Abstract
Autophagy is an evolutionarily conserved lysosomal degradation pathway that removes damaged organelles and protein aggregates from the cytoplasm. Being post-mitotic, neurons are particularly vulnerable to the accumulation of proteotoxins and are thus heavily dependent on autophagy to maintain homeostasis. In fact, CNS-specific and neuron-specific loss of autophagy is sufficient to cause neurodegeneration in mice. Further, mutations in PINK1 and Parkin, proteins that selectively remove damaged mitochondria, cause Parkinson’s disease, linking defective autophagy with neurodegenerative disease in humans. This review provides an overview of the mechanisms of autophagy in the axon and the role of neuronal autophagy in axonal homeostasis and degeneration. The pathway for autophagosome biogenesis and maturation along the axon will be discussed as well as several key insights revealing the diverse functions of axonal autophagy. Evidence linking altered autophagy with axonal degeneration and neuronal death will be presented. Appropriate manipulation of autophagy may lead to promising therapeutics for neurodegenerative diseases.
Introduction
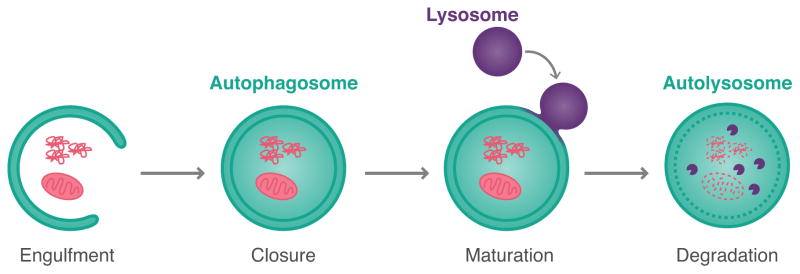
Autophagy is a lysosomal degradation pathway that maintains cellular homeostasis by destroying damaged proteins and organelles (Mizushima et al., 2008; Xie and Klionsky, 2007). In this process, a portion of the cytoplasm is engulfed and sequestered within a double membrane organelle termed an autophagosome (Fig. 1). Subsequent fusion with lysosomes enables degradation of internalized cargo by resident hydrolases. Degradation products (e.g. amino acids, lipids) can then be exported from the lysosome and recycled for new biosynthetic reactions. Thus, autophagy recycles essential biosynthetic building blocks to help sustain cell viability. Indeed, autophagy is particularly important for cell survival under conditions of starvation.
Figure 1. Schematic of the autophagy process.
Cargo is enveloped within a double membrane cisterna, which fuses onto itself to form a closed autophagosome organelle. Autophagosomes acquire hydrolases by fusing with lysosomes to form degradative autolysosomes. Degradation products can then be exported and used for new biosynthesis.
Neurons are particularly dependent on active degradation pathways such as autophagy to maintain homeostasis and viability. Neurons are post-mitotic, and thus cannot dilute out proteotoxins simply by cell division. Furthermore, the vast majority of neurons in the brain are born during embryogenesis and must survive for an entire lifetime. C14-labeling of genomic DNA indicates the age of cortical neurons as old as the human being (Spalding et al., 2005). The small percentage of neurogenesis that does occur in the adult mammalian brain is spatially restricted to the dentate gyrus of the hippocampus and the subventricular zone of the lateral ventricles; neurons generated in the subventricular zone migrate and incorporate into the olfactory bulb (Zhao et al., 2008). Therefore, there is essentially no cellular turnover or exchange of dysfunctional neurons, unlike other cell types such as intestinal epithelial cells that are replaced every few days. As a consequence, it is critical that neurons maintain robust quality control mechanisms to support their long-term viability and functionality.
The importance of autophagy in maintaining neuronal homeostasis is further underscored by various animal models in which genes required for autophagosome formation are genetically inactivated. CNS-specific and neuron-specific knockout of Atg5 or Atg7, core machinery required for autophagosome formation, is sufficient to induce axonal degeneration and neuron death in mice (Hara et al., 2006; Komatsu et al., 2006; Komatsu et al., 2007; Nishiyama et al., 2007). In the absence of autophagy, the axon terminal undergoes a swelling, followed by retraction and neuron death. This neurodegeneration occurs in the absence of any neurodegenerative disease-linked proteins, indicating that autophagy is constitutively active in neurons and basal levels of autophagy are critical for axonal homeostasis.
Neurons also face the daunting logistical challenge of executing autophagy over the extended distance of the axon. Neurons are uniquely characterized by a highly complex and polarized morphology, with extended axonal and dendritic processes. The axon serves as the highway for communication, conveying electrical and chemical information across large distances that can reach up to 1 meter in length in humans. How are homeostatic pathways adapted to maintain protein and organelle quality across such extended distances of the axon? This review will examine the fundamental mechanisms of autophagy in maintaining axonal homeostasis as well as implications of altered autophagy in neurodegenerative disease, focusing primarily on mammalian neurons.
Mechanisms of axonal autophagy
While much of the elegant work characterizing the fundamentals of autophagy has been performed in yeast and small less-polarized mammalian cells, we are only at the beginning of understanding the mechanisms of autophagy in neurons. Whereas less-polarized mammalian cells are only tens of microns wide, neurons have axons that can reach up to 1 meter in length. Therefore, how do degradative pathways respond to protein and organelle damage along the length of the axon? Where are autophagosomes generated in neurons? How do they mature into compartments capable of degradation? Does autophagy require long-range transport along the axon or is it executed within a localized region? Surprisingly, the answers to these basic questions have long remained unclear and unaddressed. Recent work, however, has provided key insights into several of these questions.
