Abstract
The minimum information required for a glycomics experiment (MIRAGE) project was established in 2011 to provide guidelines to aid in data reporting from all types of experiments in glycomics research including mass spectrometry (MS), liquid chromatography, glycan arrays, data handling and sample preparation. MIRAGE is a concerted effort of the wider glycomics community that considers the adaptation of reporting guidelines as an important step towards critical evaluation and dissemination of datasets as well as broadening of experimental techniques worldwide. The MIRAGE Commission published reporting guidelines for MS data and here we outline guidelines for sample preparation. The sample preparation guidelines include all aspects of sample generation, purification and modification from biological and/or synthetic carbohydrate material. The application of MIRAGE sample preparation guidelines will lead to improved recording of experimental protocols and reporting of understandable and reproducible glycomics datasets.
Keywords: glycobiology, glycomics, MIRAGE, sample preparation
Introduction
The minimum information required for a glycomics experiment (MIRAGE) initiative follows in the footsteps of similar reporting guidelines projects, namely the minimum information about a proteomics experiment (MIAPE, proteomics) (Taylor et al. 2007), standards for reporting enzymology data (STRENDA, enzymology) (Apweiler et al. 2005) and minimum information about a microarray experiment (MIAME, microarrays) (Brazma et al. 2001). The overarching goal of MIRAGE is to provide reporting guidelines as a tool to help facilitate clear and concise communication of glycomics datasets and experimental methods among peer reviewed journals. The concept and organization of MIRAGE were introduced in 2014 (York et al. 2014). The MIRAGE Commission consists of “working groups” in which active glycomics researchers from various areas work jointly to design guidelines encompassing mass spectrometry (MS), glycan array, capillary electrophoresis (CE), liquid chromatography (LC) and sample preparation. The MIRAGE Advisory Board, comprised of international leaders in glycoscience, are entrusted with reviewing and endorsing the guidelines prior to dissemination to the wider field. In this way MIRAGE guidelines are based on a collective agreement and represent information considered “required” to be included in published reports containing glycomics data. The MS guidelines were published in 2013 (Kolarich et al. 2013) and currently LC and glycan array guidelines are being drafted.
Glycoconjugates are inherently complex and heterogeneous biomolecules that require sophisticated analytical methods and software for interpreting and storing annotated data. By comparison, proteomics has enjoyed longstanding development of tools and software while glycomics, being a relatively young field, has not experienced the same level of development to date. Paradoxically this offers the glycobiology community greater freedom to shape the future of the field and even learn from past proteomics experiences. This outlook is exemplified by the journals Glycobiology (who recommend) and Molecular and Cellular Proteomics (MCP, who have adopted) reporting checklists to ensure completeness and accessibility of submitted glycomics data prior to peer review (Wells et al. 2013). The MCP instructions, termed “Athens Guidelines”, are complementary to the MIRAGE initiative and the developers of these guidelines are also members of the MIRAGE Commission. MIRAGE reporting guidelines will help alleviate inconsistent reporting, ensure accountable reporting of data and metadata and also help newcomers to the field in understanding the reported results. The intention of MIRAGE is to provide guidelines, not rules, to assist authors, reviewers and editors in assessing glycomics data. The guidelines serve as a framework to provide data and information as agreed by the glycobiology community to ensure reporting of understandable and reproducible scientific results.
Effective guidelines for reporting glycan structure analyses (e.g. MS, CE, LC) and glycan interaction studies (e.g. glycan arrays) require a complete description of glycan sample preparation. The starting point for both analytical and interaction studies is preparation of the glycan sample. Common examples of experimental steps may include tryptic digest of a glycoprotein, glycan release using PNGase F, glycan permethylation and purification using LC. Sample preparation steps vary and these variations can affect the carbohydrate material itself. This is particularly important for glycoconjugate types (i.e. glycolipids, glycoproteins, free oligosaccharides) and among prokaryotic/eukaryotic samples which may require complex preparation steps. Therefore a thorough report of sample preparation is fundamental to appreciate subsequent experiments. The MIRAGE sample preparation guidelines presented in this article aim to provide the metadata on these preparation steps necessary to allow readers and reviewers to understand and reproduce the presented results. In addition, these guidelines are considered as a common basis for other MIRAGE reporting guidelines in order to keep the publication requirements for data analysis short and consistent.
