Abstract
The pregnane X receptor (PXR) plays an important and diverse role in mediating xenobiotic induction of drug-metabolizing enzymes and transporters. Several protein isoforms of PXR exist, and they have differential transcriptional activity upon target genes; transcript variants 3 (PXR3) and 4 (PXR4) do not induce target gene expression, whereas transcript variants 1 (PXR1) and 2 (PXR2) respond to agonist by activating target gene expression. PXR protein variants also display differences in protein–protein interactions; PXR1 interacts with p53, whereas PXR3 does not. Furthermore, the transcript variants of PXR that encode these protein isoforms are differentially regulated by methylation and deletions in the respective promoters of the variants, and their expression differs in various human cancers and also in cancerous tissue compared to adjacent normal tissues. PXR1 and PXR4 mRNA are downregulated by methylation in cancerous tissue and have divergent effects on cellular proliferation when ectopically overexpressed. Additional detailed and comparative mechanistic studies are required to predict the effect of PXR transcript variant expression on carcinogenesis, therapeutic response, and the development of toxicity.
Abbreviations: AF, activating function; BAMCA, bacterial artificial chromosome array–based methylated CpG island amplification; CYP, cytochrome P450; GST, glutathione S-transferase; MDR, multidrug resistance protein; NHR, nuclear hormone receptor; P-gp, P-glycoprotein; PXR1, PXR transcript variant 1 (434 residues); PXR2, transcript variant 2 (473 residues); PXR3, transcript variant 3 (397 residues); PXR4, transcript variant 4 (322 residues;AK122990); RACE, 5′ rapid amplification of cDNA ends; shRNA, short hairpin RNA; siRNA, small interfering RNA; UGT, UDP-glucuronosyltransferase; UTR, untranslated region
Key words: Pregnane X receptor, Transcript variants, Drug metabolism, Therapeutic responses, Toxicity
Graphical abstract
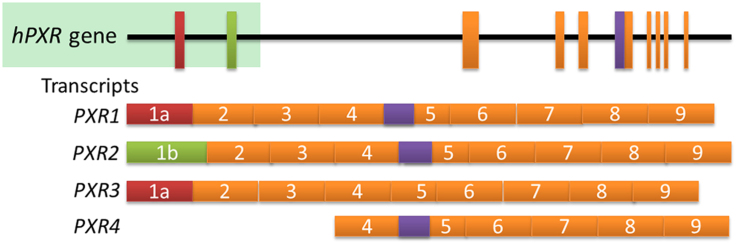
PXR induces drug-metabolizing enzymes and transporters, and has multiple transcript variants, such as PXR1, PXR2, PXR3, and PXR4, which respond to agonists, interact with other proteins, and affect cellular function differently. Further studies are required to predict the effect of PXR variants on carcinogenesis, therapeutic response, and toxicity.
1. Introduction
The pregnane X receptor (PXR), also known as NR1I2 (nuclear receptor subfamily 1, group I, member 2), SXR (steroid and xenobiotic sensing receptor), or PAR (pregnane activated receptor), regulates the expression of proteins involved in all three phases of drug metabolism and transport. PXR is a nuclear hormone receptor (NHR), one of a class of proteins characterized by a DNA-binding domain and activating function domains 1 and 2 (AF-1 and AF-2) that are relatively conserved in different species and, in PXR specifically, by a promiscuous ligand-binding domain (LBD)1, 2, 3, 4. NHRs bind to specific DNA sequences and bring the DNA molecule into the preferential steric conditions for transcription of target genes; in the case of PXR, these are predominantly genes involved in metabolizing xenobiotics. The LBD of PXR interacts with ligands to stabilize the protein (and recruit binding proteins, such as RXR, retinoic X receptor) and enable the recruitment of coactivators to the AF-2 region, resulting in further stabilization of the protein while in complex with DNA1, 2, 3, 4, 5, 6. PXR regulates the expression of phase I enzymes, including cytochrome P450 enzymes (CYPs) CYP3A4, CYP2B6, CYP2C9, and CYP2C19; phase II enzymes, including UDP-glucuronosyltransferase 1 family polypeptide A1 (UGT1A1), UGT1A2, and sulfotransferase 2 A (SULT2A); and phase III transporters, including ATP-binding cassette transporter ABCB1 (also known as MDR1 or P-gp), multiple organic anion transporters (OATs), and multidrug-resistance protein 3 (MRP3)2, 3, 4, 7, 8. PXR is mainly expressed in the liver and the intestines9. The recent identification of PXR as a potential therapeutic target in several diseases underscores the necessity of fully describing all variants of the receptor and their effect on physiologic and pathophysiologic processes10, 11, 12, 13, 14. PXR has been implicated in the pathophysiology of bone disease11, inflammatory disorders13, 14, and dyslipidemias10, in addition to its roles in hepatotoxicity and hepatic fibrosis15, 16, 17, 18. Because some PXR ligands are species-specific, a mouse model in which mouse PXR is replaced with human PXR (hPXR) enables examination of hPXR function in vivo19, 20, 21, 22; however, mouse models that are engineered to express only PXR variant 1 mRNA (PXR1) and protein fail to account for the multiple PXR transcripts. Therefore, a mouse model in which the entire hPXR gene is inserted into the mouse genome was created21, 23.
Ligand activation of PXR results in the induction of target genes by a broad range of structurally dissimilar xenobiotics, leading to the metabolism of an even greater range of compounds than those that directly activate xenobiotic metabolism via activation of PXR (Table 124, 25, 26, 27, 28, 29, 30, 31). Analgesics [NSAIDs (nonsteroidalanti- inflammatory drugs) and non-NSAIDs], protease inhibitors, antibacterials, anticonvulsants, glucocorticoids, and statins activate PXR (Table 1). PXR is also activated by a structurally and functionally diverse set of ligands implicated in a range of disease states. PXR target genes display considerable interindividual variation in expression profile and enzymatic activity32, 33, 34. Administration of xenobiotics to patients might cause adverse drug responses such as hepatotoxicity15, 17, 35, 36. Interindividual variability in drug response or nonresponse may result, in part, from variation in total PXR protein expression, single-nucleotide polymorphisms in coding or promoter regions of PXR, and variation in the relative expression of PXR isoforms9, 32. Interindividual variability in drug metabolism, therapeutic response, and the incidence and degree of dose-limiting toxicity may reflect, in part, variability in the expression levels of PXR transcript variants. There is considerable variability in PXR transcript expression in human liver and intestines; some patients express low levels of a common variant, whereas others express high levels of uncommon variants7, 9, 37, 38, 39. These variants display altered transactivation activity towards target genes7, 39. In addition, PXR1 and PXR2 have separate transcription start sites40.
Table 1.
