Abstract
Mast cells are evolutionarily ancient sentinel cells. Like basophils, mast cells express the high-affinity IgE receptor and are implicated in host defense and diverse immune-mediated diseases. To better characterize the function of these cells, we assessed the transcriptional profiles of mast cells isolated from peripheral connective tissues and basophils isolated from spleen and blood. We found that mast cells were transcriptionally distinct, clustering independently from all other profiled cells, and that mast cells demonstrated considerably greater heterogeneity across tissues than previously appreciated. We observed minimal homology between mast cells and basophils, which share more overlap with other circulating granulocytes than with mast cells. Derivation of mast cell and basophil transcriptional signatures underscores their differential capacity to detect environmental signals and influence the inflammatory milieu.
The Immunologic Genome (ImmGen) Project is a consortium of immunologists and computational biologists who seek to determine the gene expression patterns that characterize the mouse immune system through rigorously standardized cell isolation protocols and data analysis pipelines1. Tissue resident mast cells and circulating basophils are granulocytes traditionally associated with type 2 inflammation and host defense against helminthic infection2. Here, we assess the gene expression profiles associated with these populations and place them within the broader context of the immune system using the power of the ImmGen compendium.
Mast cells are evolutionarily ancient cells dating back at least as far as urochordates3, 4, predating the emergence of adaptive immunity. Mast cells are morphologically distinct tissue-resident sentinel cells densely packed with secretory granules containing pre-formed mediators including histamine, TNF-α, serotonin and a broad range of mast cell-specific serine proteases bound to a proteoglycan core with heparin glycosaminoglycans5. Granule release following mast cell activation is accompanied by the generation of pro-inflammatory leukotrienes, prostaglandins, chemokines and cytokines5, 6. This array of mediators is central to the mast cell’s sentinel function in mediating host resistance to bacteria, multicellular parasites and xenobiotic venoms7–9. Mast cells can be activated through pattern-recognition receptors9 or tissue damage10, 11 and express FcεR1 and Fcγ receptors, allowing them to respond to targets of the adaptive immune system2.
Mast cells are found in two main peripheral tissue compartments. Mucosal mast cells, absent in T cell-deficient humans and mice12, arise from bone marrow (BM)-derived agranular mast cell progenitors. These progenitors constitutively home to the intestinal mucosa13 and are further recruited to the intestine14 and lung15 during T cell-mediated inflammation, which directs their maturation into granulated mucosal mast cells16. In contrast to mucosal mast cells, connective tissue mast cells are constitutively present in most connective tissues17 and are seeded during embryogenesis by circulating progenitors derived from the fetal liver18. BM transfer experiments in adult mice show poor engraftment of donor-derived mast cells in connective tissues as compared to their recruitment to mucosal sites19, suggesting that the connective tissue mast cell compartment is maintained through longevity or self-renewal rather than replacement by BM-derived precursor cells. While studies have indicated that mast cell expression of proteases16, 20 and receptors21 is heterogeneous and regulated by the tissue microenvironment, the full degree of mast cell heterogeneity across different tissues is unknown.
Compared to mast cells, basophils are smaller circulating cells with multi-lobular nuclei and fewer, smaller cytoplasmic granules containing histamine and a restricted protease profile22, 23. Basophils infiltrate peripheral tissue during allergic inflammation24 and, like mast cells, express FcεR1. Signaling through FcεR1 induces basophil degranulation, accompanied by the rapid generation of leukotrienes and cytokines, including interleukin-4 (IL-4) and IL-1325, 26. Unlike connective tissue mast cells, circulating basophils are short-lived, with a half-life of several days in the periphery27 and are actively replenished from a progenitor cell28. Due to their FcεR1 expression and mediators produced, mast cells and basophils have been believed to be closely related.
The mast cell contribution to inflammation and immunity has been studied in mouse strains with mutations in the stem cell factor receptor c–kit, which are mast cell-deficient, in mice lacking mast cell-specific proteases and, more recently, in mice with the Cre-mediated deletion of mast cells or mast cell-associated proteins2, 29. In some cases, newer genetic approaches have supported previous findings, confirming important roles for mast cells in IgE-dependent local and systemic anaphylaxis29, uric acid crystal-induced arthritis30, sensitization to food allergen31 and resistance to animal venom32. In other models, such as contact hypersensitivity33, the Cre-mediated deletion of mast cell protease 5-expressing cells has contradicted early findings in c-kit mutant strains, by establishing a pro-inflammatory role for mast cells in sensitization to contact allergens. Discrepant findings could reflect differences in protocols, the influence of Kit mutation outside of the mast cell compartment, or differential deletion of mast cell subsets in these strains. Additionally, some mast cell-associated proteins, such as carboxypeptidase A3, used to direct Cre-expression for the generation of the mast cell-deficient “Cre-Master” and “Hello Kitty” strains, have been detected in basophils34, which are reduced in number in these strains. Thus, defining the genes and pathways uniquely or dominantly expressed in mast cells relative to other immune cells may clarify mast cell functions, identify targets for Cre-mediated disruption and provide candidate loci for the generation of novel mast cell-specific Cre-expressing strains.
Here, we isolated constitutive connective tissue mast cells from five distinct anatomical locations: the skin, the tongue, the esophagus, the trachea and the peritoneal cavity, and basophils from two locations: the spleen and peripheral blood. Our data show that the mast cell transcriptome is distinct, with mast cells clustering independently from all other analyzed lymphoid and myeloid cell populations. We find that basophils are transcriptionally closest to eosinophils and share surprisingly few distinct transcripts with mast cells. We describe the unique transcriptional signatures of mast cells and basophils and find a small shared signature between the two cell populations. Among the mast cell populations studied, we identify significant heterogeneity in gene expression and find evidence for previously unappreciated connective tissue mast cell turnover in the periphery in the absence of tissue inflammation.
Results
Mast cells are transcriptionally distinct among immunocytes
Mast cells were sorted based on co-expression of FcεR1α and CD117 from the peritoneal cavity, the ear, where they reside in the dermis, the tongue, where they reside in the muscular layer, the trachea, where they reside in the submucosa and serosal tissue and the esophagus, where they reside in the submucosa proximal to the stomach (Supplementary Fig. 1). Mast cells constituted between 0.05–10% of CD45+ cells in each compartment. Basophils were sorted based on co-expression of FcεR1α and CD49b from the spleen and peripheral blood, where they comprised 0.1% of CD45+ cells (Fig. 1a). The gating strategy used for isolating mast cells (Supplementary Fig. 2a) and basophils (Supplementary Fig. 2b) was validated through histochemical staining, indicating that the isolated cells were morphologically mast cells and basophils, respectively (Fig. 1b). Cells were enriched to high purity through multiple rounds of sorting (Supplementary Fig. 3) and final purity was assessed using parallel samples (Supplementary Table 1). RNA extracted from sorted mast cells and basophils was examined by microarray and compared to immunocytes from the ImmGen database, including blood eosinophils, peritoneal macrophages and B1a B cells; and splenic dendritic cells (DCs), neutrophils, CD4+ T cells, CD8+ T cells, γδ T cells, B2 B cells, NK cells and NK T cells.