Preliminary evidence for autophagy in neurons emerged in the 60’s and 70’s, long before the molecular basis of autophagy was established (Bunge, 1973; Dixon, 1967; Matthews and Raisman, 1972). Initial electron microscopic (EM) studies observed an increase in autophagosome-like organelles in the soma and axon terminals generated by axotomy (Dixon, 1967; Matthews and Raisman, 1972). In another study, Bunge (1973) used EM to map the ultrastructural characteristics of each compartment of a cultured sympathetic neuron from axon tip to soma. Double membrane-bound autophagic structures appeared to be enriched in growth cones, particularly within retracting regions, and were also found along the shaft of the neurite. Many autophagic structures were detected early in formation as they were apparently engulfing cytoplasmic organelles such as dense core vesicles. Furthermore, these early autophagic structures appeared to be continuous with smooth ER (SER), suggesting they were derived from the SER present within the axon terminal. However, without labeling these structures for autophagosomal markers, their identity remained unconfirmed.
Advances in our molecular understanding of autophagy (Mizushima et al., 2011; Weidberg et al., 2011) have now enabled us to track this process in real-time in live cells. Autophagy is readily followed using a GFP-tagged LC3, LC3 being a well-characterized marker for the autophagosome (Kabeya et al., 2000; Mizushima et al., 2004). Recent live-cell imaging analysis has shown that autophagosome formation in primary neurons is a constitutive process enriched in the distal axon (Fig. 2); few autophagosomes are generated in the mid-axon under basal conditions (Hollenbeck, 1993; Maday and Holzbaur, 2012; Maday et al., 2012; Maday and Holzbaur, 2014). Autophagosome biogenesis is evident by the appearance of GFP-LC3-positive puncta that grow progressively into ring structures ~800 nm in diameter (Maday et al., 2012; Maday and Holzbaur, 2014). Autophagosome formation in the distal axon proceeds via an ordered assembly of Atg (Autophagy-related) proteins (Mizushima et al., 2011; Weidberg et al., 2011) recruited with stereotypical kinetics onto the endoplasmic reticulum (Maday and Holzbaur, 2014). These results are consistent with early EM reports citing autophagosome membranes being continuous with SER (Bunge, 1973). Thus, while the fundamental core machinery required for autophagosome formation is conserved from yeast and smaller less-polarized mammalian cells, autophagy in neurons is distinct due to the spatial regulation of autophagosome biogenesis along the axon.
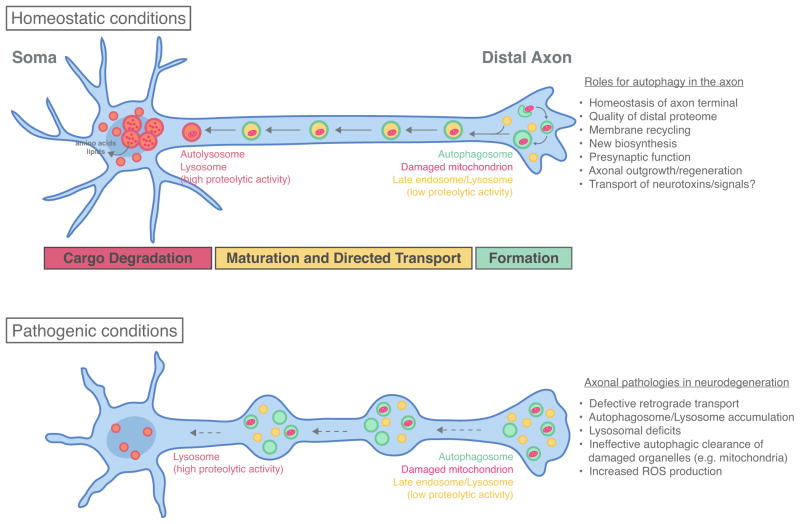
Figure 2. Model for neuronal autophagy in the axon under homeostatic versus pathogenic conditions.
Under homeostatic conditions, autophagosome formation is enriched in the distal axon. Autophagosomes then undergo retrograde transport toward the soma. As autophagosomes move toward the soma, they mature into degradative autolysosomes. The various roles for autophagy in axonal function are listed. Under pathogenic conditions, defects in retrograde transport likely impair maturation of lysosomes and autophagosomes, resulting in an accumulation of dysfunctional organelles within dystrophic axons. Failed clearance of proteotoxins such as damaged mitochondria leads to increased ROS production and axon degeneration.
Following formation in the distal axon, autophagosomes undergo robust retrograde motility along the axon toward the soma (Fig. 2), driven by the microtubule-based molecular motor dynein (Cheng et al., 2015; Hollenbeck, 1993; Lee et al., 2011; Maday et al., 2012; Maday and Holzbaur, 2014; Wang et al., 2015; Yue, 2007). Retrogradely moving autophagosomes transport engulfed soluble and organelle cargoes such as ubiquitin and mitochondrial fragments (Maday et al., 2012). As autophagosomes travel along the axon, they mature into degradative organelles (Lee et al., 2011; Maday et al., 2012; Wang et al., 2015). Autophagosomes along the mid-axon are positive for the late endosome/lysosome marker LAMP1, suggesting that exit from the distal axon is accompanied by fusion with late endosomes/lysosomes (Maday et al., 2012). Recent evidence suggests that the fusion event itself triggers retrograde transport by recruiting dynein (Cheng et al., 2015). Retrograde transport of autophagosomes is also induced by acquisition of the scaffolding proteins JIP1 and huntingtin that promote dynein-based motility (Fu et al., 2014; Wong and Holzbaur, 2014a).
