Abstract
Intra-individual tumor heterogeneity may reduce the efficacy of molecularly guided systemic therapy for cancers that have metastasized. To determine whether the genomic alterations in a single metastasis provide a reasonable assessment of the major oncogenic drivers of other dispersed metastases within an individual, we analyzed multiple tumors from men with disseminated prostate cancer by whole exome sequencing, array CGH and RNA transcript profiling and compared the genomic diversity within and between individuals. In contrast to substantial heterogeneity between men, there was limited diversity comparing metastases within an individual. Numbers of somatic mutations, the burden of genomic copy number alterations, and aberrations in known oncogenic drivers were highly concordant as were metrics of androgen receptor (AR) activity and cell cycle activity. AR activity inversely associated with cell proliferation, whereas the expression of Fanconi anemia (FA) complex genes correlated with elevated cell cycle progression, E2F1 expression and RB1 loss. Men with somatic aberrations in FA complex genes or ATM exhibited significantly longer treatment response durations to carboplatin compared to men without defects in genes encoding DNA repair proteins. Collectively, these data indicate that though exceptions exist, evaluating a single metastasis provides a reasonable assessment of the major oncogenic driver alterations present in disseminated tumors within an individual, and may be useful for selecting treatments based on predicted molecular vulnerabilities.
Personalized approaches for cancer treatment rely on the premise that defined molecular alterations in tumors will confer susceptibility to specific therapeutics, and those malignancies without the alteration will not respond1. Consequently, identifying subsets of patients with and without a particular oncogenic driver is predicted to enhance efficacy and reduce futile treatment. This concept requires that heterogeneity exist between individuals with respect to the composition of therapy vulnerability characteristics and is supported by studies demonstrating extensive diversity in genomic aberrations and treatment responses between cancers with the same histological classification2-8. To achieve full success, precision oncology also requires limited molecular heterogeneity within an individual such that the targeted vulnerability is represented in most or all tumor cells.
While striking successes exploiting vital drivers of tumor cell growth have been achieved9, deep molecular profiling has uncovered a substantial degree of intratumoral heterogeneity with divergent cancer clones existing within a primary tumor10-12. The extent of this diversity indicates the potential for misclassification using current biopsy-based strategies that may under-sample tumors, and provides clear pathways to rapid treatment resistance. To date, most studies assessing the extent of tumor heterogeneity have characterized primary tumors. However, localized carcinomas are generally managed with surgery or radiation, which are often curative. A major unmet need involves the systemic treatment of metastatic cancer, which usually involves multiple spatially distributed anatomic sites. There is limited information concerning the molecular diversity of the metastatic burden within an individual13-15. Such knowledge has considerable implications for the success of precision oncology.
Here, we analyzed the intra-individual molecular diversity of metastatic prostate cancer (mPC), a disease that accounts for more than 27,000 deaths annually in the US16. The mainstay of initial therapy for mPC, essentially unchanged for more than 50 years, relies on suppressing circulating levels of testosterone and consequently suppressing the androgen receptor (AR) signaling program in PC cells17. This therapy, conceptually one of the first precision medicine approaches for treating cancer, is highly effective with response rates exceeding 90% and durations of disease control lasting approximately 24 months. At the time of disease progression, a nearly universal event, metastatic castration resistant prostate cancers (mCRPC) exhibit a spectrum of recurrent molecular aberrations, a number of which are predicted to confer susceptibility to targeted therapeutics18-21. Our objective was to determine whether the molecular composition of a single metastatic focus at a particular point in the treatment continuum provides a reasonable assessment of the major oncogenic drivers of the other dispersed metastases in a patient. We used whole exome sequencing (WES), array CGH and transcript profiling to characterize the composition of metastatic tumors and evaluated the consistency and diversity of molecular aberrations within and between individuals.
RESULTS
Genomic landscapes of metastatic prostate cancers
We resected 176 primary or metastatic tumors from 63 men with mCPRC at the time of rapid autopsy and determined mutations by WES (n = 141 tumors from 56 men), copy number aberrations by array CGH (n = 149 tumors from 60 men), and transcript expression by microarray hybridization (n = 171 tumors from 63 men). A small subset of tumors could not be evaluated by all three methods due to limited tumor quantity. Histological assessments of each metastasis determined that the vast majority (n = 156) were adenocarcinomas while 20 tumors from 2 men were of small cell neuroendocrine histology. All men were treated with androgen deprivation therapy (ADT) and following disease progression, most also received additional treatments including at least one additional AR pathway-targeted agent and at least one systemic chemotherapy, most commonly docetaxel (Supplementary Tables 1 and 2).
As reported in previous studies of mCRPC, we found recurrent aberrations involving the AR, ERG, TP53, RB1, SPOP, CHD1, ZBTB16/PLZF and regions of chromosome 8q gain (including the MYC locus) and 8p loss (Fig. 1a). We also identified recurrent mutations in FOXA2, which encodes a transcription factor expressed in tumors exhibiting neuroendocrine features (Supplementary Fig. 1a,b)22,23. Metastases from five men exhibited hypermutated genomes with complex structural aberrations in MSH2 and MSH6 mismatch repair genes24. All metastases and the primary tumors from these men were hypermutated, suggesting that mismatch repair deficiency occurred early in the genesis of these carcinomas.
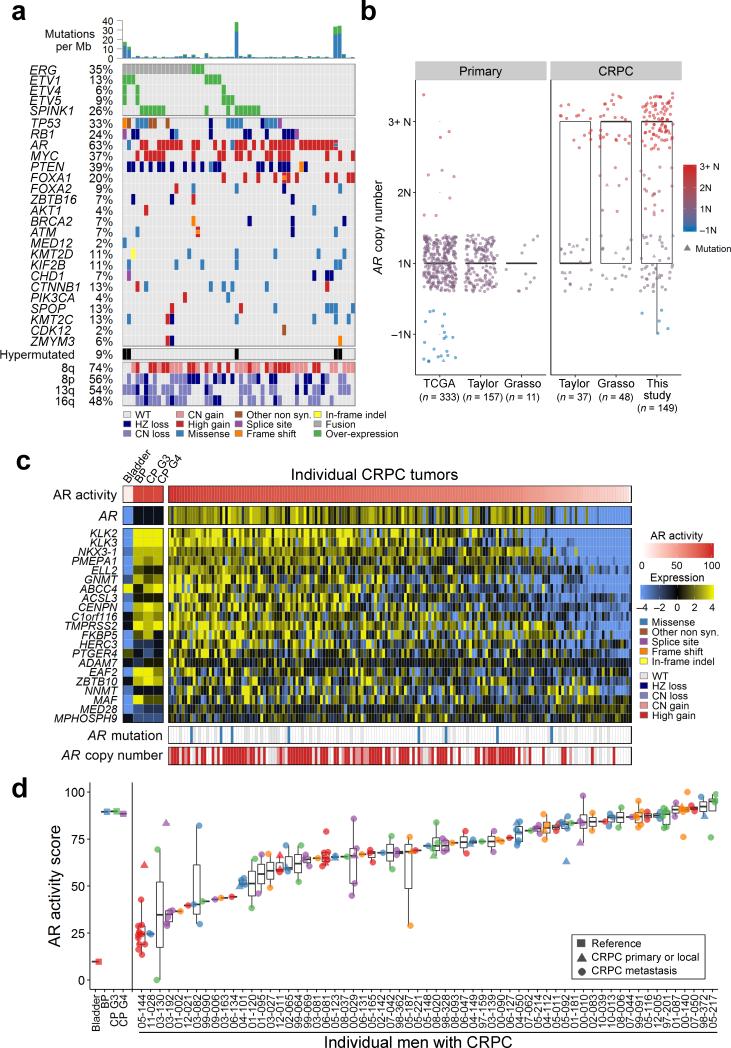
Figure 1. Integrated landscape of somatic aberrations and AR activity in mCRPC.
