Abstract
Although ribosomes are ubiquitously expressed and essential for life, recent data indicate that monogenic causes of ribosomal dysfunction can confer a remarkable degree of specificity in terms of human disease phenotype. Box C/D small nucleolar RNAs (snoRNAs) are evolutionarily conserved non-protein encoding RNAs involved in ribosome biogenesis. Here we show that biallelic mutations in the gene SNORD118, encoding the box C/D snoRNA U8, cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts (LCC), presenting at any age from early childhood to late adulthood. These mutations affect U8 expression, processing and protein binding and thus implicate U8 as essential in cerebral vascular homeostasis.
Introduction
In 1996, Pierre Landrieu and colleagues described three unrelated children with a purely neurological disorder characterized by the radiological triad of cerebral white matter disease (leukoencephalopathy), intracranial calcification and cysts (LCC)(Fig. 1)1. This disease manifests as a progressive cerebral degeneration, where a microangiopathy characterized by angiomatous-like blood vessels with gliosis and Rosenthal fiber deposition is the most striking pathological feature (see Supplementary Fig. 1)1–3. The observation of sibling pairs, including affected females, suggested that LCC is a genetic disorder, likely inherited as an autosomal recessive trait. However, uncertainty has remained over the molecular basis of this phenotype, not least because of the wide range of age at presentation that has been observed4. Here, we define LCC as an apparently single-gene disorder due to germ-line biallelic mutations in the box C/D snoRNA U8, thus indicating that LCC may represent a novel ribosomopathy. Furthermore, the genetic architecture of LCC illustrates some of the difficulties associated with the attribution of causation to sequence variants in non-protein encoding genomic DNA.
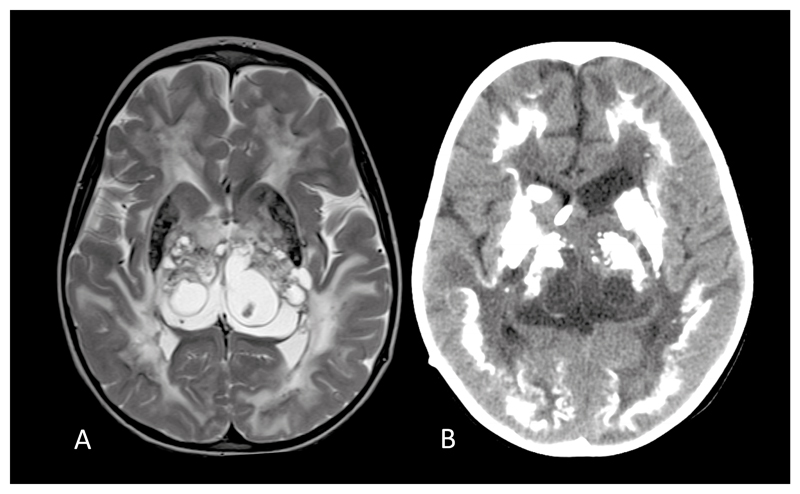
Fig. 1. Typical magnetic resonance (MR) and computed tomography (CT) appearances of LCC.
(a). Axial T2 cranial MR at 4 years of age of patient F172 demonstrating symmetrical high signal of the periventricular, deep and subcortical white matter, multiple cysts within the thalami and basal ganglia, and calcification involving the putamen. (b). Cranial CT of the same patient at age 5 years showing dense, rock-like calcification in the basal ganglia and thalami. There is also dense calcification of the deep cortex and some deep white matter calcification.
Results
Identification of SNORD118 mutations in patients with LCC
Over a period of 12 years we collected clinical data and biological samples relating to 40 patients affected with LCC (see Supplementary Table 1). Surprisingly for a rare autosomal recessive disorder there was a relative paucity of consanguineous families in our cohort (only two of 33 pedigrees), strongly suggesting that homozygous mutations might not be typically associated with this phenotype. These 40 patients, who were mainly of white European ancestry, demonstrated an age at presentation ranging from early infancy to 54 years of life. In an attempt to define the genetic basis of LCC we sequenced the exomes of 19 affected individuals, and analyzed the data both on the basis of an autosomal recessive trait and an autosomal dominant model with reduced penetrance. However, no mutations were identified (data not shown). We then pursued a different strategy, using linkage and haplotype analysis in five pairs of affected siblings born to unrelated parents, and two singletons who were the product of separate consanguineous unions. In this way, genome-wide we were able to identify a single region of > 1 Mb in size with a LOD score > 3, giving a minimal mapping locus of 1.2 Mb on chromosome 17 (genomic coordinates 7,721,931-8,930,080, GRCh37) (LOD score of 6.02), indicating that LCC disease-causing variants lie within this interval (see Supplementary Fig. 2).
Considering the absence of any obvious pathogenic variants on re-examination of our sequence data covering the coding exons and essential splice sites in this mapping region, we undertook a capture sequencing assay of 3 million base-pairs (bp) of genomic DNA on chromosome 17 (coordinates: 7,000,000 – 10,000,000) using samples from 10 unrelated patients. In each of these affected individuals we identified two rare variants (defined as a frequency of < 0.005 alleles on the Exome Aggregation Consortium (ExAC) database) lying within a 199 bp stretch of DNA (8,076,761 - 8,076,960) encompassing the gene SNORD118 (NR_033294.1). Sanger sequencing confirmed these changes, leading us to analyze a further 30 patients demonstrating typical clinical and neuroradiological characteristics of LCC. In total, we observed two rare sequence variants to segregate with phenotypic status (40 affected individuals; five unaffected full siblings) in all 33 families in our cohort (Table 1, Fig. 2, Supplementary Table 2). Where DNA was available (18 families), all parents showed appropriate heterozygosity for a single variant except in two cases: in F819, the mother carried two rare variants, and her two affected children each inherited a distinct maternal rare allele in combination with a paternally-derived genomic deletion of SNORD118 (see Supplementary Fig. 3); whilst in F906, an n.103G>A nucleotide alteration arose de novo on the paternal allele (microsatellite analysis confirming paternity, see Supplementary Table 3).
Table 1. SNORD118 variants identified in each LCC family.
Family number, structure (persons genotyped), ethnicity, variants detected, their zygosity and their frequency on ExAC.
