Abstract
The trillions of microbes that inhabit the human gut (the microbiota) together with the host comprise a complex ecosystem, and like any ecosystem, health relies on stability and balance. Some of the most important members of the human microbiota are those that help maintain this balance via modulation of the host immune system. Gut microbes, through both molecular factors (such as capsular components) and by-products of their metabolism (such as Short Chain Fatty Acids (SCFAs)), can influence both innate and adaptive components of the immune system, in ways that can drive both effector, and regulatory responses. Here we review how commensal microbes can specifically promote a dynamic balance of these immune responses in the mammalian gut.
Keywords: Th17, iTreg, MAMPS, SCFA, metabolites, PSA, IL-10, inflammatory disease
Introduction
Microbiome research has recently undergone a rapid expansion and is fundamentally changing the way we view microbial life and our relationship to that unseen world. This explosion of microbiome research has been fueled by advances in genomic technologies, such as next generation sequencing, that allows researchers to gain a better understanding of the complex communities of microbes that inhabit the human body. The use of these technologies has provided unprecedented insight into how our microbial communities shift in states of disease and health, inspiring researchers to investigate the complex relationships between the host’s health and the state of their microbiome.
Our bodies are teeming with microbial life. Some reports estimate that we host up to 1023–1024 microbial cells, vastly outnumbering human cells; these microbes encode an estimated 3.3 million genes compared to a paltry 23,000 encoded by the human genome [1,2]. To put it simply, we are the human scaffold for a dense microbial community, and this is not a simple jumble of bacteria. This population of microbes, together with the human host, comprises a complex ecosystem that consists of thousands of different microbial species that inhabit unique niches in the body and play different roles within their communities [3]. Some of these microbes help us digest our food and extract necessary vitamins and minerals from our diet, some protect us from pathogenic microbes, others may even contribute to our mood and behaviors, and some help regulate our immune system [4,5,6]. Like any ecosystem, the health of the microbiome relies on stability and balance. Thus some of the most important members of the human microbiota are those that help maintain this balance, a feat that is accomplished in part via microbial modulation of the host immune system.
The human immune system plays many complex roles in maintaining health. A key role of the effector arm of the immune system is to vigilantly patrol for and destroy pathogens and malignant host cells, while the regulatory arm maintains balance by quelling the effector responses to prevent tissue damage during pathogen clearance, prevent autoimmunity, and promote tolerance of innocuous and beneficial microbiota. There is no anatomical site where the need for these two arms of the immune system is more critical than the intestinal tract. The intestinal tract is the largest surface of the human body that is exposed to the external world. It is constantly faced with novel food antigens and it is colonized by trillions of bacteria, many of which are harmless and even beneficial, but some of which are dangerous pathogens.
Millennia of coevolution between host and microbes have driven the development of a sophisticated host immune system as well as microbial strategies to manipulate that system. The observation that germ free animals have defective and incomplete immune systems coupled with the fact that a number of pathologies are linked to dysbiosis of the gut microbiota belies the critical role of the microbiota in the development and maintenance of immune homeostasis [7,8]. Although pathogenic microbes have evolved many strategies to evade or manipulate the host immune system to avoid detection and destruction [9], much more frequently, microbes have co-evolved with the host to modulate the immune system in a manner that benefits both the microbe and the host. Here, we focus on commensal bacteria that have been found to contribute to the maintenance of immune homeostasis in the host. Two of the major ways microbes choreograph the host immune system to maintain balance is via the production of metabolites and via bacterial associated factors that directly interact with the host immune system.
General Cues: Directing Innate Immune Responses
Innate immune factors respond to some highly conserved microbial associated molecular patterns (MAMPS), such as lipopolysaccharide (LPS) and peptidoglycan, whose ubiquity in the microbial world belie the many and often opposing responses to these MAMPS by the host immune system. For example, if these MAMPS are recognized on the apical surface of the gut epithelium they can promote tolerance and balance the inflammatory response, however if they are sensed by the basolateral surface of the same cell layer it can indicate a breach in the protective gut epithelium and trigger an inflammatory response [10].
