Abstract
The size and shape of dendrite arbors are defining features of neurons and critical determinants of neuronal function. The molecular mechanisms establishing arborization patterns during development are not well understood, though properly regulated microtubule (MT) dynamics and polarity are essential. We previously found that FoxO regulates axonal MTs, raising the question of whether it also regulates dendritic MTs and morphology. Here we demonstrate that FoxO promotes dendrite branching in all classes of Drosophila dendritic arborization (da) neurons. FoxO is required both for initiating growth of new branches and for maintaining existing branches. To elucidate FoxO function, we characterized MT organization in both foxO null and overexpressing neurons. We find that FoxO directs MT organization and dynamics in dendrites. Moreover, it is both necessary and sufficient for anterograde MT polymerization, which is known to promote dendrite branching. Lastly, FoxO promotes proper larval nociception, indicating a functional consequence of impaired da neuron morphology in foxO mutants. Together, our results indicate that FoxO regulates dendrite structure and function and suggest that FoxO-mediated pathways control MT dynamics and polarity.
Keywords: FoxO, dendrites, Drosophila, EB1, microtubules, microtubule polarity
Introduction
Dendrite architecture is established during development and lays the groundwork for neuronal connectivity and function. Dendrites acquire simple or complex morphologies depending on the degree of branching and growth of their arbors. Regulation of dendrite branching and growth requires accurate integration of cell-intrinsic and cell-extrinsic factors. On the cell-intrinsic side, cohorts of transcription factors direct the expression of downstream effector molecules that together impart cell-type specific morphologies. While a number of transcription factors have been implicated in dendrite morphogenesis, the remarkable morphological diversity of dendrite arbors suggests that others remain to be identified.
Dendritic arborization (da) neurons are sensory neurons that innervate the larval epidermis and are grouped into four classes (classes I–IV) based on the size and shape of their dendrite arbors (Corty et al., 2009; Grueber et al., 2002). Work in this system has detailed cytoskeletal characteristics that distinguish dendrite morphologies of classes of da neurons (Grueber et al., 2003; Jinushi-Nakao et al., 2007). For instance, simple class I arbors and complex class IV arbors differ in the extent to which their dendrite branches are populated by stable microtubules (MTs). The MT-associated protein (MAP) Futsch/MAP1B, binds and stabilizes MTs (Halpain and Dehmelt, 2006; Hummel et al., 2000; Roos et al., 2000). In class I neurons, many branches contain Futsch, while in class IV neurons, Futsch is confined primarily to main branches (Grueber et al., 2002; Jinushi-Nakao et al., 2007). Moreover, loss of Futsch increases branching of class I neurons (Yalgin et al., 2015). Together these data suggest that dynamic MTs are particularly critical in generating the highly branched dendrite arbors in class IV neurons.
The stereotyped and superficial positions of da neurons, as well as the two-dimensional shapes of their dendrite arbors, have greatly facilitated in vivo live imaging of dendrite growth and cytoskeletal dynamics in this system (Rolls et al., 2007; Stone et al., 2008). Such studies have established that da neuron dendrites have mixed MT polarity during developmental stages characterized by rapid dendrite growth and branching (Hill et al., 2012). In other words, MT polymers are a mixture of plus-end-out (anterograde polymerizing) and minus-end-out (retrograde polymerizing) filaments. MT polarity matures over the course of larval development to an almost entirely minus-end-out orientation (Hill et al., 2012). The presence of plus-end-out MTs during stages of extensive branching suggests that anterograde MT polymerization may play a role in generating dendrite arbors. This hypothesis is supported by recent studies demonstrating a function for anterograde MT polymerization in facilitating nascent branch formation and stabilization (Ori-McKenney et al., 2012; Yalgin et al., 2015).
Transcription factor-mediated pathways play leading roles in regulating cytoskeletal assembly and organization in da neurons (Lefebvre et al., 2015; Santiago and Bashaw, 2014), suggesting that developmental competence for dendrite growth and branching is established by cell-intrinsic factors. Interestingly, a number of transcription factors selectively regulate either the MT or actin cytoskeleton in da dendrites. For example, Cut and Lola control actin organization while Abrupt, Dar1, and Knot regulate MTs (Ferreira et al., 2014; Jinushi-Nakao et al., 2007; Yalgin et al., 2015; Ye et al., 2011). Identifying the suite of transcription factors regulating da neuron dendritogenesis and defining the cytoskeletal features they regulate is key to deciphering how these factors collaborate to control neuronal morphology.
We set out to test whether the transcription factor FoxO regulates development of da neuron dendrites. FoxO proteins regulate neural stem cell homeostasis, neuronal polarity, neurite outgrowth, synaptic function, and memory consolidation (Christensen et al., 2011; la Torre-Ubieta et al., 2010; Paik et al., 2009; Renault et al., 2009; Salih et al., 2012). In addition, we previously found that the sole FoxO ortholog in Drosophila regulates MT organization in presynaptic terminals of motor neurons (Nechipurenko and Broihier, 2012). Together, these studies demonstrate that FoxO proteins are evolutionarily conserved regulators of neuronal structure and function. However, a role for FoxO proteins in dendrite arborization during neurodevelopment has not been investigated.
In this study, we demonstrate that Drosophila FoxO regulates dendrite development of da neurons. We find that FoxO is expressed in da neurons, and loss of FoxO results in decreased dendrite branching in all da neuron classes. To understand how FoxO promotes dendrite branching, we undertook a time-lapse analysis and demonstrate that FoxO stimulates initiation of new branch growth and also stabilizes existing branches. We hypothesized that these morphological defects result from aberrant MT organization. In line with this hypothesis, analyses of foxO loss-of-function (LOF) and overexpressing neurons demonstrate that FoxO regulates MT dynamics. Specifically, we find that FoxO promotes overall MT dynamics as well as anterograde MT growth. Taken with our previous study of FoxO function in motoneurons, these findings indicate that FoxO regulates MT organization in both motor axons and sensory dendrites. Lastly, we examined whether FoxO is required for da neuron function. Class IV da neurons are nociceptive, sensing noxious heat and mechanical stimuli (Hwang et al., 2007; Tracey et al., 2003). We find that nociceptive responses are attenuated in foxO mutant larvae, indicating that FoxO is required for both structure and function of da neurons. Together, these findings extend in vivo functions of neuronal FoxO proteins to include dendrite arborization and suggest that regulating MT dynamics is a core neuronal function of FoxO family members.
Results
FoxO acts cell-autonomously to regulate class IV dendrite morphology
Our previous work established that FoxO organizes presynaptic MTs at the neuromuscular junction (NMJ)(Nechipurenko and Broihier, 2012). Because MT organization and dynamics are central to dendrite growth and branching, we hypothesized that FoxO regulates dendrite morphology. Class IV da cells are the largest and most elaborate of the da neurons, providing an ideal cell type in which to explore a possible function for FoxO in dendrite morphology. Dendrite outgrowth of class IV cells begins late in embryogenesis and continues through early larval stages, when it is characterized by a rapid growth as the arbor covers its receptive field. Following this phase, dendrite growth transitions to a phase of scaling growth in third instar larvae where growth of dendrite arbors and overall animal growth are synchronized (Parrish et al., 2009).
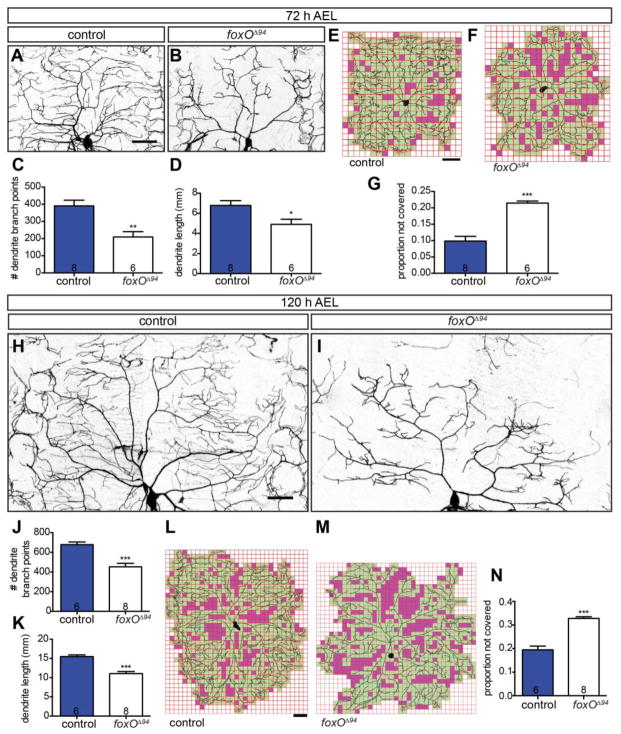
We examined dendrite growth and branching in early (72 h AEL; After Egg Laying) and late (120 h AEL) third instar larvae in ddaC, a well-characterized Class IV cell (Grueber et al., 2002). We labeled membranes of foxO nulls (foxOΔ94)(Slack et al., 2011) and controls with membrane-targeted GFP via a class IV Gal4 driver to permit morphological analyses. Consistent with previous reports (Colombani et al., 2005), we do not detect a difference in overall body size between foxO mutants and controls. We find that at 72 h AEL, foxOΔ94 animals are 2.0 ± 0.14 mm long (n=12) and control animals are 2.0 ± 0.15 mm long (n=20). At 120 h AEL, foxOΔ94 animals are 3.2 ± 0.07 mm long (n=38) and control animals are 3.2 ± 0.06 mm long (n=39). We first assessed ddaC branching at 72 h AEL. We find that loss of FoxO results in a 46.4% reduction in branch number, and a 27.7% reduction in overall dendrite length (Fig. 1A–D). We utilized Sholl analysis to quantify branching as a function of distance from the soma (Sholl, 1953). We find that relative to controls, foxO nulls display decreased branching at both proximal and medial regions of the arbor (Fig. S1 A). Decreased dendrite branching in foxO mutants leads to large regions of non-innervated epidermis within the area covered by individual class IV cells. We developed an ImageJ macro to first overlay a grid of 250 μm2 squares on dendrite arbors, and then analyze internal coverage as reflected by squares with/without a dendrite branch (Jinushi-Nakao et al., 2007; Stewart et al., 2012). We find that foxO nulls display a 2.2-fold increase in the proportion of empty squares relative to controls (Fig. 1E–G), consistent with decreased dendrite branching. Together, these analyses argue that FoxO regulates the early, rapid phase of dendrite outgrowth and branching.
Figure 1. FoxO regulates class IV dendrite morphology.