| Family | Family members tested | Ethnicity | Chromosomal position (Hg19)† | Variants detected | Zygosity | ExAC frequency |
|---|---|---|---|---|---|---|
| F172 | 1A | White European (Swedish) | g.8076761C>A | n.*10G>T | Het | 0.002056 |
| g.8076851dup | n.56dup | Het | Novel | |||
| F278 | 1A, 3U, M, F | White European (North American) | g.8076832T>C | n.75A>G | Het | Novel |
| g.8076899C>G | n.8G>C | Het | 0.00000887 | |||
| F281 | 1A, M, F | White European (Estonian) | g.8076826C>G | n.81G>C | Het | Novel |
| g.8076904G>A | n.3C>T | Het | 0.001473 | |||
| F285 | 1A | White European (British) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076850C>T | n.57G>A | Het | Novel | |||
| F309 | 1A | White European (North American) | g.8076776G>C | n.131C>G | Het | Novel |
| g.8076904G>A | n.3C>T | Het | 0.001473 | |||
| F330 | 1A | White European (British) | g.8076825T>C | n.82A>G | Het | 0.00007093 |
| g.8076899C>T | n.8G>A | Het | 0.002803 | |||
| F331 | 2A, M, F | White European (German) | g.8076761C>A | n.*10G>T | Het | 0.002056 |
| g.8076835T>C | n.72A>G | Het | 0.00007093 | |||
| F334 | 1A | White European (British) | g.8076825T>C | n.82A>G | Het | 0.00007093 |
| g.8076761C>A | n.*10G>T | Het | 0.002056 | |||
| F337 | 1A, M, F | White European (Italian) | g.8076905A>G | n.2T>C | Het | 0.0000355 |
| g.8076849dup | n.58dup | Het | Novel | |||
| F343 | 1A | White European (Dutch) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076885_8076913dup | n.-7_22dup | Het | Novel | |||
| F344 | 1A | East African (Somali) | g.8076794G>A | n.113C>T | Hom# | 0.00001773 |
| g.8076899C>G | n.8G>C | Hom# | 0.00000887 | |||
| F362 | 3A (2 sequenced), 1U, M, F | White European (British) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076887G>A | n.20C>T | Het | 0.00001774 | |||
| F414 | 1A | White European (Finnish) | g.8076770G>A | n.*1C>T | Het | 0.00473 |
| g.8076846T>C | n.61A>G | Het | 0.00007980 | |||
| F426 | 2A, M, F | White European (North American) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076826C>T | n.81G>A | Het | 0.00003546 | |||
| F433 | 1A | White European (British) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076887G>A | n.20C>T | Het | 0.00001774 | |||
| F445 | 1A | White European (British) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076780G>C | n.127C>G | Het | 0.00005322 | |||
| F446 | 1A | White European (British) | g.8076766G>C | n.*5C>G | Hom | 0.0005781 |
| F454 | 2A, M, F | White European (Canadian) | g.8076766G>C | n.*5C>G | Het | 0.0005781 |
| g.8076955_8076960del | n.-54_-49del | Het | Novel | |||
| F465 | 1A, M, F | White European (German) | g.8076825T>C | n.82A>G | Het | 0.00007093 |
| g.8076846_8076847insA | n.60_61insT | Het | 0.00001773 | |||
| F521 | 2A, M, F | White European (North American) | g.8076803C>T | n.104G>A | Het | 0.0004877 |
| g.8076776G>C | n.131C>G | Het | Novel | |||
| F551 | 1A | White European (Belgium) | g.8076762G>A | n.*9C>T | Het | 0.00202 |
| g.8076780G>C | n.127C>G | Het | 0.00005322 | |||
| F564 | 1A | White European (Belgium) | g.8076770G>A | n.*1C>T | Het | 0.00473 |
| g.8076781G>A | n.126C>T | Het | 0.00009756 | |||
| F691 | 1A, M, F | White European (British) | g.8076762G>A | n.*9C>T | Het | 0.00202 |
| g.8076849T>C | n.58A>G | Het | 0.0000266 | |||
| F730 | 1A | White European (Australian) | g.8076761C>A | n.*10G>T | Het | 0.002056 |
| g.8076826C>T | n.81G>A | Het | 0.00003546 | |||
| F766 | 1A, M, F | White European (British) | g.8076865C>T | n.42G>A | Het | 0.00102 |
| g.8076904G>T | n.3C>A | Het | 0.000008875 | |||
| F780 | 2A, 1U, M, F | White European (North American) | g.8076770G>A | n.*1C>T | Het | 0.00473 |
| g.8076776G>C | n.131C>G | Het | Novel | |||
| F819 | 2A, M | White European (British) | g.8076762G>A g.8076770G>A g.8076696_8076977del§ |
n.*9C>T n.*1C>T |
Het Het Het |
0.00202 0.00473 Novel |
| F906 | 1A, M, F | White European (Italian) | g.8076804C>T | n.103G>A (de novo) | Het | 0.00001773 |
| g.8076868C>G | n.39G>C | Het | 0.0001419 | |||
| F1127 | 1A, M, F | White European (French) | g.8076762G>A | n.*9C>T | Het | 0.00202 |
| g.8076807A>C | n.100T>G | Het | Novel | |||
| F1172 | 1A | White European (German) | g.8076762G>A | n.*9C>T | Het | 0.00202 |
| g.8076776G>C | n.131C>G | Het | Novel | |||
| F1288 | 1A, M, F | White European (North American) | g.8076848A>C | n.59T>G | Het | Novel |
| g.8076762G>A | n.*9C>T | Het | 0.00202 | |||
| F1424 | 1A, M, F | Mixed white European (German) / north African | g.8076777A>G | n.130T>C | Het | Novel |
| g.8076912C>T | n.-6G>A | Het | 0.0009584 | |||
| F1445 | 1A, M, F | White European (British) | g.8076826C>T | n.81G>A | Het | 0.00003546 |
| g.8076904G>A | n.3C>T | Het | 0.001473 |
Het = heterozygous; Hom = homozygous. ExAC = Exome Aggregate Consortium
All genomic coordinates should be preceded by Chr17(GRCh37):
Deletion extends beyond these boundaries, but boundaries have not been fully defined. One sibling carries g.8076762G>A plus g.8076696_8076977del. The other sibling carries g.8076770G>A plus g.8076696_8076977del. Mum carries g.8076762G>A plus g.8076770G>A
In F344, both of these rare variants were seen in the homozygous state. However, n.8G>C was also observed in F278, suggesting that this is the likely pathogenic variant
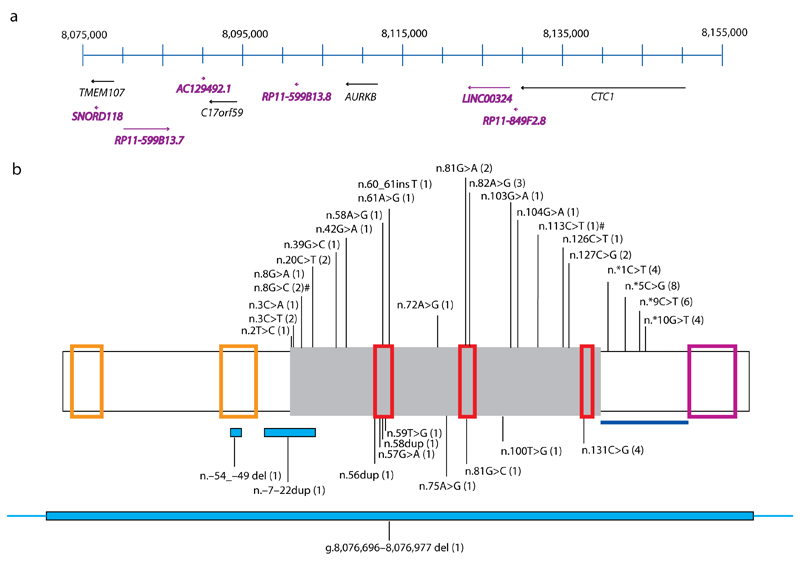
Fig. 2. Schematic of chromosome 17p13.1 and SNORD118.
(a) Genes across chromosome 17p13.1 between 8,075,000 and 8,155,000 are drawn to scale (numbered according to GRCh37). Protein encoding genes are represented in black text, whilst non-protein encoding genes (including SNORD118) are annotated in purple. SNORD118 lies within the 3’ UTR of TMEM107 and 50 kb from CTC1. (b) Positions of variants identified in SNORD118. Green box represents mature SNORD118. From 5’ to 3’, the orange boxes highlight the distal (DSE) and proximal (PSE) sequence elements. From 5’ to 3’, the C box, LSm and D box binding sites of SNORD118 are shown by the red boxes. The violet box represents the 3’ box (end of precursor transcript). The blue line represents the sequence encompassing the 3’ precursor transcripts of SNORD118 which are intermediates of the mature transcript. Variants that have been seen on the ExAC browser are shown above the box, with novel variants not seen on ExAC shown below. The number of LCC families with each variant is shown in brackets. Deletions and duplications are represented by blue boxes beneath the schematic. # In F344, both of these rare variants were seen in the homozygous state. However, n.8G>C was also observed in F278, suggesting that this is the likely pathogenic variant.
Of the total of 36 rare putative pathogenic variants observed in SNORD118, 13 were novel (i.e. they were not annotated on the ExAC database, comprising more than 112,000 annotated alleles for each of these 12 variants, or in our in-house data-set of > 5,000 exomes). In 15 and 16 of 33 families, the affected individuals were compound heterozygous for either one completely novel allele and one rare allele, or for two rare alleles (where the rarer of the two variants has an ExAC allele frequency of < 0.00009756 in all cases) respectively. Consistent with our linkage analysis, two unrelated patients born to consanguineous parents demonstrated homozygosity for a rare allele (with allele frequencies of 0.0005781 and 0.00000887 for these putative mutations and no recorded homozygotes on the ExAC database). As further proof that biallelic variants in SNORD118 are causative of LCC, we noted recurrent putative mutant alleles in our cohort. Specifically, eleven novel / rare variants were observed in more than one family, with a mutant allele shared by four or more different sets of families at five distinct nucleotide positions. One of these alleles, n.131C>G, was seen in four LCC families, but is not recorded on the ExAC database of more than 112,000 alleles at this position, whilst an n.*5C>G variant, observed in the compound heterozygous state in eight disease pedigrees (i.e. 8 of 66 alleles in affected individuals), has an ExAC frequency of 0.0005781 (1 in 1730 of control alleles)(8 in 66 versus 1 in 1730, Chi-squared p < 0.000005). Importantly, screening of a panel of 677 European controls to determine the frequency of biallelic novel / rare variants in the same person, which is not possible to derive from ExAC data, revealed only four individuals to carry two rare variants on distinct alleles (four in 677 versus 20 of 20 LCC probands where it was possible to test for / impute biallelic inheritance; p < 0.000005 Chi squared test) (see Supplementary Tables 4 and 5). Of further note, none of these alleles were novel, and only one was seen in our patient cohort. Thus, despite a remarkable variability in age at presentation between some patients, ranging from the neonatal period (e.g. F172, F344) to the sixth decade of life (e.g. F433, F1172), these data indicate that LCC is a genetically homogeneous disorder and that we have identified the disease-causing genomic variants in all 40 patients conforming to the LCC phenotype in our study.