Sample preparation guidelines—general principles
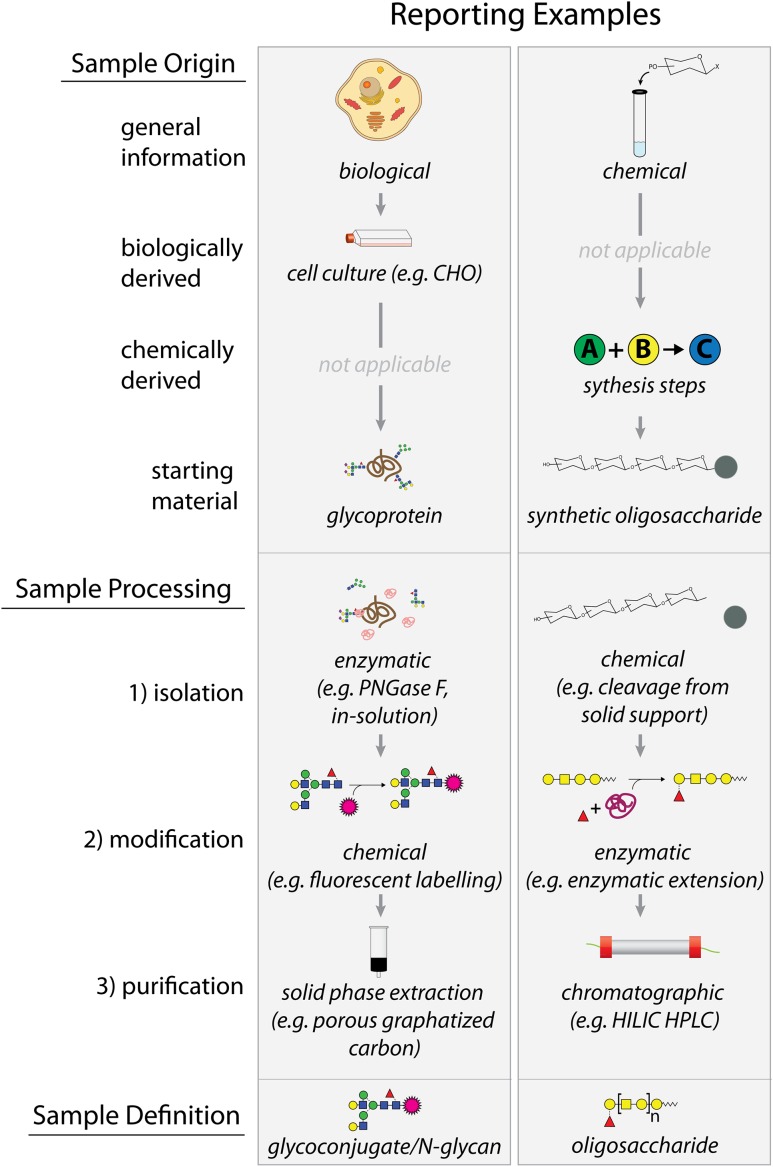
The MIRAGE sample preparation guidelines (Table I) have been designed for broad usability and application to a wide-range of oligosaccharide/glycoconjugate types produced from biological or synthetic sources. The guidelines are separated into three categories: (1) sample origin, (2) sample processing and (3) sample definition. Guidelines should be used accordingly to fit individual research experiments and therefore reporting in a given manuscript may or may not be linear as presented here. The sample origin requirements should fully explain where and how the starting material (which contains the oligosaccharide material of interest) was obtained. The reagents and methods by which starting material is produced can influence glycosylation and therefore should be reported. Within this category, general information requirements are broad but may be conceptually divided into biologically or synthetically derived material, as exemplified in Figure 1.
Table I.
