After the publication of this work [1] it was noticed that there were typographical errors in the following equations: equation 5 in column 2, equation 7 in column 2, equation 8 in column 1.
The bracket was placed incorrectly, so it should read:
\ log _2 (Y_{gij} + 0.5) rather than (\ log _2 Y_{gij} + 0.5)
It was brought to our attention that a new submission to the webtool for the eXpress algorithm for the ENCODE GM12878 dataset performs better than what is reported in the paper. While looking into the reason for this discrepancy we found two errors. First, the commands and parameter settings provided in the log information on the webtool were incorrect. Second, we realized that we ran the eXpress submission differently from the other methods for this particular dataset. One cause for the discrepancy was the accidental use of a different transcript FASTA file. We reran eXpress controlling for these differences and confirmed that better results are attained. Row 2 in Table 1 is changed, and the updated row is below.
Table 1.
Summarized metrics for analyzed pipelines based on an experimental dataset
| Method | SD low | SD medium | SD high | NE (K = 1) | NN (K = 1) | TxDiff low | TxDiff medium | TxDiff high | deFC low | deFC medium | deFC high | pAUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cufflinks | 0.62 (0.002) | 0.26 (0.001) | 0.12 (0.000) | 0.08 | 0.70 | 0.31 (0.007) | 0.08 (0.002) | 0.03 (0.001) | 2.65 (0.022) | 2.25 (0.047) | 1.01 (0.024) | 0.77 |
| eXpress | 0.53 (0.002) | 0.22 (0.001) | 0.10 (0.000) | 0.07 | 0.72 | 0.24 (0.006) | 0.06 (0.002) | 0.02 (0.001) | 2.86 (0.022) | 2.21 (0.048) | 1.00 (0.019) | 0.79 |
| Flux Capacitor | 0.62 (0.003) | 0.57 (0.003) | 0.18 (0.001) | 0.10 | 0.73 | 0.42 (0.008) | 0.15 (0.004) | 0.07 (0.003) | 2.62 (0.024) | 2.40 (0.050) | 1.01 (0.025) | 0.75 |
| kallisto | 0.53 (0.002) | 0.24 (0.001) | 0.12 (0.000) | 0.09 | 0.64 | 0.28 (0.007) | 0.08 (0.002) | 0.03 (0.0001 | 2.36 (0.024) | 2.06 (0.045) | 1.03 (0.024) | 0.76 |
| RSEM | 0.54 (0.002) | 0.22 (0.001) | 0.11 (0.000) | 0.06 | 0.73 | 0.39 (0.008) | 0.07 (0.002) | 0.02 (0.001) | 2.72 (0.022) | 2.22 (0.048) | 1.03 (0.026) | 0.78 |
| Sailfish | 0.46 (0.002) | 0.25 (0.001) | 0.13 (0.000) | 0.08 | 0.60 | 0.27 (0.006) | 0.08 (0.002) | 0.04 (0.001) | 2.30 (0.023) | 2.08 (0.044) | 0.97 (0.022) | 0.77 |
| Salmon | 0.46 (0.002) | 0.23 (0.001) | 0.12 (0.000) | 0.08 | 0.65 | 0.29 (0.007) | 0.07 (0.002) | 0.04 (0.001) | 2.30 (0.024) | 2.06 (0.045) | 1.03 (0.022) | 0.77 |
Metrics for single cell lines are averaged for both cell lines, except standard deviation is the square root of average squares. Columns 2–4 shows median standard deviation on three transcript abundance levels; column 5 shows proportions of discordant calls when K = 1; column 6 shows proportions of both non-expressed when K = 1; columns 7–9 show the mean proportion differences of transcripts in genes only having two annotated transcripts based on three transcript abundance levels; columns 10–12 show median log fold changes of true differentially expressed genes based on three abundance levels; column 13 shows standardized partial area under the curve for differential expression of genes. pAUC partial area under the receiver operating characteristic curve
The comparative figures for GM12878 change (panel A Figures 3, 4, 5, 6 and Additional file 1: Figure S5). The new figures are below.
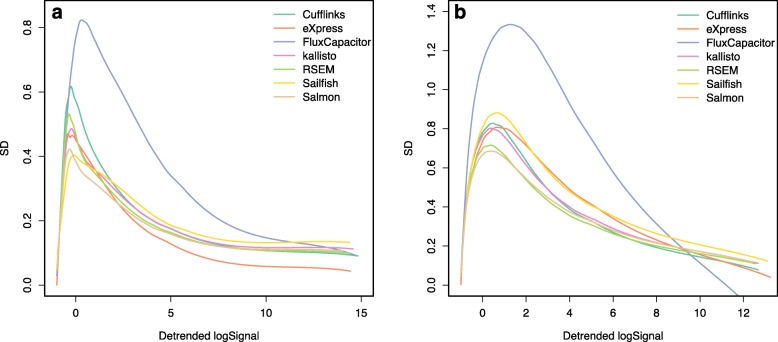
Fig. 3.