(a) Recurrent somatic molecular aberrations from an index metastasis from each of 54 men with mCRPC (columns) ascertained by transcript microarray, array CGH and WES on the same tumor. Columns represent the index tumor from each individual, and rows represent specific genes. Mutations per Mb are shown in the upper histogram. The frequency of the aberration in the cohort is shown as a percentage (%). Copy number variations (CNVs) common to mCRPC are shown in the lower matrix, with red representing gain and blue representing loss. Color legends including amplification, two-copy loss, one copy loss, copy neutral loss of heterozygosity (LOH), splice site mutation, frameshift mutation, missense mutation, in-frame indel, and gene fusion. Cases with more than one aberration in a gene are represented by split colors. Except for arm level CNVs, only high-level gain and loss are shown. The 54 patients with expression, CN, and mutations data are shown.
(b) AR copy number quantitation in three studies of untreated primary prostate cancer and three studies comprising mCRPC.
(c) AR activity as determined by transcript levels of 21 AR-regulated genes across CRPC tumors obtained at rapid autopsy. AR activity scores are also shown for microdissected cell populations of bladder urothelium (Bladder) benign prostate epithelium (BP), primary PC of Gleason grade 6 (CP G3) and primary PC of Gleason grade >6 (CP G4). AR somatic mutation and copy number status for each tumor sample are shown in the lower matrix.
(d) Consistency of the AR activity score for metastatic tumors within individuals and diversity of the AR activity score in tumors between individuals. The x-axis is the AR activity score and y-axis shows individual men. Scores for individual tumors are plotted over a range of 0–100% and colors are used to denote different patients. Circles are metastatic tumors (n = 149) and triangles are primary tumors (n = 22) resected at rapid autopsy.
Overall, the molecular landscapes of the lethal PCs exhibited a subset of aberrations observed recurrently across men, and also combinations of events unique to tumors originating within an individual. Excluding men with hyper-mutated tumors, the average number of protein coding mutations averaged 44 events per tumor, and the overall percentage of tumor genomes affected by structural changes, termed the DNA copy number alteration (CNA) burden25, averaged 38%, though there were wide ranges in these metrics. Of interest most tumors from a given man exhibited similar numbers of mutations and CNA burden, and there were no differences in overall mutation abundance, or aberrations in any specific genes that associated with specific visceral or bone metastatic sites (Supplementary Fig. 1c-g).
AR Activity is Maintained in the Majority of CRPC Metastases
Amplification or mutation of the AR gene occurred in 63% of men (Fig. 1a). Such aberrations were extremely rare in untreated primary PCs (Fisher's exact test; P < 0.0001) (Fig. 1b). Using a gene expression signature of AR transcriptional output26, we found that 88% of patients had tumors with robust AR activity, despite extensive prior treatment to suppress AR function (Fig. 1c). To evaluate this more fully, we assessed AR expression and AR output across multiple metastases from individual men. AR activity scores (Fig. 1d) and AR expression (Supplementary Fig. 2a) diverged across individuals, but were highly concordant when comparing metastasis within individuals. AR expression was positively associated with AR locus amplification (p ≤ 0.05) but did not differ between lymph node, visceral or bone metastatic sites (Supplementary Fig. 2b,c). AR genomic aberrations were generally concordant within individuals: all tumors were consistent in having no AR aberrations in 8 men, and for 24 men all tumors were found to have AR copy gain or mutation. In 9 men there was discordance with one or more tumors exhibiting an AR copy gain or mutation and one or more tumors with no aberration. We identified one man with evidence of convergent evolution where several metastases had AR mutation and other metastases had AR amplification, each potentially contributing to treatment resistance. In two men with small cell carcinomas confirmed by histology and immunohistochemical stains showing chromogranin expression, high neuroendocrine (NE) and low AR activity signatures were concordant across all metastases (Supplementary Fig. 2d).
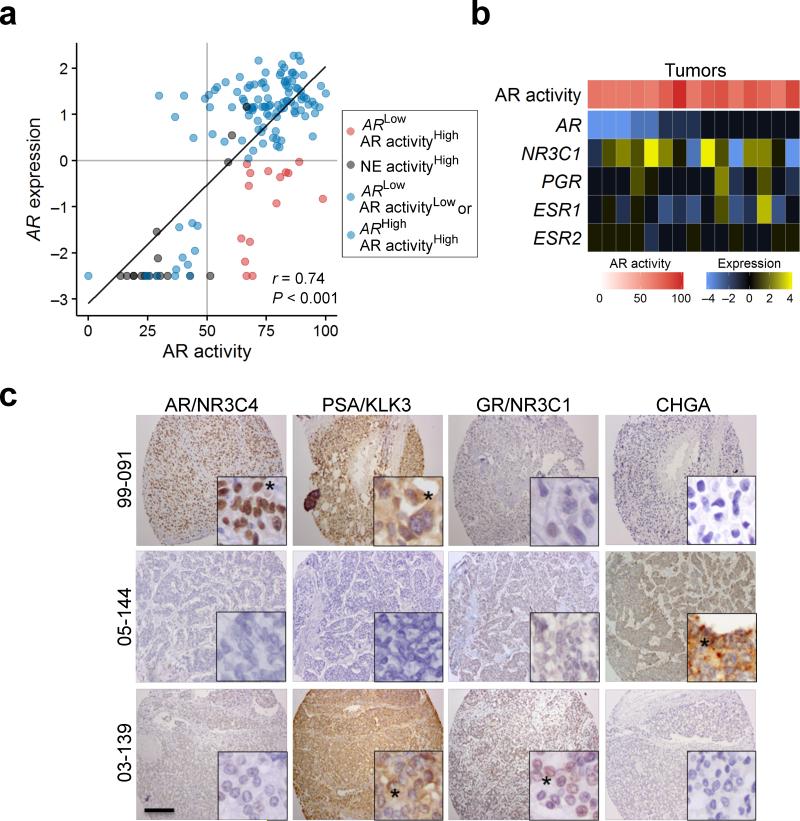
Though AR expression was usually associated with AR activity, we identified outliers with high AR activity but with absent or limited expression of the AR itself (Fig. 2a). Previous reports indicate that other nuclear hormone receptors such as the glucocorticoid receptor (GR;NR3C1), progesterone receptor (PGR), or estrogen receptor (ESR1) could activate canonical AR target genes27-30. In a small minority of tumors, GR;NR3C1 and ESR1 were expressed in the absence of AR (Fig. 2b,c), and thus could account for the maintenance of at least a component of the AR transcriptome.
Figure 2. Relationships between AR activity and the expression of AR and other nuclear hormone receptors.
(a) The relationship between AR transcript expression plotted as mean centered log2 ratio to the AR activity score for each tumor. Blue points represent adenocarcinomas where AR activity levels correlate with AR level; black points are neuroendocrine tumors; red circles are tumors with high AR activity and relatively low AR expression. There was a positive overall correlation between AR transcript levels and AR activity score (r = 0.74, P < 0.001) using Pearson's correlation coefficient,
(b) Transcript levels of nuclear hormone receptors for the 15 tumors with high AR activity and low AR expression. NR3C1, glucocorticoid receptor; PGR, progesterone receptor; ESR1, estrogen receptor alpha, ESR2, estrogen receptor beta.
(c) Immunohistochemical assessment of AR (NR3C4), GR (NR3C1), PSA (KLK3) and chromogranin (CHGA) protein in mCRPC tumors from three men with different AR expression and AR activity relationships: 99-091 expresses AR and the AR regulated protein PSA while 03-139 lacks AR expression but expresses GR (NR3C1) and PSA (KLK3). Black asterisk indicates cells with positive staining. Black scale bar is 200 micrometers.