LPS, is a cell wall component of gram-negative bacteria, which comprise much of the commensal population of the human microbiome; of course, many pathogens are also gram-negative and LPS is readily recognized by dedicated receptors of the innate immune system and can induce inflammation and sepsis [11]. Despite this, LPS levels have also been shown to contribute to immune homeostasis and have multiple beneficial effects on the host. For example, exposure of germ-free mice to LPS leads to a recovery of normal expression of mucous in the colon [12]. Recognition of LPS by Toll-like receptor 4 (TLR4) on epithelial cells initiates a signaling cascade that activates MyD88 which can promote antimicrobial activity and tissue protection and repair thus facilitating pathogen clearance and maintenance of a healthy epithelium [13]. Mice lacking MyD88 or other components of the LPS recognition pathway, or mice lacking microbiota are more susceptible to infection by a number of pathogenic bacteria and to intestinal injury suggesting levels of LPS may act as an indicator of the state of the system and enhance host defenses as needed [14].
Peptidoglycan, a component of the bacterial cell wall, has been shown to induce the formation of isolated lymphoid follicles (ILF’s), which prime the system for subsequent adaptive immune responses. The important role of peptidoglycan in immune homeostasis is underscored by the fact that the composition of the microbiome is dramatically altered when the peptidoglycan driven production of ILF’s is lost. Peptidoglycan has also been shown to enhance neutrophil function and thus promote the clearance of pathogenic bacteria [15]. Non-pathogenic strains of Salmonella as well as Bacteroides thetaiotaomicron have been shown to reduce inflammation via interference of the NF-κB signaling cascade [16,17].
The Effector T cell/Treg Axis
The balance of the immune system in the GI tract relies heavily on the relative abundance and activity of two subsets of CD4+ T-cells, the effector or helper CD4+ T cells and regulatory CD4+ T-cells (Tregs). Helper T-cells were canonically designated as either Th1, promoting cellular immunity and characterized by the production of IL-2, IFN-γ, and TFN-β or Th2, known for promoting humoral immunity and producing IL-4, IL-5, and IL-13 [18,19]. However, more recent research has defined Th17 cells as a new subset of the helper T cells. Th17s are characterized by production of cytokines interleukin (IL)-17A, IL-17F, IL21, and IL22 and are particularly important at mucosal surfaces including the GI tract, where there is a high risk of infection by pathogenic microbes.
Balancing out effector cell activity, Tregs suppress inflammation and promote immunologic tolerance [20]. There are two major subsets of Tregs; natural Tregs (nTregs) which develop in the thymus and are critical to preventing autoimmunity and controlling immune responses, and induced Tregs (iTregs) which leave the thymus as mature but naïve CD4+Tcells and differentiate upon activation in the periphery [21]. Tregs are characterized by the production of anti-inflammatory cytokines such as IL-10 that can quell an inflammatory response in a number of ways. IL-10 can reduce MHC-II expression on monocyte-macrophages thus dampening the production of pro-inflammatory cytokines, can inhibit chemokine production in dendritic cells in an autocrine fashion, and directly inhibits cytokine production and proliferation in CD4+ T cells [22]. Recent work has shown that iTreg deficient mice suffer from Th2 driven pathologies of the mucosa indicating that iTregs are critical to controlling mucosal Th2 responses [23]. Although iTregs have been canonically defined to express the cell surface marker FoxP3 [20], Tr1 cells which are CD4+ and Foxp3 negative, also contribute to immune tolerance in the gut [24] through in part, high expression of IL-10 [25]. Multiple human gut bacteria, including Bacteroides fragilis and Bifidiobacterium breve, can induce IL-10 producing Tr1 cells with B. fragilis also promoting the homing of these cells to the gut, indicating an important role for Tr1 cells in the relationship between commensals and host [26,27].
Immune cell subsets do not always fit neatly into effector versus regulatory functional bins. Although Th17s are generally considered to be effector cells for their contribution to host defense, their role varies by context. Th17 cells have been implicated in autoimmune disorders and inflammatory disease where they have been found to play both pathogenic and protective roles via IL-17 or IL-22 production [28]. Interestingly, it has been found that pro-inflammatory Th17 cells can transition to an immunosuppressive role upon recruitment to the small intestine and can in certain contexts produce IL-10 [29]. The cytokines IL-1β and IL-2 have been shown to play a role in regulating pro- versus anti- inflammatory function of Th17 in the context of exposure to different pathogens in priming and effector stages [29]. Taken together, it is clear that Th17 activity is tightly regulated to maintain protection from pathogens while avoiding auto-injury [30].