Examples of hPXR agonists.
| Compound | Description (indication, drug class, etc.) | Reference |
|---|---|---|
| 12-Ketolithocholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| 3-Keto-7α,12α-dihydroxy-5α-cholanic acid | Bile salt found in sea lamprey | Krasowski et al. (2005)24 |
| 5α-Cyprinol 27-sulfate | Bile salt found in zebrafish | Krasowski et al. (2005)24 |
| 5β-Pregnane-3,20-dione | Steroid hormone | Jones et al. (2000)25 |
| 5β-Scymnol 27-sulfate | Bile salt found in cartilaginous fish | Krasowski et al. (2005)24 |
| 7,12-Diketolithocholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| 7-Ketodeoxycholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| 7-Ketolithocholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| Carbemazepine | Epilepsy | Luo et al. (2002)26 |
| Cholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| Clotrimazole | Anti-fungal | Xie et al. (2000)27; Luo et al. (2002)26; Xie et al. (2003)28; Jones et al. (2000)25 |
| Corticosterone | Glucocorticoid | Jones et al. (2000)25 |
| Cyproterone acetate | Antineoplastic (prostate); androgen disorders, steroidal anti-androgen | Jones et al. (2000)25 |
| Deoxycholic acid | Bile salt found in rabbit | Krasowski et al. (2005)24 |
| Dexamethasone | Glucocorticoid | Xie et al. (2000)27; Luo et al. (2002)26 |
| Dexamethasone-t-butyl acetate | Glucocorticoid | Synold et al. (2001)29; Luo et al. (2002)26 |
| Estradiol | Steroid hormone | Luo et al. (2002)26 |
| Glycolithocholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| Glycolithocholic acid 3-sulfate | Bile salt found in humans | Krasowski et al. (2005)24 |
| Hyodeoxycholic acid | Bile salt found in many mammals | Krasowski et al. (2005)24 |
| Hyperforin | St. John׳s Wort; herbal supplement commonly used for depression | Luo et al. (2002)26 |
| Lithocholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| Lithocholic acid 3-sulfate | Bile salt found in humans | Krasowski et al. (2005)24 |
| Lithocholic acid acetate | Bile salt found in humans | Krasowski et al. (2005)24 |
| Lithocholic acid acetate methyl ester | Bile salt found in humans | Krasowski et al. (2005)24 |
| Mifepristone (RU486) | Pregnancy termination, steroidal antiprogesterone | Xie et al. (2000)27; Jones et al. (2000)25 |
| Nonylphenol | Anthropogenic environmental estrogen | Mota et al. (2011)30 |
| Paclitaxel (Taxol) | Anti-neoplastic | Synold et al. (2001)29; Luo et al. (2002)26 |
| Petromyzonol 24-sulfate | Bile salt found in sea lamprey | Krasowski et al. (2005)24 |
| Phenobarbital | Epilepsy | Jones et al. (2000)25; Luo et al., (2002)26 |
| Pheytoin | Seizure disorders | Luo et al. (2002)26 |
| Piperine | Component of the spice, black pepper | Wang et al. (2013)31 |
| Pregnenolone | Steroid hormone | Jones et al. (2000)25 |
| Progesterone | Steroid hormone | Jones et al. (2000)25 |
| Rifampicin | A component of the first line anti-tuberculosis therapy | Jones et al. (2000)25; Synold et al. (2001)29; Xie et al. (2000)27; Xie et al. (2003)28; Luo et al. (2002)26 |
| Ritonavir | HIV, protease inhibitor | Luo et al. (2002)26 |
| Spironolactone | Diuretic, steroidal anti-mineralocorticoid | Jones et al. (2000)25 |
| SR12813 | Dyslipidemia, HMG-CoA inhibitor | Jones et al. (2000)25; Synold et al. (2001)29 |
| Sulfadimidine | Antibiotic, sulfonamide | Luo et al. (2002)26 |
| Sulfinpyrazone | Gout | Luo et al. (2002)26 |
| Taurochenodeoxycholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| Taurohyodeoxycholic acid | Bile salt found in many mammals | Krasowski et al. (2005)24 |
| Taurolithocholic acid | Bile salt found in humans | Krasowski et al. (2005)24 |
| Trans-nonachlor (Chlordane) | Pesticide | Jones et al. (2000)25 |
| Troglitazone | Diabetes (withdrawn), thiazolidinedione | Jones et al. (2000)25 |
| Troleandomycin | Antibiotic, macrolide | Luo et al. (2002)26 |
| α-Muricholic acid | Bile salt found in rat | Krasowski et al. (2005)24 |
2. PXR and chemotherapeutic metabolism
Distinct from its less investigated direct role in cellular proliferation and senescence, PXR can play seemingly dual roles in the development of resistance to chemotherapeutic agents. For example, after treatment with a PXR agonist, an inactive anticancer prodrug is metabolized to a greater degree to an active metabolite which may confer the anticancer chemotherapeutic activity41. Conversely, PXR activation may enhance the metabolism of the active forms of a drug into less active metabolites or excreted, with a resultant increase in resistance to chemotherapy28, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60. The role of PXR in chemotherapeutic metabolism has been elegantly reviewed by Zhuo61.
Cyclophosphamide and ifosfamide are prodrugs that are converted by CYP3A4 and CYP2B6 to active 4-hydroxy metabolites41. Indeed, the treatment with rifampicin and dexamethasone, both agonists of PXR, increases the metabolism of the prodrugs to their 4-hydroxy form to confer anticancer activity41. Irinotecan is a prodrug that is metabolized by carboxylesterase to its active form SN38 and is further metabolized by UGT1A1 and, to a lesser extent, CYP3A4 to an inactive form, SN38 glucuronide (SN38G)57, 58. PXR protein overexpression, and activation by rifampicin and SN38 itself, results in increased metabolism to inactive SN38G42, 43. Tamoxifen is metabolized to 4-hydroxytamoxifen, a more active metabolite, by CYP2D662. Conversely, tamoxifen is metabolized by CYP3A4 to N-desmethyltamoxifen, a metabolite with very low activity63. PXR protein overexpression and activation by rifampicin, SR12813, tamoxifen, and 4-hydroxytamoxifen lead to increased metabolism to N-desmethyltamoxifen and efflux which may result in increased resistance to chemotherapy48, 49, 60. Paclitaxel is metabolized by CYP2C8 and CYP3A4 to inactive metabolites64, 65. PXR knockdown by small interfering RNA (siRNA) or short hairpin RNA (shRNA) results in increased sensitivity to therapy51, 59, whereas the activation of PXR by rifampicin, paclitaxel, or SR12813 leads to increased metabolism and efflux of paclitaxel, resulting in increased resistance to chemotherapy44, 50, 51, 59. Doxorubicin is metabolized directly by the Cyp3a family or by carbonyl reductases to doxorubicinol and is further metabolized by Cyp3a enzymes to inactive aglycone metabolites66. Additionally, transcriptional activation of PXR by either overexpressing a constitutively active PXR protein or binding of rifampicin to the wild-type hPXR results in decreased sensitivity to doxorubicin39, 67. Vinblastine is metabolized by CYP3A4 and CYP3A5 to inactive metabolites68. Pretreatment with a PXR agonist decreases the sensitivity of cancer cells to vinblastine treatment, whereas siRNA knockdown of PXR increases their sensitivity to vinblastine51, 56. Together, these results clearly demonstrate the roles of PXR in regulating the efficacy of chemotherapeutic agents.