Figure 1.
Identification of mast cells as distinct from other assayed cell populations. (a) Relative concentrations of mast cells (MC) and basophils (Ba) in digested tissue as a percentage of CD45+ cells. Mean ± sd. Data are combined from 3 (peritoneal mast cell, spleen basophil), 4 (skin mast cell, tongue mast cell) or 5 (trachea mast cell, esophagus mast cell, blood basophil) independent experiments. (b) Chloracetate esterase (CAE) staining of mast cells sorted from trachea, esophagus tongue, ear skin, peritoneal lavage and toluidine blue staining of basophils sorted from spleen confirming gating strategy used to isolate cell populations. Scale bar indicates 10 μm. (c) Hierarchical clustering of indicated cell populations using the top 15% most variable transcripts. Cell populations denoted using standard ImmGen abbreviations include trachea mast cells (MC.Tr), tongue mast cells (MC.To), esophagus mast cells (MC.Es), skin mast cells (MC.Sk), peritoneal mast cells (MC.PC), blood basophils (BA.Bl), spleen basophils (BA.Sp), blood eosinophils (Eo.Bl), spleen neutrophils (GN.Sp), peritoneal macrophages (MF.PC), spleen dendritic cells (DC.Sp), peritoneal B1a B cells (B1ab.PC), splenic B2 B cells (BB.Sp), splenic natural killer cells (NK.Sp), splenic NK T cells (NKT.Sp), splenic γδ T cells (Tgd.Sp), splenic CD8+ T cells (T8.Sp), splenic CD4+ T cells (T4.Sp) and splenic regulatory T cells (Treg.Sp). Bar height is inversely correlated to homology between linked populations. (d) Principal component analysis of cell populations indicated in (c) using the top 15% most variable transcripts. Numbers in parentheses indicate percentage of transcripts described by each principal component. Data (c–d) are from n = 3 independent experiments (skin, tongue, and trachea mast cells, spleen and blood basophils), from n = 5 independent experiments from peritoneal mast cells, and from n = 2 independent experiments from esophagus mast cells.
Hierarchical clustering using the top 15% most variable genes showed that the five sorted mast cell populations clustered separately from all other lymphoid and myeloid cells analyzed (Fig. 1c). Lymphoid and myeloid cells clustered independently as expected, and the myeloid cluster was further divided into one group containing granulocytes (eosinophils, neutrophils and both basophil populations) and a second containing macrophages and DCs. Mast cell distinction among immunocytes was based on both high expression of a distinct set of genes and low expression of many other transcripts associated with other cell types. Basophils showed strong expression of a smaller cluster of genes that had little overlap with the mast cell-enriched transcripts. Principal components analysis further highlighted the distinction of mast cells from the other profiled cell populations, with mast cells from different tissues grouping closely with each other and distant from other myeloid and lymphoid cells (Fig. 1d).
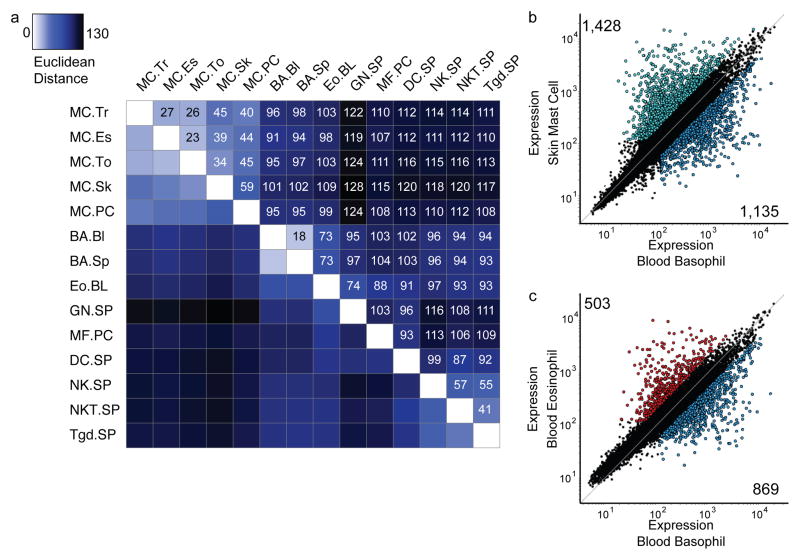
The transcriptional relationship between mast cells, basophils and the other analyzed cell populations was quantitated through Euclidean distance measurements (Fig. 2a), calculated using the top 15% most variable transcripts (Fig. 1c). Among mast cells, the trachea, esophagus and tongue mast cell subsets were the most similar, and the skin and peritoneal mast cell subsets were the most different. Mast cells as a whole were closest to basophils and eosinophils and furthest from neutrophils. Blood and spleen basophils were very similar to each other and were closest to eosinophils. The distance between basophils and mast cells was similar to the distance between basophils and neutrophils. Pairwise comparisons of dermal mast cells and blood basophils revealed 2,563 transcripts differentially expressed at an arbitrary two-fold or greater level (Fig. 2b), underscoring further their transcriptional differences. In contrast, pairwise comparisons of blood eosinophils and blood basophils revealed 1372 transcripts differentially expressed at a twofold or greater level (Fig. 2c). Thus tissue-resident mast cell populations express a gene program that distinguishes them from other immunocytes.
Figure 2.
Characterization of mast cells as transcriptionally distinct from basophils. (a) Euclidean distance matrix indicating degree of similarity between selected cell populations calculated using the top 15% most variable genes determined in Fig. 1c. Numbers in boxes indicate Euclidean distance. (b) Gene expression in skin mast cells and spleen basophils. Colored dots indicate transcripts expressed at two-fold or greater levels in skin mast cells (aqua) or spleen basophils (dark blue) and with expression values greater 120. Numbers indicate total genes enriched in each population. (c) Gene expression in blood eosinophils and spleen basophils. Colored dots indicate transcripts expressed at two-fold or greater levels in blood eosinophils (red) or spleen basophils (dark blue) and with expression values greater than 120. Numbers indicate total genes differentially expressed in each population. Data are combined from independent experiments as per Fig. 1c,d.
Transcriptional signature of tissue-resident mast cells
We next determined a 128 gene transcriptional signature whose expression was two-fold or greater in mast cells than in all other cells analyzed (Fig. 3a). Functional analysis using the PANTHER pathway classification system revealed that the mast cell signature was most significantly enriched in ‘serine proteases’ compared to transcripts encoding other functional categories (Table 1). This group included transcripts for many canonical mast cell proteases, but also Plau, encoding the urokinase-type plasminogen activator, Adamts9, encoding a metalloprotease and C2, encoding complement component C2 of the classical C3 convertase. Mast cells were also enriched in Ctsg, encoding cathepsin G, with more than five-fold higher expression than in neutrophils (Fig. 3b). Additional pathways enriched in mast cells included ‘sulfur metabolism’, which contained transcripts encoding enzymes important for heparin sulfate biosynthesis, ‘polysaccharide metabolism’ and ‘transferases’. The latter category included Hpgds, encoding hematopoietic prostaglandin D2 synthase, which is important for synthesis of the mast cell inflammatory product prostaglandin D2.
