Double-strand DNA breaks (DSBs) continuously arise and are a source of mutations and chromosomal rearrangements. Here, we present DSBCapture, a sequencing-based method that captures DSBs in situ and directly maps these at single nucleotide resolution enabling the study of DSB origin. DSBCapture shows substantially increased sensitivity and data yield compared to other methods. Employing DSBCapture, we uncovered a striking relationship between DSBs and elevated transcription within nucleosome-depleted chromatin.
Due to their mutagenic potential, their role in cancer and their exploitation in cancer therapies1,2, it is important to identify the precise location of endogenous DSBs and those induced by therapeutic drugs and environmental insults such as radiation. ChIP-seq3, GUIDE-seq4 and BLESS5, are currently used methods for the genome-wide investigation of DSBs in situ. ChIP-seq, however, is indirect and relies on the capture of specific proteins that mark certain DSBs by proxy thus compromising both accuracy and resolution. GUIDE-seq profiles off-target cleavage induced by CRISPR-Cas nucleases and relies on the transfection and successful integration of a double stranded oligonucleotide at DSBs by non-homologous end joining (NHEJ)4. BLESS captures DSBs through the ligation of a barcoded adapter to broken DNA ends5, however the nature of this adapter introduces practical constraints that preclude the elucidation of the complete DSB landscape. The BLESS adapter requires a blunt-ended ligation for DSB capture, which is known to be less efficient than cohesive-end ligation6. Furthermore, the sequential addition of the capture and sequencing adapters, results in the need for two rounds of PCR, a technique known to introduce biases7. The resulting libraries suffer from low sequence diversity (i.e. low variation in the initial bases of the sequences read) that leads to problems during the first cycles of Illumina sequencing. This is normally remedied by diluting the sample DNA by mixing in an unrelated library (typically phiX) to artificially increase the sequence diversity to enable sequencing on the Illumina platform. However, this reduces data yield of the sample under investigation by greater than 50 %8. Finally, as sequencing is not only initiated from the captured DSB (proximal end) but also from the end generated through fragmentation (distal end), the number of sequencing reads that directly identify the site of DNA damage in single-end sequencing is halved. To overcome these limitations we developed DSBCapture, a substantially improved method that is derived from BLESS but enables direct in situ capture of DSBs using a modified P5 Illumina adapter to facilitate sequencing without additional library preparation steps (Fig. 1a and Supplementary Fig. 1 a,b,c,d). Ligation of the T-tailed modified P5 Illumina adapter to the break site identifies solely the site of DSB formation in single-end sequencing and, as DSBCapture libraries do not suffer from low sequence diversity, no spike-in of another library is required for sequencing. DSBCapture displays enhanced data yield and data quality, higher reproducibility and superior sensitivity compared to BLESS. In a head-to-head comparison DSBCapture identified 4.5 × more (84,946 compared to 18,816) DSBs in normal human epidermal keratinocytes (NHEK) than BLESS, demonstrating the most comprehensive elucidation of a DSB landscape in a normal human cell line.
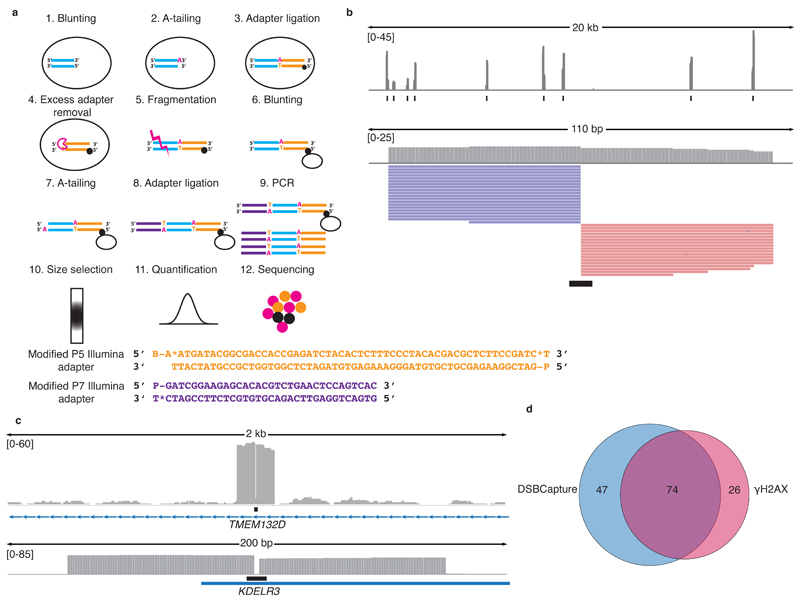
Figure 1. DSBCapture methodology and validation.
(a) DSBCapture workflow. (1) DSBs in fixed nuclei were blunt-end repaired, (2) A-tailed and (3) ligated to a biotinylated (black ball) modified P5 Illumina adapter (orange lines). (4) Excess adapters were removed by lambda exonuclease digestion; (5) DNA extracted from lysed nuclei was fragmented by sonication, (6) bead-captured (hollow ball) and blunted-end repaired, (7) A-tailed, and (8) ligated to a modified P7 Illumina adapter (purple lines). (9) Captured break sites were PCR amplified, (10) size selected, (11) quantified and (12) sequenced. Sequences of the DSBCapture adapters: modified P5 Illumina adapter and modified P7 Illumina adapter (B = biotin; P = phosphorylated; * = phosphorothioate bond). (b) DSBs created by EcoRV cleavage in fixed nuclei (N = 1). PCR duplicates have been removed. Data range is shown in square brackets and black boxes illustrate the genomic location of EcoRV sites. A 20 kb region and a 110 bp region are shown. Pink and purple lines: reads from the sense and antisense strand, respectively. As EcoRV is a blunt cutter, reads originate directly from the cleavage site. (c) AsiSI cleavage sites (black boxes) detected by DSBCapture (N = 1). Cleavage by AsiSI generates a 2 bp 3’ overhang; end processing removes this overhang generating the 2 bp gap in the center of the peak. A 2 kb and a 200 bp region are shown. (d) Venn diagram illustrating the overlap of DSBs detected at AsiSI sites by DSBCapture and γH2AX ChIP-seq9.