Targeting T-cells: Shaping Local Populations of iTreg/effector Cells
The microbiota has been found to play a role in the broad choreography of the effector/ regulatory T-cell axis to promote immune homeostasis [31] (Figure 1). A more complete understanding of which particular microbes facilitate immune homeostasis and by what mechanism, has the potential to inform the development of probiotic strategies for diseases that are driven by dysbiosis of the microbiota [10].
Figure 1.
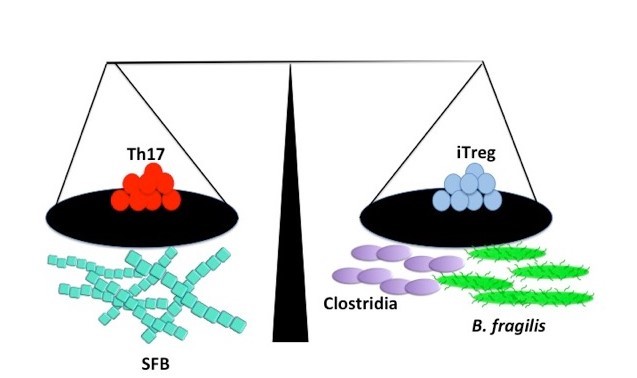
Commensal factors can promote immune homeostasis. Segmented filamentous bacteria have been shown to promote Th17 levels, while Bacteroides fragilis and some Clostridia species increase iTreg populations.
Studies in mouse models of inflammatory disease have allowed for the discovery of specific bacteria that can help to restore homeostasis in pathologic states through influence of the adaptive immune system. For instance, a consortium of 17 strains of non-pathogenic Clostridia that were derived from the stool of a healthy human has been shown to attenuate symptoms of colitis and allergic diarrhea via the induction of Tregs and IL-10 [32] and oral administration of the membranous fraction of the common gut bacterium Parabacteroides distasonis attenuated experimental murine colitis while preventing increases in several pro-inflammatory cytokines [33]. Although the precise mechanism of anti-colitic activity by bacteria is often not well understood, one exception to this case is immune-modulation by the common gut bacterium B. fragilis.
B. fragilis has been shown to promote the induction of Foxp3+ Tregs and promote IL-10 secretion, which can rebalance biased Th17/Treg levels and increase the suppressive capacity of Foxp3+ Tregs, thus mitigating pathological inflammation. This interaction is mediated by one of the 8 capsule polysaccharides produced by B. fragilis, PSA. PSA is a zwitterionic capsular polysaccharide and is reportedly presented by MHC-II following PSA activation of dendritic cells [34]. The anti-inflammatory effects of PSA benefit the bacteria by enhancing the colonization capabilities of B. fragilis, as a strain lacking PSA had a reduced capacity for colonization. PSA also confers a benefit to the host by supporting an anti-inflammatory phenotype. The significance of this effect is emphasized by the fact that B. fragilis and/or purified PSA can promote protection in colitis models but B. fragilis strains in which the PSA operon has been deleted cannot [35,36]. However, while B. fragilis PSA is known for its anti-inflammatory effects in the lamina propria, it has also been found to play a role in the systemic induction of Th1 responses [37,38].
Segmented filamentous bacteria (SFB) can also tip the scale in the other direction by promoting Th17 commitment of naïve Tregs and bolstering antimicrobial defenses, which was shown to enhance protection against some pathogenic agents [39]. The colonization of mice with whole microbiota and segmented filamentous bacteria specifically has been shown to drive the maturation of helper T-cell populations in the gut [40]. The ability of SFB to increase Th17 levels appears to be contingent upon adhesion of the SFB to the epithelium. T-cells with SFB specific antigen receptors will differentiate into Th17 cells indicating the fate of these effector cells is driven by SFB in the intestine [39,41,42].
Modifying the Immune System via the Production of Metabolites
In addition to the more direct interaction between bacteria and the immune system via molecules such as LPS and PSA, bacteria in the gut can also influence immune status through the metabolites that they produce while digesting components of our diets (Figure 2). In this case, the relationship between particular microbial community compositions and immune phenotypes may be harder to identify because they are dependent on complex, often cooperative metabolic processes that occur in a diet-dependent manner. One set of compounds that have been particularly well studied for their immune-modulatory properties are short chain fatty acids (SCFAs), a major by-product of microbial fermentation of substrates such as dietary soluble fibers. The three most prominent SCFAs produced by bacterial fermentation are butyrate, acetate and propionate.