Proximal to the soma, these compartments are fully acidified (Lee et al., 2011; Maday et al., 2012; Wang et al., 2015), consistent with formation of autolysosomes that more effectively degrade cargo (Fig. 2). Full acidification may involve additional fusion events with late endosomes/lysosomes en route as they travel through the axon, as defects in autophagosome transport impair autophagosome maturation and cargo degradation (Fu et al., 2014; Wong and Holzbaur, 2014a). Together, autophagosome biogenesis and maturation is spatially and temporally regulated along the axon, consistent with a highly compartmentalized pathway for autophagy in neurons under basal conditions.
During acute and localized axonal stress, autophagy may be executed locally within the axon. Focally-induced depolarization of mitochondria along the axon recruits LC3 and neighboring lysosomes for local degradation of damaged mitochondria (Ashrafi et al., 2014). While conditions of stress may favor local degradation, autophagy under basal conditions exhibits a pronounced retrograde flux toward the soma (Lee et al., 2011; Maday et al., 2012; Maday and Holzbaur, 2014). Since the soma is the primary site of protein synthesis, delivery of autophagosomes to the soma likely facilitates the recycling of degradation products for new and constitutive biosynthesis.
The localized mitophagy in the axon observed by Ashrafi et al. (2014) was dependent on PINK1 and Parkin. PINK1 is a serine/threonine kinase and Parkin is an E3 ubiquitin ligase that function together to ubiquitinate damaged mitochondria with decreased membrane potential and target them for degradation by autophagy (Pickrell and Youle, 2015). The role of PINK1 and Parkin in mitophagy was elegantly defined in HeLa cells (Kane et al., 2014; Lazarou et al., 2015; Narendra et al., 2008; Narendra et al., 2010), but their role in neuronal mitophagy is less clear. Ashrafi et al. (2014) observed localized recruitment of Parkin to depolarized mitochondria in the axon. Moreover, axonal mitophagy was impaired in hippocampal neurons isolated from Parkin or PINK1 −/− rodents (Ashrafi et al., 2014). In contrast, another report has shown that damaged mitochondria traffic to the soma for envelopment by Parkin-mediated mitophagy (Cai et al., 2012). Further support for the soma being the primary site of mitophagy comes from recent work showing that PINK1 mutant flies have enlarged mitochondria in the soma of motor neurons, with no change in mitochondrial density or morphology along the axon (Devireddy et al., 2015). Another group proposes that PINK1/Parkin-mediated mitophagy does not represent a dominant degradative pathway in neurons due to bioenergetic characteristics of neurons (Van Laar et al., 2011). They postulate that neurons, in contrast to HeLa cells, are less glycolytic and derive most of their energy from mitochondrial oxidative phosphorylation. Hence, neurons must invest heavily in preserving functional, healthy mitochondria and excessive PINK1/Parkin-mediated mitophagy would be highly detrimental to the neuron unless it was offset by increased mitochondrial biogenesis (Van Laar et al., 2011). Additional experiments will be needed to resolve the precise conditions under which PINK1 and Parkin play a role in mitophagy within each neuronal compartment, and further identify potential neuron-specific mechanisms of mitochondrial quality control.
What is the function of autophagy in axons?
Key insights into the physiological function of autophagy in neurons (Fig. 2) come from several knockout mouse models. CNS-specific knockout of Atg5 or Atg7 in mice results in a dramatic and progressive degeneration of Purkinje cells in the cerebellum (Hara et al., 2006; Komatsu et al., 2006). There is also loss of pyramidal cells in the cerebral cortex, although to a lesser degree (Hara et al., 2006; Komatsu et al., 2006). Animals display motor deficits by 4 weeks of age and die by 28 weeks (Komatsu et al., 2006). Thus, constitutive and basal levels of autophagy are essential for neuronal homeostasis and protect against fatal neurodegeneration.
In the absence of autophagy, ubiquitin-positive aggregates accumulate in neurons with age (Hara et al., 2006; Komatsu et al., 2006). Interestingly, different neuronal populations respond differently to the lack of autophagy, accumulating aggregates to a differing degree (Hara et al., 2006; Komatsu et al., 2006). The most vulnerable neuronal subtype, Purkinje cells, lacks aggregates, suggesting perhaps a protective effect for these structures (Hara et al., 2006; Komatsu et al., 2006). Why Purkinje cells are most susceptible to the deficiency in autophagy remains unclear. In these studies (Hara et al., 2006; Komatsu et al., 2006), the cre recombinase was under control of the nestin promoter, which is also expressed in glial cells. Thus, it was not possible to assess cell autonomous effects of autophagy deficiency. In subsequent studies (Komatsu et al., 2007; Nishiyama et al., 2007), neuron-specific knockout animals were generated to investigate these effects directly.
