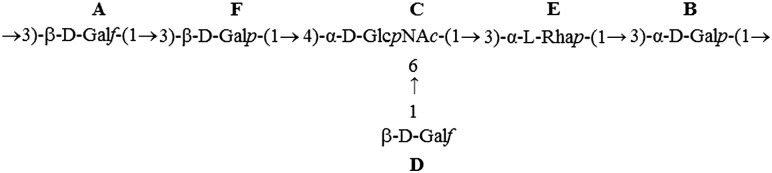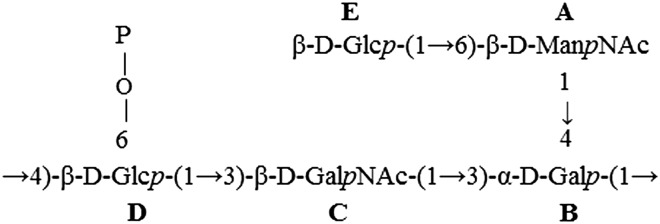Abstract
The Lactobacillus casei strain, LOCK 0919, is intended for the dietary management of food allergies and atopic dermatitis (LATOPIC® BIOMED). The use of a probiotic to modulate immune responses is an interesting strategy for solving imbalance problems of gut microflora that may lead to various disorders. However, the exact bacterial signaling mechanisms underlying such modulations are still far from being understood. Here, we investigated variations in the chemical compositions and immunomodulatory properties of the polysaccharides (PS), L919/A and L919/B, which are produced by L. casei LOCK 0919. By virtue of their chemical features, such PS can modulate the immune responses to third-party antigens. Our results revealed that L919/A and L919/B could both modulate the immune response to Lactobacillus planatarum WCFS1, but only L919/B could alter the response of THP-1 cells (in terms of tumor necrosis factor alpha production) to L. planatarum WCFS1 and Escherichia coli Nissle 1917. The comprehensive immunochemical characterization is crucial for the understanding of the biological function as well as of the bacteria–host and bacteria–bacteria cross-talk.
Keywords: immunomodulation, Lactobacillus, NMR spectroscopy, polysaccharide, probiotic
Introduction
Lactic acid bacteria (LAB) are used in many fermented foods, particularly fermented dairy products, such as cheese, buttermilk and fermented milk. Some LAB confer health benefits and are thus considered to be probiotics (Mattu and Chauhan 2013). As defined by the Food and Agriculture Organization of the United Nations, “probiotics are live microorganisms which when administrated in adequate amounts confer a health benefit on the host” (FAO/WHO 2001). The bacteria in probiotic products typically belong to Lactobacillus genera, although other microorganisms (e.g. Bifidobacterium, Gram-negative bacteria and yeasts) have also been proposed as probiotics (McFarland 2006). The beneficial effects associated with the consumption of probiotic bacteria include the prevention of antibiotic-associated, travelers’ or Clostridium difficile-associated diarrhea; necrotizing enterocolitis; colitis (Górska et al. 2009); food allergies and atopic eczema (Weston et al. 2005). Moreover, probiotic bacteria have also been shown to reduce inflammation, improve gastrointestinal tract homeostasis (Tursi et al. 2010), enhance epithelial barrier function (Luyer et al. 2005) and modulate the immune response (Taverniti and Guglielmetti 2011; Wells 2011). It has been well established that the effects of probiotics depend on the strain(s) and the dose/duration of treatment. Thus, the proposed efficacy of a particular strain cannot be extrapolated to other probiotic strains. We do not yet know precisely how lactobacilli interact with the gut bacteria or host immune cells, or the mechanisms through which they adhere to host surfaces. The growing availability of genomic sequences for various lactobacilli has highlighted the presence of various molecules involved in host–bacteria interactions (Kant et al. 2011). These molecules include the polysaccharides (PS), which are macromolecules that are used to improve the viscosity and texture of fermented products (Marshall and Rawson 1999). Recently, it has been proposed that these polymers may contribute to host colonization (Dertli et al. 2015), and promote host health by supporting immunomodulatory properties (Ciszek-Lenda et al. 2011; Fanning et al. 2012; Górska et al. 2014), antiviral activity, antioxidant activity (Zhang et al. 2013) and antihypertensive activity (Ai et al. 2008).
The Lactobacillus PS are a large group of carbohydrate molecules that may be closely or loosely associated with the bacterial surface (called capsular PS or slime PS, respectively), and some strains may release capsular PS into the environment. The PS show considerable diversity in their composition and structure. The LAB polysaccharides may be divided into two classes: homopolysaccharides that comprise a single type of monosaccharide; and heteropolysaccharides that comprise regular repeating units of three to nine different carbohydrate moieties synthesized from intercellular sugar nucleotide precursors (Górska et al. 2007; Górska-Frączek et al. 2011). Their structural diversity is largely due to the presence of different sugar monomers, variations in the modes of linkage, substitution and branching and even differences in molecular mass. PS may be recognized by a number of receptors, including C-type lectin receptors, Toll-like receptors (TLR) and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin receptors (Segers and Lebeer 2014).
The L. casei LOCK 0919 are of particular interest because are intended for the dietary management of food allergies and atopic dermatitis as a commercially available product (LATOPIC® BIOMED). The aim of this work was to perform a structural analysis and evaluate the immunomodulatory effects of PS isolated from L. casei LOCK 0919. To further assess the structure–function relationships between PS and their potential immunomodulatory activities, we examined their ability to modulate immune response of the following: mouse bone marrow-derived dendritic cells (BM-DC); human embryonic kidney (HEK) 293 cells stably transfected with vectors encoding TLR or the NOD2 receptor; and THP-1 cells. We also examined the immune response of these cells to other bacteria, including Lactobacillus plantarum WCFS1; the probiotic Escherichia coli strain, Nissle 1917; and Crohn's disease-associated E. coli strain, LF82. Finally, we determined the immunoreactivity of human sera to the PS.
Results
PS L919/A and L919/B show appreciable structural differences
Antigens L919/A and L919/B were isolated from the bacterial cell mass of L. casei LOCK 0919 and investigated for their monosaccharide composition. Gas–liquid chromatography-mass spectrometry (GLC-MS) analysis of obtained alditol acetates (sugar and methylation analysis) and acetylated 2-butyl glycosides (determination of the absolute configuration) revealed that L919/A is composed of L-Rha, D-Gal and D-GlcNAc in a molar ratio of 1:3.8:0.8. Methylation analysis revealed the presence of 3-substituted 6-deoxy hexose, 3-substituted hexofuranose, 3-substituted hexose, 4,6-disubstituted GlcNAc and t-hexofuranose in a molar ratio of 0.5:0.7:1:0.5:0.6. The average molecular mass was 9.7 × 104 Da. The same analysis showed that L919/B is composed of D-Glc, D-Gal, D-ManNAc and D-GalNAc in a molar ratio of 1:0.4:0.5:0.9. Methylation analysis revealed the presence of 6-substituted hexose, 3,4-disubstituted or 3,5-disubstituted hexose, 3-substituted GalNAc, 4 or 5-substituted hexose and t-hexose in a molar ratio of 0.5:0.4:0.9:0.4:1. The average molecular mass was 18.9 × 103 Da.