Limited genomic diversity within patients
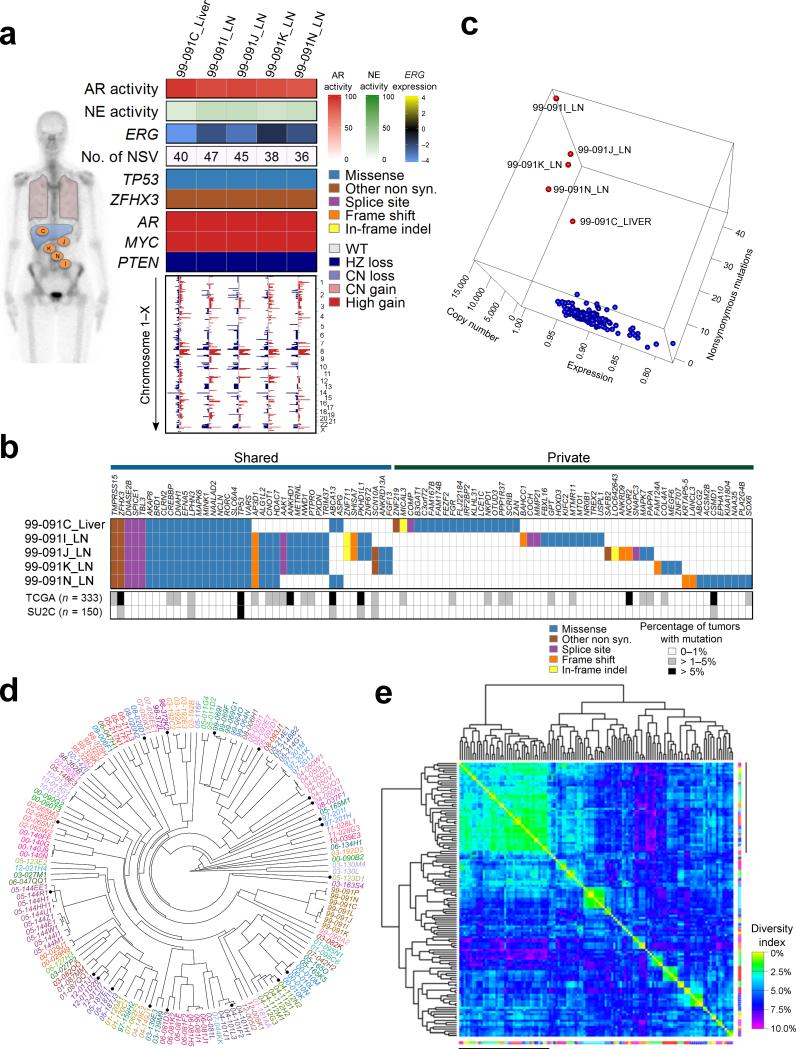
The substantial concordance of AR- or NE-associated gene expression across metastases within an individual patient prompted a further assessment of intra-individual molecular diversity. Presumptive driver aberrations identified in an index tumor from each man were generally shared in all tumors. For example, patient 99-091 had AR copy gain, high AR activity, TP53 mutation, and lacked an ERG rearrangement (Fig. 3a). The other 4 evaluable tumors from 99-091 shared these and other features. CNAs were also highly concordant (Fig. 3a). At the level of mutations, a subset of non-synonymous single nucleotide variants (NSVs) were shared, though each tumor had private mutations, of which few were predicted to be deleterious (Fig. 3b). To further assess the potential for these metastasis-private NSVs to influence pathogenesis, we examined WES data from 333 primary PCs from TCGA31 and 150 metastatic CRPC tumors from the PCF/SU2C study20. Of 51 genes with NSVs exclusive to an individual metastasis from patient 99-091, only two, CSMD1 and NCOR2, were altered at frequencies exceeding 5% in either the TCGA or SU2C tumors. Similar results were observed across multiple tumors from other men (Supplementary Fig. 3,4). Collectively, these data suggest that the vast majority of mutations unique to an individual tumor are not drivers of tumor behavior.
Figure 3. Molecular aberrations are shared between metastasis within individuals with mCRPC.
(a) Gene expression pathways and putative somatic driver aberrations are consistent across metastasis within an individual. Shown are selected features from five metastatic tumors from patient 99-091. Activity scores comprise transcripts of genes regulated by the AR (AR activity score), genes expressed in neuroendocrine carcinoma (NE activity score). ERG transcript levels are relative to the mean-centered ratio for the cohort. NSV is the number of somatic nonsynonymous nucleotide variants. Genome wide copy number losses (blue) and gains (red) are shown for each tumor ordered by chromosome.
(b) Distribution of all 91 nonsynonymous point mutations and indels identified in the five tumors from patient 99-091. The table indicates the presence of a mutation (blue/tan/purple/orange/yellow) or its absence (white) in each tumor. The grey scale bars indicate the frequency of a mutation in the specified gene in the TCGA (primary tumors) or SU2C (metastatic tumors) datasets.
(c) Relationships of tumors derived from a single patient (red points) relative to tumors from all other individuals (blue points) based on a calculated a sample similarity score that comprised single nucleotide alterations, copy number alterations, and gene expression using the 133 tumors with data on all three platforms.
(d) Unsupervised clustering of 149 tumors by correlation of genome-wide copy number variations. Tumors from the same individual are designated with the same color. Primary tumors are noted by a black circle.
(e) Heatmap of the percent of the total of 984 distinct copy number losses and gains identified in the entire cohort of tumors that differ between samples and sorted based on complete linkage dendrograms. Margin line denotes tumors with very low copy number aberration burden. The diversity index is defined as the proportion of corresponding aberrations that differed between samples.
Using an integrated analysis that incorporated CNAs, mutations, and gene expression, we found that all tumors from an individual closely congregated with other tumors from the same individual, and were distinct from tumors in other men (Fig. 3c; Supplementary Fig 3,4). Unsupervised clustering based on CNA profiles grouped tumors from the same individual together with near 100% accuracy (Fig. 3d). Of note, where the primary tumor removed at the time of rapid autopsy was analyzed, it grouped with the metastatic tumors from the same individual, rather than with primary tumors from different men.
We next evaluated the intra-individual consistency of a recurrent driver aberration, the TMPRSS2-ERG rearrangement, present in approximately 40% of primary PCs32. FISH using TMPRSS2 and ERG probes on 53 tumors from 13 men, found 100% concordance, all positive or all negative, between all metastasis within an individual (Supplementary Table 2). Using array CGH to identify chromosomal loss between the TMRPSS2 and ERG genes, 32 of 34 men including the 13 men with FISH results had 100% concordance across all metastases (94%) with divergence in two men. For one, 00-029, FISH demonstrated all tumors were positive for a TMPRSS2-ERG rearrangement.
Clustering tumors based on expression profiles also grouped tumors by patient of origin, excepting bone metastasis (Supplementary Fig. 5a). This may reflect inherent tumor biology, microenvironment effects, the overall lower tumor cell content of bone metastasis, or technical factors associated with processing for transcript quantitation. While we and others have reported differences in the expression of specific genes between bone and visceral sites33, we did not identify mutations or CNAs associated with metastasis location.
We next determined the overall molecular similarity and diversity of tumors using a statistical framework. We focused on the subset of 38 patients where multiple tumors per individual were successfully sequenced. Of the 123 tumors from this cohort, 5389 genes harbored a somatic NSV of which 2843 were from hypermutated tumors, leaving 2546 NSVs derived from 112 tumors from 34 patients. Collectively, there were 984 regions with defined CNAs in these tumors. We compared the CNAs in each tumor with every other tumor and determined that the median percent of corresponding aberrations that differed between tumors from the same individual was 6.7% compared to 22.1% from different individuals. The median inter- versus intra-individual tumor difference was 15.7% (95% CI 15.5-15.9%; P < 0.001; 2-sided 2-sample Wilcoxon rank sum test). Heatmaps with complete linkage dendrograms using copy number aberrations (Fig. 3e) or mutations (Supplementary Fig. 5b) clustered tumors according to patient of origin.