3. Structural, functional, and expression characteristics of PXR transcript variants
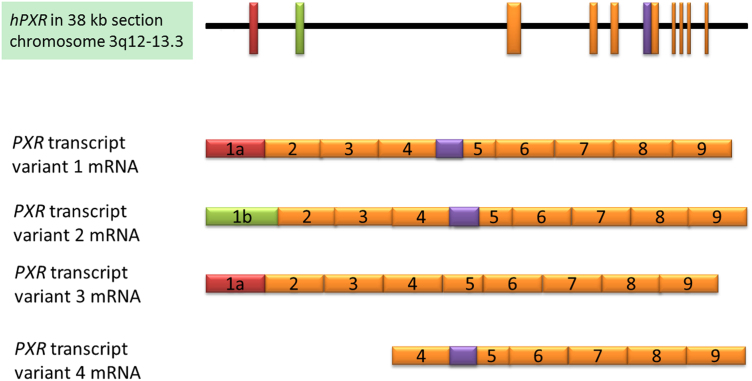
The PXR gene located on chromosome 3 encodes multiple transcript variants, resulting in structurally and functionally distinct proteins3, 7, 37, 38, 39, 69, 70. The identification of 9 splicing and transcript variants of PXR in human livers has led to the postulation that these variants, which may have different transactivation activity, contribute to interindividual variability in the expression of drug-metabolizing enzymes and efflux transporters, such as CYP3A4 and P-gp69. Recent work has focused on four of these mRNA transcript variants. The hPXR gene (NC_000003.12, Fig. 1) is located on chromosome 3q12-q13.3 and consists of approximately 38,000 base pairs69. PXR1 mRNA possesses the noncoding exon 1a of the hPXR gene and is transcribed into an mRNA of approximately 4400 nucleotides (NM_003889), which is then translated into a protein of 434 amino acids using a CTG start codon in exon 2 (NP_003880). PXR3 mRNA, a splicing variant of PXR1, originates from exon 1a of the hPXR gene, which contains a 111-base pair (bp) deletion at the 5′ end of exon 5, due to preferential usage of a cryptic splice acceptor site within exon 5, and is transcribed into an mRNA of approximately 4300 nucleotides (NM_033013), which is translated into a protein of 397 amino acids (NP_148934) with a deletion of 37 amino acids in the LBD of variant 137, 69. The 37-amino acid deletion in human PXR3 corresponds to a 41-amino acid deletion in the LBD of the corresponding mouse PXR variant3. PXR2 mRNA originates from the exon 1b of the hPXR gene and is transcribed into an mRNA of 2800 nucleotides (NM_022002), which is translated into a protein of 473 amino acids using a ATG start codon in exon 1b (NP_071285) that contains an additional 39 amino acids at the N-terminus, as compared to PXR169, 71, 72. PXR1 and PXR2 variants share exon 2 through exon 9 therefore they have identical LBD and DNA-binding domain. Many other PXR variants have been described, including PXR4, also known as short PXR (sPXR), a newly identified 37-kDa short PXR protein containing 322 amino acids that is translated from exon 4 to exon 9 (cDNA AK122990)72. The naming of different PXR transcript variants and their corresponding abbreviations, though mostly consistent, varies between authors; please refer to the “Abbreviations” section for the naming scheme used throughout this article.
Figure 1.
The human PXR gene locus and mRNA transcripts 1–4. Exons are represented as rectangles; the region of exon 5 that is not expressed in variant 3 is shown in purple, and the alternative positions of exon 1 are shown in red and green.
PXR splice and transcript variants differ in their effects on target gene transcription7, 38, 39, 70. PXR3 lacks the activity of PXR1 in terms of both gene transactivation activity and protein–protein interactions7, 38, 39, 70, 73, whereas the protein encoded by the PXR2 mRNA has comparable activity to the main protein, PXR17, 38, 70. Similarly to PXR3, PXR4 has been shown to lack gene transactivational activity, but, in contrast to PXR3, it has an intact LBD72. These observations and the lack of exhaustive study into the functions of PXR transcript variants highlight the necessity of investigating the functions of these variant proteins and transcripts and their effects on drug metabolism.
The PXR transcript variants possess distinct and overlapping functionality. PXR2 was as effective as PXR1 in mediating transcription of CYP3A77. Transcription of the UGT1A family increased in cells transfected with PXR1 and PXR2, but this was not consistent between UGT1A family members: Whereas UGT1A1, UGT1A3, UGT1A4, and UGT1A6 mRNA were induced to various extents by PXR1 and PXR2 in response to rifampicin treatments, UGT1A9 mRNA was not induced by rifampicin7. PXR3 did not activate CYP3A7 or UGT1A mRNA expression7, but when PXR3 was cotransfected with PXR1, it displayed a dominant negative effect on PXR1 activation of CYP3A4 induced by rifampicin39. Although PXR3 binds to PXR response element, it does not activate the expression of a luciferase reporter construct under the control of the CYP3A4 promoter (CYP-Luc)39. In addition, it binds to corepressors but not coactivators39. One study69 found 9 variants of PXR1, 7 of which had deletions in exon 5. PXR3 results from splicing of PXR1, whereas PXR2 is generated by an alternate transcription start site and has a first exon distinct from that of PXR1. No publications exist that further investigate the PXR variants 5–9. Stable transfection of HepG2 and LS180 cells with PXR1, but not with PXR3, resulted in an increase in CYP3A4, MDR1, CYP2B6, and UGT1A1 mRNA in both the absence and presence of the PXR agonist rifampicin38, 39.
The expression profiles of the main PXR transcript variants have not been fully elucidated, nor have the implications of the 5′ diversity among the transcripts been fully investigated. There are data available on the organism-wide expression profile for variants 1 and 3, but the expression of other variants has been quantified only in the liver9, 38, 40, 72. Quantification of PXR2 mRNA in a larger sample (n = 56) found that PXR2 represented, on average, approximately 15% of the total PXR transcripts in the liver, although this proportion was as high as 60% in some cases38. The average mRNA level of PXR3, which lacks the transactivation activity of PXR1 on inducing CYP3A470 accounts for 7% of the total PXR transcripts in the liver9. As PXR transcript variants 1 to 4 have identical 3′ regions, it would be predicted that similar regulatory mechanisms act on this region in all variants. However, the 5′ region of the PXR transcripts differs between PXR1 and PXR240, 71, 74, and this 5′ diversity may contribute to differential regulation by transregulatory factors at both the genomic (i.e., gene) and post-transcriptional (i.e., mRNA) levels74. When 5′ rapid amplification of cDNA ends (RACE) sequencing was performed to map the 5′ untranslated region (UTR) of PXR transcripts, putative transcription factor binding sites were found upstream of the PXR2 first exon that were distinct from those in the upstream region of the PXR1 first exon74. As mentioned previously, PXR1 contains a noncoding exon 1a and uses a CTG start codon located in exon 274. However, PXR2 contains exon 1b and an ATG start site that results in a protein with an N-terminal addition of 39 amino acids (as compared to variant 1)74. The alternating use of exons 1a and 1b results in distinct 5′ regions in PXR1 and PXR274. The characterization of the upstream region of PXR2 revealed a promoter region 1.5 kb upstream of the major PXR2 transcript transcription start site that displayed gene transactivation activity when placed upstream from a luciferase gene in a reporter construct75. A search of the TRANSFAC transcription factor database (http://www.gene-regulation.com/pub/databases.html) also revealed consensus binding sites for hepatocyte nuclear factor 1 (HNF1), HNF3β, and HNF4 in this 1.5-kb region that is required for activating luciferase expression75. Further characterization of this promoter region by sequential deletion revealed that a putative HNF1 response element was required for luciferase expression75. Additionally, putative TATA boxes and consensus sites for HNF-3β, octamer factor 1 (Oct-1), CCAAT/enhancer binding protein β (C/EBPβ), and glucocorticoid receptor (GR), were found upstream of the major transcription start site identified for PXR174. In another study, the region upstream of the transcription start site of PXR1 containing putative transcription factor binding sites displayed promoter activity when placed upstream of a luciferase gene and transfected into HepG2 cells71. The authors also mapped the minimal essential region for promoter activity to a 160-bp region upstream of the transcription initiation site, showing that this region also binds to nuclear proteins and that mutations of this region disrupt protein binding and reduce promoter activity. Alternative promoter use would account for the multiple 5′ ends of the transcripts for PXR1 and PXR274. The 5′ regions for PXR1 and PXR2 were later extended, and new proximal promoters were identified40. Additionally, the exon 3 region with the CpG island also displays promoter activity by luciferase reporter assay76, suggesting that transcription factors may be able to regulate PXR4 differently from the other variants, as PXR4 lacks exon 3.