During the development of DSBCapture, we validated an early version of the method using the controlled double strand cleavage of fixed HeLa nuclei by the EcoRV restriction endonuclease. EcoRV has 430,897 predicted recognition sites in the human genome with the palindromic sequence GATATC. In our pilot experiment, 99 % of the 13.4 million aligned, single-end sequencing reads mapped to EcoRV restriction sites and DSBCapture identified 93.7 % of EcoRV sites as cleaved (with a coverage of ≥ 5 reads/site), illustrating the ability of this method to efficiently capture in vitro generated DSBs (Fig. 1b and Supplementary Table 1). Furthermore, DSB capture is not influenced by the chromatin state or the DNA sequence content. 0.5 % of all predicted EcoRV sites lie within open chromatin regions, whilst 99.5 % lie within heterochromatic regions. The EcoRV sites detected as cleaved by DSBCapture reflect this distribution (of the detected sites 0.5 % are in open chromatin and 99.5 % are in heterochromatin), illustrating that DSBCapture can detect DSBs that occur in both euchromatin and heterochromatin and does not display bias towards either one. Likewise, the average GC content of 100 bp either side of the EcoRV sites detected as cleaved and those not detected as cleaved is very similar (37 % and 36 %, respectively) demonstrating that there is no evident GC bias for DSB detection.
To verify that DSBCapture is equally suited to the detection of in situ generated DSBs, we employed AID-DIvA cells - a U2OS cell line that expresses the AsiSI restriction enzyme fused to a modified estrogen receptor ligand-binding domain9. Upon 4-hydroxy tamoxifen (4OHT) treatment, the restriction enzyme translocates to the nucleus to cause sequence-specific DSBs at GCGATCGC sites. The locations of these AsiSI-induced DSBs have been extensively studied by ChIP-seq for the DNA damage markers γH2AX, RAD51 and XRCC49. DSBCapture was performed on AID-DIvA cells following 4OHT treatment (Supplementary Table 2). A total of 121 AsiSI sites were identified as DSBs (p ≤ 0.05) (Fig. 1c,d). Of the previously reported 100 ‘most cleaved’ sites identified by γH2AX ChIP-seq9, DSBCapture recovered 74, illustrating good overlap between entirely independent methods of DSB detection (Fig. 1d). DSBCapture also uncovered 47 additional AsiSI cleavage sites that were not previously identified as break sites by γH2AX ChIP-seq; 28 (60 %) of these sites overlap with XRCC4 or RAD51 ChIP-seq peaks9, illustrating that DNA repair proteins localize to the majority of these sites. It is noteworthy that the ability of DSBCapture to precisely map nuclease-induced DSBs illustrates its application for the study of CRISPR off-target cleavage sites.
In AsiSI-expressing U2OS cells, we noted that DSBCapture also recovered 2,372 endogenous DSBs (i.e. those lacking an AsiSI recognition sequence). By comparison, GUIDE-seq identified 25 endogenous DSBs in U2OS cells4 (Supplementary Fig. 2). The substantially higher number of DSBs revealed by DSBCapture is most probably a result of the direct mechanism of DSB detection. DSBCapture detects DSBs that occur at a given moment in time through the ligation of a capture oligonucleotide to DNA ends, whereas GUIDE-seq relies on a specific biological process (NHEJ) to integrate a double stranded oligonucleotide at the DSB during DNA repair. Overall, this illustrates the better sensitivity and genome-wide coverage for the detection of endogenous DSBs by DSBCapture.