Cell cycle progression inversely correlates with AR activity
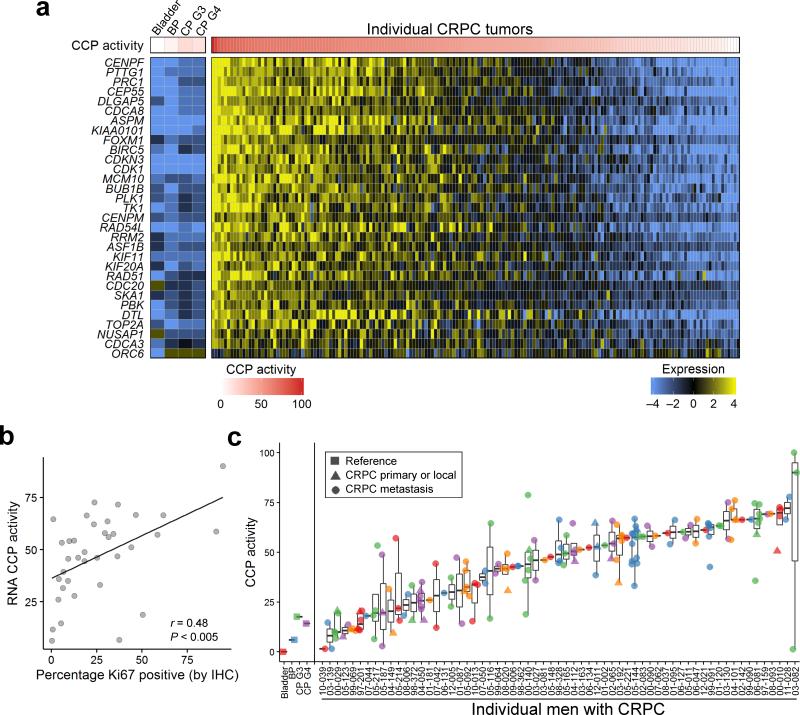
The concordance of AR activity or NE activity in tumors derived from the same patient prompted further analyses to determine if a relationship existed between these programs and a key metric of treatment resistance: tumor cell proliferation. We evaluated a 31 gene signature of cell cycle progression (CCP) shown to associate with PC mortality when assessed in primary tumors34. A wide-range of CCP activity was evident across the CRPC tumors (Fig. 4a). We confirmed that CCP activity was associated with cell proliferation determined by Ki67 immunohistochemistry on the same tumor samples (r = 0.48; P < 0.005)(Fig. 4b and Supplementary Fig 5c). CCP activity was generally concordant between tumors within an individual compared to tumors from different individuals (Fig. 4c). There were modest positive associations between CCP activity and mutation burden (r = 0.21; P = 0.023) (Supplementary Fig. 5d,e).
Figure 4. Tumor cell cycle activity within and between patients.
(a) Cell cycle progression (CCP) scores based on the expression of 31 cell cycle-associated transcripts vary across tumors. The genes comprising the CCP score are shown with expression levels colored to reflect high (yellow) or low (blue) transcript abundance. Columns represent individual tumors excepting Bladder; BP, benign prostate; CP3, primary PC with Gleason grade 6; CP4, primary PC with Gleason pattern ≥ 7, which are composites of 10 to 20 samples. The 171 CRPC tumors from 63 patients with expression data are shown.
(b) Correlation between CCP score and Ki67 IHC (r = 0.48, P < 0.005) in 36 tumors with matching protein and RNA expression data.
(c) Cell cycle progression scores (y-axis) for each of 171 tumors grouped by patient (x-axis). BP, benign prostate epithelium; CP, primary prostate cancer; G, Gleason pattern.
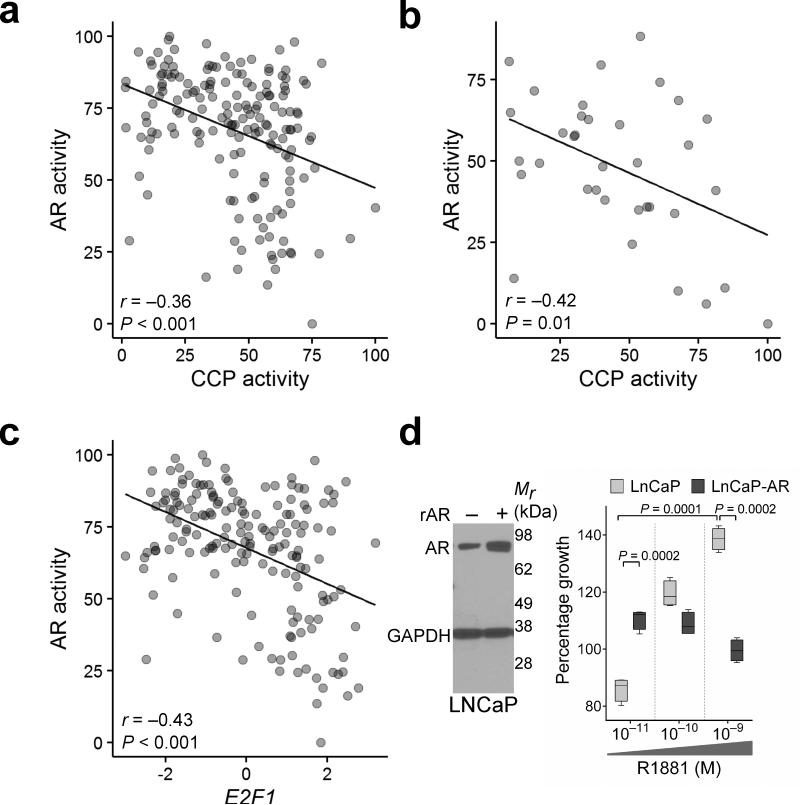
Contrary to expectations, AR activity was inversely associated with CCP activity (r = −0.33; P < 0.001) (Fig. 5a), a finding confirmed in an independent dataset comprising mCRPC tumors (Fig. 5b)19. AR activity was also inversely associated with E2F1, a key regulator of cell proliferation (P < 0.001) (Fig. 5c). To determine the functional relevance of this correlation, we compared the proliferation of LNCaPWT cells expressing endogenous AR versus LNCaPAR cells engineered to overexpress AR. LNCaPAR proliferation was significantly greater than LNCaPWT cells at very low AR ligand concentrations of 10−11 M R1881 (P = 0.002) (Fig. 5d) that approximate castrate serum testosterone concentrations achieved with androgen deprivation therapy35. However, at higher ligand levels (10−9 M), approximating concentrations in a non-castrate male, the growth of LNCaPWT cells substantially exceeded LNCaPAR cells whose proliferation was suppressed by the higher androgen concentration (Fig. 5d). Consistent with these results, after the introduction of AR into AR-null PC3 cells 10−9 M R1881 also significantly inhibited growth (P = 0.02) (Supplementary Fig. 5f). These findings support the concept that in addition to oncogenic roles, the AR may have activities that suppress aspects of tumor progression 36,37.
Figure 5. Cell cycle progression activity and E2F1 expression are inversely associated with AR activity.
(a) CCP scores are inversely associated with AR activity scores as determined by the expression levels of 21 AR-regulated genes in 171 tumors (r = −0.36, P < 0.001).
(b) Inverse association between CCP scores and AR activity scores was also observed in the Grasso et al. dataset of 35 CRPC tumors (r = −0.42, P = 0.01).
(c) AR activity is inversely associated with E2F1 transcript expression in 171 tumors (r = −0.43, P < 0.001).
(d) AR expression level influences cellular responses to the AR ligand R1881. Representative Western blot (n = 2) of AR and GAPDH protein levels in wild type LNCaP cells and LNCaP-AR cells engineered to overexpress AR (rAR) (left panel). LNCaP and LNCaP-AR cell growth measured 72 h after exposure to the indicated R1881 concentration, n = 4 in each group (right panel).