(A, B, H, I) Representative z-projections of class IV ddaC neurons marked with mCD8-GFP driven by 477-GAL4 at the indicated larval ages and backgrounds. (C) Quantification of dendrite branch point numbers at 72 h AEL in control animals: 390.6 ± 32.8, n=8 cells; foxOΔ94 animals: 209.5 ± 30.7, n=6 cells. (D) Quantification of dendrite length at 72 h AEL in control animals: 6.78 ± 0.48 mm, n=8; foxOΔ94 animals: 4.91 ± 0.50 mm, n=6 cells. (E, F, L, M) Representative analysis of internal coverage of ddaC cells with 250 μm2 squares of the indicated ages and backgrounds. Green squares mark areas covered by the dendritic arbor and soma, while magenta squares mark areas not covered. (G) Quantification of the proportion of squares not covered by the dendrite and soma at 72 h AEL in control animals: 0.10 ± 0.01, n=8 cells; foxOΔ94 animals: 0.21 ± 0.01, n=6 cells. (J) Quantification of dendrite branch point numbers at 120 h AEL in control animals: 678.5 ± 26.2, n=6 cells; foxOΔ94 animals: 453.3 ± 35.0, n=8 cells. (K) Quantification of dendrite length at 120 h AEL in control animals: 15.48 ± 0.44 mm, n=6 cells; foxOΔ94 animals: 11.04 ± 0.54 mm, n=8 cells. (N) Quantification of the proportion of squares not covered by the dendrite and soma at 120 h AEL in control animals: 0.19 ± 0.02, n=6 cells; foxOΔ94 animals: 0.33 ± 0.01, n=8 cells. Scale bars: 50 μm. Error bars are mean ± s.e.m., *, p<0.05, **, p<0.01, ***, p<0.001.
We next examined if the decrease in dendrite branching observed at 72 h AEL persists until 120 h AEL, the late third instar stage. At 120 h AEL, we find a 33.2% reduction in branch number (Fig. 1H–J) and a 28.7% reduction in overall dendrite length in foxO nulls (Fig. 1K). We again utilized Sholl analysis to quantify branching as a function of distance from the soma, and find reductions in branching throughout the arbor in foxO nulls relative to controls (Fig. S1 B). We next quantified internal coverage and find that foxO nulls display a 1.7-fold increase in the proportion of empty squares relative to controls (Fig. 1L–N). Together, these findings indicate that loss of FoxO results in a sustained decrease in dendrite branching and a corresponding increase in epidermal area lacking innervation.
To assess cell autonomy, we undertook a clonal analysis of class IV ddaC using MARCM (Lee and Luo 1999; Grueber et al. 2002). At 120 h AEL, foxO null ddaC clones display a 29.7% reduction in branch number relative to control cells (Fig. 2A–C), consistent with the phenotype observed in foxO nulls. foxO null clones also display a 24.4% reduction in total dendrite length at this stage (Fig. 2D). Sholl analysis reveals a similarly shaped arbor as observed in foxO null animals (Fig. S1 C). We again tested internal coverage using an overlaid grid and find that foxO null ddaC clones display a 1.6-fold increase in the proportion of empty squares relative to controls at 120 h AEL (Fig. 2E–G). Because neither dendrite length nor branching of ddaC are more severely disrupted in foxO null animals than in foxO mutant clones (p>0.05 for both), we conclude that FoxO acts cell-autonomously in class IV ddaC neurons to promote dendrite branching and growth.
Figure 2. FoxO acts cell-autonomously to regulate class IV dendrite morphology.
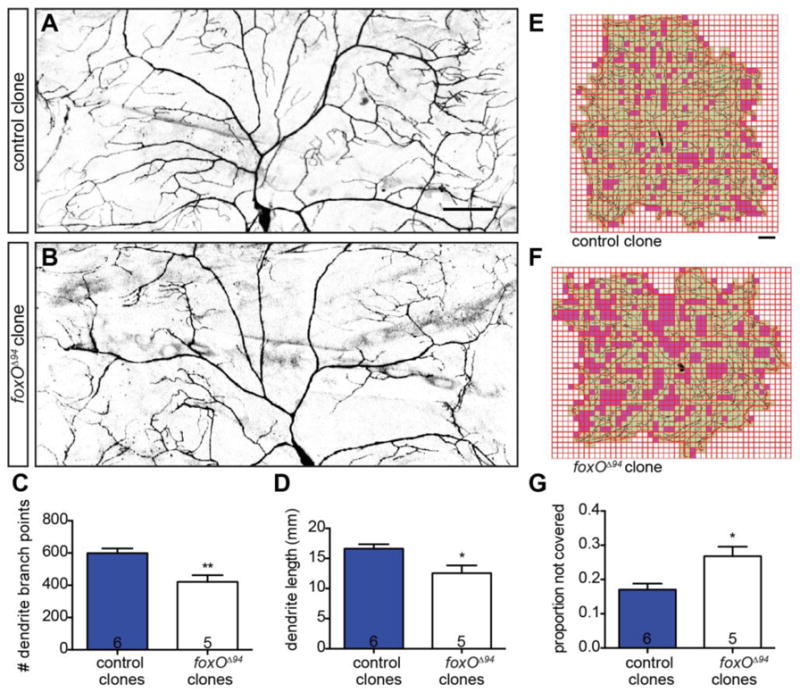
(A–B) Representative z-projections of class IV ddaC MARCM clones at 120 h AEL of the indicated backgrounds marked with mCD8-GFP driven by 109(2)80-GAL4. (C) Quantification of dendrite branch point numbers in control clones: 598.7 ± 29.5, n=6 cells; foxOΔ94 clones: 420.8 ± 42.5, n=5 cells. (D) Quantification of dendrite length in control clones: 16.62 ± 0.73 mm, n=6 cells; foxOΔ94 clones: 12.56 ± 1.28 mm, n=5 cells. (E–F) Representative analysis of internal coverage of ddaC cells with 250 μm2 squares of the indicated backgrounds. Green squares mark areas covered by the dendritic arbor and soma, while magenta squares mark areas not covered. (G) Quantification of the proportion of squares not covered by the dendrite and soma in control clones: 0.17 ± 0.02, n=6 cells; foxOΔ94 clones: 0.27 ± 0.03, n=5 cells. Scale bars: 50 μm. Error bars are mean ± s.e.m., *, p<0.05, **, p<0.01.
FoxO is expressed in da neurons and regulates class I–III dendrite morphology
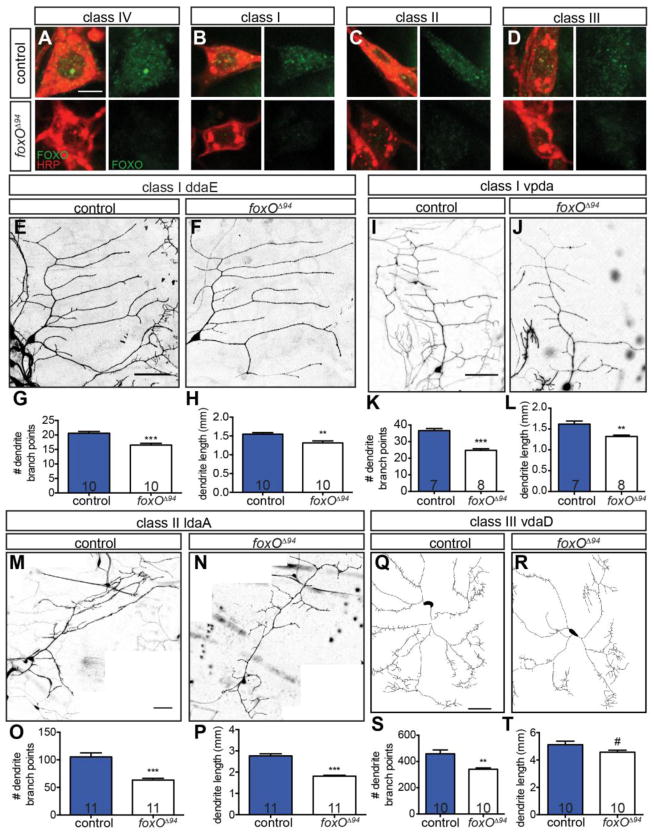
Our MARCM analysis implies that FoxO protein is expressed in class IV da neurons. In line with a cell-autonomous function, FoxO is expressed in ddaC neurons (Fig. 3A), as assessed with an anti-FoxO antibody (Nechipurenko and Broihier, 2012). We further find that FoxO is expressed in class I–III da neurons (Fig. 3B–D). The widespread expression of FoxO in da neurons raised the possibility that FoxO regulates morphology of multiple da neuron classes. To analyze morphology in class I–III cells, we labeled them using a class I–III Gal4 driver and membrane-targeted GFP. Class I cells have the simplest dendrite arbors of da neurons (Grueber et al., 2002). To test if FoxO Is necessary for branching in these cells, we asked if dendrite morphology of two distinct class I cells, ddaE and vpda, is aberrant in foxO nulls. Compared with control class I ddaE cells, foxO nulls display a 19.9% reduction in branch number and a 15.1% reduction in length (Fig. 3E–H). In class I vpda cells, foxO nulls display a 32.3% reduction in branch number and an 18.3% reduction in length (Fig. 3I–L). Thus, FoxO promotes length and branching in neurons with simple dendrite arbors.
Figure 3. FoxO is expressed in da neurons and regulates class I–III dendrite morphology.
(A–D) FoxO staining (green) of FoxO positive controls and foxOΔ94 class I–IV cells, counterstained for HRP (red). Scale bar: 5 μm. (E, F, I, J, M, N) Representative z-projections of class I and II cells of the indicated cell type and backgrounds, marked with mCD8-GFP driven by C161-GAL4. (G) Quantification of branch point numbers in class I ddaE cells in control animals: 20.6 ± 0.6, n=10 cells; foxOΔ94 animals: 16.5 ± 0.6, n=10 cells. (H) Quantification of dendrite length in class I ddaE in control animals: 1.55 ± 0.04 mm, n=10 cells; foxOΔ94 animals: 1.31 ± 0.06 mm, n=10 cells. (K) Quantification of branch point numbers in class I vpda cells in control animals: 36.6 ± 1.3, n=7 cells; foxOΔ94 animals: 24.8 ± 1.0, n=8 cells. (L) Quantification of dendrite length in class I vpda in control animals: 1.62 ± 0.07 mm, n=7 cells; foxOΔ94 animals: 1.32 ± 0.03 mm, n=8 cells. (O) Quantification of branch point numbers in class II ldaA cells in control animals: 105.4 ± 7.2, n=11 cells; foxOΔ94 animals: 63.2 ± 3.2, n=11 cells. (P) Quantification of dendrite length in class II ldaA in control animals: 2.76 ± 0.10 mm, n=11 cells; foxOΔ94 animals: 1.81 ± 0.04 mm, n=11 cells. (Q–R) Representative traces of class III vdaD cells of the indicated backgrounds, marked with mCD8-GFP driven by C161-GAL4. (S) Quantification of branch point numbers in class III vdaD cells in control animals: 458.7 ± 29.1, n=10 cells; foxOΔ94 animals: 341.4 ± 10.1, n=10 cells. (T) Quantification of dendrite length in class III vdaD in control animals: 5.12 ± 0.26 mm, n=10 cells; foxOΔ94 animals: 4.57 ± 0.14 mm, n=10 cells. Scale bars in E, I, M, and Q: 50 μm. Error bars are mean ± s.e.m., #, p<0.1, **, p<0.01, ***, p<0.001.