Figure 3.
Derivation of the mast cell transcriptional signature. (a) Mast cell-specific gene signature derived based on two-fold or greater transcript expression levels in all mast cell populations compared to all other analyzed cell populations. Highlighting indicates five-fold (purple) or ten-fold (red) higher expression levels in all mast cell subsets compared to all other cell populations. (b) Protease transcripts specifically enriched in the mast cell signature. (c) Mas-related G protein receptor transcript expression across analyzed cell populations. Data are combined from independent experiments as per Fig. 1c,d.
Table 1. Functional pathway enrichment analysis of mast cell signature genes.
Identifiers in parentheses (left column) indicate pathway module designations using the PANTHER pathway classification system. P values were calculated using the DAVID software program using a modification of the Fisher’s exact test.
| Pathway | Genes | P value | |||
|---|---|---|---|---|---|
| Serine protease (MF00216) | Ctsg | Cma1 | C2 | Cma2 | 1.30E-06 |
| Mcpt2 | Mcpt4 | Mcpt9 | Plau | ||
| Tpsab1 | Tpsb2 | Tpsg1 | |||
| Other transferase (MF00140) | Ndst2 | Chst1 | Hs3st3a1 | Hs2st1 | 2.30E-06 |
| Hs6st2 | Hpgds | ||||
| Other polysaccharide metablism (BP00009) | Ndst2 | St6galnac3 | Ext1 | Hs3st3a1 | 1.90E-03 |
| Hs2st1 | Ids | ||||
| Sulfur metabolism (BP00101) | Papss2 | Ndst2 | Hs3st3a1 | Hs2st1 | 3.80E-03 |
| Ids | |||||
| Carbohydrate metabolism (BP00001) | Ndst2 | St6galnac3 | A4galt | Chst1 | 1.10E-02 |
| Eno2 | Ext1 | Hs3st3a1 | Hs2st1 | ||
| Ids | Slc45a3 | ||||
| Signal Transduction (BP00102) | Cdc42bpa | Mrgprb1 | Mrgprb2 | Mrgprx1 | 2.60E-02 |
| Mrgprx2 | Ndst2 | Rab27b | Rapgef2 | ||
| Stard13 | Tiam2 | Bmpr2 | Cdh9 | ||
| Ccl2 | Ccl7 | Dgki | Dusp18 | ||
| Ednra | Grik2 | Gp1ba | Gnai1 | ||
| Gnaz | Hs3st3a1 | Kit | Lrrc66 | ||
| Mfge8 | Neo1 | Pde1c | Plau | ||
| Mrgpra4 | Pcdh7 | Ror1 | Rgs13 | ||
| Sgce | Socs2 | Tph1 |
Five of the genes in the ‘signal transduction’ pathway were members of the mas-related G-protein coupled receptor (Mrgpr) family: Mrgpra4, Mrpgrb1, Mrgprb2, Mrgprx1 and Mrgprx2. One of these, Mrgprb2, was previously described as the homologue to human MRGPRX2. The encoded protein mediates mast cell activation to a broad array of stimuli ranging from wasp venom to several pharmaceutical compounds associated with IgE-independent anaphylactic reactions in patients35. In addition to the five Mrgpr transcripts in the mast cell signature, Mrgprb8 and Mrgprb13 were strongly expressed specifically in skin mast cells, while Mrgpra6 was strongly expressed in basophils (Fig. 3c). Mrgpra2a and Mrgpra2b were predominantly expressed by neutrophils, as previously described36, but also showed lower expression on all mast cell populations, and Mrgpre was detected in B cells and NKT cells in addition to mast cells (Fig. 3c). Thus, the unique mast cell transcriptional program contained a broader degree of proteases, biosynthetic enzymes, and Mrgpr receptors than previously appreciated.
Distinct and shared mast cell gene expression
A basophil transcriptional signature containing 66 transcripts was similarly calculated based on two-fold or greater expression of transcripts in both basophil populations compared to all other analyzed cell populations, including mast cells (Fig. 4a). The basophil signature contained a single protease transcript, Mcpt8. The basophil signature transcripts also included several genes encoding chemokines (Ccl3, Ccl4 and Ccl9), growth factors (Hgf and Bmp4) and adhesion proteins (Cdh1, Itga1), suggesting mechanisms through which the basophil can interact with and influence the local environment.
Figure 4.
Distinct and shared transcriptional expression patterns between basophils and mast cells. (a) Basophil-specific gene signature derived based on two-fold or greater transcript expression levels in both basophil populations compared to all other analyzed cell populations. Highlighting indicates five-fold (purple) or ten-fold (red) higher expression. (b) Shared mast cell and basophil gene signature derived based on two-fold or greater transcript expression levels in all mast cell and basophil populations compared to all other analyzed cell populations. Red highlighting indicates ten-fold higher expression. (c) Transcripts involved in monoamine biosynthesis and neurotransmitter receptors expressed in mast cells or basophils. All transcripts aside from Hdc were included in either the mast cell-specific signature or the shared mast cell and basophil signature. (d) Transcription factors present in the distinct and shared mast cell and basophil gene signatures. Data are combined from independent experiments as per Fig. 1c,d.
To better understand the relationship between mast cells and basophils, a shared signature was derived based on two-fold higher expression of transcripts in all basophil and mast cell subsets, as compared to any other analyzed population. This analysis revealed a small shared transcriptional signature consisting of only 24 genes (Fig. 4b), many of which had previously been characterized in mast cells and basophils. These included Cd200r3, encoding an activating receptor, Fcer1a and Ms4a2, encoding the high affinity IgE receptor α and β chains, respectively, Slc24a3, encoding a Ca2+ transporter, and Gata2, encoding a transcription factor which directs the differentiation and function of both cell types37. The protease-encoding transcript Cpa3 was also present in the shared signature (Fig. 4b), consistent with previous reports of this transcript being highly expressed by basophils38, in addition to mast cells.
Mast cells and basophils are well-known sources of histamine2. Consistent with this, the mast cell-basophil shared profile identified here included the transcript encoding Slc18a2, a solute transporter involved in loading histamine into secretory vesicles (Fig. 4b). A further analysis of the monoamine biosynthetic pathways indicated that both mast cells and basophils strongly expressed transcripts encoding the histidine transporter (Slc3a2) and histidine decarboxylase (Hdc) (Fig. 4c). Mast cells further expressed transcripts encoding the L-tryptophan transporter (Slc7a5), tryptophan hydrolase 1 (Tph1) and dopa decarboxylase (Ddc) (Fig. 4c). The mast cell signature also contained Maob, encoding monoamine oxidase B (Fig. 4c), consistent with prior reports39. Mast cells and basophils both expressed transcripts encoding the histamine receptor Hrh4 and the serotonin re-uptake transporter Slc6a4 (Fig. 4c), while basophils expressed transcript encoding the serotonin receptor Htr1b (Fig. 4c).