Interestingly, a 6-bp deletion (−133GAGAAG−128) spanning the putative HNF1 binding sequence upstream of the PXR2 transcription start site74 was reported in a Japanese population75. This deletion was associated with a complete loss of promoter activity when conjugated to a luciferase gene and transfected into HepG2 cells. The allele frequency of the deletion variant was 28% in both healthy control subjects and patients with aspirin-induced asthma (AIA), and almost half of the population sampled had this 6-bp deletion, either heterozygously or homozygously. The same 6-bp deletion was further investigated in a Chinese population in healthy controls and in patients with hepatic carcinoma38. In this study, the allelic frequency, as determined by analyzing blood samples (n = 177), in healthy controls (22%) was significantly different from that in patients with hepatic carcinoma (38%). This research group also reported that PXR2 mRNA represented an average of 15% of the total PXR mRNA expression, with a range from 1% to 60%. The 6-bp deletion reduces the levels of PXR2 and, thus, of total PXR and of MDR1 and CYP3A4 mRNA expression38. The significantly higher allelic frequency of the 6-bp deletion in patients with hepatic carcinoma might also suggest its correlation with carcinoma formation, possibly as a result of the reduced capacity for detoxification.
4. Relation of PXR variant expression and activity in the context of chemotherapeutic response and carcinogenesis
Organism-wide expression profiling studies have revealed an incomplete correlation between the expression of PXR target genes and that of the major transcript variant, PXR1 (434 residues). PXR2 (473 residues), which regulates similar target genes to PXR1, may contribute to this incomplete correlation; however, an organism-wide expression profile for PXR2 has not been attempted9. It is possible that the interindividual and tissue variation in target gene expression may be related to the tissue-specific expression of specific PXR transcript variants. For example, PXR1 and PXR3 are, in some instances, not expressed in tissues that express CYP3A4, whereas PXR2 may be expressed in those tissues. Similarly, Caco-2 cells only express PXR1 mRNA, whereas HepG2 cells, human jejunum, and human hepatocytes express PXR1, PXR2, and PXR3 mRNA7. In addition to tissue-specific expression, the PXR transcript variants also display divergent effects on target gene transcription. PXR3 binds to the PXR response element but fails to activate the expression of PXR target genes in response to PXR ligands39. Instead, it functions as a dominant negative interfering with PXR139. PXR1 and PXR2, but not PXR3, induce CYP3A7, UGT1A1, UGT1A3, and UGT1A4 mRNA in HepG2 and Caco-2 cells in response to rifampicin7. These data suggested that both PXR1 and PXR2, although they may be differentially expressed, contribute to the overall PXR activity7. Conversely, PXR3 and PXR4 lack transcriptional activity in inducing target gene expression39, 72. Thus, the relative expression levels of the PXR transcript variants may directly affect chemotherapeutic metabolism.
A recent report describes the inverse correlation between the expression level of PXR4 and the methylation status of a CpG island upstream of exon 3, as well as the effect of overexpressed PXR4 protein in reducing cell proliferation markers72. The CpG island was reported to be methylated in various cell lines and normal colon tissue, and the methylation status of this region did not affect PXR1 mRNA expression77. PXR4 contains 322 amino acids, is homologous to the LBD of PXR1 and PXR2, retains ligand and coregulator binding capacity, and is detectable in the nucleus, but it lacks target gene transactivation activity72. These investigators also found PXR4 to represent approximately 10% of the total PXR mRNA expressed in human livers. Overexpression of PXR4 in HepG2 cells decreased cellular proliferation and rifampicin-induced expression of CYP3A4 and FGF1972. The authors postulated that PXR4 was a tumor suppressor functioning as a dominant negative against PXR1. Although no significant difference in expression of PXR4 mRNA was observed in normal tissue and hepatocellular carcinoma or adenoma, the authors did observe differences in expression of PXR4 mRNA among certain tumor subtypes. For example, PXR4 mRNA was decreased in inflammatory hepatocellular adenomas compared to normal livers or noninflammatory subtypes. Interestingly, higher levels of PXR4 mRNA expression were associated with favorable prognostic indicators and greater disease-free survival, whereas lower levels of expression in subtypes of hepatocellular carcinoma were associated with poor prognoses. The authors also observed decreased expression of PXR4 in the inflammatory subtype of hepatocellular adenoma that is believed to progress to hepatocellular carcinoma as a result of inflammatory activation via the JAK/STAT pathway. These observations suggest that the suppressive effect of PXR4 on PXR target genes may also extend to interactions with the NF-κB pathway, a pathway known to interact with PXR14, 72, 78, 79. PXR4 may thus serve as a prognostic marker in hepatic carcinoma, as well as playing a role in the malignant transformation of hepatic carcinomas.
The PXR variants may also display differential behavior in protein–protein interactions. It has been reported that p53 interacts with PXR1 and represses PXR-mediated transactivation of the CYP3A4 promoter. However, PXR3 does not interact with p5373. The p53 protein, mutated in many cancers, is a tumor suppressor that leads to cell cycle arrest, DNA repair, and apoptosis when activated by a wide array of genotoxic stresses80, 81. Interestingly, while both the wild-type and mutated forms of p53 interact with PXR, only the wild-type form inhibits PXR activity73. Therefore, cell populations with a mutant p53 protein will have higher levels of PXR transcriptional activity and drug-metabolizing enzymes, such as CYP3A4, possibly rendering these populations less responsive to chemotherapeutic agents that are metabolized by CYP3A4. Whether other PXR protein variants interact with p53 has not been investigated.
As discussed earlier, the differential regulation of PXR1 and PXR2 resulting from their different 5′ UTRs may contribute to the differential expression of the variants and the levels of total PXR and its target genes, which might be associated with differential risk of carcinoma formation and the response of cancer to chemotherapeutic agents.