To test if FoxO is also required in cells with intermediate-sized dendrite arbors, we analyzed both class II and III cells. An analysis of the class II cell ldaA reveals a 40.0% reduction in branch number and a 34.4% reduction in length in foxO nulls relative to controls (Fig. 3M–P). Finally, relative to control class III vdad cells, foxO null cells display a 25.6% reduction in branch number, and a trending, but not significant, 10.7% reduction in length (Fig. 3Q–T). These analyses demonstrate that FoxO promotes branching in cells of all da neurons classes, indicating that FoxO is broadly required for proper da neuron morphology.
FoxO promotes initiation and stabilization of new branches
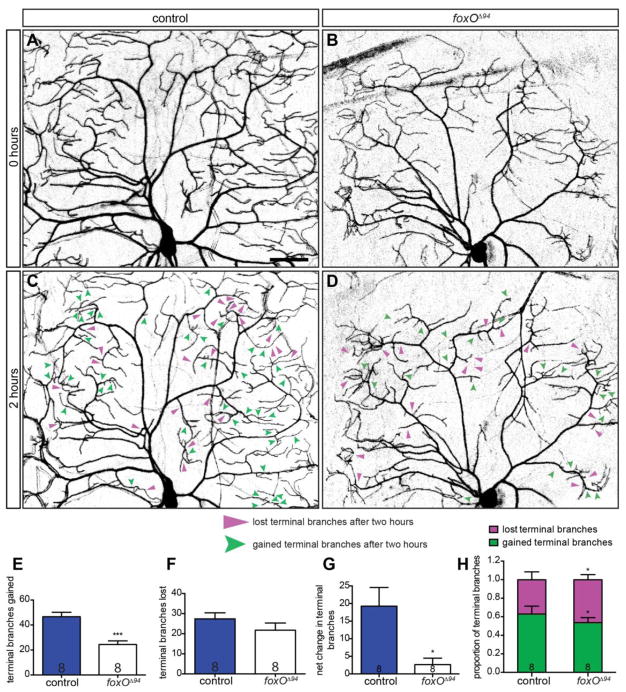
We wondered whether FoxO acts to promote new branch growth, to stabilize existing branches, or both. To explore relative functions for FoxO in branch formation and stabilization, we undertook a time-lapse analysis of the class IV ddaC cell. We analyzed the class IV ddaC neuron at 96 h AEL because these cells are highly branched and dynamic at this time point (Lee et al., 2011; Ori-McKenney et al., 2012; Parrish et al., 2009). We imaged individual ddaC neurons once, removed the animals and returned them to food, then 2 h later, re-imaged the same cells to assess branch gain and loss within a 2 h window (Fig. 4AD). We find that control ddaC cells gain 46.6. ± 3.6 branches on average, while foxO mutants gain 24.4 ± 2.9 branches over this time period, indicating decreased branch initiation in foxO mutants (Fig. 4E). Over the same period, control cells lose 27.4 ± 3.0 branches for a net gain of 19.3 ± 5.3 branches, while foxO mutants lose 21.8 ± 3.6 branches for a net gain of only 2.6 ± 1.8 branches (Fig. 4F–G). Thus, foxO mutant cells lose almost as many branches as they gain in the two-hour interval.
Figure 4. FoxO promotes initiation and stabilization of new branches.
(A–D) Representative images of class IV ddaC cells within intact, 96 h AEL animals, marked with mCD8-GFP driven by 477-GAL4, in the indicated backgrounds and time points. Filled, magenta arrows indicate lost terminal branches after a two-hour period, while notched, green arrows indicate new terminal branches. (E) Quantification of gained branch points after a two-hour period, controls: 46.6 ± 3.6, n=8 cells; foxOΔ94 animals: 24.4 ± 2.9, n=8 cells. (F) Quantification of lost branch point after a two-hour period, controls: 27.4 ± 3.0, n=8; foxOΔ94 animals: 21.8 ± 3.6, n=8 cells. (G) Quantification of net branches after a two-hour period, controls: 19.3 ± 5.3, n=8 cells; foxOΔ94 animals: 2.6 ± 1.8, n=8 cells. (H) Quantification of the proportion of gained branches compared with lost branches over a two-hour period, gained branches in controls: 0.63 ± 0.04, n=8 cells; lost branches in controls: 0.37 ± 0.04, n=8 cells; gained branches in foxOΔ94 animals: 0.54 ± 0.02, n=8 cells; lost branches in foxOΔ94 animals: 0.46 ± 0.02, n=8 cells. Scale bar: 50 μm. Error bars are mean ± s.e.m., *, p<0.05, ***, p<0.001.
We also calculated gain and lost branches as fractions of the total number of dynamic branches present in each genotype in the two-hour window. By this measure, controls exhibit a significantly greater fraction of gained branches than foxO mutants, whereas foxO mutants display a greater fraction of lost branches (Fig. 4H). If these relatively short-term changes in branch loss and growth are summed over development, they are predicted to result in the significantly smaller arbors observed in foxO mutant animals. Together, these data indicate that FoxO serves to both initiate new branch growth as well as to stabilize existing branches.
FoxO is sufficient to promote branch formation
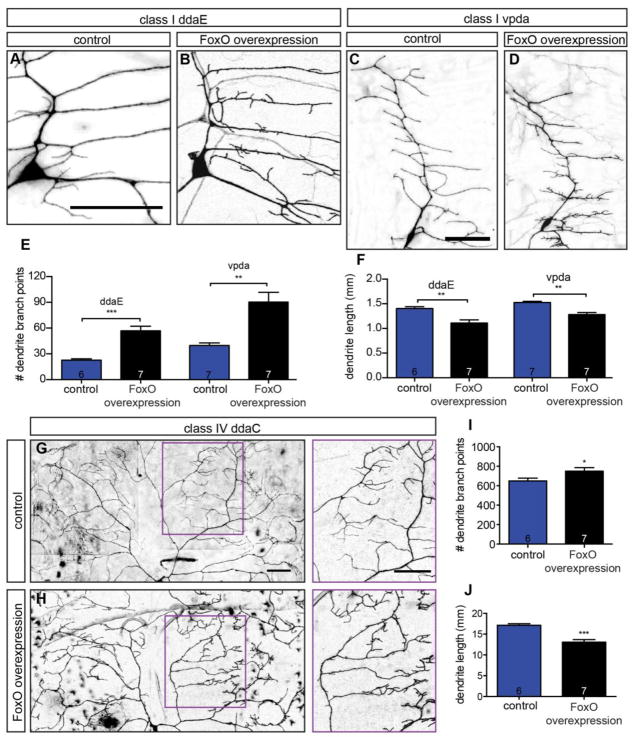
The preceding loss-of-function analysis indicates that FoxO is necessary for dendrite branching. To test if FoxO is also sufficient for branching, we tested if its overexpression drives increased branching. We began by investigating whether FoxO overexpression increases branching in class I cells, because these cells are simple with comparatively fewer branches. Thus, an increase in branching might be more apparent in class I cells than in highly branched class IV cells. Indeed, FoxO overexpression leads to dramatically increased branching in two distinct class I cells: ddE and vpda (Fig. 5A–D). Specifically, we find a 2.5-fold increase in branch number in ddaE and a 2.3-fold increase in vpda (Fig. 5E) in FoxO overexpressing neurons relative to controls. Interestingly, FoxO overexpression in these cells does not alter the overall branching patterns of primary and secondary branches, but rather drives the formation of ectopic short, spiky branches. The main branches in FoxO overexpressing cells are slightly shorter than in controls, leading to an overall reduction in total dendrite length in spite of the elevated branch number in these cells (Fig. 5F). We next assessed whether FoxO overexpression also results in increased branching in slightly more complex class II da neurons. We find that FoxO overexpression leads to a 1.3-fold increase in branch number in the class II ldaA neuron and an overall reduction in dendrite length (Fig. S2 A–D). The short, ectopic branches observed in class II are very similar in appearance to those observed with FoxO overexpression in class I. Thus, FoxO overexpression promotes dendrite branching, but not dendrite length, in neurons with simple dendrite arbors.
Figure 5. FoxO is sufficient to promote branch formation.
(A–D) Representative z-projections of class I ddaE and vpda cells of the indicated backgrounds, marked with mCD8-GFP driven by 2-21-GAL4. (E) Quantification of class I branch point numbers in animals expressing control RNAi #2 in ddaE: 22.7 ± 1.6, n=6 cells; control RNAi #2 in vpda: 39.7 ± 3.1, n=7 cells; FoxO WT #1 in ddaE: 56.9 ± 5.3, n=7 cells; FoxO WT #1 in vpda: 90.3 ± 11.4, n=7 cells. (F) Quantification of class I dendrite length in animals expressing control RNAi #2 in ddaE: 1.40 ± 0.04 mm, n=6 cells; control RNAi #2 in vpda: 1.52 ± 0.02 mm, n=7 cells; FoxO WT #1 in ddaE: 1.11 ± 0.07 mm, n=7 cells; FoxO WT #1 in vpda: 1.27 ± 0.05 mm, n=7 cells. (G–H) Representative z-projections of class IV ddaC cells of the indicated backgrounds, marked with mCD8-GFP driven by 477-GAL4. Magenta boxes in A and B are magnified in side panels. (I) Quantification of class IV ddaC branch point numbers in animals expressing control RNAi #1: 649.0 ± 28.6, n=6 cells; FoxO WT #2: 749.7 ± 35.4, n=7 cells. (J) Quantification of class IV ddaC dendrite length in animals expressing control RNAi #1: 17.12 ± 0.37 mm, n=6 cells; FoxO WT #2: 13.08 ± 0.61 mm, n=7 cells. Scale bars: 50 μm. Error bars are mean ± s.e.m., *, p<0.05, **, p<0.01, ***, p<0.001.
We next wanted to determine if FoxO overexpression increases branch number in neurons with more complex dendrite arbors. For this analysis, we again turned to the class IV ddaC cell. Similar to our findings in class I and II cells, FoxO overexpression in ddaC generates ectopic short, spiky branches (Fig. 5G–H). We quantified branch number in these cells and find a 1.2-fold increase in branch number in class IV cells ddaC relative to controls (Fig. 5I). Again, as in class I and II cells, the main branches in FoxO overexpressing neurons are shorter than in controls, leading to an overall reduction in dendrite length (Fig. 5J). The increase in short ectopic branches, coupled with the decrease in main branch length, gives these cells a compact, bushy appearance. Because we utilized class-specific drivers to overexpress FoxO in these experiments, they support the conclusion that FoxO cell-autonomously promotes dendrite branching. Furthermore, our analyses of dendrite morphology in foxO LOF and overexpression backgrounds indicate that foxO is necessary and sufficient for dendrite branch formation in multiple classes of sensory neurons.
