Abstract
Asthma is a highly heterogeneous disease characterized by inflammation of the airways, which invokes symptoms such as wheeze, dyspnea, and chest tightness. Asthma is the product of multiple interconnected immunological processes and represents a constellation of related, but distinct, disease phenotypes. The prevalence of asthma has more than doubled since the 1980s, and efforts to understand this increase have inspired consideration of the microbiome as a key player in the pathophysiology and regulation of this disease. While recent years have seen an explosion of new research in this area, researchers are only beginning to untangle to mechanisms by which the microbiome may influence asthma. This review will focus on the relationship between the microbiome and the immune system and how this influences development of asthma. This review will also highlight evidence that may point the way toward new therapies and potential cures for this ancient respiratory foe.
Keywords: asthma, microbiome, microbiota, allergy, hygiene hypothesis, immune system, immunity
Asthma: Pre-History to Modern Malady
Descriptions of the condition known as asthma have existed since the eighth century B.C., when the term was first used in reference to Hector’s attempt to catch his breath post battle (The Iliad, book XV, line 290). The word ‘asthma’ has evolved much since originally described by Homer, who simply meant it to refer to the symptom of dyspnea (difficulty breathing) rather than the disease itself. Yet it is no wonder that the condition known as modern-day asthma is rooted in the Greek language, having stemmed from the verb ‘aazein’ (to pant or exhale with an open mouth).
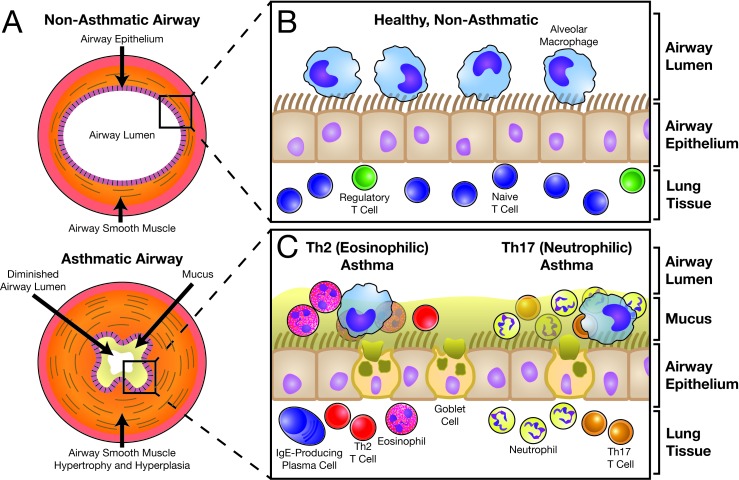
Asthma is a chronic inflammatory disorder of the airways characterized by recurrent symptoms of coughing, wheezing, dyspnea, and chest tightness [1] and affecting people of all ages, races, and sexes in every nation worldwide. This inflammation promotes thickening of the airway wall (Figure 1A) secondary to fibrotic remodeling and airway smooth muscle (ASM) hypertrophy and hyperplasia [2]. Goblet cell hyperplasia and increased mucus production augment wall thickening [3]. An overwhelming drive to expectorate this mucus contributes to the development of asthma-associated cough [3], while airway fibrosis, ASM thickening, and airway hyper-reactivity (AHR, i.e. spasms) of the thickened ASM layer impedes airflow secondary to reduced cross-sectional area of the airway lumen [1]. This airflow restriction is largely responsible for the devastating morbidity of the disorder.
Figure 1.
The Normal and Asthmatic Airway. The anatomy of a healthy, non-diseased airway (A, top) is similar to that of a hose or pipe, with a patent and unobstructed luminal space permitting free flow of air with respiration. Surrounding the airway lumen is a wall of connective tissue and airway smooth muscle. In an asthmatic airway (A, bottom), the airway smooth muscle has undergone hypertrophy and hyperplasia, obstructing the lumen through an increase in mass. Furthermore, asthmatic airway smooth muscle is classically hyperreactive to its triggering stimulus, which leads to constriction and further obstruction of airflow. Finally, heightened mucus production reduces the already diminished lumen, resulting in the sensations of dyspnea and chest tightness that are so characteristic of the disease. From a cellular perspective, the normal and asthmatic airways are dramatically different. In a healthy airway (B), a ciliated respiratory epithelium separates the airway lumen from the surrounding lung tissue. Alveolar macrophages are the dominant cell type within the airway. Within lung tissue, many cell types are present, including naive T cells and regulatory T cells (Tregs). A vigorous immune response is not present. By contrast, the asthmatic airway (C) is characterized by a robust immune response regardless of the asthma endotype. For Th2-driven, eosinophilic asthma (C, left), the airway space becomes dominated by eosinophils and Th2 cells, with more of these cells also present within the lung tissue. Plasma cells are also present and are responsible for production of allergen-specific IgE. For Th17-driven, neutrophilic asthma (C, right), the dominant cell types are Th17 cells and the neutrophils they recruit, with eosinophils, Th2 cells, and IgE being absent. Other cell types, including B cells, mast cells, and basophils, may be present depending on the endotype.
Asthma may fundamentally be a disease of airway inflammation, but this inflammation can manifest itself in multiple ways. While commonly thought of as a childhood disease, asthma may appear throughout life [1]. The severity of symptoms, frequency of severe asthma attacks (a sudden worsening of asthma symptoms), and response to treatment vary between and within age groups [3]. Some children who acquire asthma grow out of it, while others seem unable to shake the condition [4]. Today, the concept of asthma is one of multiple endotypes [5-7], or subtypes defined by “distinct pathophysiologic mechanism[s]” [8].
While many specific features separate asthma endotypes, one concept that has become central to this disorder is the microbiome, i.e. the community of microorganisms that dwell on and within its host. In this review, we will highlight the immunologic mechanisms that contribute to the pathogenesis and regulation of asthma and the means by which interactions with the microbiome may influence these mechanisms.
The Actors on Asthma’s Immunologic Stage
The cast of immunologic actors that put on the asthma “show” is complex and variable [5,7]. The classic description of allergic asthma is an inappropriate immune response against an otherwise innocuous environmental antigen (i.e. allergen), leading to eosinophilia, AHR, and IgE production [1]. Asthma may also be aggravated by non-allergen triggers, ranging from tobacco smoke and environmental pollution to emotional stress and viral or bacterial respiratory infections. Regardless of cause, Th2 cells commonly reside at the center of this process by providing cytokines that influence the function of both immune cells (e.g. eosinophils, B cells) and non-hematopoietic cells (e.g. goblet cells, fibroblasts) (Figure 1B) [5,7].
One particularly interesting development in asthma pathogenesis has been the relatively recent realization that pathways outside the traditional Th2 axis can also drive asthmatic responses, particularly in adult-onset disease [5,7]. One such alternative pathway drives a form of asthma dominated by neutrophils and lacking eosinophilia and other characteristics of Th2 immunity (Figure 1C). This nonatopic, non-IgE dependent form of asthma has been associated with increased levels of gamma interferon (INF-γ), interleukin 17 (IL-17), and Th17 cells, a subset of helper T cells known for production of IL-17 [9]. Both Th17 cells and the IL-17 they produce have been linked to the recruitment of neutrophils to airways [10], increased severity of airway inflammation [11], and the promotion of AHR [12]. In humans, airway neutrophilia negatively correlates with airway function [13], and asthmatic airways on the whole tend to have more Th17 cells than controls [14]. Intriguingly, Th17 cells in animal models have been shown to be resistant to steroid therapy [11], and human Th17 cells seem to share this property [15], potentially explaining why human adults with severe asthma are often resistant to traditional pharmacologic treatments [16].
Of particular importance to asthma are mechanisms of immune regulation, as asthma is at its heart an imbalance of inflammation and regulation tilted in favor of inflammation. One of the most studied immune regulators is the regulatory T cell (Treg), a class of T lymphocytes first identified as being protective against autoimmune inflammation [17]. Since their initial identification, evidence supporting the function of Tregs in protection against a variety of human diseases including asthma has rapidly expanded [18]. Tregs, which are often characterized by constitutive expression of both CD25 and the transcription factor forkhead box P3 (Foxp3), have been shown to protect against experimental asthma by inducing tolerance, or lack of an immune response, to inhaled allergens [19]. Tregs utilize multiple suppressor functions to regulate allergic inflammation, including secretion of inhibitory cytokines such as transforming growth factor beta (TGF-β) and interleukin 10 (IL-10), cell-contact dependent mechanisms, consumption of limiting growth factors, and modulation of dentritic cell (DC) and Th17 activity [20]. Decreased Treg levels or defective Treg function has been associated with development of hyper IgE syndrome, hypereosinophilia, and allergic airway inflammation in Foxp3 mutant mice [21]. Several studies have also shown that the suppression of experimental asthma is accompanied by accumulation of Tregs in the lung and lung-draining lymph node [22,23].
Tregs have been shown to be reduced in the blood of asthmatic individuals relative to control subjects and also possess diminished suppressive capability [24]. Interestingly, studies of Treg numbers in asthmatic lungs have been mixed. Some have shown that asthmatic patients harbor fewer Tregs in their airways [25,26] or a reduced ratio of Tregs to Th2 cells as well as decreased expression of Foxp3 in CD4+CD25+ T cells [27]. One pediatric study also reported Tregs to be functionally impaired in asthmatic children [28], and another investigation has linked a genetic polymorphism in interleukin 33 (IL-33) that is known to predispose to asthma to reduced Tregs [29]. However, earlier work in adult asthmatics reported increased numbers of Tregs in asthmatic airways [30], although they did not examine Treg function. One possible explanation is that these studies assessed patients broadly classified as having “asthma” but who may have exhibited subtly different asthma phenotypes or frankly distinct endotypes. More work comparing human Treg populations among various asthma classifications is required.