Methylation of the PXR promoter was first reported in detail in neuroblastoma76. Neuroblastoma is the most common solid tumor in children, with advanced stages having an incidence of mortality greater than 60%82, 83. Normal adrenal tissue, the tissue of origin of neuroblastoma, expresses PXR1 mRNA exclusively76. Hypermethylation in CpG-rich exons and promoter regions has been observed in many cancers and can lead to the transcriptional inactivation of tumor suppressors84. PXR mRNA expression was analyzed across a panel of 19 neuroblastoma cell lines, and no PXR was detected in 14 of the cell lines76. By using bacterial artificial chromosome array-based methylated CpG island amplification (BAMCA), the promoter of PXR was found to be methylated in several neuroblastoma cell lines and primary tumors76. Specifically, PXR mRNA was expressed in the low-grade tumors but not in the advanced neuroblastoma cell lines analyzed. However, PXR mRNA expression increased in the advanced neuroblastoma cell lines after they were treated with 5-aza-Cyd, an inhibitor of DNA methyltransferase76. The authors performed bisulfite sequencing in the exon 1a region and in a region of exon 3. Methylation of the exon 3 region was more frequently detected in advanced tumors, tumors from patients with a poor prognosis, and tumors from patients who were more than 1 year of age. The methylation status of exon 1a did not correlate with PXR mRNA expression. However, hypermethylation of the exon 3 region was detected in cell lines lacking PXR mRNA expression (IMR32 and SH-SY5Y), whereas hypomethylation, as well as PXR mRNA expression, was detected in a normal lymphoblast cell line and two neuroblastoma cell lines (SK-N-AS and SK-N-KP). The exon 3 region with the CpG island exhibited promoter activity. Additionally, there was no detectable difference in the methylation status of the exon 1a region among these cell lines. PXR1 and PXR2 mRNA were expressed in cell lines in which the PXR promoter is unmethylated, but not in cell lines with methylated PXR promoters. In cell lines with methylated PXR promoter, the expression of PXR1 mRNA, but not of PXR2 mRNA, was restored by the treatment with 5-aza-Cyd. PXR2 was detected in the hypomethylated (SJ-N-KP and SK-N-AS) cell lines and not in the hypermethylated ones (SJ-N-GC and SMS-KAN), whereas PXR3 mRNA was not expressed in any of the cell lines, regardless of their methylation status or whether they were treated with 5-aza-Cyd. Consequently, the methylation status of separate regions of the PXR gene differentially regulates the expression of specific PXR variants. In this study, the ectopic overexpression of PXR decreased cell proliferation, suggesting that PXR functions as a tumor suppressor76. However, whether this finding is specific to neuroblastoma cell lines is unclear.
In colon cancer cell lines, PXR promoter methylation was associated with PXR mRNA and CYP3A4 mRNA expression levels77. Moreover, the PXR promoter was less methylated and PXR and CYP3A4 mRNA expression levels were correspondingly higher in cancerous colon tissues than in adjacent normal tissues. In a xenograft model of colon cancer, PXR activation correlates with the growth of both human colon tumor cell lines and primary human colon cancer tissue. PXR potentially functions as an oncogene85. In colon cancer cell lines (LS180, LoVo, Caco-2, HCT116, HT29, and SW48), PXR and CYP3A4 mRNA levels were associated with hypermethylation of a CpG-rich sequence in the promoter of PXR and were increased after treatment with 5-aza-dC77. The cell lines that express high levels of PXR1 and low levels of PXR2 and PXR3 have mostly methylated CpG-rich sequences in exon 1b and in the intron of the 3′ end of exon 1b. However, a mostly unmethylated CpG-rich sequence exists in exon 1a and in the intron of the immediate 3′ end of exon 1a in the cell lines with higher expression of PXR (LS180 and LoVo)77. Thus, it would seem that the mRNA expression of different PXR variants could be differentially regulated by methylation. It would be expected that methylation of the CpG-rich sequences in and around exon 1a might specifically affect the expression of PXR1 mRNA and PXR3 mRNA, whereas the methylation status of and near exon 1b would affect the expression patterns of PXR2 mRNA. In colon cancer cells excised from colonic tissue samples, the CpG island within the exon 3 region is the most methylated region of the PXR gene, whereas the CpG-rich sequence around exon 1a is the most unmethylated region77. The CpG-rich sequence around exon 1b is methylated in these colon cancer cells, and this may correlate with low PXR2 mRNA expression. Additionally, the methylation of the CpG island within exon 3 seen in cell lines77 may be associated with low expression of PXR4 and, subsequently, with higher expression observed after treatment with a demethylating agent72. Therefore, determination of the protumor effect of PXR in colon cancer may involve a complex series of events that requires a specific relative expression pattern of multiple transcript variants. Additional analysis of the methylation of PXR variants in different tumor types is warranted to further establish the correlation between methylation and expression of PXR and tumor development, as well as tumor response to chemotherapeutic agents.
5. Conclusions
It is clinically and scientifically important to elucidate the functions of the PXR variants in order to predict the outcome of ligand activation of PXR and the role of PXR in various model systems. PXR is activated by structurally and functionally diverse ligands that are associated with and prescribed in a multitude of disease states. A greater understanding of the roles of the hPXR transcript variants would greatly contribute to predicting therapeutic failure of cotherapies arising from xenosensor-mediated decreases in drug bioavailability or increases in toxicity. For example, high overall PXR expression and low PXR3 (which functions in a dominant negative manner) expression may result in decreased drug bioavailability whereas high overall PXR expression and high PXR3 expression (compared to transcriptionally active variants) would be expected to result in increased drug bioavailability. PXR1 plays a well-described role in the detoxifying drug metabolism pathways of cancer therapeutics and can cause treatment failure at high levels of expression. PXR2 has similar effect on target gene transactivation but has not been fully characterized in terms of coregulator recruitment, ligand response, protein–protein interactions, or organism-wide tissue expression. PXR3 has been reported to recruit corepressors but does not similarly participate in the protein–protein interactions associated with PXR1 or transactivate PXR target genes. However, it is unknown whether the dominant negative effect of PXR3 plays a significant pathophysiologic or physiologic role. PXR4 has been reported to compete with PXR1 in binding to ligand and coactivators to exert a dominant negative effect, as has PXR3 with respect to PXR1 transactivational activity. Several of these transcripts have been implicated in cancer pathogenesis, with PXR1 expression being a negative prognostic factor in some but not all cancers because of its role in drug metabolism and transport. There have been many contradictory reports concerning the effects of PXR1 on cellular proliferation and apoptosis; these effects may be more clearly defined by investigating the roles of the variants in these various model systems. PXR polymorphisms, the use of alternate promoters, and splicing variations have not been fully elucidated in terms of expression across disease states and transactivation profiles of target genes. Future studies investigating the role of PXR on drug metabolism would be remiss not to evaluate multiple transcript variants and the resultant proteins.