FoxO limits the distribution of stable microtubules in dendrites
How does loss of FoxO alter dendrite morphology? To shed light on the cellular mechanism by which FoxO promotes dendrite branching, we characterized the MT cytoskeleton in foxO mutants. We examined MTs because our prior work demonstrated that loss of FoxO alters MT stability and organization at the NMJ (Nechipurenko and Broihier, 2012). Thus, a straightforward hypothesis is that FoxO-dependent pathways regulate MT stability in dendrites. To examine this possibility, we labeled dendrites with Futsch/MAP1B, a MT-associated protein that stabilizes MTs and is itself a marker of the stable MT population (Halpain and Dehmelt, 2006; Hummel et al., 2000; Roos et al., 2000).
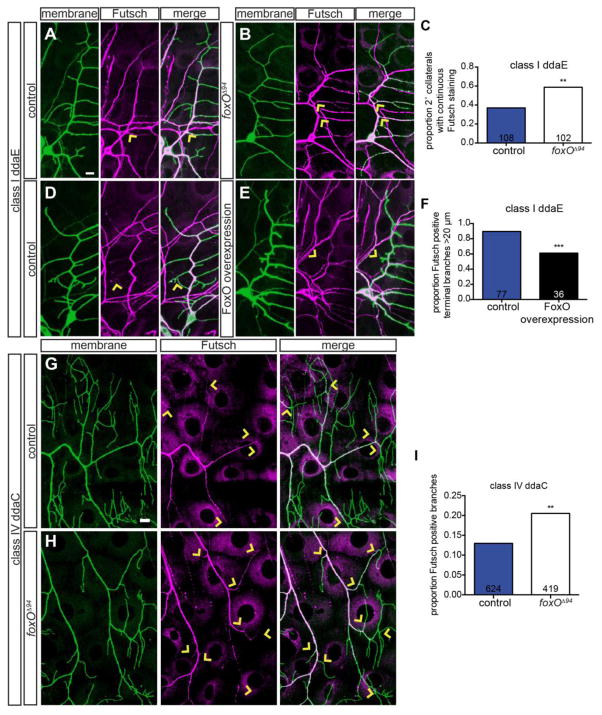
We first assessed class I ddaE neurons to ask if FoxO regulates the distribution of stable MTs in these cells. We began with these cells because of their simple morphology and stereotyped pattern of Futsch staining (Jinushi-Nakao et al., 2007). We find that in controls, while primary ddaE branches are strongly Futsch-positive, secondary branches typically have much weaker Futsch (Fig. 6A). Strikingly, in foxO nulls, we frequently find strong Futsch expression extending well into secondary ddaE branches (arrows in Fig. 6B). We quantified the proportion of secondary branches with continuous Futsch, and find a 1.6-fold increase in this proportion in foxO nulls relative to controls (Fig. 6C). Thus, loss of FoxO results in an increased distribution of Futsch in ddaE.
Figure 6. FoxO limits the distribution of stable microtubules in dendrites.
(A, B, D, E) Representative z-projections of class I ddaE cells of the indicated backgrounds, marked with mCD8-GFP, in green, driven by C161-Gal4 (A–B) or 2-21-GAL4 (D–E), counterstained for Futsch, in magenta. Arrows in A and B mark examples of continuous Futsch staining at 2° collaterals, while arrows in D and E mark examples of terminal branches with or without Futsch staining. (C) Quantification of the proportion of 2° collaterals with continuous Futsch staining in control animals: 37.0% of 108 collaterals from 9 cells; in foxOΔ94 animals: 58.8% of 102 collaterals from 11 cells. (F) Quantification of the proportion of Futsch positive, greater than 20 μm terminal branches in animals expressing control RNAi #1: 89.6% of 77 branches from 6 cells; FoxO WT #1: 61.1% of 36 branches from 6 cells. (G–H) Representative z-projections of class IV ddaC cells of the indicated backgrounds, marked with mCD8-GFP, in green, driven by 477-GAL4, counterstained for Futsch, in magenta. Arrows in G and H mark Futsch staining in terminal or near terminal branches. (I) Quantification the proportion of Futsch positive branches in control animals: 13.0% of 624 branches from 6 cells; foxOΔ94 animals: 20.5% of 419 branches from 7 cells. Scale bars: 10 μm. Comparisons made with two-tailed Fisher’s exact tests, **, p<0.01, ***, p<0.001.
To ask if FoxO is also sufficient to limit stable MTs in class I ddaE neurons, we tested the effect of FoxO overexpression on the distribution of Futsch. FoxO overexpression in ddaE results in a clear decrease in Futsch intensity throughout the arbor (Fig. 6D–E). We find that the intensity of Futsch staining is decreased in primary branches as well as in higher order branches. For purposes of quantification, we focused on Futsch expression in terminal branches greater than 20 μm in length. We wanted to exclude the short, spiky branches present only in the FoxO overexpressing cells to ensure that any change in Futsch-positive branches was not solely the result of the presence of these short branches. We observe a 1.5-fold reduction in Futsch-positive branches in FoxO overexpressing neurons (Fig. 6F). Thus, while loss of FoxO leads to an expanded Futsch distribution in ddaE dendrites, FoxO overexpression results in a reduced Futsch distribution.
We went on to analyze the more complex distribution of Futsch in foxO null class IV ddaC neurons. In controls, the primary branches of these cells contain Futsch, while higher order branches are generally Futsch-negative (Fig. 6G)(Grueber et al., 2003). We noticed that Futsch appeared to extend farther into higher order branches in foxO nulls relative to wild type (Fig. 6G–H). We quantified branches with Futsch and find a 1.6-fold increase the proportion of Futsch-positive branches in foxO nulls relative to controls (Fig. 6I). This difference is pronounced in tertiary and terminal branches, which lack Futsch in controls, but are frequently Futsch-positive in foxO mutants (arrowheads in Fig. 6G–H). Thus, loss of FoxO results in an expanded distribution of Futsch, a stable MT marker, arguing that FoxO normally limits MT stability in class IV da neurons. Together, these results raise the possibility that the observed morphological phenotypes in foxO mutants reflect alterations in underlying MT organization.
To test if increased MT stability underlies the morphological defects in foxO mutants, we examined whether otherwise decreasing MT stabilization rescues dendrite length and/or branching in foxO mutants. To this end, we analyzed genetic interactions between FoxO and Futsch. Because Futsch stabilizes MTs, we tested if decreasing MT stability by removing one copy of Futsch counteracts the loss of FoxO in class IV ddaC neurons. Relative to control dendrites, we find a 39.9% reduction in branching and a 38.3% reduction in length in foxO nulls (Fig. S3 A–B, E–F). We find that futsch dominantly suppresses deficits in branching and length observed in ddaC cells in foxO nulls (Fig. S3 A–F). Branch number is increased 1.3-fold in futschK68/+;;foxOΔ94 relative to foxOΔ94 alone, while length is increased 1.2-fold. This partial rescue is notable given that in our hands, futsch heterozygosity on its own significantly decreases both length and branching in ddaC (Fig. S3 E–F). Thus, we conclude that FoxO activity is normally balanced by Futsch activity in ddaC. The genetic interaction between foxO and futsch argues that elevated MT stability in foxO nulls contributes to the observed morphological defects. Moreover, we interpret our finding that both increased MT stability (foxO nulls) and decreased MT stability (futsch heterozygotes) result in decreased branching to suggest that MT stability and dynamics must be precisely balanced to support proper branch formation and maintenance.
FoxO is necessary for both anterograde polymerization and dynamics of microtubules
The alterations to Futsch distribution in foxO LOF and foxO overexpression backgrounds are consistent with altered underlying MT dynamics. To test this hypothesis, we utilized EB1-GFP to visualize plus-end MT growth in vivo. EB1 binds the plus end of MTs, and an EB1-GFP fusion protein is widely used to track plus-end MT growth. When EB1-GFP binds to the growing plus-end of a MT, it is visualized as an EB1-GFP comet (Baas and Lin, 2011; Rolls et al., 2007; Stone et al., 2008). EB1-GFP comet number reveals the amount of MT growth, whereas comet direction (anterograde or retrograde) indicates MT orientation. Developmental analyses of EB1-GFP dynamics have shown that da neurons gradually acquire minus-end-out polarity (Hill et al., 2012). During embryonic stages, dendrites contain roughly equal numbers of plus-end-out and minus-end-out MTs. This distribution gradually resolves to almost entirely minus-end-out polarity by the end of larval stages.
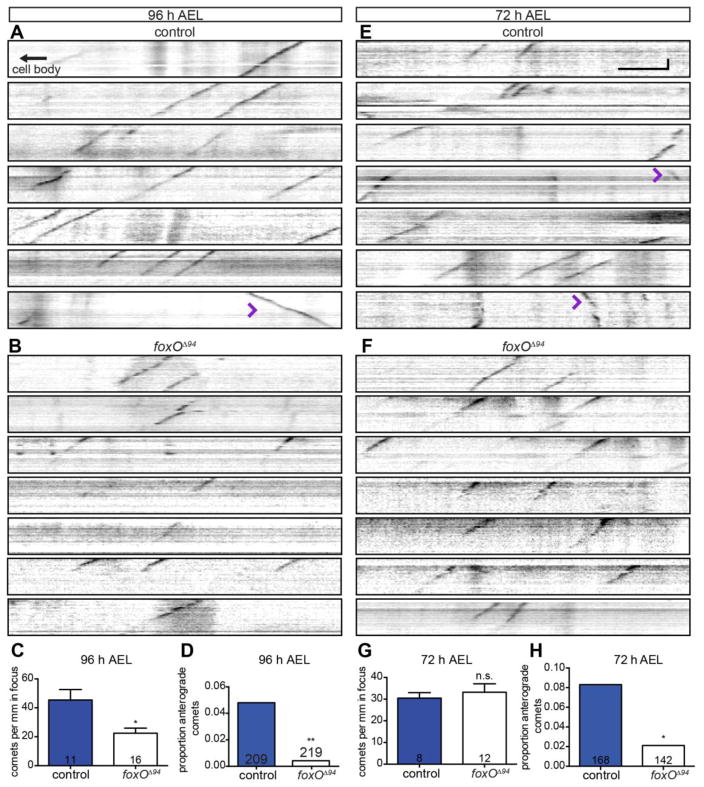
To investigate if FoxO regulates MT dynamics, we undertook a live EB1-GFP analysis in class IV ddaC neurons. We began by analyzing 96 h AEL larvae both because MTs are dynamic at this stage and because the larvae are amenable to live imaging (see Materials and Methods). In controls, we find 45.4 ± 7.2 comets/mm dendrite, in line with published reports (Fig. 7A, C) (Stewart et al., 2012). In contrast, in foxO mutants, we find 22.5 ± 3.5 comets/mm dendrite, or a 2.0-fold decrease in total comet number (Fig. 7B–C). Thus, at 96 h AEL, MTs are less dynamic in foxO mutant ddaC dendrites, consistent with the expanded distribution of Futsch in this background (Fig. 6G–I). Unexpectedly, we also find a marked 9.6-fold reduction in the percentage of anterograde comets in foxO nulls relative to controls. While 4.8% of the comets are anterograde in controls, only 0.5% of the comets are anterograde in foxO nulls (Fig. 7A–B, D; Supplemental Movie 1). These data suggest that FoxO normally promotes anterograde polymerization of MTs.