Functional analysis of a selection of SNORD118 variants
SNORD118 encodes the box C/D small nucleolar RNA (snoRNA) U8. Box C/D snoRNAs are evolutionarily conserved RNAs involved in ribosomal biogenesis and function5. The U8 snoRNA is independently transcribed, producing precursor U8 snoRNAs which are processed to mature box C/D U8 snoRNAs6. The conserved box C/D motif binds to four core proteins, namely 15.5K, NOP56, NOP58 and fibrillarin7. These proteins assemble onto the box C/D motif in a stepwise manner, with the initial binding of 15.5K necessary for recruitment of the three remaining proteins, thus facilitating stabilization, processing and trafficking. U8 also contains a well conserved LSm (like Sm) binding site8. Seven individual LSm proteins assemble into a ring structure which binds U8 to form a ribonucleoprotein complex. This U8 snoRNP is necessary for ribosomal RNA processing9.
Of the 36 mutations in U8 that we identified, one (n.-54_-49del) lies within a recognized 5’ enhancer element important for U8 expression6, 29 are within the U8 sequence, and four are situated immediately 3’ of the U8 sequence in a region which is transcribed but then removed by processing of the precursor U8 transcript10. We also identified one complete gene deletion (in F819), and a further variant just 5’ of the start of the U8 sequence (F1424) of undetermined functional consequence. Within the mature U8 sequence itself, 10 variants were observed in the C/D box and LSm binding site, affecting nucleotides which are strictly conserved across evolution (see Supplementary Fig. 4). Additionally, the variants at n.103, n.104 and n.113 lie within the stem of a hairpin loop which is very well conserved and would be predicted to decrease the stability of this structure (see Supplementary Fig. 5); whilst the three variants at n.81 and the variant at n.82 are situated within the highly conserved GAUU motif of the LSm binding site.
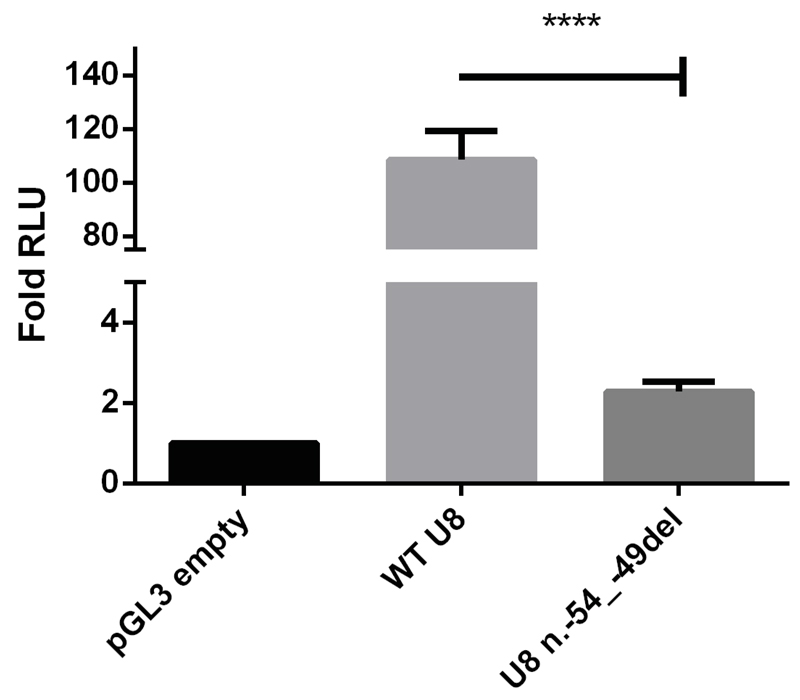
To demonstrate a functional consequence of a selection of these U8 variants, we first made a reporter construct encompassing the entire U8 promoter region both as wild type (WT) and with the n.-54_-49del variant. In keeping with a promotor function, we observed a 109-fold increase in expression with the WT sequence compared to empty vector. In contrast, the n.-54_-49del variant was associated with a profound loss of transcriptional activity (Fig. 3).
Fig. 3. Variant n.-54_-49del found in F454 reduces the activity of the PSE element in dual luciferase assays.
HeLa cells were transfected with the Promega pGL3 reporter vector carrying the wild type PSE without the deletion (WT U8), with n.-54_-49del (U8 n.-54_-49del), or the reporter vector without an insert (pGL3 empty). The WT U8 PSE vector functioned as a promoter, enhancing luciferase activity by a mean of 109-fold in comparison with empty vector. In contrast, the n.-54_-49del vector demonstrated a mean of 2 fold activity compared to empty vector. Data presented relate to the mean fold change (+/- SD) of relative light units (RLU) compared to the control vector for three independent experiments each with three technical replicates. Data were analyzed using a one way Anova with multiple comparisons where **** = p<0.0001.
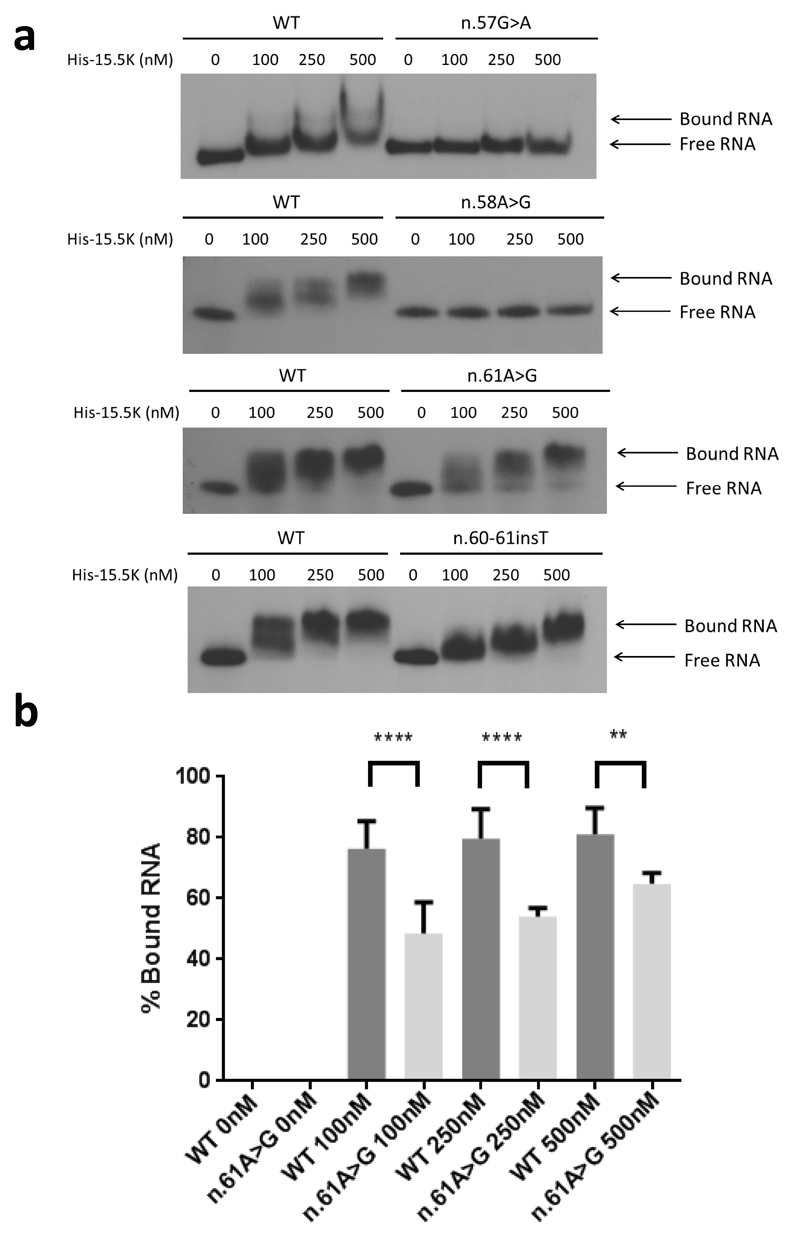
We then assessed the effect of four variants (n.57G>A; n.58A>G; n.61A>G; n.60_61insT) observed in the U8 box C region responsible for the interaction of U8 with the 15.5K protein. These patient-related putative mutations all displayed reduced binding to the 15.5K protein compared to WT (Fig. 4). Binding of His-15.5K with n.57G>A and n.58A>G was severely impaired (Fig 4a, Supplementary Fig. 6), concordant with previous studies which demonstrate that disruption of the essential GA dinucleotide within the asymmetric internal loop completely abrogates 15.5K binding11. A more subtle reduction in 15.5K binding was seen with n.61A>G and n.60-61insT (Fig 4a, 4b). The nucleotides at positions n.60 and n.61 form part of a conserved stem loop II of C/D box motifs which is necessary for the assembly of the C/D box snoRNP complex12.