Acknowledgments
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children′s Research Hospital, and the U.S. National Institutes of Health (Grants GM086415, GM110034, GM118041 and P30-CA21765). The funding sources had no involvement in the writing or the decision to submit the manuscript for publication. We thank Dr. Keith A. Laycock for comprehensively editing the manuscript.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Watkins R.E., Davis-Searles P.R., Lambert M.H., Redinbo M.R. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J Mol Biol. 2003;331:815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 2.Zollner G., Wagner M., Trauner M. Nuclear receptors as drug targets in cholestasis and drug-induced hepatotoxicity. Pharmacol Ther. 2010;126:228–243. doi: 10.1016/j.pharmthera.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Kliewer S.A., Moore J.T., Wade L., Staudinger J.L., Watson M.A., Jones S.A. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 4.Lehmann J.M., McKee D.D., Watson M.A., Willson T.M., Moore J.T., Kliewer S.A. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T. Nuclear receptor drug discovery. Curr Opin Chem Biol. 2008;12:418–426. doi: 10.1016/j.cbpa.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y.M., Ong S.S., Chai S.C., Chen T. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol. 2012;8:803–817. doi: 10.1517/17425255.2012.685237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner-Stephen D., Heydel J.M., Goyal A., Lu Y., Xie W., Lindblom T. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos. 2004;32:340–347. doi: 10.1124/dmd.32.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aleksunes L.M., Klaassen C.D. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos. 2012;40:1366–1379. doi: 10.1124/dmd.112.045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamba V., Yasuda K., Lamba J.K., Assem M., Davila J., Strom S. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–265. doi: 10.1016/j.taap.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y., Liu D. Activation of pregnane X receptor by pregnenolone 16α-carbonitrile prevents high-fat diet–induced obesity in AKR/J mice. PLoS One. 2012;7:e38734. doi: 10.1371/journal.pone.0038734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azuma K., Casey S.C., Ito M., Urano T., Horie K., Ouchi Y. Pregnane X receptor knockout mice display osteopenia with reduced bone formation and enhanced bone resorption. J Endocrinol. 2010;207:257–263. doi: 10.1677/JOE-10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helsley R.N., Sui Y., Ai N., Park S.H., Welsh W.J., Zhou C. Pregnane X receptor mediates dyslipidemia induced by the HIV protease inhibitor amprenavir in mice. Mol Pharmacol. 2013;83:1190–1199. doi: 10.1124/mol.113.085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng J., Shah Y.M., Ma X., Pang X., Tanaka T., Kodama T. Therapeutic role of rifaximin in inflammatory bowel disease: clinical implication of human pregnane X receptor activation. J Pharmacol Exp Ther. 2010;335:32–41. doi: 10.1124/jpet.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah Y.M., Ma X., Morimura K., Kim I., Gonzalez F.J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-κB target gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1114–G1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 15.Hawkins M.T., Lewis J.H. Latest advances in predicting DILI in human subjects: focus on biomarkers. Expert Opin Drug Metab Toxicol. 2012;8:1521–1530. doi: 10.1517/17425255.2012.724060. [DOI] [PubMed] [Google Scholar]
- 16.Marschall H.U., Wagner M., Zollner G., Trauner M. Clinical hepatotoxicity. Regulation and treatment with inducers of transport and cofactors. Mol Pharm. 2007;4:895–910. doi: 10.1021/mp060133c. [DOI] [PubMed] [Google Scholar]
- 17.Andrews E., Armstrong M., Tugwood J., Swan D., Glaves P., Pirmohamed M. A role for the pregnane X receptor in flucloxacillin-induced liver injury. Hepatology. 2010;51:1656–1664. doi: 10.1002/hep.23549. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y.M., Chai S.C., Brewer C.T., Chen T. Pregnane X receptor and drug-induced liver injury. Expert Opin Drug Metab Toxicol. 2014;10:1521–1532. doi: 10.1517/17425255.2014.963555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong H., Sinz M.W., Feng Y., Chen T., Venkataramanan R., Xie W. Animal models of xenobiotic receptors in drug metabolism and diseases. Methods Enzymol. 2005;400:598–618. doi: 10.1016/S0076-6879(05)00034-0. [DOI] [PubMed] [Google Scholar]
- 20.Ma X., Shah Y., Cheung C., Guo G.L., Feigenbaum L., Krausz K.W. The PREgnane X receptor gene-humanized mouse: a model for investigating drug–drug interactions mediated by cytochromes P450 3A. Drug Metab Dispos. 2007;35:194–200. doi: 10.1124/dmd.106.012831. [DOI] [PubMed] [Google Scholar]
- 21.Scheer N., Ross J., Rode A., Zevnik B., Niehaves S., Faust N. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest. 2008;118:3228–3239. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng J., Ma X., Gonzalez F.J. Pregnane X receptor- and CYP3A4-humanized mouse models and their applications. Br J Pharmacol. 2011;163:461–468. doi: 10.1111/j.1476-5381.2010.01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheer N., Ross J., Kapelyukh Y., Rode A., Wolf C.R. In vivo responses of the human and murine pregnane X receptor to dexamethasone in mice. Drug Metab Dispos. 2010;38:1046–1053. doi: 10.1124/dmd.109.031872. [DOI] [PubMed] [Google Scholar]
- 24.Krasowski M.D., Yasuda K., Hagey L.R., Schuetz E.G. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol Endocrinol. 2005;19:1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones S.A., Moore L.B., Shenk J.L., Wisely G.B., Hamilton G.A., McKee D.D. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 26.Luo G., Cunningham M., Kim S., Burn T., Lin J., Sinz M. CYP3A4 induction by drugs: correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab Dispos. 2002;30:795–804. doi: 10.1124/dmd.30.7.795. [DOI] [PubMed] [Google Scholar]