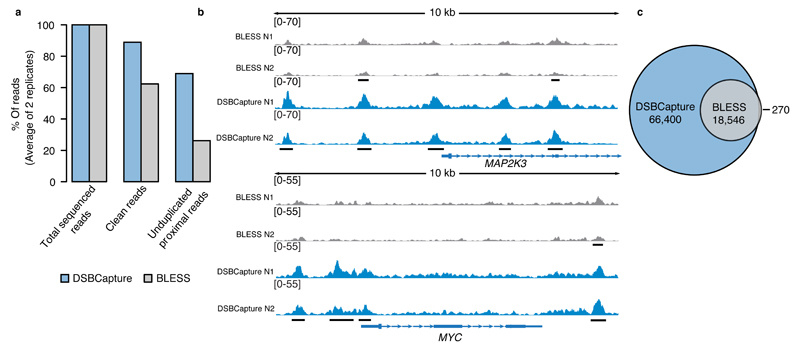
We next applied both DSBCapture and BLESS in a side-by-side comparison to study endogenous DSBs present in two biological replicates of NHEKs. While no DNA was recovered in three negative controls (DSBCapture performed without T4 DNA ligase during the first or second ligation or using a non-biotinylated modified P5 Illumina adapter), the complete DSBCapture procedure recovered DSBs successfully (Supplementary Fig. 1e). After artificially enhancing the BLESS data yield by repeated sequencing to obtain similar read numbers to those obtained by DSBCapture, we evaluated the data quality of the two methodologies. We found that the sequencing data quality of the DSBCapture libraries surpassed that of BLESS: when taking the average of the two replicates for both methods, 69 % of the DSBCapture data passed filtering, in contrast to 26 % of the BLESS data (Fig. 2a). This difference results from the enhanced alignment quality and reduced duplication frequency in DSBCapture (Supplementary Table 3). Overall, 84,946 high confidence peaks were observed common to replicates for DSBCapture versus 18,816 for BLESS (Supplementary Fig. 3a). Fig. 2b exemplifies the increased signal of the DSBs seen in DSBCapture for two representative genes: MAP2K3 and MYC. Moreover, DSBCapture showed higher reproducibility: 83 % of peaks overlapped between two independent experiments, compared to 63 % in BLESS (Supplementary Fig. 3a). Of the DSBs identified using BLESS, 99 % were observed by DSBCapture whereas 78 % of DSBs identified by DSBCapture were missed by BLESS and are therefore uniquely identified by our method (Fig. 2c, Supplementary Fig. 3b). In a DSBCapture experiment with reduced DNA input, 73 % of the original peaks were identified, illustrating good overlap irrespective of input material (Supplementary Fig. 3c). The increased number of peaks detected by DSBCapture indicates that our method has substantially higher sensitivity for DSB detection than the prior art and is therefore able to identify genomic regions containing DSBs with high statistical confidence. We next compared the sequence context of DSBs detected uniquely by DSBCapture to those shared between BLESS and DSBCapture. We found that DSBCapture detected a significantly greater proportion of DSBs in genomic regions where the GC content surpassed 70 %. When the GC content of DSBs exceeded 80 %, DSBCapture unique peaks were more than 4.5-fold enriched compared to those shared with the BLESS dataset (Supplementary Fig. 3d).
Figure 2. DSBCapture and BLESS comparison in NHEK cells.
(a) Bar plot comparing the fraction of good quality alignment reads (clean reads) and unduplicated proximal (identifies the break site) reads in DSBCapture and BLESS, shown as percentage of total sequenced reads, set to 100 %. (b) Representative genomic view of DSBs detected by BLESS (grey) and DSBCapture (blue) in two biological replicates (N1 and N2). Peaks in common between the two replicates for each method are underlined with black bars; the data range (absolute read counts) is shown in square brackets. Two 10 kb genomic regions in proximity of the MAP2K3 and MYC genes (blue tracks) are shown. (c) Venn diagram depicting the overlap of high confidence DSBCapture and BLESS peaks.
Four stranded DNA secondary structures called G-quadruplexes (G4s), form in G-rich genomic regions in human cells10. G4 structures have been implicated as fragile sites during transcription and replication3,11,12, and have been associated with somatic copy number alterations in human cells13. We found that observed G-quadruplex-forming sequences (OQs), previously mapped in human genomic DNA13, were 3-fold enriched over random within the high confidence DSBCapture peaks (Supplementary Fig. 3e). Notably, OQs with increasing GC content were found to be progressively enriched within DSBs in NHEK cells: up to 15-fold for OQs with GC content > 70 %, corresponding to 9,966 DSBs (Supplementary Fig. 3e), illustrating that OQs, particularly those with high GC content, are intrinsic sites of DSB formation in NHEKs. Due to the reduced representation of sequences with high GC content in the BLESS data, we expected a reduced representation of DSBs at OQ regions with high GC content; indeed, only 11 % (1,123) of the OQs with GC content > 70 % were shared between BLESS and DSBCapture.
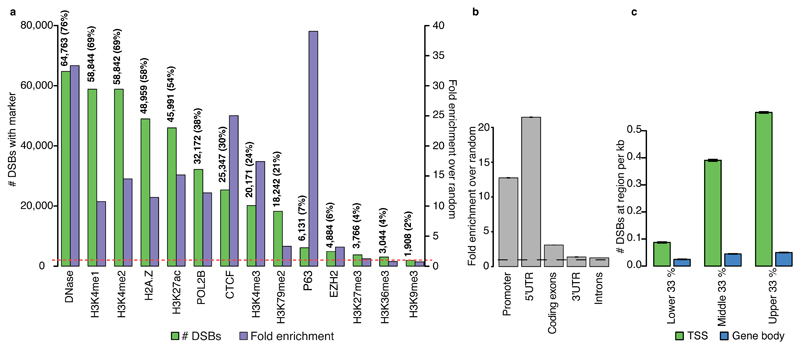
We next analysed our high confidence DSBCapture data for NHEK cells in the context of chromatin features using ChIP-seq and DNase-seq datasets from ENCODE14. Notably, 58 % (11.4-fold enrichment) of DSBs localized with the histone H2A.Z; a histone variant transiently incorporated at DSBs and known to have a role in DSB repair15 (Fig. 3a, Supplementary Fig. 4a and Supplementary Table 4). Strikingly, over 76 % (33.3-fold enrichment) of the DSBCapture peaks overlapped with regulatory, nucleosome-depleted regions (overlap with DNase-seq), revealing a relationship between regulatory chromatin and genome instability (Fig. 3a and Supplementary Table 4). This is consistent with the notion that nucleosome density mediates the susceptibility of DNA to genotoxic insults, with euchromatin showing enhanced DNA repair signaling16,17. Our finding that endogenous DSBs mainly occur in transcriptionally active regions was further supported by the observation that DSBs correlate with markers of active genes (mono-, di- and tri-methylated H3K4); enhancer regions (H3K27ac and H3K4me1; Supplementary Fig. 4b), the architectural protein CTCF and the transcription factor P63 (Fig. 3a and Supplementary Table 4). 