Nuclear magnetic resonance analysis of L919/A
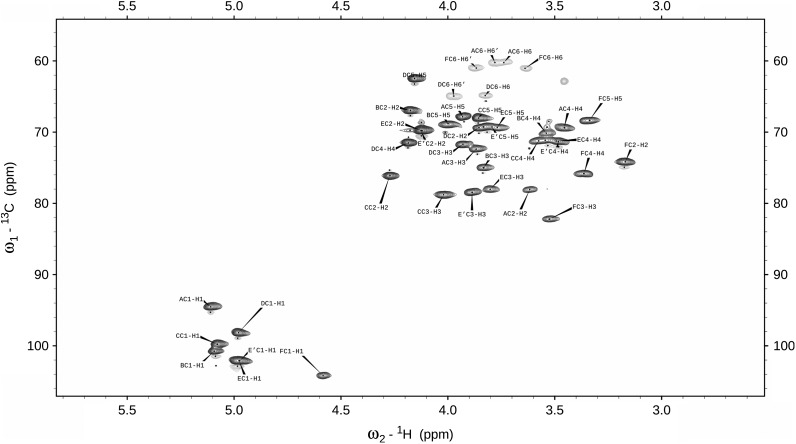
The 1D 1H-NMR spectrum of L919/A obtained at 600 MHz revealed six anomeric proton signals (at 5.15, 4.99, 4.96, 4.94, 4.94 and 4.24 ppm) correlated with six carbon resonances in the 1H-13C HSQC spectrum (at 109.2, 99.3, 94.2, 107.4, 102.0 and 102.6 ppm, respectively), as shown in Figure 1. The 1H-NMR spectrum also contained signals for acetyl groups at 1.93 ppm. The six monosaccharides of L919/A designated as residues A–F, according to their proton chemical shifts. The resonances corresponding to residues C, D and E were heavily but not intractably overlapped. The anomeric carbon resonances of residues A and D were characteristic of the furanosidic ring form (Katzenellenbogen et al. 2009; Shibata and Okawa 2011). Consistent with this, a correlation between H1 and C4 was observed in the 1H,13C HMBC spectrum. The chemical shifts of the anomeric protons and carbons of residues B, C, E and F were characteristic of the pyranose form (Jansson et al. 1989). The complete 1H and 13C chemical shifts for all components of L919/A were assigned by two-dimensional COSY, TOCSY and HSQC experiments, as well as by comparison with previously published 1H-and 13C-NMR data (Gorin and Mazurek 1975; Bock and Pedersen 1983; Lipkind et al. 1988). The chemical shifts are reported in Table I. Disentangling of the spin system showed that residues A, B, D and F were assigned to the galacto-configuration, whereas residue C was recognized as an N-acetyl amino sugar. The latter finding was confirmed by correlation of protons at N-bearing carbons to the corresponding carbons at 53.2 ppm (C2 of C), as revealed by the 1H-13C HSQC experiment. Residue E, which exhibited the characteristic chemical shifts for the H6 and C6 resonances, is a 6-deoxy hexose that, according to the above chemical analysis, must be rhamnose. The sequence of the monosaccharide residues within the repeating unit of L919/A was obtained by assignment of the inter-residue connectivities observed in the two-dimensional NOESY and HMBC spectra, which showed cross-peaks between the anomeric protons and the carbons at the linkage positions (Table I). The structure of the hexasaccharide repeating unit of L919/A, as determined herein, is shown in Figure 2. This structure is identical to that of the PS isolated from L. rhamnosus GG (GG) (Landersjö et al. 2002), but the two differ in that the molecular mass of L919/A was calculated to be 9.7 × 104 Da, whereas that of GG was reported to be 1.4 × 106 Da (Lebeer et al. 2009). Therefore, in our further experiments, assessing the immunomodulatory properties of L919/A, we included GG for comparison.
Fig. 1.
Selected parts of the 1H-13C HSQC nuclear magnetic resonance (NMR) spectrum of L919/A.
Table I.
1H-and 13C-NMR chemical shifts and selected inter-residue (HMBC) connectivities obtained for the anomeric protons of L919/A from Lactobacillus casei strain LOCK 0919
| Sugar residue | Chemical shifts 1H, 13C (ppm) | Selected connectivities (ppm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H6′ | CH3CO | NOESY | HMBC | ||
| C1 | C2 | C3 | C4 | C5 | C6 | ||||||
| A | →3)-β-D-Galf-(1→ | 5.15 | 4.31 | 4.00 | 4.15 | 3.78 | 3.61 | — | A1-F3 (3.62) | ||
| 109.2 | 79.5 | 84.4 | 81.6 | 70.6 | 62.6 | ||||||
| B | →3)-α-D-Galp-(1→ | 4.99 | 3.85 | 3.83 | 3.96 | 4.04 | 3.65 | B1-A3 (4.00) | B1-A3 (84.4) | ||
| 99.3 | 67.0 | 77.2 | 68.9 | 71.2 | 60.9 | ||||||
| C | →4,6)-α-D-GlcpNAc-(1→ | 4.96 | 3.92 | 3.85 | 3.75 | 4.12 | 3.76 | 3.94 | 1.93 | C1-E2 (4.06) | C1-E3 (75.5) |
| 21.8 | C1-E3 (3.78) | ||||||||||
| 94.2 | 53.2 | 69.3 | 78.1 | 69.3 | 65.5 | ||||||
| D | β-D-Galf-(1→ | 4.94 | 4.06 | 3.98 | 3.91 | 3.74 | 3.58 | D1-C6 (3.76, 3.94) | D1-C6 (65.5) | ||
| 107.4 | 80.8 | 76.4 | 82.1 | 70.3 | 62.6 | ||||||
| E | →3)-α-L-Rhap-(1→ | 4.94 | 4.06 | 3.78 | 3.42 | 3.73 | 1.22 | E1-B3 (3.83) | E1-B3 (77.2) | ||
| 102.0 | 66.9 | 75.5 | 70.2 | 69.2 | 17.0 | ||||||
| F | →3)-β-D-Galp-(1→ | 4.24 | 3.54 | 3.62 | 3.99 | 3.63 | 3.64 | F1-C4 (3.75) | F1-C4 (78.1) | ||
| 102.6 | 69.8 | 79.9 | 68.4 | 75.1 | 60.9 | ||||||
Spectra were obtained for 2H2O solutions at 25°C, with acetone (δH 2.225, δC 31.05 ppm) used as an internal reference.
Fig. 2.
The structure of the hexasaccharide repeating unit of L919/A.
NMR analysis of L919/B
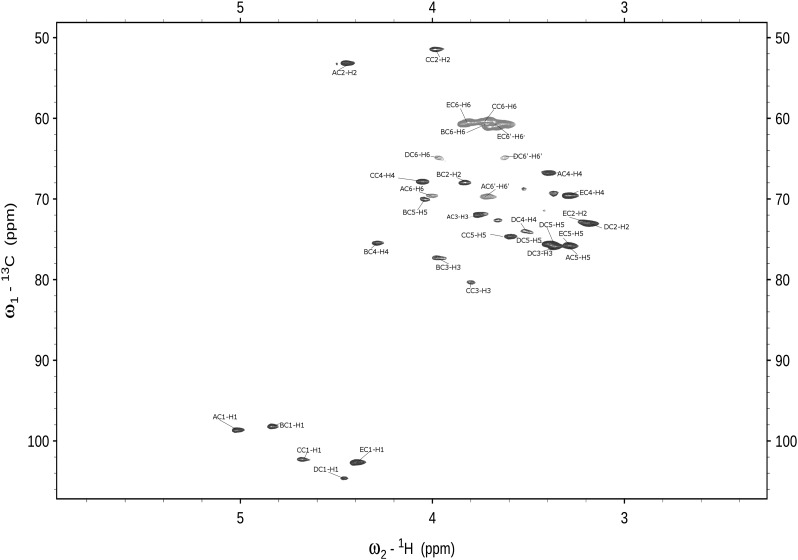
The 1D 1H-NMR spectrum of L919/B obtained at 600 MHz revealed five anomeric proton signals (at 5.02, 4.81, 4.65, 4.46 and 4.37 ppm) that correlated with five carbon resonances (at 98.6, 98.2, 103.3, 104.6 and 102.5 ppm, respectively), as shown in the 1H-13C HSQC spectrum of L919/B presented in Figure 3. The five monosaccharides of L919/B were designated as residues A–E, according to their proton chemical shifts. The anomeric carbon resonances of all residues were found to be characteristic of the pyranose ring form (Jansson et al. 1989). The 31P-NMR spectrum contained one signal for a phosphomonoester group at 2.6 ppm. The 1H-NMR spectrum also contained signals of acetyl groups at 1.93–1.95 ppm. The complete 1H-and 13C-NMR resonances were obtained using different two-dimensional nuclear magnetic resonance experiments and comparison to previously published 1H-and 13C-NMR data sets (Gorin and Mazurek 1975; Bock and Pedersen 1983; Lipkind et al. 1988). The chemical shifts are reported in Table II. Residues A and C were recognized as the N-acetyl amino sugars; this was confirmed by the correlation of protons at N-bearing carbons to the corresponding carbons at 53.0 ppm (C2 of A) and 51.5 ppm (C2 of C), as revealed by the 1H-13C HSQC experiment. Based on our analysis of TOCSY spectra and 1H and 13C chemical shifts, residue B was assigned as having the galacto-configuration, whereas residues D and E have the gluco-configuration. The chemical shifts were used to identify residues A, B, C, D and E as N-acetylmannosamine, galactose, N-acetylgalactosamine and two glucose residues, respectively. The sequence of the monosaccharide residues within the repeating unit of the L919/B was obtained by assignment of the inter-residue connectivities observed in the two-dimensional NOESY and HMBC spectra, which showed cross-peaks between the anomeric protons and the carbons at the linkage positions (Table II). The phosphate substitution position was revealed using a 1H,31P-correlation experiment (HMQC and HSQC). The 31P resonance at δ 2.6 ppm showed connectivity to the H6 signal at δ 3.95 ppm (H6 of residue D) and the carbon signal at δ 64.8 ppm (C6 of residue D). The structure of the pentasaccharide repeating unit of L919/B, as determined herein, is shown in Figure 4. The molecular mass was calculated to be 18.9 × 103 Da.