Knockout of Atg5 or Atg7 specifically in Purkinje cells in mice results in strikingly similar phenotypes (Komatsu et al., 2007; Nishiyama et al., 2007). The earliest pathology is swelling and dystrophy of the axon terminal, followed by degeneration (Komatsu et al., 2007; Nishiyama et al., 2007). Despite the elaborate nature of the Purkinje cell dendritic arbor, little effect was observed on dendrites and spines (Komatsu et al., 2007), suggesting that axons may be more vulnerable to the absence of autophagy. Ultimately, the progressive axon degeneration induced by the loss of autophagy leads to Purkinje cell death and deficits in motor coordination (Komatsu et al., 2007; Nishiyama et al., 2007). These effects are cell-autonomous as the loss of Atg7 or Atg5 expression is exclusively in Purkinje cells. Thus, autophagy is critical to maintain homeostasis of the axon in vivo. Evidence also suggests that autophagy plays a role in neural development and well as axonal outgrowth (Ban et al., 2013; Fimia et al., 2007), with alterations in autophagy linked to neurodevelopmental disorders (Lee et al., 2013; Tang et al., 2014), however, precise mechanisms remain elusive.
Interestingly, both Atg5 and Atg7 Purkinje cell knockout animals displayed an accumulation of membranous structures, reminiscent of SER, in the axon terminal (Komatsu et al., 2007; Nishiyama et al., 2007). In control animals, double membrane vacuoles were present within axon terminals, some of which are likely to be autophagosomes whereas the majority are likely derived from neighboring oligodendrocytes (Komatsu et al., 2007). These structures, however, were absent in the Atg7 knockout animals (Komatsu et al., 2007). Knockout animals displayed, rather, an accumulation of SER and cup-shaped structures potentially representing nascent autophagosomes arrested during development; these structures were not observed in dendrites (Komatsu et al., 2007; Nishiyama et al., 2007). Thus, autophagy may play an essential role in membrane turnover and recycling in the distal axon.
In addition to maintaining axonal homeostasis, autophagy also regulates presynaptic function (Fig. 2). Knockout of Atg7 specifically in dopaminergic neurons in mice increased the amplitude of neurotransmitter release and presynaptic recovery (Hernandez et al., 2012). In control tissue, induction of autophagy by mTOR-inhibition increased the number of autophagosomes at presynaptic sites, and also decreased the number of synaptic vesicles (Hernandez et al., 2012). Thus, autophagy modulates neurotransmission, by potentially sequestering synaptic vesicles. In addition to its role in presynaptic function, autophagy also regulates synapse development by promoting formation of the neuromuscular junction in Drosophila (Shen et al., 2015; Shen and Ganetzky, 2009).
The autophagosome journey to the soma provides a pathway for the relay and flow of information from the distal axon to the soma (Fig. 2). Wang et al. (2015) show that neurotoxins hitch a ride with retrogradely moving autophagosomes. This pathway is enhanced with presynaptic activity as neuronal depolarization increases autophagosome formation in the axon terminal and subsequent retrograde trafficking to the soma (Wang et al., 2015). These findings raise several key questions as to whether autophagosomes also carry signaling information to effect changes in the soma, in addition to their immediate function as degradative organelles.
Autophagy in the distal axon likely also plays an important role in maintaining the integrity of the distal proteome (Fig. 2). Since proteins and organelles in the distal axon are further from primary sites of protein synthesis in the soma, they may be more susceptible to aging and damage. In fact, mitochondria residing in the distal axon are older than mitochondria located in the proximal axon (Ferree et al., 2013), and accumulate more mutations in mtDNA (Lehmann et al., 2011). Furthermore, the vast majority of newly synthesized cytosolic proteins are generated in the soma and travel toward the distal axon via slow axonal transport (Scott et al., 2011). These cytosolic proteins travel slowly at ~10 mm per day and will age ~120 days before arriving at the tip of a meter-long human motor axon. Enhanced degradative or chaperone activities in the distal axon may counteract the accumulation of aged and damaged protein in the distal axon.
Autophagy in axon degeneration
Defects in autophagy have been linked to neuronal dysfunction and degeneration (Table 1) (Kiriyama and Nochi, 2015; Rubinsztein et al., 2005; Yamamoto and Yue, 2014; Yang et al., 2013; Yue et al., 2009). Mutations in WDR45 (encodes WIPI4, a component of the core machinery that generates the autophagosome (Mizushima et al., 2011; Weidberg et al., 2011)), are directly linked with neurodegenerative disease in humans. Mutations in WDR45 cause autophagy defects and lead to static encephalopathy of childhood with neurodegeneration in adulthood (SENDA), a neurodegenerative disease affecting the substantia nigra (Saitsu et al., 2013). Cell lines derived from SENDA patients display an accumulation of immature autophagic structures and inefficient autophagic flux (Saitsu et al., 2013). Mutations in genes that encode PINK1 and Parkin, machinery that remove damaged mitochondria from the cell, lead to a juvenile onset form of Parkinson’s Disease (Kitada et al., 1998; Valente et al., 2004). Mutations in genes that encode autophagy receptors optineurin, p62, and ubiquilin-2 cause ALS (Deng et al., 2011; Fecto et al., 2011; Majcher et al., 2015; Maruyama et al., 2010). Optineurin and p62 recruit ubiquitinated cargoes to autophagosomes via their LC3-interacting regions (LIR), whereas ubiquilin-2 lacks a characterized LIR and likely interacts with LC3 indirectly (Majcher et al., 2015; Rothenberg et al., 2010). These autophagy receptors function to target misfolded proteins and damaged mitochondria for degradation (Bjorkoy et al., 2005; N’Diaye et al., 2009; Pankiv et al., 2007; Rothenberg et al., 2010; Wong and Holzbaur, 2014b). Mutations in VCP lead to ALS (Johnson et al., 2010); VCP (valosin-containing protein, also known as p97) regulates mitophagy (Kim et al., 2013), granulophagy (Buchan et al., 2013), and autophagosome maturation into degradative organelles (Ju et al., 2009; Tresse et al., 2010). Exome sequencing revealed several risk genes within the autophagic pathway involved in ALS progression, including a novel ALS gene TBK1 (Cirulli et al., 2015). TBK1 (TANK-binding kinase 1) promotes mitophagy by enhancing the functions of optineurin and p62 through direct phosphorylation (Heo et al., 2015; Matsumoto et al., 2015; Wild et al., 2011). Thus, genetic evidence directly links components of the autophagic pathway with neurodegenerative disease in humans.