Beyond Tregs, many other cell types can regulate asthma. Among these are a population of B cells known as regulatory B cells (Bregs) [31]. Bregs have been found to suppress symptoms of experimental asthma [32] through various mechanisms including induction of Tregs [33]. This induction may occur in the lung-draining lymph node, where Bregs have been shown to accumulate during tolerance development [34]. Unfortunately, the role of Bregs in human asthma is currently unclear. One recent investigation reported that B cells from asthmatic patients produced less IL-10 than those from controls, suggesting that a qualitative difference in Bregs may exist in human asthma [35].
A further cell type with regulatory activity is the DC. A subset of DCs has been shown to be protective in animal models of asthma [36], with subsequent work extending this finding to report that this protection is mediated through DC induction of Tregs [37] and proteolytic modification of DC surface markers [38]. Additionally, evidence has shown that mice lacking CD103+ DCs have diminished ability to resolve asthma-associated inflammation [39]. However, the DC-asthma story is complex, and there is equally good evidence that subsets of DCs help promote Th2 inflammation [40]. Which role DCs play may depend on their interactions with other immune cell types, including Tregs [41].
Asthma: A Growing Epidemic
Asthma has been a scourge of humanity since ancient times, but the disease truly became what has been dubbed the “asthma epidemic” [42] only in the late 20th century. Between the 1980s and 2009, childhood asthma rates more than doubled [42-46]. While there is some recent evidence that overall prevalence may be stabilizing, rates in certain populations, including poor children and adolescents, continue to rise [46].
Heterogeneity of asthma presentation and diagnosis based largely on clinical features with lack of a gold-standard test to confirm or rule out disease have long made estimates of asthma prevalence challenging for epidemiologists [6,42]. To what degree prevalence numbers are over- or underestimated is unknown, but that they are imprecise is certain. For example, some types of asthma that are not accompanied by a Th2 response may be missed by surveys that utilize IgE or pinprick testing as key components of diagnosis.
Regardless of whether prevalence has stabilized, asthma continues to produce millions of missed school and work days [44], representing a significant drain on the national economy and a hindrance to learning. Asthma remains the third leading cause of hospitalization in children [47] and sends millions of victims of all ages to the emergency room annually. From a health disparities perspective, asthma is a challenging disease for many minority populations in the United States. Both African American and Puerto Rican children have significantly higher prevalence of asthma than the general population; the rate for Puerto Rican children is the highest of any group, approaching 20 percent [46]. Unfortunately, efforts to staunch the rising tide of asthma are hampered by a lack of understanding of why certain populations are more prone to asthma development than others [44].
Cleaner Is Not Always Better: the Hygiene Hypothesis
Many theories have been offered to explain the rise in asthma rates in the decades since the upward trend in asthma prevalence first became apparent. Among the most compelling is the hygiene hypothesis of Strachan, originally raised in a study of hay fever [48]. The hypothesis states that the increasingly clean and sterile environment of modern life has promoted the development of many diseases, including asthma, and evidence in support of this has grown exponentially since Strachan’s initial proposal [49].
What, then, is the driving force? Interestingly, the answer may have been proposed over twenty years prior to Strachan’s hygiene hypothesis by a Canadian team comparing native and white communities. In the 1970s, this team observed that native communities suffered far less from asthma and allergic disease than their white counterparts. Years ahead of their time, the researchers opined that the higher frequency of asthma and allergy was merely “the price paid by some members of the white community for their relative freedom from diseases due to viruses, bacteria and helminths” [50]. Microbes, or lack thereof, could be the source of the asthma epidemic [51].
Bring on the Bugs: the Microbiome
The term “microbiome”, popularized by Joshua Lederberg in the beginning of this century [52], refers to the microorganisms that live on and inside another living organism and the interactions they share with their host (for a discussion of the term microbiome and its conflicting definitions, refer to footnote 1).
While recognition of the role of the microbiome has grown primarily in the last two decades, the idea that humans co-exist with microorganisms is far from new. Antonie van Leeuwenhoek first described “animalcules” in his own stool in a letter to Robert Hooke of the Royal Society in 1681 (Letter 34, [57,58]). The explosion in science and knowledge related to the microbiome has suggested that we may be able to harness the microbiome to advance human health, reviving a concept first proposed by Élie Metchnikoff over a century ago [59].
Unfortunately, this increased understanding has also revealed a disturbing trend. Humans have likely coexisted with their microbiomes since the dawn of the species, but opportunities to acquire these microorganisms have dwindled with increasing societal emphasis on cleanliness and sterility, leading to the loss of loss of many so-called microbial friends [60]. According to this “old friends” hypothesis [61], it is the loss of exposure to microorganisms, many of whom humans co-evolved with, that underlies the increasing prevalence of many diseases of the modern age from diabetes and obesity to asthma and allergy.
The Benefits of Dirty Living
The idea that microbial exposure can protect against asthma has a long history, particularly with respect to farm living. Residing on a farm (where microorganisms are commonplace) has long been linked to protection from asthma [62], with a veritable host of studies finding farm living [63-68] and exposure to animals and dirt [69,70] to be protective against the development of wheeze and asthma. Suggestive of a role for microbes as a causative factor for the aforementioned findings is the fact that exposure to endotoxin, a component of gram negative bacterial cell walls, negatively correlated with the development of asthma in children on farms [71]. More recently, the association has extended beyond bacterial endotoxin to encompass a number of bacterial and even fungal products, which together may provide protection from asthma development [72]. Interestingly, this study found the greatest protection came in the middle of the range of exposure levels, with those exposed to the highest levels of microbial products actually at higher risk of developing asthma. As is often true, too much of a good thing can prove problematic.
However, assessing exposure to microbial products does not indicate which specific organisms may be responsible. A growing number of studies have sought to change that, and today researchers know of multiple specific microbial populations that may protect from or predispose to asthma [73-83]. One of the best studied has been the Gram positive phylum Firmicutes. Increased contact with Firmicutes conveys asthma protection [75,77,80,81,84], although at least one Firmicutes species, the enteric pathogen Clostridium difficile, may predispose to asthma [83]. The importance of Firmicutes is illustrative of the need to identify specific microorganisms rather than just microbial products such as endotoxin. Gram positive bacteria, including Firmicutes, lack the endotoxin-containing outer membrane characteristic of Gram negative bacteria, and the influence of such organisms would have escaped notice in earlier studies solely assessing endotoxin.
Perhaps unsurprisingly, there appears to be no single microorganism or group of microorganisms common amongst all studies, suggesting that it is the presence of many different microorganisms that is protective. Indeed, several of the aforementioned microbiome assessments have found that the greater the diversity of microorganisms that one encounters as a child, the lower the lifetime risk of asthma [75,78]. However, other studies have shown no link between overall microbial diversity and asthma development [76,79-81] or even a tendency toward asthma development with increased microbial diversity [82]. The inconsistency may have multiple roots, including varying sources of microbial DNA and the use of both differing methods of microbial DNA extraction and multiple microbial DNA assessment technologies between studies. Overall understanding of how microbial diversity can impact asthma is clearly incomplete, and whether more or less exposure is protective remains to be determined.
Several studies have pointed to fungi as opposed to bacteria as being responsible for protection against asthma. Individual fungal genera, such as Eurotium [75] and Cryptococcus [79], as well as overall fungal diversity [79] have been linked to protection from asthma. There is a relative paucity of information available on the fungal microbiome, particularly in its relationship to asthma, and more study in this area is needed to determine which host-fungal relationships drive asthma protection.
The Dangers of Very Clean Living: Early-Life Antibiotics
Antibiotics have saved countless lives, and one need only look to growing panic over rising antibiotic resistance to understand the necessity of antibiotic use [85]. Unfortunately, antibiotics may affect the development of asthma even as they protect us from truly deadly pathogenic microorganisms. The microbiome changes rapidly in early life [86-88], and antibiotic treatment can have lasting effects on both the microbiome and the host [87,89,90]. Many studies have linked early-life antibiotic exposure, particularly in the first year of life, to the development of asthma [91-95], including one meta-analysis of 20 studies that found antibiotic use in the first year of life was associated with greater than 50 percent higher odds of asthma development [96]. However, there are some caveats. In the aforementioned meta-analysis, prospective studies tended to show no or minimal association between asthma and antibiotic use, while retrospective studies tended to show a clear association [96]. The discrepancy may center on childhood infections, as several groups have suggested that it may be the infections the antibiotics are prescribed to treat rather than the antibiotics themselves that predisposes to asthma [97,98]. The association is likely further complicated, particularly for meta-analyses, by the lack of a gold-standard diagnostic test, with some tests potentially identifying only one or a few asthma endotypes [6,42].
In experimental models of asthma, the relationship between antibiotic use and asthma is decidedly less controversial. Antibiotic administration heightens severity of experimental asthma in mice [99-103], and two studies have reported that neonatal, but not adult, antibiotic exposure produces more severe allergic airway inflammation [101,103]. Determining the link between early antibiotic exposure and asthma development is critical to the reversal of the asthma epidemic, and the answer to this question is likely to be tied to how the microbiome interacts with the host immune system.
The Microbiome Governs the Immune System
The intricate relationship between the microbiome and the host immune system is one of the most important revelations of microbiome research. Today, it is clear that the microbiome touches all aspects of immunity, from development of specific immune cell types to formation of immune organs and tissues [104-108]. Importantly, most of what is known about microbiome-immune interactions comes from work in mouse models of asthma; unless otherwise indicated, all reported relationships in the following section are from studies of laboratory mice. Figure 2 summarizes many of these relationships.