Cell cycle progression correlates with FA gene expression
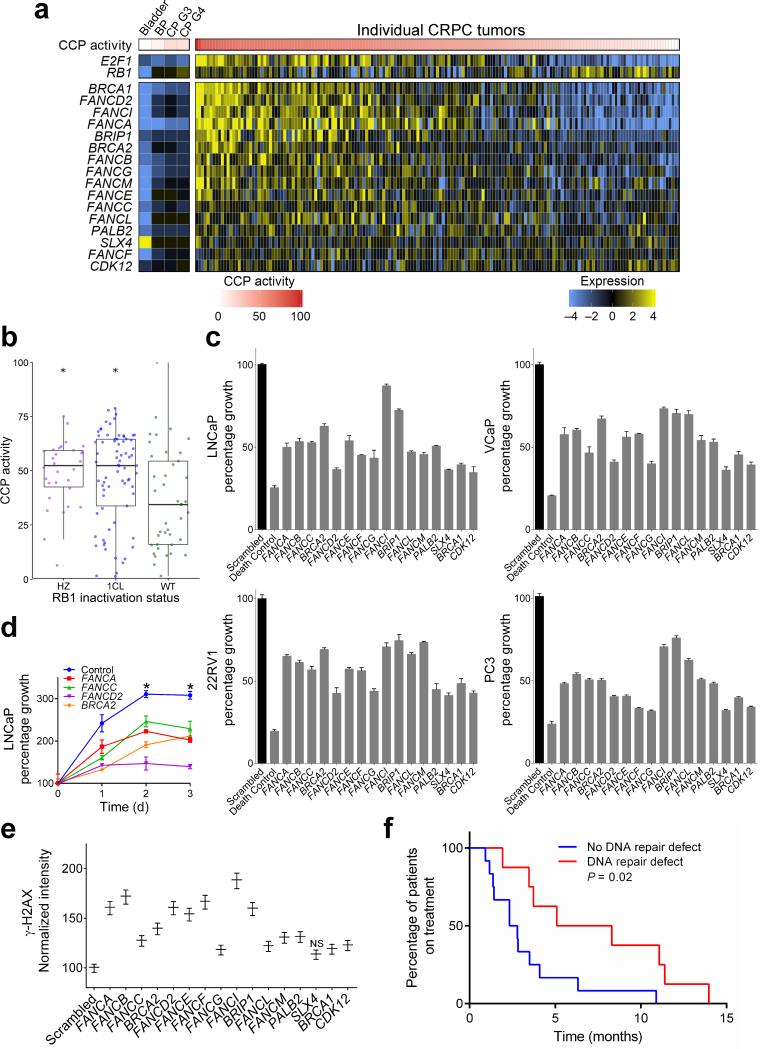
We next sought to identify genes with expression levels positively associated with elevated cell proliferation. We computed genome-wide correlations of CCP score to each gene and found that elevated expression of Fanconi Anemia (FA) complex components including FANCA, FANCI, FANCD2, BRCA1 and BRCA2 associated with those tumors exhibiting high CCP activity (Fig. 6a). Genes comprising the FA pathway were expressed at low levels in untreated primary PCs, which also had low CCP scores (Fig. 6a). For several FA genes, including FANCA, BRCA2 and PALB2 we found significant relationships between copy number and transcript levels though most tumors did not have alterations in genes encoding FA components (Supplementary Fig 6a-c). A subset of FA genes are positively regulated by E2F transcription factors, primarily E2F138,39. We found a significant relationship between high CCP activity and loss of the retinoblastoma (RB1) tumor suppressor locus (P < 0.01) (Fig 6b), which also associated with E2F1 expression and remained significant after removing the highly proliferative NE metastases40. Tumors with high E2F1 expression also had elevated CCP scores (r = 0.8; P < 0.001) and elevated activity of a signature comprising 15 FA pathway genes (r = 0.78; P < 0.001) (Supplementary Fig 6d-g).
Figure 6. Expression levels of Fanconi Anemia complex genes are associated with cell cycle progression and E2F1 expression.
(a) CCP scores for each tumor are ordered from high (left) to low (right). Corresponding Fanconi Anemia complex genes expressed in each tumor are colored to reflect high (yellow) or low (blue) transcript abundance. Columns represent individual tumors excepting Bladder; BP, benign prostate; CP G3, primary PC with Gleason pattern 3; CP G4, primary PC with Gleason pattern 4, which are composites of 10 to 20 samples. The 171 CRPC tumors from 63 patients with expression data are shown.
(b) Higher cell cycle progression score is associated with heterozygous and homozygous RB1 inactivation by copy loss and/or mutation (asterisk indicates P < 0.01 by pairwise t-test to WT group). HZ, homozygous loss; 1CL, heterozygous loss, WT, wild-type.
(c) Suppression of individual Fanconi anemia complex genes reduces the proliferation of multiple prostate cancer cell lines. Cell numbers were measured 5 d after introducing siRNAs targeting the specified genes relative to scrambled control siRNAs (n = 3 biological replicates per gene). All gene knockdowns produced a significant reduction in growth compared to scrambled control (2 sample t-test P < 0.05).
(d) Time course of LNCaP growth following the suppression of individual Fanconi Anemia complex genes. Growth curves measured over 3 days after introducing siRNAs targeting the specified genes. Asterisk indicates a significant difference in percent growth compared to control for each time point using two-sample t-tests with P < 0.05).
(e) Assessment of DNA damage in LNCaP cells by γ–H2AX assays following the knockdown of individual FA genes by siRNA and exposure to 50μM carboplatin. All FA knockdowns produced significantly greater γ–H2AX compared to scrambled siRNA controls (P < 0.05) excepting SLX4.
(f) Kaplan-Meier plot comparing the duration of carboplatin treatment (months) for 21 men with metastatic CRPC with or without a germline or somatic alteration in genes involved in DNA repair: BRCA2, PALB2, or ATM (P = 0.02 by logrank test).
To determine if FA components can influence cell proliferation, we suppressed expression of individual FA genes using siRNAs in LNCaP, 22Rv1, VCaP and PC3 cells. Suppression of each FA gene reduced the proliferation of each cell line (Fig. 6c,d; Supplementary Fig 6h,i). Consistent with the known role of the FA complex in facilitating DNA repair, suppression of individual FA complex genes significantly increased phosphorylation of H2AX (γ–H2AX), a marker of DNA double strand breaks, in LNCaP cells following exposure to the genotoxic drug carboplatin (Fig 6e). Of the men comprising this rapid autopsy cohort, 20 received carboplatin chemotherapy during their clinical course. Those with a somatic aberration in a DNA repair pathway gene, defined as a homozygous deleterious event in any FA pathway component, or heterozygous inactivating event in the ATM gene, events recently shown to associate with responses to the PARP inhibitor olaparib41, had significantly greater durations of response to carboplatin chemotherapy as determined by time on drug treatment (P = 0.02) (Fig. 6f).
DISCUSSION
Comprehensive molecular assessments of PC have identified numerous recurrent genomic and epigenomic aberrations in genes implicated in driving cancer development, of which a substantial fraction have clinical implications for directing treatment12,19,20,42-45. These findings provide rationale for tumor sequencing to identify facets of tumor biology that prioritize and individualize therapeutics. However, deep sequencing of tumor genomes has also identified mutational heterogeneity, particularly within primary tumors. Branching evolution and the coexistence of multiple cancer lineages occur in a spectrum of carcinomas which have implications with respect to accurately sampling tumors, and for evolutionary mechanisms that may rapidly promote treatment resistance11,12,46.
In the present study of mCRPC, we determined that constellations of genomic events were unique to a given patient from which a tumor was derived, demonstrating substantial inter-patient heterogeneity. In contrast, the molecular aberrations identified in multiple spatially-distinct metastases derived from a single individual and sampled at a single point in time were highly concordant, though a limited number of unique events specific to any particular tumor were evident of which very few represent known oncogenic drivers. We found that overall mutation burden, CNA burden, AR activity signature, and cell cycle output were generally concordant when comparing tumors within an individual. Our results provide further evidence for monoclonal seeding of metastatic sites, though our methods precluded assessments of their polyclonal composition or inter-mixing of metastases as reported by others15. It is interesting to consider that the ability of tumor clones from one site to seed other metastatic foci and intermix would result in a degree of homogenization, leading to the practical consequence that sampling one site has the potential to interrogate the extent of mutational heterogeneity present within an individual. Further studies are required to ascertain the influence of intra-tumoral heterogeneity with respect to treatment responses of tumors with variable frequencies of clones that possess druggable alterations.
Though the analysis of a single metastatic tumor site clearly does underestimate the total burden of molecular aberrations found in the totality of all metastases, the vast majority of drivers and actionable features are represented as common roots across all tumors or result from convergent evolution conferred by therapeutic pressure15, and most molecular differences between metastases do not appear to influence tumor phenotypes. Though there are exceptions, these findings lead to the practical conclusion that clinical decision-making based on a biopsy from a single metastatic site is reasonable. This assumption may not be relevant when sampling a primary tumor where greater molecular diversity appears to occur12, and must be tempered as only three men in our study were treated with the potent AR pathway antagonists abiraterone or enzalutamide that could induce diverse resistance mechanisms that include AR amplification, AR mutation, and expression of AR splice variants that may differ across metastases within an individual. In some situations, simply determining the activity status of a particular pathway will be sufficient for directing therapy, whereas in other instances the mechanism by which the pathway is activated, for example a specific AR ligand binding site mutation, will have treatment implications.