Table 1.
Examples of genes mutated in human neurodegenerative disease that encode autophagy proteins
| Protein | Gene with mutation | Neurodegenerative Disease | Protein function | References |
|---|---|---|---|---|
| WIPI4 | WDR45 | SENDA | Core autophagosome formation machinery | Mizushima et al., 2011, Weidberg et al., 2011, Saitsu et al., 2013 |
| PINK1 | PINK1 | PD | Serine/Threonine Kinase; Mitophagy | Valente et al., 2004, Narendra et al., 2010, Lazarou et al., 2015 |
| Parkin | PARK2 | PD | E3 Ubiquitin Ligase; Mitophagy | Kitada et al., 1998, Narendra et al., 2008, Lazarou et al., 2015 |
| Optineurin | OPTN | ALS | Autophagy/mitophagy receptor | Maruyama et al., 2010, Wild et al., 2011, Wong and Holzbaur, 2014b |
| p62/SQSTM1 | SQSTM1 | ALS | Autophagy/mitophagy receptor | Bjorkoy et al., 2005, Pankiv et al., 2007, Fecto et al., 2011 |
| Ubiquilin-2 | UBQLN2 | ALS | Autophagy receptor | Rothenberg et al., 2010, Deng et al., 2011 |
| VCP/p97 | VCP | ALS IBMPFD |
AAA(+)-ATPase; Regulates mitophagy, granulophagy, and autophagosome maturation | Ju et al., 2009, Johnson et al., 2010, Tresse et al., 2010, Buchan et al., 2013, Kim et al., 2013 |
| TBK1 | TBK1 | ALS | Serine/Threonine Kinase; Mitophagy | Cirulli et al., 2015, Heo et al., 2015, Matsumoto et al., 2015 |
Neuronal autophagy is also altered in a variety of axonopathies and axonal stress. Levels of autophagy are elevated in the Lurcher mouse model for excitotoxic neurodegeneration, which has a constitutively active glutamate receptor, leading to death of Purkinje cells (Wang et al., 2006). Autophagosomes accumulate specifically in dystrophic and swollen distal axons of degenerating Purkinje cells and subsequently appear in the soma and dendrites but at lower frequency (Wang et al., 2006). This enrichment of autophagosomes in dying-back distal axons in vivo (Wang et al., 2006) is similar to earlier observations in which autophagosomes were enriched in retracting neurites observed in vitro (Bunge, 1973).
Autophagosomes also accumulate in human neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and familial Amyotrophic Lateral Sclerosis (fALS) that are characterized by a widespread accumulation of protein aggregates and misfolded species (Rubinsztein et al., 2005; Yamamoto and Yue, 2014; Yang et al., 2013; Yue et al., 2009). Cortical biopsies from Alzheimer’s disease patients display elevated levels of autophagy in cortical neurons (Nixon et al., 2005). Autophagosomes accumulate throughout the neuron, but are enriched in dystrophic neurites, particularly within dendrites and synaptic terminals, as compared with the soma (Nixon et al., 2005).
The underlying basis for autophagosome accumulation in neurodegenerative disease is unclear. While genetic inactivation of autophagy leads to neurodegeneration, indicating a neuroprotective role for neuronal autophagy, the significance of elevated autophagy on neuronal viability is unclear. Is elevated autophagy a cause or effect of neurodegenerative disease? Since autophagic flux is a balance between autophagosome formation and degradation, an accumulation of autophagosomes could result from either an increase in autophagosome biogenesis or a defect in clearance. Autophagy may be elevated in response to increased protein aggregation in effort to save the axon. Conversely, high levels of autophagy may be destructive and toxic. Alternatively, defects downstream in autophagy could impair efficient clearance of proteotoxins, contributing to axonal degeneration. The fact that many models of neurodegenerative disease have defects in retrograde transport (Maday et al., 2014; Millecamps and Julien, 2013) and impaired lysosome function (Gowrishankar et al., 2015; Lee et al., 2010; Xie et al., 2015) suggests that downstream steps in autophagy may be blocked (Gowrishankar et al., 2015; Lee et al., 2010; Lee et al., 2011; Nixon et al., 2005; Xie et al., 2015), leading to failed degradation of protein aggregates and damaged organelles.