Sample Preparation Guidelines descriptors
| Classification | Guidelines |
|---|---|
| 1. Sample Origin | |
| Here “sample” is defined as any carbohydrate-containing material; polysaccharide, oligosaccharide or glycoconjugate that originates from any given starting material. The starting material may be a compound, mixture or cell product used to produce the oligosaccharide sample of interest. The source and/or methods used to produce the starting sample material can vary considerably but minimum information that describes its origin is outlined. | |
| General information | Describe how original starting sample material was generated or where it was obtained. Descriptions may include but are not limited to material produced in the laboratory (e.g. chemically, enzymatically or chemoenzymatically synthesized), acquired from natural sources (e.g. human tissues) or purchased from a commercial manufacturer. In the case of commercial material, provide vendor and applicable item information. Starting material descriptions are further delineated by biologically or chemically derived material. In all cases reagents and methods used to generate starting material should be reported. |
| Biologically derived material |
|
| Chemically derived material | Describe how material was generated. If samples were synthetically derived, provide information detailing synthesis steps or specify where the equivalent reaction protocol is available. |
| Description of starting material | Define the type of starting material used or produced that contains the oligosaccharide to be used/analysed in subsequent experiments. These may include glycoprotein(s), proteoglycan, glycolipid, GPI-anchored, free-oligosaccharides, sugar-nucleotides or synthetically derived material but are not limited to these definitions. |
| 2. Sample Processing | |
| The sample processing is divided into 1) isolation, 2) modification and 3) purification subcategories. Required information for each is described below. | |
| 2.1 Sample Processing - Isolation | |
| Processing may include methods to remove the oligosaccharide from the starting material prior to downstream experiments or conversely the starting material may also be altered so the oligosaccharide remains conjugated to non-carbohydrate material such as chemical (e.g. linker) or biological (e.g. peptides) components. | |
| Enzymatic treatments | Describe any enzymes used to for the purpose of oligosaccharide removal (e.g. PNGase F) or for modification of the starting material (e.g. trypsin protease). Specify where it was obtained (vendor) or for enzymes produced in-house, describe expression and purification procedure. Also state if sample material was treated in-solution or immobilized (SDS-PAGE, PVDF etc.) as well as temperature, duration, volume, enzyme concentration. |
| Chemical treatments | Define the technique for oligosaccharide release (hydrazinolysis, β- elimination etc.) or other chemical modifications. Provide details of reaction conditions (temperature, duration, volume and chemical concentrations). |
| 2.2 Sample Processing - Modification | |
| Isolated material may be subject to a wide variety of chemical or enzymatic modifications. The type of modification and reaction protocols should be documented. If new protocols are used provide a thorough description. | |
| Chemical modifications | Describe any treatments made to the isolated material. Explain the type of modification employed (e.g. hydrolysis, sample tagging (including fluorescent labels), isotopic labelling, permethylation/peracetylation, etc.). Include source of materials, description of kits used, reaction conditions and detailed workflow. |
| Enzymatic modifications | Document enzyme concentration, supplier, biological source, incubation time and temperature. If novel glycosidase was used, provide information indicating the origin (i.e. species) of the enzyme. |
| 2.3 Sample Processing - Purification | |
| The processing steps encompass any type of refining or clean-up strategies used to produce the purified final sample product. | |
| Purification step(s) | Specify all steps used to purify starting material after isolation/modification steps. Examples of procedures include solid phase extraction (SPE), liquid-liquid extraction or other chromatographic methods. For each method describe the all experimental materials (e.g. stationary phase) and methods (e.g. flow rates, fractionation etc.). |
| 3. Sample Definition | |
| Provide a designation for the final sample in accordance with conventional terminology. Resources for the symbol nomenclature for graphical representation of glycans (SNFG) are available (Varki, A., Cummings, R.D., et al. 2015). | |
| Sample name | Name or specify the type of sample material to be analysed or used in other experiments. These may include but are not limited to glycoconjugates, glycosaminoglycans, N- or O-glycans, glycopeptides, glycolipids, monosaccharides, poly- and oligosaccharides |
Fig. 1.
Examples of MIRAGE Sample Preparation Reporting Guidelines. Two types of starting “sample material” are shown: (1) a recombinant glycoprotein produced from mammalian cell expression (biological) and (2) a synthetic oligosaccharide derived from chemical production (chemical). These Sample Origin descriptors provide background information for the sample starting material prior to further preparation. Sample Processing guidelines help explain how the starting material (e.g. glycoprotein and synthetic oligosaccharide) was treated. These include all purification and modification steps used to produce a final sample that is to be defined by the researcher (Sample Definition).
Sample processing steps provide essential information on how the starting material was prepared to create the sample that is used for further analysis. These sample processing steps include (1) isolation, (2) modification and (3) purification which can be iterated in any way. Isolation is defined as treatments to the “starting material” which yields an unmodified, non-purified sample. These may include glycan/oligosaccharide release, protease treatments or any technique aimed to produce an oligosaccharide sample for further processing. Secondly, modification describes any further alteration(s) made to the sample and is not restricted to a single action. Thirdly, purification includes the steps employed to produce the final refined sample material. Sample preparation may entail multiple and/or various processing steps and therefore the sample processing guidelines should be used accordingly to reflect all experimental procedures. For example, a given glycoprotein (sample material) may be treated with a protease, most often trypsin (isolation of glycopeptides) prior to PNGase F release of N-glycans (isolation of N-glycans). Furthermore, the released N-glycans may be fluorescently labeled (chemical modification) and then sequentially digested with a panel of exoglycosidases (enzymatic modification). Finally, the excess fluorescent dye may be removed by normal phase solid phase extraction (purification, normal phase SPE).