Fig. 4. Protein binding of U8 variants.
(a). Electrophoretic mobility shift assay (EMSA) using wild type (WT) and mutant 5’ end-labeled in vitro transcribed U8 snoRNA with increasing concentrations of recombinant 6His-tagged 15.5K protein (His-15.5K). The concentration of the recombinant protein is given in nM above the panels. Binding of WT RNA resulted in a shift in mobility at 100 nM. Binding of His-15.5K with n.57G>A, and n.58A>G was severely impaired. A shift in mobility could not be observed for n.57G>A or n.58A>G at protein concentrations up to 500 nM. Binding between His-15.5K and n.61A>G demonstrated a shift in mobility at 100 nM; however, this shift was less than observed in WT RNA at the same concentration, and excess free RNA can be seen at all concentrations up to 500 nM indicating that binding is impaired. Similarly binding between His-15.5K and n.60_61insT demonstrated a shift in mobility at 100 nM. This shift was less than observed in WT RNA and equivalent shift was only achieved at the highest concentration tested 500nM, indicating that binding is impaired. (b). Quantification of binding between His 15.5K and n.61A>G compared to WT RNA. The percentage of protein bound RNA is significantly decreased at all concentrations. Data are given as the mean +/- SD; n= 4 independent experiments. Data were analyzed using a one way Anova with multiple comparisons where **** = p<0.0001.
Finally, to explore the effect of variants in the region immediately downstream of U8, including the n.*5C>G sequence alteration seen in eight families, we examined the in vitro processing of precursor U8 snoRNAs with extended 3’ regions encompassing four distinct variants. Previous work has defined a highly stereotyped pattern of processing of the SNORD118 transcript10, and we observed disturbed processing of the precursor U8 RNAs with four mutations compared to WT (Fig. 5, Supplementary Fig. 7a). Additionally, we examined in vitro processing of precursor U8 with the C box mutation n.58A>G located away from the extended 3’ region. The processing pattern observed with the n.58A>G mutant was indistinguishable from WT U8 (Supplementary Fig. 7b) indicating that variants located in the 3’ extension of U8 specifically effect the processing of the precursor U8 snoRNA.
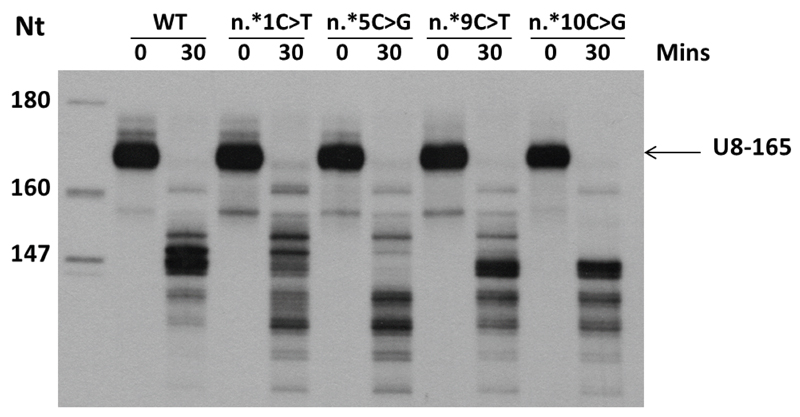
Fig. 5. 3’ end precursor processing of U8 variants.
In vitro 3’ end processing of 5’ end labeled in vitro transcribed precursor U8 snoRNA (U8-165) in HeLa nuclear extracts. At 30 minutes, multiple pre-U8 snoRNA processing intermediates can be seen with the wild-type (WT) pre-U8 snoRNA. Four mutants (n.*1C>T, n.*5C>G, n.*9C>T and n.*10C>G) in the pre-U8 snoRNA at 1, 5, 9 and 10 nucleotides downstream of the mature 3’ end of U8 were assessed. All four mutants exhibited a difference in processing intermediates compared to WT. Furthermore, all mutant RNAs displayed additional shorter intermediates which are not present with the WT RNA.. Data are representative of 3 independent experiments.
Together, these data provide compelling evidence that the disease-associated variants that we identified are likely to have a functional effect on U8 activity.
LCC patient fibroblasts demonstrate a cellular phenotype
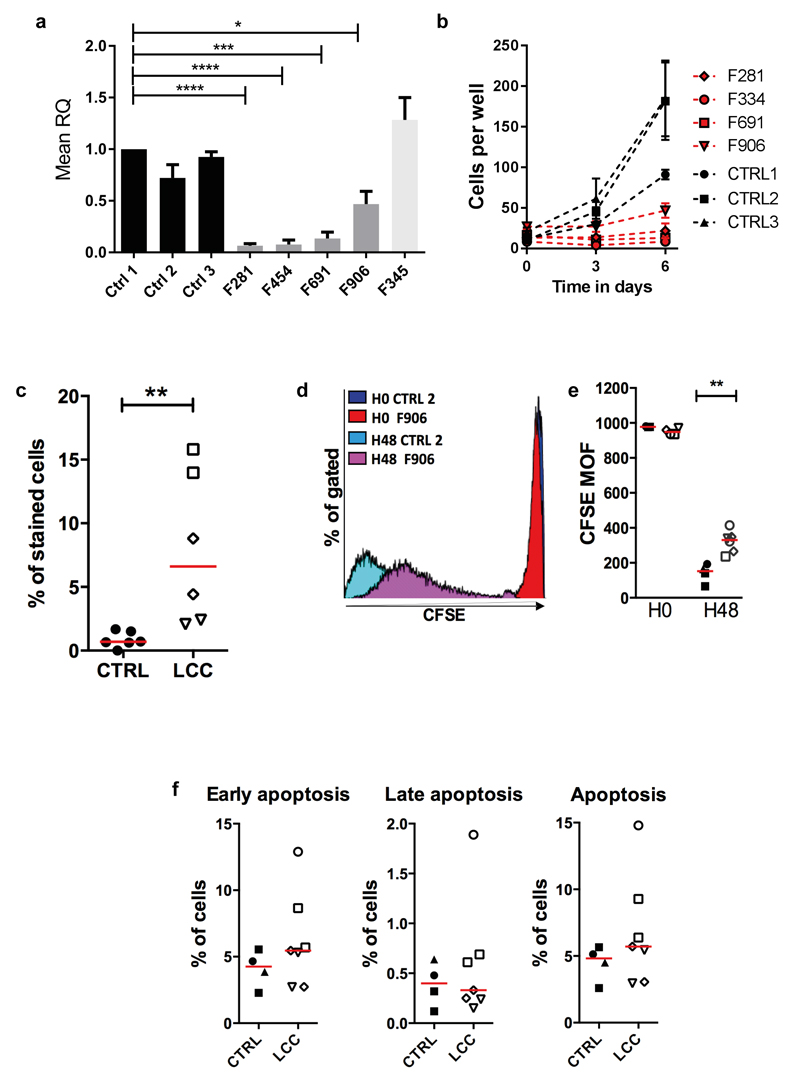
Although the clinical features of LCC are apparently limited to the central nervous system, we observed a marked reduction in expression of U8 in patient fibroblasts compared to control cells (Fig. 6a). Furthermore, primary fibroblasts from patients with LCC did not grow as well as cells from controls (Fig. 6b). A higher percentage of LCC fibroblasts were senesced compared to control cells (Fig. 6c), and a proliferation defect was confirmed by carboxyfluorescein succinimidyl ester (CFSE) labeling (Fig. 6d and e). This proliferative defect was not associated with increased apoptosis (Fig. 6f), and we did not observe a disturbance in cell cycling under basal conditions or after treatment with the DNA cross-linking agent mitomycin C (see Supplementary Fig. 8). Coats plus (CP) patient fibroblasts also demonstrate proliferative defects (secondary to a problem with telomere homeostasis)13,14, but qPCR data from fibroblasts of a CP patient (F345) showed levels of U8 comparable to controls. Due to an inability to grow enough cells, we were unable to investigate translation efficiency in fibroblasts. EBV transformed patient-derived lymphoblasts, which grow normally (data not shown), did not demonstrate any abnormality on polyribosome (polysome) fractionation analysis (see Supplementary Fig. 9).