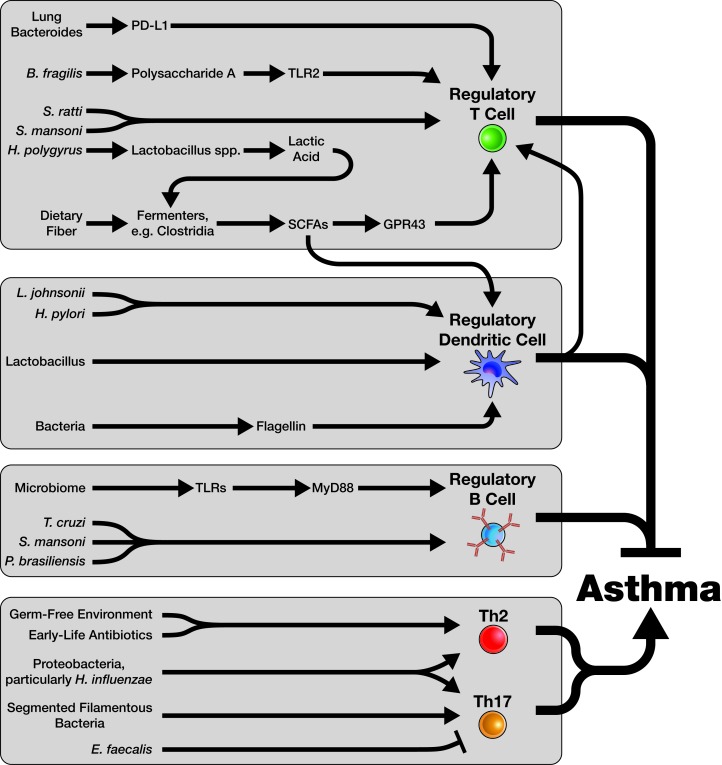
Figure 2.
Immune-Microbiome Interactions in Asthma. The immune system is influenced by the microbiome in a myriad of ways, many of which we are only beginning to understand. With respect to asthma, four key areas of microbiome-immune interactions are particularly relevant. Regulatory T cells (Tregs) play a critical role in regulating asthma pathogenesis and preventing asthma in healthy individuals. The microbiome has a large role in the generation and maintenance of Tregs. Microbial products, particularly short-chain fatty acids (SCFAs); microbe-microbe interactions; and microbial sensing by elements of the immune system (such as through TLRs) all contribute to Treg formation and function. Dendritic cells (DCs) can also play key regulatory roles in the context of asthma. Multiple microbiome-related mechanisms drive regulatory DC formation, including sensing of bacterial products such as flagellin and production of SCFAs by certain microorganisms. Regulatory DCs may also promote the formation of Tregs. The role of the microbiome in generating Regulatory B cells (Bregs) is less clear, but similar mechanisms (e.g. sensing of microbes by TLRs) seem to play a role in this process. Specific microbial products which can induce Bregs are less understood, particularly when compared to Tregs. Tregs, Bregs, and regulatory DCs all act to inhibit asthma pathogenesis. However, pathogenic Th2 and Th17 immune responses can also be driven by the microbiome. An absence of microorganisms, particularly early in life, can drive a Th2 response to allergen, as in a germ free (GF) mouse. Similarly, early-life antibiotic use can promote Th2 responses. Proteobacteria, particularly Haemophilus influenzae, can induce either a Th2 or Th17 response depending on the timing of infection. Other specific organisms can either inhibit or promote Th17 responses. It is likely that many more such relationships will be discovered for both Th2 and Th17 responses as the science of the microbiome advances.
Immune Regulation: Tregs and DCs
The ability of the microbiome to influence regulatory arms of the immune system, particularly Tregs, has fundamentally changed our view of what the microbiome means for asthma control. Multiple studies have shown that increased severity of experimental asthma is associated with a loss of colonic Tregs [99,101,103], and the links between the microbiome and Tregs are increasingly well-described. One of the earliest specific links was made to Bacteroides fragilis, which is able to induce Tregs in the mouse gut [109] through a mechanism mediated by toll-like receptor 2 (TLR2) detection of B. fragilis polysaccharide A (PSA) [110]. Interestingly, loss of PSA leads to a Th17 rather than a Treg response to B. fragilis [111], potentially indicating that loss of Tregs may contribute to the development of Th17-dominated, neutrophilic asthma.
Five years ago, another group demonstrated that a mixture of organisms in the class Clostridia was also capable of inducing Tregs not only in the gut but systemically [112]. Shortly thereafter, the mechanism was elucidated: Clostridial metabolism of dietary fiber leads to production of short chain fatty acids (SCFAs), including butyric acid and propionic acid, which then act through the receptor GPR43 to induce Tregs [113-115]. Further work narrowed down the responsible Clostridia to a specific group of 17 organisms within clusters IV, XIVa, and XVIII [116]. Interestingly, this is one instance where mechanism preceded organism, as the role of SCFAs and GPR43 in Treg induction was identified in an earlier report demonstrating more severe asthma in mice lacking GPR43 [117]. Since these initial reports, the importance of SCFA production to asthma has become increasingly evident, with multiple reports showing that a high fiber diet [118,119] or directly feeding mice SCFAs [119] is protective against experimental asthma. One of these studies provided important insight into how gut microbial metabolism can impact the lung, demonstrating that SCFAs travel to the bone marrow and promote the development of DCs that are highly phagocytic but poor inducers of Th2 responses [118]. This represents a similar phenotype to that of tolerogenic DCs, known Tregs inducers [37].
Other bacteria linked to Treg induction include Bifidobacterium infantis [120], Helicobacter pylori [121-123], and Lactobacillus johnsonii [124]. Interestingly, the protective mechanism of both H. pylori and L. johnsonii also appears to impact induction of Treg-promoting regulatory DCs [121-124]. L. johnsonii’s protective effects may depend on other microorganisms, as the lactic acid they produce may promote beneficial SCFA production by other bacteria [125].
Over the past decade, there has been an explosion of research showing that parasitic infection can influence Treg development [126]. Parasites including Heligmosomoides polygyrus [127-131], Schistosoma mansoni [132], and Strongyloides ratti [133] are all known inducers of Tregs. H. polygyrus infection has also been shown to attenuate the severity of asthma development, possibly through parasite-induced Treg induction [127,128,131]. The mechanism by which H. polygyrus induces Tregs is indirect, as H. polygyrus promotes growth of Lactobacillus species [130] and increases SCFA production by the overall gut microbiome [131]. The rationale behind parasite-mediated Treg induction appears to be entirely evolutionary in nature: without Tregs, parasites such as S. ratti are quickly killed by the host immune system [133].
Unfortunately, the available human data on microbiome Treg induction are mixed. One study lends support to the regulatory DC axis, as human blood DCs cultured with two Lactobacillus species switch phenotype to one that drives development of Tregs [134]. However, B. fragilis and Clostridial species in cluster XIVa represent a more complicated story, as one clinical study linked abundance of these microorganisms at infancy to increased asthma at age three [84]. However, these data may not be as discordant as they first appear. In a study of the effect of day care on asthma development, children in day care had more severe wheeze as toddlers but less asthma by age six [135], suggesting that early promotion of asthma may belie later protection.
For SCFAs, human clinical data are supportive of a protective role. Maternal intake of acetic acid but not butyric or propionic acid was associated with less wheeze and physician office visits in the first 12 months of infant life [119]. The SCFA discrepancy with respect to mice may simply reflect that the SCFAs that drive mouse Treg development differ from those in humans. This area of microbiome research is constantly advancing, and it is likely that answers to some of these open questions will be yielded in the near future.
The Balance Between Th2 and Th17 Responses
The microbiome may also play a role in driving asthma endotype polarization. Germ-free (GF) mice, born and raised without a microbiome, develop immune systems primed to produce Th2 responses that cannot be suppressed [136], and similar Th2-skewing results have been reported with early-life antibiotic exposure [84,99,102]. This immunologic shift results in more severe disease if experimental asthma is induced [99,102,137]. Together, these studies suggest that the rise in asthma seen with a poorly-diverse microbiome may arise from polarization of the human immune system in a Th2 direction.
As previously noted, removing PSA from B. fragilis turns a Treg inducer into a Th17 driver [111], and similar species which lack PSA may be drivers of Th17 themselves. Indeed, in 2008, it was reported that certain microorganisms were required for induction of Th17 cells in the mouse gut [138], with the culprit later identified as segmented filamentous bacteria (SFB), a member of class Clostridia, family Clostridiaceae [139].
While the evidence for SFB influencing asthma phenotype remains minimal, there is evidence of a role for other organisms. Haemophilus influenzae, a common respiratory pathogen, has been found in the lungs of patients with severe neutrophilic asthma [140]. Infecting mice with H. influenzae during the induction of experimental asthma shifts the phenotype of disease, reducing markers of Th2 immunity and driving accumulation of neutrophils and production of IL-17 [141]. However, if H. influenzae is introduced very early in life, prior to asthma induction, it can result in increased severity of disease through elevated Th2 immunity (e.g. IL-5) and reduced Tregs [142]. Finally, one enteric pathogen, an Enterococcus faecalis subspecies, can suppress Th17 immunity and symptoms of allergic airway disease simultaneously [143], although specifically Th2-driven features of asthma (eosinophils, IgE) were not explored in that study. E. faecalis represents an organism worth evaluating to develop a therapeutic that targets both asthma and Th17 immunity.
B Cells and Immunoglobulin
Unlike for Tregs, the Breg-microbiome relationship remains very poorly understood, despite the first evidence in support of such a relationship predating similar Treg findings. In 1996, a pioneering study reported that B10 cells, a population of Bregs characterized by production of the anti-inflammatory cytokine IL-10, are induced by the parasite S. mansoni, although the authors did not examine the phenotype of those Bregs much beyond IL-10 production [144]. Later work confirmed this link in patients suffering from the autoimmune disease multiple sclerosis, showing that B10 cell induction accompanied either Trypanosoma cruzi or Paracoccidioides brasiliensis infection [145]. More recently, two studies have shown that recognition of microbial antigens through TLRs [146] and the TLR signal mediator MyD88 [147] produces B cells which can suppress inflammation during microbial infection. One experimental study demonstrated that S. mansoni can induce B10 cells (together with Tregs) and that transfer of these B10 cells can protect the recipient against experimental asthma [132]. While more work remains before this parasite-B10 axis can be targeted for asthma therapies, it is an exciting area of research and one further potential explanation for why a “dirty” childhood full of infections can protect against asthma.