Figure 7. FoxO is necessary for anterograde polymerization and dynamics of microtubules.
(A, B, E, F) Representative kymographs from ddaC cells from live, intact larvae of the indicated ages and backgrounds expressing EB1-GFP driven by 477-GAL4. Retrograde EB1 comets move down and to the left, anterograde comets down and to the right (purple arrowheads). (C) Quantification of comets per mm in focus at 96 h AEL in control animals: 45.4 ± 7.2, n=11 movies; foxOΔ94 animals: 22.5 ± 3.5, n=16 movies. (D) Quantification of the proportion of anterograde comets at 96 h AEL in control animals: 10 of 209, 4.8%; foxOΔ94 animals: 1 of 219, 0.5%. (G) Quantification of EB1 comets per mm in focus at 72 h AEL in control animals: 30.4 ± 2.6, n=8 movies; foxOΔ94 animals: 33.3 ± 3.9, n=12 movies. (H) Quantification of the proportion of anterograde comets at 72 h AEL in control animals: 14 of 168, 8.3%; foxOΔ94 animals: 3 of 142, 2.1%. Vertical scale bar: 20 seconds; horizontal scale bar: 5 μm. Comparisons in D and H made with two-tailed Fisher’s exact tests. Error bars are mean ± s.e.m., n.s., not significantly different; *, p<0.05, **, p<0.01.
To further test this hypothesis, we analyzed EB1-GFP comets in ddaC cells at an earlier developmental time point when a higher proportion of MTs are expected to have anterograde orientation. Thus, we adapted our live imaging protocol for younger, 72 h AEL larvae. At this stage, we do not detect a difference in overall comet number in foxO nulls relative to controls (Fig. 7E–G), arguing that FoxO does not regulate overall MT dynamics at 72 h AEL. In contrast, we find a 4.0-fold decrease in the percentage of anterograde comets (Fig. 7E–F, H). In control animals, 8.3% of the comets move anterogradely, while only 2.1% of the comets in foxO nulls are anterograde. Together, these data indicate that FoxO promotes overall MT polymerization (anterograde and retrograde) at 96 h AEL, while it is necessary for normal levels of anterograde MT polymerization at both 72 h and 96 h AEL.
FoxO drives anterograde microtubule polymerization
While the loss of plus-end-out, anterograde polymerizing, MTs in foxO mutant dendrites was unexpected, it is consistent with recent studies that have revealed a link between plus-end-out MTs and dendrite branching (Ori-McKenney et al., 2012; Yalgin et al., 2015). These studies demonstrate that anterograde polymerizing MTs are important for nascent dendrite branch growth and/or stability. Does decreased dendrite branching in foxO nulls result, at least in part, from reduced anterograde MT polymerization? To investigate whether FoxO regulates anterograde MT polymerization to promote dendrite branching, we investigated the effect of FoxO overexpression on MT polymerization. Because FoxO overexpression drives dendrite branching (Fig. 5), we predicted that anterograde MT polymerization would be increased in this background.
To test this hypothesis, we investigated whether FoxO overexpression in the class IV ddaC cell alters MT dynamics at 96 h AEL. We do not detect a difference in total comet number between foxO overexpressing neurons and controls at 96 h AEL (Fig. 8A–C), indicating that FoxO is not sufficient to alter overall MT dynamics. However, FoxO overexpression results in a 2.8-fold increase in the percentage of anterograde comets at 96 h AEL: 18.4% of comets are anterograde in FoxO overexpressing neurons, relative to 6.7% of comets in controls. (Fig. 8A–B, D; Supplemental Movie 2). In wild-type animals, comets in short, nascent branches are more frequently anterograde, while comets in long, main branches are more frequently retrograde (Ori-McKenney et al., 2012). To interrogate the relationship between anterograde comets and branching in FoxO overexpressors, we asked if the excess anterograde comets are found in higher-order branches, or rather in main branches. In controls, 74.5% (n=51) of anterograde comets are in thin, higher-order branches, consistent with the established link between anterograde comets and nascent branches (Ori-McKenney et al., 2012). Similarly, 79.3% (n=82) of anterograde comets in FoxO overexpressing neurons are in higher-order branches. Thus, the excess anterograde comets in FoxO overexpressing neurons arise in higher-order branches—and are thus spatially positioned to contribute to increased branching.
Figure 8. FoxO drives anterograde microtubule polymerization.
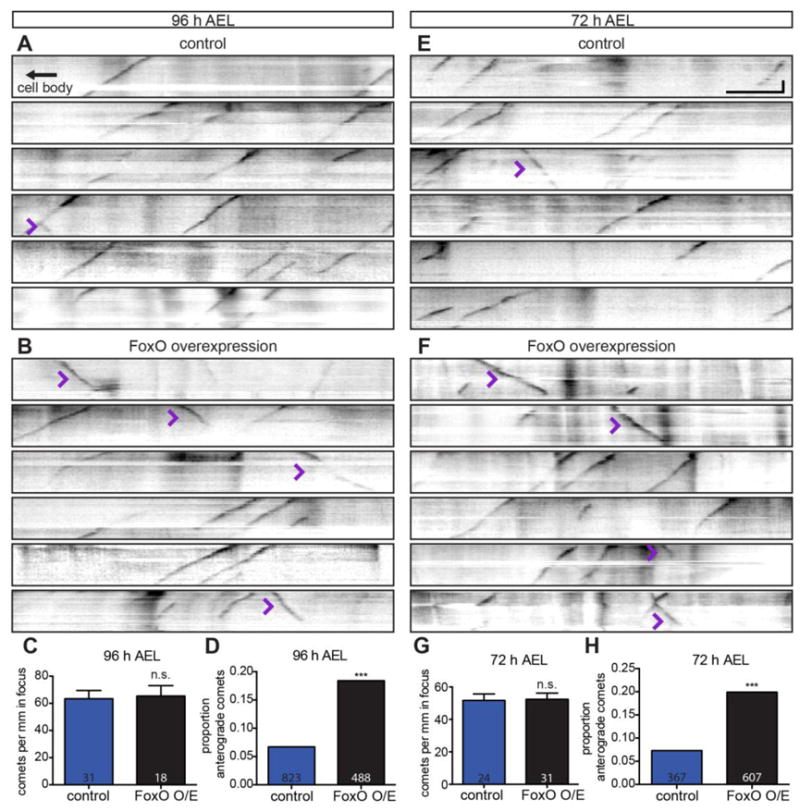
(A, B, E, F) Representative kymographs from ddaC cells from live, intact larvae of the indicated ages and backgrounds expressing EB1-GFP driven by 477-GAL4. Retrograde EB1 comets move down and to the left, anterograde comets down and to the right (purple arrowheads). (C) Quantification of comets per mm in focus at 96 h AEL in animals expressing control RNAi #1: 63.4 ± 6.0, n=31 movies; FoxO WT #2: 65.3 ± 8.1, n=18 movies. (D) Quantification of the proportion of anterograde comets at 96 h AEL in animals expressing control RNAi #1: 55 of 823, 6.7%; FoxO WT #2: 90 of 488, 18.4%. (G) Quantification of comets per mm in focus at 72 h AEL in animals expressing control RNAi #1: 51.7 ± 4.0, n=24 movies; FoxO WT #2: 52.4 ± 3.8, n=31 movies. (H) Quantification of the proportion of anterograde comets at 72 h AEL in animals expressing control RNAi #1: 27 of 367, 7.4%; FoxO WT #2: 121 of 607, 19.9%. Vertical scale bar: 20 seconds; horizontal scale bar: 5 μm. Comparisons in D and H made with two-tailed Fisher’s exact tests. Error bars are mean ± s.e.m., n.s., not significantly different; ***, p<0.001.
We next asked if FoxO overexpression increases the percentage of anterograde MT polymerization at 72 h AEL. Similar to 96 h AEL, we do not detect a difference in total comet number in FoxO overexpressing neurons relative to controls at 72 h AEL (Fig. 8E–G). However, FoxO overexpression in ddaC leads to a 2.7-fold increase in the proportion of anterograde comets at 72 h AEL. In FoxO overexpressing neurons, 19.9% of comets are anterograde, relative to 7.4% of comets in controls (Fig. 8E–F, H). Together, these analyses demonstrate that FoxO is sufficient to promote anterograde polymerization of MTs in dendrites.
FoxO is necessary for proper nociceptive response
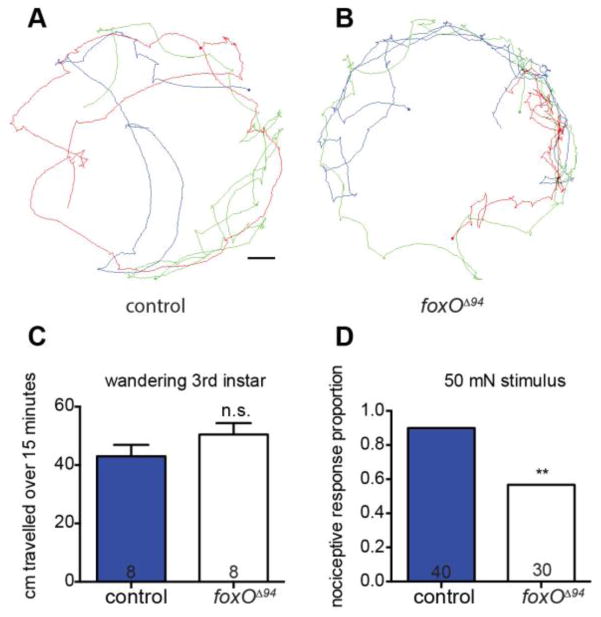
Class IV neurons are nociceptive, responding to both noxious heat and strong touch stimuli, and elicit a stereotyped 360° rolling behavior when activated (Hwang et al., 2007; Tracey et al., 2003). Reduced class IV cell complexity correlates with reduced nociceptive responses (Ferreira et al., 2014; Stewart et al., 2012). We therefore hypothesized that foxO null animals would have impaired responses to noxious touch stimuli. To quantify overall crawling behavior, we recorded the behavior of wandering 3rd instar larvae over a 15-minute period and analyzed total distance traveled. We find that foxO nulls and controls crawl similar distances, indicating that loss of FoxO does not result in a gross deficit in movement (Fig. 9A–C). To test for nociception, we calibrated 50 mN Von Frey filaments and stimulated larvae once on hemisegment 4, 5, or 6 (Tracey et al., 2003; Zhong et al., 2010). Animals were scored for whether or not they rolled 360° at least once in response to a single stimulus. We find that control animals respond 90.0% of the time, in line with previous studies (Tracey et al., 2003). In contrast, foxO mutants respond only 56.7% of the time (Fig. 9D). Animals otherwise paused upon stimulation, similar to reports of light touch sensation. Therefore, reduced dendrite complexity seen in foxO nulls correlates with a reduced nociceptive response.