Fig. 3.
Selected parts of the 1H-13C HSQC nuclear magnetic resonance (NMR) spectrum of L919/B.
Table II.
1H- and 13C-NMR chemical shifts and selected inter-residue (HMBC) connectivities obtained for the anomeric protons of L919/B from Lactobacillus casei LOCK 0919
| Sugar residue | Chemical shifts 1H, 13C (ppm) | Selected connectivities (ppm) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H6′ | CH3CO | NOESY | HMBC | ||
| C1 | C2 | C3 | C4 | C5 | C6 | ||||||
| A | →6)-β-D-ManpNAc-(1→ | 5.02 | 4.43 | 3.74 | 3.46 | 3.26 | 4.01 | 3.72 | 1.93 | A1-B4 (4.26) | A1-A2 (intra) |
| 98.6 | 53.0 | 71.8 | 66.8 | 75.6 | 69.4 | 22.1 | A1-B4 (75.5) | ||||
| B | →3,4)-α-D-Galp-(1→ | 4.81 | 3.80 | 3.95 | 4.26 | 4.06 | 3.68 | B1-D4 (3.52) | B1-B5 (intra) | ||
| B1-B5 (4.06) | |||||||||||
| C | →3)-β-D-GalpNAc-(1→ | 98.2 | 67.9 | 77.4 | 75.5 | 69.9 | 61.0 | B1- B6 (3.68) | |||
| 4.65 | 3.97 | 3.77 | 4.02 | 3.56 | 3.70 | 1.95 | C1-B3 (3.95) | C1-B3 (77.4) | |||
| 21.9 | |||||||||||
| D | →4,6)-β-D-Glcp-(1→ | 103.3 | 51.5 | 80.3 | 67.9 | 74.7 | 60.3 | ||||
| 4.46 | 3.28 | 3.36 | 3.52 | 3.39 | 3.95 | 3.65 | D1-C3 (3.77) | D1-C3 (80.3) | |||
| 104.6 | 72.8 | 75.7 | 73.8 | 75.3 | 64.8 | ||||||
| E | β-D-Glcp-(1→ | 4.37 | 3.26 | 3.37 | 3.27 | 3.38 | 3.63 | 3.80 | E1-A6 (4.01 and 3.72) | E1-A6 (69.4) | |
| 102.5 | 72.9 | 75.5 | 69.2 | 75.9 | 60.6 | ||||||
| P 2.6 ppm | P-D6 (64,8) | ||||||||||
Spectra were obtained for 2H2O solutions at 25°C, with acetone (δH 2.225, δC 31.05 ppm) used as an internal reference.
Fig. 4.
The structure of the pentasaccharide repeating unit of L919/B.
L919/A and L919/B modulate the immune response of mouse BM-DC to L. plantarum WCFS1, but not E. coli Nissle 1917, or Crohn's disease-associated E. coli LF82
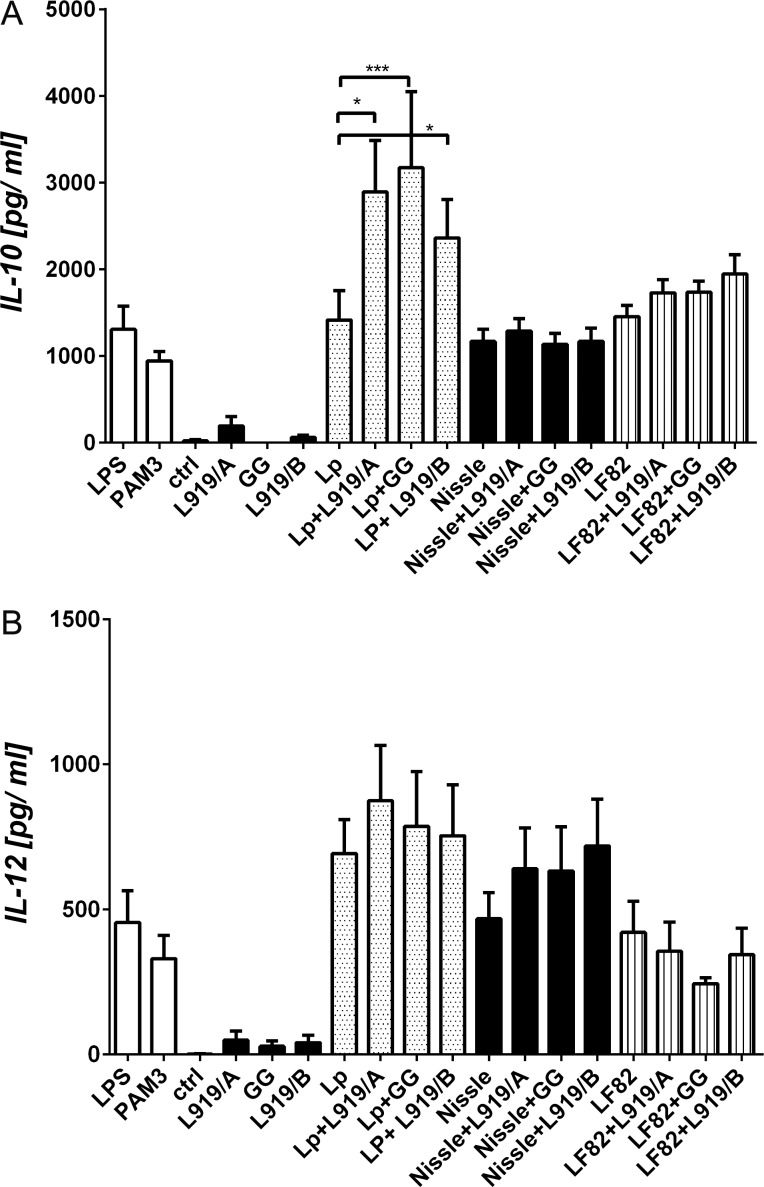
When BM-DC derived from BALB/c mice were stimulated with PS L919/A or L919/B, we observed very low (near control levels) expression of the costimulatory molecules, CD40, CD80 and CD86 (data not shown). Similarly, the PS-triggered inductions of IL-10 and IL-12p70 were low and nonsignificant (p > 0.05; Figure 5). In contrast, cocultivation of BM-DC with L. plantarum WCFS1 (Lp), E. coli Nissle 1917 (Nissle) or E. coli LF82 (LF82) cells induced both of the tested cytokines (Figure 5). Interestingly, stimulation of BM-DC with Lp plus L919/A, L919/B or GG significantly increased IL-10 to different levels (Figure 5A) but did not alter IL-12p70 production compared to the levels in cells treated with Lp alone (Figure 5B). In contrast, admixing L919/A, L919/B or GG with Nissle did not trigger any detectable change in these cytokines, nor did admixture of L919/A or L919/B with LF82. Admixture of LF82 and GG decreased the level of IL-12p70 in LF82 cells, but not to a significant degree (p > 0.05).
Fig. 5.