What About the Lung Microbiome?
While discussion has thus far centered on the gut microbiome, it is critical to consider the role of the microbiome most “proximal” to asthma, the lung microbiome. Despite the obvious potential relevance of the lung microbiome to asthma, this relationship, as with so many things related to the lung microbiome, remains poorly understood. This deficit is, to a large extent, an accident of history. While humans have known for well over three centuries of the existence of gut microorganisms [57,58], the lung microbiome was long considered to be sterile in the absence of active disease [148-152] and was correspondingly excluded from the sites explored in the first Human Microbiome Project [153]. Today, it is widely accepted that even healthy lungs harbor a microbiome, although how it interacts with the pulmonary immune system and what constitutes a “healthy” lung microbiome remain matters of great debate [154,155].
Despite our limited understanding of the lung microbiome, some patterns are beginning to emerge with respect to asthma. Phylum Proteobacteria has been linked to increased severity of asthma in human asthmatics [156-159], with the explanation possibly returning to Th17 cells, as one of these studies reported that Th17-related genes were upregulated in individuals harboring abundant Proteobacteria [159]. It may also return to H. influenzae, as that microorganism is a Proteobacteria. While one recent study has somewhat contradicted this evidence by suggesting that Proteobacteria are associated with less severe asthma, this report did not comment on non-Th2 forms of asthma [160]. When it comes to diversity of the lung microbiome, limited evidence suggests that lung microbiome diversity may have the opposite effect as in the gut, with at least one large clinical study reporting higher diversity and bacterial burden in asthmatic lungs when compared to controls [157].
Just as early life gut microbiome exposures and perturbations appear to affect lifelong asthma risk, so too goes the lung. Two cohort studies of the human infant respiratory microbiome reported that the infant lung microbiome, like the gut microbiome, changes rapidly [161,162], but that colonization with Streptococcal organisms was a strong predictor of wheeze or asthma [161,162]. One of these studies also mirrored results from adult asthmatics, reporting an association between two Proteobacteria, Moraxella catarrhalis and H. influenzae, and development of wheeze and asthma at five years of age [161].
The lung microbiome is one area where the body of experimental work is even more limited than the clinical evidence. However, available experimental evidence does support some clinical findings, particularly the relevance of Proteobacteria. In one recent study, newborn mice tended to produce exaggerated Th2 responses in an experimental asthma model, even in the face of high numbers of thymically-derived Tregs [163]. However, as the lung microbiome bacterial load increased, Gammaproteobacteria were replaced by Bacteroidetes, and inducible Tregs (iTregs), the very Treg population also generated by gut Treg inducers, increased, evidently driven by interactions involving the cell surface receptor PD-L1. When challenged with allergen after this shift, mice were more resistant, and the exaggerated Th2 response disappeared [163]. Perhaps a lack of Bacteroidetes or persistence of Proteobacteria in asthmatic lungs drives human disease.
In the past year, two elegant studies have shown that the lung immune system can be manipulated through application of inhaled bacterial products. In one, endotoxin or endotoxin- and microbial product-containing farm dust was administered intranasally to mice, and either treatment suppressed subsequent experimental asthma through a mechanism mediated by the immunosuppressive protein A20 [164]. Interestingly, this result conflicts with at least one published clinical study, where higher levels of endotoxin were noted in broncho-alveolar lavage (BAL) fluid of asthmatics [158], although it could simply be that endotoxin burden in the human asthmatics was beyond that level at which it was capable of providing benefit. In the second study, a different bacterial product, flagellin, was found to provide protection from experimental asthma induced by either ovalbumin (OVA) or the common human allergen house dust mite (HDM) when administered concurrently with the allergen [165]. The mechanisms of action, while not fully defined, may rely on regulatory DCs and Tregs, as administration of flagellin to human DCs polarized them toward a regulatory, Treg-inducing phenotype [165].
The Changing Landscape of Asthma Therapy
The increasing prevalence and soaring cost of asthma has inspired a search for cures through therapies with immunomodulatory potential. The critical role of Th2 cytokines in asthma has led to the development of several immunobiologic agents for the disease, although results from clinical trials have been largely disappointing. Despite experimental evidence that IL-4 blockade reduces allergic inflammation, phase 2 trials have found no efficacy of anti-IL-4 therapy [166]. Similarly, clinical trials of blockade of either IL-5 [167,168] or IL-13 [169] demonstrated promising reductions in some aspects of asthma pathophysiology but ultimately no meaningful disease symptom improvement. Such studies raise doubts about the efficacy of treating such a complex disorder by targeting single molecules and illustrate the need for more effective immunologic strategies for managing asthma. One unusual proposed therapy is the macrolide class of antibiotics, which were hypothesized to be beneficial due to their polymodal anti-inflammatory functions, including reduction of mucus hypersecretion [170], as opposed to their bacteriostatic effects. However, clinical trials have been contentious to say the least, with no current recommendations supporting the use of macrolides for patients with asthma [171,172]. At present, allergen-specific immunotherapy (ASIT), recognized for more than a century, is the only etiologic therapy for asthma [173]. While human trials largely support the benefits of ASIT for treatment of atopic asthma [174-178], variable safety and efficacy profiles of ASIT regimens [179-182] has limited widespread clinical application. Importantly, the potential for substantial side effects including life-threatening anaphylaxic reactions [183] has led to practice guidelines recommending administration of ASIT only in supervised medical facilities with thirty minutes of post-injection monitoring [179,184,185]. These significant impediments to widespread adoption of ASIT emphasize the need for new therapies with improved safety profiles.
Given the long recognition of the relationship between asthma and the microbiome, it is perhaps surprising that no effective microbiome-based therapies exist to treat or prevent asthma. This paucity of therapies is not for lack of effort. Consider probiotics, or live microorganisms administered to provide a health benefit [186]. A number of experimental studies have fed mice probiotic microorganisms or their products, particularly Lactobacillus species, and found protection from experimental asthma [187-192]. This protection has been variously attributed to increased TLR signaling [187], increased systemic regulatory T cell numbers [189,191], and increased macrophage production of anti-inflammatory substances including IL-10 and TGF-β [190]. However, a meta-analysis of 25 clinical trials of probiotics failed to show any beneficial effect of probiotics on asthma development [193]. While the reasons for the lack of translation from the mouse to the human remain unclear, one possibility is that the effect of adding a single microorganism or product is too small. It is worth noting that the only clinical microbiome therapy in widespread use today is fecal microbiota transplantation (FMT), which involves treatment with an entire microbial community and represents the new gold-standard therapy for antibiotic-resistant C. difficile colitis [194-196]. Successful microbiome asthma therapy may need to look beyond the addition of just one or even a few organisms at a time.
That said, perhaps live organisms are not the answer. Consider the deep pool of evidence reviewed herein that endotoxin exposure prevents asthma development. Administration of endotoxin, or even a cocktail of microbial products including but not limited to endotoxin, may provide protection from asthma development. One active clinical trial is investigating this very therapeutic strategy [197]. Unfortunately, such efforts may correct one problem and cause another, as at least one study has shown that endotoxin and beta glucan shift the profile of asthma from Th2 to Th17 [198].
Another potential option is the prebiotic, or a substance that is not alive but which modifies the microbiome or is utilized by the microbiome to produce a health effect [199]. Dietary fiber is a potent modulator of both the microbiome and (through microbial metabolism) the immune system. At least one active clinical trial is assessing whether increased intake of dietary fiber can modulate asthma pathology [200]. Several studies have looked at another prebiotic, oligosaccharides [201-203], by administering them to infants and assessing asthma symptoms later in life. Unfortunately, results are mixed, with one study reporting that wheeze was reduced later in life [201] and others reporting no effect on wheeze [202,203]. All three studies administered prebiotics during the first six months of life, although the means of administration differed (formula supplementation [201,202] versus probiotic and prebiotic pills [203]). Interestingly, two of these studies assessed the same cohort at different times, two years [201] and five years of age [202], suggesting that a relatively short course of prebiotics very early in life provides protection only for a limited time. Indeed, the second study showing no benefit also looked at asthma and allergy at five years of age [203].
A final point to consider is the relationship between early-life antibiotics and asthma. Avoiding all antibiotics in childhood is illogical, as they are a necessary cure for many childhood illnesses. However, physicians should carefully consider use of antibiotics in all patients and only administer them if truly deemed necessary. Administering antibiotics for an infection that is likely to be viral, for example, is a costly, unnecessary, and potentially harmful endeavor [85]. However, if an antibiotic is absolutely required, it may be reasonable to restore the microbiome to a “healthier” state after the pathogenic organism has been eradicated (i.e. via FMT or similar strategies), as this may reduce the ill effects of antibiotics on rates of asthma.
Conclusion
Although more is learned about the association between asthma and the microbiome on a daily basis, the precise details of this relationship require continued investigation. Advances in the realm of microbiome-immune interactions have fundamentally changed our view of how microorganisms can regulate asthma (Figure 3), but a host of questions remains to be answered. Can microbial modulation of Tregs be harnessed to control asthma? Is the microbiome responsible for the switch from Th2-driven, eosinophilic asthma to the severe, Th17-driven endotype? Although observations from epidemiological studies that have examined the influences of early life exposures (e.g. farming, antibiotic use) on asthma development have certainly been fundamental to our current understanding of the pathophysiological links between asthma and the microbiome, this knowledge has yet to be translated into improved therapeutic strategies. The rapid increase in our understanding of the microbiome suggests that these mysteries may not remain unsolved for long, and the day may soon come when asthma guidelines include one or more microbiome-derived therapies for the prevention or treatment of this widespread and disabling disorder.