38 % (12.2-fold enrichment) of DSBs overlapped with peaks of RNA polymerase II (POL2B), confirming an association of DSBs with transcriptional activity (Fig. 3a and Supplementary Table 4). Crucially, no enrichment of DSBs was found in heterochromatic regions of the genome (H3K9me3 and H3K27me3), or in regions featuring the transcriptional repressor EZH2 (Fig. 3a and Supplementary Table 4). Transcription has been linked to DNA damage18 and DSBs have been found to occur in the proximity of transcription start sites (TSSs) of highly expressed genes19,20. Indeed, we found DSBs to be enriched in genic compared to intergenic regions (Supplementary Fig. 4c), in particular at 5’UTRs (21.5-fold) and promoters (12.8-fold) (Fig. 3b) and upon further examination, we discovered a striking relationship between DSB formation and elevated gene expression in NHEK cells (Fig. 3c). By assigning genes into three categories, according to their gene expression level, we analyzed the average number of DSBs present ± 1 kb of the TSS and within gene bodies at different transcriptional levels. Increasing levels of gene expression correlated well with increased DNA damage around the TSS, whereas damage within gene bodies showed little association with transcription (Fig. 3c). Furthermore, we found that genes that do not contain DSBs ± 1 kb of the TSS are generally not expressed (Supplementary Fig. 4d). Four of the five most highly expressed genes in NHEKs are keratin genes (KRT5, KRT14, KRT6A and KRT17); keratins form filaments in epithelial cells to give the physical resilience that is characteristic of keratinocytes. DSBCapture detects DSBs ± 1 kb of the TSS in all of these genes; KRT5 and KRT17 even show two DSBs within this region; BLESS, however, did not identify DSBs within ± 1 kb of the TSS of any of these genes. In contrast to DSBCapture, which identified 93 % of the most highly expressed genes (top 5 %) to contain a DSB ± 1 kb of the TSS, BLESS only found 32 % of these genes to contain a DSB within this region. Thus, owing to reduced sensitivity, BLESS can miss observations of biological relevance. Taken together, these data clearly link elevated gene expression and sites of regulatory, nucleosome-depleted chromatin to DSB formation in NHEKs.
Figure 3. Genomic location and epigenetic context of endogenous DSBs in NHEK cells.
(a) Composite bar plot showing the overlap of high confidence DSBs detected by DSBCapture with epigenetic marks and DNA-binding proteins taken from ENCODE14. Green bars: number of DSBs overlapping each mark, ordered from highest (left) to lowest (right); corresponding y-axis on the left. Purple bars: fold change of DSBs overlapping each mark over random, corresponding y-axis on the right. A signal above 1 (dashed red line) is indicative of enrichment. The number and percentages of DSBs that overlap with each mark are shown above their respective bars. (b) Fold enrichment of high confidence DSBs in different genomic regions, calculated as the number of peaks divided by the number of randomly shuffled peaks overlapping to each region. Error bars: standard deviation of fold enrichment over random. (c) Number of high confidence DSBs detected ± 1 kb of the TSS (green bars) or within gene bodies (i.e., exons and introns excluding ± 1 kb of the TSS; blue bars) for the lower, middle and upper third of gene expression values, split according to the rpkm values (lower: < 0.041, middle: 0.041-6.08 and upper: > 6.08), where rpkm is: reads per kilo base (of the total exon length) per million sequenced reads. Error bars: SEM.
Finally, we computationally estimated the average number of DSBs per cell and found this to be 38; this may be an over-estimate as telomeres are recognized and detected as DSBs by DSBCapture.
Overall, DSBCapture enables the identification of DSBs with high sensitivity at single nucleotide resolution, without the need of signaling proteins as proxies and is therefore independent of the biological repair state. DSBCapture will be a valuable methodology for the study of DNA damage and repair, where it will enable the elucidation of sites of endogenous and drug-induced DNA damage and has the potential to elucidate DNA repair processes by uncoupling break formation from repair signaling.
Online Methods
Cell culture
HeLa cells (CRUK-CI Biorepository and Cell Services Core) were cultured in DMEM (Sigma, D6429) supplemented with 10 % heat inactivated FBS. U2OS AID-DIvA cells were cultured in DMEM supplemented with 10 % heat inactivated FBS and 800 μg/mL G418 Sulfate (Gibco, 10131). To induce nuclear localization of AsiSI, AID-DIvA cells were treated with 300 nM 4OHT (Sigma, H7904) for 4 h. Normal human epidermal keratinocytes (NHEK), pooled from multiple donors, were purchased from Thermofisher (A13401) and cultured in EpiLife medium (Thermofisher, M-EPI-500-CA) supplemented with human keratinocyte growth supplement (HKGS) (Thermofisher, S-001-5). NHEKs were detached using accutase (Sigma, A6964). HeLa and U2OS AID-DIvA cells were STR genotyped and mycoplasma tested. NHEK cells were obtained from Thermofisher and were certified as mycoplasma negative.
BLESS
The method published on http://breakome.utmb.edu was followed; with the exception that DNA was solely fragmented by sonication and amplified using Phusion HF polymerase (NEB, M0530S) as published by Crosetto, et al5. The TruSeq Nano DNA Library Prep Kit (Illumina, FC-121-4001) was used to generate libraries for sequencing (size selection for 550 bp) and all centrifugation steps were carried out at the speeds described for DSBCapture in the step by step method on the protocol exchange (Insert URL).