Figure 2.
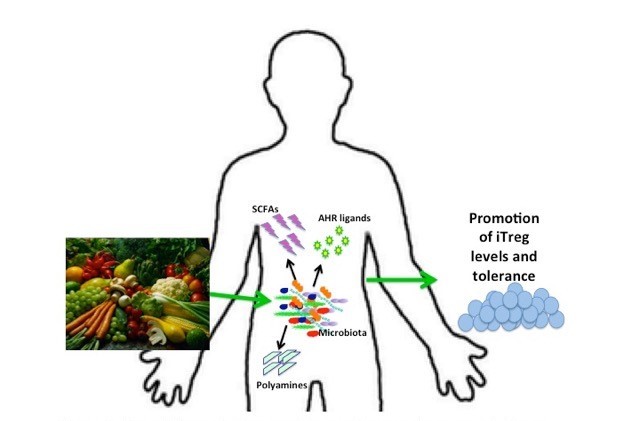
Bacterial metabolites contribute to immune homeostasis. Some microbiota metabolize complex carbohydrates into short chain fatty acids that can promote anti-inflammatory immune responses.
Butyrate, and also propionate to a lesser degree, have been shown to induce the differentiation of colonic/extra-thymic (peripheral) Treg cells in mice [43,44], indicating an anti-inflammatory effect. One potential mechanism for the induction of colonic Treg differentiation by butyrate is through histone modification of the Foxp3 locus [43]. In contrast, acetate and propionate have been shown to promote colonic Treg accumulation by activating a specific G Protein Coupled Receptor (GPCR), GPCR43 [45].
Multiple research studies suggest that immune modulation by microbially produced SCFAs can modulate disease. For instance, giving mice a diet of butyrylated high-amylose maize starch ameliorated the development of colitis induced by adoptive transfer of CD4+ CD45RBhi T cells in Rag 1 knockout mice [43]. Interestingly, SCFAs can also affect health status through immune-modulation at extra-colonic sites. As an example, in one study, feeding dietary fermentable fiber to mice resulted in increased circulating levels of SCFAs which conferred protection against allergic airway disease [46]. This protective effect was dependent on interaction between propionate and a G Protein Coupled Receptor in the bone marrow (GPCR41). In another study, circulating SCFAs were shown to directly influence the pancreatic immune environment and development of Type I diabetes in mice [47].
Another important class of microbiota-produced metabolites that influence mucosal immune response is aryl hydrocarbon receptor (AHR) ligands. AHR ligands can promote IL-22 production via the activation of innate lymphoid cells (ILC3s), and increased levels of IL-22 can protect from colonization by pathogenic microbes [48]. Mice deficient for AHR or AHR ligands were found to have altered microbial communities and pathogenic inflammation, and AHR deficient mice suffered from reduced numbers of ILCs and altered development of intestinal tissue [49]. Specific microbes such as species of Lactobacilli can produce AHR ligands from tryptophan precursors, implicating both diet and microbial metabolism as drivers of AHR activity [48,50].
Polyamines, such as putrescine, cadaverine, spermidine, and spermine are another class of microbial metabolites that can influence the host immune system, although their levels can also be influenced by diet or de novo synthesis by the host. Due to the high biodiversity and biodensity in the GI tract there are very high levels of available polyamines in the gut, which have been found to be critical to the maintenance of the health of the epithelium in the intestine by facilitating the constant and rapid turnover of intestinal epithelial cells and enhancing the structural and functional integrity of the epithelial barrier by increasing levels of junction proteins, mucous secretions, and antimicrobial factors [51,52].
Therapies that utilize knowledge of microbial metabolites on the immune system must consider the dynamic interplay between diet, gut microbial composition, and the generation of metabolites. There appears to be a high correlation between the composition of an individual’s microbiome and the diet of that individual. While this might not come as a surprise, the fact that a dietary change leads to a rapid and predictable shift in the gut microbiota suggests that dietary interventions could provide promising and effective strategies to modulate the gut microbiota. In addition to diet interventions, the use of prebiotics, compounds that may not be digestible by the host but promote the health of beneficial commensal bacteria, may be a promising treatment strategy for dysbiosis. For example, arbinooxylooligosaccharides, a component of wheat bran, has been shown to protect against Salmonella infection in chickens, and it has been suggested that these strategies could hold promise for prevention or interventions in human disease as well [53]. Ingestion of fructooligosaccharides has been shown to promote the expansion of endogenous bifidobacteria [54]. These finding suggest prebiotics can promote the growth of specific microbes and may be effective as targeted therapies to correct dysbiosis.