Several transcription factors were more highly expressed in either mast cells or basophils compared to other immunocytes. Mast cells were specifically enriched for Creb3l1, Mitf, Smarca1 and Zfp9 (Fig. 4d). Of these, to the best of our knowledge, only Mitf has previously been described in mast cell biology, regulating expression of kit and mast cell proteases40. Basophils were specifically enriched for Sncaip, Cebpa, Supt3h and Nfil3 transcripts (Fig. 4d). Of these, only Cebpa has been previously described to play an important role in basophil biology28, where it directs progenitor commitment to the basophil lineage. Gata2 was the only transcription factor common to both cells. The diverse transcription factors, cell surface receptors, and inflammatory cell proteins expressed by mast cells and basophils extend our earlier cluster analysis (Fig. 1c) and pairwise analysis (Fig. 2b), indicating these cell types are not closely related in function.
Comparison of mast cell and basophil signatures across species
Next, we used a FANTOM consortium dataset that defined the resting transcriptome of human dermal mast cells and blood basophils41 to evaluate the mast cell and basophil signatures across species. Human mast cells were significantly enriched in the murine mast cell signature, with 55 of the 82 mast cell signature genes found in both datasets expressed two-fold higher in human skin mast cells compared to human blood basophils (Fig. 5). The transcripts conserved across species included those encoding proteases, hematopoietic prostaglandin D2 synthase, Mrgpr proteins and kit (Supplementary Table 2). Other transcripts conserved across species have less well-defined roles, including Maob and Gnai1, encoding the G protein.
Figure 5.
Enrichment of human mast cells in the murine mast cell signature. Relative expression (log2fold) for all 10,773 transcripts represented in both the ImmGen consortium data set (mouse) and the FANTOM consortium data sets (human). Expression (log2fold) in human skin mast cells relative to blood basophils, X axis; expression (log2fold) in mouse skin mast cells relative to blood basophils, Y axis. Blue line indicates two-fold relative expression. Human mast cells are statistically enriched (P=5.5e–16) in the murine mast cell signature (82 transcripts, red) and in the shared mast cell and basophil signature (17 transcripts, green) (P=0.0028, hypergeometric cumulative distribution upper tail). Human blood basophils are not enriched in the murine basophil signature (44 transcripts, blue) (P=0.33). Data from murine skin mast cells are from 3 independent experiments. Data from human skin mast cells was derived from 3 independent donors41.
In contrast, human basophils were not significantly enriched in the murine basophil signature, with only 10 of the 44 signature genes present in both data sets expressed twofold higher in human blood basophils compared to human skin mast cells (Supplementary Table 3). Among the transcripts conserved in both human and mouse basophils are those encoding the chemokines Ccl3 and Ccl4 (Fig. 5), suggesting a shared role for basophils across species in recruiting other leukocytes to sites of inflammation. Compared to human basophils, human mast cells were enriched in the signature shared in murine mast cells and basophils, with transcripts such as Cpa3 expressed 7.6-fold higher and Gata2 expressed 5.2 fold higher in human mast cells relative to human basophils (Supplementary Table 4), again demonstrating the conserved nature of the mast cell transcriptional program across species.
Tissue-specific genetic programs among mast cell populations
Next, we assessed the diversity among mast cell subsets through pairwise comparisons. Because peritoneal mast cells were the only mast cell population derived from non-digested tissue, we first assessed the effect of digestion enzymes on mast cell transcription. Enzymatic treatment of peritoneal mast cells increased expression of 137 genes by two-fold or more compared to untreated cells, including 17 genes that increased 5–10 fold and 7 genes that increased more than 10-fold such as Ccl3, Il13, and the transcription factor Egr2 (Supplementary Fig. 4a). Enzymatic digestion decreased expression of 26 genes by two-fold or more, including one transcript at 5–10 fold, Myl1, encoding the myosin light chain protein (Supplementary Fig. 4b). None of these genes were mast cell signature genes, and mast cell hierarchical clustering using enzymatically treated peritoneal mast cells again demonstrated that peritoneal mast cells were the most transcriptionally distinct subset (Supplementary Fig. 5). However, because a subset of genes was transcriptionally altered, we used enzymatically treated peritoneal mast cells for subsequent comparisons to other mast cell populations.
Mast cells from the tongue showed high homology with mast cells from both the trachea and esophagus, with only 110 genes differentially expressed two-fold or greater in tongue relative to trachea mast cells (Fig. 6a), and only 122 genes differentially expressed in tongue relative to esophagus mast cells (Fig. 6b). In contrast, tongue and peritoneal mast cells differentially expressed 612 transcripts (Fig. 6c). 957 genes were differentially expressed two-fold or greater in peritoneal relative to skin mast cells (Fig. 6c), indicating these two mast cell subsets were the most distinct.
Figure 6.
Tissue-specific mast cell gene expression. (a–c) Differential gene expression between mast cell subsets. Colored dots indicate transcripts expressed at two-fold or greater levels and at expression levels greater than 120 in tongue mast cells (light green), trachea mast cells (dark green), esophagus mast cells (pink), peritoneal mast cells (turquoise) or skin mast cells (aqua). Numbers indicate total genes enriched in each population. (d–g) Transcripts expressed at least four-fold higher or lower levels in (d) esophagus mast cells, (e) tracheal mast cells, (f) peritoneal mast cells, or (g) skin mast cells than in any other mast cell population. (h) Flow cytometric validation of differential gene expression suggested by transcript data. Grey solid histogram indicates isotype control staining, black histogram indicates cell surface protein expression in the indicated mast cell population. Flow plots are representative of three independent experiments. Data (a–g) are combined from independent experiments as per Fig. 1c,d.
We next analyzed transcripts specifically enriched or downregulated four-fold or more in single mast cell subsets compared to all other mast cell populations. Consistent with the transcriptional similarity between the trachea, esophagus and tongue, tongue mast cells showed no transcriptional enrichment. Esophagus mast cells showed at least 4-fold enrichment for five transcripts, including the protease transcripts Mcpt1, which was limited to this subset, and Mcpt2 (Fig. 6d). Tracheal mast cells showed four-fold enrichment for a single transcript, Lipf (Fig. 6e). No transcript was downregulated more than four-fold in trachea, esophagus or tongue mast cells compared to other mast cell populations. Three transcripts were enriched more than four-fold in peritoneal mast cells, including Itgb2, encoding β2 integrin and Bmp2, encoding bone morphogenic protein 2 (Fig. 6f). Peritoneal mast cells showed more than four-fold decreased expression of 10 transcripts, including Cd59a, encoding a membrane attack complex inhibitor and Olr1, encoding oxidized lipoprotein receptor 1(Fig. 6f).