Our findings are supported by prior studies reporting substantially greater similarities in the genomic and epigenomic composition between metastases relative to the more expansive diversity found within regions of primary tumors10-12,45-49. In addition to the severe bottlenecks imposed by the metastatic process, the time scales involved in the evolutionary stages of cellular transformation to invasion and metastasis may underlie the disparity in molecular heterogeneity within primary tumors and between metastases. It has been estimated that decades of chronological time are required for cells with initiating events to acquire sufficient additional mutations that endow subclones with characteristics enabling invasion and metastatic spread10. In contrast, intervals between the acquisition of metastatic capability and the ability to grow in distant organs span only months to a few years10. Indeed, prostatic intraepithelial neoplasia and primary PCs are readily evident in men aged 30-40 years, decades before metastatic PC is commonly diagnosed50. These years of intra-organ tumor growth allow for substantial evolutionary diversity, clonal expansion, clonal mixing, and also for the genesis of completely independent primary tumors.
In the present study, an additional bottleneck limiting tumor diversity may be treatment pressures that likely eliminate sensitive tumor clones and allow the expansion of resistant clones. Extreme diversity across metastases would likely result in the rapid expansion of one or a few metastases and regression or stasis of others. Our finding that most metastatic sites within a given individual exhibited concordant proliferation rates suggests that there are limited differences across metastases with respect to treatment responses to androgen deprivation therapy or taxane chemotherapy. This finding may reflect the fact that most successful therapies in mPC are directed toward the AR which functions as a core lineage oncogene51 or else toward fundamental cellular processes involved in cell division such as microtubule dynamics6. In this context, the observation that FA pathway activity is associated with the subset of tumors with high proliferation rates, likely reflecting important roles in replication stress52, coupled with the high frequency of somatic events involving DNA repair proteins supports targeting this pathway in selected patients41.
ONLINE METHODS
Patient Enrollment and Tissue Acquisition
All procedures involving human subjects were approved by the Institutional Review Board (IRB) of the University of Washington and of the Fred Hutchinson Cancer Research Center. Samples were obtained from patients who died of metastatic CRPC and who signed written informed consent for a rapid autopsy performed within 6 hours of death, under the aegis of the Prostate Cancer Donor Program at the University of Washington53. The inclusion criteria for the Prostate Cancer Donor Program included a diagnosis of prostate cancer, informed consent to perform an autopsy and informed consent to use biospecimens for biomedical research. Visceral metastases were identified at a gross level. Biopsies of all bone sites, except ribs (a dyke cutter was used for rib biopsies) were obtained using a drill with an attached trephine (11 mm diameter coring drill bit). All specimens were either flash frozen and embedded in Optimal Cutting Temperature compound (OCT; Tissue-Tek) or formalin fixed and embedded in paraffin respectively (bone metastases were decalcified in 10% formic acid before paraffin embedding). This study included 176 localized and metastatic tumors from 63 men with CRPC and a matched benign tissue sample from each individual to facilitate the identification of somatic mutations. For the molecular studies, the investigators were blinded as to sample identities and samples were randomly assigned to exome sequencing runs and CGH analyses. Samples were included for the molecular studies if histological analysis estimated >50% neoplastic cells, DNA quantity ≥ 3 μg and the presence of high molecular weight DNA determined by gel electrophoresis.
Transcript Profiling
A total of 176 tumors from 63 patients were collected for profiling by expression microarray. For five tumors, RNA was of insufficient RNA quality or quantity leaving 171 tumors from 63 patients that were successfully profiled. A Leica CM3050S cryostat was used to cut 8 micron sections, which were collected on PEN Membrane Frame Slides (Life Technologies) and immediately fixed in 95% ethanol. Sections were briefly stained (5–10 seconds) with Hematoxylin solu tion according to Mayer (Sigma-Aldrich) then dehydrated in 100% ethanol. Laser capture microdissection was performed with an Arcturus Veritas instrument, using both UV cutting and infrared capture lasers, with CapSure® Macro LCM Caps (Life Technologies) collecting 5,000-10,000 tumor cells per sample. Digital photographs were taken of tissue sections before, during, and after LCM and assessed by a pathologist to confirm the tumor content. Following LCM, captured cells were lysed in Arcturus RNA Extraction Buffer. RNA was isolated using the Arcturus PicoPure RNA Isolation Kit and DNAse treated using the Qiagen RNase-Free DNase Set. The bone metastases were sampled using a 1 mm diameter tissue punch of the frozen OCT-embedded tumors in a −20°C cryostat. The sample was obtained from the region of the block where there was tumor based upon a section of an adjacent decalcified FFPE block. RNA was isolated from the tissue cores using the RNeasy Plus Micro Kit (Qiagen Inc.). Subsequently, total RNA from all tumors was amplified for two rounds using the Ambion MessageAmp aRNA Kit. Probe labeling and hybridization to Agilent 44K whole human genome expression oligonucleotide microarray slides (Agilent Technologies, Inc., Santa Clara, CA) was performed following the manufacturer's suggested protocols and fluorescent array images were collected using the Agilent DNA microarray scanner G2565BA. Agilent Feature Extraction software was used to grid and extract the data. Data was loess normalized within arrays (normexp background correction with offset 50) and quantile normalized between arrays in R using the Limma Bioconductor package by batch. Control probes were removed, duplicate probes averaged, and spots flagged by Agilent Feature Extraction software as being foreground feature nonuniformity or population outliers were assigned a value of “NA”. An additional normalization step was applied to remove systematic batch effects due to date of RNA-amplification by application of the ComBat function within the sva Bioconductor package to the log2 Cy3 signal intensities. Expression microarray data are deposited in the Gene Expression Omnibus database under the accession number GSE74685
Copy Number Profiling
A total of 176 tumors from 63 patients were collected for profiling by microarray comparative genomic hybridization (CGH). For 27 tumors, DNA was of insufficient quality or quantity leaving 149 tumors from 60 patients that were successfully profiled. Genomic DNA was isolated from OCT sections of localized and metastatic tissues with ≥50% tumor content, 1mm punches from frozen bone metastases as well as patient-matched normal tissues using the Qiagen QiaAmp DNA Micro kit incorporating RNAse-treatment. DNA quality was assessed by agarose gel electrophoresis. One microgram of high molecular weight genomic DNA from each sample was labeled by random priming using the Agilent Genomic DNA Enzymatic Labeling Kit (Cy3-dUTP). A pool of reference normal DNA (Promega) was labeled with Cy5-dUTP. Cy3 and Cy5 probes were combined and hybridized to Agilent 2×400K SurePrint G3 CGH Microarrays and washed following the manufacturer's specifications. Fluorescent array images were collected using the Agilent DNA microarray scanner G2505C and Agilent Feature Extraction software. Control probes were removed, duplicate probes averaged, and log2 ratios defined as gProcessed-Signal/rProcessedSignal. Recurrent germline copy number variations (CNVs) were identified across benign tissues uninvolved with cancer obtained from 56 patients where DNA of sufficient quality and quantity was obtained for this study, as well as the Grasso, et. al copy number dataset of 28 benign samples, and a list of CNV regions defined by the Broad Institute and markers within these regions were removed from further analysis. The log2 ratios were corrected for the GC content wave effect using 1Mb windows for genome GC content. Outliers were smoothed and the normalized log2 ratios were segmented using the Circular Binary Segmentation (CBS) algorithm implemented with the DNAcopy Bioconductor package using default parameters and undo.splits = “sdundo” (undo.sd=1). The mode of the histogram of segment means was set to zero. To determine significantly recurrent regions of SCNA, the GISTIC 2.0 version 6.2 GenePattern module was applied to the segmented data using amplifications and deletions thresholds of 0.2, join.segment.size of 4 probes, a copy-ratio cap of 2.5, arm-level peel-off enabled, broad length cutoff of 0.7 chromosome arms, and confidence level of 95%. CGH array data are deposited in the Gene Expression Omnibus database under the accession number GSE74685.
Clustering
Unsupervised clustering of expression and CN profiles was performed in R on the 5000 most variable genes for each dataset using Pearson correlation as the distance metric and average-linkage hierarchical clustering. Clusters were visualized using the ape R package. Circular plots of expression and CN profiles by chromosome location were created with the OmicCircos R package version 1.8.0.