Figure 9. FoxO is necessary for proper nociceptive response.
(A, B) Representative traces of wandering 3rd instar larvae of the indicated backgrounds and their movement over a 15-minute period. Scale bar: 1 cm. (C) Quantification of movement over a 15-minute period by controls: 43.0 ± 3.9 cm, n=8 animals; foxOΔ94 animals: 50.5 ± 3.9 cm, n=8 animals. (D) Quantification of the proportion of nociceptive response of animals given a 50 mN Von Frey filament stimulation, analyzed with two-tailed Fisher’s exact test, of control animals: 36 of 40, 90.0%; foxOΔ94 animals: 17 of 30, 56.7%. Error bars are mean ± s.e.m., n.s., not significantly different; **, p<0.01.
Discussion
Here we demonstrate a role for FoxO in arborization of sensory neuron dendrites during development. Both loss-of-function and overexpression analyses indicate that FoxO broadly promotes dendrite branching in da neurons. A time-lapse analysis provides insight into FoxO function and indicates that FoxO promotes initiation and stabilization of new branches. Moreover, we find that FoxO limits the distribution of Futsch/MAP1B in multiple classes of da neuron dendrites, indicating an expansion of the stable MT pool. Moreover, analysis of dynamic MTs in foxO LOF and overexpressing neurons indicates that FoxO promotes anterograde polymerization of MTs. Lastly, loss of FoxO leads to a reduced larval nociceptive response, arguing that FoxO is also necessary for function of class IV da neurons. We conclude that FoxO is necessary and sufficient for dendrite branching, at least in part, by promoting anterograde MT polymerization.
Anterograde MT growth and dendrite branching
Mature da neurons contain largely minus-end-out, retrograde polymerizing, MTs (Hill et al., 2012; Rolls and Jegla, 2015; Stone et al., 2008). In contrast, during development as da dendrites grow and branch, their MTs have mixed polarity (Hill et al., 2012). The presence of anterograde polymerizing MTs during dendrite extension suggests that this MT population is linked to growth/branching. Consistent with this hypothesis, differences in MT polarity are observed in different types of branches in class IV neurons (Ori-McKenney et al., 2012). These authors found that longer, established branches have mostly retrograde comets, while shorter, nascent branches have mostly anterograde comets. Moreover, following dendrite severing, nascent dendrites initially contain both retrograde and anterograde polymerizing MTs, which resolves to the mature pattern of minus-end-out (Song et al., 2012; Stone et al., 2014).
Further analyses support a direct link between anterograde polymerizing MTs and dendrite branching. Ori-McKenney et al. (2012) find a striking difference between stable and retracting terminal branches with respect to anterograde MT growth. They demonstrate that the majority of stable branches contain anterograde EB1 comets, while the majority of retracting branches do not contain comets. If anterograde MT polymerization is involved in branching, then one might expect the relatively simple class I cells to have a mechanism to limit anterograde polymerization. Indeed, Yalgin et al. (2015) have recently demonstrated that the class I–specific transcription factor Abrupt limits branching in class I neurons by promoting Centrosomin expression. They find that Centrosomin represses dendrite branching by orienting MT nucleation to repress anterograde polymerization. Centrosomin is proposed to execute this function by tethering MT nucleation events to one face of Golgi outposts and biasing the direction of MT growth away from dendrite tips.
In the present study, we demonstrate that FoxO promotes branching in all classes of da neurons. We also find that FoxO is necessary and sufficient for anterograde MT polymerization. Based on the established role of anterograde polymerizing MTs in branching, we propose that FoxO drives branching, at least in part, by regulating MT orientation. Moreover, the link between MT polarity and dendrite maturity suggests that by stimulating plus-end-out MTs, FoxO promotes a more immature, dynamic MT environment that is well suited for branching.
Our genetic analyses in foxO LOF and overexpression backgrounds indicate that FoxO drives anterograde MT polymerization. Our results also indicate that this is unlikely to be the only function of FoxO in MT regulation. Several lines of evidence also indicate that FoxO promotes MT dynamics. First, we find that the distribution of Futsch/MAP1B is expanded in dendrites of both class I and class IV neurons in foxO nulls. Second, the reduced length and branching observed in ddaC in foxO nulls are partially suppressed by removing one copy of futsch. Third, we find an approximate two-fold reduction in EB1 comets in foxO nulls at 96 h AEL. These LOF analyses indicate that foxO plays a role in regulating overall dynamics.
Moreover, it will be important to determine if the actin cytoskeleton is altered in foxO LOF or overexpression backgrounds. The ectopic short, spiky branches observed with FoxO overexpression are Futsch-negative and resemble the actin-rich branches in class III dendrites (Jinushi-Nakao et al., 2007; Nagel et al., 2012). We propose that the presence of these actin-rich branches in FoxO overexpressing neurons is indirectly caused by alterations to the MT network. It is alternately possible that FoxO more directly regulates the actin cytoskeleton. Identification of FoxO’s transcriptional targets will clarify the mechanism(s) through which FoxO controls these interrelated cytoskeletal components.
FoxOs in neurodevelopment
FoxO family members have recently emerged as key regulators of neuronal processes such as neural stem cell homeostasis, neuronal polarity, neurite outgrowth, synaptic function, and memory consolidation (Christensen et al., 2011; la Torre-Ubieta et al., 2010; Paik et al., 2009; Renault et al., 2009; Salih et al., 2012). Of particular interest, simultaneous RNAi-mediated knockdown of FoxO1, 3 and 6 interferes with neuronal polarization in hippocampal and cerebellar neurons (la Torre-Ubieta et al., 2010). Pak1, a kinase known to regulate neuronal MT dynamics and neuronal polarization (Jacobs et al., 2007), was shown to be a critical for this function of FoxO. Based on this link between mammalian FoxOs and Pak1, we tested if Pak1 might be a FoxO effector in da neurons. However, RNAi-mediated knockdown of Pak1 does not yield phenotypes consistent with an essential role for Pak1 downstream of FoxO in regulating da neuron arborization (JCS and HTB, data not shown).
FoxO family members serve key functions in neuronal development and function subsequent to initial polarization. Knockdown of mammalian FoxOs after neurons have polarized reveals defects in axon and dendrite outgrowth in vitro (Christensen et al., 2011). Arguing that FoxO function in neurite outgrowth is evolutionarily conserved, the C. elegans foxO homolog, daf-16, is likewise required for axon outgrowth of the AIY interneuron (Christensen et al., 2011). We previously examined the role of Drosophila FoxO in motoneurons and found that FoxO is required for proper MT architecture in presynaptic terminals at the NMJ, though we did not find defects in initial axon outgrowth or guidance (Nechipurenko and Broihier, 2012). Genetic and molecular analyses argued that MT stability at the NMJ is enhanced in foxO LOF and attenuated in foxO overexpressing neurons. These findings are in good agreement with the present study and together indicate that Drosophila FoxO limits MT stability in both axons and dendrites in multiple neuronal populations.
Loss of mammalian FoxO6 results in decreased spine density in hippocampal neurons both in vitro and in vivo (Salih et al., 2012). Spines are actin-rich protrusions on dendrites that house postsynaptic components of excitatory synapses in the mammalian CNS. Intriguingly, MT entry into spines is linked to aspects of spine development and function, including density and morphology (Gu et al., 2008; Hu et al., 2008; Jaworski et al., 2009), raising the possibility that aberrant MT behavior could underlie spine defects in FoxO6 mutants. It will be important to investigate whether loss of mammalian FoxO family members results in defects in neuronal MT dynamics or polarity.
The pathways upstream of FoxO proteins in neurons are not well understood. FoxOs can be regulated by post-translational modifications including phosphorylation, acetylation, and ubiquitylation, which together direct their subcellular localization and transcriptional activity (Calnan and Brunet, 2008). Akt kinase phosphorylates FoxO and inhibits its transcriptional activity by retaining it in the cytoplasm (Huang and Tindall, 2007). Thus, if an Akt-FoxO axis is central to FoxO function in da neurons, loss of Akt is predicted to result in increased branching similar to overexpression of FoxO (Fig. 4). However, loss of Akt results in strongly reduced dendrite growth and branching (Parrish et al., 2009), suggesting that Akt is not a critical regulator of FoxO function in da neuron dendrites. Thus, upstream regulation of FoxO in da neurons is likely distinct from FoxO regulation in motoneurons, which appears to depend on Akt-dependent inhibition (Nechipurenko and Broihier, 2012).
In the future, it will be important to define effector(s) of FoxO in da neurons. FoxO can now be considered a member of a small group of transcription factors, including Abrupt, Dar1, and Knot, that regulate dendrite morphology via MT dynamics in da neurons (Jinushi-Nakao et al., 2007; Li et al., 2004; Sugimura et al., 2004; Ye et al., 2011). Critical transcriptional targets of Abrupt, Dar1, and Knot have recently been identified that mediate the MT regulatory functions of these proteins (Jinushi-Nakao et al., 2007; Wang et al., 2015; Yalgin et al., 2015; Ye et al., 2011). Of these transcription factors, Knot is most similar to FoxO in that it drives branching (Jinushi-Nakao et al., 2007). However, FoxO is expressed in, and promotes branching of, all da neuron classes, These results suggest that FoxO does not differentially regulate the neuronal subtype fate of a particular da neuron class, but rather promotes branching of all da neuron subtypes.
Based on the reciprocal changes we observe in the proportion of plus-end-out MTs in foxO LOF and overexpressing neurons, we predict that FoxO’s downstream transcriptional targets include proteins regulating MT polarity in dendrites. Possible targets include Kinesin-2 subunits, EB1, and APC, as RNAi-mediated knockdown of these proteins results in shifts in MT polarity similar to phenotypes described here for FoxO overexpression (Mattie et al., 2010). Further investigation of the molecular mechanism by which FoxO directs MT polarity will elucidate cell-intrinsic programs controlling dendrite branching during neurodevelopment.