In contrast to the other mast cell subsets, skin mast cells showed a four-fold increase in 28 genes and a four-fold decrease in 18 genes (Fig. 6g). In addition to Mrgprb8 and Mrgprb13, skin mast cells showed increased expression of transcripts encoding the metalloproteases Adamts1 and Adamts5, the cytokine and mast cell growth factor IL-3 and the transcription factor Sox7. Skin mast cells also showed enhanced expression of CD59a (Fig. 6g), suggesting strong differential expression of this gene between skin and peritoneal mast cells. Transcripts downregulated in skin mast cells compared with other subsets included CD34, encoding a canonical mast cell marker and Alox5 and Alox5ap, encoding 5-lipoxygenase and 5-lipoxygenase activating protein, respectively (Fig. 6g).
In support of the transcriptional data, flow cytometric analysis indicated CD34 was expressed on all mast cell subsets except for skin mast cells, CD59a expression was strongest in the skin mast cells and undetectable on peritoneal mast cells and ItgB2 expression was only detected on peritoneal mast cells (Fig. 6h). Enzymatically treated peritoneal mast cells showed no decrease in either CD34 or ItgB2 surface staining (Supplementary Fig. 6).
Because the skin and peritoneal mast cell populations showed the greatest degree of differential gene expression, these populations were compared using Gene Set Enrichment Analysis (GSEA). Among the most enriched Gene Ontology (GO) Consortium terms in peritoneal mast cells were Mitosis and M phase (Fig. 7a), suggesting that peritoneal mast cells might be undergoing cellular turnover. Thus, we evaluated peritoneal mast cell expression of Ki67, a nuclear protein present during mitosis but rapidly degraded during the G-0 phase. Ki67 staining was increased in peritoneal mast cells relative to skin mast cells, which also expressed Ki67 (Fig. 7b). In total, 16% of peritoneal mast cells were positive for Ki67 compared to only 4% of skin mast cells (Fig. 7c), indicating a significantly higher rate of mitosis in the peritoneal mast cell population and notable Ki67 expression in both populations in the absence of inflammation.
Figure 7.
Transcriptional analysis predicts peritoneal mast cell turnover. (a) GSEA Identification of Mitosis and M phase GO Terms as significantly enriched in digest enzyme-treated peritoneal cavity mast cells compared to skin mast cells. Both terms enriched with a nominal P-value <0.001 with a false discovery rate Q-value<0.005. (b) Intracellular Ki67 expression in peritoneal and skin mast cells. Results representative of three independent experiments with a total of n-9 mice per group. (c) Quantification of Ki67+ mast cells found in peritoneum and skin. * indicates P=0.0000062 (two-tailed unpaired t test with Welch’s correction). Data are combined from three independent experiments with a total of n=9 mice per group. Dots show individual data points, lines indicate mean ± sd.
Discussion
Heparin-containing mast cell-like cells are found as far back as urochordates3, and although mast cells were first identified over 100 years ago their contribution to immune defense and disease has been poorly defined. Here we provide the first comprehensive transcriptional analysis of murine mast cells in comparison to 14 other lymphoid and myeloid cell populations. We identify mast cells as the most transcriptionally distinct cell type, clustering independently from all other populations including basophils. We describe a shared mast cell transcriptional signature and further recognize tissue-specific regulation of the mast cell transcriptome. We find that mast cells are enriched in distinct pathways for sensing and responding to environmental cues, providing a framework for understanding their sentinel function.
Mast cells from various tissues share a transcriptional signature of 128 genes, of which serine proteases are a significant contributor. Mast cells are also enriched for metabolic pathways required for the generation of a broad range of other preformed mediators, including histamine, serotonin and heparin sulfate. Furthermore, mast cells express transcripts allowing the acute generation of eicosanoids such as prostaglandin D2 and rapid production of cytokines and chemokines. Together, these findings indicate the capacity to generate a unique repertoire of mediators. The murine mast cell signature is also highly enriched in human mast cells, suggesting evolutionary pressures to retain a core mast cell functionality. These highly conserved genes include well-known mast cell genes such as proteases and hpgds, but also several that are poorly understood in the context of mast cells, including Maob, Gnai1 and Mrgpr family members.
The array of Mrgprs expressed in mast cells is broader than previously appreciated. Originally discovered in sensory neurons42, eight members of this family are expressed in skin mast cells and six are expressed in the other mast cell populations. Further expression of Mrgpra6 in basophils and Mrgpra2a and Mrgpra2b in neutrophils suggests that Mrgprs may play a significant role in innate immune function. Human MRGPRX2 was recently shown to mediate mast cell degranulation in response to the classical mast cell activating compound 48/80 in human cord blood derived mast cells43 and the transformed human LAD2 mast cell line44. The murine homologue of MRGPRX2, Mrgprb2, mediates degranulation in response to wasp venom, 48/80 and a diverse array of other basic compounds, including therapeutic agents that induce IgE-independent mast cell degranulation in humans 35. Thus, members of this family may play a critical role in mediating the innate activation of mast cells to both pharmacologic agents and as-yet unidentified native ligands.
The low homology observed between murine mast cells and basophils in this study is similar to that previously observed in human cells, as is the closer relationship between basophils and eosinophils41. While murine mast cells and basophils share expression of transcripts encoding several activating receptors and histamine biosynthetic enzymes, basophils lack the diversity of proteases seen in mast cells and express different combinations of soluble mediators and receptors. Thus, transcriptional analysis of mast cells and basophils suggests that these cells play independent roles in regulating homeostasis and host defense rather than serving similar roles in different tissue compartments. The basophil signature contained Ccl3, Ccl4 and Ccl9. Two of these transcripts, Ccl4 and Ccl4, were also enriched in human basophils compared to human mast cells, suggesting a conserved role in directing cellular recruitment. However, the poor conservation of other basophil signature genes between human and mouse basophil may reflect evolutionary pressures driving divergence of this cell type between species.
Comparative analysis of mast cell populations revealed considerable tissue-specific gene expression, consistent with mast cell maturation in peripheral tissue and with studies demonstrating mast cell regulation by neighboring fibroblasts21, 45. Unlike other mast cell populations, peritoneal mast cells are not embedded in the tissue but rather line the serosal gut wall. We observed that they are enriched for transcripts associated with cellular turnover, leading to the finding that a substantial fraction of peritoneal mast cells stain positive for Ki67. Thus, the profound transcriptional differences between peritoneal mast cells and other mast cell compartments may reflect both cell maturation and differential signaling from neighboring cells. Notably, Ki67 staining was also detectable at low levels in skin mast cells, suggesting that local proliferation may play a role in the renewal and maintenance of this compartment.
In conclusion, mast cells are extraordinarily distinct cells at the transcriptional level. Their core signature is enriched for a diverse array of proteases and biosynthetic pathways, allowing for the generation of a broad range of mediators, and includes several novel gene families whose function is not yet understood. Analysis of mast cell heterogeneity reveals three distinct connective tissue mast cell subsets and varying capacity for in situ proliferation in the absence of tissue inflammation. These findings provide an important framework for better defining the role of these evolutionarily ancient cells in homeostasis, host defense and disease.