Cytokine production by bone marrow-derived dendritic cells (BM-DC) exposed individually to L919/A, L919/B, GG, Lactobacillus plantarum WCFS1 cells, Escherichia coli Nissle 1917 cells or E. coli LF82 cells, or to mixtures of each PS and bacterial cells. BM-DC from naive BALB/c mice were cultured for 18 h with media alone (ctrl), ultrapure lipopolysaccharide from E. coli (LPS, 1 µg/ml), Pam3Cys (PAM3, 1 µg/ml), 10 µg/ml of PS, 107 CFU/ml of formaldehyde-inactivated bacteria or with their indicated mixtures. The levels of IL-10 (A) and IL-12p70 (B) in culture supernatants were determined by enzyme-linked immunosorbent assay (ELISA). Pooled results from three independent experiments are shown, and are expressed as mean ± SEM; *p < 0.05, and ***p < 0.001.
TLR2, TLR4 and NOD2 are not involved in the recognition of L919/A or L919/B
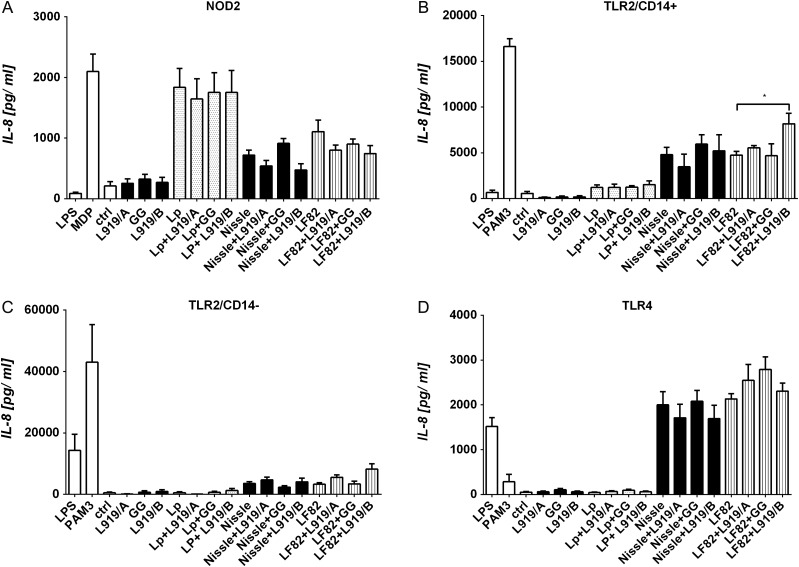
To specify the pattern recognition receptors involved in the relevant PS signaling pathways, each PS alone, Lp, Nissle or LF82 cells alone, or mixtures of each PS and bacterium were incubated with HEK293 cells stably expressing TLR2, TLR4 or NOD2. As positive controls, we used Pam3Cys, lipopolysaccharide (LPS) and muramyl-dipeptide (MDP) for the cells overexpressing TLR2, TLR4 and NOD2, respectively. The level of the cytokine, IL-8, was measured as an indicator of specific receptor-mediated antigen stimulation. Our results showed that the tested PS, with or without admixture with Lp, Nissle or LF82 cells, failed to induce the production of IL-8 in HEK293 cells expressing TLR2, TLR4 or NOD2 (Figure 6).
Fig. 6.
Activation of TLR and NOD2 receptors by PS. HEK293 cells stably transfected with vectors encoding human NOD2 (A), TLR2 (293-hTLR2/CD14) (B), TLR2 (293-hTLR2/CD14-) (C) or TLR4 (293-hTLR4/MD2/CD14) (D) were cultured for 20 h with or without 10 µg/ml of the indicated PS. Pam3Cys (PAM3, 1 µg/ml), muramyl-dipeptide (MDP, 1 µg/ml) and lipopolysaccharide (LPS, 1 µg/ml) were used as positive controls for TLR2, TLR4 and NOD2, respectively. Unstimulated cells (ctrl) were used as the negative control. Stimulation was evaluated by measurement of IL-8 production. The results are expressed as mean ± SEM, and pooled results from three independent experiments are shown.
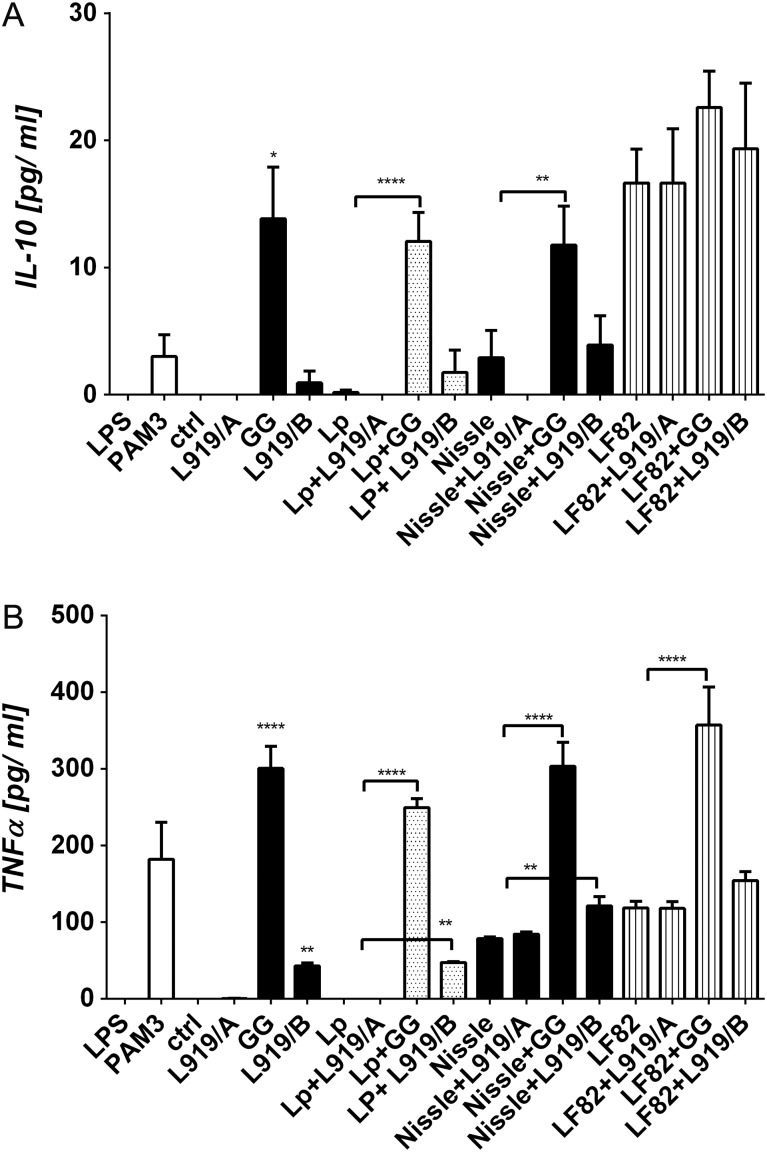
Polymer L919/B stimulates THP-1 cells to produce tumor necrosis factor alpha
The THP-1 cell line was stimulated with each PS alone, with Lp, Nissle or LF82 cells alone, or with PS/bacteria mixtures, and the levels of IL-10 and tumor necrosis factor alpha (TNF-α) were measured as indicators of antigen stimulation. L919/A, Lp cells or Nissle cells alone failed to induce the production of IL-10 or TNF-α; L919/B alone increased the level of TNF-α; and GG or LF82 cells alone triggered the productions of IL-10 and TNF-α (Figure 7). Mixtures of L919/A plus Lp, Nissle or LF82 cells did not trigger any change in the levels of IL-10 or TNF-α. Interestingly, however, exposure of THP-1 cells to mixtures of GG or L919/B plus Lp or Nissle cells significantly increased cytokine production compared to treatment with Lp or Nissle cells alone, but not with LF82 cells alone (Figure 7).
Fig. 7.
Differentiation of THP-1 cell treated individually with L919/A, L919/B, GG, L. plantarum WCFS1, E. coli Nissle 1917, or E. coli LF82 or the indicated mixtures of PS and bacteria. Unstimulated cells (ctrl) were used as a negative control. The levels of IL-10 (A) and TNF-α (B) in culture supernatants were determined by enzyme-linked immunosorbent assay (ELISA). Pooled results from three independent experiments are shown as mean ± SEM are shown; **p < 0.01, and ****p < 0.0001.