Standard deviations of transcript quantifications based on a an experimental dataset (GM12878) and b a simulation dataset (one of the cell lines). Seven quantification methods are shown here
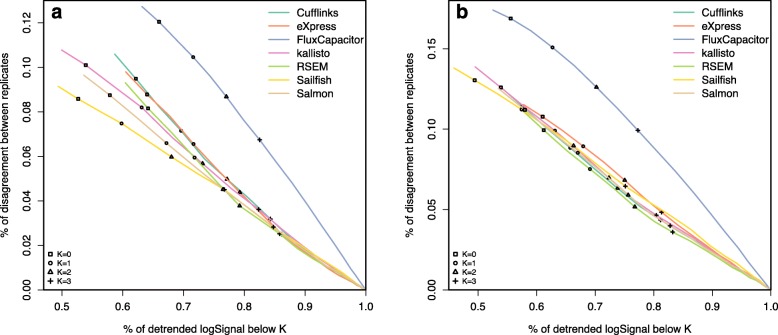
Fig. 4.
Proportions of discordant expression calls based on a an experimental dataset (GM12878) and b a simulation dataset (one of the cell lines). Seven quantification methods are shown here
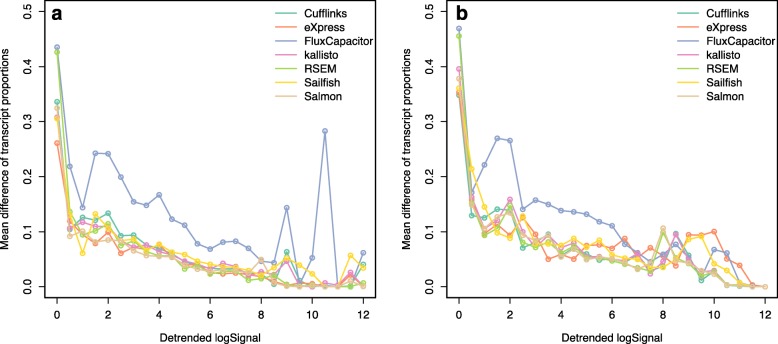
Fig. 5.
Proportion differences of transcript quantifications in genes with only two annotated transcripts based on a an experimental dataset (GM12878) and b a simulation dataset (one of the cell lines). Seven quantification methods are shown
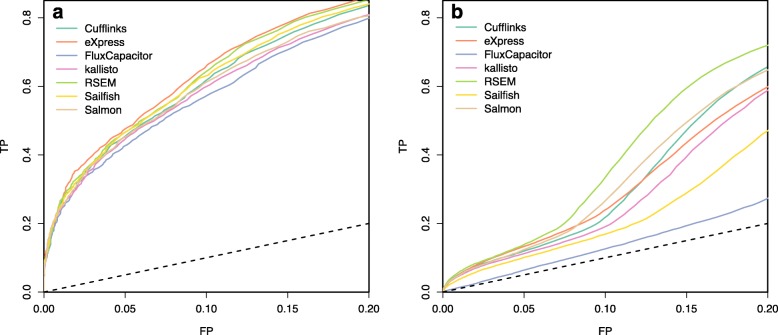
Fig. 6.
ROC curves indicating performance of quantification methods based on differential expression analysis of a an experimental dataset and b a simulation dataset. Seven quantification methods are shown. FP false positive, TP true positive
The following statements should now read:
Performance was generally poor, with one method clearly underperforming and RSEM slightly outperforming the rest.
In the first dataset, Flux Capacitor clearly underperform s compared with the other methods in the regions with most data (A between 3 and 8).
Here we see Flux Capacitor underperforming and RSEM slightly outperforming the other methods in the simulation dataset.
With the exception of the underperforming Flux Capacitor, we found that the other algorithms performed similarly.
The eXpress entry in the webtool, including the log-file entry which includes the scripts, has also been updated. You can see this in the ENCODE: 2 reps, high depth tab here: http://rafalab.rc.fas.harvard.edu/rnaseqbenchmark
The authors apologize for this error.
Additional file
Log fold changes of true differential expression fitted by losses. (a) Plot based on experimental dataset from cell lines GM12878 and K562. True differentially expressed genes are estimated using microarray data. (b) Plot based on simulation dataset with true differentially expressed transcripts predefined. (PDF 100 kb)
Footnotes
The online version of the original article can be found under doi:10.1186/s13059-016-0940-1.
Reference
- 1.Teng M, Love MI, Davis CA, Djebali S, Dobin A, Graveley BR, et al. A benchmark for RNA-seq quanitification pipelines. Genome Biol. 2016;17:74. doi: 10.1186/s13059-016-0940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]