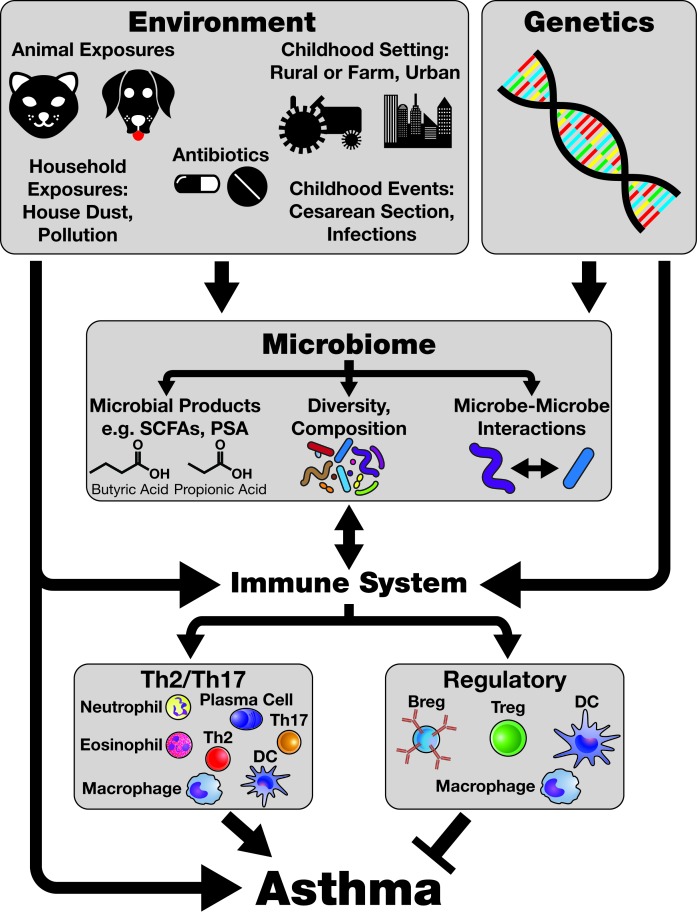
Figure 3.
The Environmental-Microbiome-Immune Axis and Asthma. The relationships between the environment, the microbiome, the immune system, and asthma are extremely complex. Key points discussed in this review are highlighted in Figure 3. The exposures humans encounter in the Environment are particularly important in shaping the microbiome. Antibiotics can eliminate some microorganisms from the microbiome, while the setting in which humans live, such as on a farm as compared to a city apartment, can determine which microorganisms are acquired in childhood. These exposures influence and impact the Microbiome and can determine which microbial products, such as short-chain fatty acids (SCFAs) or polysaccharide A (PSA), are produced. Two SCFAs, butyric acid and propionic acid, are represented here. Factors which affect the microbiome can broadly influence microbiome diversity and composition, including down to the specific organismal level (e.g. antibiotic elimination of Helicobacter pylori). The microbiome can in turn influence the Immune System, which may also be altered by the environment. Many cells responsible for regulating immune activity (from regulatory T cells to dendritic cells, or DCs) and producing effector immune responses (Th2 cells, Th17 cells, eosinophils, and so on) are induced by or otherwise influenced through interaction with the microbiome and its products. This influence can impact the type of immune response generated in the context of asthma, promoting Th2-driven responses, Th17-driven responses, or even a combination thereof. Activity of the immune system is also capable of feeding back on the microbiome through, for example, production of antimicrobial proteins. The activity of the immune system, in conjunction with Genetic influences (which may also impact the microbiome), then determines Asthma pathophysiology, including the specific endotype present, steroid resistance, and how susceptible an individual is to developing asthma in the first place. All of these factors converge to create the constricted, obstructed asthmatic airway that produces such profound morbidity in this condition.
Acknowledgments
The authors wish to thank Travis Bracken for critical comments and suggestions on the manuscript and figures. This work was supported by NHLBI grants F30 HL-126324 to AJA and F30 HL-122018 to SJB.
Abbreviations
- AHR
Airway Hyper-Reactivity
- ASIT
Allergen-Specific Immunotherapy
- AM
Alveolar Macrophage
- ASM
Airway Smooth Muscle
- Breg
Regulatory B Cell
- BAL
Broncho-Alveolar Lavage
- DC
Dendritic Cell
- FMT
Fecal Microbiota Transplant
- Foxp3
Forkhead Box P3
- GPR43
G-Protein Coupled Receptor 43
- GF
Germ Free
- SPF
Specific Pathogen Free
- IgE
Immunoglobulin E
- INF-γ
Interferon Gamma/Gamma Interferon
- IL-4
Interleukin 4
- IL-5
Interleukin 5
- IL-10
Interleukin 10
- IL-17
Interleukin 17
- IL-33
Interleukin 33
- iTreg
Inducible Regulatory T Cell
- LPS
Lipopolysaccharide
- NGS
Next Generation Sequencing
- OVA
Ovalbumin
- PD-L1
Programmed Death-Ligand 1
- PSA
Polysaccharide A
- rRNA
Ribosomal RNA
- SCFA
Short Chain Fatty Acid
- SFB
Segmented Filamentous Bacteria
- Th1
T Helper Type 1
- Th2
T Helper Type 2
- Th17
T Helper Type 17
- TLRs
Toll-like Receptors
- TGF-β
Transforming Growth Factor β
- Treg
Regulatory T Cell
Footnotes
1Microbiome, Microbiota, or Metagenome? The field of microbiome research utilizes many specific terms, and unfortunately some of these have no universally accepted definition, particularly microbiome [53]. Some researchers consider the word “microbiome” to be derived from a joining of the word “microbe” and suffix “ome”, implying all the genomes of all the microorganisms in a community. A second definition is broader, pairing the prefix “micro” with the word “biome” and implying the sum of all organisms, their activity, and their genes. As originally popularized by Joshua Lederberg, microbiome was closer to the latter, defined as “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space” [52]. Such a definition seemingly encompasses the organisms, their interactions with each other, and their interactions with the host. Furthermore, this definition fits with classic use of the term. In work dating to 1988 [54], 1952 [55], and even 1949 [56], the microbiome referred to the entire ecological community of microorganisms and their interactions with their environment. The authors prefer to use microbiome in its classical sense, to refer to the sum of all the organisms, their genomes, and their environmental interactions. When discussing only the organisms, the authors shall use the term microbiota. To speak of the genomes of the microorganisms, the authors shall use the term metagenome.
References
- Postma DS, Rabe KF. The Asthma-COPD Overlap Syndrome. N Engl J Med. 2015;373(13):1241–1249. doi: 10.1056/NEJMra1411863. [DOI] [PubMed] [Google Scholar]
- Berair R, Saunders R, Brightling CE. Origins of increased airway smooth muscle mass in asthma. BMC Med. 2013;11:145. doi: 10.1186/1741-7015-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanta CH. Asthma. N Engl J Med. 2009;360(10):1002–1014. doi: 10.1056/NEJMra0804579. [DOI] [PubMed] [Google Scholar]
- Bisgaard H, Bønnelykke K. Long-term studies of the natural history of asthma in childhood. J Allergy Clin Immunol. 2010;126(2):187–199. doi: 10.1016/j.jaci.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18(5):716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- Sears MR. Trends in the prevalence of asthma. Chest. 2014;145(2):219–225. doi: 10.1378/chest.13-2059. [DOI] [PubMed] [Google Scholar]
- Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2015;16(1):45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- Lötvall J. et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- Cosmi L, Liotta F, Maggi E. et al. Th17 cells: new players in asthma pathogenesis. Allergy. 2011;66(8):989–998. doi: 10.1111/j.1398-9995.2011.02576.x. [DOI] [PubMed] [Google Scholar]
- Liang SC. et al. An IL-17F/A heterodimer protein is produced by mouse Th17 cells and induces airway neutrophil recruitment. J Immunol. 2007;179(11):7791–7799. doi: 10.4049/jimmunol.179.11.7791. [DOI] [PubMed] [Google Scholar]
- McKinley L. et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181(6):4089–4097. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M. et al. IL-17A produced by αβ T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. 2012;18(4):547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw DE. et al. Association between neutrophilic airway inflammation and airflow limitation in adults with asthma. Chest. 2007;132(6):1871–1875. doi: 10.1378/chest.07-1047. [DOI] [PubMed] [Google Scholar]
- Barczyk A. et al. Decreased percentage of CD4(+) Foxp3(+) TGF-β(+) and increased percentage of CD4(+)IL-17(+) cells in bronchoalveolar lavage of asthmatics. J Inflamm Lond Engl. 2014;11:22. doi: 10.1186/1476-9255-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R. et al. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J Exp Med. 2014;211(1):89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesné J. et al. IL-17 in severe asthma. Where do we stand? Am J Respir Crit Care Med. 2014;190(10):1094–1101. doi: 10.1164/rccm.201405-0859PP. [DOI] [PubMed] [Google Scholar]