- 27.Xie W., Barwick J.L., Simon C.M., Pierce A.M., Safe S., Blumberg B. Reciprocal activation of xenobiotic response genes by nuclear receptors SXR/PXR and CAR. Genes Dev. 2000;14:3014–3023. doi: 10.1101/gad.846800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie W., Yeuh M.F., Radominska-Pandya A., Saini S.P., Negishi Y., Bottroff B.S. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc Natl Acad Sci U S A. 2003;100:4150–4155. doi: 10.1073/pnas.0438010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Synold T.W., Dussault I., Forman B.M. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 30.Mota L.C., Barfield C., Hernandez J.P., Baldwin W.S., Nonylphenol-mediated C.Y.P. induction is PXR-dependent: the use of humanized mice and human hepatocytes suggests that hPXR is less sensitive than mouse PXR to nonylphenol treatment. Toxicol Appl Pharmacol. 2011;252:259–267. doi: 10.1016/j.taap.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y.M., Lin W., Chai S.C., Wu J., Ong S.S., Schuetz E.G. Piperine activates human pregnane X receptor to induce the expression of cytochrome P450 3A4 and multidrug resistance protein 1. Toxicol Appl Pharmacol. 2013;272:96–107. doi: 10.1016/j.taap.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Kuehl P., Green E.D., Touchman J.W., Watkins P.B., Daly A. The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics. 2001;11:555–572. doi: 10.1097/00008571-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Lamba J.K., Lin Y.S., Schuetz E.G., Thummel K.E. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 34.Lamba V., Panetta J.C., Strom S., Schuetz E.G. Genetic predictors of interindividual variability in hepatic CYP3A4 expression. J Pharmacol Exp Ther. 2010;332:1088–1099. doi: 10.1124/jpet.109.160804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts R.A., Ganey P.E., Ju C., Kamendulis L.M., Rusyn I., Klaunig J.E. Role of the Kupffer cell in mediating hepatic toxicity and carcinogenesis. Toxicol Sci. 2007;96:2–15. doi: 10.1093/toxsci/kfl173. [DOI] [PubMed] [Google Scholar]
- 36.James L.P., Mayeux P.R., Hinson J.A. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos. 2003;31:1499–1506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- 37.Dotzlaw H., Leygue E., Watson P., Murphy L.C. The human orphan receptor PXR messenger RNA is expressed in both normal and neoplastic breast tissue. Clin Cancer Res. 1999;5:2103–2107. [PubMed] [Google Scholar]
- 38.Liu Y., Ji W., Yin Y., Fan L., Zhang J., Yun H. The effects of splicing variant of PXR PAR-2 on CYP3A4 and MDR1 mRNA expressions. Clin Chim Acta. 2009;403:142–144. doi: 10.1016/j.cca.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y.S., Yasuda K., Assem M., Cline C., Barber J., Li C.W. The major human pregnane X receptor (PXR) splice variant, PXR.2, exhibits significantly diminished ligand-activated transcriptional regulation. Drug Metab Dispos. 2009;37:1295–1304. doi: 10.1124/dmd.108.025213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tompkins L.M., Sit T.L., Wallace A.D. Unique transcription start sites and distinct promoter regions differentiate the pregnane X receptor (PXR) isoforms PXR 1 and PXR 2. Drug Metab Dispos. 2008;36:923–929. doi: 10.1124/dmd.107.018317. [DOI] [PubMed] [Google Scholar]
- 41.Chang T.K., Yu L., Maurel P., Waxman D.J. Enhanced cyclophosphamide and ifosfamide activation in primary human hepatocyte cultures: response to cytochrome P-450 inducers and autoinduction by oxazaphosphorines. Cancer Res. 1997;57:1946–1954. [PubMed] [Google Scholar]
- 42.Raynal C., Pascussi J.M., Leguelinel G., Breuker C., Kantar J., Lallemant B. Pregnane X Receptor (PXR) expression in colorectal cancer cells restricts irinotecan chemosensitivity through enhanced SN-38 glucuronidation. Mol Cancer. 2010;9:46. doi: 10.1186/1476-4598-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basseville A., Preisser L., de Carne Trecesson S., Boisdron-Celle M., Gamelin E., Coqueret O. Irinotecan induces steroid and xenobiotic receptor (SXR) signaling to detoxification pathway in colon cancer cells. Mol Cancer. 2011;10:80. doi: 10.1186/1476-4598-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta D., Venkatesh M., Wang H., Kim S., Sinz M., Goldberg G.L. Expanding the roles for pregnane X receptor in cancer: proliferation and drug resistance in ovarian cancer. Clin Cancer Res. 2008;14:5332–5340. doi: 10.1158/1078-0432.CCR-08-1033. [DOI] [PubMed] [Google Scholar]
- 45.Yonemori K., Takeda Y., Toyota E., Kobayashi N., Kudo K. Potential interactions between irinotecan and rifampin in a patient with small-cell lung cancer. Int J Clin Oncol. 2004;9:206–209. doi: 10.1007/s10147-004-0394-4. [DOI] [PubMed] [Google Scholar]
- 46.Miki Y., Suzuki T., Kitada K., Yabuki N., Shibuya R., Moriya T. Expression of the steroid and xenobiotic receptor and its possible target gene, organic anion transporting polypeptide-A, in human breast carcinoma. Cancer Res. 2006;66:535–542. doi: 10.1158/0008-5472.CAN-05-1070. [DOI] [PubMed] [Google Scholar]
- 47.Choi H.K., Yang J.W., Roh S.H., Han C.Y., Kang K.W. Induction of multidrug resistance associated protein 2 in tamoxifen-resistant breast cancer cells. Endocr Relat Cancer. 2007;14:293–303. doi: 10.1677/ERC-06-0016. [DOI] [PubMed] [Google Scholar]
- 48.Nagaoka R., Iwasaki T., Rokutanda N., Takeshita A., Koibuchi Y., Horiguchi J. Tamoxifen activates CYP3A4 and MDR1 genes through steroid and xenobiotic receptor in breast cancer cells. Endocrine. 2006;30:261–268. doi: 10.1007/s12020-006-0003-6. [DOI] [PubMed] [Google Scholar]
- 49.Sane R.S., Buckley D.J., Buckley A.R., Nallani S.C., Desai P.B. Role of human pregnane X receptor in tamoxifen- and 4-hydroxytamoxifen-mediated CYP3A4 induction in primary human hepatocytes and LS174T cells. Drug Metab Dispos. 2008;36:946–954. doi: 10.1124/dmd.107.018598. [DOI] [PubMed] [Google Scholar]
- 50.Nallani S.C., Goodwin B., Maglich J.M., Buckley D.J., Buckley A.R., Desai P.B. Induction of cytochrome P450 3A by paclitaxel in mice: pivotal role of the nuclear xenobiotic receptor, pregnane X receptor. Drug Metab Dispos. 2003;31:681–684. doi: 10.1124/dmd.31.5.681. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y., Tang Y., Wang M.T., Zeng S., Nie D. Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res. 2007;67:10361–10367. doi: 10.1158/0008-5472.CAN-06-4758. [DOI] [PubMed] [Google Scholar]
- 52.Masuyama H., Suwaki N., Tateishi Y., Nakatsukasa H., Segawa T., Hiramatsu Y. The pregnane X receptor regulates gene expression in a ligand- and promoter-selective fashion. Mol Endocrinol. 2005;19:1170–1180. doi: 10.1210/me.2004-0434. [DOI] [PubMed] [Google Scholar]
- 53.Zhou J., Liu M., Zhai Y., Xie W. The antiapoptotic role of pregnane X receptor in human colon cancer cells. Mol Endocrinol. 2008;22:868–880. doi: 10.1210/me.2007-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandanaraj E., Lal S., Selvarajan V., Ooi L.L., Wong Z.W., Wong N.S. PXR pharmacogenetics: association of haplotypes with hepatic CYP3A4 and ABCB1 messenger RNA expression and doxorubicin clearance in Asian breast cancer patients. Clin Cancer Res. 2008;14:7116–7126. doi: 10.1158/1078-0432.CCR-08-0411. [DOI] [PubMed] [Google Scholar]
