 10.1037/a0022695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinall J. et al. Neonatal pain in relation to postnatal growth in infants born very preterm. Pain. 2012;153(7):1374–1381. doi: 10.1016/j.pain.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Bonaz BL, Bernstein CN. Brain-Gut Interactions in Inflammatory Bowel Diseases. Gastroenterology. 2012;144(1):36–49. doi: 10.1053/j.gastro.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Sudo N. et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558(Pt. 1):263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater R. et al. Evoked potentials generated by noxious stimulation in the human infant brain. Eur J Pain. 2010;14(3):321–326. doi: 10.1016/j.ejpain.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Grunau RE. et al. Altered basal cortisol levels at 3, 6, 8 and 18 months in infants born at extremely low gestational age. J Pediatr. 2007;150(2):151–156. doi: 10.1016/j.jpeds.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouei J, Moussavi M, Hodgson DM. Effect of maternal probiotic intervention on HPA axis, immunity and gut microbiota in a rat model of irritable bowel syndrome. PLoS One. 2012;7(10):e46051. doi: 10.1371/journal.pone.0046051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy ME, Shi HN, Walker WA. The long-term health effects of neonatal microbial flora. Curr Opin Allergy Clin Immunol. 2009;9(3):197–201. doi: 10.1097/ACI.0b013e32832b3f1d. [DOI] [PubMed] [Google Scholar]
- Turco F. et al. Enteroglial-derived S100B protein integrates bacteria-induced Toll-like receptor signalling in human enteric glial cells. Gut. 2014;63(1):105–115. doi: 10.1136/gutjnl-2012-302090. [DOI] [PubMed] [Google Scholar]
- Dabritz J. et al. Fecal S100A12: identifying intestinal distress in very-low-birth-weight infants. J Pediatr Gastroenterol Nutr. 2013;57(2):204–210. doi: 10.1097/MPG.0b013e3182946eb2. [DOI] [PubMed] [Google Scholar]
- Boschetti G. et al. Accuracies of serum and fecal S100 proteins (calprotectin and calgranulin C) to predict the response to TNF antagonists in patients with Crohn's disease. Inflamm Bowel Dis. 2015;21(2):331–336. doi: 10.1097/MIB.0000000000000273. [DOI] [PubMed] [Google Scholar]
- Serpero LD. et al. Next generation biomarkers for brain injury. J Matern Fetal Neonatal Med. 2013;26(2):44–49. doi: 10.3109/14767058.2013.829688. [DOI] [PubMed] [Google Scholar]
- Douglas-Escobar M, Weiss MD. Biomarkers of brain injury in the premature infant. Front Neurol. 2012;3:185. doi: 10.3389/fneur.2012.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Theis KR. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015;13(8):e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Dinan TG, Cryan JF. Microbial genes, brain & behaviour - epigenetic regulation of the gut-brain axis. Genes Brain Behav. 2014;13(1):69–86. doi: 10.1111/gbb.12109. [DOI] [PubMed] [Google Scholar]
- Goodrich JK. et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54(Pt. 12):1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- Blekhman R. et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM. et al. Microbes & neurodevelopment - Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun. 2015;50:209–220. doi: 10.1016/j.bbi.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Putignani L. et al. The human gut microbiota: a dynamic interplay with the host from birth to senescence settled during childhood. Pediatr Res. 2014;76(1):2–10. doi: 10.1038/pr.2014.49. [DOI] [PubMed] [Google Scholar]
- Borre YE. et al. The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv Exp Med Biol. 2014;817:373–403. doi: 10.1007/978-1-4939-0897-4_17. [DOI] [PubMed] [Google Scholar]