Fig. 6. Defective proliferation of LCC fibroblasts.
(a). Quantitative reverse transcription PCR (qPCR) of SNORD118 expression in three control (CTRL1, 2, 3), four LCC (F281, F454, F691, F906), and one CP patient (F345) primary fibroblast cell lines, normalized to two housekeeping genes, RNU24 and U6. RQ is equal to 2-ΔΔCt i.e. normalized fold change relative to CTRL1. Data given as mean +/- SEM; n=3 independent experiments. Data analyzed using one way Anova with multiple comparisons **** = p<0.0001. (b). Proliferation of patient (F281, F334, F691, F906) and control (CTRL1, 2, 3) fibroblasts. The passage number of patient cells was the same or lower than controls, except for F691 which had 3 more passages. Data given as mean +/- SEM; n=2 independent experiments. (c). Percentage of beta galactosidase positive control (n=3) and LCC (n=3) fibroblasts. Red bar represents median value for each group. Mann Whitney U test **p<0.01. (d). Representative histogram of fibroblasts from one patient (F906) and one control (CTRL2). Mean of Fluorescence (MOF) assessed at 30 minutes (H0) and 2 days (H48) after carboxyfluorescein succinimidyl ester (CFSE) labeling. (e). Quantification of mean CFSE fluorescence in fibroblasts from patients (n=4) and controls (n=3). Red bar represents median value for each group. Mann Whitney U test **p<0.01; n=2 independent experiments. (f). Percentage cells in early, late and total apoptosis for four patients and three controls. Red bar represents median value for each group. No significant difference by Mann Whitney U testing; n=2 independent experiments. ●CTRL1, ■CTRL2, ▲CTRL3, ◊F281, ○F334, □F691, ▿F906.
Although the phenotype of the exclusively neurological disease LCC is clearly distinctive, it is not pathognomonic, since a similar radiological association can also be seen in the context of the multisystem disorder CP (see Supplementary Fig. 10)15. CP is most frequently caused by mutations in CTC1, encoding conserved telomere maintenance component 1, but such mutations were not found in patients without systemic involvement, indicating that CP and LCC are genetically distinct entities13,16. Noting that U8 is situated 50 kb upstream of CTC1 on chromosome 17p, we considered if the LCC disease-associated variants across SNORD118 might be affecting a control element for the expression of CTC1. In the absence of a functional antibody against CTC1, expression was tested in patient fibroblasts using quantitative reverse transcription PCR (qPCR) and was similar to controls (see Supplementary Fig. 11). As expected, we observed increased DNA damage and numbers of telomere dysfunction-induced foci (TIF) in primary fibroblasts from patients with biallelic CTC1 hypomorphic mutations. In contrast, primary fibroblasts from LCC patients displayed a similar number of DNA damage foci compared to controls, and no evidence of disturbed telomeric integrity (see Supplementary Fig. 12). We also looked at telomere length in four families using Flow-FISH, but saw no consistent association with genotype status (data not shown). Taken in the context of the data relating to U8 per se, these results indicate that the physical proximity of SNORD118 and CTC1 is likely not causally relevant to LCC.
LCCfibroblasts do not demonstrate dysfunction of TMEM107
Finally, SNORD118 is located within the 3’ UTR of the protein encoding gene TMEM107 (NM_032354.3). Although dysfunction of TMEM107 could, in theory, be causal of the LCC phenotype, we did not observe any exonic variants in this gene. Moreover, expression of TMEM107 at the RNA and protein level was similar between patients and controls in primary fibroblasts (see Supplementary Fig. 13). An ENU-mutagenesis derived mouse with a homozygous loss of function mutation in Tmem107 demonstrates a defect in ciliogenesis leading to polydactyly, neural tube defects and microphthalmia - none of which are seen in LCC17, and mutations in TMEM107 have been reported to result in a typical human ciliopathy phenotype18. These observations lead us to conclude that LCC is not consequent to TMEM107 dysfunction.
Discussion
Here we demonstrate that mutations in SNORD118 cause the cerebral microangiopathy LCC. We identified completely novel or rare biallelic sequence variants in all 40 radiologically stereotyped patients belonging to every one of the 33 families in our cohort. Moreover, the additional observation of novel and rare allele sharing across multiple pedigrees, and the results of screening of a large panel of control samples to determine the frequency of biallelic rare variants in unaffected individuals, provide unequivocal evidence of genetic causality.
Of particular note, although LCC is rare and inherited as an autosomal recessive trait, only 2 of 33 mutation-positive families that we identified are consanguineous. In keeping with this, the affected individuals in 31 families were compound heterozygotes for two different SNORD118 variants, likely comprising one ‘severe’ and one ‘mild’ mutation. Nineteen families in our cohort carry one of six putative pathogenic variants with an overall allelic frequency of > 0.001 in the ExAC control database (akin to, for example, the known pathogenic mutations p.Ala177Thr in RNASEH2B and p.Pro193Ala in ADAR1, which demonstrate allele frequencies of 0.001 and 0.002 respectively). In each of these 19 families, the second allele is either not present at all on ExAC (i.e. is completely novel; 9 families), or is only present at a very low frequency (< 0.00009756 i.e. 1 in 10,250 alleles in all cases)(Supplementary Table 6). We suggest that these more frequent, but still rare, variants are likely hypomorphic, whilst the still rarer variants confer a greater deleterious effect on U8. Similar to the situation observed in certain other Mendelian autosomal recessive diseases13,19,20, these data lead to the conclusion that biallelic null mutations in SNORD118 are likely incompatible with development, whereas homozygosity for (most) hypomorphic variants may be associated with no disease, later-onset pathology, or a different phenotype not ascertained here. Such a possibility would explain the presence on the ExAC database of a small number of homozygotes for five of the 36 putative mutations that we report (Supplementary Tables 2 and 6). Related to this understanding, it is of possible relevance that one, F446, of the two individuals in our cohort with homozygous SNORD118 mutations died at the youngest age of any of our patients (13 years), and we also note that two of our patients were completely asymptomatic until the sixth decade of life. Such variability in age at presentation likely relates to the overall functional effect of the two molecular lesions combined. Furthermore, it is interesting that the mother in F819 carries two rare variants (each one on a different allele), both of which were seen in affected patients. However, she has not demonstrated any disease features to the age of 60 years. In contrast, both of her children, each carrying a different maternally-derived rare allele in combination with a paternally-inherited whole-gene deletion, experienced the onset of symptoms in their teenage years.
As the apparatus of protein synthesis, the ribosome is one of the most precisely constructed and regulated molecular machines in the cell21. The ribosome consists of ribosomal RNA (rRNA) and ribosomal proteins (RPs), with snoRNAs being an evolutionarily conserved group of non-protein encoding RNAs involved in the modification and processing of rRNAs. U8 is a vertebrate-specific factor, which is the only known snoRNA essential for maturation of the 60S large ribosomal subunit RNAs, 5.8S and 28S22–24. It is thought that U8 snoRNA binds to newly transcribed pre-rRNA and thereby facilitates its proper folding, but later needs to be displaced for further processing to occur. We provide evidence that a variant in the promoter region of U8 affects expression of the snoRNA. Furthermore, alterations in the C box disturb the association of U8 with the snoRNA-binding protein 15.5K, whilst variants in the 3’ end of the gene confer aberrant processing of the precursor U8 snoRNA. In total, we recorded seven putative mutations in the invariant C/D box motifs, three within the stem of a hairpin loop which is very well conserved and would be predicted to decrease the stability of this structure, and three within the highly conserved GAUU motif of the LSm binding site. Taken together, these data indicate that the variants we describe are pathogenic, and likely act as loss of function mutations.
LCC, the first example of a human disease to be associated with point mutations in a C/D box snoRNA, illustrates some of the difficulties associated with the attribution of causation to sequence variants in a non-protein encoding portion of genomic DNA, where it is not possible to use conventional in silico algorithms to assess the pathogenicity of sequence variants. This fact, together with the high degree of sequence variation across SNORD118 (Supplementary Table 7), and the finding that the disease occurs almost invariably in the context of compound heterozygosity, leads us to suggest that the identification of the genetic basis of LCC would likely only have been possible with a very large cohort of patients such as presented here. The degree of SNORD118 sequence variability in the general population is notable, perhaps indicating that this variation confers important biological effects worthy of further study.