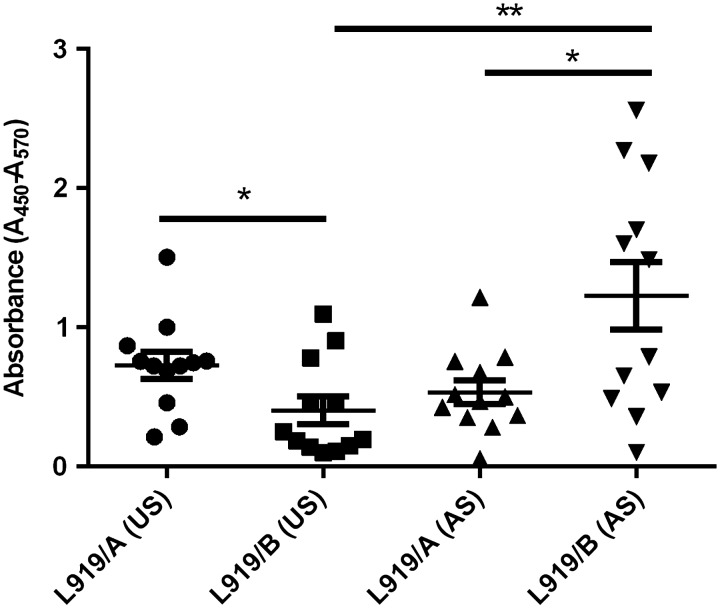
L919/A and L919/B differ in their reactivity with human sera
The reactivity of PS with human sera obtained from healthy adult volunteers (AS) and umbilical cord blood sera (US) was examined by enzyme-linked immunosorbent assay (ELISA; Figure 8). All studied human sera were found to contain antibodies toward both PS, at various titers. The reactivity of L919/B was much higher with AS than with US (p < 0.01), and that of L919/B with AS was much higher than that of L919/A with AS (p < 0.05). In contrast, the reactivity of L919/A with US was much higher than that of L919/B with US (p < 0.05).
Fig. 8.
Enzyme-linked immunosorbent assay (ELISA) examining the reactivity between L919/A or L919/B and human sera obtained from healthy adults volunteers (AS) and umbilical cord blood (US); * p< 0.05, and ** p < 0.01.
Discussion
The study of PS isolated from probiotic bacteria is relatively new, and there is a growing interest in understanding the precise mechanisms that underlie the biological activities of PS. In the future, we need to elucidate the means through which PS interact with host immune systems, as well as their potential for use as therapeutic agents. We must also seek to determine the compositions, chemical structures and immunomodulatory properties of PS. Here, our group sought to determine the relationship between the biological properties and chemical structure/molecular mass of PS antigens isolated from the probiotic bacterium, L. casei LOCK 0919. The structures of these PS were determined by classical chemical analysis, NMR spectroscopy and mass spectrometry, which collectively revealed that (1) polymer L919/A has a relatively small molecular mass of 9.7 × 104 Da, is a branched heteropolysaccharide with a repeating six-residue unit (Rha, four Gal and GalNAc), and has a structure identical to that of PS GG isolated from L. rhamnosus GG (Landersjö et al. 2002); and (2) polymer L919/B contains five monosaccharide residues (two Glc, Gal, ManNAc and GalNAc) and one phosphate group in its repeating unit.
Although L919/A and GG were found to be identical in their chemical structures, they exhibited molecular masses of 9.7 × 104 and 1.4 × 106 Da, respectively. We thus decided to compare their modulatory properties, in order to determine if this difference in molecular mass affected their immunomodulatory features. Despite the existing high-quality research, we are still far from completely understanding the biological effects of Lactobacillus PS and the relationship between their chemical structure and function. As PS molecules can vary widely, their potential immunomodulatory properties may differ with respect to the exact chemical features of the molecules. We herein report that L919/A, L919/B and GG do not show any evidence of acting through the TLR2, TLR4 or NOD2 receptors, altering the maturation of BM-DC, or stimulating the production of IL-10 or IL-12p70. This is interesting given that whole-cell L. casei LOCK 0919 has been shown to act through NOD2 and induce the productions of IL-10 and IL-12p70 (Kozakova et al. 2015). In many cases, whole bacteria (as opposed to their bacterial compounds) are necessary for the stimulation of immune cells (Shida et al. 2006).
Our results show that particular structure can display different effects itself or in a complex multicomponent context. We found that the tested PS could effectively modulate the immune response elicited by L. plantarum WCFS1 (Lp), but not by the probiotic E. coli strain, Nissle 1917, or Crohn's disease-associated E. coli strain, LF82. L919/A, GG and L919/B all triggered the synergistic induction of IL-10 (but not IL-12p70) when coadministered with Lp cells, compared to the results obtained with the bacterium alone. Moreover, GG, but not L919/A, stimulated the production of TNF-α and IL-10 by THP-1 cells and affected the immunomodulation of Lp and Nissle cells, suggesting that the difference in molecular weight between these two PS appeared to have some functional effect. It is likely that subtle differences in the structures and action modes of Gram-positive (Lp) and Gram-negative (Nissle, LF82) bacteria could explain some of the differences in the immune responses obtained in the presence of the L. casei LOCK 0919 PS.
Few studies have examined whether given PS act directly or indirectly on the immune system, and/or if their triggered immunomodulations are transient or permanent. Furthermore, the potential immunomodulatory activity of a specific bacterial compound can be masked by various structures (e.g. the cell wall) or influenced by additional bioactive substances. Our observation, that PS antigens triggered the synergistic induction of IL-10 when coadministered with Lp cells, is consistent with a previous report by Kaji et al. (2010), who demonstrated that cell-wall teichoic acid and lipoteichoic acid induced macrophages to produce IL-10 only weakly when applied alone, but triggered a strong induction when applied in the presence of L. casei strain Shirota. In another study, Kim et al. (2011) found that lipoteichoic acid isolated from Lactobacillus planatarum K8 could attenuate the proinflammatory signaling induced by the peptidoglycan from Shigella flexneri KCTC 2517 in THP-1 cells. Recently, Gao et al. (2015) found that the PS from L. rhamnosus GG reduced the mRNA levels of inflammatory cytokines in LPS-induced cells by inhibiting p38MAPK and NF-κB signaling, and further indicated that PS exerts immunomodulatory effects in intestinal epithelial cells to alleviate inflammation. Moreover, Lebeer et al. (2011) reported that the PS of L. rhamnosus GG forms a protective shield against innate immune factors in the intestine. If we understood the developmental windows during which the host is most responsive to PS, we could more rationally apply beneficial microbes or their compounds to treat diseases during such periods. A better understanding of the action mechanisms of these bacterial compounds could help us define specific and efficient therapeutics, perhaps at an individual and customized level.
Some Lactobacillus strains have been shown to stimulate the production of secretory antibodies such as immunoglobulin A and immunoglobulin G (IgG). For instance, Maassen et al. (2003) described that oral administration of individual Lactobacillus strains could differentially affect IgG1 versus IgG2a depending on the phase of bacterial growth. Gill et al. (2000) reported that probiotic strains (e.g. L. rhamnosus HN001) potentiated the antibody response to both systemically and orally administered T-dependent antigens in mice, and identified increases in the specific antibody titers of both serum and intestinal tract secretions. Here, we tested the immunoreactivity of purified PS with human sera, and found that the physiological sera of healthy adult humans contain relatively high titers of antibodies that reacted to the PS. The reactivity of PS L919/A with adult human sera could reflect its structural similarity to GG, whose source (L. rhamnosus GG) is a main probiotic strain widely used in the food and pharmaceutical industries. Reactive antibodies were also present in human umbilical cord blood sera. L919/A exhibited higher reactivity than L919/B with umbilical cord blood sera, whereas L919/B exhibited higher reactivity than L919/A with adult human sera, suggesting that the reactivity reflects the chemical properties of a given PS.