Annealing of oligonucleotides
To anneal the modified P5 and P7 Illumina adapters; oligonucleotides (modified P5 and modified P5 complement / modified P7 and modified P7 complement) were made up to 10 μM in 1 × T4 DNA ligase reaction buffer (NEB, B0202S). Oligonucleotides were heated to 95 °C for 10 minutes. Tubes were removed form the heat source and gradually cooled to room temperature.
DSBCapture (EcoRV cleavage in HeLa cells)
During the development of the DSBCapture method, pilot experiments were performed in which permeabilized nuclei were treated with the restriction enzyme EcoRV to generate DSBs. This pilot method was essentially comparable to the final DSBCapture protocol described below with the following significant differences: Cells were fixed in formaldehyde for 10 min. After re-suspension in nucleus break buffer the nuclei were washed 2 × with 1 × NEBuffer4 (NEB, B7004S) + 1 % TritonX-100 followed by re-suspension in 500 μL 1 × CutSmart buffer (NEB, B7204S). 500 units of EcoRV (NEB, R0195T) were added and the nuclei were incubated over night at 37 °C with shaking at 950 rpm. The 8 min Proteinase K digest was performed at a final concentration of 50 μg/mL. The first blunting step was missed out before A-tailing, as EcoRV is a blunt cutting enzyme. No treatment with Lambda Exonuclease was performed before subsequent DNA isolation. 20 μg of extracted DNA was bound to beads. PCR reactions was performed each using 5 μL beads, 1 μL 20 μM PCR F primer, 1 μL 20 μM PCR R primer, 10 μL 5 × Phusion HF buffer, 1 μL 10 mM dNTPs and 0.5 μL Phusion HF DNA polymerase (NEB, M0530S) in a final reaction volume of 50 μL with the following cycling parameters: 94 °C 5 min, 94 °C 1 min, 60 °C 45 s, 72 °C 1 min, 72 °C 10 min, 4 °C hold, repeating steps 2-4, for a total of 18 cycles. The DNA was size selected (400-500 bp) by gel electrophoresis and was subsequently extracted using the MinElute gel extraction kit (Qiagen, 28604). The DNA library was subjected to paired-end sequencing on an Illumina MiSeq platform.
DSBCapture (U2OS and NHEK)
A detailed protocol can be found on the protocol exchange (Insert URL). Briefly: cells were detached, counted and fixed in complete medium with 2 % formaldehyde (Pierce, 28908) at a density of 1 million cells/1.5 mL for 30 min at room temperature whilst gently rotating. Formaldehyde was quenched by adding glycine to a final concentration of 125 mM. Cells were washed twice in ice cold PBS and lysed in lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.2 % NP40 substitute (Sigma, 74385), cOmplete Roche proteinase inhibitors (REF 11873580001), 1 mM DTT) by gently rotating at 4 °C for 90 min. Nuclei were subsequently re-suspended in nucleus break buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.3 % SDS, 1 mM DTT) and incubated at 37 °C for 45 min whilst gently rotating. Nuclei were then re-suspended in 1 × NEBuffer 2 + 0.1 % TritonX-100 (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, pH 7.9 at 25 °C) and transferred into 2 mL eppendorf tubes (10 million nuclei/tube). Proteinase K (Ambion, AM2546) was added to a final concentration of 100 μg/mL on ice and nuclei were incubated at 37 °C for 8 min before adding an equal volume of 1 × NEBuffer 2 + 0.1 % TritonX-100 + 1:50 PMSF (Sigma, 93482) to kill the reaction. Nuclei were then washed twice in 1 × NEBuffer 2 + 0.1 % TritonX-100 at 4 °C. To repair DNA ends, nuclei were washed once in 1 × Blunting Buffer (NEB, E1201L) + 100 μg/mL BSA (NEB, B9000S) and were blunt-end repaired using the NEB Quick Blunting Kit (NEB, E1201L) + 100 μg/mL BSA in a final volume of 100 μL at 25 °C for 45 min, shaking for 10 s at 800 rpm, every 5 min. Nuclei were then washed three times with 1 × NEBuffer 2 + 0.1 % TritonX-100 at 4 °C and were subsequently A-tailed using Klenow Fragment 3’-5’ exo- (NEB, M0212L) and dATP (Promega, U120D) in a final volume of 50 μL at 37 °C for 45 min, shaking for 10 s at 800 rpm, every 10 min. Following A-tailing, nuclei were washed 3 × with 1 × NEBuffer 2 + TritonX-100 at 4 °C, once with T4 DNA ligase buffer (NEB, B0202S) + 0.1 % TritonX-100 at 4 °C and once with T4 DNA ligase buffer at 4 °C. The modified P5 Illumina adapter, previously annealed in 1 × T4 DNA ligase buffer, was then ligated to DNA ends using T4 DNA ligase (NEB, M0202M) for 15-20 h at 16 °C in a final reaction volume of 50 μL whilst shaking at 350 rpm for 15 s every 45 min during the ligation. To remove excess adapters, nuclei were washed 2 × in WB buffer + 0.1 % TritonX-100 (5 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 M NaCl) and once in Lambda Exonuclease Reaction Buffer (NEB, M0262L) and were then treated with 50 units Lambda Exonuclease (NEB, M0262L) in a final reaction volume of 50 μL for 30 min at 37 °C. Following washing in 1 × NEBuffer 2 + 0.1 % TritonX-100 nuclei were then re-suspended in 1 × NEBuffer 2 + 0.5 % TritonX-100 at 4 °C. To extract genomic DNA, nuclei were lysed with Proteinase K (200 μg/ml) for 30 min at 55 °C, shaking at 800 rpm followed by 30 min at 65 °C, shaking at 800 rpm and the DNA was precipitated using isopropanol. The DNA was then re-suspended in 90 μL H20 and incubated at 55 °C shaking