Statistical Analyses
Pearson's correlation coefficient was used to study the relationships between variables shown in scatterplots using the cor.test function in R. RB1 inactivation status was determined as homozygous if there was a two copy loss or 1 copy loss accompanied by mutation. Single copy loss or mutation only was categorized as heterozygous loss. The CCP activity in each of these groups was compared to the RB1 wild-type group using both pairwise t-tests and pairwise Wilcoxon rank sum tests, using the pairwise.t.test and pairwise.wilcox.test functions in R. The pairwise t-test and pairwise Wilcoxon rank sum test were also used to compare the number of nonsynonymous mutations and percent genome altered in different metastatic tissue sites, and all copy number vs. expression comparisons. Assessments of the proportions of bone or soft tissue metastases with homozygous PTEN cn loss or high gain of AR were performed by Fisher's Exact test using GraphPad Prism version 6.02, GraphPad Software, La Jolla California USA. Duration on carboplatin treatment was compared for percentages of patients harboring a DNA repair defect with those who did not by Kaplan-Meier plot with logrank test using GraphPad Prism version 6.02. Patients with a DNA repair defect were defined as those with any combination of the following criteria: homozygous or heterozygous loss in ATM by copy number loss and/or mutation, homozygous loss of one of the following 15 Fanconi anemia associated genes (FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, BRIP1, FANCL, FANCM, PALB2, SLX4, BRCA1) or germline mutation in BRCA2 and/or ATM.
Exome sequencing
A total of 176 tumors from 63 patients were collected for whole exome sequencing. For 35 tumors, DNA was of insufficient quality or quantity leaving 141 tumors from 56 patients that were successfully sequenced using the Nimblegen V2 or V3 platforms as previously described18 with the following modification: in a subset of tumors, individually barcoded libraries were pooled in pairs prior to capture. Genomic DNA was isolated as described above. Sequencing was per formed using the Illumina Hiseq 2000 with either 50 bp or 100 bp paired end sequences. Reads were mapped to the human reference genome sequence (hg19) with bwa v0.7.154. After remov al of PCR duplicate pairs we performed local realignment around indels using the Genome Analysis Toolkit (GATK)47. We subsequently called mutations using the Mutect software package with the following parameters: “--minimum_normal_allele_fraction 0.02 -- max_alt_alleles_in_normal_count 12 --intervals poscont.list --fraction_contamination 0.02”. To remove common polymorphisms and enrich for likely somatic mutations, we imposed a number of additional requirements, including requiring variants to be observed in at least 10% of reads at a position and removing variants present within a modified database of SNPs (dbSNP v137) that had first been stripped of all COSMIC variants. We investigated mutations for significance with MutSigCV using standard parameters, and inspected mutations in the top 50 significant genes manually using the Integrated Genomics Viewer (IGV) to remove sequence artifacts. The data are available for visualization and analysis in the cBioPortal for Cancer Genomics at: http://www.cbioportal.org/study.do?cancer_study_id=prad_fhcrc”.
Immunohistochemistry
One hundred and fifty-five metastatic PCa tissues (including 73 soft tissue metastases and 82 bone metastases) from a total of 50 autopsy patients (up to 4 sites per patient) were embedded in paraffin. A human tissue microarray (TMA) was constructed with duplicate 1 mm cores using the aforementioned tissues. Due to insufficient tissue, not all tumors subjected to molecular profiling were included on the TMA. Formalin-fixed paraffin-embedded TMA sections (5 μm) were deparaffinized and rehydrated with 3 changes of xylene and graded ethanol. Antigen retrieval was performed with 10 mM citrate buffer (pH 6.0) in a high pressure cooker for 30 min. Endogenous peroxidase and avidin/biotin were blocked respectively (Vector Laboratories Inc.). Sections were then blocked with 5% normal goat-horse-chicken serum at room temperature for 1h, and incubated with primary antibody at 4°C overnight. After washing 3 times with 1X PBS, slides were incubated with biotinylated secondary antibody (Vector Laboratories Inc.), followed by ABC reagent (Vector Laboratories Inc.) and stable DAB (Invitrogen Corp.). All sections were lightly counterstained with hematoxylin and mounted with Cytoseal XYL (Richard Allan Scientific). Mouse or rabbit IgG were used as appropriate for negative controls. Antibodies sources and dilutions: ER beta was from Biogenex cat # PU385-UP and used at 1:100; ER alpha was from Abcam, cat# ab173467, used at 1:500; GR/NR3C1 was from Santa Cruz, cat# sc-8992, used at 1:75; Androgen Receptor was from Biogenex, cat# F39.4.1, used at 1:60; Chromogranin A was from Dako, cat# DAK-A3, used at 1:100, and Ki-67 was from Dako, cat# M7240, used at 1:100.
Assessments of Inter- and Intra-Individual Tumor Molecular Heterogeneity
Mutation and copy number aberrations for 141 tumor samples from 56 patients that had successful sequencing and CGH analyses performed were initially examined separately. We identified 5389 mutations across tumor samples. Based on visualization of individual profiles, we flagged 5 patients as hypermutated; these patients together contributed 2843 (53%) unique mutations. To summarize heterogeneity across samples from non-hypermutated patients, we removed these patients and all mutations that were present only for these patients. This left 2546 mutations from 129 tumor samples from 51 patients. Next, we identified 984 regions with defined chromosome copy gain or loss across tumor samples from all 56 patients. We collapsed copy numbers with 1 or 2 gains into a single category (and similarly for 1 or 2 losses). The resulting mutation and copy number aberrations were then combined to create aggregate tumor samples. Finally, to compare heterogeneity across samples from the same individual vs between individuals, we restricted attention to individuals with at least 2 samples. This left a total of 3530 mutations or copy number aberrations from 133 tumors from 39 patients.
We quantified heterogeneity between two tumor samples using a “diversity index,” defined as the proportion of corresponding aberrations that differed between samples. For statistical inference, we randomly drew 10,000 pairs of tumor samples from the same individual and, separately, drew another 10,000 random pairs of tumor samples from different patients. We summarized distributions of the resulting diversity index using kernel density estimates and tested for equality of distributions using a 2-sided 2-sample Wilcoxon rank sum test.
As an alternative visualization of heterogeneity across tumor samples, we created heatmaps of the same diversity index calculated for all possible pairs of tumor samples. The heatmaps were sorted based on results of a complete linkage cluster analysis using the diversity index as the Euclidean distance between samples.
For the 3D plot of tumor molecular relationships (e.g. Figure 2C), we calculated a sample similarity score between the primary tumor of each patient and all other tumors using (a) single nucleotide alterations, (b) copy number alterations, and (c) gene expression. Similarity in single nucleotide alterations was measured by the number of impacted genes that are shared between two tumors. Similarity in copy number alterations was calculated as the inner (dot) product of the vectors of GISTIC-score per-gene for each tumor. Similarity in expression was measured by Pearson correlation across all genes.
We generated one plot for each patient. Each coordinate axis in a plot represents one of the above three similarity measures. Plots were interactively rotated in 3D to obtain the clearest viewing angle. In the example plot of Figure 2C, the vertex at the top marks the location of an index tumor. The red plot points mark samples from the same patient as the index tumor. The blue plot points mark all other tumor samples.
Gene Manipulation and Cell Growth Assessments
LNCaP, PC3, VCaP and 22Rv1 cells were obtained from ATCC and used within six passages of growth after receipt or validated using STR (DNA Diagnostics) and confirmed to be mycoplasma negative. Cell lines were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS). siRNAs were obtained from either Sigma (FANC F, L, M and BRIP1) or Qiagen (all others and the Allstars negative control). Target sequences are shown below. Cells were transfected with the siRNAs using Liopofectamine RNAiMAX (Invitrogen) following the manufacturer's protocol. Briefly, for gene expression analyses, 250,000 cells were plated in a 6 well dish with 50nM RNAi and 5μl of RNAiMAX in Optimem. After 8 hours the media was replaced with RPMI 1640 with 10%FBS. After 5 days RNA was extracted using the RNeasy kit (Qiagen). cDNA was made using SuperScript II (Invitrogen) and oligo dT primers. Gene expression was determined using qRT-PCR. Each assay comprised three biological replicates. Reactions were performed using Power SybrGreen on an Applied Biosystems (Foster City, CA) 7900HT Fast Real-Time PCR System. Cycling conditions included a 10 minute incubation at 95°C followed by 40 cycles of 95°C for 15 seconds with a one minute extension at 60°C. Disassociation curves were generated using the machines default conditions. Expression was normalized to RPL13A expression. Gene specific primer sequences are shown below.