Materials and Methods
Fly stocks, alleles, and driver lines
We used 477GAL4, UAS-mCD8-GFP; 477-Gal4, UAS-mCD8RFP; 477-Gal4, UAS-EB1-GFP; and 2-21-Gal4, UAS-mCD8-GFP for visualization of class IV morphology, class IV EB1 comets, and class I morphology (gifts from Melissa Rolls [Pennsylvania State University, University Park, PA, US]). We used foxOΔ94 as a foxO genetic null allele (a gift from Linda Partridge [University College London, London, England, UK]). For MARCM, we used hsFLP, 109(2)80-Gal4, UAS-mCD8-GFP, and FRT82b, TubP-Gal80 from the Bloomington Stock Center (BDSC 8862, BDSC 8768, BDSC 5135). For visualization of classes I–III we used C161-Gal4 (BDSC 27893). For overexpression of FoxO, we used UAS-FoxOWT (listed as FoxO WT #1; a gift from Robert Tjian [University of California, Berkeley, Berkeley, CA, US]) in class I and II and UAS-FoxOWTf19-5 (listed as FoxO WT #2; a gift from Marc Tatar [Brown University, Providence, RI, US]) in class IV. For control RNAi we used Vienna Drosophila RNAi Center lines 25271 (gamma-tubulin 37C RNAi, listed as control RNAi #1) and 33320 (Rtnl2 RNAi, listed as control RNAi #2) (Chen et al., 2012; Hill et al., 2012). For FoxO/Futsch interaction, we used Oregon R, futschk68; a gift from Christian Klämbt (University of Muenster, Muenster, Germany; Hummel et al., 2000), futschk68;; foxOΔ94, and ppk-CD4-GFP (BDSC 35842). For this study, we generated recombinants FRT82b, foxOΔ94 from FRT82b, Sb (a gift from Jocelyn McDonald [Kansas State University, Manhattan, KS, US]), and C161-Gal4, foxOΔ94 from stocks listed above. Recombinants were generated via standard genetic techniques.
Aging, imaging, and analysis
Size and age of larvae were controlled by length. Classification of h AEL was determined by the rostral to caudal length of larvae. Around 2 mm larvae were counted as 72 h AEL, or early 3rd instar larvae. Around 3 mm larvae were counted as 96 h AEL. Around 4 mm larvae were counted as 120 h AEL, or late 3rd instar larvae.
Images were taken using either a Zeiss Axioplan 2 widefield microscope with Colibri.2 LED light system, a Zeiss LSM 510 confocal system, or a Leica SP8 confocal system. For native CD8-GFP and CD4-GFP fluorescence, larvae were flattened beneath a coverslip in 60% glycerol with pressure applied to the caudal end of the animal, such that the guts were pressed out to reduce background fluorescence. For stained preps, larval filets were fixed in 4% PFA for approximately 25 minutes, then stained with chicken anti-GFP primary antibody (Abcam, ab15769) at 1:1000 and labeled with goat anti-chicken 488 secondary antibody at 1:750 (Invitrogen, A-11039). Widefield or confocal z-stacks were converted to 2D projections with Zeiss extended focus and maximum projection, respectively. Projections of larger cells that required multiple image fields were stitched together with either Adobe Photoshop, or the ImageJ FIJI plugins Pairwise Stitching or Grid/Collection Stitching (Preibisch et al., 2009). For clarity in figures, background fluorescence outside the plane of focus of marked cells was at times reduced. Images were traced with a Wacom graphics pad and either Adobe Illustrator or ImageJ. Traces were then analyzed for branch points and length in NeuronStudio (Wearne et al., 2005), while Sholl was analyzed with the FIJI analysis tool Sholl Analysis (Ferreira et al., 2014). Classes with smaller cells were analyzed for length in ImageJ, and branch points were counted in ImageJ with the Cell Counter plugin. Overlays and analysis of 250 μm2 squares to determine internal coverage of class IV cells was accomplished with an ImageJ macro of our own design. The macro defines a grid of Regions of Interest (ROI) based on a defined selection around the cell. The macro then checks each ROI for the presence of the neuron (displayed in green) and tallies it. Afterwards, the macro checks each ROI for the absence of a neuron (displayed in magenta), but does not count ROI outside the defined selection around the cell (displayed in white). The macro will be available at http://imagej.net/User:JamesSears.
For clonal analysis, we followed established heat shock protocols and timings (Grueber et al., 2002; Shrestha and Grueber, 2011). Briefly, crosses were allowed to lay over a 3-hour period onto molasses filled, yeast covered caps at 25 C. Caps were removed and placed at 25 C for an additional 4–5 hours. Each cap was then sealed onto another cap with Parafilm, then placed under a floating foam device and sufficient weight to submerge the caps in a 38 C water bath for a one hour period of heat shock. Caps were removed from the water bath and then returned to 25 C until larvae had developed to the desired stage.
For time-lapse imaging, 96 h AEL larvae were measured and mounted, intact, in 60% glycerol under a coverslip. To prevent damage to the animals, layers of tape were placed between the slide and the coverslip as spacers. Native GFP fluorescence in class IV ddaC cells was imaged with a Zeiss LSM 510 confocal system. Then the larvae were washed in 1X PBS, dried, and allowed to roam freely with food for two hours until the next imaging period.
For EB1-GFP comet imaging, 72 h and 96 h AEL larvae were measured and mounted, intact, in 80% glycerol under a coverslip. To prevent damage to the animals, layers of tape were placed between the slide and the coverslip as spacers. Extra care was taken with 72 h AEL larvae, whose smaller size and shape make imaging throughout the ddaC arbor more challenging. Comets were imaged on a Zeiss Axioplan 2 with a Colibri.2 LED light system, a 100×1.3 NA oil immersion objective, an 800 ms exposure time, and 25% light strength at 2-second intervals. Length in focus was determined in imageJ, and comet direction was counted with the ImageJ plugin Cell Counter. Comets were counted in both higher-order and main branches. Kymographs were generated with the ImageJ FIJI plugin KymoResliceWide.
For antibody staining for Futsch and FoxO, larval filets were fixed in 4% PFA for 25 minutes, dorsal muscles were removed, and preparations were stained. To mark neuronal membranes with HRP, fluorescently conjugated goat anti-HRP-594 was used at 1:500 (Jackson ImmunoResearch Laboratories, Inc., 123-585-021). To stain for FoxO, guinea pig anti-FoxO (Nechipurenko and Broihier, 2012) was used at 1:20 with goat anti-guinea pig 488 secondary antibody at 1:300 (Invitrogen, A-11073). To stain for Futsch, the primary antibody 22C10 (Developmental Studies Hybridoma Bank) was used at 1:8 or 1:10 depending on the aliquot, with goat anti-mouse 568 secondary antibody at 1:300 (Invitrogen, A-11031). For continuous Futsch staining, the first 20 μm of 2° collaterals was assessed for any breaks in Futsch staining.
Behavior
For behavior we modified a larval learning paradigm in order to assess free movement of animals (Gerber et al., 2013). At least one day prior to imaging, Petri dishes were prepared, each with a thin layer of 1% agarose. Wandering third instar larvae were placed in the middle of the Petri dishes, given 10 minutes to acclimate, then recorded for 15 minutes. Movies were converted to 1 Hz, and then analyzed with the FIJI plugin Manual Tracking. For nociceptive responses, we calibrated 30 mN and 50 mN von Frey filaments from 6lb test, 0.23 mm diameter, Omniflex monofilament fishing line (Tracey et al., 2003; Zhong et al., 2010). Larvae were stimulated with a single, quick depression on the dorsal side, until the von Frey filament visibly bent. If at least one 360° roll was observed, it was counted as a positive response. Each larva was stimulated only one time, and if a stimulus glanced the animal, it was not counted.
Statistical Analysis
Statistical analysis was performed in GraphPad Prism. When two groups were compared, unpaired, two-tailed t-tests with Welch’s correction were performed. When more than two groups were compared, one-way ANOVA was performed with multiple comparisons between each group with Tukey’s correction. For categorical data, two-tailed Fisher’s exact tests were performed. For significance, * denotes p<0.05, ** denotes P<0.01, and *** denotes p<0.001. For trending, # denotes p<0.10. No significant difference is n.s.
Supplementary Material
(AC) Class IV ddaC Sholl analysis of controls or control MARCM clones (blue with circles) and foxOΔ94 animals or foxOΔ94 MARCM clones (black with squares) at the indicated ages, displaying the numbers of dendrite intersections at 10 μm intervals from the cell body. Cells in A and B are marked with mCD8-GFP driven by 477-GAL4, while cells in C are marked with mCD8-GFP driven by 109(2)80-GAL4. In A, n=8 control cells from 72 h AEL animals, n=6 foxOΔ94 cells from 72 h AEL animals. In B, n=6 cells from 120 h AEL animals, n=8 foxOΔ94 cells from 120 h AEL animals. In C, n=6 control clones from 120 h AEL animals, n=5 foxOΔ94 clones from 120 h AEL animals.
(A–B) Representative z-projections of class II IdaA cells of the indicated backgrounds marked with mCD8-GFP driven by C161-GAL4. (C) Quantification of the number of dendrite branch points in control animals: 105.4 ± 7.2, n=11 cells; in animals expressing FoxO WT #1: 135.3 ± 4.5 mm, n=6 cells. (D) Quantification of the total dendrite length in control animals: 2.76 ± 0.10 mm, n=11 cells; in animals expressing FoxO WT #1: 2.22 ± 0.09 mm, n=6 cells. Scale bar: 50 μm. Error bars are mean ± s.e.m., **, p<0.01, ***, p<0.001.
(A–D) Representative z-stacks of class IV ddaC cells of the indicated background, marked with CD4-GFP driven by the ppk promoter. (E) Quantification of the number of dendrite branch points in control animals: 926.3 ± 37.4, n=8 cells; foxOΔ94 animals: 556.3 ± 37.6, n=8 cells; futschk68/+;; foxOΔ94 animals: 708.5 ± 28.3, n=6 cells; futschk68/+ animals: 736.4± 46.5, n=5 cells. (F) Quantification of the total dendrite length in control animals: 26.18 ± 0.42 mm, n=8 cells; foxOΔ94 animals: 16.14 ± 0.71 mm, n=8 cells; futschk68/+;; foxOΔ94 animals: 19.64 ± 0.41 mm, n=6 cells; futschk68/+ animals: 22.40 ± 0.82 mm, n=5 cells. Scale bar: 50 μm. Error bars are mean ± s.e.m., n.s., not significantly different; *, p<0.05, **, p<0.01, ***, p<0.001.
Representative movies of class IV ddaC cells expressing EB1-GFP driven by 477-GAL4. A control cell is presented in the left panel, and a foxOΔ94 cell is presented in the right panel, both at 96 h AEL. The arrows mark the retrograde direction, toward the cell body. Scale bar: 5 μm.