Application of beneficial metabolites of bacteria directly to the site of disease can also be an effective treatment strategy. For instance, introducing butyrate and/or SCFA cocktails to the colon via enema has been shown to lessen colonic inflammation in Inflammatory Bowel Disease (IBD) patients [55,56].
Conclusion
Human immunity is an incredibly powerful and dynamic system that has the ability to mount a robust response to insults, but it’s greatest power may lie in the ability to maintain homeostasis. While homeostasis suggests a static state of being, immune homeostasis is anything but; it is a dynamic state that keeps the system primed to respond rapidly to pathogenic microbes while avoiding autoimmunity and maintaining tolerance for beneficial commensals.
Here we’ve reviewed how the microbiota contributes to the maintenance of immune homeostasis. While it is clear that dysbiosis of the microbiota is tightly associated with an imbalance of the immune system and many human diseases, in many scenarios it remains unclear if the dysbiosis is causal or results from a shift in the immune balance. While not without complications, studies using humanized gnotobiotic mice (i.e. mice originally devoid of microbes that are colonized with the microbiota of humans of different pathologic and non-pathologic states) can help to differentiate between cause and effect and allow for further exploration of the role of microbiota in promoting a homeostatic immune system in the host [57,58]. Understanding the etiology of these diseases and the mechanisms by which dysbiosis of the microbiota may cause or contribute to their progression is critically important to understanding the epidemic incidences of diseases that are characterized by chronic inflammation of the gut, as well as to informing effective and targeted treatment strategies.
Acknowledgments
We would like to thank Dr. Brent Palmer for his input and valuable comments.
Abbreviations
- SCFAs
Short Chain Fatty Acids
- GI
gastrointestinal
- TH17
helper T 17 cells
- Tregs
regulatory T cells
- iTregs
inducible regulatory T cells
- nTreg
natural regulatory T cells
- MAMPS
microbial associated molecular patterns
- LPS
lipopolysaccharide
- ILF's
isolated lymphoid follicles
- PSA
polysaccharide A
- SFB
Segmented filamentous bacteria
- GPCR
G Protein Coupled Receptor
- IBD
Inflammatory Bowel Disease
Author Contributions
KA and CL wrote the article together. Both are funded by R01 DK104047 and CL is also funded by R01 DK108366.
References
- Ley RE, Peterson DA, Gordon JI. Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell. 2006;124(4):837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy M, Bouladoux N, Belkaid Y. Intestinal Microbiota: Shaping local and systemic immune responses. Semin Immunol. 2012;24(1):58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW. et al. Human nutrition, the gut microbiome, and immune system: envisioning the future. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N. et al. Control of Pathogens and Pathobionts by the Gut Microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabotti M, Scirocco A, Maselli MA. et al. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol Q Publ Hell Soc Gastroenterol. 2015;28(2):203–209. [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh PJ, Balart LA, Johnson DA. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin Transl Gastroenterol. 2015;6(6):e91. doi: 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, McFadden G. Anti-Immunology: Evasion of the Host Immune System by Bacterial and Viral Pathogens. Cell. 2006;124(4):767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Peterson CT, Sharma V, Elmén L. et al. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015;179(3):363–377. doi: 10.1111/cei.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. J Endotoxin Res. 2001;7(3):167–202. [PubMed] [Google Scholar]
- Petersson J, Schreiber O, Hansson GC. et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 211;300(2):G327–G333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DL, Ma C, Bergstrom KS. et al. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10(3):618–631. doi: 10.1111/j.1462-5822.2007.01071.x. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F. et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ES. et al. Recognition of Peptidoglycan from the Microbiota by Nod1 Enhances Systemic Innate Immunity. Nat Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier-Hyams LS, Sloane V, Batten BC. et al. Cutting Edge: Bacterial Modulation of Epithelial Signaling via Changes in Neddylation of Cullin-1. J Immunol. 2005;175(7):4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP. et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5(1):104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Romagnani S. Th1/Th2 cells. Inflamm Bowel Dis. 1999;5(4):285–294. doi: 10.1097/00054725-199911000-00009. [DOI] [PubMed] [Google Scholar]