at 800 rpm for 1 h. The isolated DNA was fragmented by sonication to obtain an average fragment size of 200-500 bp and then 50 μg (20 μg only for data depicted in Supplementary Fig. 3c) of DNA was bound to 5 μl MyOne strepdavidin C1 Dynabeads (Invitrogen, 65001) at 4 °C for 45 min, whilst gently rotating. Beads were washed 3 × with WB buffer + 0.1 % TritonX-100 and captured DNA was blunt end-repaired using the Quick Blunting Kit in a final reaction volume of 50 μL for 45 minutes at 25 °C, shaking at 800 rpm every 5 min for 10 s. The beads were then washed 3 × in WB buffer + 0.1 % TritonX-100 and DNA ends were once more A-tailed using Klenow Fragment 3’-5’ exo- in a final reaction volume of 25 μL at 37 °C for 45 min, shaking at 800 rpm for 10 s every 10 min. After washing beads 3 × with WB buffer + 0.1 % TritonX-100, the modified P7 Illumina adapter, previously annealed in 1 × T4 DNA ligase buffer, was ligated to DNA ends in a final reaction volume of 50 μL for 15-20 h at 16 °C, every 45 minutes samples were mixed for 1 min at 1200 rpm. Beads were then washed 3 × in WB buffer + 0.1 % TritonX-100 before re-suspending in 25 μL nuclease free water. The DNA DSBCapture library was then amplified by PCR with PCR F and PCR R primers and NEB Next PCR mix for 15 cycles, following the manufacturers recommendations (annealing temperature: 65 °C; NEB, M0541L). The amplified DNA library was then purified using a MinElute PCR Purification kit (Qiagen, 28004) and the library was size-selected (250-1200 bp) using a BluePippin (Sage Science). Samples were run on the Bioanalyzer (Agilent, 5067-4626) to determine the average library size, and the library was quantified using the KAPA library quantification kit (Kapa Biosystems, kk4824), following the manufacturers recommendations. Libraries were subsequently sequenced paired-end on an Illumina NextSeq 500 platform.
RNA-seq
Total RNA for RNA-seq experiments was extracted using the RNeasy kit (Qiagen, cat. no. 74104), following the manufacturer’s instructions. RNA-seq libraries were generated using the Illumina Truseq RNA HT (stranded mRNA) kit (RS-122-2103).
Epigenome analysis
The ENCODE project NHEK epigenome ChIP-seq data14 was retrieved from the NCBI’s GEO repository as follows: H3K4me3 (GSM945175); H3K4me2 (GSM733686); H3K27ac (GSM733674); H3K4me1 (GSM733698); H3K79me2 (GSM1003527); H3K36me3 (GSM945174); H2A.Z (GSM1003488); POLR2B (GSM733671); H3K9me3 (GSM1003528); H3K27me3 (GSM733701); EZH2 (GSM1003489); CTCF (GSM822271); TP63 (GSE32061), DNase-seq (GSM736556); Transcription (GSE76688).
Sequencing
For the EcoRV enzyme cleavage experiment, 50 bp paired-end sequencing was conducted on the Illumina MiSeq instrument. For the AID-DIvA U2OS and NHEK cell experiments, 75 bp paired-end sequencing was performed on the Illumina NextSeq 500 instrument. Despite DSB detection with DSBCapture requiring only single-end sequencing, we sequenced all DSBCapture libraries in paired-end mode in order to be able to distinguish DSBs at restriction sites from PCR duplicates and to be able to compare our method to BLESS in NHEK cells.
Sequencing analysis
DSBCapture libraries: fastq files containing sequencing reads were pre-processed to remove the Illumina adapters and to trim low quality tails using trim_galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/); reads were aligned to the human reference genome (hg19) using the bwa mem aligner (http://sourceforge.net/projects/bio-bwa/files/) and cleaned for low quality alignments (mapQ < 10) using samtools (clean reads) (http://samtools.sourceforge.net). Duplicates were identified using picard tools (https://github.com/broadinstitute/picard/) and removed (unduplicated reads). Only reads from read 1, which are proximal to the break site, were retained for downstream analysis and are shown in the coverage plots.
BLESS libraries: when performing the Illumina adapter trimming, the BLESS linker sequences are trimmed (TCGAGGTAGTA and TCGAGACGACG for proximal and distal linkers, respectively), and only read pairs containing both linkers are retained. Trimmed fastq files then undergo the same processing pipeline as for DSBCapture libraries. Finally, only sequencing reads that originally contained the proximal linker (originating either from read 1 or read 2) are retained for subsequent analysis and are shown in the coverage plots.
Peak calling analysis
Regions with enriched reads over the background were called as peaks using the MACS2 software (https://github.com/taoliu/MACS/) using default parameters for the statistical threshold (i.e., corrected p-value q ≤ 0.05) and the options “no-model” and “no control”. For the NHEK cell analysis, peaks where called independently for each replicate on the clean bam files containing only the proximal linker, and the two peaks files were then intersected using the bedtools package (http://bedtools.readthedocs.org/) to generate the high-confidence peak files for both BLESS and DSBCapture. The sample size was constrained by experimental considerations. For the endogenous DSBs mapping in primary NHEK cells, we observed good reproducibility with 2 biological replicates for both BLESS and DSBCapture.