Ultimately, the sample preparation guidelines describe the step-by-step actions used to produce a final sample as defined by the researcher (sample definition). The type of oligosaccharide/glycoconjugate should be designated in accordance with existing and acceptable terminology (Lindhorst 2003; Varki et al. 2009) and not by ambiguous vocabulary. Additionally, the Symbol Nomenclature for Glycans system is recommended for illustrating glycan structures (Varki et al. 2015). This defined sample may or may not be used for additional downstream assays or analysis, but such reporting of subsequent experiments, such as MS, is recommended to be compliant with the specific MIRAGE guidelines for these experimental techniques.
Discussion
The goal, as with all MIRAGE reporting guidelines, is to outline and provide the community with a framework for reporting glycomics data. Here we present sample reporting guidelines that are considered essential information to be included in published reports as determined by the broader glycomics community. The implementation of reporting guidelines is not only welcomed by glycoscientists but is already recommended in two leading journals in the field and as such the establishment of MIRAGE guidelines is especially timely. In due course, the use of MIRAGE guidelines will establish transparency and reproducibility in communicating glycomics research worldwide and in so doing advance the field as a whole.
Availability
This manuscript describes the sample preparation guidelines as of March 2016. The current versions of all MIRAGE guidelines and examples are available on the MIRAGE project web site: http://www.beilstein-institut.de/en/projects/mirage/guidelines#sample_preparations (http://dx.doi.org/10.3762/mirage.1).
Funding
We thank the Beilstein-Institut for funding the MIRAGE initiative. W.S. acknowledges a research grant from Against Breast Cancer [www.againstbreastcancer.org; UK Charity 1121258]. J.Z and C.C acknowledge funding from the National Institutes of Health [P41GM104603]. M.N. acknowledges funding from the National Institutes of Health [RO1GM024349]. The participation of T.F. and Y.L. is supported by the Wellcome Trust [Grant No. WT099197MA]. N.K. acknowledge support from the Swedish Foundation for International Cooperation in Research and Higher Education [STINT IB2015-5931] and the Swedish Research Council [621-2013-5895]. A.D. and S.M.H. acknowledge funding from the Biotechnology and Biological Sciences Research Council [Grant No. BB/K016164/1]. This work was supported in part by the NIGMS/NIH funded [P41GM103490] National Center for Biomedical Glycomics [R.R., W.S.Y. and L.W.].
Fig. 1:
Examples of MIRAGE Sample Preparation Reporting Guidelines. Two types of starting “sample material” are shown: (1) a recombinant glycoprotein produced from mammalian cell expression (biological) and (2) a sythetic oligosaccharide derived from chemical production (chemical). These sample origin descriptors provide background information for the sample starting material prior to further preparation. Sample processing guidelines help explain how the starting material (e.g. glycoprotein and synthetic oligosaccharide) was treated. These include all purification and modificaiton steps used to produce a final sample that is to be defined by the researcher (sample definition).
References
- Apweiler R, Cornish-Bowden A, Hofmeyr JH, Kettner C, Leyh TS, Schomburg D, Tipton K. 2005. The importance of uniformity in reporting protein-function data. Trends Biochem Sci. 30:11–12. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 29:365–371. [DOI] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R, et al. 2013. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting mass-spectrometry-based glycoanalytic data. Mol Cell Proteomics. 12:991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhorst TK. 2003. Essentials of Carbohydrate Chemistry and Biochemistry, 2nd ed.; Wiley-VCH: Weinheim, Germany.
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, et al. 2007. The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol. 25:887–893. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, et al. 2015. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi C, Hart G, Etzler M. 2009. Essentials of Glycobiology, 2nd ed. Cold Spring Harbor, N.Y, Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Wells L, Hart GW, Athens Guidelines for the Publication of Glycomics D . 2013. Glycomics: Building upon proteomics to advance glycosciences. Mol Cell Proteomics. 12:833–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Agravat S, Aoki-Kinoshita KF, McBride R, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson N, et al. 2014. MIRAGE: The minimum information required for a glycomics experiment. Glycobiology. 24:402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]