- 55.Huang R., Murry D.J., Kolwankar D., Hall S.D., Foster D.R. Vincristine transcriptional regulation of efflux drug transporters in carcinoma cell lines. Biochem Pharmacol. 2006;71:1695–1704. doi: 10.1016/j.bcp.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 56.Smith N.F., Mani S., Schuetz E.G., Yasuda K., Sissung T.M., Bates S.E. Induction of CYP3A4 by vinblastine: role of the nuclear receptor NR1I2. Ann Pharmacother. 2010;44:1709–1717. doi: 10.1345/aph.1P354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haaz M.C., Rivory L., Riche C., Vernillet L., Robert J. Metabolism of irinotecan (CPT-11) by human hepatic microsomes: participation of cytochrome P-450 3A and drug interactions. Cancer Res. 1998;58:468–472. [PubMed] [Google Scholar]
- 58.Mathijssen R.H., van Alphen R.J., Verweij J., Loos W.J., Nooter K., Stoter G. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11) Clin Cancer Res. 2001;7:2182–2194. [PubMed] [Google Scholar]
- 59.Masuyama H., Nakatsukasa H., Takamoto N., Hiramatsu Y. Down-regulation of pregnane X receptor contributes to cell growth inhibition and apoptosis by anticancer agents in endometrial cancer cells. Mol Pharmacol. 2007;72:1045–1053. doi: 10.1124/mol.107.037937. [DOI] [PubMed] [Google Scholar]
- 60.Desai P.B., Nallani S.C., Sane R.S., Moore L.B., Goodwin B.J., Buckley D.J. Induction of cytochrome P450 3A4 in primary human hepatocytes and activation of the human pregnane X receptor by tamoxifen and 4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:608–612. doi: 10.1124/dmd.30.5.608. [DOI] [PubMed] [Google Scholar]
- 61.Zhuo W., Hu L., Lv J., Wang H., Zhou H., Fan L. Role of pregnane X receptor in chemotherapeutic treatment. Cancer Chemother Pharmacol. 2014;74:217–227. doi: 10.1007/s00280-014-2494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dehal S.S., Kupfer D. CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res. 1997;57:3402–3406. [PubMed] [Google Scholar]
- 63.Jacolot F., Simon I., Dreano Y., Beaune P., Riche C., Berthou F. Identification of the cytochrome P450 IIIA family as the enzymes involved in the N-demethylation of tamoxifen in human liver microsomes. Biochem Pharmacol. 1991;41:1911–1919. doi: 10.1016/0006-2952(91)90131-n. [DOI] [PubMed] [Google Scholar]
- 64.Harris J.W., Rahman A., Kim B.R., Guengerich F.P., Collins J.M. Metabolism of taxol by human hepatic microsomes and liver slices: participation of cytochrome P450 3A4 and an unknown P450 enzyme. Cancer Res. 1994;54:4026–4035. [PubMed] [Google Scholar]
- 65.Rahman A., Korzekwa K.R., Grogan J., Gonzalez F.J., Harris J.W. Selective biotransformation of taxol to 6α-hydroxytaxol by human cytochrome P450 2C8. Cancer Res. 1994;54:5543–5546. [PubMed] [Google Scholar]
- 66.Lee H.J., Lee M.G. Effects of dexamethasone on the pharmacokinetics of adriamycin after intravenous administration to rats. Res Commun Mol Pathol Pharmacol. 1999;105:87–96. [PubMed] [Google Scholar]
- 67.Harmsen S., Meijerman I., Febus C.L., Maas-Bakker R.F., Beijnen J.H., Schellens J.H. PXR-mediated induction of P-glycoprotein by anticancer drugs in a human colon adenocarcinoma-derived cell line. Cancer Chemother Pharmacol. 2010;66:765–771. doi: 10.1007/s00280-009-1221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leveque D., Jehl F. Molecular pharmacokinetics of catharanthus (vinca) alkaloids. J Clin Pharmacol. 2007;47:579–588. doi: 10.1177/0091270007299430. [DOI] [PubMed] [Google Scholar]
- 69.Fukuen S., Fukuda T., Matsuda H., Sumida A., Yamamoto I., Inaba T. Identification of the novel splicing variants for the hPXR in human livers. Biochem Biophys Res Commun. 2002;298:433–438. doi: 10.1016/s0006-291x(02)02469-5. [DOI] [PubMed] [Google Scholar]
- 70.Hustert E., Zibat A., Presecan-Siedel E., Eiselt R., Mueller R., Fuss C. Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab Dispos. 2001;29:1454–1459. [PubMed] [Google Scholar]
- 71.Kurose K., Ikeda S., Koyano S., Tohkin M., Hasegawa R., Sawada J. Identification of regulatory sites in the human PXR (NR1I2) promoter region. Mol Cell Biochem. 2006;281:35–43. doi: 10.1007/s11010-006-0167-7. [DOI] [PubMed] [Google Scholar]
- 72.Breuker C., Planque C., Rajabi F., Nault J.C., Couchy G., Zucman-Rossi J. Characterization of a novel PXR isoform with potential dominant-negative properties. J Hepatol. 2014;61:609–616. doi: 10.1016/j.jhep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 73.Elias A., Wu J., Chen T. Tumor suppressor protein p53 negatively regulates human pregnane X receptor activity. Mol Pharmacol. 2013;83:1229–1236. doi: 10.1124/mol.113.085092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurose K., Koyano S., Ikeda S., Tohkin M., Hasegawa R., Sawada J. 5ʹ diversity of human hepatic PXR (NR1I2) transcripts and identification of the major transcription initiation site. Mol Cell Biochem. 2005;273:79–85. doi: 10.1007/s11010-005-7757-7. [DOI] [PubMed] [Google Scholar]
- 75.Uno Y., Sakamoto Y., Yoshida K., Hasegawa T., Hasegawa Y., Koshino T. Characterization of six base pair deletion in the putative HNF1-binding site of human PXR promoter. J Hum Genet. 2003;48:594–597. doi: 10.1007/s10038-003-0076-5. [DOI] [PubMed] [Google Scholar]
- 76.Misawa A., Inoue J., Sugino Y., Hosoi H., Sugimoto T., Hosoda F. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 2005;65:10233–10242. doi: 10.1158/0008-5472.CAN-05-1073. [DOI] [PubMed] [Google Scholar]
- 77.Habano W., Gamo T., Terashima J., Sugai T., Otsuka K., Wakabayashi G. Involvement of promoter methylation in the regulation of pregnane X receptor in colon cancer cells. BMC Cancer. 2011;11:81. doi: 10.1186/1471-2407-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu X., Ke S., Liu D., Sheng T., Thomas P.E., Rabson A.B. Role of NF-κB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem. 2006;281:17882–17889. doi: 10.1074/jbc.M601302200. [DOI] [PubMed] [Google Scholar]
- 79.Zhou C., Tabb M.M., Nelson E.L., Grun F., Verma S., Sadatrafiei A. Mutual repression between steroid and xenobiotic receptor and NF-κB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest. 2006;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muller P.A., Vousden K.H. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 81.Appella E., Anderson C.W. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 82.Brodeur G.M. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 83.Westermann F., Schwab M. Genetic parameters of neuroblastomas. Cancer Lett. 2002;184:127–147. doi: 10.1016/s0304-3835(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 84.Baylin S.B., Herman J.G., Graff J.R., Vertino P.M., Issa J.P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 85.Wang H., Venkatesh M., Li H., Goetz R., Mukherjee S., Biswas A. Pregnane X receptor activation induces FGF19-dependent tumor aggressiveness in humans and mice. J Clin Invest. 2011;121:3220–3232. doi: 10.1172/JCI41514. [DOI] [PMC free article] [PubMed] [Google Scholar]