Alterations in ribosomal components, structure or function can cause a heterogeneous class of diseases referred to as the ribosomopathies25, with this diversity in clinical phenotype informing a developing understanding of the multiple specialized roles of the ribosome in normal physiology26,27. Despite being ubiquitously expressed, germ-line mutations in the snoRNA U8 cause an exclusively neurological, progressive microangiopathy, thus suggesting further subtleties in ribosomal activity directly relevant to human health and disease. That being said, the precise link between U8 and cerebral vascular homeostasis awaits elucidation, and may conceivably involve a direct effect on translation, or other yet to be defined functions of U8.
Online Methods
Subject ascertainment
We ascertained subjects demonstrating a characteristic neuroradiological picture of white matter disease (leukoencephalopathy), intracranial calcification and brain cysts internationally through colleagues in pediatric neurology and clinical genetics. All subjects had been previously screened negative for mutations in CTC1. Written informed consent was obtained for all participants. The study had ethical approval from the Leeds Multicentre Research Ethics Committee (07/Q1206/7).
Exome sequencing
Genomic DNA was extracted from lymphocytes from affected individuals by standard techniques. For whole-exome analysis, targeted enrichment and sequencing were performed on DNA extracted from peripheral blood from 19 patients F281, F330, F331 (2 affected individuals), F343, F344, F362 (2 affected individuals), F426 (2 affected individuals), F433, F446, F451, F454 (2 affected individuals), F521 (2 affected individuals), F551 and F564. Enrichment was undertaken using the SureSelect Human All Exon kits following the manufacturer’s protocol (Agilent Technologies), and samples were paired-end sequenced on either an Illumina HiSeq 2000 or SOLiD platform. Sequence data were mapped using BWA (Burrows-Wheeler Aligner) and the hg18 (NCBI36) human genome as a reference. Variants were called using SOAPsnp and SOAPindel (from the Short Oligonucleotide Analysis Package) with medium stringency.
Linkage analysis
We used the Merlin package28 to perform nonparametric linkage analysis in 5 pairs of affected siblings (F331, F426, F454, F521, F780) born to unrelated parents (with the inclusion of one unaffected sibling in F780), and two singletons (F344 and F446) who were the product of independent consanguineous unions (first and second cousin parents respectively). We assumed allele frequencies as provided by Affymetrix for the Caucasian population, and calculated a LOD score using the Kong and Cox exponential model29. Data were annotated according to the hg19 human genome.
Genomic capture
Genomic capture, next generation sequencing and bioinformatic analysis was performed as a service at BGI Tech (Hong Kong) on 10 affected individuals from 10 separate families (F330, F331, F362, F426, F433, F446, F454, F521, F691 and F780). A 3 Mb interval (ch17:7,000,000-10,000,000) was captured using Agilent Sure select technology, and was then sequenced using the Illumina HiSeq2000 platform. Genomes were aligned to hg19 with BWA, and all subsequent analyses were performed with hg19.
Sanger sequencing
Mutation analysis was performed by direct sequencing of purified genomic PCR products using the BigDye™ Terminator v3.1 cycle sequencer system (Applied Biosystems) and an ABI 3130 DNA sequencer. Primers were designed for a 544 bp region around SNORD118. Primers used are listed in Supplementary Table 8. Mutation description is based on the reference sequence SNORD118 NR_033294.1.
Copy number analysis
Copy number analysis of SNORD118 in F819 was performed using DNA from the two affected individuals and their mother (paternal DNA not available). Parental samples from F426, F780 and F619 were used as controls. Copy number analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems), and 40ng of DNA. Copy number probes chosen were Hs01424421_cn (76bp upstream of U8) and Hs01021672_cn (72bp downstream of U8). F619 Mum was chosen as the calibrator sample, and copy number was assessed with the Applied Biosystems StepOne Software v2.1 and Applied Biosystems CopyCaller software V2.0.
Microsatellite genotyping
To confirm maternity and paternity in F906, informative polymorphic microsatellite markers Penta E, D18S51, D21S11, TH01, D3S1358, FGA, TPOX, D8S1179, vWA, Amelogenin, Penta D, CSF1PO, D16S539, D7S820, D13S317 and D5S818 were genotyped using DNA from the affected child and parents using the PowerPlex 16HS System (Promega).
Control panel sequencing
Human Random Control DNA Panels (UK Caucasian blood donors) HCR-1 to HCR-5 were purchased from Public Health England (Porton Down, Salisbury). DNA panels NDPT099 and NDPT095 were acquired from the NINDS Human Genetics Resource Center DNA and Cell Line Repository.All panels were sequenced as above for the 544 bp region around SNORD118. Where two rare variants were identified, the PCR products were cloned into the pGEM T easy vector (Promega) so that each allele could be sequenced separately.
In silico analysis
In silico analysis of variants identified in LCC patients was carried out using RNAfold (version 2.1.9), from the ViennaRNA suite.
U8 Cloning
A 525 bp region of chromosome 17 encompassing the entire U8 coding region was subcloned into the pGEM T easy vector (Promega). Site directed mutagenesis was carried out using the QuikChange Site-Directed Mutagenesis protocol (Stratagene). Primers used for the cloning and site directed mutagenesis are listed in Table S6.
In Vitro transcription of U8 snoRNA
RNA was transcribed from PCR templates (primer sequences are listed in table S6) amplified from pGEM constructs containing WT and mutant U8 snoRNA sequence, using the T7 Ribomax Express large scale RNA Kit (Promega) in accordance with the manufacturer’s instructions. Transcribed RNAs were purified on a 6% acrylamide/7M urea gel. Following passive elution from the gel, RNA was precipitated and re-suspended in 20 μl water. Quantification of RNA was carried out on the Boeco S-22 UV/Vis spectrophotometer.
De-phosphorylation and labeling of transcribed RNA
De-phosphorylation of transcribed RNA was carried out using 10 U of Calf Intestinal Alkaline phosphatase (NEB) in Cut Smart buffer. Reactions were incubated for 1 hour at 37°C, followed by phenol extraction and re-suspension in water to give a 1 μM final concentration. De-phosphorylated RNA (at 1 μM concentration) was end-labeled with 32P-γATP 3000 Ci/mmole (Hartmann Analytic) using T4 Polynucleotide kinase in PNK buffer (NEB). Labeled RNA was phenol extracted, precipitated and resuspended in water. The amount of labeled RNAs was then determined using the Bioscan QC 2000 radioactivity counter.
15.5K cloning, expression and purification
The 15.5K open reading frame was amplified from human cDNA (see table S6 for primer sequences) then cloned into the NdeI and XhoI sites of pET28a (Novagen) to produce the pET28a-TEV-15.5K plasmid. Positive clones were sequenced to confirm the correct fusion of the human 15.5K open reading frame with an N-terminal 6-His tag and TEV protease cleavage site. The pET28a-TEV-15.5K plasmid was transformed into Rosetta 2 (DE3) (Novagen) and expression of 6His-TEV-15.5K induced by using Overnight Express Instant TB medium (Novagen). The 6His-TEV-15.5K protein was purified as previously described for Snu13p30.
Electrophoretic mobility shift assay (EMSA)
For electrophoretic mobility shift assays, recombinant His-15.5K was incubated with 50,000 dpm 32P end labeled U8 snoRNA in EMSA buffer (20 mM HEPES-KOH, 150 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100) for 30-45 minutes on ice. Resulting RNA-protein complexes were resolved on a native 7% acrylamide gel for 8.5 hours at 4°C. Gels were dried and exposed to an X-ray film for approximately 8 hours at -80°C in the presence of an intensifying screen. Binding between His-15.5K and 32P end-labeled RNA was quantified using the Typhoon FLA 7000IP phosphorimager. For the super-shift assay recombinant His-15,5K was incubated for 30 minutes on ice with 6XHis antibody ab18184 (Abcam) prior to addition of 50,000 dpm 32P end labeled U8 snoRNA. For the competition assay recombinant His-15,5K was incubated for 30 minutes on ice with 2.5ug of unlabeled U8 snoRNA prior to addition of 50,000 dpm 32P end labeled U8 snoRNA.
3’ Processing assay
For 3’ processing assays 32P end-labeled U8 snoRNA was incubated with HeLa nuclear extract (CIL biotech) at 30°C in buffer containing 0.25 mM ATP, 10 mM phosphocreatine, 3.2 mM MgCl2, 20 mM HEPES KOH pH 7.9, 2.6% PVA and 240 U RNasin (Promega). At 0 and 30 minute time points 10 μl of the reaction was removed and added to a tube containing 4 μl stop solution (Stop solution: 1 mg/ml proteinase K, 50 mM EDTA, 1% SDS). Reactions were then incubated at 37°C for 15 minutes, phenol extracted, precipitated and resolved on a 6% acrylamide/7M Urea gel. Gels were dried and exposed to an X-ray film overnight at -80°C in the presence of an intensifying screen.