Online Methods
Mice
All cells used for transcriptional and flow cytometric analyses were obtained from male six-week-old C57BL/6J mice and tissue used for histology was obtained from male six- to ten-week-old C57BL/6J mice from the Jackson Laboratory. Mice were housed (4 mice per cage) in specific pathogen-free facilities at the Dana Farber Cancer Institute (DFCI) under a 12 hour light/12 hour dark cycle. The use of all mice for these studies was in accordance with institutional guidelines with review and approval by the Animal Care and Use Committee of DFCI.
Cell isolation and sorting
Cells were purified according to the standardized ImmGen standard operations protocol (http://www.immgen.org/Protocols/ImmGen%20Cell%20prep%20and%20sorting%20SOP.pdf) using the indicated antibodies (below) with modifications for increased digestion time as noted below. Peritoneal cell suspensions were obtained by lavaging the peritoneal cavity with 7 mL HBSS containing 1 mM EDcell TA. Single-cell suspensions were obtained from tongue, esophagus, and trachea by finely mincing tissue between two scalpel blades and incubating for 30 minutes at 37° C with 600 U/mL collagenase IV (Worthington), 0.1% dispase (Gibco) and 20 μg/mL DNAse 1 (Roche) in RPMI supplemented with 10% fetal bovine serum at 500 RPM. Ear digests were obtained using modifications of a previously described protocol46. Briefly, dorsal and ventral halves of the ear were separated and incubated for 20 minutes in HBSS with 2.5 μg/mL dispase at 300 RPM to separate the epidermis. After pulling away the epidermis, remaining tissue was finely minced between two scalpel blades and incubated for 30 minutes with 600 U/mL collagenase IV and 20 μg/mL DNAse 1 in RPMI supplemented with 10% fetal bovine serum at 500 RPM. Spleen suspensions were obtained through mechanical disruption of the spleen followed by erythrocyte lysis using ACK buffer (Sigma). Following lysis, lymphocytes were depleted using Dynal beads directed against B220 and Thy1.2 (Invitrogen). Blood was obtained through cardiac puncture and erythrocytes were depleted using a 44%/67% Percol gradient (Sigma). Mast cells were identified as CD45+ CD11b− CD11c− CD19− CD4− CD8− FcεR1α+ CD117+. Basophils were identified as CD3− CD19− NK1.1− CD117− FcεR1α+ CD49b+. Cells were sorted at the Brigham and Women’s Human Immunology Flow Core using a BD FACSAria Fusion cell sorter. For surface marker and intracellular analysis, data was acquired on a BD FACSCanto II and analyzed with FlowJo software (Treestar). The following monoclonal antibodies (clone, concentration) were used: Anti-FcεR1α (MAR-1, 1:250), anti-CD117 (2B8, 1:250), anti-CD45 (30-F11, 1:250), anti-CD11b (M1/70, 1:250), anti-CD11c (N418, 1:250), anti-CD19 (6D5, 1:250), anti-CD4 (GK1.5, 1:250), anti-CD8 (53–6.7, 1:250), anti-CD49b (DX5, 1:250), anti-NK1.1 (PK136, 1:250), anti-CD34 (MEC14.7, 1:250), anti-CD59b (mCD59.3, 1:250), anti-ItgB2 (M18/2, 1:250), and isotype-matched control monoclonal antibodies (mAbs) were obtained from Biolegend. Anti-IgE (23G3 1:250), anti-Ki67 (SolA15, 1:100), isotype-matched control mAbs, and FoxP3 staining buffer set used for Ki67 staining were obtained from eBioscience.
Cytospins and microscopy
For histochemical evaluation of mature mast cells in peripheral tissues, tissue sections were fixed overnight in 4% paraformaldehyde and embedded in glycolmethacrylate. For cytospin evaluation, sorted cells were spun onto charged glass slides and dried overnight. Cut section and cytospins were stained for CAE reactivity for the identification of mast cells, and cytospins were stained with toluidine blue for the identification of basophils.
Cells and animals per microarray replicate
Mast cells were collected from the skin (n=3, each replicate was 25,000 cells pooled from 8 mice), peritoneal cavity (n=5, each replicate was 30,000 cells pooled from 4 mice), tongue (n=3, each replicate was 10,000 cells pooled from 10 mice), esophagus (n= 2, each replicate was 10,000 cells pooled from 24 mice), and trachea (n= 3, each replicate was 15,000 cells pooled from 8 mice). Basophils were collected from the blood (n=3, each replicate was 10,000 cells pooled from 5 mice) and spleen (n=3, each replicates was 25,000 cells pooled from 4 mice). Whenever possible, multiple tissues were harvested from each mouse to minimize total number of animals used. Sample sizes were determined based on ImmGen standard protocols targeting a minimum of 10,000 cells per microarray sample.
Microarray analysis and data evaluation
Samples were sorted twice and collected directly into TRIzol. RNA was amplified and hybridized to the Affymetrix Mouse Gene 1.0 ST array by ImmGen according to the consortium’s standard protocols (https://www.immgen.org/Protocols/Total%20RNA%20Extraction%20with%20Trizol.pdf ) with modification. To improve microarray success rate, RNA was treated with heparinase as previously described47, 48. Briefly, following an initial round of chloroform extraction, RNA was incubated in 5μm Tris buffer containing 50U of RNAsin plus (Promega) and 0.02 U of heparinase (Sigma) for 2h at room temperature, and then subjected to a second round of TRIzol extraction. Comparison of peritoneal mast cell RNA treated with heparinase (n=3) or control showed that 4 transcripts among the 21,775 assayed were reduced by a 2-fold statistically significant (p<0.05) degree, demonstrating minimal impact on detected transcript levels. Data generation and quality-control documentation was also conducted by ImmGen according to the consortium’s standard protocols (https://www.immgen.org/Protocols/ImmGen%20QC%20Documentation_ALL-DataGeneration_0612.pdf). Transcripts identified through multiple probes were collapsed based on median values and differential gene expression was characterized using the Multiplot Studio module of GenePattern software (Broad Institute). Tracheal mast cells were found to be enriched for several B cell genes, including immunoglobulin genes, suggesting B cell contamination. Contaminating B cell genes in tracheal mast cells were identified by comparing fold changes in expression between tracheal mast cells and esophagus mast cells to fold changes in expression between esophagus mast cells and splenic B cells. All transcripts with greater than 16-fold increased expression in splenic B cells compared to tongue mast cells also showed increased expression in tracheal mast cells compared to tongue mast cells and were excluded from all pairwise comparisons. Hierarchical clustering for transcripts was conducted using Gene-E (http://broadinstitute.org/cancer/software/GENE-E) based on the top 15% most variable transcripts using Pearson’s correlation and cell population clustering was calculated using Spearman’s correlation. Euclidean distance matrix and all transcript heat maps were also constructed using Gene-E. Principal component analysis was visualized using MatLab software (MathWorks) using principal components calculated using the PopulationDistances PCA program (S. Davis, Harvard Medical School) based on the top 15% most variable transcripts across all analyzed cell populations. The skin and enzyme-treated peritoneal mast cell transcriptomes were further compared using the Gene Set Enrichment Analysis software program (Broad Institute)49, 50 using Gene Ontology Consortium (www.geneontology.org) gene sets.