- Hall BM. T Cells: Soldiers and Spies—The Surveillance and Control of Effector T Cells by Regulatory T Cells. Clin J Am Soc Nephrol. 2015;10(11):2050–2064. doi: 10.2215/CJN.06620714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S, Wing K, Onishi Y. et al. Regulatory T cells: how do they suppress immune responses? Int Immunol. 2009;21(10):1105–1011. doi: 10.1093/intimm/dxp095. [DOI] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N. et al. Conversion of Peripheral CD4+CD25− Naive T Cells to CD4+CD25+ Regulatory T Cells by TGF-β Induction of Transcription Factor Foxp3. J Exp Med. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couper KN, Blount DG, Riley EM. IL-10: The Master Regulator of Immunity to Infection. J Immunol. 2008;180(9):5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY. et al. Extrathymically generated regulatory T cells control mucosal Th2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Zeng H, Zhang R, Jin B. et al. Type 1 regulatory T cells: a new mechanism of peripheral immune tolerance. Cell Mol Immunol. 2015;12(5):566–571. doi: 10.1038/cmi.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SG, Kayama H, Ueda Y. et al. Probiotic Bifidobacterium breve Induces IL-10-Producing Tr1 Cells in the Colon. Plos Pathog. 2012;8(5):e1002714. doi: 10.1371/journal.ppat.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisman LSC, Cobb BA. Glycoantigens induce human peripheral Tr1 cell differentiation with gut-homing specialization. J Biol Chem. 2011;286(11):8810–8818. doi: 10.1074/jbc.M110.206011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Kolls JK, Zheng Y. The Biological Functions of T Helper 17 Cell Effector Cytokines in Inflammation. Immunity. 2008;28(4):454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski CE, Mele F, Aschenbrenner D. et al. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484(7395):514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- Omenetti S, Pizarro TT. The Treg/Th17 Axis: A Dynamic Balance Regulated by the Gut Microbiome. Mucosal Immun. 2015;6:639. doi: 10.3389/fimmu.2015.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM. et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Kverka M, Zakostelska Z, Klimesova K. et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol. 2011;163(2):250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesford KM, Yan W, Ochoa-Reparaz J. et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes. 2015;6(4):234–242. doi: 10.1080/19490976.2015.1056973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J. et al. The Toll-like receptor pathway establishes commensal gut colonization. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO. et al. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lécuyer . et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M. et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163(2):367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M. et al. Focused Specificity of Intestinal Th17 Cells towards Commensal Bacterial Antigens. Nature. 2014;510(7503):152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N. et al. The microbial metabolites, short chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145) doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Sun J, Furio L, Mecheri R. et al. Pancreatic β-Cells Limit Autoimmune Diabetes via an Immunoregulatory Antimicrobial Peptide Expressed under the Influence of the Gut Microbiota. Immunity. 2015;43(2):304–317. doi: 10.1016/j.immuni.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S. et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334(6062):1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buts JP, De Keyser N, Kolanowski J. et al. Maturation of villus and crypt cell functions in rat small intestine. Role of dietary polyamines. Dig Dis Sci. 1993;38(6):1091–1098. doi: 10.1007/BF01295726. [DOI] [PubMed] [Google Scholar]
- Chen J, Rao JN, Zou T. et al. Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2007;293(3):G568–G576. doi: 10.1152/ajpgi.00201.2007. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V, Van Immerseel F, Dewulf J. et al. Arabinoxylooligosaccharides from wheat bran inhibit Salmonella colonization in broiler chickens. Poult Sci. 2008;87(11):2329–2334. doi: 10.3382/ps.2008-00193. [DOI] [PubMed] [Google Scholar]
- Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125(6):1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- Scheppach W, Sommer H, Kirchner T. et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103(1):51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- Harig JM, Soergel KH, Komorowski RA. et al. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320(1):23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ. et al. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci Transl Med. 2009;1(6):6ra14–6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TLA, Vieira-Silva S, Liston A. et al. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]