Luciferase assay
A 298 bp region of chromosome 17 encompassing the entire U8 promoter region from control DNA was cloned into the pGEM T easy vector (Promega). Site directed mutagenesis was carried out to create the n.-54_-49del mutation. The DNA from WT and mutant clones were then subcloned into the pGL3 basic vector (Promega). Primers used for the cloning are listed in table S6.
HeLa-M cells were maintained in 75 cm2 flasks in Dulbecco's Modified Eagle Medium (DMEM) with GlutaMAX (Life Technologies 10566-016) supplemented with 10% fetal bovine serum (Sigma F7524). Cells for transfection were plated out in triplicate wells in Corning 96 well clear bottom plates at 1 × 104 cells per well in a 100 μl volume of culture medium. Transfections were carried out using the Effectene kit (Qiagen 301425) according to the manufacturer’s instructions. Cells in each well were co-transfected with 200 ng pGL3 vector and 20 ng pRL-TK control vector (10:1 ratio). Plates were incubated for 24 hours at 37°C 5% CO2. Twenty four hours after transfection, medium containing transfection reagent was removed from all wells and replaced with 75 μl of fresh culture medium. Cells were assayed for firefly and renilla luciferase activity using the Dual-GLO Luciferase Assay System (Promega E2920) in accordance with manufacturer’s instructions. Luciferase activity was measured on the BMG Labtech Fluostar Omega microplate reader. The firefly/renilla luciferase ratio was calculated for each well after subtraction of average background values from untransfected cells. The mean firefly/renilla ratio was calculated from triplicate values and used to calculate the fold increase in relative light units (RLU) compared to empty pGL3 vector.
Polysome fractionation and profiling
Human lymphoblastoid cell lines (LCLs) from control (CTRL4) and patient (F433, F446) samples were used for polysome profiling. Approximately 2x107 cells were incubated in RPMI medium with 100 µg/ml cycloheximide for 5 minutes at 37°C 5% CO2, followed by a wash in 1 x PBS containing 100 µg/ml cycloheximide. Cells were collected by centrifugation at 200 x g for 5 minutes at 4°C and re-suspended in 425 µl hypotonic buffer (Hypotonic buffer: (5 mM Tris-HCl (pH 7.5), 2.5 mM MgCl2, 1.5 mM KCl and 1x protease inhibitor cocktail-EDTA-free). The re-suspended cell mixture was supplemented with 5 μl of 10 mg/ml cycloheximide, 1 μl of 1M DTT and 100 units RNasin. The cell mixture was incubated on ice for 10 minutes followed by 5 second vortex. The mixture was further supplemented with 25 μl of 10% Triton X-100 and 25 μl of 10% sodium deoxycholate, and vortexed again for 5 seconds. To pellet debris the lysates were centrifuged at 16,000 x g for 7 min at 4°C and the supernatant transferred to a new pre-chilled tube. The OD at 260 nm was measured for all lysates and adjusted so that all samples contained the same OD. Lysates were loaded onto sucrose gradients (15-50%) and centrifuged for 2.5 hrs at 40,000 rpm. Polysome profiles were recorded using the Isco UA-6 UV/Vis detector.
Cell culture
Human primary fibroblasts and HeLa-M cells were cultured in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% heat-inactivated fetal calf serum and 1% penicillin/streptomycin. Human lymphoblastoid cell lines (LCLs) were cultured in RPMI (Gibco) supplemented with 10% heat-inactivated fetal calf serum and 1% penicillin/streptomycin. The control fibroblast line CTRL1 is a commercially available line purchased from ATCC. All other fibroblasts were generated by the referring institutes and shipped as growing cultures. All LCLs were generated as a service at St Mary’s Hospital, Manchester. HeLa-M cells were a kind gift from the Manson Lab at the University of Manchester. All cells are routinely tested for mycoplasma contamination. All putative patient cell lines were genotyped for relevant variants in SNORD118 to confirm authenticity.
Cell growth / proliferation
Proliferation of primary fibroblast cells from patients (F281, F334, F691, F906) and healthy controls (CTRL 1, 2, 3). Cells were plated in triplicate for each patient and control at 25 cells per well in a 96 well plate. Every cell from each well was counted under the microscope at 12 hours after plating (D0), and then at day 3 and day 6. Cell proliferation was measured in patient and healthy control primary fibroblasts using the Cell Trace CFSE (carboxyfluorescein succinimidyl ester) Cell Proliferation Kit (Life Technologies). Cells were labeled following the manufacturer’s protocol, and then plated as 5000 cells per well in a 24 well plate. CFSE content was analyzed by flow cytometry at 30 minutes and 2 days after staining by flow cytometry on a Gallios flow cytometer with Kaluza acquisition software.
Apoptosis
Apoptosis was measured in patient and healthy control primary fibroblasts using an ANNEXIN V – FITC Kit (Beckman Coulter). Briefly, cells were seeded in 24-well plates and incubated for 24 hours. Cells were harvested, washed twice in PBS, and then stained with Annexin V-FITC and propidium iodide (PI) according to the manufacturer’s instructions. The resulting fluorescence was detected by flow cytometry on a Gallios flow cytometer with Kaluza acquisition software.
Senescence
Senescence was measured in patient and healthy control primary fibroblasts using the Senescence beta-galactosidase staining kit from Cell Signaling. Briefly, cells were seeded in 24-well plates and incubated for 24 hours. Cells were labeled following manufacturer instructions. The percentage of senescence was calculated with the following formula: (number of beta-galactosidase positive cells in 5 fields/ total number of cells in these 5 fields) X 100.
DNA Content
DNA content was measured in patients and healthy control primary fibroblasts. Briefly, cells were plated out as 5000 cells per well in 24 well plates and treated with 50 ng/ml of Mitomicin C (Sigma Aldrich). Cells from a patient with Fanconi anemia were used as a positive control. After 5 days of treatment, cells were harvested, washed twice in PBS, and then incubated for 30 minutes in 70% ethanol. Cells were washed 3 times in PBS, and incubated in PBS 1 µg/ml DAPI and 0.1% Triton X100 (Sigma Aldrich) for 30 minutes on ice, and analyzed by flow cytometry on a Gallios flow cytometer with Kaluza acquisition software.
ImmunoFISH for telomere dysfunction-induced foci
Primary fibroblasts from healthy controls, CTC1 mutation positive patients and LCC patients were fixed for 5 minutes in 4% vol/vol formaldehyde in H2O and permeabilized in PBS with 1% BSA 0.1% Triton X100. Cells were incubated with primary anti-53BP1 antibody (SantaCruz) for 1 hour at room temperature, and then with Alexa 488 labeled secondary antibody (Life Technology). Samples were fixed for 5 minutes in 4% vol/vol paraformaldehyde and dehydrated in successive 5 minutes baths of 70% Ethanol, 95% ethanol and 100% ethanol. PNA-cy3-Telo-C probes (DAKO) were hybridized according to the supplier’s recommendations. Briefly, probes were incubated with the samples for 5 minutes at 80°C, and left in the dark at room temperature for 90 minutes. Samples were then washed twice in 70% formamide, 10 mM Tris-HCL, and PBS, and mounted with DAPI mounting media (Vectashield).
Protein analysis
Whole cell lysates were prepared from patient primary fibroblasts (5x106 cells per sample) using 10mM EDTA / RIPA buffer containing protease inhibitors (Roche) and Halt phosphatase inhibitor (Thermo scientific). For western blot analysis, 30 µg of total protein was loaded onto 15% SDS-PAGE gels and electrophoresis was performed using the Mini-PROTEAN system (Biorad Laboratories Ltd). Following wet-blotting transfer of the protein onto nitrocellulose membrane (Li-cor), non-specific antibody binding was blocked using Odyssey Blocking Buffer (PBS) for a minimum of 1 hour at room temperature. Rabbit TMEM107 primary antibody (Abcam; ab181396) was incubated with the membrane for 1 hour at room temperature with agitation, using a dilution of 1:1000 in blocking buffer. As a loading control, membranes were incubated with a 1:3000 dilution of mouse anti- beta actin primary antibody (Abcam; ab125248). Following washes with PBS, membranes were incubated with IRDye® 800CW Donkey anti-Mouse IgG (Li-cor; 925-32212) and IRDye® 680RD Donkey anti-Rabbit IgG (Li-cor; 925-68073) diluted 1:10000 in blocking buffer for 45 minutes at room temperature. Following further washes, signal was detected using Odyssey digital infrared imaging system (Li-cor) and images were analyzed using Image Studio v5 (Li-Cor).
rtPCR
RNA was isolated from patient and control primary fibroblasts using Ambion RNAqueous Micro kit (Life technologies) according to the manufacturer’s instructions. Reverse transcriptase PCR was performed with the High Capacity cDNA reverse transcription Kit and the Taqman MicroRNA reverse transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Quantitative reverse transcription PCR analysis was performed using the TaqMan Universal PCR Master Mix (Applied Biosystems), and cDNA derived from 40 ng total RNA (for CTC1 and TMEM107) or 10ng RNA (for SNORD118). The relative abundance of target transcripts, measured using TaqMan probes for CTC1 (Hs01558648) and TMEM107 (Hs00766060) was normalized to the expression level of HPRT1 (Hs03929096_g1) and 18s (Hs999999001_s1). The relative abundance of target transcripts, measured using a custom TaqMan small RNA probe for SNORD118 (CSS07G9) was normalized to the expression level of RNU24 (4427975) and U6 snRNA (4427975). All data were assessed with the Applied Biosystems StepOne Software v2.1 and DataAssist Software v.3.01.