Comparison of DSBs identified by DSBCapture with 50 μg and 20 μg of input material
DSBCapture was performed using 50 μg and 20 μg of input material from the same biological sample (N=1). The two DSBCapture experiments were processed in the same way, as previously described in the paragraphs “Sequencing analysis” and “Peak calling analysis”. The 20 μg library was sub-sampled in order to have the same number of unduplicated, proximal reads as the 50 μg library, i.e. 124 million reads. Peak calling was performed on the subsampled library. Peak intersection was calculated by using the bedtools, as previously explained.
GC content analysis of the EcoRV data set
The 430,897 predicted EcoRV sites were split into two subsets: 403,905 (94 % of total) sites displaying a coverage of at least 5 reads, marked as detected, and the remaining 26,992 sites (6 % of the total) displaying coverage below 5, marked as not detected. The GC content around the EcoRV restriction site was calculated, by considering 100 base pairs either side of the restriction site.
Analysis of EcoRV restriction sites at DNase hypersensitivity sites
DNase hypersensitivity sites in HeLa cells were downloaded from the ENCODE project (https://www.encodeproject.org/; GEO sample accession numbers GSM736564 and GSM736510), high confidence sites were calculated as the intersection of the genomic regions from the two replicates (bedtools intersect), resulting in 96,541 DNase sites. The overlap of the 430,897 predicted EcoRV sites and the cleaved subset (403,905; coverage ≥ 5 reads) with the high confidence DNase sites was calculated. The percentage of total and cleaved sites within DNase sites is reported. The non-parametric Chi-squared test for proportions was used for statistical testing.
AsiSI-induced DSB analysis in AID-DIvA U2OS cells
AsiSI restriction sites were identified as DSBs by peak calling as described above. The list of 100 most cleaved AsiSI sites identified by γH2AX ChIP-seq signal, and the RAD51 and XRCC4 ChIP-seq data, were obtained from Aymard, et al9. Peaks on RAD51 and XRCC4 were called using the MACS2 peak caller with default parameters (q ≤ 0.05) using the respective input files.
Comparison of DSBCapture to GUIDE-seq
The 25 endogenous DSB hotspots identified in U2OS cells by GUIDE-seq4 were compared (bedtools intersect) to the 2,372 endogenous DSBCapture peaks identified by MACS2 in AID-DIvA U2OS cells.
GC content and G-quadruplex analysis
The high confidence (~ 85,000) peaks detected in both replicates of DSBCapture were split into two subsets: DSBs uniquely identified by DSBCapture (~ 66,000 peaks) and peaks common to BLESS (~ 19,000). GC content was measured within the peak regions and each peak was assigned to a GC % category. Independently, for both the DSBCapture unique and BLESS-DSBCapture common peak subsets, the number of peaks in each GC % category were divided by the total number of peaks in the subset, resulting in a fraction of peaks within each GC % category for both subsets. The two fractions of each GC % category were divided one by each other to obtain the fold enrichment of DSBCapture unique peaks over the BLESS-DSBCapture common peaks. Values > 1 indicate that a given GC % category is enriched among the DSBCapture unique peaks, or otherwise is depleted in the BLESS-DSBCapture common peaks. Similarly for the G-quadruplex analysis, the fraction of reads from the DSBCapture high confidence peaks overlapping to observed G-quadruplex-forming sequences (OQs13) was calculated for each GC % content group and compared to random (fold enrichment). The random sets were generated by random shuffling of DSB genomic intervals throughout the entire genome and calculating the random overlap to OQs (N = 3).
Comparison to the ENCODE data sets in NHEK cells
Data sets for epigenetic marks (histone modifications), DNase hypersensitivity sites (DNase) and DNA binding proteins (CTCF, P63, POL2B) were downloaded from ENCODE (see Epigenome analysis, Methods). The number of high confidence DSBCapture peaks overlapping each mark (command intersectBed of the bedtools package) was calculated for each mark (independently of all other marks) and visualized on a bar plot (green bars in Fig. 3a and Supplementary Fig. 4b). Additionally, the DSBCapture peak files were randomly shuffled across the genome (command shuffleBed of the bedtools package) three independent times and the overlap of the random sets with each mark was calculated. The ratio of the DSB overlap divided by the average (N = 3) random overlap for each mark was computed (fold enrichment over random) and visualized (purple bars in Fig. 3a and Supplementary Fig. 4b). Fold enrichment values > 1 indicates that a mark overlaps to DSBs more often than random. Standard deviations from the 3 independent randomizations are not indicated in the bar plot as they lay in the range 0.004-0.12 for all marks, and are therefore negligible when compared to the fold change values.
Genomic regions analysis
Gene annotation files for the human genome (hg19) were downloaded from the Illumina iGenomes support website (https://support.illumina.com/sequencing/sequencing_software/igenome.html, and different gene features were calculated as follows: Promoter = 1 kb upstream of the transcription start site (TSS); 5’UTR = sequence from the TSS to the annotated translation start codon; Coding exons = all exons that are translated (coding sequences) from the start until the stop codon; 3’UTR = sequence from the annotated end of translation (stop codon) to the end of the last exon; introns = all the regions spliced out during transcript processing. The number of high confidence DSB sites overlapping each region was calculated by intersecting DSBCapture high confidence peaks to each feature separately. The DSBCapture peak files were then randomly shuffled across the genome (command shuffleBed of the bedtools package) three independent times and the overlap of the random sets to each feature was calculated. The ratio of the DSB overlap divided by the average (N = 3) random overlap for each region was calculated (fold enrichment over random). Similarly, the fold enrichment over random for genic regions (i.e. within gene bodies) versus intergenic features was assessed.