The continued study of immunomodulatory molecules is needed to increase our understanding of the complex nature of bacteria–bacteria and bacteria–host interactions. Our knowledge of PS-elicited immune changes is improving, but further experiments are needed if we hope to develop better probiotic-derived therapeutics aimed at preventing and attenuating human disorders. To draw general conclusions about the molecular mechanisms of probiotic effects, we need to conduct comprehensive immunochemical characterizations of bacterial compounds in terms of both their structures and molecular masses. PS from probiotic bacteria could potentially be used to influence the immunomodulatory properties of other bacteria, such as by enhancing their beneficial effects. Moreover, the administration of molecules with well-defined structures instead of whole bacteria could reduce the potential risks of sepsis and bacteremia that currently accompany the administration of probiotics.
Materials and methods
Microorganisms and growth conditions
Lactobacillus casei LOCK 0919 (formerly Lactobacillus paracasei LOCK919; patent no. 209986) was originally isolated from a fecal sample collected from an healthy 5-year-old boy (Aleksandrzak-Piekarczyk et al. 2013). For this work, cells were obtained from the Pure Culture Collection of Technical University, Lodz, Poland. Lactobacillus plantarum WCFS1 is a single-colony isolated from L. plantarum NCIMB8826, originally isolated from human saliva (National Collection of Industrial and Marine Bacteria, Aberdeen, United Kingdom) (Hayward and Davis 1956). Lactobacillus rhamnosus GG was obtained from the ATCC (Cat. #53103; American Type Culture Collection). Lactobacillus strains were stored at −75°C in MRS broth (Difco) supplemented with 20% glycerol; prior to use, they were subcultured twice in MRS broth (Biocorp, Poland) under anaerobic conditions at 37°C for 48 h. Escherichia coli Nissle 1917 (DMS 6601, serotype O6:K5:H1, Nissle; Mutaflor) and Crohn's disease-associated E. coli LF82 (LF82; serotype O83:H1, was obtained from Institute of Microbiology, Academy of Sciences of the Czech Republic, Czech Republic) were cultivated overnight in LB broth (Fluka Biochemika, Switzerland) at 37°C. Prior to inactivation, cells were cultivated on a shaker for 4 h at 37°C under aerobic conditions, to the end of the exponential growth phase. For in vitro experiments, bacteria were inactivated in 1% formaldehyde in phosphate-buffered saline (PBS) for 3 h at room temperature, washed twice with sterile PBS and stored at −20°C.
Isolation of PS antigens
The PS antigens of L. casei LOCK 0919 were isolated as previously described (Górska-Frączek et al. 2013). Briefly, cells were harvested by centrifugation at 5000 × g (4°C, 20 min), washed twice with PBS, washed once with MilliQ water and lyophilized. The freeze-dried bacterial mass was extracted with 50 ml of 10% trichloroacetic acid (25°C, 2.5 h) and centrifuged at 13,000 × g for 20 min. The pellet was discarded and the supernatant was collected. PS antigens were precipitated with five volumes of cold 96% ethanol (4°C, overnight), the mixture was centrifuged and the pellet was suspended in water. The recovered PS antigens were dialyzed for 48 h against water, and then lyophilized.
PS purification
The freeze-dried crude PS antigen preparations (20 mg) were dissolved in 1 ml of buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2), treated with DNase (210 µg, Sigma) and RNase (210 µg, Sigma) at 37°C for 6 h, and then treated with Streptomyces griseus protease (447 µg, 37°C, overnight; Sigma). The PS were dialyzed against water at 4°C for 24 h, and then purified by ion exchange chromatography on a diethylaminoethyl-Sephadex A-25 column (1.6 × 20 cm; Pharmacia). The neutral fractions were eluted with 20 mM Tris buffer, pH 8.2. The charged fractions were eluted with a NaCl gradient (0–2 M in 20 mM Tris buffer, pH 8.2) at a flow rate of 0.5 ml/min. All the elutions were monitored at 220 nm using an UV–VIS absorbance detector (contaminant detection; Amersham Pharmacia Biotech) and a differential refractometer (Knauer, Poland). The carbohydrate content was analyzed by the phenol/sulfuric acid method (Dubois et al. 1956). The phosphate content was determined as previously described (Chen et al. 1956). The PS-containing fractions were pooled, desalted by dialysis against water at 4°C for 24 h, and lyophilized. The PS were then further purified by gel permeation chromatography (GPC) on a Toyopearl HW-55S column (1.6 × 100 cm; Tosoh Bioscience, Belgium), fitted to an fast protein (or fast performance) liquid chromatography system (Amersham Pharmacia Biotech) and eluted with 0.1 M ammonium acetate buffer. The column effluents were monitored by absorbance at λ = 280 nm (protein contamination detection), with a differential refractometer (Knauer, Poland), and for the carbohydrate content, as described earlier. The purified PSs were subjected to the Limulus Amebocyte Lysate assay (PyroGene™ Recombinant Factor C Endotoxin Detection Assay; Lonza), which revealed that the endotoxin levels were below 0.1 EU per 1 µg of pure PS.
Analysis of sugars, methylation, molecular mass and the absolute configuration of monosaccharides
Each PS sample (0.5 mg) was hydrolyzed with 10 M HCl at 80°C for 25 min and subjected to evaporation under a stream of N2. The resulting monosaccharides were converted into alditol acetates, as described previously (Sawardeker et al. 1956). For methylation analysis, the PS samples were permethylated as previously described (Ciukanu and Kerek 1984), and the product was purified by water–chloroform extraction, hydrolyzed with 2 M trifluoroacetic acid at 120°C for 2 h and subjected to evaporation under N2. Finally, methylated monosaccharides were reduced with NaBD4 and acetylated for GLC-MS analysis. GLC was used to determine the absolute configurations of the acetylated (S)-2-octyl glycosides, as previously described (Gerwig et al. 1979). Alditol acetates and acetylated 2-butyl glycosides were analyzed by GLC-MS using a Thermo Focus ITQ 700 instrument (Thermo Scientific) equipped with a TR-5 ms glass capillary column (0.2 mm × 12 m; Thermo Scientific). The program was set to increase the temperature from 150 to 270°C at 12°C/min. The average molecular mass of each PS was determined by GPC (Dionex Ultimate 3000) on an OHpak SB-806M HQ column (8 × 300 mm, maximum pore size 15,000 Å; Shodex); the run was calibrated with dextran standards (Mw 12, 25, 50, 80, 150 and 270 kDa) and 0.1 M ammonium acetate buffer was used as the eluent. The flow rate was 0.5 ml/min and the column eluate was monitored with a refractive index detector (RI 102; Shodex). The working detector temperature and sensitivity were adjusted to 35°C and 512×, respectively. System control, data acquisition and treatments were performed using the Chromeleon software (Dionex).
NMR spectroscopy
The NMR spectra were obtained with a Bruker 600 MHz Avance III spectrometer, using a 5-mm QCI 1H/13C/15N/31P probe equipped with a z-gradient (Bruker). The NMR spectra were obtained for each 2H2O PS solution at 25°C, using acetone (δH 2.225, δC 31.05 ppm) as an internal reference. The PS (10 mg) was repeatedly exchanged with 2H2O (99.9%) via intermediate lyophilization. The data were acquired and processed using the Topspin software package (version 3.1; Bruker) and the SPARKY program (Goddard and Kneller 2001). The signals were assigned using 1D and 2D experiments, COSY, TOCSY, NOESY, HSQC with and without carbon decoupling, and HMBC. The TOCSY experiments were carried out with mixing times of 30, 60 and 100 ms, while NOESY was performed with mixing times of 100 and 300 ms. The 1H,13C HMBC spectrum was recorded with a 60-ms delay for the evolution of long-range spin couplings.
Preparation and activation of BM-DC
Bone marrow precursors were isolated from the femurs and tibias of conventional (CV) BALB/c mice. Cells were cultured at 4 × 105/ml in bacteriological Petri dishes in 10 ml RPMI-1640 complete supplemented with granulocyte macrophage colony-stimulating factor (20 ng/ml; eBioscience, United States of America). Fresh medium was added on days 3 and 6, and BM-DC were used on day 8 of culture. The purity of the obtained BM-DC, which was assessed by CD11c staining and flow cytometry, ranged from 69 to 75%. For activation BM-DC were harvested, diluted to 2 × 106 cells/ml, and stimulated for 20 h with 10 µg/ml of PS antigens, 107 CFU/ml formalin-inactivated Lp, Nissle or LF82 cells or mixtures of each PS and each cell type. As controls, BM-DC were incubated with PAM3 (Pam3CSK4, 1 µg/ml; InvivoGen, San Diego, United States of America), or ultrapure LPS from E. coli (LPS, 1 µg/ml; InvivoGen). The levels of IL-10 and IL-12p70 in culture supernatants were determined using ELISA Ready-Set-Go! kits (eBioscience) according to the manufacturer's instructions. These experiments were approved by the 1st Local Committee for Experiments with the Use of Laboratory Animals, Wroclaw, Poland (no. 38/2012).