Representative movies of class IV ddaC cells expressing EB1-GFP driven by 477-GAL4. A control RNAi #1-expressing cell is presented in the left panel, and a FoxO WT #2-expressing cell is presented in the right panel, both at 96 h AEL. The arrows mark the retrograde direction, toward the cell body. Scale bar: 5 μm.
Highlights.
-FoxO promotes dendrite branching in all classes of Drosophila dendritic arborization (da) neurons
-FoxO is required for initiating growth of new branches and for maintaining existing branches
-FoxO is necessary and sufficient for anterograde microtubule polymerization in dendrites
-FoxO promotes proper larval nociception
Acknowledgments
We are grateful to Melissa Rolls (Pennsylvania State University, University Park, PA, US), for her support and guidance. We thank Melissa Rolls, Jocelyn McDonald (Kansas State University, Manhattan, KS, US), Linda Partridge (University College London, London, England, UK), Robert Tjian (University of California, Berkeley, Berkeley, CA, US), the Bloomington Drosophila Stock Center, and the Vienna Drosophila RNAi Center for fly strains. We thank the Developmental Studies Hybridoma Bank for antibodies, the ImageJ community for insight and inspiration, Nan Liu and Priya Tumuluru for technical assistance, the Case Western Reserve University Neurosciences/Genetics Imaging facility for providing confocal microscopes and support, and members of the Broihier lab for helpful discussion and comments on the manuscript. This work was supported by NIH T32AG00271 to JCS and NIH R56NS055245 and R21NS090369 to HTB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baas PW, Lin S. Hooks and comets: The story of microtubule polarity orientation in the neuron. Dev Neurobiol. 2011;71:403–418. doi: 10.1002/dneu.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Chen L, Stone MC, Tao J, Rolls MM. Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proc Natl Acad Sci USa. 2012;109:11842–11847. doi: 10.1073/pnas.1121180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen R, de la Torre-Ubieta L, Bonni A, Colón-Ramos DA. A conserved PTEN/FOXO pathway regulates neuronal morphology during C. elegans development. Development. 2011;138:5257–5267. doi: 10.1242/dev.069062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carré C, Noselli S, Léopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, Ou Y, Li S, Giniger E, van Meyel DJ. Dendrite architecture organized by transcriptional control of the F-actin nucleator Spire. Development. 2014;141:650–660. doi: 10.1242/dev.099655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B, Biernacki R, Thum J. Odor-taste learning assays in Drosophila larvae. Cold Spring Harb Protoc. 2013;2013 doi: 10.1101/pdb.prot071639. pdb.prot071639. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Gu J, Firestein BL, Zheng JQ. Microtubules in dendritic spine development. J Neurosci. 2008;28:12120–12124. doi: 10.1523/JNEUROSCI.2509-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006;7:224. doi: 10.1186/gb-2006-7-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SE, Parmar M, Gheres KW, Guignet MA, Huang Y, Jackson FR, Rolls MM. Development of dendrite polarity in Drosophila neurons. Neural Dev. 2012;7:34. doi: 10.1186/1749-8104-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Viesselmann C, Nam S, Merriam E, Dent EW. Activity-dependent dynamic microtubule invasion of dendritic spines. J Neurosci. 2008;28:13094–13105. doi: 10.1523/JNEUROSCI.3074-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, Tracey WD. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T, Causeret F, Nishimura YV, Terao M, Norman A, Hoshino M, Nikolić M. Localized activation of p21-activated kinase controls neuronal polarity and morphology. J Neurosci. 2007;27:8604–8615. doi: 10.1523/JNEUROSCI.0765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, Spangler SA, Di Stefano P, Demmers J, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- la Torre-Ubieta de L, Gaudillière B, Yang Y, Ikeuchi Y, Yamada T, DiBacco S, Stegmüller J, Schüller U, Salih DA, Rowitch D, et al. A FOXO-Pak1 transcriptional pathway controls neuronal polarity. Genes Dev. 2010;24:799–813. doi: 10.1101/gad.1880510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Sanes JR, Kay JN. Development of dendritic form and function. Annu Rev Cell Dev Biol. 2015;31:741–777. doi: 10.1146/annurev-cellbio-100913-013020. [DOI] [PubMed] [Google Scholar]
- Li W, Wang F, Menut L, Gao FB. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43:823–834. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM. Directed microtubule growth, +TIPs, and kinesin-2 are required for uniform microtubule polarity in dendrites. Curr Biol. 2010;20:2169–2177. doi: 10.1016/j.cub.2010.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel J, Delandre C, Zhang Y, Förstner F, Moore AW, Tavosanis G. Fascin controls neuronal class-specific dendrite arbor morphology. Development. 2012;139:2999–3009. doi: 10.1242/dev.077800. [DOI] [PubMed] [Google Scholar]
- Nechipurenko IV, Broihier HT. FoxO limits microtubule stability and is itself negatively regulated by microtubule disruption. J Cell Biol. 2012;196:345–362. doi: 10.1083/jcb.201105154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DAM, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Jegla TJ. Neuronal polarity: an evolutionary perspective. J Exp Biol. 2015;218:572–580. doi: 10.1242/jeb.112359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, Doe CQ. Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, Hummel T, Ng N, Klämbt C, Davis GW. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26:371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Salih DAM, Rashid AJ, Colas D, la Torre-Ubieta de L, Zhu RP, Morgan AA, Santo EE, Ucar D, Devarajan K, Cole CJ, et al. FoxO6 regulates memory consolidation and synaptic function. Genes Dev. 2012;26:2780–2801. doi: 10.1101/gad.208926.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C, Bashaw GJ. Transcription factors and effectors that regulate neuronal morphology. Development. 2014;141:4667–4680. doi: 10.1242/dev.110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha BR, Grueber WB. Generation and staining of MARCM clones in Drosophila. Cold Spring Harb Protoc. 2011;2011:973–979. doi: 10.1101/pdb.prot5659. [DOI] [PubMed] [Google Scholar]
- Slack C, Giannakou ME, Foley A, Goss M, Partridge L. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell. 2011;10:735–748. doi: 10.1111/j.1474-9726.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Ori-McKenney KM, Zheng Y, Han C, Jan LY, Jan YN. Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev. 2012;26:1612–1625. doi: 10.1101/gad.193243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Tsubouchi A, Rolls MM, Tracey WD, Sherwood NT. Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. J Neurosci. 2012;32:11631–11642. doi: 10.1523/JNEUROSCI.0729-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Albertson RM, Chen L, Rolls MM. Dendrite injury triggers DLK-independent regeneration. Cell Rep. 2014;6:247–253. doi: 10.1016/j.celrep.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Roegiers F, Rolls MM. Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell. 2008;19:4122–4129. doi: 10.1091/mbc.E07-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Satoh D, Estes P, Crews S, Uemura T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron. 2004;43:809–822. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang MW, Kim JH, Macara AM, Sterne G, Yang T, Ye B. The Krüppel-Like Factor Dar1 Determines Multipolar Neuron Morphology. J Neurosci. 2015;35:14251–14259. doi: 10.1523/JNEUROSCI.1610-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- Yalgin C, Ebrahimi S, Delandre C, Yoong LF, Akimoto S, Tran H, Amikura R, Spokony R, Torben-Nielsen B, White KP, et al. Centrosomin represses dendrite branching by orienting microtubule nucleation. Nat Neurosci. 2015;18:1437–1445. doi: 10.1038/nn.4099. [DOI] [PubMed] [Google Scholar]
- Ye B, Kim JH, Yang L, McLachlan I, Younger S, Jan LY, Jan YN. Differential regulation of dendritic and axonal development by the novel Krüppel-like factor Dar1. J Neurosci. 2011;31:3309–3319. doi: 10.1523/JNEUROSCI.6307-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(AC) Class IV ddaC Sholl analysis of controls or control MARCM clones (blue with circles) and foxOΔ94 animals or foxOΔ94 MARCM clones (black with squares) at the indicated ages, displaying the numbers of dendrite intersections at 10 μm intervals from the cell body. Cells in A and B are marked with mCD8-GFP driven by 477-GAL4, while cells in C are marked with mCD8-GFP driven by 109(2)80-GAL4. In A, n=8 control cells from 72 h AEL animals, n=6 foxOΔ94 cells from 72 h AEL animals. In B, n=6 cells from 120 h AEL animals, n=8 foxOΔ94 cells from 120 h AEL animals. In C, n=6 control clones from 120 h AEL animals, n=5 foxOΔ94 clones from 120 h AEL animals.
(A–B) Representative z-projections of class II IdaA cells of the indicated backgrounds marked with mCD8-GFP driven by C161-GAL4. (C) Quantification of the number of dendrite branch points in control animals: 105.4 ± 7.2, n=11 cells; in animals expressing FoxO WT #1: 135.3 ± 4.5 mm, n=6 cells. (D) Quantification of the total dendrite length in control animals: 2.76 ± 0.10 mm, n=11 cells; in animals expressing FoxO WT #1: 2.22 ± 0.09 mm, n=6 cells. Scale bar: 50 μm. Error bars are mean ± s.e.m., **, p<0.01, ***, p<0.001.
(A–D) Representative z-stacks of class IV ddaC cells of the indicated background, marked with CD4-GFP driven by the ppk promoter. (E) Quantification of the number of dendrite branch points in control animals: 926.3 ± 37.4, n=8 cells; foxOΔ94 animals: 556.3 ± 37.6, n=8 cells; futschk68/+;; foxOΔ94 animals: 708.5 ± 28.3, n=6 cells; futschk68/+ animals: 736.4± 46.5, n=5 cells. (F) Quantification of the total dendrite length in control animals: 26.18 ± 0.42 mm, n=8 cells; foxOΔ94 animals: 16.14 ± 0.71 mm, n=8 cells; futschk68/+;; foxOΔ94 animals: 19.64 ± 0.41 mm, n=6 cells; futschk68/+ animals: 22.40 ± 0.82 mm, n=5 cells. Scale bar: 50 μm. Error bars are mean ± s.e.m., n.s., not significantly different; *, p<0.05, **, p<0.01, ***, p<0.001.
Representative movies of class IV ddaC cells expressing EB1-GFP driven by 477-GAL4. A control cell is presented in the left panel, and a foxOΔ94 cell is presented in the right panel, both at 96 h AEL. The arrows mark the retrograde direction, toward the cell body. Scale bar: 5 μm.
Representative movies of class IV ddaC cells expressing EB1-GFP driven by 477-GAL4. A control RNAi #1-expressing cell is presented in the left panel, and a FoxO WT #2-expressing cell is presented in the right panel, both at 96 h AEL. The arrows mark the retrograde direction, toward the cell body. Scale bar: 5 μm.