- Asano M, Toda M, Sakaguchi N. et al. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184(2):387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Khare A, Krishnamoorthy N. et al. Regulatory T cells in many flavors control asthma. Mucosal Immunol. 2010;3(3):216–229. doi: 10.1038/mi.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroukhova M. et al. Tolerance induced by inhaled antigen involves CD4(+) T cells expressing membrane-bound TGF-beta and FOXP3. J Clin Invest. 2004;114(1):28–38. doi: 10.1172/JCI20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead GS. et al. IL-35 production by inducible costimulator (ICOS)-positive regulatory T cells reverses established IL-17-dependent allergic airways disease. J Allergy Clin Immunol. 2012;129(1):207–215. doi: 10.1016/j.jaci.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkoc T, Akdis M, Akdis CA. Update in the mechanisms of allergen-specific immunotheraphy. Allergy Asthma Immunol Res. 2011;3(1):11–20. doi: 10.4168/aair.2011.3.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson WF. et al. Accumulation of regulatory T cells in local draining lymph nodes of the lung correlates with spontaneous resolution of chronic asthma in a murine model. Int Arch Allergy Immunol. 2008;145(3):231–243. doi: 10.1159/000109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken SJ. et al. Long-Term Exposure to House Dust Mite Leads to the Suppression of Allergic Airway Disease Despite Persistent Lung Inflammation. Int Arch Allergy Immunol. 2015;166(4):243–258. doi: 10.1159/000381058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamessier E. et al. T-cell activation during exacerbations: a longitudinal study in refractory asthma. Allergy. 2008;63(9):1202–1210. doi: 10.1111/j.1398-9995.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- Akdis M. et al. Immune responses in healthy and allergic individuals are characterized by a fine balance between allergen-specific T regulatory 1 and T helper 2 cells. J Exp Med. 2004;199(11):1567–1575. doi: 10.1084/jem.20032058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DS. Regulatory T cells and asthma. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2009;39(9):1314–1323. doi: 10.1111/j.1365-2222.2009.03301.x. [DOI] [PubMed] [Google Scholar]
- Lin Y-L, Shieh C-C, Wang J-Y. The functional insufficiency of human CD4+CD25 high T-regulatory cells in allergic asthma is subjected to TNF-alpha modulation. Allergy. 2008;63(1):67–74. doi: 10.1111/j.1398-9995.2007.01526.x. [DOI] [PubMed] [Google Scholar]
- Hartl D. et al. Quantitative and functional impairment of pulmonary CD4+CD25hi regulatory T cells in pediatric asthma. J Allergy Clin Immunol. 2007;119(5):1258–1266. doi: 10.1016/j.jaci.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Schröder PC. et al. IL-33 polymorphisms are associated with increased risk of hay fever and reduced Regulatory T cells in a birth cohort. Pediatr Allergy Immunol. 2016;00:00. doi: 10.1111/pai.12597. [DOI] [PubMed] [Google Scholar]
- Smyth LJC, Eustace A, Kolsum U. et al. Increased airway T regulatory cells in asthmatic subjects. Chest. 2010;138(4):905–912. doi: 10.1378/chest.09-3079. [DOI] [PubMed] [Google Scholar]
- Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- Singh A. et al. Regulatory Role of B Cells in a Murine Model of Allergic Airway Disease. J Immunol. 2008;180(11):7318–7326. doi: 10.4049/jimmunol.180.11.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amu S. et al. Regulatory B cells prevent and reverse allergic airway inflammation via FoxP3-positive T regulatory cells in a murine model. J Allergy Clin Immunol. 2010;125(5):1114–1124. doi: 10.1016/j.jaci.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Natarajan P. et al. Regulatory B cells from hilar lymph nodes of tolerant mice in a murine model of allergic airway disease are CD5(+), express TGF-β, and co-localize with CD4(+)Foxp3(+) T cells. Mucosal Immunol. 2012;5(6):691–701. doi: 10.1038/mi.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlugt LEPM. et al. CD24(hi)CD27(+) B cells from patients with allergic asthma have impaired regulatory activity in response to lipopolysaccharide. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2014;44(4):517–528. doi: 10.1111/cea.12238. [DOI] [PubMed] [Google Scholar]
- Fujita S. et al. Regulatory dendritic cells protect against allergic airway inflammation in a murine asthmatic model. J Allergy Clin Immunol. 2008;121(1):95–104. doi: 10.1016/j.jaci.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Huang H, Dawicki W, Zhang X. Tolerogenic dendritic cells induce CD4+CD25hiFoxp3+ regulatory T cell differentiation from CD4+CD25-/loFoxp3- effector T cells. J Immunol. 2010;185(9):5003–5010. doi: 10.4049/jimmunol.0903446. [DOI] [PubMed] [Google Scholar]
- Secor ER. et al. Bromelain Inhibits Allergic Sensitization and Murine Asthma via Modulation of Dendritic Cells. Evid-Based Complement Altern Med. 2013;2013:702196. doi: 10.1155/2013/702196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez E. et al. Pulmonary CD103 expression regulates airway inflammation in asthma. Am J Physiol Lung Cell Mol Physiol. 2015;308(8):L816–L826. doi: 10.1152/ajplung.00319.2014. [DOI] [PubMed] [Google Scholar]
- Plantinga M. et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38(2):322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Lewkowich IP. et al. CD4+CD25+ T cells protect against experimentally induced asthma and alter pulmonary dendritic cell phenotype and function. J Exp Med. 2005;202(11):1549–1561. doi: 10.1084/jem.20051506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355(21):2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- Akinbami LJ, Moorman JE, Garbe PL. Status of childhood asthma in the United States, 1980-2007. Pediatrics. 2009;123(3):S131–S145. doi: 10.1542/peds.2008-2233C. [DOI] [PubMed] [Google Scholar]
- Akinbami LJ, Moorman JE, Liu X. Asthma prevalence, health care use, and mortality: United States, 2005-2009. . Natl Health Stat Rep. 2011;32:1–14. [PubMed] [Google Scholar]
- Moorman JE. et al. National surveillance of asthma: United States, 2001-2010. . Vital Health Stat 3. 2012;35:1–58. [PubMed] [Google Scholar]
- Akinbami LJ, Simon AE, Rossen LM. Changing Trends in Asthma Prevalence Among Children. Pediatrics. 2016;137(1):1–7. doi: 10.1542/peds.2015-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Health Statistics. Health, United States, 2014: With Special Feature on Adults Aged 55–64. Centers for Disease Control. Available from: http://www.cdc.gov/nchs/hus.htm . [PubMed]
- Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan DP. Family size, infection and atopy: the first decade of the “hygiene hypothesis.”. Thorax. 2000;55(1):S2–S10. doi: 10.1136/thorax.55.suppl_1.s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard JW, Geddes CA, Reggin PL. et al. Serum IgE levels in white and metis communities in Saskatchewan. Ann Allergy. 1976;37(2):91–100. [PubMed] [Google Scholar]
- Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1(1):69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- Lederberg J, McCray A. ’Ome Sweet ’Omics—a genealogical treasury of words. The Scientist. 2001;15(7):8. [Google Scholar]
- Huss J. Methodology and Ontology in Microbiome Research. Biol Theory. 2014;9(4):392–400. doi: 10.1007/s13752-014-0187-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge MN. Fungi in Biological Control Systems. Manchester: Manchester University Press; 1988. [Google Scholar]
- Mohr JL. Protozoa as Indicators of Pollution. Sci Mon. 1952;74(1):7–9. [Google Scholar]
- Revue odontologique. Paris: Société syndicate odontologique de France; 1949. [Google Scholar]
- van Leeuwenhoek A. Ontledingen en ontdekkingen van levende dierkens in de teel-deelen van verscheide dieren, vogelen en visschen: van het hout met der solver meenigvuldige vaaton ... Leiden: C. Boutesteyn; 1686. [Google Scholar]
- Dobell C, van Leeuwenhoek A. Antony van Leeuwenhoek and his “Little animals”; being some account of the father of protozoology and bacteriology and his multifarious discoveries in these disciplines. [cited 2016 May 21]. Availabel from: http://archive.org/details/antonyvanleeuwen00dobe .
- Metchnikoff E. The Prolongation of Life: Optimistic Studies. [cited 2015 Jan 17]. Available from: http://archive.org/details/prolongationofli00metciala .
- Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook GA. et al. Mycobacteria and other environmental organisms as immunomodulators for immunoregulatory disorders. Springer Semin Immunopathol. 2004;25(3-4):237–255. doi: 10.1007/s00281-003-0148-9. [DOI] [PubMed] [Google Scholar]
- von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10(12):861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- Von Ehrenstein OS. et al. Reduced risk of hay fever and asthma among children of farmers. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2000;30(2):187–193. doi: 10.1046/j.1365-2222.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- Riedler J, Eder W, Oberfeld G. et al. Austrian children living on a farm have less hay fever, asthma and allergic sensitization. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2000;30(2):194–200. doi: 10.1046/j.1365-2222.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- Braun-Fahrländer C. et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347(12):869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- Sordillo JE. et al. Multiple microbial exposures in the home may protect against asthma or allergy in childhood. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2010;40(6):902–910. doi: 10.1111/j.1365-2222.2010.03509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illi S. et al. Protection from childhood asthma and allergy in Alpine farm environments-the GABRIEL Advanced Studies. J Allergy Clin Immunol. 2012;129(6):1470–1477. doi: 10.1016/j.jaci.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Feng M. et al. Associations of early life exposures and environmental factors with asthma among children in rural and urban areas of Guangdong, China. Chest. 2016;149(4):1030–1041. doi: 10.1016/j.chest.2015.12.028. [DOI] [PubMed] [Google Scholar]
- Riedler J. et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet Lond Engl. 2001;358(9288):1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- Ege MJ. et al. Not all farming environments protect against the development of asthma and wheeze in children. J Allergy Clin Immunol. 2007;119(5):1140–1147. doi: 10.1016/j.jaci.2007.01.037. [DOI] [PubMed] [Google Scholar]
- Karvonen AM. et al. Exposure to microbial agents in house dust and wheezing, atopic dermatitis and atopic sensitization in early childhood: a birth cohort study in rural areas. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2012;42(8):1246–1256. doi: 10.1111/j.1365-2222.2012.04002.x. [DOI] [PubMed] [Google Scholar]
- Karvonen AM. et al. Quantity and diversity of environmental microbial exposure and development of asthma: a birth cohort study. Allergy. 2014;69(8):1092–1101. doi: 10.1111/all.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167(8):821–827. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- Penders J. et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ege MJ. et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- Bisgaard H. et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128(3):646–655. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- Lynch SV. et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134(3):593–601. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson TR. et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2004;44(6):842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- Dannemiller KC. et al. Next-generation DNA sequencing reveals that low fungal diversity in house dust is associated with childhood asthma development. Indoor Air. 2014;24(3):236–247. doi: 10.1111/ina.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M-C. et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- Ciaccio CE. et al. Home dust microbiota is disordered in homes of low-income asthmatic children. J Asthma Off J Assoc Care Asthma. 2015;52(9):873–880. doi: 10.3109/02770903.2015.1028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemiller KC, Gent JF, Leaderer BP. et al. Indoor microbial communities: Influence on asthma severity in atopic and nonatopic children. J Allergy Clin Immunol. 2016;138(1):76–83. doi: 10.1016/j.jaci.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen FA. et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128(5):948–955. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Vael C, Vanheirstraeten L, Desager KN. et al. Denaturing gradient gel electrophoresis of neonatal intestinal microbiota in relation to the development of asthma. BMC Microbiol. 2011;11:68. doi: 10.1186/1471-2180-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Cars O. Antibiotic Resistance — Problems, Progress, and Prospects. N Engl J Med. 2014;371(19):1761–1763. doi: 10.1056/NEJMp1408040. [DOI] [PubMed] [Google Scholar]
- Koenig JE. et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulfer A, Blaser MJ. Risks of Antibiotic Exposures Early in Life on the Developing Microbiome. PLoS Pathog. 2015;11(7):e1004903. doi: 10.1371/journal.ppat.1004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone KA, Dowd SE, Stamatas GN. et al. Diversity of the Human Skin Microbiome Early in Life. J Invest Dermatol. 2011;131(10):2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney KM, Yurist-Doutsch S, Arrieta M-C. et al. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68:2017–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- Droste JH. et al. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease? Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2000;30(11):1547–1553. doi: 10.1046/j.1365-2222.2000.00939.x. [DOI] [PubMed] [Google Scholar]
- Sobko T. et al. Neonatal sepsis, antibiotic therapy and later risk of asthma and allergy. Paediatr Perinat Epidemiol. 2010;24(1):88–92. doi: 10.1111/j.1365-3016.2009.01080.x. [DOI] [PubMed] [Google Scholar]
- Mai X-M, Kull I, Wickman M. et al. Antibiotic use in early life and development of allergic diseases: respiratory infection as the explanation. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2010;40(8):1230–1237. doi: 10.1111/j.1365-2222.2010.03532.x. [DOI] [PubMed] [Google Scholar]
- Kwon J-W. et al. Changes in the Prevalence of Childhood Asthma in Seoul from 1995 to 2008 and Its Risk Factors. Allergy Asthma Immunol Res. 2011;3(1):27–33. doi: 10.4168/aair.2011.3.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risnes KR, Belanger K, Murk W. Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. Am J Epidemiol. 2011;173(3):310–318. doi: 10.1093/aje/kwq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk W, Risnes KR, Bracken MB. Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics. 2011;127(6):1125–1138. doi: 10.1542/peds.2010-2092. [DOI] [PubMed] [Google Scholar]
- Lapin B. et al. The Relationship of Early-Life Antibiotic Use with Asthma in At-Risk Children. J Allergy Clin Immunol. 2014;134(3):728–729. doi: 10.1016/j.jaci.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örtqvist AK. et al. Antibiotics in fetal and early life and subsequent childhood asthma: nationwide population based study with sibling analysis. BMJ. 2014;349:g6979. doi: 10.1136/bmj.g6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Noggle RM, Toews GB. et al. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72(9):4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr MC, Falkowski NR, McDonald RA. et al. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73(1):30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL. et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA. et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18(4):538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL. et al. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4(2):158–164. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol. 2015;6:1085. doi: 10.3389/fmicb.2015.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159(2):122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Arpaia N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Maslowski KM. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2012;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Thorburn AN. et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- O’Mahony C. et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4(8):e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold IC. et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121(8):3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertli M. et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J Clin Invest. 2012;122(3):1082–1096. doi: 10.1172/JCI61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler DB. et al. Effective treatment of allergic airway inflammation with Helicobacter pylori immunomodulators requires BATF3-dependent dendritic cells and IL-10. Proc Natl Acad Sci U S A. 2014;111(32):11810–11815. doi: 10.1073/pnas.1410579111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE. et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111(2):805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T, Hashizume K, Koyama H. et al. Stimulation of butyrate production through the metabolic interaction among lactic acid bacteria, Lactobacillus acidophilus, and lactic acid-utilizing bacteria, Megasphaera elsdenii, in porcine cecal digesta. Anim Sci J. 2006;77(4):454–461. [Google Scholar]
- Reynolds LA, Finlay BB. Worming Their Way into the Picture: Microbiota Help Helminths Modulate Host Immunity. Immunity. 2015;43(5):840–842. doi: 10.1016/j.immuni.2015.10.025. [DOI] [PubMed] [Google Scholar]
- Wilson MS. et al. Suppression of allergic airway inflammation by helminth-induced regulatory T cells. J Exp Med. 2005;202(9):1199–1212. doi: 10.1084/jem.20042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagaki K. et al. Intestinal helminths protect in a murine model of asthma. J Immunol. 2006;177(3):1628–1635. doi: 10.4049/jimmunol.177.3.1628. [DOI] [PubMed] [Google Scholar]
- Grainger JR. et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-β pathway. J Exp Med. 2010;207(11):2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LA. et al. Commensal-pathogen interactions in the intestinal tract: lactobacilli promote infection with, and are promoted by, helminth parasites. Gut Microbes. 2014;5(4):522–532. doi: 10.4161/gmic.32155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss MM. et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity. 2015;43(5):998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlugt LEPM. et al. Schistosomes induce regulatory features in human and mouse CD1d(hi) B cells: inhibition of allergic inflammation by IL-10 and regulatory T cells. PloS One. 2012;7(2):e30883. doi: 10.1371/journal.pone.0030883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhaus B. et al. Strongyloides ratti infection induces expansion of Foxp3+ regulatory T cells that interfere with immune response and parasite clearance in BALB/c mice. J Immunol. 2011;186(7):4295–4305. doi: 10.4049/jimmunol.1001920. [DOI] [PubMed] [Google Scholar]
- Smits HH. et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J Allergy Clin Immunol. 2005;115(6):1260–1267. doi: 10.1016/j.jaci.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Ball TM. et al. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med. 2000;343(8):538–543. doi: 10.1056/NEJM200008243430803. [DOI] [PubMed] [Google Scholar]
- Sudo N. et al. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159(4):1739–1745. [PubMed] [Google Scholar]
- Herbst T. et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184(2):198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- Ivanov II. et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LG, Simpson JL, Hansbro PM. et al. Potentially pathogenic bacteria cultured from the sputum of stable asthmatics are associated with increased 8-isoprostane and airway neutrophilia. Free Radic Res. 2010;44(2):146–154. doi: 10.3109/10715760903362576. [DOI] [PubMed] [Google Scholar]