Supplementary Material
Acknowledgments
We are very grateful to the affected families for their involvement in our research. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. DNA panels from the NINDS Human Genetics Resource Center DNA and Cell Line Repository were used in this study, as well as clinical data. The submitters that contributed samples are acknowledged in detailed descriptions of each panel: NDPT099, NDPT095. YJC acknowledges funding from the Newlife Foundation (14-15/15), the Great Ormond Street Hospital Children’s Charity (V1212) and a state subsidy managed by the National Research Agency (France) under the “Investments for the Future” (ANR-10-IAHU-01). RTO acknowledges the Wellcome Trust (104981). This work was supported by Wellcome Trust funding (097820/Z/11/B) to YJC and RTO. PR has received research support from La Ligue (Equipe lab elis ée) and Centre National de la Recherche Scientifique (CNRS). This paper is dedicated to the memory of Dr. John L Tolmie.
Footnotes
URLs
UCSC Human Genome Browser, http://genome.ucsc.edu/; Ensembl, http://www.ensembl.org/; dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/; Exome Variant Server, NHLBI Exome Sequencing Project (ESP), http://snp.gs.washington.edu/EVS/; Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org (accessed 29th May 2015); Clustal Omega, https://www.ebi.ac.uk/Tools/msa/clustalo/; Alamut, http://www.interactive-biosoftware.com/; GraphPad, http://www.graphpad.com/. University of Michegan Center for Statistical Genetics, http://csg.sph.umich.edu/abecasis/merlin/reference.html; GTEx gene expression portal, http://www.gtexportal.org/home/gene/SNORD118; NINDS Human Genetics Resource Center DNA and Cell Line Repository, https://catalog.coriell.org/1/ninds
Author contributions
Exome sequencing was performed by JEU, JO’S, SGW and SB. Exome and genomic capture data were analyzed by EMJ. Linkage analysis was undertaken by JEU. Sanger sequencing and cloning were performed by EMJ with assistance from AO and LCG. Copy number analysis and microsatellite genotyping were undertaken by EMJ. Cell lines were maintained by EMJ, AO, LCG, MPR and YR. In vitro transcription of U8 snoRNA, EMSAs, 3’ processing assays, luciferase assays and polysome assays were performed by EMJ, CJK and RTO with assistance from GDP. Cell proliferation, senescence and apoptosis assays were performed by MPR. DNA content and ImmuoFISH assays were undertaken by MPR with assistance from PR. Western blots were performed by PRK. rtPCR was performed by MPR with assistance from GIR. RNA modeling studies were performed by SG-J. Telomere analysis was undertaken by GMB, MH and GA. YJC and RTO designed and supervised the project and wrote the manuscript supported by GIR and EMJ. MMvdK, JHL and YJC reviewed the patient scans. KB, AJB, RB, AB, JEB-H, JAB, DMCas, RRCC, DMCor, LMdW, AJF, PF, NAF, AEF, HG, CAH, IH, RJJ, RK, GK, LL, EL, CML, SL, TJM, SGM, IM, SN, KO, PP, RS, EHS, CS, HS, JS, CU, HvE, CEGvM, AV, ELW and AW identified affected patients or assisted with related clinical and laboratory studies.
The authors declare that they have no competing financial interests.
References
- 1.Labrune P, et al. Extensive brain calcifications, leukodystrophy, and formation of parenchymal cysts: a new progressive disorder due to diffuse cerebral microangiopathy. Neurology. 1996;46:1297–301. doi: 10.1212/wnl.46.5.1297. [DOI] [PubMed] [Google Scholar]
- 2.Nagae-Poetscher LM, et al. Leukoencephalopathy, cerebral calcifications, and cysts: new observations. Neurology. 2004;62:1206–9. doi: 10.1212/01.wnl.0000119341.59445.cf. [DOI] [PubMed] [Google Scholar]
- 3.Corboy JR, Gault J, Kleinschmidt-DeMasters BK. An adult case of leukoencephalopathy with intracranial calcifications and cysts. Neurology. 2006;67:1890–2. doi: 10.1212/01.wnl.0000244470.06748.18. [DOI] [PubMed] [Google Scholar]
- 4.Livingston JH, et al. Leukoencephalopathy with calcifications and cysts: a purely neurological disorder distinct from coats plus. Neuropediatrics. 2014;45:175–82. doi: 10.1055/s-0033-1364180. [DOI] [PubMed] [Google Scholar]
- 5.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 6.Peculis BA, DeGregorio S, McDowell K. The U8 snoRNA gene family: identification and characterization of distinct, functional U8 genes in Xenopus. Gene. 2001;274:83–92. doi: 10.1016/s0378-1119(01)00596-0. [DOI] [PubMed] [Google Scholar]
- 7.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–17. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 8.Tomasevic N, Peculis BA. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol Cell Biol. 2002;22:4101–12. doi: 10.1128/MCB.22.12.4101-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kufel J, Allmang C, Petfalski E, Beggs J, Tollervey D. Lsm Proteins are required for normal processing and stability of ribosomal RNAs. J Biol Chem. 2003;278:2147–56. doi: 10.1074/jbc.M208856200. [DOI] [PubMed] [Google Scholar]
- 10.Watkins NJ, Lemm I, Luhrmann R. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol Cell Biol. 2007;27:7018–27. doi: 10.1128/MCB.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins NJ, et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–66. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 12.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol. 2002;22:8342–52. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson BH, et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet. 2012;44:338–42. doi: 10.1038/ng.1084. [DOI] [PubMed] [Google Scholar]
- 14.Takai H, et al. A POT1 mutation implicates defective telomere end fill-in and telomere truncations in Coats plus. Genes Dev. 2016;30:812–26. doi: 10.1101/gad.276873.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Briggs TA, et al. Cerebroretinal microangiopathy with calcifications and cysts (CRMCC) Am J Med Genet A. 2008;146A:182–90. doi: 10.1002/ajmg.a.32080. [DOI] [PubMed] [Google Scholar]
- 16.Polvi A, et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet. 2012;90:540–9. doi: 10.1016/j.ajhg.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christopher KJ, Wang B, Kong Y, Weatherbee SD. Forward genetics uncovers Transmembrane protein 107 as a novel factor required for ciliogenesis and Sonic hedgehog signaling. Dev Biol. 2012;368:382–92. doi: 10.1016/j.ydbio.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaheen R, et al. Identification of a novel MKS locus defined by TMEM107 mutation. Hum Mol Genet. 2015;24:5211–8. doi: 10.1093/hmg/ddv242. [DOI] [PubMed] [Google Scholar]
- 19.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 20.Scheper GC, et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet. 2007;39:534–9. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 21.Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol. 2015;22:11–9. doi: 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- 22.Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–9. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peculis BA, Steitz JA. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–55. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- 24.Peculis BA. The sequence of the 5' end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol. 1997;17:3702–13. doi: 10.1128/mcb.17.7.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst. 2010;6:481–93. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramagopal S. Are eukaryotic ribosomes heterogeneous? Affirmations on the horizon. Biochem Cell Biol. 1992;70:269–72. doi: 10.1139/o92-042. [DOI] [PubMed] [Google Scholar]
- 27.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–69. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 29.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–88. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobbyn HC, O'Keefe RT. Analysis of Snu13p mutations reveals differential interactions with the U4 snRNA and U3 snoRNA. RNA. 2004;10:308–20. doi: 10.1261/rna.5970404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.