Flow cytometry
BM-DC were labeled with monoclonal antibodies against CD11c (fluorescein isothiocyanate), MHCII (APC), CD40, CD80 or CD86 (PE) (eBioscience). Appropriate isotype antibodies were used as controls to determine nonspecific binding. Cells were analyzed using a FACSCalibur flow cytometer (Becton-Dickinson) and the obtained data were analyzed with the FlowJo 7.6.2 software (TreeStar).
Stimulation of HEK 293 cells stably transfected with vectors encoding TLRs and NOD2
HEK293 cells stably transfected with plasmids encoding human (h)TLR2/CD14 were kindly provided by M. Yazdanbakhsh (Leiden, Netherlands), those stably expressing hTLR4/MD2/CD14 were a gift from B. Bohle (Vienna, Austria) and those stably expressing hNOD2 were purchased from InvivoGen. Cells were stimulated for 20 h with PS antigens (10 µg/ml), 107 CFU/ml of formalin-inactivated Lp, Nissle or LF82 cells or mixtures of each PS and cell type. The TLR2 ligand, PAM3 (Pam3CSK4, 1 µg/ml; InvivoGen), the TLR4 ligand, ultrapure LPS from E. coli (LPS, 1 µg/ml; InvivoGen) and the NOD2 ligand, MDP (muramyl-dipeptide, 1 µg/ml; InvivoGen) were used as positive controls. Culture supernatants were assessed for their levels of human IL-8 using an ELISA kit (Thermo Scientific, United States of America) according to the manufacturer's instructions.
Stimulation of THP-1 cells
THP-1 cells (a human acute monocytic leukemia cell line) were cultured in RPMI-1640 supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM l-glutamine (all from Sigma–Aldrich) at 37°C with 5% CO2. The cells were seeded to 48-well tissue culture plates (2 × 106/ml in a volume of 250 µl per well) and incubated for 18 h with each PS antigen (10 µg/ml), 107 CFU/ml of formalin-inactivated Lp, Nissle or LF82 cells or mixtures of each PS and cell type. PAM3 (Pam3CSK4, 1 µg/ml; InvivoGen) and LPS from E. coli (LPS, 1 µg/ml; InvivoGen) were used as controls. Culture supernatants were measured for their levels of human tumor necrosis factor alpha (TNF-α) and IL-10 using ELISA Ready-Set-Go! kits (eBioscience) according to the manufacturer's instructions.
Human antisera
Serum samples were obtained from healthy adult volunteer blood donors (n = 12), and human umbilical cord sera were obtained from healthy women (n = 12) at the Obstetric Clinic of the Medical University of Wroclaw. Patients provided written informed consent. The use of umbilical cord serum samples was approved by the Medical Ethics Committee of the Medical University of Wroclaw (number KB-882-2012), and was conducted in accordance with the Helsinki Declaration of 1975.
Enzyme-linked immunosorbent assay
ELISA experiments were performed with PS L919/A and L919/B. Flat-bottom 96-well MaxiSorp plates (Nunc) were coated overnight at 4°C with 100 µl of a 0.01 mg/ml PS solution and stored overnight at 4°C. The plates were washed three times with PBS-T (PBS + 0.05% Tween 20) and blocked at 25°C for 1 h with 0.2% casein in PBS (w/v). The plates were again washed three times with PBS-T, and then incubated with 100 µl of human serum (geometric dilutions from 1:400 to 1:6400 in PBS) at 37°C for 2 h. The plates were subjected to five washes, and then incubated at 25°C for 1 h with secondary horseradish peroxidase-conjugated goat antihuman IgG (1:10,000). After a further five washes with PBS-T, the plates were treated with 100 µl/well of 3,3′,5,5′-tetramethylbenzidine (Sigma) substrate and incubated for 30 min at 25°C. Each reaction was stopped with 50 µl of 2 M H2SO4, and the plates were read at 450 and 570 nm on a Biotek instrument (Tekane).
Statistical analysis
Data are expressed as means ± SEM. Statistical analysis was performed by one-way analysis of variance followed by Sidak's multiple comparison test, as applied using the GraphPad software (GraphPad Prism 5.04; San Diego, CA); p values < 0.05 were considered significant.
Acknowledgements
We would like to thank Dr Scott Butler from English Manager Science Editing for proof reading of the manuscript.
Funding
National Science Centre of Poland (UMO-2012/05/D/NZ7/02494).
Conflict of interest statement
None declared.
Abbreviations
BM-DC, bone marrow-derived dendritic cells; COSY, COrrelation SpectroscopY; GLC-MS, gas–liquid chromatography-mass spectrometry; GPC, gel permeation chromatography; HEK293, human embryonic kidney cell line 293; HMBC, 1H-detected Heteronuclear Multiple-Bond Correlation spectroscopy; HSQC, 1H-detected Heteronuclear Single Quantum Coherence Spectroscopy; IgG, immunoglobulin G; IL, interleukin; LAB, lactic acid bacteria; LF82, Crohn's disease-associated Escherichia coli LF82; Lp, Lactobacillus plantarum WCFS1; LPS, lipopolysaccharide; Nissle, Escherichia coli strain Nissle 1917; NOESY, Nuclear Overhauser Effect SpectroscopY; PBS, phosphate-buffered saline; PS, polysaccharide; THP-1, a human monocytic cell line derived from an acute monocytic leukemia; TLR, Toll-like receptor; TOCSY, TOtal Correlation SpectroscopY.
References
- Ai L, Zhang H, Guo B, Chen W, Wu Z, Wu Y.. 2008. Preparation, partial characterization and bioactivity of exopolysaccharides from Lactobacillus casei LC2W. Carbohydr Polym. 74:353–357. [DOI] [PubMed] [Google Scholar]
- Aleksandrzak-Piekarczyk T, Koryszewska-Bagińska A, Bardowski J.. 2013. Complete genome sequence of the probiotic strain Lactobacillus casei (formerly Lactobacillus paracasei) LOCK919. Genome Announc. doi:10.1128/genomeA.00758–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock K, Pedersen C.. 1983. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv Carbohydr Chem Biochem. 41:27–66. [Google Scholar]
- Chen PS, Toribara TV, Warner H.. 1956. Microdetermination of phosphorus. Anal Chem. 28:1756–1758. [Google Scholar]
- Ciszek-Lenda M, Strus M, Górska-Frączek S, Targosz-Korecka M, Śróttek M, Heczko PB, Gamian A, Szymoński M, Marcinkiewicz J.. 2011. Strain specific immunostimulatory potential of lactobacilli-derived exopolysaccharides. Cent Eur J Immunol. 36:121–129. [Google Scholar]
- Ciukanu I, Kerek FA.. 1984. Simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 131:209–217. [Google Scholar]
- Dertli E, Mayer MJ, Narbad A.. 2015. Impact of the exopolysaccharide layer on biofilms, adhesion and resistance to stress in Lactobacillus johnsonii FI9785. BMC Microbiol. 15:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Giller KA, Rebers PA, Smith FA.. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem. 28:350–356. [Google Scholar]
- Fanning S, Hall LJ, van Sinderen D.. 2012. Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection. Gut Microbes. 3:420–425. [DOI] [PubMed] [Google Scholar]
- FAO/WHO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. www.fao.org.