Neurodegenerative diseases are characterized by defective cargo transport along the axon (Maday et al., 2014; Millecamps and Julien, 2013). Since burgeoning evidence supports the hypothesis that axonal transport may be coupled to organelle function (Fu et al., 2014; Lee et al., 2011; Maday et al., 2012; Wong and Holzbaur, 2014a), impaired cargo motility could accumulate dysfunctional organelles along the axon. Recent reports indicate decreased transport of late endosomal/lysosomal cargoes along the axon of motor neurons expressing SOD1G93A, an ALS-linked SOD1 variant (Xie et al., 2015). Consequently, proteolytic lysosomes are depleted from the soma and autophagosomes containing abnormal mitochondria accumulate along the axon in vivo in SOD1G93A transgenic mice (animal model for fALS) (Xie et al., 2015), similar to the autophagosome accumulation reported by others in models of fALS (Li et al., 2008; Morimoto et al., 2007). These events occur early in disease progression, during asymptomatic stages of disease. Rescuing transport defects by expression of a late endosomal adaptor for dynein restores the distribution of degradative lysosomes in the soma and promotes the clearance of damaged mitochondria from the axon (Xie et al., 2015).
Recent evidence demonstrates the accumulation of protease-deficient lysosomes along the axon in AD mouse models (Gowrishankar et al., 2015). Interestingly, lysosomes accumulate in dystrophic regions of the axon proximal to extracellular amyloid plaques; accumulations are specific for lysosomes and devoid of endosomes, mitochondria and the Golgi (Gowrishankar et al., 2015). In vitro studies have shed light on the mechanisms potentially underlying these phenotypes observed in AD. Pharmacologically blocking lysosomal proteolytic activity inhibits autophagosome and lysosome transport within the axon, resulting in autophagosomes arrested within axon swellings, reminiscent of AD phenotypes (Boland et al., 2008; Lee et al., 2011; Nixon et al., 2005). Thus, impaired lysosomal function may contribute to disease pathogenesis in AD.
Taken together, information gained from studying neurodegenerative disease models sheds insights into the fundamental processes of the autophagic-lysosomal pathway in neurons. These data support a model whereby retrograde transport is important for the efficient maturation of lysosomes and autophagosomes into degradative organelles (Fig. 2). As a result, a gradient of lysosome function is generated along the axon, with proteolytic activity concentrated in the soma (Gowrishankar et al., 2015; Lee et al., 2011; Xie et al., 2015). Thus, organelles need to reach the soma for effective degradation, suggesting a highly compartmentalized pathway for autophagy in neurons. Being the primary site of protein synthesis, delivery of these organelles to the soma can ensure effective recycling of degradation products back into essential biosynthetic reactions.
Under pathogenic conditions, impaired retrograde transport arrests immature and/or dysfunctional autophagosomes/lysosomes in the axon (Fig. 2). Consequently, damaged organelles and proteins are not effectively removed from the axon. Mutations in the dynein motor itself prevent maturation of autophagosomes and degradation of polyQ-huntingtin aggregates (Ravikumar et al., 2005). Furthermore, polyQ-huntingtin also directly impairs autophagosome transport along the axon, resulting in undigested mitochondrial contents within autophagosomes (Wong and Holzbaur, 2014a). In some models of Huntington’s disease, autophagosomes are devoid of cargo including mitochondria (Martinez-Vicente et al., 2010). In SOD1G93A-expressing motor neurons, defective mitochondria are engulfed within autophagosomes, but arrested along the axon (Xie et al., 2015). Loss of autophagy in the CNS also leads to accumulation of Parkinson’s disease-linked proteins alpha-synuclein and LRRK2 in the distal axon (Friedman et al., 2012). Thus, a network of factors contributes to disease pathogenesis including defects in transport, organelle function, and cargo degradation. The accumulation of dysfunctional organelles such as mitochondria in the axon is another hallmark of disease pathogenesis for many neurodegenerative diseases, leading to oxidative damage and apoptotic cascades, thus rendering the axon more vulnerable to degeneration.
Interestingly, the efficiency of the autophagic-lysosomal pathway as well as other proteostasis networks (e.g. chaperones and proteasomes) declines with age (Cuervo, 2008; Labbadia and Morimoto, 2014). Furthermore, autophagy genes required for autophagosome formation (e.g. Atg5 and Atg7) are transcriptionally downregulated in aging human brain (≥70 years old as compared with ≤40 years old) (Lipinski et al., 2010). Thus, compromised quality control pathways may contribute to the late age-onset that is typical of neurodegenerative disease progression. Upon disease progression, however, positive regulators of autophagy are transcriptionally upregulated in AD brain, potentially as a mechanism to compensate for increased protein aggregation (Lipinski et al., 2010).
Outstanding questions
Autophagy was initially identified in yeast as a response to starvation (Tsukada and Ohsumi, 1993), however, autophagy is not upregulated in the brain during nutrient deprivation (Mizushima et al., 2004), suggesting that the primary function of autophagy in neurons may not be a response to starvation. To date, autophagy serves many non-canonical roles in axonal homeostasis, growth, and presynaptic function. Future work will further define the primary physiological functions of autophagy in neurons during homeostasis and its contribution to degeneration. Future studies will elucidate precisely how autophagy is regulated and tuned to the compartment-specific needs of the neuron to facilitate normal neuronal function, and how these processes are altered in response to various modalities of stress. Recent evidence implicates autophagy proteins in learning and memory (Zhao et al., 2015), but the mechanisms underlying these phenotypes are largely unknown. Another key outstanding question is whether the autophagic pathway is exploited to deliver critical signals from the distal axon to the soma to perhaps relay information concerning the integrity of the axonal compartment. Perhaps autophagosomes merge with other retrogradely moving cargo such as signaling endosomes (Chowdary et al., 2012; Harrington and Ginty, 2013). In effect, are autophagosomes more than just garbage trucks? That is, are they also critical conduits relaying important signals to the cell soma?