Controlling for the effects of collagenase treatment on peritoneal mast cells
Peritoneal cell suspensions obtained by lavaging the peritoneal cavity with 7 mL HBSS containing 1 mM EDTA were incubating for 30 minutes at 37° C with 600 U/mL collagenase IV (Worthington), 0.1% dispase (Gibco) and 20 μg/mL DNAse 1 (Roche) in RPMI supplemented with 10% fetal bovine serum. Following enzymatic treatment, peritoneal mast cells were either isolated for microarray analysis or stained for cell surface marker expression.
Derivation of mast cell and basophil transcriptional signatures
The mast cell signature was generated in comparison to all cell populations analyzed. Multiple replicates for each cell population were collapsed based on median values. Transcripts in the mast cell signature were expressed at least two-fold higher in all mast cell populations than in any non- mast cell population, including basophils. All transcripts expressed below 120 relative units in more than two mast cell subsets were excluded, as were all in which there was no statistically significant difference between mast cell and non- mast cell expression by student’s t-test. The mast cell signature was calculated using non-digested peritoneal mast cells to exclude any genes induced by collagenase and dispase treatment. The basophil signature was calculated similarly, and the shared mast cell and basophil signature was calculated by determining all transcripts expressed at least two fold higher in both mast cell and basophil than in any non-mast cell and non-basophil cell. After calculating the signatures, enriched pathways were determined using DAVID software51, 52 based on the PANTHER classification system with P < 0.05. Mast cell and basophil-specific transcription factors were determined by identifying transcripts in the individual and shared mast cell and basophil signatures that also appeared in the Riken institute transcription factor database (http://genome.gsc.riken.jp/TFdb/)
Comparison of human and mouse mast cells and basophils
All 10,773 transcripts identified in both the Affymetrix Mouse 1.0 array and in human cells via CAGE sequencing were visualized on a fold change vs fold change plot. To allow for fold change comparisons in the CAGE sequencing dataset, in which numerous transcript levels had a value of zero, a value of 1 was added to each datapoint. Genes found in the murine mast cell, basophil and combined signatures were then highlighted.
Statistics
There was no randomization, blinding, or exclusion of data. Sample size was not predetermined statistically. Significance of PANTHER pathway enrichment was determined using a modified Fisher’s exact test in DAVID. Enrichment of human mast cells and basophils for the murine mast cell and basophil signatures was evaluated using the hypergeometric cumulative distribution upper tail in Matlab (Mathworks). Differences in intracellular Ki67 levels were evaluated using Prism 6.0 (GraphPad) with a two-tailed unpaired T test with Welch’s correction after determining that the samples represented a gausian distribution using the D’Agostino & Pearson omnibus normality test. P values of <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
The authors would like to thank the other members of the ImmGen consortium for many helpful discussions, especially C. Benoist and T. Shay; the core ImmGen team for technical assistance, especially A. Rhodes and K. Rothamel; and A. Chicoine of the Brigham and Women’s Human Immunology Flow Core for assistance with isolating cells. Supported by the US National Institutes of Health (R24AI072073 to the ImmGen Consortium, R01 HL120952 to N.A.B, AI095219 to N.A.B, T32 AI007306 to D.F.D) and the Steven and Judy Kaye Young Innovators Award to N.A.B.
Footnotes
The authors have no competing financial interest
GEO accession
Microarrays have been deposited in the gene expression omnibus under Series GSE37448. Microarray results are also accessible through www.ImmGen.org
Author Contributions
D.F.D. wrote the manuscript, conceived of and conducted experiments and analyzed the data. N.A.B. and K.F.A. wrote the manuscript and supervised the experimental design. The ImmGen Project Consortium contributed to data collection and assisted in experimental design.
References
- 1.Heng TS, Painter MW Immunological Genome Project, C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 2.Voehringer D. Protective and pathological roles of mast cells and basophils. Nat Rev Immunol. 2013;13:362–375. doi: 10.1038/nri3427. [DOI] [PubMed] [Google Scholar]
- 3.Cavalcante MC, et al. Occurrence of heparin in the invertebrate styela plicata (Tunicata) is restricted to cell layers facing the outside environment. An ancient role in defense? J Biol Chem. 2000;275:36189–36186. doi: 10.1074/jbc.M005830200. [DOI] [PubMed] [Google Scholar]
- 4.Wong GW, et al. Ancient origin of mast cells. Biochem Biophys Res Commun. 2014;451:314–318. doi: 10.1016/j.bbrc.2014.07.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 6.Razin E, et al. IgE-mediated release of leukotriene C4, chondroitin sulfate E proteoglycan, beta-hexosaminidase, and histamine from cultured bone marrow-derived mouse mast cells. J Exp Med. 1983;157:189–201. doi: 10.1084/jem.157.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metz M, et al. Mast cells can enhance resistance to snake and honeybee venoms. Science. 2006;313:526–530. doi: 10.1126/science.1128877. [DOI] [PubMed] [Google Scholar]
- 8.Schneider LA, Schlenner SM, Feyerabend TB, Wunderlin M, Rodewald HR. Molecular mechanism of mast cell mediated innate defense against endothelin and snake venom sarafotoxin. J Exp Med. 2007;204:2629–2639. doi: 10.1084/jem.20071262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10:440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abonia JP, et al. Mast cell protease 5 mediates ischemia-reperfusion injury of mouse skeletal muscle. J Immunol. 2005;174:7285. doi: 10.4049/jimmunol.174.11.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Younan G, et al. The inflammatory response after an epidermal burn depends on the activities of mouse mast cell proteases 4 and 5. J Immunol. 2010;185:7681–7690. doi: 10.4049/jimmunol.1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurish MF, Austen KF. Developmental origin and functional specialization of mast cell subsets. Immunity. 2012;37:25–33. doi: 10.1016/j.immuni.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Crapper RM, Schrader JW. Frequency of mast cell precursors in normal tissues determined by an in vitro assay: antigen induces parallel increases in the frequency of P cell precursors and mast cells. J Immunol. 1983;131:923–928. [PubMed] [Google Scholar]