Growth assays were performed using CellTiter 96 Aqueous One Solution (Promega). For each assay/condition, 10,000 cells per well were transfected as above using 10nM RNAi and 0.2μl of RNAiMAX in 100ul of Optimem. After 8 hours 100μl of RPMI 1640 with 20% FBS was added to each well. Each day, 10μl of CellTiter 96 Aqueous One Solution was added to designated wells, incubated at 37°C for 2 hours and the absorbance was measured at 490nM.
LNCaP-AR cells were the kind gift of Dr. Charles Sawyers (Memorial Sloan Kettering Cancer Center). PC3-AR were created by cloning the androgen receptor from a normal prostate cDNA library obtained from Clontech (Mountain View, CA) into the pBABE retroviral expression vector. After virus production and infection cells were selected using puromycin and clones selected and screened for AR expression. AR expression was determined by immunoblot using an antibody against AR (GTX62599, Genetex, Irvine, CA).
AR expressing cells were assayed for their response to varying concentrations of the synthetic androgen R1881. Five thousand cells were plated in charcoal stripped growth media for one day prior to exposure to androgen. Cell growth was determined as above using CellTiter 96 Aqueous One Solution. Percent cell growth was normalized to day 0 or to vector controls. Percent growth in each group was compared to the control using two-sample t-tests. Variances in each group were compared and not found to be significantly different by F test.
DNA Damage and γ-H2AX Assessment
Cells were transfected in 96 well plates with siRNAs as described above. Three days after transfection half of the plates were dosed with 50 μM of Carboplatin (Sigma) or vehicle. After 16 hours of exposure to Carboplatin or vehicle, the level of phosphorylated γ-H2AX was determined using the H2A.X (pSer139) Human In-Cell ELISA Kit (Abcam). The manufactures procedures were followed precisely. PCR primer sequences and siRNA sequences are in the Supplemental Tables 5 and 6.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all of the patients and their families that participated in this study.
We thank A. Mckenna and other members of the Nelson, Shendure and Vessella laboratories for helpful advice and assistance. We thank T. Taniguchi for helpful discussions and the provision of reagents. We thank M. Roudier, J. Noteboom, J. Kho and all other members of the tissue acquisition necropsy team for their hard work and dedication.
This work was supported by National Institutes of Health awards: Pacific Northwest Prostate Cancer SPORE grant P50 CA097186 (P.S.N., B.M., E.M., L.T., R.V., C.M.), P01 CA163227 (P.S.N., R.V. C.M.), P01 CA85859 (R.V., C.M., M.F, P.S.N.), Department of Defense awards PC140519 and PC140794 (P.S.N, R.D., I.C., R.C., N.S.) a Fred Hutchinson Cancer Research Center Solid Tumor Translational Research award (P.S.N., H.B., C.P., B.M.), and the Prostate Cancer Foundation. C.P. J.S. and N.S. are supported by Prostate Cancer Foundation Young Investigator Awards.
Footnotes
AUTHOR CONTRIBUTIONS
A.K., T.W., and I.C. coordinated overall sequencing and bioinformatics analysis.
I.C. and N.S. coordinated data deposition, assembly, figures, and tables.
R.G., R.E., I.C., and H.B. performed statistical analyses.
A.K., I.C., R.C., R.F.D., T.W., C.P., N.DS., and J.S. performed sequencing and analyses.
L.D.T., M.F., and X.Z. coordinated central pathology review, FISH, and IHC studies.
J.M.L. conducted gene manipulation studies and growth assays.
R.L.V., L.G.B. and C.M. coordinated clinical enrollment and tissue procurement
P.H.L., C.H, E.Y.Y, P.S.N., and B.M. enrolled patients and provided clinical insights.
P.S.N. I.C., J.S. and A.K. wrote the manuscript which all authors reviewed.
ACCESSION CODES
Molecular profiling data are deposited in the Gene Expression Omnibus database under the accession number GSE74685
COMPETING FINANCIAL INTERESTS.
The authors declare that they have no competing financial interests of relevance to this manuscript.
REFERENCES
- 1.National Research Council (U.S.) Toward precision medicine : building a knowledge network for biomedical research and a new taxonomy of disease. National Academies Press; Washington, D.C.: 2011. Committee on A Framework for Developing a New Taxonomy of Disease. [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RG, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tannock IF, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 7.de Bono JS, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scher HI, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 10.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper CS, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47:367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009 doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haffner MC, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–4922. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 17.Huggins C, Hodges CV. Studies on prostate cancer 1: the effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Research. 1941;1:293–297. [Google Scholar]
- 18.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson D, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosquera JM, et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia. 2013;15:1–10. doi: 10.1593/neo.121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi J, et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18:23–38. doi: 10.1016/j.ccr.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, et al. Mash1 expression is induced in neuroendocrine prostate cancer upon the loss of Foxa2. Prostate. 2013;73:582–589. doi: 10.1002/pros.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard CC, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun. 2014;5:4988. doi: 10.1038/ncomms5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hieronymus H, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A. 2014;111:11139–11144. doi: 10.1073/pnas.1411446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahu B, et al. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res. 2013;73:1570–1580. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- 28.Arora VK, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155:1309–1322. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai J, Myers SA, Lawrence MG, Odorico DM, Clements JA. Direct progesterone receptor and indirect androgen receptor interactions with the kallikrein-related peptidase 4 gene promoter in breast and prostate cancer. Mol Cancer Res. 2009;7:129–141. doi: 10.1158/1541-7786.MCR-08-0218. [DOI] [PubMed] [Google Scholar]
- 30.Setlur SR, et al. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Electronic address, s.c.m.o. Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 33.Morrissey C, et al. Differential expression of angiogenesis associated genes in prostate cancer bone, liver and lymph node metastases. Clin Exp Metastasis. 2008;25:377–388. doi: 10.1007/s10585-007-9116-4. [DOI] [PubMed] [Google Scholar]
- 34.Cuzick J, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedelaar JP, Isaacs JT. Tissue culture media supplemented with 10% fetal calf serum contains a castrate level of testosterone. Prostate. 2009;69:1724–1729. doi: 10.1002/pros.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokontis JM, Hay N, Liao S. Progression of LNCaP prostate tumor cells during androgen deprivation: hormone-independent growth, repression of proliferation by androgen, and role for p27Kip1 in androgen-induced cell cycle arrest. Mol Endocrinol. 1998;12:941–953. doi: 10.1210/mend.12.7.0136. [DOI] [PubMed] [Google Scholar]
- 37.Schweizer MT, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med. 2015;7:269ra262. doi: 10.1126/scitranslmed.3010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tategu M, Arauchi T, Tanaka R, Nakagawa H, Yoshida K. Systems biologybased identification of crosstalk between E2F transcription factors and the Fanconi anemia pathway. Gene Regul Syst Bio. 2007;1:1–8. [PMC free article] [PubMed] [Google Scholar]
- 39.Meier D, Schindler D. Fanconi anemia core complex gene promoters harbor conserved transcription regulatory elements. PLoS One. 2011;6:e22911. doi: 10.1371/journal.pone.0022911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan HL, et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res. 2014;20:890–903. doi: 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mateo J, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindberg J, et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol. 2013;63:702–708. doi: 10.1016/j.eururo.2012.11.053. [DOI] [PubMed] [Google Scholar]
- 43.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature Genetics. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robbins CM, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aryee MJ, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5:169ra110. doi: 10.1126/scitranslmed.3005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakr WA, et al. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20-69: an autopsy study of 249 cases. In-vivo. 1994;8:439–443. [PubMed] [Google Scholar]
- 51.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 52.Jones MJ, Huang TT. The Fanconi anemia pathway in replication stress and DNA crosslink repair. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roudier MP, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34:646–653. doi: 10.1016/s0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 54.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.