While the mechanisms of autophagy in axons are being investigated in model systems in vitro, less is known about the dynamics of autophagy in mammalian neurons in vivo. Autophagic flux may be lower in vivo as compared with in vitro systems (Mizushima et al., 2004; Nixon et al., 2005). The fact that protein half-lives in the brain are 2–5 fold longer than corresponding proteins in the liver is consistent with slower autophagic turnover in brain tissue (Price et al., 2010). Future work will need to explore the mechanisms of autophagy in vivo, particularly in disease models to discern the contribution of autophagy during axon degeneration.
Lastly, is there a limited capacity for autophagy in the axon? The axon has long been considered the Achilles heel of the neuron and is particularly vulnerable to damage and injury. Few autophagosomes form in the mid-axon under basal conditions (Maday et al., 2012; Maday and Holzbaur, 2014) and autophagosomes that are generated under pathological conditions appear to be dysfunctional (Lee et al., 2011; Xie et al., 2015). Therefore, is autophagy a viable target for developing drugs to treat neurodegenerative diseases? Since many neurodegenerative diseases exhibit defects in autophagosome-mediated degradation, simply increasing the number of autophagosomes may not be sufficient to restore axon viability. Nevertheless, small molecules that upregulate autophagy have, in fact, been shown to hold promise for the treatment of neurodegenerative disease models (Barmada et al., 2014). Indeed, a combinatorial approach that targets multiple deficits in autophagosome biogenesis, transport, and maturation may powerfully counter fatal neurodegenerative disease phenotypes. If disease-associated defects in autophagy can be restored, how do we specifically target disease-linked misfolded proteins for destruction and avoid digestion of healthy proteins? Since many neurodegenerative diseases have a slow accumulation of protein aggregation with time, can autophagy levels be finely tuned to compensate accordingly? Perhaps increasing autophagy modestly (perhaps even only by 10%?) would be sufficient to counteract protein aggregation over time without deleterious side effects on selectively vulnerable neurons. Answers to these questions can only be achieved with a more accurate understanding of the precise roles of autophagy in degeneration and disease pathogenesis. This understanding, in turn, will likely empower the development of urgently needed therapeutics for a number of invariably fatal and increasingly prevalent neurodegenerative disorders.
Highlights.
Autophagy is an essential lysosomal degradation pathway in neurons
Autophagosome biogenesis and maturation is spatiotemporally regulated in the axon
Autophagy maintains axonal homeostasis and regulates presynaptic function
Neuronal autophagy is altered in axonopathies and neurodegenerative diseases
Manipulating autophagy may lead to therapeutics for neurodegenerative diseases
Acknowledgments
I thank Swathi Ayloo and James Shorter for helpful comments on this manuscript. This work was funded by NIH grants K99NS082619 and R00NS082619 to S.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206:655–70. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban BK, Jun MH, Ryu HH, Jang DJ, Ahmad ST, Lee JA. Autophagy negatively regulates early axon growth in cortical neurons. Mol Cell Biol. 2013;33:3907–19. doi: 10.1128/MCB.00627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–85. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–74. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge MB. Fine structure of nerve fibers and growth cones of isolated sympathetic neurons in culture. J Cell Biol. 1973;56:713–35. doi: 10.1083/jcb.56.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–52. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XT, Zhou B, Lin MY, Cai Q, Sheng ZH. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol. 2015;209:377–86. doi: 10.1083/jcb.201412046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdary PD, Che DL, Cui B. Neurotrophin signaling via long-distance axonal transport. Annu Rev Phys Chem. 2012;63:571–94. doi: 10.1146/annurev-physchem-032511-143704. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, Ren Z, Keebler J, Han Y, Levy SE, Boone BE, Wimbish JR, Waite LL, Jones AL, Carulli JP, Day-Williams AG, Staropoli JF, Xin WW, Chesi A, Raphael AR, McKenna-Yasek D, Cady J, Vianney de Jong JM, Kenna KP, Smith BN, Topp S, Miller J, Gkazi A, Al-Chalabi A, van den Berg LH, Veldink J, Silani V, Ticozzi N, Shaw CE, Baloh RH, Appel S, Simpson E, Lagier-Tourenne C, Pulst SM, Gibson S, Trojanowski JQ, Elman L, McCluskey L, Grossman M, Shneider NA, Chung WK, Ravits JM, Glass JD, Sims KB, Van Deerlin VM, Maniatis T, Hayes SD, Ordureau A, Swarup S, Landers J, Baas F, Allen AS, Bedlack RS, Harper JW, Gitler AD, Rouleau GA, Brown R, Harms MB, Cooper GM, Harris T, Myers RM, Goldstein DB Consortium FS. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–12. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, Jiang H, Hirano M, Rampersaud E, Jansen GH, Donkervoort S, Bigio EH, Brooks BR, Ajroud K, Sufit RL, Haines JL, Mugnaini E, Pericak-Vance MA, Siddique T. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy S, Liu A, Lampe T, Hollenbeck PJ. The Organization of Mitochondrial Quality Control and Life Cycle in the Nervous System In Vivo in the Absence of PINK1. J Neurosci. 2015;35:9391–401. doi: 10.1523/JNEUROSCI.1198-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JS. “Phagocytic” lysosomes in chromatolytic neurones. Nature. 1967;215:657–8. doi: 10.1038/215657a0. [DOI] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, Donkervoort S, Ajroud-Driss S, Sufit RL, Heller SL, Deng HX, Siddique T. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- Ferree AW, Trudeau K, Zik E, Benador IY, Twig G, Gottlieb RA, Shirihai OS. MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy. 2013;9:1887–96. doi: 10.4161/auto.26503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–93. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu MM, Nirschl JJ, Holzbaur EL. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell. 2014;29:577–90. doi: 10.1016/j.devcel.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci U S A. 2015;112:E3699–708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harrington AW, Ginty DD. Long-distance retrograde neurotrophic factor signalling in neurons. Nat Rev Neurosci. 