- Gao K, Wang C, Liu L, Dou X, Liu J, Yuan L, Zhang W, Wang H.. 2015. Immunomodulation and signaling mechanism of Lactobacillus rhamnosus GG and its components on porcine intestinal epithelial cells stimulated by lipopolysaccharide. J Microbiol Immunol Infect. doi:10.1016/j.jmii.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Gerwig GJ, Kamerling JP, Vliegenthart JFG.. 1979. Determination of the absolute configuration of monosaccharides in complex carbohydrates by capillary G.L.C. Carbohydr Res. 77:10–17. [DOI] [PubMed] [Google Scholar]
- Gill HS, Rutherfurd KJ, Prasad J, Gopal PK.. 2000. Enhancement of natural and acquired immunity by Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019). Br J Nutr. 83:167–176. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG.. 2001. SPARKY. 3rd ed. San Francisco, CA: University of California. [Google Scholar]
- Gorin PAJ, Mazurek M.. 1975. Further studies on the assignment of signals in 13C magnetic resonance spectra of aldoses and derived methyl glycosides. Can J Chem. 53:1212–1223. [Google Scholar]
- Górska S, Grycko P, Rybka J, Gamian A.. 2007. Exopolysaccharides of lactic acid bacteria: Structure and biosynthesis. Postepy Hig Med Dosw. 61:805–818. [PubMed] [Google Scholar]
- Górska S, Jarząb A, Gamian A.. 2009. Probiotic bacteria in the human gastrointestinal tract as a factor stimulating the immune system. Postepy Hig Med Dosw. 63:653–667. [PubMed] [Google Scholar]
- Górska S, Schwarzer M, Jachymek W, Srutkova D, Brzozowska E, Kozakova H, Gamian A.. 2014. Distinct immunomodulation of bone marrow-derived dendritic cell responses to Lactobacillus plantarum WCFS1 by two different polysaccharides isolated from Lactobacillus rhamnosus LOCK 0900. Appl Environ Microbiol. 80:6506–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska-Frączek S, Sandström C, Kenne L, Paściak M, Brzozowska E. Strus M, Heczko PB, Gamian A.. 2013. The structure and immunoreactivity of exopolysaccharide isolated from Lactobacillus johnsonii strain 151. Carbohydr Res. 378:148–53. [DOI] [PubMed] [Google Scholar]
- Górska-Frączek S, Sandström C, Kenne L, Rybka J, Strus M, Heczko PB, Gamian A.. 2011. Structural studies of the exopolysaccharide consisting of a nonasaccharide repeating unit isolated from Lactobacillus rhamnosus KL37B. Carbohydr Res. 346:2926–2932. [DOI] [PubMed] [Google Scholar]
- Hayward AC, Davis GHG.. 1956. The isolation and classification of Lactobacillus strains from Italian saliva. Br Dent J. 101:43–46. [Google Scholar]
- Jansson PE, Kenne L, Widmalm G.. 1989. Computer-assisted structural analysis of polysaccharides with an extended version of CASPER using 1H and 13C-NMR data. Carbohydr Res. 188:169–191. [DOI] [PubMed] [Google Scholar]
- Kaji R, Kiyoshima-Shibata J, Nagaoka M, Nanno M, Shida K.. 2010. Bacterial teichoic acids reverse predominant IL-12 production induced by certain lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol. 184:3505–3513. [DOI] [PubMed] [Google Scholar]
- Kant R, Blom J, Palva A, Siezen RJ, de Vos WM.. 2011. Comparative genomics of Lactobacillus. Microbial Biotechnol. 4:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenellenbogen E, Kocharova NA, Toukach PV, Górska S, Korzeniowska-Kowal A, Bogulska M, Gamian A, Knirel YA.. 2009. Structure of an abequose-containing O-polysaccharide from Citrobacter freundii O22 strain PCM 1555. Carbohydr Res. 344:1724–1728. [DOI] [PubMed] [Google Scholar]
- Kim HG, Lee SY, Kim NR, Lee HY, Ko MY, Jung BJ, Kim CM, Lee JM, Park JH, Han SH, et al. . 2011. Lactobacillus plantarum lipoteichoic acid down-regulated Shigella flexneri peptidoglycan-induced inflammation. Mol Immunol. 48:382–391. [DOI] [PubMed] [Google Scholar]
- Kozakova H, Schwarzer M, Tuckova L, Srutkova D, Czarnowska E, Rosiak I, Hudcovic T, Schabussova I, Hermanova P, Zakostelska Z, et al. . 2015. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell Mol Immunol. doi:10.1038/cmi.2015.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landersjö C, Yang Z, Huttunen E, Widmalm G.. 2002. Structural studies of the exopolysaccharide produced by Lactobacillus rhamnosus strain GG (ATCC 53103. Biomacromolecules. 3:880–884. [DOI] [PubMed] [Google Scholar]
- Lipkind GM, Shashkov AS, Knirel YA, Vinogradov EV, Kochetkov NK.. 1988. A computer-assisted structural analysis of regular polysaccharides on the basis of carbon-13 NMR data. Carbohydr Res. 175:59–75. [DOI] [PubMed] [Google Scholar]
- Lebeer S, Verhoeven TL, Francius G, Schoofs G, Lambrichts I, Dufrêne Y, Vanderleyden J, De Keersmaecker SC.. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol. 75:3554–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S, Claes IIJ, Verhoeven TLA, Vanderleyden J, De Keersmaecker SCJ.. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microbial Biotechnol. 4:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyer MD, Buurman WA, Hadfoune M, Speelmans G, Knol J, Jacobs JA, Dejong CHC, Vriesema AJM, Greve JWM.. 2005. Strain-specific effects of probiotics on gut barrier integrity following hemorrhagic shock. Infect Immun. 73:3686–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maassen CBM, Boersma WJA, van Holten-Neelen C, Claassen E, Laman JD.. 2003. Growth phase of orally administered Lactobacillus strains differentially affects IgG1/IgG2a ratio for soluble antigens: Implications for vaccine development. Vaccine. 21:2751–2757. [DOI] [PubMed] [Google Scholar]
- Marshall VM, Rawson HL.. 1999. Effects of exopolysaccharide - Producing strains of thermophilic lactic acid bacteria on the texture of stirred yoghurt. Int J. Food Sci Technol. 34:137–143. [Google Scholar]
- Mattu B, Chauhan A.. 2013. Lactic acid bacteria and its use in probiotics. J Bioremed Biodeg. doi:10.4172/2155-6199.1000e140 [Google Scholar]
- McFarland LV. 2006. Meta‑analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol. 101:812–822. [DOI] [PubMed] [Google Scholar]
- Sawardeker JS, Sloneker JH, Jeanes A.. 1956. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal Chem. 37:1602–1603. [Google Scholar]
- Segers ME, Lebeer S.. 2014. Towards a better understanding of Lactobacillus rhamnosus GG - Host interactions. Microb Cell Fact. doi:10.1186/1475-2859-13-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Okawa Y.. 2011. Chemical structure of β-galactofuranose-containing polysaccharide and O-linked oligosaccharides obtained from the cell wall of pathogenic dematiaceous fungus Fonsecaea pedrosoi. Glycobiology. 21:69–81. [DOI] [PubMed] [Google Scholar]
- Shida K, Kiyoshima-Shibata J, Nagaoka M, Watanabe K, Nanno MJ.. 2006. Induction of interleukin-12 by Lactobacillus strains having a rigid cell wall resistant to intracellular digestion. J Dairy Sci. 89:3306–3317. [DOI] [PubMed] [Google Scholar]
- Taverniti V, Guglielmetti S.. 2011. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes Nutr. 6:261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursi A, Brandimarte G, Papa A, Giglio A, Elisei W, Giorgetti GM, Forti G, Morini S, Hassan C, Pistoia MA, et al. . 2010. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: A double-blind, randomized, placebo-controlled study. Am J Gastroenterol. 105:2218–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JM. 2011. Immunomodulatory mechanisms of lactobacilli. Microb Cell Fact. doi:10.1186/1475-2859-10-S1-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S, Halbert A, Richmond P, Prescott SL.. 2005. Effects of probiotics on atopic dermatitis: A randomised controlled trial. Arch Dis Child. 90:892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu C, Li D, Zhao Y, Zhang X, Zeng X, Yang Z, Li S.. 2013. Antioxidant activity of an exopolysaccharide isolated from Lactobacillus plantarum C88. Int J Biol Macromol. 54:270–275. [DOI] [PubMed] [Google Scholar]