- 14.Friend DS, et al. Mast cells that reside at different locations in the jejunum of mice infected with Trichinella spiralis exhibit sequential changes in their granule ultrastructure and chymase phenotype. J Cell Biol. 1996;135:279–290. doi: 10.1083/jcb.135.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones TG, et al. Antigen-induced increases in pulmonary mast cell progenitor numbers depend on IL-9 and CD1d-restricted NKT cells. J Immunol. 2009;183:5251–5260. doi: 10.4049/jimmunol.0901471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing W, Austen KF, Gurish MF, Jones TG. Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc Natl Acad Sci U S A. 2011;108:14210–14215. doi: 10.1073/pnas.1111048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gersch C, et al. Mast cells and macrophages in normal C57/BL/6 mice. Histochem Cell Biol. 2002;118:41–49. doi: 10.1007/s00418-002-0425-z. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi C, Sonoda T, Nakano T, Nakayama H, Kitamura Y. Mast-cell precursors in the skin of mouse embryos and their deficiency in embryos of Sl/Sld genotype. Dev Biol. 1985;109:234–241. doi: 10.1016/0012-1606(85)90363-x. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura Y, Shimada M, Hatanaka K, Miyano Y. Development of mast cells from grafted bone marrow cells in irradiated mice. Nature. 1977;268:442–443. doi: 10.1038/268442a0. [DOI] [PubMed] [Google Scholar]
- 20.Friend DS, et al. Reversible expression of tryptases and chymases in the jejunal mast cells of mice infected with Trichinella spiralis. J Immunol. 1998;160:5537–5545. [PubMed] [Google Scholar]
- 21.Kurashima Y, et al. The enzyme Cyp26b1 mediates inhibition of mast cell activation by fibroblasts to maintain skin-barrier homeostasis. Immunity. 2014;40:530–541. doi: 10.1016/j.immuni.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Dvorak AM, et al. Ultrastructural identification of the mouse basophil. Blood. 1982;59:1279–1285. [PubMed] [Google Scholar]
- 23.Poorafshar M, Helmby H, Troye-Blomberg M, Hellman L. MMCP-8, the first lineage-specific differentiation marker for mouse basophils. Elevated numbers of potent IL-4-producing and MMCP-8-positive cells in spleens of malaria-infected mice. Eur J Immunol. 2000;30:2660–2668. doi: 10.1002/1521-4141(200009)30:9<2660::AID-IMMU2660>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 24.Siracusa MC, Perrigoue JG, Comeau MR, Artis D. New paradigms in basophil development, regulation and function. Immunol Cell Biol. 2010;88:275–284. doi: 10.1038/icb.2010.1. [DOI] [PubMed] [Google Scholar]
- 25.Gibbs BF, et al. Purified human peripheral blood basophils release interleukin-13 and preformed interleukin-4 following immunological activation. Eur J Immunol. 1996;26:2493–2498. doi: 10.1002/eji.1830261033. [DOI] [PubMed] [Google Scholar]
- 26.Warner JA, et al. Differential release of mediators from human basophils: differences in arachidonic acid metabolism following activation by unrelated stimuli. J Leukoc Biol. 1989;45:558–571. doi: 10.1002/jlb.45.6.558. [DOI] [PubMed] [Google Scholar]
- 27.Ohnmacht C, Voehringer D. Basophil effector function and homeostasis during helminth infection. Blood. 2009;113:2816–2825. doi: 10.1182/blood-2008-05-154773. [DOI] [PubMed] [Google Scholar]
- 28.Arinobu Y, et al. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galli SJ, et al. Approaches for analyzing the roles of mast cells and their proteases in vivo. Adv Immunol. 2015;126:45–127. doi: 10.1016/bs.ai.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reber LL, et al. Contribution of mast cell-derived interleukin-1beta to uric acid crystal-induced acute arthritis in mice. Arthritis Rheumatol. 2014;66:2881–2891. doi: 10.1002/art.38747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burton OT, et al. Immunoglobulin E signal inhibition during allergen ingestion leads to reversal of established food allergy and induction of regulatory T cells. Immunity. 2014;41:141–151. doi: 10.1016/j.immuni.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marichal T, et al. A beneficial role for immunoglobulin E in host defense against honeybee venom. Immunity. 2013;39:963–975. doi: 10.1016/j.immuni.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dudeck A, et al. Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity. 2011;34:973–984. doi: 10.1016/j.immuni.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 34.Feyerabend TB, et al. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 2011;35:832–844. doi: 10.1016/j.immuni.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 35.McNeil BD, et al. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 2015;519:237–241. doi: 10.1038/nature14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ericson JA, et al. Gene expression during the generation and activation of mouse neutrophils: implication of novel functional and regulatory pathways. PLoS ONE. 2014;9:e108553. doi: 10.1371/journal.pone.0108553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Qi X, Liu B, Huang H. The STAT5-GATA2 Pathway Is Critical in Basophil and Mast Cell Differentiation and Maintenance. J Immunol. 2015;194:4328–4338. doi: 10.4049/jimmunol.1500018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 39.Vitalis T, et al. Developmental expression pattern of monoamine oxidases in sensory organs and neural crest derivatives. J Comp Neurol. 2003;464:392–403. doi: 10.1002/cne.10804. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura Y, Morii E, Jippo T, Ito A. Effect of MITF on mast cell differentiation. Mol Immunol. 2002;38:1173–1176. doi: 10.1016/s0161-5890(02)00058-5. [DOI] [PubMed] [Google Scholar]
- 41.Motakis E, et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood. 2014;123:e58–67. doi: 10.1182/blood-2013-02-483792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 43.Tatemoto K, et al. Immunoglobulin E-independent activation of mast cell is mediated by Mrg receptors. Biochem Biophys Res Commun. 2006;349:1322–1328. doi: 10.1016/j.bbrc.2006.08.177. [DOI] [PubMed] [Google Scholar]
- 44.Kashem SW, et al. G protein coupled receptor specificity for C3a and compound 48/80-induced degranulation in human mast cells: roles of Mas-related genes MrgX1 and MrgX2. Eur J Pharmacol. 2011;668:299–304. doi: 10.1016/j.ejphar.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levi-Schaffer F, Austen KF, Gravallese PM, Stevens RL. Coculture of interleukin 3-dependent mouse mast cells with fibroblasts results in a phenotypic change of the mast cells. Proc Natl Acad Sci U S A. 1986;83:6485–6488. doi: 10.1073/pnas.83.17.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogunovic M, et al. Identification of a radio-resistant and cycling dermal dendritic cell population in mice and men. J Exp Med. 2006;203:2627–2638. doi: 10.1084/jem.20060667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai M, Miyamoto M, Tam SY, Wang ZS, Galli SJ. Detection of mouse mast cell-associated protease mRNA. Heparinase treatment greatly improves RT-PCR of tissues containing mast cell heparin. Am J Pathol. 1995;146(2):335–343. [PMC free article] [PubMed] [Google Scholar]
- 48.Gilchrist M, MacDonald AJ, Neverova I, Ritchie B, Befus AD. Optimization of the isolation and effective use of mRNA from rat mast cells. J Immunol Methods. 1997;201(2):207–214. doi: 10.1016/s0022-1759(96)00230-x. [DOI] [PubMed] [Google Scholar]
- 49.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 52.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.