- Essilfie A-T. et al. Haemophilus influenzae infection drives IL-17-mediated neutrophilic allergic airways disease. PLoS Pathog. 2011;7(10):e1002244. doi: 10.1371/journal.ppat.1002244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann JR, Mason SN, Auten RL. et al. Early life intranasal colonization with nontypeable Haemophilus influenzae exacerbates juvenile airways disease in mice. Infect Immun. 2016;84(7):2022–2030. doi: 10.1128/IAI.01539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, An J, Shimada T. et al. Oral administration of Enterococcus faecalis FK-23 suppresses Th17 cell development and attenuates allergic airway responses in mice. Int J Mol Med. 2012;30(2):248–254. doi: 10.3892/ijmm.2012.1010. [DOI] [PubMed] [Google Scholar]
- Palanivel V. et al. B-cell outgrowth and ligand-specific production of IL-10 correlate with Th2 dominance in certain parasitic diseases. Exp Parasitol. 1996;84(2):168–177. doi: 10.1006/expr.1996.0102. [DOI] [PubMed] [Google Scholar]
- Correale J, Farez M, Razzitte G. Helminth infections associated with multiple sclerosis induce regulatory B cells. Ann Neurol. 2008;64(2):187–199. doi: 10.1002/ana.21438. [DOI] [PubMed] [Google Scholar]
- Sayi A. et al. TLR-2-activated B cells suppress Helicobacter-induced preneoplastic gastric immunopathology by inducing T regulatory-1 cells. J Immunol. 2011;186(2):878–890. doi: 10.4049/jimmunol.1002269. [DOI] [PubMed] [Google Scholar]
- Neves P. et al. Signaling via the MyD88 adaptor protein in B cells suppresses protective immunity during Salmonella typhimurium infection. Immunity. 2010;33(5):777–790. doi: 10.1016/j.immuni.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Johanson WG, Pierce AK, Sanford JP. Changing pharyngeal bacterial flora of hospitalized patients. Emergence of gram-negative bacilli. N Engl J Med. 1969;281(21):1137–1140. doi: 10.1056/NEJM196911202812101. [DOI] [PubMed] [Google Scholar]
- Higuchi JH, Coalson JJ, Johanson WG. Bacteriologic diagnosis of nosocomial pneumonia in primates. Usefulness of the protected specimen brush. Am Rev Respir Dis. 1982;125(1):53–57. doi: 10.1164/arrd.1982.125.1.53. [DOI] [PubMed] [Google Scholar]
- Baughman RP, Thorpe JE, Staneck J. et al. Use of the protected specimen brush in patients with endotracheal or tracheostomy tubes. Chest. 1987;91(2):233–236. doi: 10.1378/chest.91.2.233. [DOI] [PubMed] [Google Scholar]
- Thorpe JE, Baughman RP, Frame PT. et al. Bronchoalveolar lavage for diagnosing acute bacterial pneumonia. J Infect Dis. 1987;155(5):855–861. doi: 10.1093/infdis/155.5.855. [DOI] [PubMed] [Google Scholar]
- Chastre J. et al. Diagnosis of nosocomial bacterial pneumonia in intubated patients undergoing ventilation: comparison of the usefulness of bronchoalveolar lavage and the protected specimen brush. Am J Med. 1988;85(4):499–506. doi: 10.1016/s0002-9343(88)80085-8. [DOI] [PubMed] [Google Scholar]
- Peterson J. et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal LN, Blaser MJ. A Brave New World: The Lung Microbiota in an Era of Change. Ann Am Thorac Soc. 2014;11(1):S21–S27. doi: 10.1513/AnnalsATS.201306-189MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Martinez FJ. The Microbiome and the Respiratory Tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty M. et al. Disordered microbial communities in asthmatic airways. PloS One. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ. et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127(2):372–383. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goleva E. et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188(10):1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ. et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015;136(4):874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PloS One. 2016;11(4):e0152724. doi: 10.1371/journal.pone.0152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard H. et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357(15):1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- Teo SM. et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollwitzer ES. et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20(6):642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- Schuijs MJ. et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349(6252):1106–1110. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- Shim J-U. et al. Flagellin suppresses experimental asthma by generating regulatory dendritic cells and T cells. J Allergy Clin Immunol. 2016;137(2):426–435. doi: 10.1016/j.jaci.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Hart TK. et al. Preclinical efficacy and safety of pascolizumab (SB 240683): a humanized anti-interleukin-4 antibody with therapeutic potential in asthma. Clin Exp Immunol. 2002;130(1):93–100. doi: 10.1046/j.1365-2249.2002.01973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckie MJ. et al. Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response. Lancet Lond Engl. 2000;356(9248):2144–2148. doi: 10.1016/s0140-6736(00)03496-6. [DOI] [PubMed] [Google Scholar]
- Flood-Page P. et al. A study to evaluate safety and efficacy of mepolizumab in patients with moderate persistent asthma. Am J Respir Crit Care Med. 2007;176(11):1062–1071. doi: 10.1164/rccm.200701-085OC. [DOI] [PubMed] [Google Scholar]
- Gauvreau GM. et al. Effects of interleukin-13 blockade on allergen-induced airway responses in mild atopic asthma. Am J Respir Crit Care Med. 2011;183(8):1007–1014. doi: 10.1164/rccm.201008-1210OC. [DOI] [PubMed] [Google Scholar]
- Kanoh S, Rubin BK. Mechanisms of Action and Clinical Application of Macrolides as Immunomodulatory Medications. Clin Microbiol Rev. 2010;23(3):590–615. doi: 10.1128/CMR.00078-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EHC, Porter JD, Edwards MR. et al. The role of macrolides in asthma: current evidence and future directions. Lancet Respir Med. 2014;2(8):657–670. doi: 10.1016/S2213-2600(14)70107-9. [DOI] [PubMed] [Google Scholar]
- Kew KM, Undela K, Kotortsi I. et al. Macrolides for chronic asthma. Cochrane Database Syst Rev. 2015;9:CD002997. doi: 10.1002/14651858.CD002997.pub4. [DOI] [PubMed] [Google Scholar]
- Noon L. Originally published as Volume 1, Issue 4580 PROPHYLACTIC INOCULATION AGAINST HAY FEVER. The Lancet. 1911;177(4580):1572–1573. [Google Scholar]
- Barnes PJ. New therapies for asthma: is there any progress? Trends Pharmacol Sci. 2010;31(7):335–343. doi: 10.1016/j.tips.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Durham SR. et al. Long-term clinical efficacy of grass-pollen immunotherapy. N Engl J Med. 1999;341(7):468–475. doi: 10.1056/NEJM199908123410702. [DOI] [PubMed] [Google Scholar]
- Eng PA, Reinhold M, Gnehm HPE. Long-term efficacy of preseasonal grass pollen immunotherapy in children. Allergy. 2002;57(4):306–312. doi: 10.1034/j.1398-9995.2002.1o3264.x. [DOI] [PubMed] [Google Scholar]
- Ali I, Goksal K, Ozan B. et al. Long-term allergen-specific immunotherapy correlates with long-term allergen-specific immunological tolerance. Adv Ther. 2008;25(1):29–36. doi: 10.1007/s12325-008-0004-3. [DOI] [PubMed] [Google Scholar]
- Hankin CS, Cox L, Bronstone A. et al. Allergy immunotherapy: reduced health care costs in adults and children with allergic rhinitis. J Allergy Clin Immunol. 2013;131(4):1084–1091. doi: 10.1016/j.jaci.2012.12.662. [DOI] [PubMed] [Google Scholar]
- Greenberg MA. et al. Late and immediate systemic-allergic reactions to inhalant allergen immunotherapy. J Allergy Clin Immunol. 1986;77(6):865–870. doi: 10.1016/0091-6749(86)90385-4. [DOI] [PubMed] [Google Scholar]
- Roy SR. et al. Increased frequency of large local reactions among systemic reactors during subcutaneous allergen immunotherapy. Ann Allergy Asthma Immunol Off Publ Am Coll Allergy Asthma Immunol. 2007;99(1):82–86. doi: 10.1016/S1081-1206(10)60626-6. [DOI] [PubMed] [Google Scholar]
- Cox L. Accelerated immunotherapy schedules: review of efficacy and safety. Ann Allergy Asthma Immunol Off Publ Am Coll Allergy Asthma Immunol. 2006;97(2):126–202. doi: 10.1016/S1081-1206(10)60003-8. [DOI] [PubMed] [Google Scholar]
- Mellerup MT, Hahn GW, Poulsen LK. et al. Safety of allergen-specific immunotherapy. Relation between dosage regimen, allergen extract, disease and systemic side-effects during induction treatment. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2000;30(10):1423–1429. doi: 10.1046/j.1365-2222.2000.00910.x. [DOI] [PubMed] [Google Scholar]
- Caminati M. et al. Incidence and risk factors for subcutaneous immunotherapy anaphylaxis: the optimization of safety. Expert Rev Clin Immunol. 2015;11(2):233–245. doi: 10.1586/1744666X.2015.988143. [DOI] [PubMed] [Google Scholar]
- Huggins JL, Looney RJ. Allergen immunotherapy. Am Fam Physician. 2004;70(4):689–696. [PubMed] [Google Scholar]
- Cox L. et al. Allergen immunotherapy: a practice parameter third update. J Allergy Clin Immunol. 2011;127(1):S1–S55. doi: 10.1016/j.jaci.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Sanders ME. Probiotics: definition, sources, selection, and uses. Clin Infect Dis Off Publ Infect Dis Soc Am. 2008;46(2):S58–S151. doi: 10.1086/523341. [DOI] [PubMed] [Google Scholar]
- Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am J Respir Crit Care Med. 2007;175(6):561–569. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- Ezendam J, van Loveren H. Lactobacillus casei Shirota administered during lactation increases the duration of autoimmunity in rats and enhances lung inflammation in mice. Br J Nutr. 2008;99(1):83–90. doi: 10.1017/S0007114507803412. [DOI] [PubMed] [Google Scholar]
- Karimi K, Inman MD, Bienenstock J. et al. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med. 2009;179(3):186–193. doi: 10.1164/rccm.200806-951OC. [DOI] [PubMed] [Google Scholar]
- Harb H. et al. Neonatal supplementation of processed supernatant from Lactobacillus rhamnosus GG improves allergic airway inflammation in mice later in life. Clin Exp Allergy J Br Soc Allergy Clin Immunol. 2013;43(3):353–364. doi: 10.1111/cea.12047. [DOI] [PubMed] [Google Scholar]
- Jang S-O. et al. Asthma Prevention by Lactobacillus Rhamnosus in a Mouse Model is Associated With CD4(+)CD25(+)Foxp3(+) T Cells. Allergy Asthma Immunol Res. 2012;4(3):150–156. doi: 10.4168/aair.2012.4.3.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacSharry J. et al. Immunomodulatory effects of feeding with Bifidobacterium longum on allergen-induced lung inflammation in the mouse. Pulm Pharmaco. Ther. 2012;25(4):325–334. doi: 10.1016/j.pupt.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Elazab N. et al. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 2013;132(3):e666–e676. doi: 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- Bakken JS. et al. Treating Clostridium difficile Infection with Fecal Microbiota Transplantation. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2011;9(12):1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt LJ. et al. Long-Term Follow-Up of Colonoscopic Fecal Microbiota Transplant for Recurrent Clostridium difficile Infection. Am J Gastroenterol. 2012;107(7):1079–1087. doi: 10.1038/ajg.2012.60. [DOI] [PubMed] [Google Scholar]
- Youngster I. et al. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA. 2014;312(17):1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- Martinez FD, Firmage L. National Library of Medicine. 2000. Available from: https://clinicaltrials.gov/ct2/show/NCT02148796 .
- Hadebe S. et al. Microbial Ligand Costimulation Drives Neutrophilic Steroid-Refractory Asthma. PloS One. 2015;10(8):e0134219. doi: 10.1371/journal.pone.0134219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavin J. Fiber and Prebiotics: Mechanisms and Health Benefits. Nutrients. 2013;5(4):1417–1435. doi: 10.3390/nu5041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig D, Hron B, Ebbeling C. Asthma (and Dietary) Inflammation Reduction (AIR). Available from: ClinicalTrials.gov .
- Arslanoglu S. et al. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr. 2008;138(6):1091–1095. doi: 10.1093/jn/138.6.1091. [DOI] [PubMed] [Google Scholar]
- Arslanoglu S. Early neutral prebiotic oligosaccharide supplementation reduces the incidence of some allergic manifestations in the first 5 years of life. J Biol Regul Homeost Agents. 2012;26(3):49–59. [PubMed] [Google Scholar]
- Kukkonen AK. Airway inflammation in probiotic-treated children at 5 years. Pediatr Allergy Immunol Off Publ Eur Soc Pediatr Allergy Immunol. 2011;22(2):249–251. doi: 10.1111/j.1399-3038.2010.01079.x. [DOI] [PubMed] [Google Scholar]