2013;14:177–87. doi: 10.1038/nrn3253. [DOI] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, Torres CA, Setlik W, Cebrian C, Mosharov EV, Tang G, Cheng HC, Kholodilov N, Yarygina O, Burke RE, Gershon M, Sulzer D. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 2012;74:277–84. doi: 10.1016/j.neuron.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol. 1993;121:305–15. doi: 10.1083/jcb.121.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ Consortium I. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–64. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187:875–88. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NC, Tresse E, Kolaitis RM, Molliex A, Thomas RE, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, Winborn BJ, Moore J, Lee JY, Yao TP, Pallanck L, Kundu M, Taylor JP. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama Y, Nochi H. The Function of Autophagy in Neurodegenerative Diseases. Int J Mol Sci. 2015;16:26797–26812. doi: 10.3390/ijms161125990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–94. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. Proteostasis and longevity: when does aging really begin? F1000Prime Rep. 2014;6:7. doi: 10.12703/P6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–58. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Hwang SK, Lee JA. Neuronal autophagy and neurodevelopmental disorders. Exp Neurobiol. 2013;22:133–42. doi: 10.5607/en.2013.22.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci. 2011;31:7817–30. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann HC, Chen W, Borzan J, Mankowski JL, Hoke A. Mitochondrial dysfunction in distal axons contributes to human immunodeficiency virus sensory neuropathy. Ann Neurol. 2011;69:100–10. doi: 10.1002/ana.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–3. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2010;107:14164–9. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Holzbaur EL. Autophagosome assembly and cargo capture in the distal axon. Autophagy. 2012;8:858–60. doi: 10.4161/auto.20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–17. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Holzbaur EL. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev Cell. 2014;30:71–85. doi: 10.1016/j.devcel.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majcher V, Goode A, James V, Layfield R. Autophagy receptor defects and ALS-FTLD. Mol Cell Neurosci. 2015;66:43–52. doi: 10.1016/j.mcn.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–42. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- Matthews MR, Raisman G. A light and electron microscopic study of the cellular response to axonal injury in the superior cervical ganglion of the rat. Proc R Soc Lond B Biol Sci. 1972;181:43–79. doi: 10.1098/rspb.1972.0040. [DOI] [PubMed] [Google Scholar]
- Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–76. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, Abe K. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–7. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–9. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 2007;3:591–6. doi: 10.4161/auto.4964. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–22. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–73. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci U S A. 2010;107:14508–13. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–6. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–32. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A, Ryujin F, Yoshioka S, Nishiyama K, Kondo Y, Tsurusaki Y, Nakashima M, Miyake N, Arakawa H, Kato M, Mizushima N, Matsumoto N. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet. 2013;45:445–9. 449e1. doi: 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- Scott DA, Das U, Tang Y, Roy S. Mechanistic logic underlying the axonal transport of cytosolic proteins. Neuron. 2011;70:441–54. doi: 10.1016/j.neuron.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DN, Zhang LH, Wei EQ, Yang Y. Autophagy in synaptic development, function, and pathology. Neurosci Bull. 2015;31:416–26. doi: 10.1007/s12264-015-1536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–9. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–43. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–43. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–27. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Van Laar VS, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–40. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP, Chait BT, Zhong Y, Heintz N, Yue Z. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–68. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Martin S, Papadopulos A, Harper CB, Mavlyutov TA, Niranjan D, Glass NR, Cooper-White JJ, Sibarita JB, Choquet D, Davletov B, Meunier FA. Control of autophagosome axonal retrograde flux by presynaptic activity unveiled using botulinum neurotoxin type a. J Neurosci. 2015;35:6179–94. doi: 10.1523/JNEUROSCI.3757-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–56. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur EL. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014a;34:1293–305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014b;111:E4439–48. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhou B, Lin MY, Wang S, Foust KD, Sheng ZH. Endolysosomal Deficits Augment Mitochondria Pathology in Spinal Motor Neurons of Asymptomatic fALS Mice. Neuron. 2015;87:355–70. doi: 10.1016/j.neuron.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Yue Z. Autophagy and its normal and pathogenic states in the brain. Annu Rev Neurosci. 2014;37:55–78. doi: 10.1146/annurev-neuro-071013-014149. [DOI] [PubMed] [Google Scholar]
- Yang Y, Coleman M, Zhang L, Zheng X, Yue Z. Autophagy in axonal and dendritic degeneration. Trends Neurosci. 2013;36:418–28. doi: 10.1016/j.tins.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3:139–41. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- Yue Z, Friedman L, Komatsu M, Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim Biophys Acta. 2009;1793:1496–507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–60. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhao YG, Sun L, Miao G, Ji C, Zhao H, Sun H, Miao L, Yoshii SR, Mizushima N, Wang X, Zhang H. The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy. 2015;11:881–90. doi: 10.1080/15548627.2015.1047127. [DOI] [PMC free article] [PubMed] [Google Scholar]