Gene expression analysis
RNA-seq sequencing reads were pre-processed by the trim galore software (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) for removal of Illumina sequencing adapters and low quality read tails. Trimmed data were then aligned to the human reference genome (hg19) using tophat2 (https://ccb.jhu.edu/software/tophat/index.shtml/). Reads overlapping unambiguously to each gene feature were assessed and counted by the htseq-count (http://www-huber.embl.de/users/anders/HTSeq/). Gene expression values were split into three groups containing the same number of genes (lower, middle and upper third) according to their rpkm gene expression values (rpkm < 0.041; rpkm 0.041-6.08 and rpkm > 6.08), where rpkm is the reads per kilo base (of the total exon length) per million sequenced reads. The average number of high confidence DSBs per kb overlapping ± 1 kb of the TSS (green bars in Fig. 3c) and to gene bodies, i.e. all exons and introns excluding ± 1 kb around the TSS (Gene body, blue bars in Fig. 3c), were calculated. Genes with length less than 2 kb were excluded from the gene body category. Additionally, all genes were split into two categories according to the presence (n=12,984) / absence (n=9,272) of DSBs at ± 1 kb around the TSS and the rpkm for all genes in the two groups was inspected and visualized as box plots.
Theoretical estimation of DSBs per cell
The amount of DNA used for DSB mapping is 50 µg; the average DNA content per cell is 6 pg, therefore the captured breaks come from 50 µg / 6 pg = 8.3 million cells. The MarkDuplicates function of the Picard software tool (https://broadinstitute.github.io/picard/) allows the calculation of the fraction of duplicated reads (either optical or PCR duplicates) and to infer the estimated library size, i.e. the estimated number of unique (unduplicated) molecules in the library based on PE duplication. According to Picard, the DSBCapture libraries display an average number of 318 million reads as library size. This allows the estimation of the average number of DSBs per cell: the total number of estimated reads in the library divided by the estimated number of cells used for DSB mapping. This is equal to 318 million / 8.3 million ≈ 38 DSBs / cell.
Code availability
The computer code used to analyze all data in this work, including sequencing processing and data comparisons, can be obtained on request by contacting the corresponding author.
Supplementary Material
Acknowledgements
We thank G. Legube, LBCMCP, Center for Integrative Biology (CBI), Université de Toulouse, Toulouse, France for providing U2OS AID-DIvA cells. We thank the genomic core facility at Cancer Research UK Cambridge Institute. R.H.H. acknowledges EMBO for support (EMBO Long-Term Fellowship to R.H.H.). We acknowledge support from the University of Cambridge and the Cancer Research UK program. The Balasubramanian laboratory is supported by core funding from Cancer Research UK (C14303/A17197 to S.B.) and by an ERC Advanced Grant (S.B.). S.B. is a Senior Investigator of the Wellcome Trust.
Footnotes
Accession code
The data reported in this paper are available at the NCBI’s GEO repository, accession number GSE78172 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qzonkauajryvtod&acc=GSE78172).
Author Contributions
S.V.L. developed the DSBCapture method, conceived the study, conducted experiments, interpreted results and wrote the manuscript. G.M. conceived the study, performed bioinformatics analyses, interpreted results and wrote the manuscript. R.H.H. contributed to the development of the DSBCapture method, conceived the study, conducted experiments, contributed to bioinformatics analyses, interpreted results and wrote the manuscript. E.Y.L. contributed to the development of the DSBCapture method, conceived the study and conducted experiments. D.T. conceived the study, interpreted results and wrote the manuscript. S.B. conceived the study, interpreted results and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Srivastava M, Raghavan SC. DNA double-strand break repair inhibitors as cancer therapeutics. Chem Biol. 2015;22:17–29. doi: 10.1016/j.chembiol.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchuk D, Drumm M, Saulino A, Collins FS. Construction of T-vectors, a rapid and general sytem for direct cloning of unmodified PCR products. Nucleic Acids Res. 1990;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitra A, Skrzypczak M, Ginalski K, Rowicka M. Strategies for achieving high sequencing accuracy for low diversity samples and avoiding sample bleeding using illumina platform. PLoS One. 2015;10:e0120520. doi: 10.1371/journal.pone.0120520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aymard F, et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribeyre C, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers VS, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 14.Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gursoy-Yuzugullu O, Ayrapetov MK, Price BD. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc Natl Acad Sci USA. 2015;112:7507–7512. doi: 10.1073/pnas.1504868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storch K, et al. Three-dimensional cell growth confers radioresistance by chromatin density modification. Cancer Res. 2010;70:3925–3934. doi: 10.1158/0008-5472.CAN-09-3848. [DOI] [PubMed] [Google Scholar]
- 17.Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52:291–302. doi: 10.1016/j.molcel.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Kemp CJ, Henikoff S. Anthracyclines induce double-strand DNA breaks at active gene promoters. Mutat Res. 2015;773:9–15. doi: 10.1016/j.mrfmmm.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwer B, et al. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc Natl Acad Sci USA. 2016;113:2258–2263. doi: 10.1073/pnas.1525564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.